Summary
Background
Tuberculosis, a major cause of death in people living with HIV, remains challenging to diagnose. Diagnostic accuracy data are scarce for promising triage and confirmatory tests such as C-reactive protein (CRP), sputum and urine Xpert MTB/RIF Ultra (Xpert Ultra), and urine Determine TB LAM Ag (a lateral flow lipoarabinomannan [LF-LAM] test), without symptom selection. We evaluated novel triage and confirmatory tests in ambulatory people with HIV initiating antiretroviral therapy (ART).
Methods
897 ART-initiators were recruited irrespective of symptoms and sputum induction offered. For triage (n=800), we evaluated point-of-care blood-based CRP testing, compared with the WHO-recommended four-symptom screen (W4SS). For sputum-based confirmatory testing (n=787), we evaluated Xpert Ultra versus Xpert MTB/RIF (Xpert). For urine-based confirmatory testing (n=732), we evaluated Xpert Ultra and LF-LAM. We used a sputum culture reference standard.
Findings
463 (52%) of 897 participants were female. The areas under the receiver operator characteristic curves for CRP was 0·78 (95% CI 0·73–0·83) and for number of W4SS symptoms was 0·70 (0·64–0·75). CRP (≥10 mg/L) had similar sensitivity to W4SS (77% [95% CI 68–85; 80/104] vs 77% [68–85; 80/104]; p>0·99] but higher specificity (64% [61–68; 445/696] vs 48% [45–52; 334/696]; p<0·0001]; reducing unnecessary confirmatory testing by 138 (95% CI 117–160) per 1000 people and number-needed-to-test from 6·91 (95% CI 6·25–7·81) to 4·87 (4·41–5·51). Sputum samples with Xpert Ultra, which required induction in 49 (31%) of 158 of people (95% CI 24–39), had higher sensitivity than Xpert (71% [95% CI 61–80; 74/104] vs 56% [46–66; 58/104]; p<0·0001). Of the people with one or more confirmatory sputum or urine test results that were positive, the proportion detected by Xpert Ultra increased from 45% (26–64) to 66% (46–82) with induction. Programmatically done haemoglobin, triage test combinations, and urine tests showed comparatively worse results.
Interpretation
CRP is a more specific triage test than W4SS in those initiating ART. Sputum induction improves diagnostic yield. Sputum samples with Xpert Ultra is a more accurate confirmatory test than with Xpert.
Funding
South African Medical Research Council, EDCTP2, US National Institutes of Health–National Institute of Allergy and Infectious Diseases.
Introduction
Tuberculosis is the single biggest cause of death among people living with HIV. Such individuals are most immunocompromised before antiretroviral therapy (ART) initiation1 and, at HIV diagnosis, are already captured within HIV care cascades. ART initiation clinics represent a valuable opportunity to rapidly screen and test for tuberculosis. However, the point-of-care feasibility, accuracy, and potential use of new and repurposed tools is unclear, especially in the absence of symptomatic pre-selection.2
The WHO-recommended four-symptom screen (W4SS) is used to identify people requiring confirmatory tuberculosis testing. However, in addition to being subjective, stigmatising, and poorly implemented,3,4 prevalence surveys show that most bacteriologically positive tuberculosis is in W4SS-negative individuals.5 Consequently, WHO emphasises that, as an alternative or adjunct to symptoms, an inexpensive and rapid triage test with sensitivity of 90% or more and specificity of 70% or more on accessible non-sputum specimens is needed to better focus relatively expensive confirmatory testing.6
C-reactive protein (CRP) is a blood biomarker with evidence to assist in tuberculosis diagnosis in people with HIV.7–10 CRP platforms are commercially-available;11 however, few are evaluated at point-of-care9 or alongside other biomarkers such as haemoglobin.12 For the implementation of CRP triage, point-of-care testing is required to enable real-time decision making within the same encounter that symptoms are ascertained. A systematic review and individual participant data meta-analysis, which focussed on aggregate data in people with HIV but not specifically ART-initiators, W4SS-negatives, or point-of-care feasibility, showed CRP to have similar sensitivity versus W4SS, but higher specificity.13 This meta-analysis informed a WHO guideline update, in which CRP was recommended as an adjunct to W4SS triage for HIV-associated tuberculosis.14
CRP performance data in concert with the new Xpert MTB/RIF Ultra (Xpert Ultra; Cepheid, Sunnyvale, CA, USA) sputum test are also scarce. Xpert Ultra has improved limit of detection for Mycobacterium tuberculosis complex versus its predecessor Xpert MTB/RIF (Xpert; Cepheid, Sunnyvale, CA, USA). Critically, this sensitivity increment is probably the largest in people with paucibacillary early-stage disease.15 Such individuals might not self-present to care due to tuberculosis symptoms (if any) but still do so, for example, to initiate ART. Xpert Ultra at this point could therefore represent a hitherto unavailable opportunity to detect tuberculosis early in a key risk group; however, data are scarce.
Individuals with early-stage tuberculosis disease or HIV, or both, might not be able to naturally expectorate sputum.16 Individuals who cannot provide a sputum sample are often excluded from rapid confirmatory testing—both programmatically and in research studies—because sputum induction is frequently unavailable in high-burden primary care settings. Obtaining a diagnostic reference standard is challenging in individuals who do not produce sputum. As a result, those who have difficulty producing sputum are often excluded from sensitivity calculations, leading to missed cases. Furthermore, the proportion of people who would have needed induction is seldom reported, meaning that studies that include only sputum expectorators potentially overestimate real-world test performance. Lastly, due to these and other challenges associated with sputum,17 it is important to evaluate confirmatory tests on easily accessible fluids such as urine.
We therefore sought to evaluate the performance of novel triage and confirmatory testing approaches among people with HIV who were selected independent of symptoms and visiting a facility in Cape Town, South Africa, to start ART. For triage, we compared point-of-care CRP to W4SS as the current standard-of-care. For confirmatory testing, we evaluated sputum tests (with Xpert and Xpert Ultra), the incremental diagnostic yield of sputum induction, and urine tests (with Xpert Ultra, and Determine TB LAM Ag, Abbott, Chicago, IL, USA).
Methods
Study design and participants
897 ambulatory adults (age ≥18 years) newly diagnosed with HIV and referred to Kraaifontein Community Health Centre, an urban referral centre serving the Northern subdistrict in Cape Town, South Africa, to start ART were eligible and prospectively enrolled from March 15, 2017, to March 17, 2020, irrespective of the presence or absence of W4SS symptoms (eg, current cough, fever, night sweats, or weight loss)14 and evaluated at a single visit (figure 1). Participants were excluded if they had received any tuberculosis treatment within the past 2 months, were of unknown treatment status, did not provide written informed consent, or had been or were currently on ART.
Figure 1: Flowchart showing participant enrolment, specimen collection and processing, and tests done.
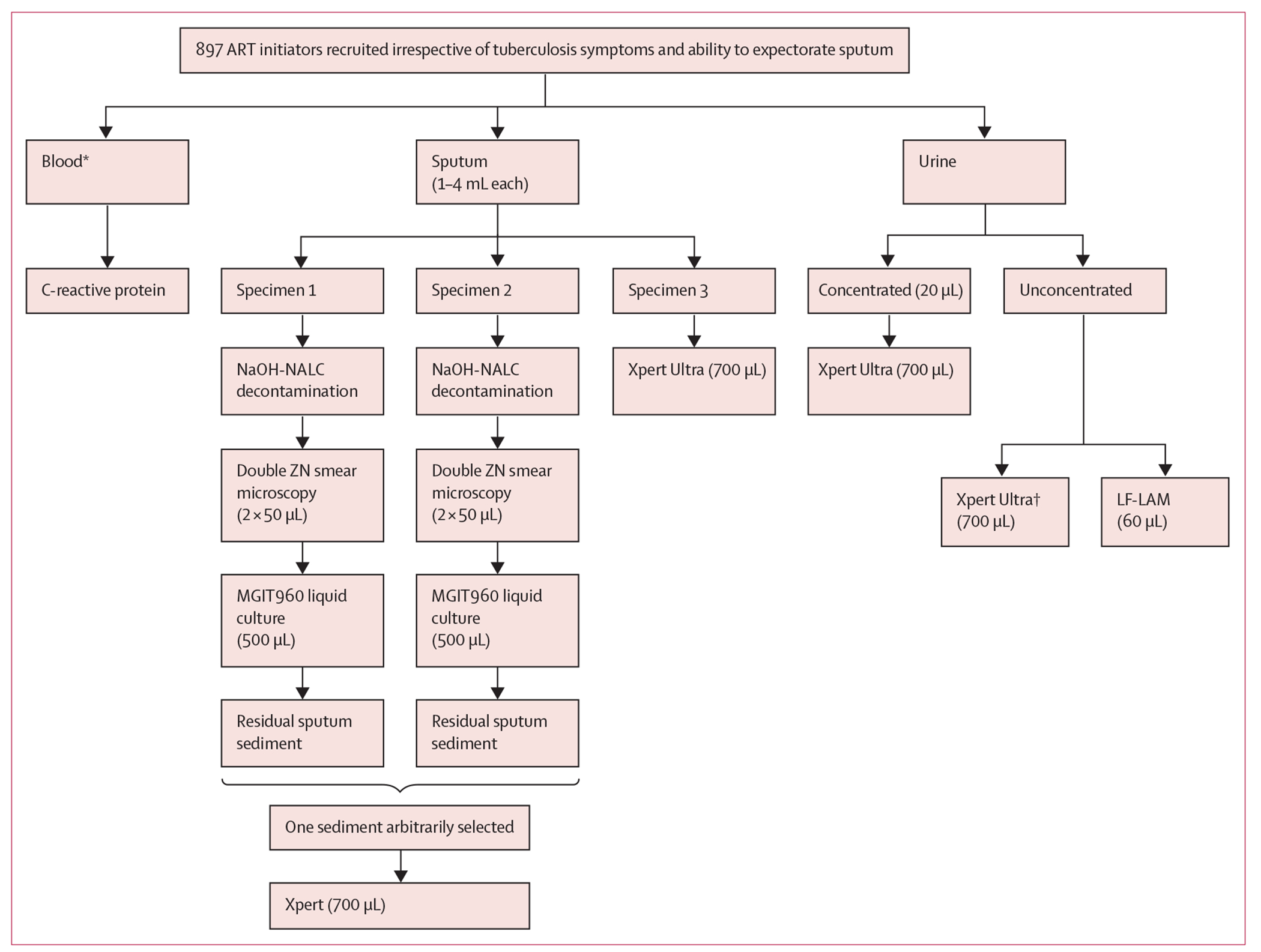
ART=antiretroviral treatment. LF-LAM=lateral flow lipoarabinomannan (Determine TB LAM Ag test). MGIT960=mycobacterial growth indicator tube. NaOH-NALC=sodium hydroxide-N-acetyl-L-cystein. Xpert=Xpert MTB/RIF. Xpert Ultra=Xpert MTB/RIF Ultra. ZN=Ziehl-Neelsen. *Haemoglobin was done when programmatically indicated outside of the study. †Unconcentrated Xpert Ultra done if concentrated Xpert Ultra non-actionable or positive.
People with at least one W4SS symptom were classified as W4SS-positive. Four to five drops of capillary blood were used by a health worker (nurse, nurse assistant, or community health worker) for point-of-care CRP measurement (iChromaII platform, Boditech, South Korea;18 appendix 1 p 2). CD4 counts were measured outside the study but captured (counts more than 3 months before or after recruitment were excluded) as well as haemoglobin (when medically indicated by routine staff per local guidelines close to HIV diagnosis).19 CRP positivity thresholds of 5 mg/L or more (CRP5) or 10 mg/L or more (CRP10), and less than 10 g/dL haemoglobin, were pre-selected on the basis of the literature.9,13 Different combinations of triage tests were evaluated.
This study was approved by the Stellenbosch University Faculty of Health Sciences Research Ethics Committee (N14/10/136) and the Western Cape Department of Health, South Africa (WC_2016RP38_944). This study is registered on ClinicalTrials.gov, NCT03187964.
Procedures
Three sputa (≥1 mL each) were required from participants, who first attempted expectoration before induction was offered (whether specific individual sputa made were expectorated or induced was, due to a database error [field staff were initially not correctly prompted in the electronic case report form to capture which sputa were a result of expectoration or induction], only successfully recorded for the last 158 participants). Induction used a nebuliser (Ultrasonic Hospital Grade WH-802, Hitech Therapy, Johannesburg, South Africa) with 5% NaCl (Ysterplaat Medical Supplies, Cape Town, South Africa) for 7–10 min.16 Two sputa, per participant, were arbitrarily selected to each undergo 1% NaOH-NALC decontamination, Ziehl-Neelsen smear microscopy, and Mycobacteria Growth Indicator Tube 960 culture (Becton Dickinson Diagnostic Systems, Sparks, NV, USA). The other sputum underwent Xpert Ultra testing and Xpert was done on the sputum sediment remnant remaining after culture inoculation. Further information on specimen storage, sputum, urine, and isolate testing is in appendix 1 (pp 2–3). Study staff had access to all clinical, index test, and reference standard data, but were masked to the reference standard results when triage and index tests were done. Sputum Xpert Ultra and microscopy (both available ≤48 h), culture, and MTBDRplus (version 2.0, Hain Lifescience, Nehren, Germany) results (typically available within 35 days) were reported for potential participant management to the programme. No adverse events occurred.
If at least one culture was M tuberculosis complex positive, participants were classified as having tuberculosis. Participants negative for two cultures or negative for culture and the other contaminated, were classified as not having tuberculosis. Participants were excluded if both cultures were contaminated.
Statistical analysis
Self-reported demographic data including but not limited to self-reported sex (male or female) and ethnicity, as well as clinical data were captured on RedCap.20 Head-to-head analyses (only people with actionable results from the relevant tests) are presented unless stated otherwise. Sensitivity, specificity, and predictive values for tests and algorithms were calculated using two-by-two tables with 95% CIs (binomial proportion method). Yield was calculated as, of the people who had a test attempted, those with a positive result (sputum Xpert Ultra, sputum Xpert, sputum culture, urine Xpert Ultra [concentrated or unconcentrated], or urine lateral flow lipoarabinomannan [LF-LAM]). If people could not make a specimen, they were still included in the yield denominator (however, with the availability of sputum induction, all could make sputum as well as urine) as were people who had non-actionable results (not positive or negative;21,22 appendix 1 p 2). People who did not have a test attempted due to human error for example (and not a lack of samples) were excluded from yield calculations.
Statistical tests included McNemar’s chi squared, Mann-Whitney, two-sample proportion, Kruskal-Wallis, and Spearman’s coefficient using Stata (version 15; StataCorp) and GraphPad Prism (version 7.0; GraphPad Software). We followed STARD analysis and reporting criteria23 and did sample size calculations using a 95% CI approach (appendix 1 p 4).
Role of the funding source
The funders of the study had no role in study design, data collection, data analysis, data interpretation, or writing of the report.
Results
Of the 897 enrolled participants, 855 (95%) had a valid culture result (22 had no cultures done, 16 had both cultures contaminated, and 4 had one culture contaminated and the second culture not done). Of those with valid results, 107 (13%) of 855 were positive for culture (36 [34%] of 107 were positive by one culture), comprising 104 (13%) of 800 participants in our CRP and W4SS triage comparison, 104 (13%) of 787 in our sputum confirmatory test comparison, and 97 (13%) of 732 in our urine test comparison (figure 2A–C). Compared with W4SS-positives, W4SS-negative participants were more likely to be younger, female, non-smokers, culture-negative, and have lower CRP but higher CD4 and haemoglobin levels (table 1). Compared with participants with culture-negative results, participants with culture-positive results were more likely to be older, of mixed ancestry, smokers, have lower CD4 counts and haemoglobin, and higher CRP levels. 26 (25%) of 104 participants with culture-confirmed tuberculosis were W4SS-negative.
Figure 2: Flow diagrams showing the number of people in head-to-head comparisons of triage (A), confirmatory (B), and urine (C) tests.
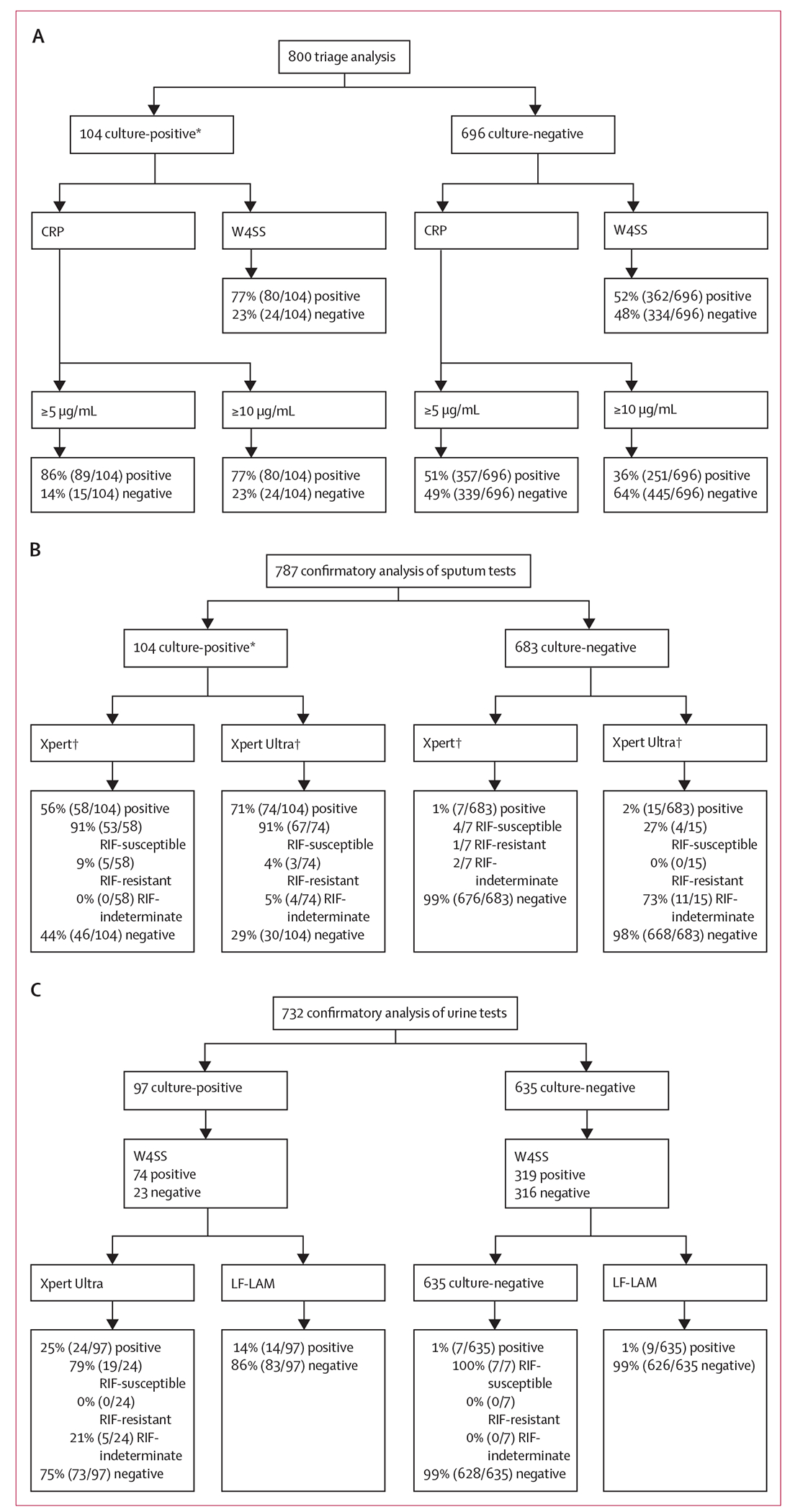
Triage tests are CRP and W4SS. Confirmatory tests are Xpert and Xpert Ultra on sputum. Urine tests are concentrated Xpert Ultra and LF-LAM. CRP correctly classified more people without tuberculosis compared with W4SS, Xpert Ultra detected more tuberculosis than Xpert on sputum, and Xpert Ultra detected more tuberculosis than LF-LAM on concentrated urine. Reasons for people excluded from each head-to-head analysis: (A–C) no culture (n=42: 22 no sputum and 20 contaminated); (A) no CRP results (n=55); (B) no actionable Xpert (n=91: 4 no result, 1 error, and 86 no or under volume specimen) and Xpert Ultra results (n=16: 2 error and 14 no sputum); and (C) no actionable concentrated urine Xpert Ultra (n=118: 3 no result, 23 invalid, and 92 error), insufficient urine (n=7: 4 for both LF-LAM and concentrated Xpert Ultra, 3 for concentrated Xpert Ultra only, and 1 for LF-LAM only), and LF-LAM unavailable (n=1). CRP=C-reactive protein. LF-LAM=lateral flow lipoarabinomannan (Determine TB LAM Ag test). RIF=rifampicin. Xpert=Xpert MTB/RIF. Xpert Ultra=Xpert MTB/RIF Ultra. W4SS=WHO-recommended four-symptom screen. *69 (66% of 104 people were culture-positive for both sputa; 35 (34%) of 104 were positive for one sputum and negative for the other (30 [86%]), contaminated for the other (4 [11%]), or the other not done (1 [3%]). †Xpert detected three Xpert Ultra-negative cases (3 [10%] of 30 [95% CI 2–27]). Xpert Ultra detected 19 Xpert-negative cases (19 [41%] of 46 [95% CI 27–57]).
Table 1:
Demographic and clinical characteristics by W4SS, culture, and smear statuses for participants in the Xpert Ultra and Xpert head-to-head comparison
| Overall (n=787) | W4SS status |
Culture and smear status |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| W4SS-positive (n=430) | W4SS-negative (n=357) | p value* | Culture positive |
Culture negative |
||||||
| All (n=104) | Smear-positive (n=16) | Smear-negative (n=88) | p value† | All (n=683) | p value‡ | |||||
|
Demographics
| ||||||||||
| Age, years | 32 (26–39) | 33 (28–40) | 31 (25–37) | <0·0001 | 34 (29–39) | 37 (33–42) | 34 (29–39) | 0·17 | 31 (26–38) | 0·010 |
| Sex | ||||||||||
| Female | 463 (59%) | 233 (54%) | 230 (64%) | .. | 43 (41%) | 4 (25%) | 39 (44%) | .. | 420 (61%) | .. |
| Male | 324 (41%) | 197 (46%) | 127 (36%) | 0·0037 | 61 (59%) | 12 (75%) | 49 (56%) | 0·15 | 263 (39%) | <0·0001 |
| Ethnicity§ | ||||||||||
| Mixed ancestry | 158 (21%) | 93 (22%) | 70 (20%) | 0·49 | 31 (30%) | 4 (25%) | 27 (31%) | 0·65 | 132 (19%) | 0·014 |
| Black | 618 (79%) | 334 (78%) | 284 (80%) | 0·52 | 72 (69%) | 11 (69%) | 61 (69%) | 0·96 | 546 (80%) | 0·013 |
| Tobacco smoker, past or current | 372 (47%) | 239 (56%) | 133 (37%) | <0·0001 | 64 (62%) | 9 (56%) | 55 (63%) | 0·64 | 308 (45%) | <0·0001 |
|
| ||||||||||
| Clinical | ||||||||||
|
| ||||||||||
| Number of W4SS symptoms (0–4) | 1 (0–2) | 2 (1–2) | .. | .. | 2 (1–3) | 3 (2–3) | 2 (1–3) | 0·17 | 1 (0–2) | <0·0001 |
| Able to expectorate at least one sputum sample of ≥1 mL | 80/121¶ (66%) | 47/79 (59%) | 33/42 (79%) | 0·035 | 16/24 (67%) | 3/5 (60%) | 13/19 (68%) | 0·72 | 64/97 (66%) | 0·95 |
| Culture-positive | 104 (13%) | 78 (18%) | 26 (7%) | <0·0001 | 104 (100%) | 16 (100%) | 88 (100%) | >0·99 | .. | .. |
| Time-to-positivity, days | 13 (9–18) | 12 (9–15) | 15 (12–21) | 0·013 | 13 (9–18) | 6 (5–7) | 13 (11–19) | <0·0001 | .. | .. |
| Previous tuberculosis | 112 (14%) | 69 (16%) | 43 (12%) | 0·11 | 16 (15%) | 3 (19%) | 13 (15%) | 0·69 | 96 (14%) | 0·72 |
| CD4, cells/μL | 299 (170–483) | 263 (110–428) | 348 (227–521) | <0·0001 | 193 (60–285) | 199 (45–257) | 192 (61–285) | 0·89 | 326 (197–506) | <0·0001 |
| C-reactive protein, mg/L | 7 (3–34) | 14 (3–77) | 4 (3–11) | <0·0001 | 72 (14–150) | 125 (101–286) | 45 (10–139) | <0·0001 | 6 (3–20) | <0·0001 |
| Haemoglobin, g/dL | 13 (12–14) | 13 (11–14) | 14 (12–15) | <0·0001 | 12 (10–13) | 10 (10–13) | 12 (10–13) | 0·31 | 13 (12–14) | <0·0001 |
Data are median (IQR) or n (%). W4SS=WHO-recommended four-symptom screen. p values for comparisons by
W4SS
smear, or
culture statuses.
Eight participants self-reported as other ethnicity.
121 participants had it recorded whether their sputum was expectorated or induced. Missing or not done (overall): C-reactive protein (n=62), CD4 count (n=4; n=47 older than 3 months), and haemoglobin (n=190).
The sensitivity and specificity for cough (for any duration), cough for 2 weeks or more, and W4SS were recorded: cough (any): 53% sensitivity (95% CI 42–62; 54/104) and 73% specificity (72–79; 524/696); cough for 2 weeks or more: 42% sensitivity (33–53; 44/104) and 83% specificity (81–86; 579/696); W4SS: 77% sensitivity (68–85; 80/104) and 48% specificity (45–52; 334/696; table 2). Point-of-care CRP testing was feasible: 5 (1%) of 835 tests were non-actionable, all resolved upon re-testing, and all prospective results were generated within 3 min of a finger prick. Sensitivity and specificity for CRP5 were 86% (95% CI 78–92; 89/104) and 49% (46–53; 339/696), and for CRP10 were 77% (68–85; 80/104) and 64% (61–68; 445/696). Sensitivity at both thresholds was decreased in participants with CD4 counts of more than 350 cells/μL versus 350 cells/μL or less, and specificity increased. When comparisons were limited to smear-negatives or included participants that did not have head-to-head data, similar patterns were observed (appendix 1 pp 12–17). Haemoglobin (<10 g/dL) had 31% (95% CI 22–43; 25/81) sensitivity and 88% (86–91; 487/553) specificity, displaying similar trends to CRP across CD4 count strata (appendix 1 p 19).
Table 2:
Head-to-head diagnostic accuracy of cough, W4SS, and CRP alone or in combination (parallel or sequential algorithms) stratified by CD4 count
| All participants (n=800) |
CD4 count of >350 cells/μL (n=312) |
CD4 count of ≤350 cells/μL (n=441) |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sensitivity | Specificity | PPV | NPV | Sensitivity | Specificity | PPV | NPV | Sensitivity | Specificity | PPV | NPV | |
| Individual biomarkers | ||||||||||||
|
| ||||||||||||
| Cough (any) | 53% (42–62); 54/104 | 73% (72–79); 524/696 | 22% (18–29); 54/226 | 91% (89–94); 524/574 | 39% (18–65); 7/18 | 80% (75–85); 235/294 | 11% (5–21); 7/66 | 96% (93–98); 235/246 | 54% (43–66); 41/76; p=0·25* | 67% (63–72); 245/365; p<0·0001* | 25% (19–33); 41/161; p=0·013* | 88% (84–92); 245/280; p<0·0001* |
| Cough for ≥2 weeks | 42% (33–53); 44/104 | 83% (81–86); 579/696 | 27% (21–35); 44/161 | 91% (89–93); 579/639 | 28% (10–54); 5/18 | 88% (84–92); 258/294 | 12% (5–27); 5/41 | 95% (92–98); 258/271 | 43% (33–56); 33/76; p=0·22* | 81% (77–85); 294/365; p=0·013* | 32% (23–42); 33/104; p=0·016* | 87% (84–91); 294/337; p=0·001* |
| Cough (any) vs cough for ≥2 weeks | p=0·13 | p<0·0001 | p=0·26 | p=0·74 | p=0·48 | p=0·010 | p=0·80 | p=0·86 | p=0·19 | p<0·0001 | p=0·27 | p=0·93 |
| W4SS | 77% (68–85); 80/104 | 48% (45–52); 334/696 | 18% (15–23); 80/442 | 93% (91–96); 334/358 | 50% (27–74); 9/18 | 54% (49–61); 160/294 | 6% (3–12); 9/143 | 95% (91–98); 160/169 | 82% (72–90); 62/76; p=0·0051* | 44% (39–50); 161/365; p=0·0085* | 23% (19–29); 62/266; p<0·0001* | 92% (87–96); 161/175; p=0·32* |
| CRP5 | 86% (78–92); 89/104 | 49% (46–53); 339/696 | 20% (17–24); 89/446 | 96% (94–98); 339/354 | 67% (41–87); 12/18 | 60% (55–66); 176/294 | 9% (5–16); 12/130 | 97% (93–99); 176/182 | 89% (81–96); 68/76; p=0·089* | 41% (36–46); 148/365; p=0·75* | 24% (20–30); 68/285; p<0·0001* | 95% (91–98); 148/156; p=0·33* |
| Cough (any) vs CRP | p<0·0001 | p<0·0001 | p=0·44 | p=0·0095 | p=0·095 | p<0·0001 | p=0·76 | p=0·54 | p<0·0001 | p<0·0001 | p=0·72 | p=0·013 |
| W4SS vs CRP | p=0·10 | p=0·79 | p=0·48 | p=0·15 | p=0·31 | p=0·18 | p=0·36 | p=0·35 | p=0·67 | p<0·0001 | p=0·11 | p=0·28 |
| CRP10 | 77% (68–85); 80/104 | 64% (61–68); 445/696 | 24% (20–30); 80/331 | 95% (93–97); 445/469 | 39% (18–65); 7/18 | 72% (67–77); 211/294 | 8% (4–16); 7/90 | 95% (92–98); 211/222 | 84% (75–92); 64/76; p<0·0001* | 59% (54–64); 214/365; p<0·0001* | 30% (24–37); 64/215; p<0·0001* | 95% (91–98); 214/226; p=0’94* |
| Cough (any) vs CRP | p<0·0001 | p<0·0001 | p=0·63 | p=0·021 | p>0·99 | p=0·021 | p=0·54 | p=0·81 | p<0·0001 | p=0·018 | p=0·36 | p=0·0056 |
| W4SS vs CRP | p>0·99 | p<0·0001 | p=0·039 | p=0·33 | p=0·50 | p<0·0001 | p=0·66 | p=0·87 | p=0·17 | p=0·33 | p=0·88 | p=0·29 |
| CRP5 vs CRP10 | p=0·39 | p<0·0001 | p=0·16 | p=0·56 | p=0·095 | p=0·0023 | p=0·71 | p=0·41 | p=0·34 | p<0·0001 | p=0·14 | p=0·93 |
|
| ||||||||||||
| Triage positive for each Algorithm if † | ||||||||||||
|
| ||||||||||||
| Algorithm 1: W4SS-positive or CRP10-positive, or both | 85 (77–91); 88/104 | 36 (33–40); 251/696 | 17 (14–20); 88/533 | 95 (91–97); 251/267 | 56 (31–79); 10/18 | 36 (38–50); 127/294 | 6 (3–11); 10/177 | 94 (89–98); 127/135 | 91 (82–97); 69/76 | 36 (28–37); 116/365 | 22 (18–27); 69/318 | 94 (89–98); 116/123 |
| Algorithm 2: W4SS-positive or CRP5-positive, or both | 90 (84–96); 94/104 | 28 (25–32); 196/696 | 16 (13–20); 94/594 | 95 (92–98); 196/206 | 72 (47–91); 13/18 | 36 (31–43); 107/294 | 7 (4–11); 13/200 | 96 (90–99); 107/112 | 95 (88–99); 72/76 | 23 (19–28); 84/365 | 20 (17–25); 72/353 | 95 (89–99); 84/88 |
| Algorithm 3: if W4SS-positive, CRP10-positive | 69 (60–78); 72/104 | 76 (73–79); 528/696 | 30 (25–37); 72/240 | 94 (93–97); 528/560 | 33 (14–60); 6/18 | 83 (79–88); 244/294 | 11 (5–22); 6/56 | 95 (92–98); 244/256 | 75 (64–85); 57/76 | 71 (67–76); 259/365 | 35 (28–43); 57/163 | 93 (90–96); 259/278 |
| Algorithm 4: if W4SS-positive, CRP5-positive | 72 (63–81); 75/104 | 69 (65–72); 477/696 | 26 (21–31); 75/294 | 94 (92–97); 477/506 | 44 (22–70); 8/18 | 78 (73–83); 229/294 | 11 (5–21); 8/73 | 96 (93–98); 229/239 | 76 (66–86); 58/76 | 62 (57–67); 225/365 | 29 (24–37); 58/198 | 93 (89–96); 225/243 |
Data are %, 95% CI, and n/N. Cough (for any duration or for ≥2 weeks) had decreased sensitivity versus W4SS, but W4SS had sub-optimal specificity. In participants with CD4 count of more than 350 cells/μL, only the CRP10 threshold (and not CRP5) had increased specificity versus W4SS, whereas in participants with CD4 counts of 350 cells/μL or less, CRP5 (and not CRP10) had increased specificity versus W4SS. CRP10 had increased specificity versus CRP5 within each CD4 count stratum without a sensitivity decrement. A sequential W4SS-CRP approach (Algorithm 3) resulted in a more balanced sensitivity and specificity, whereas a parallel approach (Algorithm 1) resulted in higher sensitivity but lower specificity. Each algorithm’s sensitivity increased at lower CD4 counts compared with higher counts, whereas specificity decreased. Participants who, in the triage positive for each algorithm section, do not meet the definition of triage-positive are triage-negative. CRP=C-reactive protein. CRP5=CRP≥5mg/L. CRP10=CRP≥10mg/L. NPV=negative predictive value. PPV=positive predictive value. W4SS=WHO-recommended four-symptom screen.
p values comparing data across CD4 count strata; missing data: no CD4 within 3 months (n=47).
Other algorithms including Algorithm 5 (W4SS-positive or, if W4SS-negative, CRP10-positive) and Algorithm 7 (W4SS-positive or, if W4SS-negative, CRP5-positive) had the same performances as Algorithm 1, whereas Algorithms 6 (CRP10-positive or, if CRP10-negative, W4SS-positive) and Algorithm 8 (CRP5-positive or, if CRP5-negative, W4SS-positive) had the same performance as Algorithm 2.
W4SS and CRP receiver operating characteristic (ROC) curves are in appendix 1 (pp 7–8). Areas under the ROC curves (AUROCs) were 0·78 (95% CI 0·73–0·83) for CRP, 0·70 (0·64–0·75) for W4SS, and 0·70 (0·64–0·75) for haemoglobin. Among participants with CD4 counts of 350 cells/μL or less, higher AUROCs occurred for each biomarker than in those with more than 350 cells/μL. AUROCs were also higher for those that were W4SS-positive versus those that were W4SS-negative. We next assessed biomarker performance under different scenarios and identified corresponding thresholds. At a rule-out threshold with sensitivity prioritised (about 95%), CRP (>3 mg/L; CRP3) sensitivity was similar to the WHO-recommended CRP5 (89% [95% CI 83–94] vs 86% [78–92]) and specificity diminished (39% [36–42] vs 49% [45–53]), suggesting CRP3 offers small sensitivity improvements at large specificity costs. CRP and haemoglobin had small variations in rule-out thresholds across CD4 cell count and W4SS strata. Sensitivities and specificities under rule-in and Youden’s index scenarios are in appendix 1 (p 20).
Each biomarker’s sensitivity decreased in W4SS-negatives compared with W4SS-positives, whereas specificity increased (appendix 1 pp 12, 15). Triage test combinations were assessed as part of algorithms (table 2). Algorithm 3 (triage positive if W4SS-positive first, then CRP10-positive) had similar sensitivity (69% [95% CI 60–78]; 72/104) to W4SS (77% [68–85]; 80/104) and CRP10 (77% [68–85]; 80/104) individually, but specificity improved compared with each biomarker (76% [74–81; 528/696] vs 48% [45–52; 334/696] for W4SS or 64% [61–68; 445/696] for CRP10; table 2, 3). We did not include haemoglobin in algorithms as it was done in a programmatically selected subset.
Table 3:
Head-to-head diagnostic accuracy of Xpert and Xpert Ultra on sputum for tuberculosis stratified by W4SS status
| All participants (n=787) | W4SS-negative (n=357) | W4SS-positive (n=430) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sensitivity | Specificity | PPV | NPV | Sensitivity | Specificity | PPV | NPV | Sensitivity | Specificity | PPV | NPV | |
| Smear microscopy | 15% (9–24); 16/104 | 99% (98–100); 678/683 | 76% (53–92); 16/21 | 89% (86–91); 678/766 | 12% (3–31); 3/26 | 100% (99–100); 331/331 | 100% (30–100); 3/3 | 94% (91–96); 331/354 | 17% (10–27); 13/78; p=0·53* | 99% (97–100); 347/352; p=0·030* | 73% (47–91); 13/18; p=0·29* | 85% (81–88); 347/412; p<0·0001* |
| Xpert | 56% (46–66); 58/104 | 99% (98–100); 676/683 | 89% (79–96); 58/65 | 94% (92–95); 676/722 | 27% (12–48); 7/26 | 100% (98–100); 328/331 | 70% (35–94); 7/10 | 95% (92–97); 328/347 | 66% (54–76); 51/78; p=0·0010* | 99% (98–100); 348/352; p=0·77* | 93% (83–98); 51/55; p=0·033* | 93% (90–96); 348/375; p=0’34* |
| Smear vs Xpert | p<0·0001 | p=0·56 | p=0·13 | p=0·001 | p=0·16 | p=0·083 | p=0·28 | p=0·57 | p<0·0001 | p=0·74 | p=0·022 | p<0·0001 |
| Ultra-negative | 10% (2–27); 3/30 | 99% (98–100); 663/668 | 38% (9–76); 3/8 | 96% (94–97); 663/690 | 0% (0–24); 0/14 | 100% (98–100); 324/327 | 0% (0–71); 0/3 | 96% (94–98); 324/338 | 19% (5–46); 3/16; p=0·088* | 100% (98–100); 339/341; p=0·62* | 60% (15–95); 3/5; p=0·090* | 97% (94–99); 339/352; p=0·76* |
| Xpert Ultra | 71% (61–80); 74/104 | 98% (96–99); 668/683 | 83% (74–90); 74/89 | 96% (94–97); 668/698 | 47% (27–67); 12/26 | 99% (97–100); 327/331 | 75% (48–93); 12/16 | 96% (94–98); 327/341 | 80% (69–88); 62/78; p=0·001* | 97% (95–99); 341/352; p=0·088* | 85% (75–93); 62/73; p=0·337* | 96% (93–98); 341/357; p=0·807* |
| Xpert vs Xpert Ultra | p<0·0001 | p<0·0001 | p=0·29 | p=0·083 | p=0·15 | p=0·70 | p=0·78 | p=0·40 | p=0·049 | p=0·44 | p=0·18 | p=0·12 |
| Xpert-negative | 41% (27–57); 19/46 | 98% (97–99); 663/676 | 59% (41–76); 19/32 | 96% (94–97); 663/690 | 27% (10–52); 5/19 | 99% (97–100); 324/328 | 56% (22–87); 5/9 | 96% (94–98); 324/338 | 52% (32–72); 14/27; p=0·083* | 98% (96–99); 339/348; p=0·20* | 61% (39–81); 14/23; p=0·78* | 97% (94–99); 339/352; p=0·76* |
Data are %, 95% CI, and n/N. Xpert Ultra’s sensitivity was higher than Xpert, with Xpert Ultra detecting W4SS-negatives missed by Xpert. Xpert Ultra specificity decreased compared with Xpert. Sensitivities were higher in W4SS-positives than W4SS-negatives. NPV=negative predictive value. PPV=positive predictive value. W4SS=WHO-recommended four-symptom screen. Xpert Ultra=Xpert MTB/RIF Ultra. Xpert=Xpert MTB/RIF.
p values comparing data across W4SS strata.
No point estimate had the minimum WHO triage test sensitivity target of 90%; however, the CRP5 and Algorithm 2 95% CIs (86% [95% CI 78–92; 89/104] and 90% [84–96; 94/104], respectively) overlapped with 90%. Algorithms 3 and 4 specificity estimates (76% [73–79; 528/696] and 69% [65–72; 477/696], respectively) overlapped with the WHO minimum triage test specificity target (70%; figure 3A; table 2).
Figure 3: Summaries of triage and confirmatory test performance and effect on NNT.
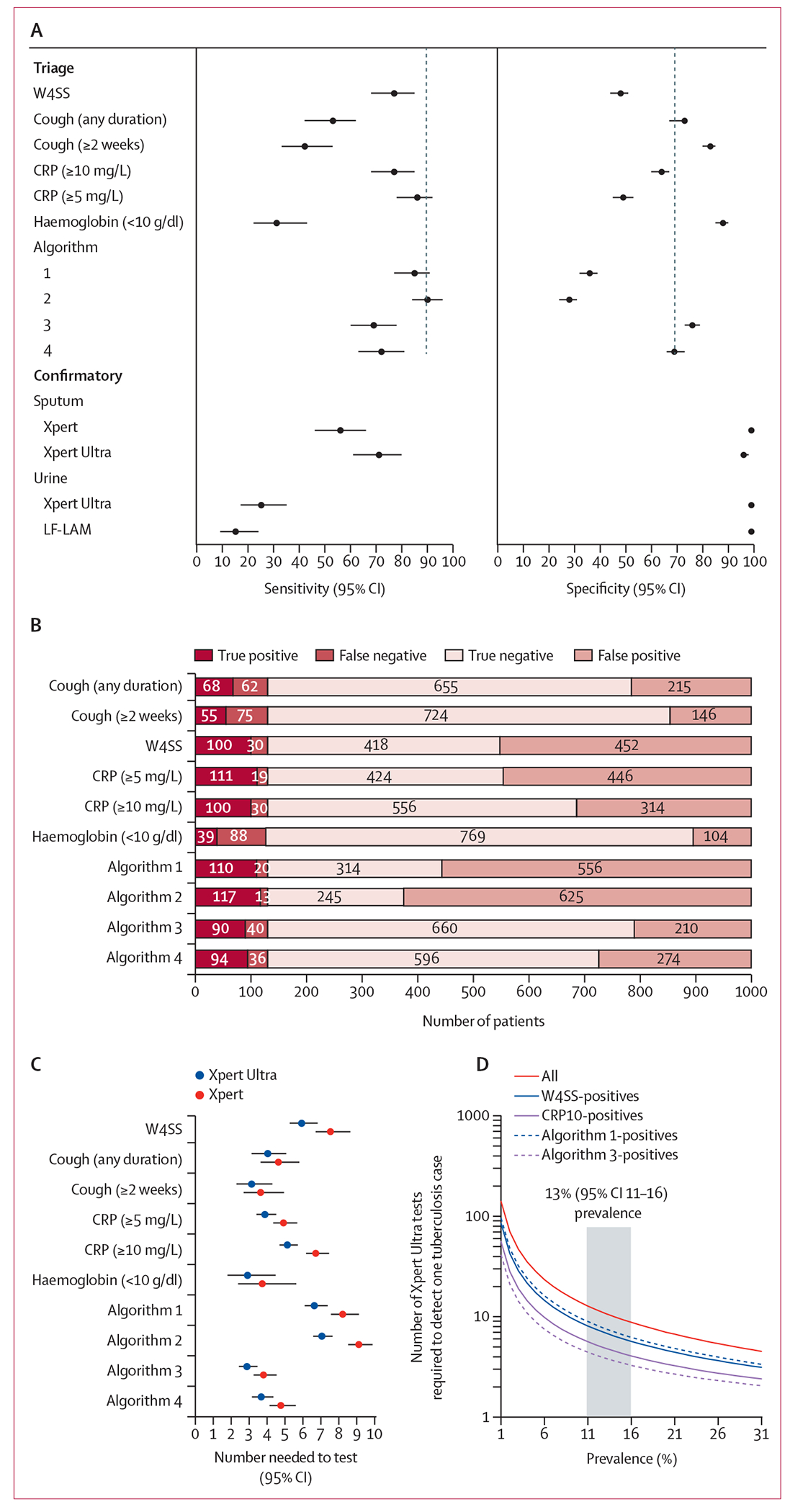
(A) Forrest plots comparing sensitivity and specificity point estimates (with 95% CIs) of triage tests and algorithms as well as sputum and urine confirmatory tests. Black dashed vertical lines indicate WHO target product profile estimates. (B) Effect of different triage tests and algorithms (table 2) on participant classification, showing CRP10 alone or in combination with W4SS (Algorithms 3 and 4) to result in fewer onward unnecessary referrals (false positives). CRP10 had 138 fewer false positives per 1000 people than W4SS (452 vs 314). The number needed to test to detect one culture-positive case when different triage methods are used in our cohort (C) and for Xpert Ultra only (D), modelled against different tuberculosis prevalences versus an Xpert Ultra-in-all scenario. Use of CRP10 would result in a tuberculosis case correctly detected every five rather than every seven people as for W4SS. Other triage methods had lower NNTs but would be offset by diminished sensitivities. The grey column in D shows our prevalence. CRP=C-reactive protein. LF-LAM=lateral flow lipoarabinomannan (Determine TB LAM Ag test). Xpert=Xpert MTB/RIF. Xpert Ultra=Xpert MTB/RIF Ultra. W4SS=WHO-recommended four-symptom screen.
The effect of individual tests and algorithms on people classified as positive or negative in a hypothetical cohort of 1000 people at the prevalence in our cohort is shown (figure 3B), as well as how these translate into different numbers of people needing downstream confirmatory testing (number needed to test [NNT]) as a function of prevalence (figure 3C–D). Haemoglobin resulted in the smallest number of unnecessary referrals or false-positives, but this was at the expense of few people with tuberculosis correctly referred (true-positives; figure 3B). CRP10 reduced unnecessary testing more than W4SS (138 [117–161] fewer false-positive referrals), and this was enhanced further by a combination strategy also involving W4SS (Algorithms 3 and 4), but this resulted in fewer people with tuberculosis correctly referred. Briefly, compared with W4SS, CRP10 would reduce Xpert Ultra NNT from 6·91 (95% CI 6·25–7·81) to 4·87 (4·41-5·51). Improvements offered by CRP10 versus W4SS remained relatively consistent across prevalences, with the NNT of triage strategies plateauing beyond 30% prevalence.
Xpert Ultra and Xpert non-actionable results rates were low (1% [5/811] for Xpert Ultra and 1% [2/883] for Xpert). Xpert Ultra had higher sensitivity than Xpert overall (71% [95% CI 61–80]; 74/104) vs 56% [46–66]; 58/104) and in smear-negatives (67% [56–77; 59/88] vs 50% [39–61; 44/88]), and lower specificity (98% [96–99; 668/683] vs 99% [98–100; 676/683]; table 3; appendix 1 p 22). The NNT for Xpert Ultra to detect a person with culture-confirmed tuberculosis was less than Xpert (4·87 [95% CI 4·41–5·51] vs 5·90 [5·34–6·67] in CRP10-positives, for example). Forest plots that include all confirmatory tests are provided (figure 3A), and Euler diagrams showing overlap between confirmatory tests are in appendix 1 (p 10).
Xpert Ultra had decreased sensitivity in people with CD4 counts of more 350 cells/μL versus those with counts of 350 cells/μL or less (42% [95% CI 22–70; 8/18] vs 77% (66–86; 61/79]) and similar specificity (99% [97–100; 305/309] vs 97% (95–99; 381/393]; appendix 1 p 21). Within CD4 count strata, Xpert Ultra sensitivity was higher than Xpert in people with counts of 350 cells/μL or less (77% [66–86; 61/79] vs 60% [48–71; 46/77]) and specificity was (97% [95–99; 730/760] vs 99% [98–100; 677/684]). Similar trends were observed in non-head-to-head data (appendix 1 p 23).
Xpert Ultra’s sensitivity differed from Xpert’s in W4SS-positives (80% [69–88; 62/78] vs 66% [54–76; 51/78]; table 3). Trends in Xpert Ultra and Xpert comparative accuracy were similar in smear-negative people and non-head-to-head data (appendix 1 pp 22, 25). Specificities between Xpert Ultra and Xpert were similar (irrespective of W4SS status) and Xpert Ultra specificity did not differ by previous tuberculosis status (97% [91–99; 93/96] vs 98% [97–99; 575/587]; p=0·503; appendix 1 p 25). Xpert specificity among people with no previous tuberculosis was 90% (79–97) vs 99% (94–100); p=0·99.
Reclassifying Xpert Ultra trace results from positive-to-negative resulted in similar sensitivity (71% [95% CI 61–80; 74/104) vs 66% [57–76; 69/104]; difference of −5% [−10 to 0]; p=0·45]) and increased specificity (98% [96–99; 668/683] vs 99% [99–100; 679/683]; difference of 1% [1 to 3]; p=0·011; appendix 1 p 26). Trace exclusion had a similar effect. Conclusions were unchanged when stratified by previous tuberculosis status.
Xpert Ultra results on concentrated urine were frequently non-actionable (118 [13%] of 890 participants). When an unconcentrated aliquot was tested, 117 (99%) of 118 of non-actionables were resolved and 13 (11%) of 117 were positive (appendix 1 pp 6, 11).
Of the concentrated urine Xpert Ultra positives, 23 (73%) of 33 had a corresponding unconcentrated urine test Xpert Ultra positive (appendix 1 p 11). Among the 732 participants with HIV in the confirmatory urine test analysis, Xpert Ultra and LF-LAM had low sensitivities (25% [95% CI 17–35] vs 15% [9–24]; p=0·070) and high specificities (99% [98–100] vs 99% [98–100]; p=0·25; figure 2C; appendix 1 p 27).
Xpert Ultra and LF-LAM overlap was high, with each test infrequently giving the only positive non-sputum result (5 [3%] of 129 for Xpert and 9 [7%] of 129 for LF-LAM; appendix 1 p 9). Five participants with culturenegative results were urine Xpert Ultra-positive and, of these, four had a later sputum-based programmatic diagnosis of tuberculosis within a year of recruitment (one had no data).
Among participants in whom it was known whether sputum induction was necessary, 49 (31%) of 158 were sputum scarce (11 W4SS negative and 38 W4SS positive), of which 10 (20%) of 49 were culture positive (all W4SS positive); similar to the culture positivity rate among participants who could expectorate (15 [14%] of 109; appendix 1 p 29).
152 (17%) of 897 participants had at least one positive sputum (culture, Xpert, and Xpert Ultra) or urine (Xpert Ultra and LF-LAM) result. When limited to participants in which each test result was available and the absence of a result was not due to human error or stock outs, the proportion of any-test-positive was 18% (145/804; 95% CI 15–21). Of those with any positive results, sputum tests had highest yields: 71% (103/145; 63–78) for culture (34% 36/107; 25–43] for single culture-positive), 61% (89/145; 53–69) for Xpert Ultra, and 44% (64/145; 36–53) for Xpert. Urine tests had lower yields with 30% (43/145; 22–38) and 17% (25/145; 12–24) positive by Xpert Ultra and LF-LAM, respectively.
Of the 158 participants in whom it was known whether expectorated sputum could be produced before induction was done, 126 (80%) had all the above tests attempted. 29 (23%) of 126 had one or more positive results. Of these, some had sputum (expectorated or induced) positive by culture (24 [83%] of 29), Xpert Ultra (19 [66%] of 29), and Xpert (17 [59%] of 29). Some had urine positive by Xpert Ultra (10 [34%] of 29) and LF-LAM (7 [24%] of 29). 42 (33%) of 126 were unable to naturally expectorate and induction was their only source of sputum. If induction was not done, yields of the aforementioned sputum tests would decrease to 55% (16/29; 36–74) for culture, 45% (13/29; 26–64) for Xpert Ultra, and 38% (11/29; 21–58) for Xpert. Comparisons of tests did not show yield and sensitivity and specificity differences when results from expectorated or induced sputum were compared (appendix 1 p 30).
Discussion
In our prospective, cross-sectional, diagnostic accuracy study, the key findings are: (1) CRP testing is feasible at point-of-care and superior to W4SS for triaging those initiating ART, reducing unnecessary referrals, improving NNT, and approaching but not capable of meeting the WHO target product profile minimum sensitivity and specificity benchmarks; (2) sequential triage algorithms combining W4SS and CRP (both positive) approach the WHO-recommended optimal specificity target but result in more missed tuberculosis than CRP alone; (3) sputum Xpert Ultra is more sensitive than Xpert for culture-confirmed tuberculosis, which is common in both W4SS-negative and sputum-scarce people; (4) offering sputum induction enhances diagnostic yield (beyond that possible using urine tests); and (5) urine testing with Xpert Ultra and LF-LAM have similar performance and urine testing with Xpert Ultra is hampered by high non-actionable result rates. These data can inform triage and confirmatory testing strategies, including specimen acquisition, in those initiating ART.
People with HIV should be screened for tuberculosis, a process most efficient at ART initiation when patients are immunosuppressed and have relatively high pre-test probability of tuberculosis. ART initiators are already within HIV treatment cascades, representing a population captured in a setting in which that we now show point-of-care CRP is technologically highly feasible and a better alternative to symptom based triage, significantly reducing unnecessary onward referrals (from 452 [for W4SS] to 314 people per 1000 using CRP10, translating into an NNT reduction of about 7 to about 5). In line with WHO guidance,14 our data also informs CRP’s use at this threshold of 10 mg/L or more, with lower thresholds negating benefits CRP has over W4SS (fewer unnecessary referrals). In contrast to community-based evaluations of CRP,24 we show little added benefit of combining W4SS with CRP in those initiating ART, unless large tuberculosis case detection reductions are acceptable, and that haemoglobin should not be considered further due to low sensitivity.
Xpert Ultra, despite being WHO-recommended, is unevaluated in those initiating ART without syndromic preselection. We now provide the first data to support the use of Xpert Ultra in this diagnostically challenging population versus the previous generation Xpert, in which sensitivity was higher for Xpert Ultra (71% vs 56%), including in people who did not meet the W4SS criteria and in people with advanced HIV (CD4 counts <350 cells/μL). This supports Xpert Ultra’s use as part of recent efforts to move beyond symptom criteria in high burden settings.25 Encouragingly, we did not observe drastically reduced Xpert Ultra specificity in contrast to previous research in our setting; however, this previous research was in self-reporting people with presumptive tuberculosis, who have a higher rate of previous tuberculosis than ART initiators without preselection based on symptoms.22
Although non-sputum tests are a priority for tuberculosis control, there are, unfortunately, unlikely to be alternatives available in the short-term for bacteriological confirmatory testing.2 Furthermore, countries are heavily invested in sputum-based testing infrastructure and might wish to consider induction, which our data support the use of: 31% of people required induction to make at least one sputum and, without induction, the proportion of people with a positive confirmatory test result detected by Xpert Ultra would reduce (66% to 45%). We therefore recommend that programmes consider making sputum induction available at ART-initiation sites, which is currently not standard-of-care in most high-burden settings, including South Africa. This is especially important given the relatively poor performance of urine tests (although LF-LAM has low diagnostic yield, it should still be available in line with current guidance, due to its low cost, point-of-care nature and, unlike urine Xpert Ultra, absence of non-actionable results).26 We also suggest that the use of sputum induction as a diagnostic intervention, and associated implementation challenges (eg, biosafety,27 space, and effect on clinic flows), require further evaluation, because, based on our data, induction improved case detection, compared to no induction, more than Xpert Ultra did relative to Xpert.
This study has strengths and limitations. Although the study was large and included all consenting ambulatory people irrespective of symptoms and ability to expectorate sputum (and is hence representative of those initiating ART in our setting), we did not have information of, and the reasons for, the relatively small number of people who declined participation. Furthermore, the study was at a single centre. This is partly mitigated as it is not a discovery cohort, but a validation of existing design-locked tools, and the centre itself is a referral node for a catchment area serving hundreds of thousands of people. Other strengths include the fact that CRP was done at point-of-care, a two-sputa culture was used as the reference standard, and that we describe sputum production in detail in people with HIV, which informs on the potential effect of induction facilities and non-sputum tests, even though we could only confidently establish if induction was necessary in a consecutive subset of people due to a database error.16,28 Other limitations include haemoglobin being done in a programmatically selected subset, which might bias accuracy estimates. We did not analyse factors other than culture-positive tuberculosis associated with CRP nor extrapulmonary tuberculosis, both of which are important future research questions. The effect of diagnostic strategies on person-important outcomes, cost, cost-effectiveness, and affordability are also important, including in lower-resource settings, and our data now provide a justification to evaluate them. We also suggest that the use of sputum induction as a diagnostic intervention, and associated implementation challenges (eg, biosafety,27 space, and effect on clinic flows), require further evaluation, because, based on our data, induction improved case detection more than Xpert Ultra did relative to Xpert. Lastly, although ours was a diagnostic accuracy study and not an implementation science exercise, shifting away from decades of entrenched W4SS-based triage to point-of-care CRP presents significant implementation challenges,4,29 including promoting the concept that people need testing despite an absence of symptoms, the necessary infrastructure requirements to do point-of-care testing and associated task-shifting (frontline health workers would now do a tuberculosis test). These and other implementation barriers and facilitators require future evaluation.
In summary, point-of-care CRP is an alternative tuberculosis triage tool that shows improved specificity compared with self-reported W4SS in people with HIV initiating ART that reduces unnecessary testing. Xpert Ultra has improved sensitivity compared with Xpert and detects W4SS-negative tuberculosis, and Xpert Ultra’s diagnostic yield is significantly enhanced through sputum induction provision. These tools should be pursued for implementation in this key risk group, together with further evaluations in different populations and settings.
Supplementary Material
Research in context.
Evidence before this study
We searched PubMed, Google Scholar, and Cochrane Library for publications published before June 13, 2023, using the search terms “diagnosis” and “tuberculosis”, “ART initiators”, “people living with HIV”, “CRP”, “Xpert MTB/RIF”, or “Xpert MTB/RIF Ultra”. The identified publications suggested that better triage and confirmatory tests are urgently needed for tuberculosis, especially in key risk groups such as people living with HIV, because many tuberculosis cases do not meet the WHO-recommended four-symptom screen (W4SS) criteria. W4SS had suboptimal specificity and alternative triage approaches, such as C-reactive protein (CRP), showed initial promise but had comparatively little data in those initiating antiretroviral therapy (ART), especially without syndromic preselection, and using point-of-care platforms. We also identified WHO-endorsed rapid molecular tests, such as Xpert MTB/RIF Ultra (Xpert Ultra), to have little supporting data in ART-initiators, who can have sputum scarce and paucibacillary early-stage disease. There was also a scarcity of data on the added value of sputum induction to augment diagnostic sampling for confirmatory testing. Lastly, the performance of urine tests (Xpert Ultra and Determine TB LAM Ag) in this population required more data on diagnostic yield and accuracy.
Added value of this study
We evaluated repurposed and new tests for triage and confirmatory testing using a rigorous microbiological reference standard in a highly vulnerable high-priority patient population (those initiating ART) regardless of symptoms and ability to naturally expectorate sputum. We showed point-of-care CRP triage is feasible, performs better than W4SS, and that combinations of different triage approaches offer no advantages over CRP alone. Sputum samples tested with Xpert Ultra has superior sensitivity to Xpert; often detecting W4SS-negative tuberculosis. Furthermore, without induction, confirmatory sputum-based testing would not be possible in a third of people. Urine tests provided poor results. This study contributed unpublished data to systematic reviews and metaanalyses used by WHO to inform global policy supporting use of CRP triage and Xpert Ultra in people with HIV.
Implications of all the available evidence
Point-of-care CRP triage testing is feasible and superior to W4SS and should be considered for roll-out in those initiating ART in high burden settings, together with sputum induction in people who triage CRP-positive, and after appropriate cost and implementation research. Such people should be offered Xpert Ultra, which outperforms Xpert.
Acknowledgments
The authors thank the participants, Fikiswa Seti, Charmaine Van der Walt, Kim Stanley, and Gian van der Spuy. GT reports funding from the EDCTP2 programme supported by the EU (RIA2018D-2509, PreFIT; RIA2018D-2493, SeroSelectTB; RIA2020I-3305, CAGE-TB) and the National Institutes of Health (D43TW010350, U01AI152087, U54EB027049, and R01AI136894). BWPR and HM acknowledge funding from the Faculty of Medicine and Health Sciences, Stellenbosch University. GN acknowledges funding from the South African Medical Research Council (SAMRC). The work reported herein was made possible through funding by the SAMRC through its Division of Research Capacity Development under the SAMRC Internship Scholarship Programme. The content hereof is the sole responsibility of the authors and does not necessarily represent the official views of the SAMRC.
Declaration of interests
GT received in-kind donations from Cepheid and Boditech.
BWPR received travel support from Cepheid to attend a conference and present unrelated data. RMW declares a salary paid by the South African Medical Research Council. MN declares a research grant funding by the Wellcome Trust and research funding by the UK National Institute for Health and Care Research Biomedical Research Centre at the University College London Hospitals NHS Foundation Trust. CCN declares grants or contracts from the US National Institutes of Health (K43TW012303) and the European and Developing Countries Clinical Trials Partnership (TMA2017CDF-1914) for career development fellowship. The authors have no financial involvement with any organisation or entity with a financial interest in, or financial conflict with, the subject matter or materials discussed in the manuscript apart from those disclosed.
Footnotes
Equitable partnership declaration
The authors of this paper have submitted an equitable partnership declaration (appendix 2). This statement allows researchers to describe how their work engages with researchers, communities, and environments in the countries of study. This statement is part of The Lancet Global Health’s broader goal to decolonise global health.
See Online for appendix 1
See Online for appendix 2
Data sharing
Data will be made available upon request directly from the corresponding author, and will include de-identified participant data (including data dictionaries) and computed variables. The corresponding author and other people involved in the study will examine and review data requests. Ethical and legal implications of data sharing will be considered. Data will be shared based on the outcome of the review.
References
- 1.Kaufmann GR, Furrer H, Ledergerber B, et al. Characteristics, determinants, and clinical relevance of CD4 T cell recovery to <500 cells/μL in HIV type 1-infected individuals receiving potent antiretroviral therapy. Clin Infect Dis 2005; 41: 361–72. [DOI] [PubMed] [Google Scholar]
- 2.Abdulgader SM, Okunola AO, Ndlangalavu G, et al. Diagnosing tuberculosis: what do new technologies allow us to (not) do? Respiration 2022; 101: 797–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Christian CS, Gerdtham U-G, Hompashe D, Smith A, Burger R. Measuring quality gaps in TB screening in South Africa using standardised patient analysis. Int J Environ Res Public Health 2018; 15: 729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yoon C, Dowdy DW, Esmail H, MacPherson P, Schumacher SG. Screening for tuberculosis: time to move beyond symptoms. Lancet Respir Med 2019; 7: 202–04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wong EB. It is time to focus on asymptomatic tuberculosis. Clin Infect Dis 2021; 72: e1044–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.WHO. High-priority target product profiles for new tuberculosis diagnostics: report of a consensus meeting. Geneva: World Health Organization, 2014. [Google Scholar]
- 7.Hanifa Y, Fielding KL, Charalambous S, et al. Tuberculosis among adults starting antiretroviral therapy in South Africa: the need for routine case finding. Int J Tuberc Lung Dis 2012; 16: 1252–59. [DOI] [PubMed] [Google Scholar]
- 8.Shapiro AE, Hong T, Govere S, et al. C-reactive protein as a screening test for HIV-associated pulmonary tuberculosis prior to antiretroviral therapy in South Africa. AIDS 2018; 32: 1811–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yoon C, Semitala FC, Atuhumuza E, et al. Point-of-care C-reactive protein-based tuberculosis screening for people living with HIV: a diagnostic accuracy study. Lancet Infect Dis 2017; 17: 1285–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Calderwood CJ, Reeve BW, Mann T, et al. Clinical utility of C-reactive protein-based triage for presumptive pulmonary tuberculosis in South African adults. J Infect 2023; 86: 24–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Griensven J, Cnops L, De Weggheleire A, Declercq S, Bottieau E. Point-of-care biomarkers to guide antibiotic prescription for acute febrile illness in sub-Saharan Africa: promises and caveats. Open Forum Infect Dis 2020; 7: ofaa260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gelaw Y, Getaneh Z, Melku M. Anemia as a risk factor for tuberculosis: a systematic review and meta-analysis. Environ Health Prev Med 2021; 26: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dhana A, Hamada Y, Kengne AP, et al. Tuberculosis screening among ambulatory people living with HIV: a systematic review and individual participant data meta-analysis. Lancet Infect Dis 2022; 22: 507–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.WHO. WHO consolidated guidelines on tuberculosis: module 2: screening: systematic screening for tuberculosis disease. Geneva: World Health Organization, 2021. [PubMed] [Google Scholar]
- 15.Chakravorty S, Simmons AM, Rowneki M, et al. The new Xpert MTB/RIF Ultra: improving detection of Mycobacterium tuberculosis and resistance to rifampin in an assay suitable for point-of-care testing. M Bio 2017; 8: e00812–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peter JG, Theron G, Singh N, Singh A, Dheda K. Sputum induction to aid diagnosis of smear-negative or sputum-scarce tuberculosis in adults in HIV-endemic settings. Eur Respir J 2014; 43: 185–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nathavitharana RR, Garcia-Basteiro AL, Ruhwald M, Cobelens F, Theron G. Reimagining the status quo: how close are we to rapid sputum-free tuberculosis diagnostics for all? EBioMedicine 2022; 78: 103939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boditech. iChromax CRP (package insert). May 12, 2021. http://web.labindustrias.com/wp-content/uploads/2022/02/ichroma-CRP-Rev.24.pdf (accessed April 4, 2022).
- 19.Provincial Government of the Western Cape. The Western Cape consolidated guidelines for HIV treatment: prevention of mother-to-child transmission of HIV (PMTCT), children, adolescents and adults (amended version 2018). Cape Town: Department of Health, 2020. [Google Scholar]
- 20.Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG. Research electronic data capture (REDCap)—a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform 2009; 42: 377–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pillay S, de Vos M, Derendinger B, et al. Non-actionable results, accuracy, and effect of the first- and second-line line probe assays for diagnosing drug-resistant tuberculosis, including on smear-negative specimens, in a high-volume laboratory. Clin Infect Dis 2023; 76: e920–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mishra H, Reeve BWP, Palmer Z, et al. Xpert MTB/RIF Ultra and Xpert MTB/RIF for diagnosis of tuberculosis in an HIV-endemic setting with a high burden of previous tuberculosis: a two-cohort diagnostic accuracy study. Lancet Respir Med 2020; 8: 368–82. [DOI] [PubMed] [Google Scholar]
- 23.Cohen JF, Korevaar DA, Altman DG, et al. STARD 2015 guidelines for reporting diagnostic accuracy studies: explanation and elaboration. BMJ Open 2a016; 6: e012799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ruperez M, Shanaube K, Mureithi L, et al. Use of point-of-care C-reactive protein testing for screening of tuberculosis in the community in high-burden settings: a prospective, cross-sectional study in Zambia and South Africa. 2023; 11: e704–14. [DOI] [PubMed] [Google Scholar]
- 25.Martinson NA, Nonyane BAS, Genade LP, et al. Evaluating systematic targeted universal testing for tuberculosis in primary care clinics of South Africa: a cluster-randomized trial (The TUTT Trial). PLoS Med 2023; 20: e1004237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.WHO. Lateral flow urine lipoarabinomannan assay (LF-LAM) for the diagnosis of active tuberculosis in people living with HIV: policy update 2019. Geneva: World Health Organization, 2019. [Google Scholar]
- 27.Nivin B, O’Flaherty T, Leibert E, Zhao BY, Driscoll J. Sputum induction problems identified through genetic fingerprinting. Infect Control Hosp Epidemiol 2002; 23: 580–83. [DOI] [PubMed] [Google Scholar]
- 28.Peter JG, Theron G, Pooran A, Thomas J, Pascoe M, Dheda K. Comparison of two methods for acquisition of sputum samples for diagnosis of suspected tuberculosis in smear-negative or sputum-scarce people: a randomised controlled trial. Lancet Respir Med 2013; 1: 471–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.WHO. Rapid communication on systematic screening for tuberculosis. Geneva: World Health Organization, 2020. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available upon request directly from the corresponding author, and will include de-identified participant data (including data dictionaries) and computed variables. The corresponding author and other people involved in the study will examine and review data requests. Ethical and legal implications of data sharing will be considered. Data will be shared based on the outcome of the review.


