Abstract
In every latently Epstein-Barr virus-infected cell the viral genes EBER-1 and EBER-2 are transcribed by polymerase III. In lytically infected cells in vivo the EBER genes could not be detected. However, in cell culture downregulation could not be confirmed, and hence the relevance of this shutdown to the replication of the virus was not clear. We assayed the transcriptional activity of the EBER genes by nuclear run-on assays with enriched lytically infected cells and demonstrated that EBER-1 and EBER-2 are differentially downregulated on the transcriptional level during the switch to lytic viral replication. This downregulation was an early event during the lytic replication of the virus.
During latency of the Epstein-Barr virus (EBV), up to 11 viral genes are expressed that encode up to nine proteins. Two of these genes, EBER-1 and EBER-2 (for a review, see reference 3), are transcribed by polymerase III (9). The transcripts are 167 and 172 bp in length, respectively, have neither a 5′ cap nor a 3′ poly(A) tail, show extensive secondary structure, and obviously do not encode proteins. Due to the large number (up to 107) of copies per cell (11), these RNAs are the most abundant transcripts in latently EBV-infected cells.
The function of these viral RNAs is unclear. In different reports on the localization of the EBERs, these RNAs were found either in the nucleus (10) or near the nuclear membrane in the cytoplasm (18), obviously tightly associated with the polyribosomes. On the basis of the homology of these RNAs to the small untranslated RNAs VAI and VAII of adenovirus (15), a highly plausible role for the EBERs is the modulation of interferon-mediated antiviral responses by binding to and inactivation of the interferon-induced DAI kinase (19–21). Furthermore, binding of the EBERs to ribosomal protein L22 was shown (23), suggesting a role in the regulation of translation. Finally, a mitogenic effect of the EBERs was described when these RNAs were transiently expressed in primary B cells, leading to an increase in protein synthesis (26).
The EBERs were found to be transcribed in every latently infected cell. Reports on expression during lytic replication, however, are controversial. In all cases of oral hairy leukoplakia (5) and in some cases of nasopharyngeal carcinoma (25) in which lytic replication of EBV was detected, no EBERs could be demonstrated by in situ hybridization. Similar results were reported in some cases of well-differentiated squamous cell carcinoma with EBV latency type II (13). Recently, we reported a case of chronic active EBV infection in which the EBV-positive lymphocytes in the peripheral blood were EBER negative (12). However, in cell culture induction of viral lytic replication by phorbol 12-myristate 13-acetate (TPA) and butyric acid (BA) did not result in a reduction of the EBERs when assayed by Northern blot analysis (8, 24). Therefore, it remained unclear whether the absence of the EBERs in some situations in vivo was related to pathogenesis or to a principal mechanism of the lytic cycle.
In this study we employed a new experimental approach in cell culture to clarify whether the expression of the EBER genes is regulated during the switch from latency to lytic viral replication. We performed nuclear run-on assays to measure the actual activity of the EBER genes in order to preclude a masking effect of stable EBERs derived from the latent phase. To avoid falsification of the results by contaminating latent cells, which transcribe vast amounts of EBERs, we enriched the population of lytically infected cells.
Induction of lytic replication of EBV.
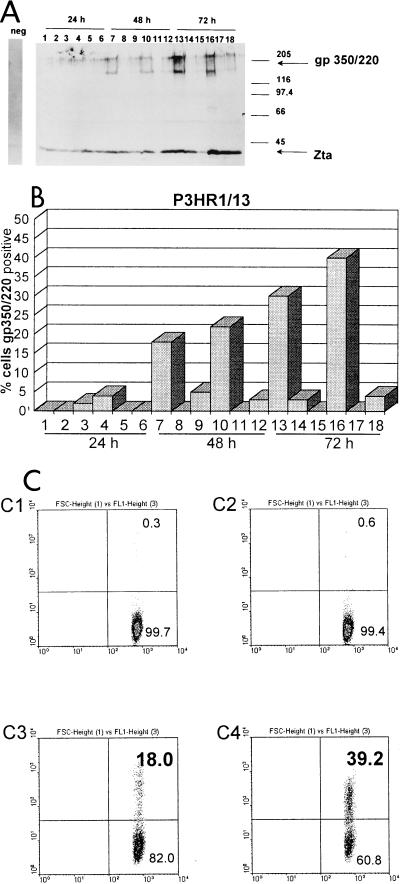
We tested several methods of chemical treatment for their abilities to induce lytic replication of EBV in cell culture. The cells were diluted with fresh medium to a concentration of 5 × 106/ml, and 24 h later TPA (final concentration, 40 ng/ml) and BA (final concentration, 3 mM) or transforming growth factor β (TGF-β) (final concentration, 50 μl/ml, activated from fetal calf serum by incubation with 1/10 volume of 2 N NaOH and neutralization with 2 N HCl) or 5-iodo-2′-deoxyuridine (IUdR) (final concentration, 50 μg/ml), either individually or in combination (1), were added and incubated for up to 72 h. In Fig. 1 the abilities of the different chemicals to induce lytic replication of EBV in the Burkitt’s lymphoma-derived cell line P3HR1/13 are shown. The proportion of cells entering lytic replication of EBV was measured with immunohistochemistry (Fig. 1A), immunofluorescence assays (Fig. 1B), and fluorescence-activated cell sorting (FACS) (Fig. 1C) with the monoclonal antibody BZ1 (Dako, Hamburg, Germany), specific for Zta (ZEBRA, gene product of BZLF-1), and the monoclonal antibody mab-64D7 (cell culture supernatant, ammonium sulfate precipitated and concentrated 10-fold in phosphate-buffered saline [PBS]), specific for the late-lytic-cycle membrane protein gp350/220 (the product of the BLLF-1 gene). Western blotting and immunohistochemistry detected increasing expression of both viral proteins 24 to 72 h after induction (Fig. 1A). Expression of gp350/220 was strongest after 72 h of treatment with TPA, BA, and TGF-β (lanes 13 and 16). Immunofluorescence assays (Fig. 1B, lanes 13 and 16) and FACS (Fig. 1C) confirmed these results, demonstrating approximately 40% of the cells in the late lytic cycle expressing gp350/220 (upper right quadrant of each panel). Hence, for the experiments described below the cells were treated for 72 h with a combination of TPA, BA, and TGF-β.
FIG. 1.
Percentage of lytically infected P3HR1/13 cells induced by various chemical treatments. P3HR1/13 cells were treated with TPA-BA (lanes 1, 7, and 13), IUdR (lanes 2, 8, and 14), TGF-β (lanes 3, 9, and 15), TPA–BA–TGF-β (lanes 4, 10, and 16), IUdR–TGF-β (lanes 5, 11, and 17), or TPA–BA–IUdR–TGF-β (lanes 6, 12, and 18), and the percentage of lytically infected cells was assayed after 24, 48, and 72 h. (A) 5 × 105 cells were harvested at each time point, protein extracts were examined by polyacrylamide gel electrophoresis and Western blotting, and Zta and gp350/220 were detected with monoclonal antibodies. The gp350/220-specific monoclonal antibody mab-64D7 yielded a double band that always migrated more quickly than the standard, probably because of heavy glycosylation. (B) Immunofluorescence assay. The percentage of gp350/220-positive cells is indicated. (C) 5 × 105 cells were harvested at each time point and analyzed by FACS with the gp350/220-specific monoclonal antibody mab-64D7. The cells in the upper right quadrant of each panel were considered positive; those in the lower right quadrant were considered negative. Left panels: C1, untreated DG75; C3, latent P3HR1/13; right panels: C2, chemically treated DG75; C4, treated P3HR1/16. The percentage of gp350/220-positive cells is indicated in the panels. The x axis indicates forward light scatter (FSC); the y axis indicates the intensity of gp350/220-specific fluorescence (FL1).
Purification of cells with lytic replication of EBV.
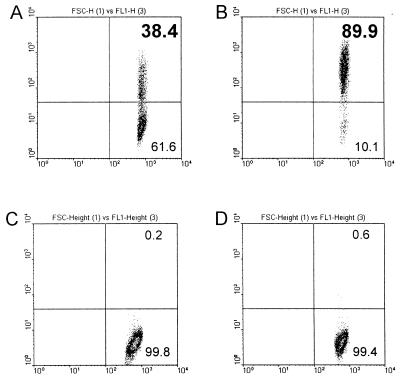
In order to enrich the population of cells with lytically replicating EBV, the viral glycoprotein gp350/220 was used as a physical marker on the cell surface for purification. Separation and purification by FACS or by use of magnetic Dynabeads were not suitable owing to great physical stress leading to damage of the cells, which had already been stressed by the chemical treatment and the lytic replication of the virus. Finally, we succeeded with the MACS system (Miltenyi Biotech, Bergisch Gladbach, Germany) and adjusted the protocol of the manufacturer to our particular situation of working with fragile lytically replicating EBV-infected cells. A quantity (5 × 107) of cells, treated as described above to induce lytic replication of EBV, was pelleted at 350 × g for 10 min at room temperature (RT) and washed twice with PBS-MACS (PBS [without Ca2+ or Mg2+] with 0.5% bovine serum albumin and 2 mM EDTA, pH 7.2). The cells were resuspended in 20 ml of PBS-MACS and incubated for 20 min at RT with 40 μl of the gp350/220-specific mouse monoclonal antibody mab-64D7. After the cells were washed twice with PBS-MACS, they were resuspended in 800 μl of PBS-MACS and incubated for 15 min at RT with 200 μl of a suspension of magnetic beads (MACS system) conjugated with a mouse-immunoglobulin-specific monoclonal antibody. The cells were washed again, and 200 μl of a fluorescein isothiocyanate-conjugated mouse-specific antibody was added and incubated for 10 min at RT in the dark. Finally, the cells were applied to a separation column (type RS+; Miltenyi Biotech) in the magnetic field. The separation column was washed, and the gp350/220-positive cells were eluted according to the manufacturer’s instructions. An aliquot of the purified cells could be used directly for FACS analysis to check the purity. Chemical treatment of the cells and subsequent purification resulted in an approximately 90% pure population of cells in the late phase of lytic replication, positive for the viral gp350/220 glycoprotein (Fig. 2B, upper right quadrant). Prior to the purification, 62% of the cells in the culture were negative for this late-lytic-cycle marker (Fig. 2A, lower right quadrant). A control experiment with the EBV-negative cell line DG75 did not show any significant difference in the signal for gp350/220 prior to and after chemical treatment (Fig. 2C and D, respectively).
FIG. 2.
Purification of lytically infected gp350/220-positive P3HR1/13 cells. P3HR1/13 cells were treated with TPA–BA–TGF-β for 72 h. Lytically infected cells were labeled with the gp350/220-specific mouse monoclonal antibody mab-64D7 and purified with paramagnetic beads (MACS system), coated with a secondary anti-mouse antibody, according to the manufacturer’s instructions. The cells in the upper right quadrant of each panel were considered positive; the cells in the lower right quadrant were considered negative. (A) P3HR1 cells after 72 h of induction, prior to purification and (B) after purification of gp350/220-positive cells. (C) DG75 cells after 72 h of induction, prior to purification and (D) after purification of gp350/220-positive cells. The percentage of gp350/220-positive cells is indicated in the panels. The x axis indicates forward light scatter (FSC); the y axis indicates the intensity of gp350/220-specific fluorescence (FL1).
Transcription of the EBER genes is downregulated during lytic replication.
Regulation of gene expression is achieved by different mechanisms affecting transcription, posttranscriptional processing, degradation, and stability of the mRNA. Previous reports investigating the expression of the EBERs in lytically infected cultured cells by Northern blotting could not confirm the downregulation observed in vivo. To avoid two problems that could mask a regulation of the EBERs, the contamination of the lytically infected cell population with latently infected cells and the presence of stable EBERs produced during the preceding viral latency, we measured the actual transcriptional activity of both EBER genes directly with nuclear run-on experiments according to the methods of Greenberg and Ziff (6) and Groudine et al. (7), with some modifications (16, 17) (Fig. 3). Cells were treated to induce viral lytic replication, and the gp350/220-positive ones were enriched and used for in vitro nuclear run-on experiments. The radioactivity-labeled RNA was hybridized to gene-specific DNA probes immobilized on membranes by Southern transfer. The gene-specific probes were synthesized by PCR using the Powerscript polymerase (PAN Systems, Nürnberg, Germany). The probes for histone, EBNA-2, LMP-1, Rta (BRLF-1), and Zta (BZLF-1) were designed to retain the intron-exon structure of the respective genes since cDNA probes of the spliced transcripts showed significantly lower hybridization signals. The genes for EBER-1, EBER-2, EA (BALF-2), and VCA (BcLF-1) were free of introns owing to their genomic organization. The sequences of primers were as follows: EBER-1 and EBER-2, reference 22; Zta (BZLF-1), 5′-TGT CCA TGA ACC GGT CGG ATC-3′ and 5′-GCG GTA AAC AAT GGC ACC CTC-3′; Rta (BRLF-1), 5′-GGC TTG GCT AAG TGC AAG GAT-3′ and 5′-GGA GGA GGC AGT TTT CAG AAG T-3′; EA (BALF-2), reference 14; VCA (BcLF-1), reference 14; EBNA-2, 5′-CAA GCT GCT TTG ATT CTT GGG-3′ and 5′-GAG CTA CCT ACC ATG CTA TAA G-3′; LMP1, 5′-GGT TCA TCG CTC AGC TCC TCC-3′ and 5′-CCT GAA TCC GCC ACC TCA TTC-3′; histone, reference 4.
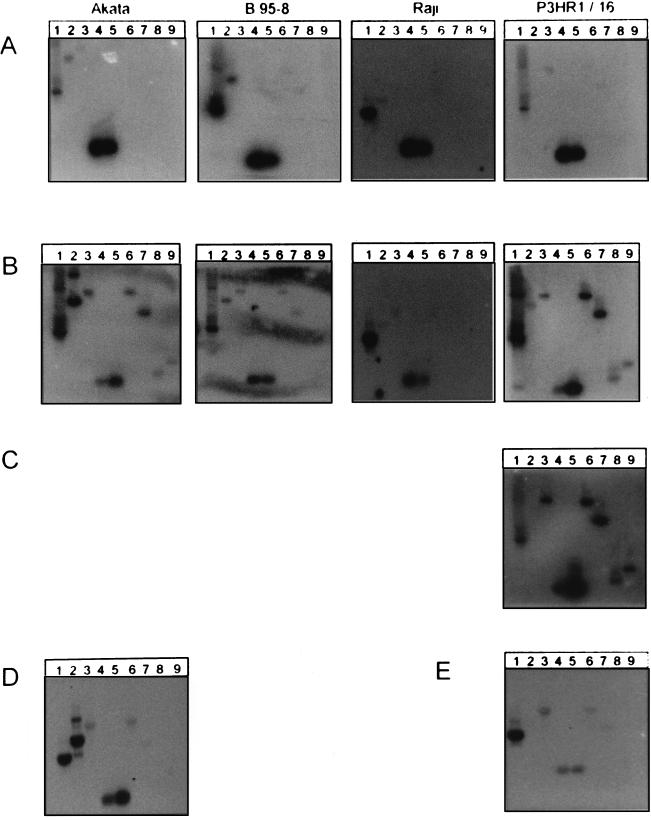
FIG. 3.
Analysis of viral gene activity in latent and lytically infected cell lines by nuclear run-on experiments. The cell lines Akata, B95-8, Raji, P3HR1/16, and P3HR1/13 (not shown here) were treated for 72 h with TPA–BA–TGF-β to induce lytic replication of EBV. After 72 h, the nuclei were purified and nuclear run-on experiments were performed. The radiolabeled RNA, reflecting the actual gene activity, was hybridized to several immobilized gene-specific DNA probes, and finally the membrane was exposed to X-ray film. Gene-specific probes: lane 1, histone H3; lane 2, EBNA-2; lane 3, LMP-1; lane 4, EBER-1; lane 5, EBER-2; lane 6, BRLF-1 (Rta); lane 7, BZLF-1 (Zta); lane 8, BALF-2 (EA); lane 9, BcLF-1 (VCA). (A) Cells prior to chemical induction. (B) Cells after 72 h of induction, prior to purification. (C) Cells after 72 h of induction, following purification of gp350/220-positive cells. (D) Akata cells after 72 h of induction with anti-immunoglobulin G, without purification. (E) P3HR1/16 cells after 72 h of induction with TPA–BA–TGF-β in the presence of PAA to inhibit viral DNA replication.
In order to compare the variable transcriptional activities of the genes between different experiments, the concentration of gene-specific probe(s), which quantitatively bind to and detect the specific transcripts, must be kept constant. In contrast to Northern blot assays, in run-on assays the constant parameter (probes) was unlabelled and was immobilized to a membrane. The variable parameter (different concentrations of various transcripts) was labeled and was contained in the hybridization solution. Since in each run-on experiment a constant number of nuclei (2 × 107) was used, the variable amounts of 32P incorporated in the individual run-on reactions reflected the sum of variable transcriptional activities of a vast number of genes. This 32P-labeled RNA was quantitatively used for hybridization to ensure that the amount of hybridization to the gene-specific probes directly reflected the genes’ transcriptional activity. Differences in hybridization due to different efficiencies in individual run-on experiments were precluded by performing assays in parallel.
In the latently infected cell lines Akata, B95-8, Raji, and P3HR1/16 (Fig. 3A), we detected transcription of the latent genes EBNA-2 (lane 2), LMP-1 (lane 3), and both EBER-1 and EBER-2 (lanes 4 and 5, respectively). After treatment with TPA, BA, and TGF-β (Fig. 3B) in all the cell lines except Raji, transcription of both the immediate-early genes BRLF-1 and BZLF-1 (lanes 6 and 7, respectively) and of early BALF-2 (lane 8) and late BcLF-1 (lane 9) was detected, demonstrating a switch to lytic replication. A significant reduction in the rate of transcription, standardized to the activity of the histone gene as our internal control, was detected for both EBER-1 (lane 4) and EBER-2 (lane 5) in the cell lines Akata (16% and 33%, respectively), P3HR1/16 (9% and 23%, respectively), and Raji (41% and 46%, respectively). In addition, downregulation of EBER-1 was more stringent than that of EBER-2. This was apparent from the EBER-1/EBER-2 ratio in the cell lines Akata (latent, 1.2; lytic, 0.6) and P3HR1/16 (latent, 1.2; lytic, 0.5), which showed the strongest overall repression of the EBERs compared to the histones. After purification of the gp350/220-positive P3HR1/16 cells in the late phase of lytic replication, the effect on the selective downregulation of EBER-1 and on the EBER-1/EBER-2 ratio was nearly the same (latent, 1.2; lytic, 0.5; purified lytic, 0.7) (Fig. 3C). At first glance, the downregulation of EBER-1 and EBER-2 relative to the expression of the histones was not as pronounced as that in unpurified cells (72% and 84%, respectively). However, this effect was only relative, owing to the downregulation of the histone itself, probably because of the host shutoff in the late lytic phase of replication. The absolute intensities of the lytic cycle transcripts (lanes 6 to 9) and of the EBERs (lanes 4 and 5) remained rather constant, suggesting that downregulation of the EBERs and of histone happens independently.
Akata and B95-8 cells could not be enriched by the protocol described above, although they showed a good induction of lytic replication in Western blot assays. This was probably a result of the particular property of the gp350/220-specific monoclonal antibody mab-64D7, recognizing the native membrane-bound glycoprotein of the EBV strains Akata and B95-8 less efficiently than that of P3HR1. In contrast, recognition of the partially denatured protein by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blotting was comparable, demonstrating similar induction of lytic replication in Akata cells (not shown).
In order to show that downregulation of the EBERs did not depend on TPA treatment, Akata cells were incubated for 72 h with anti-immunoglobulin G antibodies (Fig. 3D). These cells showed a comparable pattern of downregulation of both EBER-1 and EBER-2 (31% and 60%, respectively; EBER-1/EBER-2 ratio, 0.6).
The amount of EBER transcripts in the cell remained constant after induction of lytic virus replication.
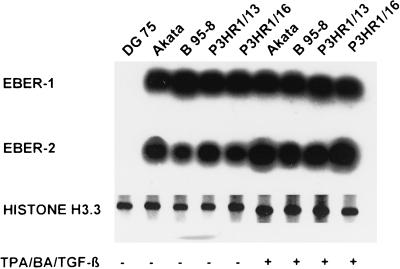
Our nuclear run-on experiments clearly demonstrated a downregulation of the transcription of the EBER genes. Therefore we investigated whether the difference from earlier reports demonstrating continuous expression of both EBERs in cell culture was due to a different experimental design, detecting the rather stable EBERs with a long half-life synthesized during viral latency. We asked whether the downregulation of EBER could also be detected at the level of RNA transcripts or whether the amount of EBERs would remain unaffected within the first 72 h after induction of lytic replication. From the same batch of stimulated P3HR1 cells used for the nuclear run-on experiments described above, RNA was prepared prior to the separation of the lytically infected cells and used for Northern blot and reverse transcriptase PCR (RT-PCR) analysis. For Northern blot analysis the gene-specific probes for EBER-1, EBER-2, and histone were synthesized by RT-PCR with total RNA from B95-8 cells. The probes for Northern blots were radiolabeled with [α-32P]dATP during the reamplification PCR. As shown in Fig. 4, there was no difference in the concentration of EBERs between the latent and the stimulated cell lines Akata (lanes 2 and 6), B95-8 (lanes 3 and 7), P3HR1/16 (lanes 4 and 8), and P3HR1/13 (lanes 5 and 9), assayed by either Northern blotting (Fig. 4) or RT-PCR (data not shown). With the same unfractionated population of cells, however, nuclear run-on assays showed significant downregulation of transcription of the EBER-1 and EBER-2 genes (Fig. 3). Thus, these experiments demonstrated that regulation of the EBERs during the initial 72 h after induction of lytic virus replication could be detected only at the level of transcription, due to the long half-life of the EBERs.
FIG. 4.
Quantification of EBER transcripts in latent and lytically infected P3HR1/13 cells by Northern blot analysis. The cell lines Akata, B95-8, P3HR1/16, and P3HR1/13 were treated for 72 h with TPA–BA–TGF-β to induce lytic replication of EBV. After 72 h, total RNA was purified and Northern blot experiments were performed. Hybridization of the immobilized RNA was done with (A) EBER-1-specific, (B) EBER-2-specific, or (C) histone H3-specific (control for RNA) radiolabeled probes. Cell lines: lanes 1 and 6, Akata; lanes 2 and 7, B95-8; lanes 3 and 8; P3HR1/13; lanes 4 and 9, P3HR1/16. Lanes 1 to 5: latent cells; lanes 6 to 9: chemically treated cells.
Downregulation of EBER-1 and EBER-2 are early events during lytic replication of EBV.
Detection of downregulation of the EBERs in unfractionated cell populations was unexpected due to the large percentage of gp350/220-negative cells, which were supposed to be latently infected and produce huge quantities of EBER-1 and EBER-2. Therefore we tested whether the downregulation may be an early event during the switch from latency to lytic replication of EBV. With nuclear run-on experiments we assayed transcription of the EBERs in P3HR1 cells, which were treated with TPA, BA, and TGF-β in combination with phosphonoacetic acid (PAA) to induce the lytic phase but block viral DNA replication (Fig. 3E). It could be shown that both EBER genes were downregulated (13% EBER-1 and 13% EBER-2) in this experimental approach when their activity was compared with the activity of the histone gene. The same results were obtained with the cell line Raji (Fig. 3B), which is blocked in the early phase of replication by a mutation (41% EBER-1 and 46% EBER-2). Transcripts of the immediate-early genes BZLF-1 and BRLF-1 were induced, whereas the late gene BcLF-1 was kept silent. The EBER-1/EBER-2 ratios remained unaltered in both P3HR-1/16 (latent, 1.2; lytic, 1.0) and Raji (latent, 1.3; lytic, 1.1) cells. These experiments clarified that the downregulation of EBER-1 is an early event in the lytic replication of EBV, probably directly regulated by cellular factors.
With this paper we demonstrate that during the phase of lytic replication transcription of EBER-1 is dramatically downregulated, as is, to a minor extent, transcription of EBER-2. This could not be reproduced experimentally in cell culture so far. Furthermore, we demonstrate that, in contrast to transcription, the amount of the EBERs remains unaltered within 72 h after induction of lytic replication, which may explain the previous incongruent reports. The difference in the detection of the EBERs in induced cultured cells and the absence in epithelial cells observed in vivo may be explained by the half-life of the EBERs. Seventy-two hours is obviously not sufficient time to degrade the EBERs produced during latency, showing that the half-lives of the EBERs are significantly prolonged in the lytic phase of viral replication compared with 8 to 9 h in latent infection (2). A study by Pathmanathan et al. (13) showed the absence of EBER expression in vivo in areas of tissue differentiation. A study of an EBV-positive lymphoma that developed in a salivary gland showed permissive replication in epithelial cells adjacent to the lymphoma with expression of BZLF-1 without EBERs and expression of EBERs and LMP-1 in the lymphoma tissue (25). These studies suggest that differentiation may negatively regulate EBER expression. Differences in epithelial infection versus lymphoid infection and differences in differentiation status may explain the lack of EBER expression in in vivo infection compared with the situation after reactivation of EBV replication in lymphocytes.
The observed differential regulation of EBER-1 and EBER-2 did not directly reveal a new role of these RNAs in the life cycle of the virus. However, these results suggested that EBER-1 may have a function different from that of EBER-2. Previous reports with recombinant EBER-negative mutant EBV demonstrated that neither gene is necessary for latency nor for the lytic replication of the virus in cell culture. Now, the downregulation of the EBERs is suggestive of a function that may be incompatible with lytic replication and thus may be involved in stabilizing viral latency, which is in accordance with the early regulation during the lytic cycle. Recently, we have described a case of chronic active infection with EBV. With a semiquantitative RT-PCR we detected expression of the immediate-early gene BZLF-1 in the B lymphocytes of the patient in an amount characteristic of primary infection, which is associated with complete lytic replication of EBV in the B cells of the peripheral blood (14). Remarkably, with in situ hybridization no EBER transcripts were detected in those B cells although EBV DNA was found (12).
Finally, the direct effect of downregulation of the transcription of the EBER genes still remains unclear, since the concentration of the EBER-1 RNA and probably of EBER-2 RNA is unaffected. One possible explanation is that the EBERs detected during the lytic replication of EBV are not functional anymore. However, as long as the function of the EBERs is not clear, this will be difficult to test. Several parameters that may influence the function of an RNA are under investigation.
Acknowledgments
We are grateful to Sabine Richter for critically reading the manuscript.
This work was supported by the Deutsche Forschungsgemeinschaft (grants Wo227/6 and Wo227/7 V).
REFERENCES
- 1.Bauer G, Hofler P, zur Hausen H. Epstein-Barr virus induction by a serum factor. I. Induction and cooperation with additional inducers. Virology. 1982;121:184–194. doi: 10.1016/0042-6822(82)90128-3. [DOI] [PubMed] [Google Scholar]
- 2.Clarke P A, Sharp N A, Clemens M J. Expression of genes for the Epstein-Barr virus small RNAs EBER-1 and EBER-2 in Daudi Burkitt’s lymphoma cells: effects of interferon treatment. J Gen Virol. 1992;73:3169–3175. doi: 10.1099/0022-1317-73-12-3169. [DOI] [PubMed] [Google Scholar]
- 3.Clemens M J. Functional significance of the Epstein-Barr virus-encoded small RNAs. Epstein-Barr Virus Rep. 1994;5:107–111. [Google Scholar]
- 4.Futscher B W, Blake L L, Gerlach J H, Grogan T M, Dalton W S. Quantitative polymerase chain reaction analysis of mdr1 mRNA in multiple myeloma cell lines and clinical specimens. Anal Biochem. 1993;213:414–421. doi: 10.1006/abio.1993.1440. [DOI] [PubMed] [Google Scholar]
- 5.Gilligan K, Rajadurai P, Resnick L, Raab Traub N. Epstein-Barr virus small nuclear RNAs are not expressed in permissively infected cells in AIDS-associated leukoplakia. Proc Natl Acad Sci USA. 1990;87:8790–8794. doi: 10.1073/pnas.87.22.8790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Greenberg M E, Ziff E B. Stimulation of 3T3 cells induces transcription of the c-fos proto-oncogene. Nature. 1984;311:433–438. doi: 10.1038/311433a0. [DOI] [PubMed] [Google Scholar]
- 7.Groudine M, Peretz M, Weintraub H. Transcriptional regulation of hemoglobin switching in chicken embryos. Mol Cell Biol. 1981;1:281–288. doi: 10.1128/mcb.1.3.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hironaka T, Nagasaki M, Morikawa S, Hirai K. Detection of Epstein-Barr virus transcripts in chemically or immunologically-activated cells and in a null cell-line (HLN-STL-C) by in situ hybridization with alkaline phosphatase-linked oligonucleotide probes. J Virol Methods. 1993;44:141–154. doi: 10.1016/0166-0934(93)90050-2. [DOI] [PubMed] [Google Scholar]
- 9.Howe J G, Shu M-D. Epstein-Barr virus small RNA (EBER) genes: unique transcription units that combine RNA polymerase II and III promoter elements. Cell. 1989;57:825–834. doi: 10.1016/0092-8674(89)90797-6. [DOI] [PubMed] [Google Scholar]
- 10.Howe J G, Steitz J A. Localization of Epstein-Barr virus-encoded small RNAs by in situ hybridization. Proc Natl Acad Sci USA. 1986;83:9006–9010. doi: 10.1073/pnas.83.23.9006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lerner M R, Andrews N C, Miller G, Steitz J A. Two small RNAs encoded by Epstein-Barr virus and complexed with protein are precipitated by antibodies from patients with systemic lupus erythematosus. Proc Natl Acad Sci USA. 1981;78:805–809. doi: 10.1073/pnas.78.2.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mitterer M, Pescosta N, Fend F, Larcher C, Prang N, Schwarzmann F, Coser P, Huemer H P. Chronic active Epstein-Barr virus disease in a case of persistent polyclonal B-cell lymphocytosis. Br J Haematol. 1995;90:526–531. doi: 10.1111/j.1365-2141.1995.tb05579.x. [DOI] [PubMed] [Google Scholar]
- 13.Pathmanathan R, Prasad U, Chandrika G, Sadler R, Flynn K, Raab-Traub N. Undifferentiated, nonkeratinizing, and squamous cell carcinoma of the nasopharynx. Variants of Epstein-Barr virus-infected neoplasia. Am J Pathol. 1995;146:1355–1367. [PMC free article] [PubMed] [Google Scholar]
- 14.Prang N S, Hornef M W, Jäger M, Wagner W J, Wolf H, Schwarzmann F. Lytic replication of Epstein-Barr virus in the peripheral blood: analysis of viral gene expression in B-lymphocytes during infectious mononucleosis and in the normal carrier state. Blood. 1997;89:1665–1677. [PubMed] [Google Scholar]
- 15.Rosa M D, Gottlieb E, Lerner M R, Steitz J A. Striking similarities are exhibited by two small Epstein-Barr virus-encoded ribonucleic acids and the adenovirus-associated ribonucleic acids VAI and VAII. Mol Cell Biol. 1981;1:785–796. doi: 10.1128/mcb.1.9.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schwarzmann F, Jäger M, Prang N, Wolf H. The control of lytic replication of Epstein-Barr virus in B lymphocytes. Int J Mol Med. 1998;1:137–142. doi: 10.3892/ijmm.1.1.137. [DOI] [PubMed] [Google Scholar]
- 17.Schwarzmann F, Prang N, Reichelt B, Rinkes B, Haist S, Marschall M, Wolf H. Negative cis-acting elements in the distal part of the promoter of Epstein-Barr virus trans-activator gene BZLF1. J Gen Virol. 1994;75:1999–2006. doi: 10.1099/0022-1317-75-8-1999. [DOI] [PubMed] [Google Scholar]
- 18.Schwemmle M, Clemens M J, Hilse K, Pfeifer K, Troster H, Muller W E, Bachmann M. Localization of Epstein-Barr virus-encoded RNAs EBER-1 and EBER-2 in interphase and mitotic Burkitt lymphoma cells. Proc Natl Acad Sci USA. 1992;89:10292–10296. doi: 10.1073/pnas.89.21.10292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sharp T V, Schwemmle M, Jeffrey I, Laing K, Mellor H, Proud C G, Hilse K, Clemens M J. Comparative analysis of the regulation of the interferon-inducible protein kinase PKR by Epstein-Barr virus RNAs EBER-1 and EBER-2 and adenovirus VAI RNA. Nucleic Acids Res. 1993;21:4483–4490. doi: 10.1093/nar/21.19.4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Swaminathan S, Huneycutt B S, Reiss C S, Kieff E. Epstein-Barr virus-encoded small RNAs (EBERs) do not modulate interferon effects in infected lymphocytes. J Virol. 1992;66:5133–5136. doi: 10.1128/jvi.66.8.5133-5136.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Swaminathan S, Tomkinson B, Kieff E. Recombinant Epstein-Barr virus with small RNA (EBER) genes deleted transforms lymphocytes and replicates in vitro. Proc Natl Acad Sci USA. 1991;88:1546–1550. doi: 10.1073/pnas.88.4.1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tierney R J, Steven N, Young L S, Rickinson A B. Epstein-Barr virus latency in blood mononuclear cells: analysis of viral gene transcription during primary infection and in the carrier state. J Virol. 1994;68:7374–7385. doi: 10.1128/jvi.68.11.7374-7385.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Toczyski D P, Matera A G, Ward D C, Steitz J A. The Epstein-Barr virus (EBV) small RNA EBER1 binds and relocalizes ribosomal protein L22 in EBV-infected human B lymphocytes. Proc Natl Acad Sci USA. 1994;91:3463–3467. doi: 10.1073/pnas.91.8.3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weigel R, Fischer D K, Heston L, Miller G. Constitutive expression of Epstein-Barr virus-encoded RNAs and nuclear antigen during latency and after induction of Epstein-Barr virus replication. J Virol. 1985;53:254–259. doi: 10.1128/jvi.53.1.254-259.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wen S, Mizugaki Y, Shinozaki F, Takada K. Epstein-Barr virus (EBV) infection in salivary gland tumors: lytic EBV infection in nonmalignant epithelial cells surrounded by EBV-positive T-lymphoma cells. Virology. 1997;227:484–487. doi: 10.1006/viro.1996.8352. [DOI] [PubMed] [Google Scholar]
- 26.Zeuthen J. Epstein-Barr virus (EBV), lymphocytes and transformation. J Cancer Res Clin Oncol. 1983;106:1–11. doi: 10.1007/BF00399890. [DOI] [PMC free article] [PubMed] [Google Scholar]