Abstract
The yeast two-hybrid system and in vitro binding assays were used to characterize 54 potential interactions between the proteins of Tf1, an LTR-retrotransposon found in Schizosaccharomyces pombe. The Tf1 integrase (IN) protein was found to interact strongly with itself and not with other control proteins. In addition, the IN core domain interacted strongly with itself and full-length IN. Interestingly, the two-hybrid analysis detected an interaction between the RNase H domain of reverse transcriptase and IN. The biological implications of these interactions are discussed.
Retroviruses and long terminal repeat (LTR)-containing retrotransposons possess coding sequences for Gag, protease (PR), reverse transcriptase (RT), and integrase (IN) proteins. The activities of these proteins have been well characterized, and it is clear that many function as multimeric complexes composed of either homomeric or heteromeric components. In many cases, the specific nature and role of these interactions are under study.
There is both genetic and biochemical evidence indicating that the IN protein functions as a multimer that coordinates the integration of reverse transcripts (11, 22) (for a review, see reference 2). The IN protein of retroelements consists of three functionally distinct domains: an N-terminal HHCC (zinc finger-like) domain, a central catalytic core, and a carboxy-terminal domain thought to have a role in nonspecific DNA binding (7, 11, 36) (for a review, see reference 2). Crystal and solution structures of the HHCC domain, catalytic core, and carboxy-terminal domain of IN have provided evidence that these three subdomains are each individually capable of homomeric interactions (8, 10, 29).
Work on several retroviral RTs indicates that they also function as dimers. As a result of biochemical and structural studies, the RTs of avian sarcoma-leukosis virus (ASLV) and human immunodeficiency virus type 1 (HIV-1) are known to exist as heterodimers (for a review, see reference 33). The ASLV RT subunits both contain the polymerase and RNase H (RH) domains, but one of the subunits possesses additional sequence encoding IN at its carboxy terminus (33). This heterodimer results from a failure in processing at the carboxy terminus of RT, and it is unclear what role this additional IN domain contributes to the activity of ASLV RT. Although the RT of Moloney murine leukemia virus is isolated from virions as a monomer, biochemical studies indicate that it may function as a homodimer (19, 34, 35).
Tf1 is an LTR-retrotransposon that exists in Schizosaccharomyces pombe. Retrotransposons serve as effective retroviral model systems because they are closely related to retroviruses. Tf1 transposition can be measured in strains of S. pombe that contain a plasmid copy of Tf1 fused to the inducible promoter nmt1 (27). Results from this in vivo assay demonstrate that each of the Tf1-encoded proteins is essential for transposition (3, 4, 25, 26, 28).
Although much is known about the formation of homomeric interactions in proteins of LTR-retroelements and their role in forming functional structures or enzymes, little is known about whether heteromeric interactions are required to coordinate the processes of particle assembly, reverse transcription, and integration. In addition, little is known about whether the Tf1 proteins form homomeric interactions. To address these questions, we developed an extensive map of the potential interactions between Tf1-encoded proteins. Similar methods have been used to generate a protein linkage map for the Escherichia coli bacteriophage T7 (5), but no comprehensive approach has been taken to study the interactions that exist between proteins of a retroelement. To characterize the function of Tf1-encoded proteins, it is important to identify the homomeric interactions of Gag, PR, RT, and IN. It is also valuable to explore systematically what heteromeric interactions occur between the proteins of retroelements to evaluate what role they may play in the propagation of these elements.
We used the interaction trap yeast two-hybrid system to screen for interactions between the Tf1-encoded proteins (9, 12, 14, 17, 20) (for a review, see reference 15). In this system, the two proteins to be studied are expressed in Saccharomyces cerevisiae as fusion proteins. One of the proteins is fused to a known DNA binding domain (BD) such as LexA, and the second is fused with a transcriptional activation domain (AD). An association between the two proteins can be detected by activation of a reporter gene that possesses a promoter with binding sites for the DNA BD.
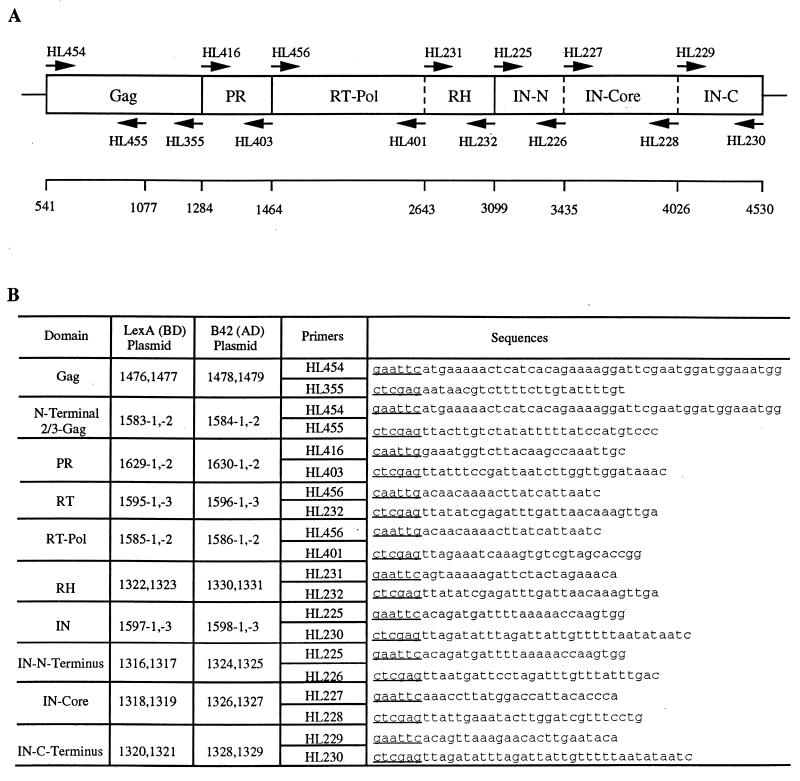
To test for interactions between the Tf1 proteins, DNA segments encoding entire proteins or subdomains were amplified by PCR and subsequently cloned into AD and BD plasmids for two-hybrid analysis. The plasmid pHL414 (28) was used as the DNA template for all PCRs. The boundaries of the protein domains were estimated based on sequence alignments with a related transposon for which the PR cleavage sites were previously determined (23). Protein domains of Tf1 analyzed in this study included the full-length Gag and the N-terminal two-thirds of Gag. The domain predicted to represent PR was also used. IN was broken down into three subdomains (N terminal [HHCC], core, and C terminal), based on the characterization of many retroviral integrases, as previously described. Finally, the RT was separated into a polymerase region and a region that consisted of the RH domain. DNA fragments encoding these domains of the Tf1 proteins were amplified by PCR with oligonucleotides that are shown in Fig. 1. In order to clone these fragments into the two-hybrid vectors, primers for each PCR product were designed to create restriction sites at the 5′ and 3′ ends. Independent duplicates of each PCR product were cloned into both BD and AD plasmids for two-hybrid analysis. pEG202 (17) was used as the expression vector for LexA-fused proteins. This plasmid constitutively expressed proteins as C-terminal fusions to 202 amino acids of the bacterial LexA DNA BD. The plasmid pJG4-5 (17) was used as the expression vector for all AD fusions. This plasmid expressed proteins under the control of the galactose-inducible GAL1 promoter as C-terminal fusions to a simian virus 40 (SV40) nuclear localization sequence, the hemagglutinin epitope tag, and the B42 AD. The various combinations of plasmids were cotransformed into S. cerevisiae EGY48 (17), which has the upstream activating sequences of the chromosomal LEU2 gene replaced with six lexA operators. Potential interactions were then scored by testing for growth on synthetic complete (SC) medium lacking leucine. Several independent transformants of each strain were screened.
FIG. 1.
Construction of two-hybrid fusion proteins. (A) Schematic diagram of the Tf1 proteins tested in the two-hybrid analysis. The coding sequences of the mature proteins are illustrated by solid vertical lines which represent predicted N-terminal and C-terminal PR processing sites. Dashed vertical lines represent subdomains used in the two-hybrid analysis. Sequence numbers on the line below represent base pairs starting from the 5′ LTR. The positions and names of the primers used in PCR amplification of various regions are indicated. (B) PCR primers used in the construction of BD and AD fusion proteins. Two independent bacterial transformants for both the BD and AD constructs were examined. The sequence is listed 5′ to 3′, and the restriction site in each primer is underlined. Thirteen additional nucleotides 5′ of the restriction site are not shown.
The results of the two-hybrid analysis are shown in Fig. 2. In some cases, fusion proteins were undetectable by immunoblotting when probed with either anti-LexA (generous gift of Erica Golemis, Fox Chase Cancer Institute) or anti-HA epitope (BAbCo) antibodies and with the ECL detection system (Amersham). As a result, these fusions are not represented in the figure (e.g., RT-B42, RT-Pol-B42, and PR-B42). All other fusions represented in the figure showed very similar high levels of expression (data not shown). The one exception to this was the RT fusion with LexA, which produced approximately 10-fold less protein than the others. Additionally, all fusions were tested for intrinsic or nonspecific activation. The AD plasmids were cotransformed with LexA fused to the human laminin C protein to test for specificity of interaction with each of the BD fusions while all the BD plasmids were cotransformed with an empty AD to test for intrinsic activation. The BD fusion with the full-length or N-terminal two-thirds of the Gag protein resulted in significant intrinsic activation and therefore could not be used in this study. All other fusions used did not intrinsically or nonspecifically activate expression of the LEU2 reporter. Plasmids containing p53 fused to LexA and SV40 large T antigen fused to B42 (Clontech) were cotransformed into EGY48 used as a positive control. Strains containing these fusion proteins showed visible growth in approximately 1 to 2 days.
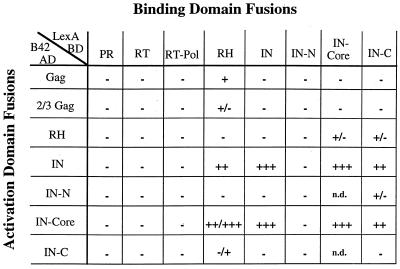
FIG. 2.
Interactions between Tf1 proteins. S. cerevisiae EGY48 was cotransformed with plasmids encoding various LexA-BD fusion proteins, together with plasmids encoding B42-AD fusions. Master plates with these transformants were grown on SC-glucose leucine-plus plates and subsequently replica plated to SC-galactose leucine-minus plates. The strength of interaction was indicated by the time required for growth on medium lacking leucine. A strain with LexA-p53 and B42-SV40 T antigen was a positive control and showed clear growth after approximately 1 to 2 days (++++). +++, very strong interaction (growth after ∼2 days); ++, strong interaction (growth after ∼3 days); +, weak interaction (growth after ∼4 days); +/− and −/+, very weak interaction (growth after ∼4 to 5 days); −, no interaction (no growth after 5 days); n.d., not determined.
Using the two-hybrid system, we observed very strong homomeric interactions within the IN protein. Full-length IN was found to interact with full-length IN and with the IN-core domain (growth on Leu− medium after ∼2 days). These interactions were of the same strength and were found to occur in either combination of the BD or AD plasmids. A very strong homomeric interaction was also found for the IN-core domain. This combination was found to be as strong as the full-length IN interactions. Additionally, the C-terminal domain of IN in the BD plasmid showed a strong interaction with both full-length IN and IN-core in the AD vector. The C-terminal domain was not found to interact with itself, however. The lack of any significant interactions of the C-terminal domain of IN in the AD fusions with domains of IN in the BD vector may reflect a masking of the AD by the C-terminal domain of IN. Particularly interesting is the IN-core homomeric interaction. This has been well described for the HIV IN catalytic core, which has been shown to dimerize or multimerize based on crystallization and biochemical studies (1, 2, 10). Solution structures of the C-terminal domain of HIV IN lead authors to speculate that the C terminus could potentially make contacts with either the catalytic core or the N-terminal domain (29). This evidence may help explain the surprising interaction observed between the C terminus and the catalytic core of Tf1 IN. The interaction between the C-terminal domain of Tf1 IN and IN-core may reflect either intramolecular or intermolecular contacts within native IN protein. Based on the two-hybrid results for the homomeric interactions, it appears that the domains of Tf1 IN are likely to possess intermolecular contacts similar to those of the well-characterized retroviral INs, perhaps requiring multimerization for activity.
Another intriguing two-hybrid interaction was found between the RH domain of RT in the BD plasmid and either full-length IN or IN-core fused to the AD. These were both strong interactions (growth after ∼3 days on Leu− medium). The fact that this interaction is not observed in the opposite orientation may be due to the individual structure of the fusions. When RH is fused to B42, the protein may be folding into a different conformation which may not allow the interaction to occur. Additionally, the LexA protein is known to dimerize (18, 32), and this dimerization of RH, which would be absent in the B42 fusion of RH, may be required for the interaction with IN. The RH and IN-core domains of HIV have a strong structural similarity (10), which could result in a heterodimerization of these two proteins. Indeed, interactions between RT and IN of other retroviruses have been observed. One example of an interaction between retroviral RT and IN is the heterodimeric RT of ASLV (33), as previously mentioned. Again, the importance of the IN domain in the ASLV RT is not clear, but it is easy to imagine that these two proteins may interact and play a mutual role in either reverse transcription or integration. Indeed, there are other reports of RT-IN complexes. Our laboratory has recently identified mutations in the RH domain of Tf1 RT that result in a severe defect in transposition without causing any defect in reverse transcription (3). It is possible that a single-amino-acid substitution in RH may disrupt an unknown function of RH which is required for optimal levels of integration, perhaps by specifically disrupting the formation of a heterodimer with IN. Consistent with this idea, there is a report that a large nucleoprotein complex defined as the preintegration complex purified from HIV-infected cells contains RT (6, 31). Based on in vitro integration assay data, this HIV preintegration complex (with RT as a member) produces double-ended insertions with a much higher efficiency than purified IN alone (13). In addition, the immunoprecipitation of murine leukemia virus particles that are boiled or treated with 2-mercaptoethanol indicates that disulfide bonds can exist between RT and IN (21). There are also several reports that describe mutations in IN that result in a defect in reverse transcription. Carboxyl-terminal deletions of the Ty3 IN cause a dramatic reduction in the amount of Ty3 DNA and a decrease in reverse transcriptase activity in vitro, without appearing to affect the size or amount of RT in viruslike particles (24). In addition, a series of point mutations generated in the zinc finger-like domain of HIV-1 IN caused a decrease in the levels of viral DNA after infection (30). Further characterization of these mutants showed that they had wild-type levels of RT within the mutant virions as well as wild-type levels of RT activity. All of this evidence suggests that interactions may exist between RT and IN.
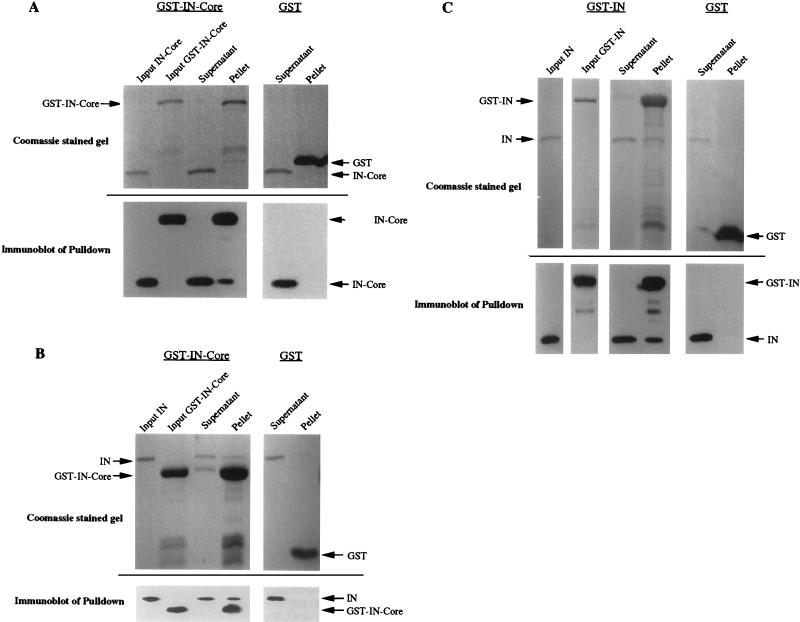
To examine further the abilities of these domains to interact, an in vitro binding assay was developed to detect interactions between IN proteins as well as interactions between IN and RH. Sequences corresponding to full-length IN, the IN core domain, and RH were amplified by PCR and cloned into two sets of pGEX vectors (Pharmacia Biotech) to direct the synthesis of glutathione S-transferase (GST) fusion proteins in bacteria (16). Independent PCR duplicates of each sequence were cloned into pGEX vectors containing either a factor Xa or Prescission protease (Pharmacia Biotech) cleavage site, allowing cleavage of the protein of interest from GST. Bacterial lysates were prepared, and the GST fusion proteins were recovered on glutathione-agarose beads (Pharmacia Biotech). One set of IN and IN-core fusion proteins bound to beads were cleaved from GST with the Prescission protease. The cleaved products were then isolated and used in pulldown experiments with GST-IN, GST-IN-core, GST-RH, or GST alone bound to beads. After extensive washing, the proteins bound to the agarose beads were analyzed by sodium dodecyl sulfate–10 or 14% polyacrylamide gel electrophoresis and Coomassie blue staining directly or transferred to nitrocellulose membranes for immunoblotting with anti-IN antibodies and ECL for detection (Fig. 3). The GST-IN-core fusion bound to agarose beads was able to pull down both full-length IN and IN-core (Fig. 3A and B). Although the IN-core pulldown (Fig. 3A) is not apparent on the Coomassie blue-stained gel, it is clearly visible on the immunoblot. Because there are breakdown products of the GST-IN-core bound to agarose beads, the pulldown experiment with full-length IN also served as a control to indicate that the IN-core result was a true pulldown and not the result of the inadvertent cleavage of the GST-IN-core by contaminating Prescission protease. The pulldown with full-length IN showed only IN in the pellet and no IN-core, based on immunoblots, and demonstrated that there was no Prescission cleavage of the GST-IN-core. Additionally, GST-IN was able to pull down IN (Fig. 3C). Importantly, the control GST protein alone didn’t bind IN or IN-core. These results confirm the yeast two-hybrid results that IN possesses strong homomeric interactions with IN and that the IN-core domain interacts with itself and IN. After several attempts, no pulldown of IN-core by GST-RH was observed. Attempts were made to cleave RH from GST and to pull down RH with GST-IN-core, but the majority of the RH was degraded after cleavage of GST.
FIG. 3.
In vitro binding between IN and IN-core proteins. IN and IN-core proteins were expressed as GST fusion proteins in bacteria and were purified by binding to glutathione-Sepharose 4B beads (Pharmacia Biotech). One set of proteins bound to resin was cleaved from GST with Prescission protease (Pharmacia Biotech) at 4°C for 2 to 4 h in the presence of cleavage buffer containing 50 mM Tris-HCl (pH 7.0), 150 mM NaCl, 1 mM EDTA, and 1 mM dithiothreitol. For binding experiments, cleaved proteins were isolated and incubated with Sepharose 4B beads bound to GST-fused IN or IN-core (containing a factor Xa cleavage site) at 22°C for 45 min in the presence of binding buffer containing 1× phosphate-buffered saline, 0.1% Tween 20, and 0.1% Casamino Acids. The binding buffer was used to wash the beads four times to remove unbound proteins. The beads were then boiled in 2× sample buffer, and the proteins bound to the GST fusion proteins were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoblotting with anti-IN antisera (28). (A) Pulldown of IN-core by GST-IN-core. Five percent of the input of IN-core and GST-IN-core and 10% of the supernatant and pellet were loaded on the Coomassie blue-stained gel. Approximately one-fourth of each of these samples was then run on a separate gel and transferred to a nitrocellulose membrane for immunoblotting with anti-IN antisera. (B) Pulldown of IN by GST-IN-core. The same amounts of samples as described above were loaded. (C) Pulldown of IN by GST-IN. Again, 5% of the input of cleaved IN was loaded along with 10% of the supernatant and pellet from each pulldown.
In summary, we have analyzed the interactions between several Tf1 proteins by the yeast two-hybrid system and in vitro binding assays with purified proteins. These results define domains of Tf1 proteins that participate in direct protein-protein interactions, both homomeric and heteromeric. There is a wealth of genetic, biochemical, and structural evidence for the role of these types of interactions in the life cycle of other retroelements. Clearly, IN is known to require the formation of a dimer or multimer to function (2), and the two-hybrid results for Tf1 IN showed that IN can have homomeric interactions. The IN-core alone was sufficient for homomeric interactions, both in the two-hybrid assay and in pulldown experiments. This may indicate that the catalytic core is the primary domain mediating the interaction, even though it appears that there are other regions involved as well. Previous studies have shown that the N-terminal domain and the C-terminal domain of IN can have homomeric interactions (8, 29). While we didn’t observe these interactions, an interaction was found between the C-terminal domain and either full-length IN or the core domain.
Another heteromeric interaction observed was found between RH and either IN or the catalytic core. It is interesting to imagine that RT may interact with IN and potentially have a role in the integration reaction. Unfortunately, difficulties in obtaining soluble, stable RT protein have hampered efforts to gain in vitro support for this interaction.
It is also interesting to consider which proteins don’t interact in this system. Obviously, it is difficult to draw conclusions from these results, but it is surprising that the Gag protein didn’t have a significant interaction with any of the polymerase (Pol) proteins tested. This leaves open the question of how Gag and Pol proteins pack within particle structures. Again, homomeric interactions for Gag could not be tested, since LexA fused to Gag caused intrinsic activation.
Generating interaction maps in this way may prove to be a valuable first tool for the analysis of potential interactions between proteins of genetic elements.
Acknowledgments
We thank Erica Golemis for providing anti-LexA antiserum and the members of the Levin and Hinnebusch laboratories for helpful discussions. We also thank Beth Agresta for reading the manuscript and providing helpful suggestions.
The NIH Intramural AIDS Targeted Antiviral Program contributed support for this research.
REFERENCES
- 1.Andrake M D, Skalka A M. Multimerization determinants reside in both the catalytic core and C terminus of avian sarcoma virus integrase. J Biol Chem. 1995;270:29299–29306. doi: 10.1074/jbc.270.49.29299. [DOI] [PubMed] [Google Scholar]
- 2.Andrake M D, Skalka A M. Retroviral integrase, putting the pieces together. J Biol Chem. 1996;271:19633–19636. doi: 10.1074/jbc.271.33.19633. [DOI] [PubMed] [Google Scholar]
- 3.Atwood A, Choi J, Levin H L. The application of a homologous recombination assay revealed amino acid residues in an LTR-retrotransposon that were critical for integration. J Virol. 1998;72:1324–1333. doi: 10.1128/jvi.72.2.1324-1333.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Atwood A, Lin J, Levin H. The retrotransposon Tf1 assembles virus-like particles that contain excess Gag relative to integrase because of a regulated degradation process. Mol Cell Biol. 1996;16:338–346. doi: 10.1128/mcb.16.1.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartel P L, Roecklein J A, SenGupta D, Fields S. A protein linkage map of Escherichia coli bacteriophage T7. Nat Genet. 1996;12:72–77. doi: 10.1038/ng0196-72. [DOI] [PubMed] [Google Scholar]
- 6.Bukrinsky M I, Sharova N, McDonald T L, Pushkarskaya T, Tarpley W G, Stevenson M. Association of integrase, matrix, and reverse transcriptase antigens of human immunodeficiency virus type 1 with viral nucleic acids following acute infection. Proc Natl Acad Sci USA. 1993;90:6125–6129. doi: 10.1073/pnas.90.13.6125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bushman F D, Engelman A, Palmer I, Wingfield P, Craigie R. Domains of the integrase protein of human immunodeficiency virus type 1 responsible for polynucleotidyl transfer and zinc binding. Proc Natl Acad Sci USA. 1993;90:3428–3432. doi: 10.1073/pnas.90.8.3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cai M, Zheng R, Caffrey M, Craigie R, Clore G M, Gronengorn A M. Solution structure of the N-terminal zinc binding domain of HIV-1 integrase. Nat Struct Biol. 1997;4:567–577. doi: 10.1038/nsb0797-567. [DOI] [PubMed] [Google Scholar]
- 9.Chien C-T, Bartel P L, Sternglanz R, Fields S. The two-hybrid system: a method to identify and clone genes for proteins that interact with a protein of interest. Proc Natl Acad Sci USA. 1991;88:9578–9582. doi: 10.1073/pnas.88.21.9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dyda F, Hickman A B, Jenkins T M, Engelman A, Craigie R, Davies D R. Crystal structure of the catalytic domain of HIV-1 integrase: similarity to other polynucleotidyl transferases. Science. 1994;266:1981–1986. doi: 10.1126/science.7801124. [DOI] [PubMed] [Google Scholar]
- 11.Engelman A, Bushman F D, Craigie R. Identification of discrete functional domains of HIV-1 integrase and their organization within an active multimeric complex. EMBO J. 1993;12:3269–3275. doi: 10.1002/j.1460-2075.1993.tb05996.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Estojak J, Brent R, Golemis E A. Correlation of two-hybrid affinity data with in vitro measurements. Mol Cell Biol. 1995;15:5820–5829. doi: 10.1128/mcb.15.10.5820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Farnet C M, Bushman F D. HIV-1 cDNA integration: requirement of HMG I(Y) protein for function of preintegration complexes in vitro. Cell. 1997;88:483–492. doi: 10.1016/s0092-8674(00)81888-7. [DOI] [PubMed] [Google Scholar]
- 14.Fields S, Song O-K. A novel genetic system to detect protein-protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 15.Fields S, Sternglanz R. The two-hybrid system: an assay for protein-protein interactions. Trends Genet. 1994;10:286–292. doi: 10.1016/0168-9525(90)90012-u. [DOI] [PubMed] [Google Scholar]
- 16.Frangioni J V, Neel B G. Solubilization and purification of enzymatically active glutathione S-transferase (pGEX) fusion proteins. Anal Biochem. 1993;210:179–187. doi: 10.1006/abio.1993.1170. [DOI] [PubMed] [Google Scholar]
- 17.Golemis E, Gyuris J, Brent R, editors. Interaction trap/two-hybrid system to identify interacting proteins. New York, N.Y: John Wiley & Sons, Inc.; 1996. [Google Scholar]
- 18.Golemis E A, Brent R. Fused protein domains inhibit DNA binding by LexA. Mol Cell Biol. 1992;12:3006–3014. doi: 10.1128/mcb.12.7.3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo J, Wu W, Yuan Z Y, Post K, Crouch R J, Levin J G. Defects in primer-template binding, processive DNA synthesis, and RNase H activity associated with chimeric reverse transcriptases having the murine leukemia virus polymerase domain joined to Escherichia coli RNase H. Biochemistry. 1995;34:5018–5029. doi: 10.1021/bi00015a013. [DOI] [PubMed] [Google Scholar]
- 20.Gyuris J, Golemis E, Chertkov H, Brent R. Cdi1, a human G1- and S-phase protein phosphatase that associates with Cdk2. Cell. 1993;75:791–803. doi: 10.1016/0092-8674(93)90498-f. [DOI] [PubMed] [Google Scholar]
- 21.Hu S C, Court D L, Zweig M, Levin J G. Murine leukemia virus pol gene products: analysis with antisera generated against reverse transcriptase and endonuclease fusion proteins expressed in Escherichia coli. J Virol. 1986;60:267–274. doi: 10.1128/jvi.60.1.267-274.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones K S, Coleman J, Merkel G W, Laue T M, Skalka A M. Retroviral integrase functions as a multimer and can turn over catalytically. J Biol Chem. 1992;267:16037–16040. [PubMed] [Google Scholar]
- 23.Kirchner J, Sandmeyer S. Proteolytic processing of Ty3 proteins is required for transposition. J Virol. 1993;67:19–28. doi: 10.1128/jvi.67.1.19-28.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kirchner J, Sandmeyer S B. Ty3 integrase mutants defective in reverse transcription or 3′-end processing of extrachromosomal Ty3 DNA. J Virol. 1996;70:4737–4747. doi: 10.1128/jvi.70.7.4737-4747.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Levin H L. A novel mechanism of self-primed reverse transcription defines a new family of retroelements. Mol Cell Biol. 1995;15:3310–3317. doi: 10.1128/mcb.15.6.3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Levin H L. An unusual mechanism of self-primed reverse transcription requires the RNase H domain of reverse transcriptase to cleave an RNA duplex. Mol Cell Biol. 1996;16:5645–5654. doi: 10.1128/mcb.16.10.5645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Levin H L, Boeke J D. Demonstration of retrotransposition of the Tf1 element in fission yeast. EMBO J. 1992;11:1145–1153. doi: 10.1002/j.1460-2075.1992.tb05155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Levin H L, Weaver D C, Boeke J D. Novel gene expression mechanism in a fission yeast retroelement: Tf1 proteins are derived from a single primary translation product. EMBO J. 1993;12:4885–4895. doi: 10.1002/j.1460-2075.1993.tb06178.x. . (Erratum, 13:1494, 1994.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lodi P L, Ernst J A, Kuszewski J, Hickman A B, Engelman A, Craigie R, Clore G M, Grononborn A M. Solution structure of the DNA binding domain of HIV-1 integrase. Biochemistry. 1995;34:9826–9833. doi: 10.1021/bi00031a002. [DOI] [PubMed] [Google Scholar]
- 30.Masuda T, Planelles V, Krogstad P, Chen I S Y. Genetic analysis of human immunodeficiency virus type 1 integrase and the U3 att site: unusual phenotype of mutants in the zinc finger-like domain. J Virol. 1995;69:6687–6696. doi: 10.1128/jvi.69.11.6687-6696.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller M D, Farnet C M, Bushman F D. Human immunodeficiency virus type 1 preintegration complexes: studies of organization and composition. J Virol. 1997;71:5382–5390. doi: 10.1128/jvi.71.7.5382-5390.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schnarr M, Pouyet J, Granger-Schnarr M, Daune M. Large-scale purification, oligomerization equilibria, and specific interaction of the LexA repressor of Escherichia coli. Biochemistry. 1985;24:2812–2818. doi: 10.1021/bi00332a032. [DOI] [PubMed] [Google Scholar]
- 33.Skalka A M, Goff S P, editors. Reverse transcriptase. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1993. [PubMed] [Google Scholar]
- 34.Telesnitsky A, Goff S P. RNase H domain mutations affect the interaction between Moloney murine leukemia virus reverse transcriptase and its primer-template. Proc Natl Acad Sci USA. 1993;90:1276–1280. doi: 10.1073/pnas.90.4.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Telesnitsky A, Goff S P. Two defective forms of reverse transcriptase can complement to restore retroviral infectivity. EMBO J. 1993;12:4433–4438. doi: 10.1002/j.1460-2075.1993.tb06128.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Gent D C, Elgersma Y, Bolk M W, Vink C, Plasterk R H. DNA binding properties of the integrase proteins of human immunodeficiency viruses types 1 and 2. Nucleic Acids Res. 1991;19:3821–3827. doi: 10.1093/nar/19.14.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]