Shareable abstract
Bedaquiline resistance is a major threat to drug-resistant tuberculosis control strategies. This analysis found a pooled prevalence of baseline bedaquiline resistance of 2.4% and a pooled prevalence of treatment-emergent bedaquiline resistance of 2.1%. https://bit.ly/3FC6yio
To the Editor:
Bedaquiline is a novel antimycobacterial agent for drug-resistant tuberculosis (TB) and is classified as a World Health Organization (WHO) group A drug due to its excellent clinical efficacy, high bactericidal activity, and potent sterilising effect [1]. The introduction of bedaquiline into treatment regimens has enabled short-course all-oral multidrug-resistant TB (MDR-TB) regimens and the shortening of drug-susceptible TB treatment [2, 3].
Bedaquiline targets F1F0-ATP synthase to impair Mycobacterium tuberculosis (Mtb) ATP synthesis and exerts other incompletely characterised bactericidal effects [4]. Variants in the target atpE and atpB genes and off-target mutations in mmpR5, mmpL5 and pepQ have been associated with bedaquiline resistance [5, 6]. We performed a systematic review and meta-analysis to estimate the frequency of, and mutations associated with, baseline and acquired (treatment-emergent) bedaquiline resistance in clinical Mtb isolates.
The study protocol was registered in PROSPERO (CRD42022346547) and the PRISMA guidelines were followed for reporting of the review methods and findings. Systematic searches of MEDLINE/PubMed, Cochrane Central Register of Clinical Trials, and EMBASE were conducted through February 2023 for publications on phenotypic resistance of bedaquiline. We included studies which reported clinical Mtb isolates with bedaquiline resistance via minimum inhibitory concentration (MIC) values from patients with at least rifampicin-resistant TB. Given the suboptimal positive predictive value of resistance-associated variants for phenotypic resistance, our study only evaluated phenotypic resistance as defined by MIC thresholds. We excluded studies with MIC cut-offs inconsistent with WHO cut-offs, in vitro Mtb isolates not obtained from patients, or ⩽3 patients/isolates. Phenotypic bedaquiline resistance was defined by critical concentrations of 1 μg·mL−1 by MGIT method or 0.25 μg·mL−1 by broth microdilution or 7H11 agar proportion method. Acquired bedaquiline resistance was defined as the absence of phenotypic resistance before treatment and demonstration of phenotypic resistance on at least one occasion during bedaquiline treatment [7]. Publication bias was evaluated using funnel plots and methodological quality was assessed by the tool proposed by Hoy et al. [8]. We performed a proportional meta-analysis in R version 4.2.2 using dmetar, metafor and meta packages. Paired MIC and resistance-associated variant (RAV) data were presented by scatterplots. Detailed methods are available online [7].
The systematic search identified 180 articles for assessment: 37 were duplicates, 47 did not meet inclusion criteria, and 82 were excluded because they were in vitro studies, review papers, case reports, used non-standardised MIC thresholds, did not distinguish between baseline and treatment-emergent resistance, contained overlapping data with another study, or had no retrievable full text [7]. The remaining 14 studies were included, comprising four randomised controlled trials and 10 cohort studies, emanating from five continents [5, 6, 9–20]. In both baseline and during-treatment analyses there was evidence of significant positive publication bias. For the baseline analysis, 13 studies were rated as low risk of bias, and one study as moderate risk of bias. For the during-treatment analysis, three studies were rated as low risk of bias, and six studies as moderate risk of bias [7].
14 studies, with a pooled sample of 9975 isolates, contributed to the estimate of baseline bedaquiline resistance. Nine studies, with a pooled sample of 1912 isolates, contributed to the estimate of treatment-emergent bedaquiline resistance. The pooled prevalence of baseline bedaquiline resistance was 2.4% (95% CI 1.7–3.5), with significant heterogeneity across all studies (I2 66%, p<0.01) (figure 1a). The pooled prevalence of treatment-emergent bedaquiline resistance was 2.1% (95% CI 1.4–3.0), with no significant heterogeneity across the included studies (I2 0%, p=0.97) (figure 1b). In the sensitivity analyses, no between-group differences were seen based on the quality of the studies (low versus moderate risk of bias).
FIGURE 1.
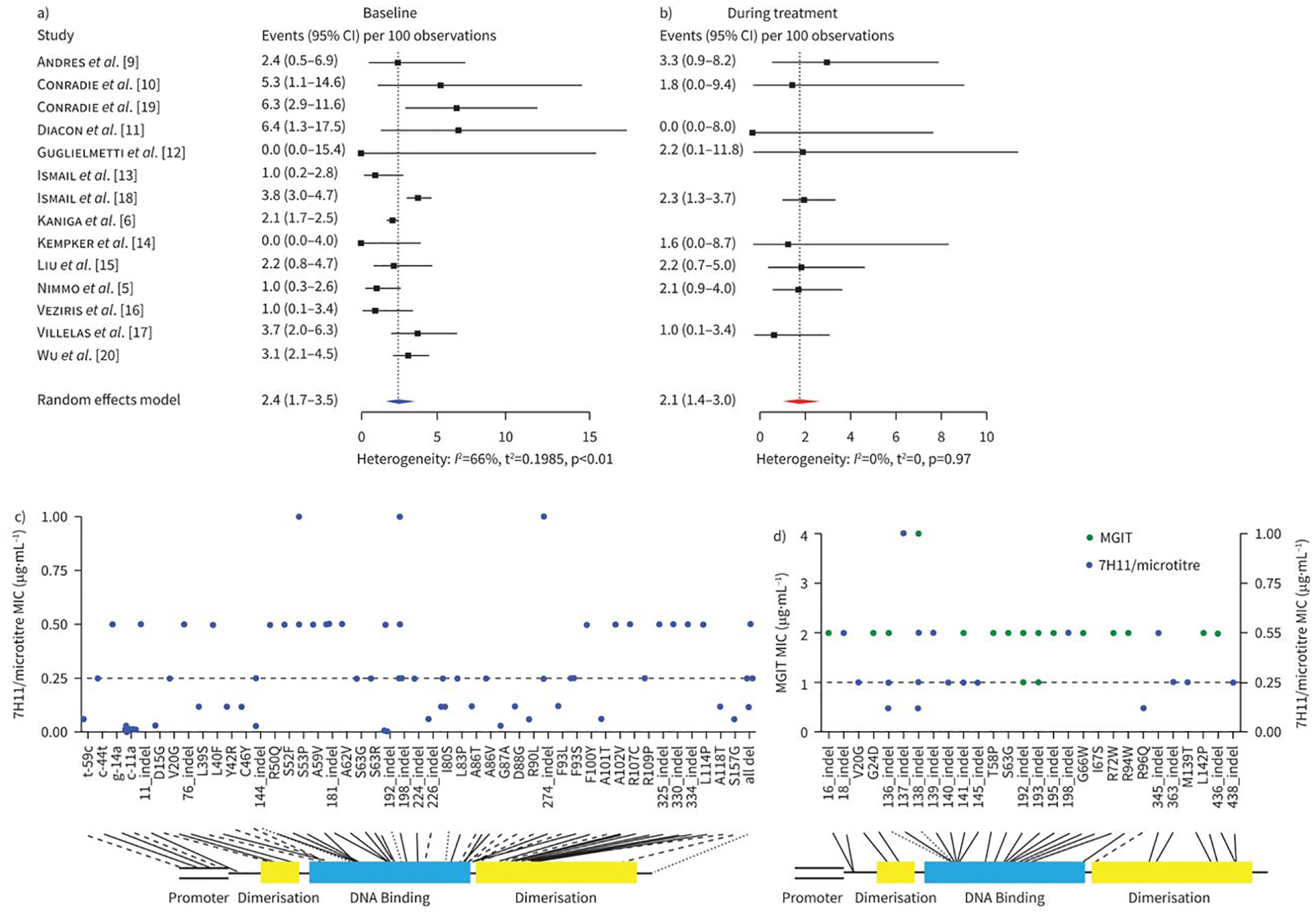
Pooled prevalence of a) baseline and b) treatment-emergent bedaquiline resistance in patients with drug-resistant tuberculosis. c) Baseline and d) treatment-emergent resistance-associated variants (RAVs) and associated minimum inhibitory concentrations (MICs). Solid black lines represent intermediate or resistant MICs, dashed lines represent susceptible MICs, and dotted lines are RAVs that are associated with both susceptible and intermediate/resistant MICs.
We identified 141 RAVs across all isolates in the included studies, comprising 71 unique RAVs at 58 unique sites. Four RAVs were mapped to the promoter region of the Mtb genome, 19 were mapped to the dimerisation domains, and 42 were mapped to the DNA binding region. The complete list of RAVs and associated MICs is presented by scatterplot (figure 1c and d). Treatment-emergent RAVs were significantly more likely to be associated with phenotypic resistance than pre-treatment RAVs (21/35 (60.0%, 95% CI 42.9–77.7) versus 24/97 (24.7%, 95% CI 15.8–33.7)).
The high degree of heterogeneity and publication bias suggests that our pooled estimate of baseline bedaquiline resistance (2.4%, 95% CI 1.7–3.5) may overestimate the true prevalence in clinical settings. However, the two included studies which involved comprehensive national surveillance of all patients initiating treatment for MDR-TB produced reliable estimates of pre-treatment bedaquiline resistance of 3.2% and 3.6% in Germany and South Africa, respectively [9, 18]. These levels of pre-treatment bedaquiline resistance likely reflect high rates of person-to-person community transmission of bedaquiline-resistant TB.
We found a concerning pooled prevalence of acquired bedaquiline resistance during treatment of 2.1% (95% CI 1.4–3.0%). Our estimate is concordant with a previous systematic review which reported a frequency of phenotypic acquired bedaquiline resistance of 2.2% [21]. The low heterogeneity among included studies reflects the systematic evaluation of serial cultures from all patients treated for MDR-TB, ranging from weekly to alternate months until culture conversion. This approach avoided the spuriously high estimates of acquired bedaquiline resistance reported in studies that only performed serial phenotypic DST in patients with evidence of treatment failure [22].
As mmpR5 (Rv0678) mutations are associated with cross-resistance to clofazimine and bedaquiline, prior exposure to clofazimine may have contributed to the high levels of pre-treatment bedaquiline resistance identified in settings with limited prior use of bedaquiline. As a WHO group B drug, clofazimine has been increasingly used as part of shortened MDR-TB regimens globally. Bedaquiline has a relatively long elimination half-life, estimated at around 5.5 months, rendering it particularly vulnerable to acquired resistance when treatment is interrupted. For this reason, high levels of adherence support, patient tracking, and robust novel treatment regimens with a high barrier to resistance are necessary to mitigate the risk of acquired resistance. In addition, bedaquiline exhibits suboptimal penetration into caseous necrotic lesions, exposing it to intralesional pharmacokinetic–pharmacodynamic mismatch [23]. Few MDR-TB drugs offer adequate protection against bedaquiline resistance, which is an increasingly important consideration when constructing an MDR-TB regimen [9].
Bedaquiline received US Food and Drug Administration accelerated approval in 2012 and by 2014 the first case of bedaquiline-resistant TB was reported in a Tibetan refugee following treatment with a bedaquiline-containing regimen [24]. Between 2015 and 2019, there were an estimated 300 cases of bedaquiline-resistant TB cases in South Africa alone [18]. We are reminded that the prevalence of rifampicin resistance was initially ~12 per 1000 patients shortly after roll out of rifampicin, but amplified to ~300 per 1000 patients currently [25, 26]. A Markov decision modelling exercise estimated the prevalence of bedaquiline resistance increasing to 588 per 1000 patients with more widespread bedaquiline rollout [27].
As we prepare for the mass rollout of bedaquiline-containing short-course regimens for the treatment of MDR-TB, National TB Programmes will require substantial strengthening to avoid escalating levels of bedaquiline resistance, ensuing surges in treatment failure and relapse, a growing dependence on complex salvage regimens, and treatment destitution. Central to reducing the threat of a bedaquiline-resistant TB epidemic is the need for a rapid diagnostic test to detect bedaquiline resistance. Research to better understand the genetics of resistance as well as investment in rapid phenotypic methods, such as microscopic observation of drug susceptibility and reporter mycobacteriophages, may be required. To date, the development of a rapid molecular diagnostic assay has been severely constrained by the poor phenotypic–genotypic concordance of resistance profiling for bedaquiline. In parallel, as bedaquiline-based regimens are implemented, national and supranational bedaquiline resistance surveillance activities must be escalated as a critical early warning system.
Footnotes
The study protocol was registered in PROSPERO (CRD42022346547). Materials related to this review are available from the authors upon request.
Conflict of interest: The authors declare no conflicts of interest.
References
- 1.Olayanju O, Limberis J, Esmail A, et al. Long-term bedaquiline-related treatment outcomes in patients with extensively drug-resistant tuberculosis from South Africa. Eur Respir J 2018; 51: 1800544. [DOI] [PubMed] [Google Scholar]
- 2.World Health Organization. Rapid communication: key changes to the treatment of drug-resistant tuberculosis. Geneva, World Health Organization, 2022. [Google Scholar]
- 3.Paton NI, Cousins C, Suresh C, et al. Treatment strategy for rifampin-susceptible tuberculosis. N Engl J Med 2023; 388: 873–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mackenzie JS, Lamprecht DA, Asmal R, et al. Bedaquiline reprograms central metabolism to reveal glycolytic vulnerability in Mycobacterium tuberculosis. Nat Commun 2020; 11: 6092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nimmo C, Millard J, van Dorp L, et al. Population-level emergence of bedaquiline and clofazimine resistance-associated variants among patients with drug-resistant tuberculosis in southern Africa: a phenotypic and phylogenetic analysis. Lancet Microbe 2020; 1: e165–e174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaniga K, Hasan R, Jou R, et al. Bedaquiline drug resistance emergence assessment in multidrug-resistant tuberculosis (MDR-TB): a 5-year prospective in vitro surveillance study of bedaquiline and other second-line drug susceptibility testing in MDR-TB isolates. J Clin Microbiol 2022; 60: e0291920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Perumal R, Bionghi N, Nimmo C, et al. Baseline and treatment-emergent bedaquiline resistance in drug-resistant tuberculosis: a systematic review and meta-analysis. medRxiv 2023; preprint [ 10.1101/2023.08.07.23293687]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoy D, Brooks P, Woolf A, et al. Assessing risk of bias in prevalence studies: modification of an existing tool and evidence of interrater agreement. J Clin Epidemiol 2012; 65: 934–939. [DOI] [PubMed] [Google Scholar]
- 9.Andres S, Merker M, Heyckendorf J, et al. Bedaquiline-resistant tuberculosis: dark clouds on the Horizon. Am J Respir Crit Care Med 2020; 201: 1564–1568. [DOI] [PubMed] [Google Scholar]
- 10.Conradie F, Diacon AH, Ngubane N, et al. Treatment of highly drug-resistant pulmonary tuberculosis. N Engl J Med 2020; 382: 893–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Diacon AH, Donald PR, Pym A, et al. Randomized pilot trial of eight weeks of bedaquiline (TMC207) treatment for multidrug-resistant tuberculosis: long-term outcome, tolerability, and effect on emergence of drug resistance. Antimicrob Agents Chemother 2012; 56: 3271–3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guglielmetti L, Jaspard M, Le Dû D, et al. Long-term outcome and safety of prolonged bedaquiline treatment for multidrug-resistant tuberculosis. Eur Respir J 2017; 49: 1601799. [DOI] [PubMed] [Google Scholar]
- 13.Ismail NA, Omar SV, Joseph L, et al. Defining bedaquiline susceptibility, resistance, cross-resistance and associated genetic determinants: a retrospective cohort study. EBioMedicine 2018; 28: 136–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kempker RR, Mikiashvili L, Zhao Y, et al. Clinical outcomes among patients with drug-resistant tuberculosis receiving bedaquiline- or delamanid-containing regimens. Clin Infect Dis 2020; 71: 2336–2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Y, Gao M, Du J, et al. Reduced susceptibility of Mycobacterium tuberculosis to bedaquiline during antituberculosis treatment and its correlation with clinical outcomes in China. Clin Infect Dis 2021; 73: e3391–e3397. [DOI] [PubMed] [Google Scholar]
- 16.Veziris N, Bernard C, Guglielmetti L, et al. Rapid emergence of Mycobacterium tuberculosis bedaquiline resistance: lessons to avoid repeating past errors. Eur Respir J 2017; 49: 1601719. [DOI] [PubMed] [Google Scholar]
- 17.Villellas C, Coeck N, Meehan CJ, et al. Unexpected high prevalence of resistance-associated Rv0678 variants in MDR-TB patients without documented prior use of clofazimine or bedaquiline. J Antimicrob Chemother 2017; 72: 684–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ismail NA, Omar SV, Moultrie H, et al. Assessment of epidemiological and genetic characteristics and clinical outcomes of resistance to bedaquiline in patients treated for rifampicin-resistant tuberculosis: a cross-sectional and longitudinal study. Lancet Infect Dis 2022; 22: 496–506. [DOI] [PubMed] [Google Scholar]
- 19.Conradie F, Bagdasaryan TR, Borisov S, et al. Bedaquiline-pretomanid-linezolid regimens for drug-resistant tuberculosis. N Engl J Med 2022; 387: 810–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu SH, Chan HH, Hsiao HC, et al. Primary bedaquiline resistance among cases of drug-resistant tuberculosis in Taiwan. Front Microbiol 2021; 12: 754249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mallick JS, Nair P, Abbew ET, et al. Acquired bedaquiline resistance during the treatment of drug-resistant tuberculosis: a systematic review. JAC Antimicrob Resist 2022; 4: dlac029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ghodousi A, Rizvi AH, Baloch AQ, et al. Acquisition of cross-resistance to bedaquiline and clofazimine following treatment for tuberculosis in Pakistan. Antimicrob Agents Chemother 2019; 63: e00915–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sarathy JP, Zuccotto F, Hsinpin H, et al. Prediction of drug penetration in tuberculosis lesions. ACS Infect Dis 2016; 2: 552–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Somoskovi A, Bruderer V, Homke R, et al. A mutation associated with clofazimine and bedaquiline cross-resistance in MDR-TB following bedaquiline treatment. Eur Respir J 2015; 45: 554–557. [DOI] [PubMed] [Google Scholar]
- 25.Loutet MG, Davidson JA, Brown T, et al. Acquired resistance to antituberculosis drugs in England, Wales, and Northern Ireland, 2000–2015. Emerg Infect Dis 2018; 24: 524–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stottmeier KD. Emergence of rifampin-resistant Mycobacterium tuberculosis in Massachusetts. J Infect Dis 1976; 133: 88–90. [DOI] [PubMed] [Google Scholar]
- 27.Kunkel A, Cobelens FG, Cohen T. Tradeoffs in introduction policies for the anti-tuberculosis drug bedaquiline: a model-based analysis. PLoS Med 2016; 13: e1002142. [DOI] [PMC free article] [PubMed] [Google Scholar]


