Abstract
Oligodendrocytes (OL) are myelin-forming glial cells in the central nervous system. In vitro primary OL culture models offer the benefit of a more readily controlled environment that facilitates the examination of diverse OL stages and their intricate dynamics. Although conventional methods for primary OL culture exist, their performance in terms of simplicity and efficiency can be improved. Here, we introduce a novel method for primary OL culture, namely the E3 (easy, efficient, and effective) method, which greatly improves the simplicity and efficiency of the primary OL culture procedure using neonatal rodent brains. We also provided the optimal media composition for the augmentation of oligodendrocyte progenitor cell (OPC) proliferation and more robust maturation into myelin-forming OLs. Overall, E3 offers an undemanding method for obtaining primary OLs with high yield and quality. Alongside its value as a practical tool, in vitro characteristics of the OL lineage additionally identified during the development of the E3 method have implications for advancing research on OL physiology and pathophysiology.
1. Introduction
Oligodendrocytes (OL) are the myelin-forming cells of the central nervous system (CNS), facilitating fast neuronal signal transmission by enabling saltatory conduction, as well as providing metabolic support to the neurons they enwrap [1,2]. OL death and demyelination cause neurological deficits, and insufficient remyelination eventually leads to axonal degeneration [3,4]. Demyelination is implicated in various neurological disorders, the most renowned being multiple sclerosis (MS) and ischemic white matter injury [5]. Considerable efforts have been made to understand OL development, characteristics, and involvement in demyelinating disorders, and strategies to enhance remyelination [6]. The complexity of the OL lineage makes it challenging to study its dynamics solely in an in vivo system, where the diversity of OL stages spanning from oligodendrocyte progenitor cells (OPCs) to mature OLs, signals, and interactions with other cell types may complicate the analysis of individual components, necessitating an in vitro cell culture model to perform detailed experiments.
In vitro OL culture models offer the advantage of a strictly regulated environment in which OL lineage cells are isolated from other elements of the brain, albeit at the expense of the structures, signals, and interactions preserved in an in vivo system. While a few established OL cell lines for culture do exist, they fall short of fully recapitulating OL morphology and function and do not cover the entire OL lineage [7,8]. Consequently, a large proportion of in vitro studies on OLs, developmental myelination, and de/remyelination in disease states have been conducted using primary cultured OLs. Various methods for isolating OPCs from rodent brains are available; the most widely used being the shaking method, which exploits the different surface adhesion capacities of glial cells to shake off OPCs from a stratified monolayer of astrocytes in a mixed glial culture [9,10]. Other techniques, such as immunopanning or magnetic-associated cell sorting (MACS) that are based on antigen-antibody reactions, also exist [[11], [12], [13], [14]]. However, such approaches may be costly, labor-intensive, and dependent on the experimenter's skills to accomplish the desired outcomes. Recently, novel and simpler methods for OL culture have been devised using OL-favoring growth factors or density gradient centrifugation to isolate OPCs from other brain cells [15,16]. Nonetheless, these methods have limitations in terms of the cell yield or purity of the obtained cell population.
Here, we introduce a novel method for primary OL culture, referred to as the E3 (easy, efficient, and effective) method. Using the E3 method, we present a simple physical method to isolate OPCs from neonatal rodent brains and provide an optimal media composition for the proliferation of OPCs and their subsequent differentiation into mature myelinating OLs.
2. Materials and methods
2.1. OPC isolation
A detailed protocol containing information on the reagents, equipment, and instructions for the full culture procedure with optimal media composition for each step is available in the Supplementary Information. Postnatal day 1 (P1) Sprague-Dawley rat pups were placed on ice for anesthetization and decapitated. The heads were collected in a 50 mL conical tube containing ice-cold Hank's Balanced Salt Solution (HBSS), transferred to another tube containing ice-cold 70 % ethanol, and immersed again in a new tube containing ice-cold HBSS. The cerebral cortices were dissected, and after removing the meninges, the hemispheres were moved to a 15 mL conical tube containing 1 mL of ice-cold Accumax™ (Merck Millipore, #SCR006) per tube, and the hemispheres were gently triturated 10 times with a 1 mL pipette tip. Four milliliters of dissection media which is composed of Hibernate-A medium (Thermo Fisher Scientific, A1247501), 1 % GlutaMAX™ (Thermo Fisher Scientific, #35050-061), and 1 % penicillin/streptomycin (Cytiva, #SV30010), plus approximately 200 IUs of papain (Worthington, #LS003126) were added to each tube. The tissues were then dissociated for 30 min at 30 °C. The dissociated cell suspension was then centrifuged at 200 g, 5 min at room temperature (RT), after which the supernatant was discarded and the remaining pellet was resuspended in 1 mL of dissection media+12 % Optiprep™ (Sigma Aldrich, #D1556). To overcome the limitation on the amount of processable tissue associated with traditional density gradient centrifugation methods, we adopted the principle of differential centrifugation, a method utilizing varying sedimentation speeds of particles commonly used for cellular organelle fractionation or exosome isolation [17,18]. The cell suspension was gently triturated 20 times with a 10 μL pipette tip attached to a 1 mL tip. Three milliliters of dissection media+12 % Optiprep™ was added, and the resulting 4 mL cell suspension was centrifuged at 200 g, 15 min at RT. The 4 mL supernatant was transferred to a different 15 mL tube, diluted with 4 mLs of dissection media, and the resulting 8 mL of 6 % Optiprep™ cell suspension was centrifuged at 200 g, 15 min at RT. The resulting pellet was resuspended in phosphate-buffered saline (PBS), and cells were counted and finally seeded on poly-d-lysine (PDL, Sigma Aldrich, #P6407) coated culture surfaces (working concentration: 10 μg/mL diluted in distilled water (DW), coated for 1 h, washed with DW three times and air dried) at a density of 1*104 cells/cm2 in OPC proliferation media (Table 1a) and incubated in a 37 °C, 5 % CO2 incubator. The same procedure was performed for P1 and P9 C57BL/6 mice, except that for P1 mice, the whole brain, excluding the hindbrain, was used, and three brains were processed per 1 column.
Table 1.
Media compositions.
| a. OPC proliferation media compositions. | |||||
|---|---|---|---|---|---|
| Base supplements: 2 % B27 + 1 % GlutaMAX™ + 1 % Penicillin/Streptomycin | |||||
|
Base growth factors: 30 ng/mL PDGF-AA + 10 ng/mL FGF | |||||
| Media composition | |||||
| 1 | 2 | 3 | 4 | 5 | 6 |
| DMEM |
DMEM + EGF |
DMEM/F12 |
DMEM/F12+EGF |
Neurobasal |
Neurobasal + EGF |
| EGF concentration: 10 ng/mL | |||||
| b. OL differentiation media compositions. | |||||
|---|---|---|---|---|---|
| Base supplements: 2 % B27 + 1 % GlutaMAX™ + 1 % Penicillin/Streptomycin +1 % N2 | |||||
|
Base growth factors: 40 ng/mL Triiodothyronine (T3) | |||||
| Media composition | |||||
| 1 | 2 | 3 | 4 | 5 | 6 |
| DMEM |
DMEM + CNTF |
DMEM/F12 |
DMEM/F12+CNTF |
Neurobasal |
Neurobasal + CNTF |
| CNTF concentration: 10 ng/mL | |||||
All animal experiments were reviewed and approved by the Institutional Animal Care and Use Committee of Ajou University School of Medicine (IACUC number; 2023-0021) and complied with the National Institutes of Health (NIH) Guide for the Care and Use of Laboratory Animals. P1 Sprague-Dawley rats were purchased from Orient Bio Korea, Inc., and P1 C57BL/6 mice were purchased from KOATECH, Korea.
2.2. OPC proliferation, passaging, and differentiation
Isolated OPCs were proliferated in an OPC proliferation medium under varying conditions (Table 1a) for 5–6 days, with a full medium change on day 3 of proliferation. For further passaging and differentiation, the OPC proliferation medium, specifically condition 4 of Table 1a, was chosen. Following the proliferation period, the media was aspirated and 5 mLs per T75 flask of Accutase® (Thermo Fisher Scientific, #A11105-01) was added, and the flask(s) were incubated in a 37 °C, 5 % CO2 incubator for 5 min. The flasks (s) were gently tapped to detach the cells, and the cell suspension was transferred to a 15 mL tube and centrifuged at 200 g, 3 min RT. The resulting pellet was resuspended in OPC proliferation media (Table 1a), counted, seeded on appropriate culture surfaces at a density of 1*104 cells/cm2, and allowed to proliferate for 2 days. For further differentiation, passaged OPCs cultured under condition 3 of Table 1a were selected. Full media exchange with OL differentiation media (Table 1b) was performed and the cells were allowed to differentiate for 2 or 4 days.
2.3. Oligodendrocyte/neuron co-culture
Primary neurons were obtained from the P1 rat cortices using the following method: Using the OPC isolation method described in Section 2.2, cells pelleted from the second centrifuge in 8 mL dissection media+6 % Optiprep™ were seeded on culture surfaces at a density of 5*104 cells/cm2 in a media composition of Neurobasal medium (NBM, Thermo Fisher Scientific, #21103049), 2 % B27 Supplement (Thermo Fisher Scientific, #17504-044), 1 % N2 supplement (Thermo Fisher Scientific, #17502048), 1 % GlutaMAX™, and 1 % penicillin/streptomycin. The cells were maintained for 14 days with half-medium exchange every 3 days, yielding a primary neuron culture. On the 14th day, the media was exchanged with OL differentiation media, specifically condition 5 of Table 1b, and passaged OPCs were seeded onto the neurons at a density of 3*104 cells/cm2. The co-culture was maintained for 4 days, with a half-medium change on day 2.
2.4. Myelinating OL culture on aligned nanofibers
Passaged OPCs were seeded on aligned nanofiber inserts (Sigma Aldrich, #Z694614-12 EA, fiber diameter = 700 nm) at a density of 3*104 cells/cm2 in OPC proliferation media, condition 3 of Table 1a. The medium was changed to the OL differentiation medium (condition 5 of Table 1b after 2 days of proliferation. Differentiation was maintained for 4 days with half-medium exchange on the second day.
2.5. WST-8 cell viability and proliferation assay
WST-8 assays were conducted with OPCs cultured in 96 well plates and the Quantimax™ cell viability kit (BioMax, #QM1000) according to the manufacturer's protocol. After isolation and proliferation for 3/4/5 days, the media was aspirated, cells were washed with pre-warmed PBS, and 100 μL of Dulbecco's Modified Eagle Medium (DMEM) without phenol red (Thermo Fisher Scientific, #21063029) was added. Ten microliters of Quantimax™ was promptly added to each well, and the plate was incubated for 30 min in a 37 °C, 5 % CO2 incubator. The OD at 450 nm was measured with a microplate reader. Proliferative capacity was calculated as a relative value using the following formula: (OD value of each well – average of OD values of the blank)/(average of OD values of wells under condition 1 of Table 1a. – average of OD values of the blank).
2.6. Immunofluorescence
For immunofluorescence, cells cultured on 9 mm coverslips and nanofiber inserts were used. After designated culture periods, the cells were fixed for 20 min with 4 % paraformaldehyde, washed three times with PBS, and stored at 4 °C until use. The cells were blocked with 10 % normal goat serum (NGS) and 0.1 % Triton X-100 in PBS for 1 h at room temperature. Cells were washed with PBS and were incubated overnight at 4 °C with the following primary antibodies in the blocking solution; anti-NG2 (1:500, Millipore, #AB5320), anti-NG2 (1:500, Santa Cruz Biotechnology, #sc-33666), anti-MBP (1:500, Abcam, #AB7349), anti-Olig2 (1:500, Merck Millipore, #MABN50), anti-GFAP (1:2000, Dako, #Z033429), anti-GFAP (1:2000, Abcam, #AB4674), anti-Tuj1 (1:1000, Dako, #G7121), anti-Iba1 (1:500, Wako, #019–19741), anti-cleaved-caspase-3 (1:200, Cell Signaling, #9664S), anti-Ki67 (1:500, Thermo Fisher Scientific, #MA514520), anti-BCAS1 (1:500, Synapse Systems, #445-003). The cells were washed with PBS and incubated for 1hr, RT with the appropriate Alexa Fluor 488-, 594-, and 680-conjugated secondary antibodies (Thermo Fisher Scientific). Subsequently, the cells were counterstained with 4′,6-diamidino-2-phenylindole, dihydrochloride (DAPI, Sigma Aldrich, #D9542) for 10 min at RT and mounted onto glass slides. The cells were imaged using a Zeiss LSM800 confocal microscope.
2.7. EdU proliferation assay
EdU proliferation assays were performed using the EdU Assay/EdU Staining Proliferation Kit (iFluor 488) (Abcam, ab219801) according to the manufacturer's protocol. Isolated OPCs were proliferated for 4.5 days and incubated with EdU for 2 h. The cells were fixed with 4 % PFA and followed by immunofluorescence staining for Ki67. An additional EdU labeling step with the EdU reaction mix, including iFluor 488 azide, was performed between the secondary antibody incubation and the DAPI counterstaining step, with an incubation period of 30 min at RT.
2.8. Statistics and image quantification
Statistical analyses, including independent t-tests, one-way analysis of variance (ANOVA) with Tukey's post hoc tests, simple linear regression, spline/LOWESS fitting, and frequency distribution analysis, were performed using GraphPad Prism, version 9.5.1 (GraphPad Software, Boston, Massachusetts USA). P-values were two-sided, and a value of < 0.05 was considered statistically significant. Cell counting was performed with Fiji/ImageJ using the Analyze particles function when applicable. Measurements of nearest neighbor distances (NND) were made by first obtaining the XY coordinates of the images of cell nuclei stained with DAPI in Fiji/ImageJ and converting the data table into a Poisson point process using the spatial statistics (spatstat) package in R, version 4.2.0 (R Foundation for Statistical Computing, Vienna, Austria). Nearest neighbor distances and spatial G-function results were obtained using spatstat. Sholl analysis was performed using the Neuroanatomy plugin in the Fiji/ImageJ. Myelin processes in OLs were visualized by immunostaining for myelin basic protein (MBP), and binary masks were obtained from thresholding the images. The center point at the location of the DAPI + nuclei of the cell being analyzed was marked, and Sholl analysis was performed by drawing circles of increasing radius from the center point and quantifying the number of intersections between each circle and the binary mask. The morphological complexity was determined by comparing the total number of cell intersections. A schematic illustration of the culture procedure was generated using Bio-Render.com.
3. Results
3.1. Isolation of OPCs from the neonatal rat brain
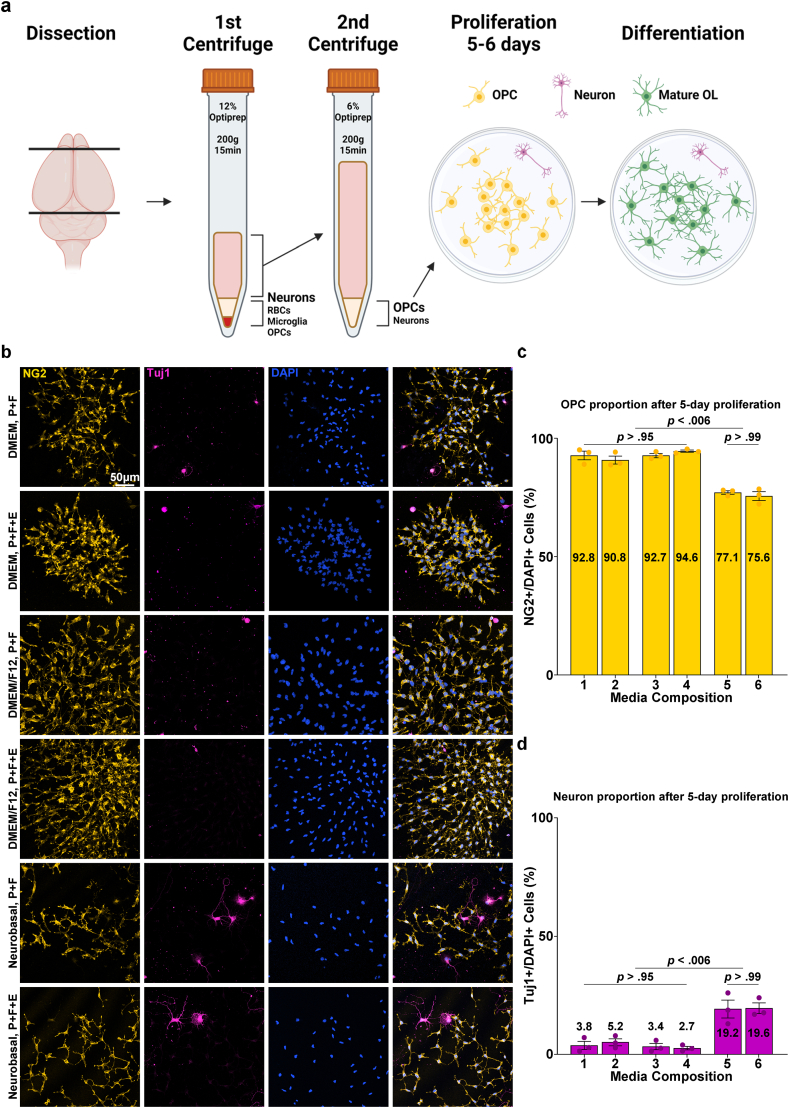
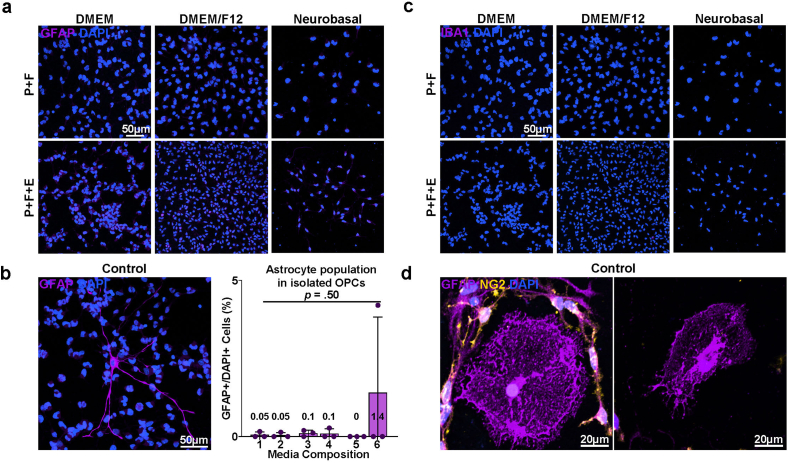
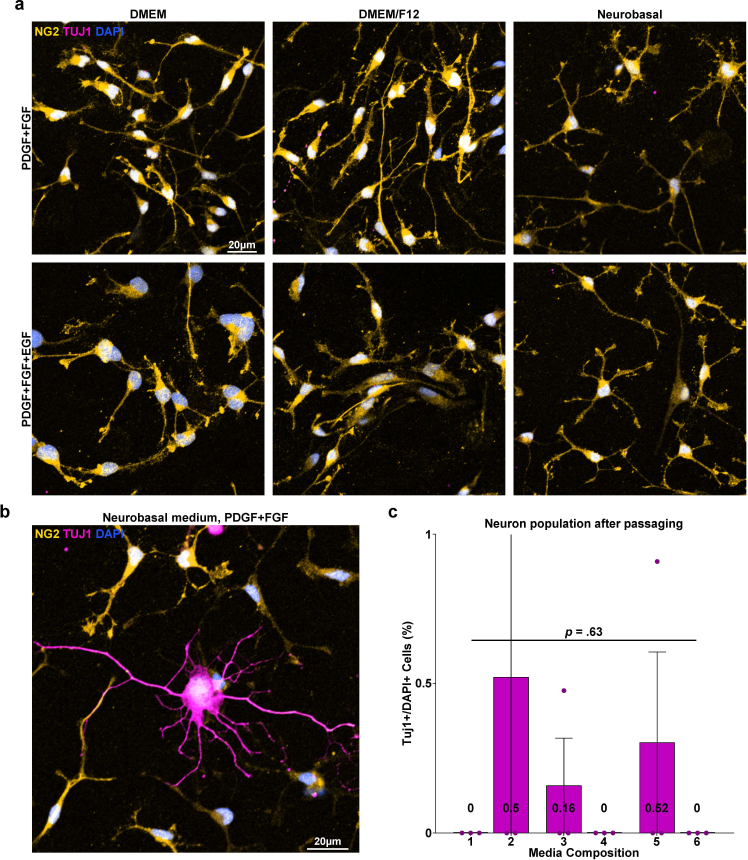
The development of the E3 method for primary OL culture commenced with an attempt to improve a density gradient centrifugation-based method for isolating adult OPCs from the rodent cerebral cortex [16]. A significant limitation of density gradient centrifugation is its restricted tissue loading capacity in a single column due to the concentration of cells and debris in one phase during the centrifugation process. To circumvent this problem, we adopted the principle of differential centrifugation, a technique widely employed for cellular organelle fractionation and exosome isolation that exploits the differential sedimentation speeds of particles [17,18] (Fig. 1a). Following the dissection and dissociation of the P1 rat cerebral cortices, the resulting cell suspension was passed through two sequential steps of centrifugation with the density gradient medium Optiprep™ to exclude unwanted cells and debris. First, the cell suspension was centrifuged in 4 mLs of dissection medium containing 12 % Optiprep™. After centrifugation, the supernatant was transferred to another 15 mL tube, diluted with dissection media to 6 % Optiprep™ (8 mL in volume), and centrifuged again. The cell population from the pellet obtained through the 2nd centrifuge was seeded at a low density of 1*104 cells/cm2 and cultured for 5 days in six compositions of the OPC proliferation medium (Table 1a). B27 supplement, known for its inclusion of various nutritional factors and is frequently used in serum-free oligodendrocyte culture media [10,14,19], was a constant component across all media compositions, as well as the growth factors platelet-derived growth factor (PDGF) and fibroblast growth factor (FGF), which are known to induce OPC proliferation and inhibit their spontaneous differentiation [[20], [21], [22], [23], [24]]. Six conditions were compared based on the differences in the base medium commonly used in primary OL cultures: DMEM, DMEM/F12, NBM, and the presence or absence of epidermal growth factor (EGF), which has been documented to promote OPC proliferation in vitro [25,26]. When cultured in DMEM or DMEM/F12-based OPC proliferation media, cells from the 2nd centrifuge grew into colonies of OPCs, with NG2+ OPCs comprising 90–95 % of the total population and 3–5% proportion of the background Tuj1+ neurons (Fig. 1b, c and 1d). GFAP + astrocytes accounted for approximately 0.5 % of the total population (Figs. S1a and S1b), and Iba1+ microglia were not observed (Figs. S1b and S1c). In NBM-based proliferation media, OPC colonies displayed reduced size and a lower proportion of OPCs, approximately 75–77 %. Furthermore, Tuj1+ neurons account for approximately 20 % of the total population. The cell pellet from the first centrifuge at 12 % Optiprep™ contained red blood cells, and when cultured using the same procedure, it showed a similar cell population except for the discernible presence of Iba1+ microglia (Fig. S1d). Thus, through two sequential density centrifugations, a cell population enriched in OPCs was obtained after 5–6 days of proliferation.
Fig. 1.
Cell composition of isolated OPC-enriched culture. a Scheme of the culture procedure. b Representative images of isolated cells (after 5 days of proliferation). Immunostaining for NG2 and Tuj1. P: PDGF, F: FGF, E: EGF. c Quantification of NG2+ OPC proportions after 5 days of proliferation according to media compositions corresponding to Table 1a (n = 3 biologically independent samples/experiments, 6 groups per sample, 5 images per group, approximately 300 DAPI + cells per image. One-way ANOVA and Tukey's post hoc test, two-sided. Data presented as mean values ± SEM.). d Quantification of Tuj1+ Neuron proportions after 5 days of proliferation according to media compositions corresponding to Table 1a (n = 3 biologically independent samples/experiments, 6 groups per sample, 5 images per group, approximately 300 DAPI + cells per image. One-way ANOVA and Tukey's post hoc test, two-sided. Data presented as mean values ± SEM.).
3.2. Media composition and growth factors for OPC proliferation
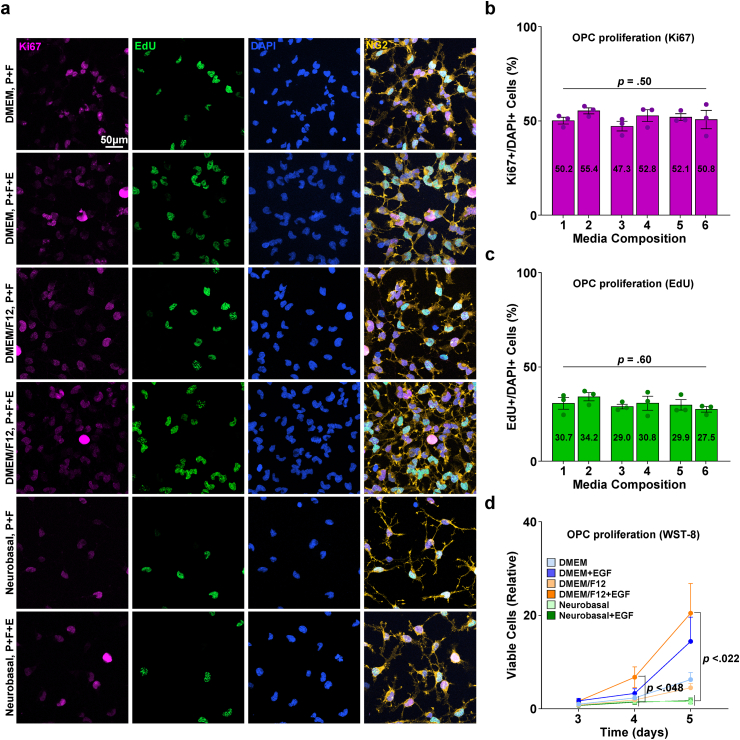
After the isolation of OPCs, various OPC proliferation media compositions were compared to determine the most effective formula for inducing OPC proliferation. Isolated OPCs were proliferated for 3–5 days in six designated OPC proliferation medium compositions. While immunostaining for the proliferation markers EdU and Ki67 showed no difference in the proportion of cells proliferating between media conditions, WST-8 proliferation/viability assays revealed that at both the 4- and 5-day time points, condition 4 (DMEM/F12 with EGF) showed an approximately 10–15 fold higher viable cell number than the NBM-based formulae (Fig. 2a, b, 2c and 2d). Therefore, we selected condition 4, DMEM/F12 with EGF, as the optimal OPC proliferation medium, and the following steps were performed throughout the culture procedure with OPCs cultured in this specific medium composition.
Fig. 2.
Proliferation of isolated primary OPC enriched culture. a Representative images of isolated cells (after 4.5 days of proliferation). Staining for NG2, EdU, Ki67. P: PDGF, F: FGF, E: EGF. b Quantification of Ki67+/DAPI + cells (%) according to media compositions corresponding to Table 1a (n = 3 biologically independent samples/experiments, 6 groups per sample, 10 images per group, approximately 500 DAPI + cells per image. One-way ANOVA. Data presented as mean values ± SEM.). c Quantification of EdU+/DAPI + cells (%) according to media compositions corresponding to Table 1a (n = 3 biologically independent samples/experiments, 6 groups per sample, 10 images per group, approximately 500 DAPI + cells per image. One-way ANOVA. Data presented as mean values ± SEM.). d Graph of WST-8 viability assay results, values relative to the DMEM (media composition 1) group cultured for 3 days (n = 3 biologically independent samples/experiments, 18 groups per sample, 3 replicates (wells) per group. One-way ANOVA and Tukey's post hoc test. Data presented as mean values ± SEM.).
3.3. Differentiation of OPCs into mature OLs and the passaging of proliferated OPCs
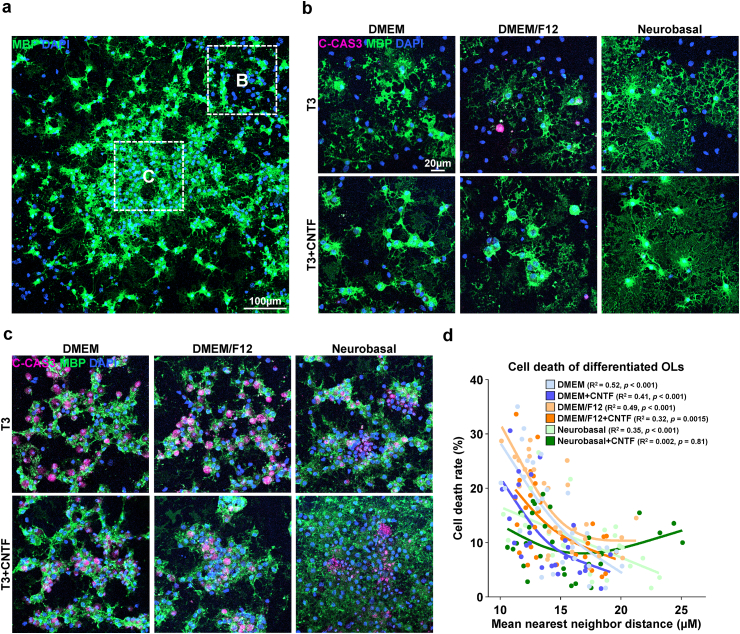
After the successful proliferation of the isolated OPCs, the culture medium was transitioned to an OL differentiation medium to induce their maturation into myelinating OLs. Different medium compositions were tested for the differentiation of OPCs into mature OLs (Table 1b). The use of B27 and N2 supplements, which incorporate insulin, which is known to be essential for the survival of mature OLs in vitro [27], and the thyroid hormone T3, a well-established OL differentiation-promoting factor, was uniform across all conditions [[28], [29], [30], [31], [32]]. As with the OPC proliferation media conditions, another six conditions were compared, which could be distinguished by their base medium: DMEM, DMEM/F12, NBM, and the presence or absence of ciliary neurotrophic factor (CNTF), a growth factor known to promote OL maturation and thus commonly included in OL differentiation media formulae [33,34]. After 4 d of differentiation, MBP + mature oligodendrocytes with web-like myelin sheets were observed in all groups (Fig. 3b). However, aside from the successful differentiation of OPCs into mature OLs, we discovered areas of clumped oligodendrocytes with entangled MBP + processes that were indistinguishable from one another, showing nuclear condensation and immunoreactivity for cleaved caspase 3 (C-Cas3), indicative of apoptosis (Fig. 3c). Notably, these apoptotic OL clusters were most prominent in the center of the colonies with high focal cell density (Fig. 3a). Correlation analysis revealed an inverse correlation between the mean nearest-neighbor distance and cell death rate, showing that overcrowding of OPCs during proliferation resulted in apoptotic cell death during the induction of maturation (Fig. 3d). To address this problem, an additional procedure to redistribute the proliferating OPCs and acquire more space was deemed necessary.
Fig. 3.
Differentiation of isolated OPC-enriched culture. a Representative image of isolated cells after 4 days of differentiation. Immunostaining for myelin basic protein (MBP). b Representative images of isolated cells after 4 days of differentiation, in the periphery of colonies (Area B of Fig. 3a). Immunostaining for cleaved-caspase 3, MBP. c Representative images of isolated cells after 4 days of differentiation, in the center of colonies (Area C of Fig. 3a). Immunostaining for cleaved-caspase 3, MBP. d Scatter plot of the mean nearest neighbor distance of cells in immunostained images and cell death rate measured as cleaved caspase-3+/DAPI + cells (%), and spline/LOWESS fitting. The correlation between mean nearest neighbor distance and cell death rate was analyzed with simple linear regression for each media condition (n = 3 biologically independent samples/experiments, 6 groups per sample, 10 images per group, approximately 300 DAPI + cells per image).
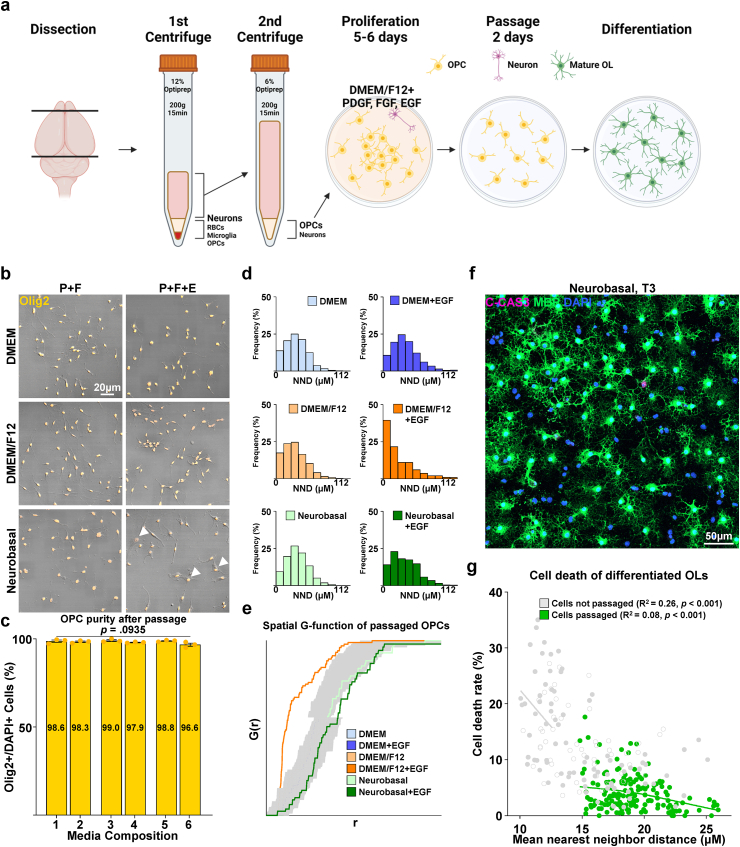
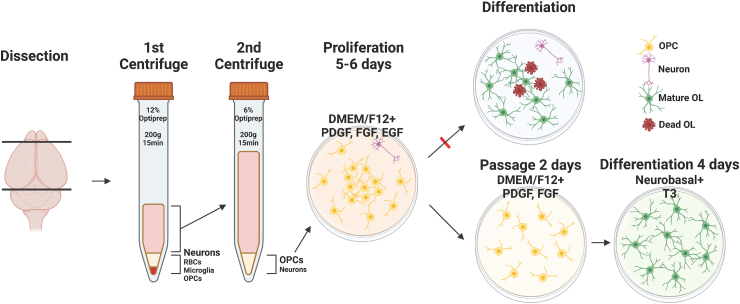
For the redistribution and spacing of OPCs to avoid cell death during differentiation, proliferated OPCs were passaged and redistributed with the synthetic enzyme Accutase®, which revealed a cell yield of approximately 3–5*106 cells/brain, and seeded at a density of 1*104 cells/cm2 (Fig. 4a). Passaged OPCs were stabilized for 2 days in OPC proliferation media. The six previously tested proliferation media compositions were reassessed to confirm which formula was most adequate for the 2-day stabilization period. After passaging, the background Tuj1+ neurons, which occupied 5 % of the total population at initial OPC isolation and proliferation, were reduced to a near-nonexistent state, and the purity of Olig2-positive OL lineage cells, OPCs judging from their bi/tripolar morphology and immunoreactivity for NG2, increased from the prior 90–95 % to approximately 98–99 % (Fig. S2a and Fig. 4c). The average nearest neighbor distance of passaged cells after 4 days of differentiation was increased from approximately 15 μm–19 μm compared with cells differentiated without passaging (P < 0.001, nested independent t-test, n = 3, two-sided), and the mean cell death rate after 4 days of differentiation, measured as the percentage of cleaved caspase-3 positive cells was reduced from 13 % to 4 % (P < 0.001, nested independent t-test, n = 3, two-sided) (Fig. 4f and g). We observed that the use of DMEM/F12 with EGF, which was the formula of choice during the OPC proliferation period preceding passaging, appeared to cause cells to have a more inhomogeneous spatial distribution compared with other media conditions. To quantify the “randomness” of the spatial distribution, the coordinates of the cell nuclei shown by DAPI in a square image were converted to Poisson point processes, and histograms of nearest-neighbor distances and graphs of the G-function, a cumulative distribution function of nearest-neighbor distances, were drawn. While cells stabilized after passaging in the other five media compositions showed a distribution within the boundaries of complete spatial randomness, cells stabilized in DMEM/F12 with EGF showed a left shift in both the histogram of the NND frequency distribution and the graph of the G function, indicating cell clustering (Fig. 4d and e). Furthermore, cells showing a morphology different from typical OPCs were also visible in media containing EGF, especially when combined with NBM (Fig. 4b); therefore, DMEM/F12 without EGF, condition 3 of Table 1a was selected as the medium for the post-passage stabilization period.
Fig. 4.
Characterization of passaged OPC culture. a Scheme of revised culture procedure, involving the passaging of expanded OPCs. b Representative images of OPCs passaged and redistributed. Immunostaining for Olig2. Arrowheads; Olig2+ cells with atypical morphology. c Quantification of Olig2+/DAPI + cells (%) according to media compositions corresponding to Table 1a (n = 3 biologically independent samples/experiments, 6 groups per sample, 10 images per group, approximately 50 DAPI + cells per image. One-way ANOVA. Data presented as mean values ± SEM.). d Representative histograms of nearest neighbor distances (NND) of OPCs after passaging (n = 3 biologically independent samples/experiments, 6 groups per sample, 16 images per group, approximately 200 DAPI + cells per image). e Graph of spatial G-function (cumulative distribution of nearest neighbor distances) of passaged OPCs by proliferation media composition. Gray zones demarcate boundaries of complete spatial randomness. f Representative low-magnification image of passaged OPCs after 4 days of differentiation into mature OLs in a Neurobasal-based differentiation media (Condition 5 of Table 1b). Immunostaining for cleaved caspase 3, MBP. g Scatter plot of the mean nearest neighbor distance of cells in immunostained images and cell death rate measured as cleaved-caspase 3+/DAPI + cells (%), and spline/LOWESS fitting. The correlation between mean nearest neighbor distance and cell death rate was analyzed with simple linear regression for each condition (n = 3 biologically independent samples/experiments, 6 groups per sample, 9 images per group, approximately 200 DAPI + cells per image).
3.4. Differentiation of passaged OPCs into mature OLs
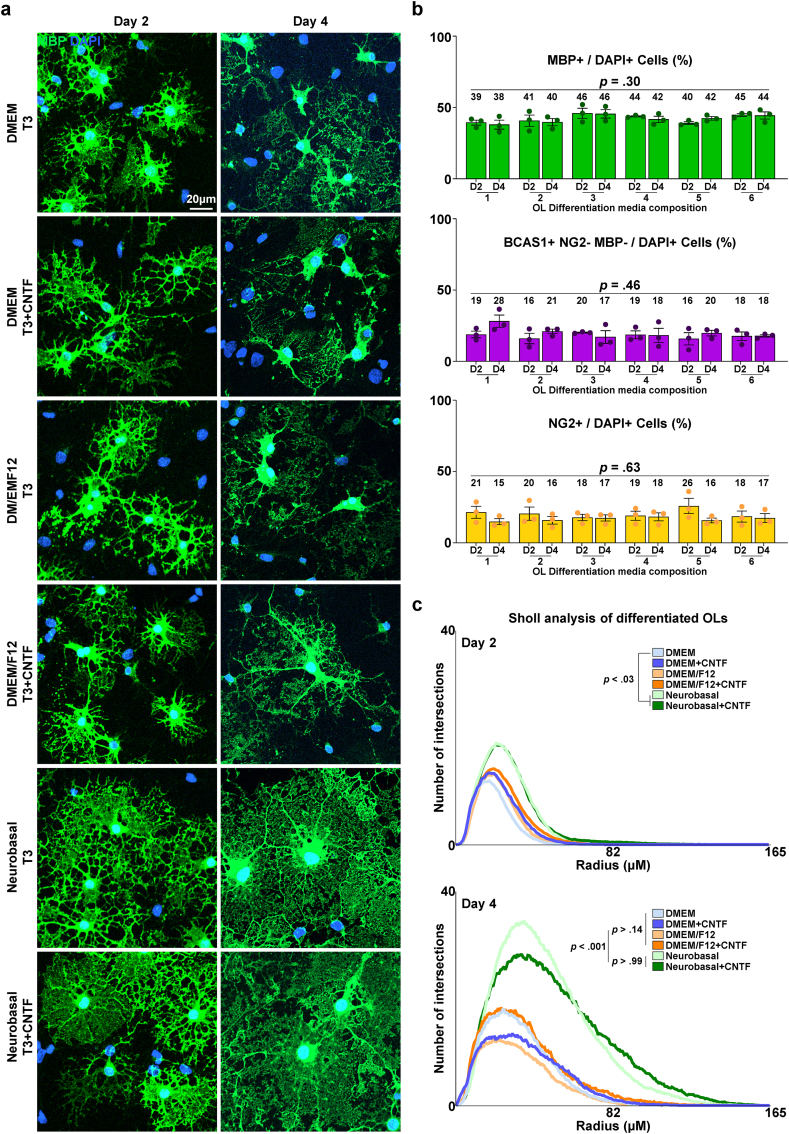
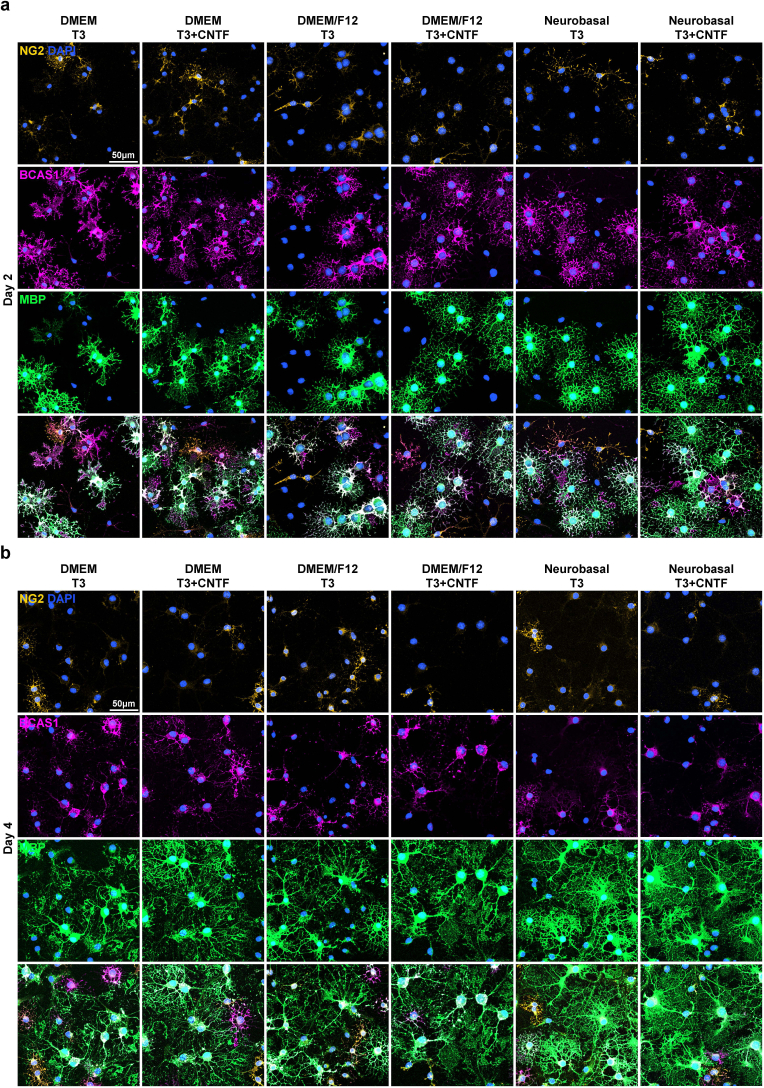
After passaging, the OPCs were stabilized and proliferated for 2 d. Again, we compared the maturation-inducing capacity of the six OL differentiation media. After 2 days of differentiation, MBP + mature OLs with branch-like processes were observed, constituting 35–50 % of the population, regardless of the media used (Fig. 5a and b). By the fourth day, web-like myelin sheets were evident in all groups. Despite the variation in morphological appearance, the actual proportion of MBP + OLs was not significantly different between cultures, regardless of the media composition and duration of differentiation (Fig. 5a and b). The proportions of NG2+ OPCs and breast carcinoma amplified sequence (BCAS1) + intermediate-stage OLs were consistent across all conditions (Fig. 5b and Fig. S3). Morphological complexity was assessed by Sholl analysis to compare the extent of differentiation. The results revealed that the NBM-based formulae produced OLs with significantly more complex processes on day 4 (Fig. 5a and c). The inclusion of CNTF did not influence OL maturation in terms of quantity or morphological complexity (Fig. 5a, b, and 5c). Overall, we successfully induced the differentiation of OPCs into mature OLs and discovered that the NBM-based differentiation media greatly enhanced the morphological complexity of mature OLs.
Fig. 5.
Differentiation of passaged OPC culture. a Representative images of differentiated OLs, differentiated after passaging for 2 or 4 days in designated OL differentiation media compositions. Immunostaining for MBP. b Quantification of MBP+/DAPI + cells, BCAS1+ NG2- MBP-/DAPI + cells, NG2+/DAPI + cells (%) of passaged OPCs after designated differentiation periods and media compositions corresponding to Table 1b (n = 3 biologically independent samples/experiments, 12 groups per sample, 10 images per group, approximately 50 DAPI + cells per image. One-way ANOVA and Tukey's post hoc test, two-sided. Data presented as mean values ± SEM.). c Results of Sholl analysis, of differentiated OLs. Comparison of morphological complexity was performed by comparing the total number of intersections per cell.. (n = 3 biologically independent samples/experiments, 12 groups per sample, 10 images per group, approximately 10 MBP + OLs per image. One-way ANOVA and Tukey's post hoc test, two-sided. Data presented as mean values ± SEM.)
3.5. Applicability of the E3 method to C57BL/6 mice
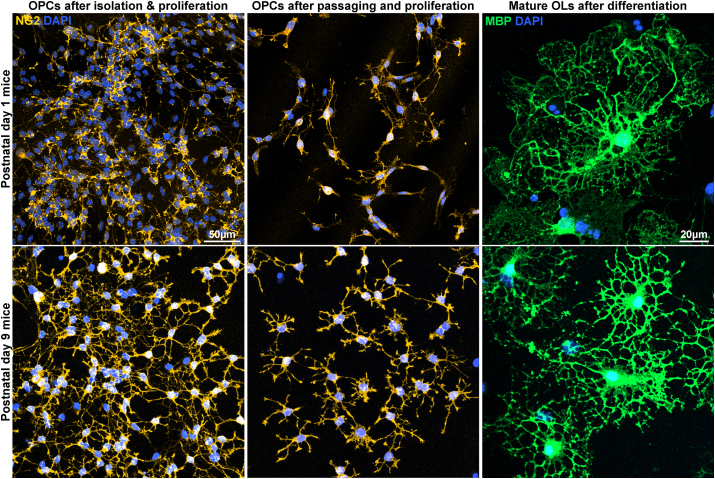
After confirming successful OL differentiation, we tested the applicability of the E3 method in C57BL/6 mice, a species that is widely used in research and constitutes the majority of transgenic animal lines. We first tested P1 mice with minor modifications to the isolation procedure to adjust for smaller brain sizes. We observed successful proliferation, passaging, and differentiation of OL-lineage cells in P1 mouse OPCs using the E3 method (Fig. 6). We also noted that during the proliferation of isolated OPC colonies, P1 mouse OPCs tended to cluster together more strongly than P1 rat OPCs and that this tendency persisted even after passaging when EGF was no longer present in the culture environment (Fig. 6). To circumvent this problem, we used mice of older age typically P9 mice. We observed that the potency of OPCs to cluster together was diminished in P9 mice and that P9 mouse OPCs showed a more ramified appearance upon staining for NG2 (Fig. 6).
Fig. 6.
Applicability of the E3 method for C57BL/6 mice. Representative images of postnatal day 1 and 9 mouse oligodendrocytes cultured through the E3 method. Immunostaining for NG2, MBP.
3.6. Myelin sheath formation of OLs culture through the E3 method
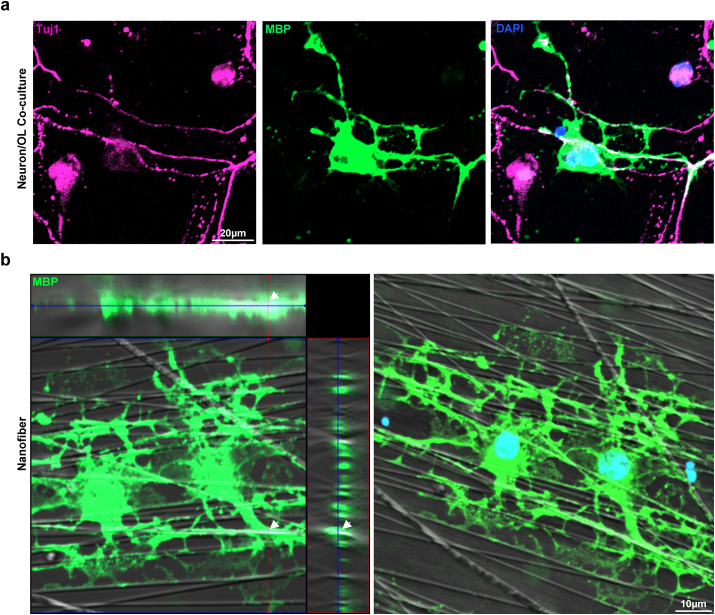
After confirmation of successful OL differentiation, we validated the capacity of cultured and maturated OLs produced with the E3 method to enwrap neurons or neuron-like structures and form myelin sheaths [10,15,19,35]. After observing the presence of background neurons during OPC proliferation after isolation, primary neuron cultures were obtained by seeding isolated cells from the second centrifuge step with 6 % Optiprep™ at a high density of 5*104 cells/cm2, in NBM supplemented with B27 and N2 without the OPC-favoring growth factors, PDGF, FGF, and EGF. Robust axonal processes were observed after culturing for 14 days. Passaged OPCs were seeded on neuron cultures at a density of 3*104 cells/cm2, and the medium was exchanged to differentiation medium condition 5, NBM-based medium without CNTF. On day 4 of differentiation, we observed areas of overlapping Tuj1+ axons and MBP + myelin sheaths (Fig. 7a). Furthermore, we examined the ability of cultured OLs to enwrap the aligned nanofibers. Passaged OPCs were seeded on 12-well aligned nanofiber inserts with a diameter of 700 nm, and Z-stack images of MBP + myelin sheaths on the aligned fibers were visualized using a differential interference contrast (DIC) filter; we noted the efficient formation of myelin sheaths in 3D reconstructions, which were wrapped around the aligned fibers (Fig. 7b). Thus, we confirmed that OLs cultured using the E3 method could form myelin sheaths and enwrap neurons in vitro. The final procedure for the E3 method is illustrated in Fig. 8.
Fig. 7.
Myelination capacity of cultured oligodendrocytes. a Representative image of neuron/oligodendrocyte co-culture. Immunostaining for MBP, Tuj1. b Representative images of differentiated oligodendrocytes cultured on aligned nanofibers. Immunostaining for MBP.
Fig. 8.
Final schematic for the E3 method for primary oligodendrocyte culture.
4. Discussion
Research on oligodendrocytes and myelination has greatly benefited from in vitro culture models, which allow the deconvolution of numerous dynamic stages of the OL lineage. All stages of the OL lineage, including OPCs, are continuous in the brain throughout life, and the stage of a typical OL lineage cell is subject to changes. This may make even simple observations, such as counting of cells, challenging and dependent on the use of multiple stage-specific reporters. Such considerations emphasize the need for an efficient method for in vitro primary OL culture. The E3 method introduced in this study significantly improves both the simplicity and efficiency of the procedure.
The shaking method, pioneered by McCarthy & de Vellis, is the oldest and most widely used method for isolating primary OPCs [9,10,[36], [37], [38], [39]]. This method exploits the different adhesion capacities of glial cell types to shake off OPCs from mixed glial cultures containing animal serum. Despite the advantages of a simple procedure, the shaking method has its drawbacks. The E3 method presents a greater cell yield, a shorter period for the acquisition of pure OPCs, and the absence of an overnight shaking step, which can cause potential damage to cells (Table 2). Immunopanning enables the direct acquisition of OPCs from brain tissue without a separate expansion process and a high purity above 99.5 %, but a prolonged dissection time due to multiple panning steps, and expensive requirements such as multiple antibodies for the negative and positive selection of cells, and hybridomas for the generation of those antibodies, are necessary to achieve results [[11], [12], [13],27,40] (Table 2). Magnetic-associated cell sorting (MACS), another method based on the exploitation of antibody-antigen reactions, also has the advantage of direct acquisition of OPCs, but the disadvantage of numerous expensive materials, such as antibodies, microbeads, tissue dissociation kits, dissociators, magnetic columns, and separators [14,41] (Table 2). The E3 method excels in terms of cost-effectiveness and simplicity of the procedure compared with these methods. A recent method developed by Yoshida et al. demonstrated that seeding dissociated brain cells from neonatal rats at a low seeding density (6.7*103 cells/cm2) in an OPC proliferation medium is sufficient to produce OL populations. While this method enables the acquisition of a high yield of OPCs using a very simple process, a significant number of non-OLs emerge during culture. The E3 method employs the principle of expanding OPCs from a low-density cell population; with the addition of two differential centrifugation steps and passaging, greater purity has been attained [15]. Another recent method, based on density gradient centrifugation, was devised for the acquisition of OPCs from adult rats. Although this method does allow for OL culture from adult rats which is not possible with conventional methods, density gradient centrifugation causes cells and debris to concentrate into a single phase during the centrifugation, and this places a limit on the amount of tissue that one column can process due to the “clogging” which can be caused by aggregated cells and debris [16,42]. OPC isolation using the E3 method is based on two sequential differential centrifugations that do not cause the particles to aggregate into a single phase, which greatly increases the amount of tissue processable per column and, as a result, results in a high cell yield. Collectively, the E3 method is a cost-effective and simple method with few requirements for producing high-purity, high-yield primary OL cultures (Table 2). The high yield, purity, and cost-effectiveness of the procedure may make the E3 method a competitive choice for high-throughput screening of drugs that promote remyelination after damage induced by noxious inflammatory stimuli.
Table 2.
Comparison of the E3 method with conventional primary OL culture methods.
| Parameter |
Method |
|||
|---|---|---|---|---|
| Shaking | Immunopanning | MACS | E3 | |
| Animal used | P1-4 rat | P6-8 rat/mouse | P5-8 rat/mouse | P1 rat/mouse |
| Dissection time (Sacrifice ∼ incubation) |
Approx. 1.5 h 3min/pup dissection 30–60min dissociation |
Approx. 4 h 3min/pup dissection 30–60min dissociation 15–45min panning*3 |
Approx. 2 h 3min/pup dissection 30–60min dissociation 35–45min MACS |
Approx. 2 h 3min/pup dissection 30–60min dissociation 15min centrifuge*2 |
| Time required for acquisition of OPCs |
7–10 days Time until shaking |
Approx. 4 h Dissection time |
Approx. 2 h Dissection time |
5–6 days Time until passage |
| Yield of OPCs | 0.2–1*106/rat |
2–2.5*106/rat 0.8–1*106/mouse |
1*106/rat 0.37*106/mouse |
3–5*106/rat 0.8–1*106/mouse |
| Purity of OPCs |
90∼98 % Contaminant = Astrocytes, Microglia |
>99.5 % Contaminant = Fibroblast-like cells |
>90 % |
98–99 % Contaminant = Neurons |
| Exclusive materials required |
Fetal Bovine Serum Shaking Incubator |
Antibodies (anti-RAN2, anti-BSL1, anti-GalC, anti-O4, anti-A2B5, anti-PDGFRα) Hybridomas |
Anti-O4 antibody or microbeads Dissociation kit MACS dissociator Magnetic columns & separator |
Iodixanol (OptiprepTM) |
| References | McCarthy & de Vellis, 1980 Yang et al., 2005 O'Meara et al., 2011 Zhu et al., 2014 Choi et al., 2017 Choi et al., 2023 |
Barres et al., 1992 Mayer-Proschel, 2001 Dugas & Emery, 2013a Dugas & Emery, 2013b Emery & Dugas, 2013 |
Dincman, Beare, Ohri, & Whittemore, 2012 Weil et al., 2019 |
|
For OL culture, rats have been used more often than mice due to their larger brain size and the fact that mouse OPCs exhibit characteristics that make their isolation more complicated with conventional methods such as the shaking method or immunopanning. In detail, mouse OPCs do not abundantly express certain surface antigens such as A2B5 compared to rat OPCs which may complicate immunopanning, and they have been shown to spontaneously differentiate in mixed glial cultures used for the shaking method [43]. When tested for applicability to C57BL/6 mice, we were able to successfully reproduce the culture procedure of the E3 method for mouse OLs. We noted subtle differences in mouse OPCs compared to their rat counterparts, such as their higher capacity to cluster together even in the absence of EGF in the environment. The use of an older animal ameliorated this tendency. A specific age between P1–P9 may be the most appropriate for the E3 method.
During the development of the E3 method, we unraveled a few intriguing in vitro characteristics of OL-lineage cells that may have implications for both in vitro and in vivo research on OL physiology and pathology. The first was the dual effect of NBM on OPC proliferation and OL differentiation. We found that NBM showed significantly inferior performance compared to DMEM or DMEM/F12 in terms of OPC proliferation; however, when it came to the differentiation of OPCs into mature OLs, NBM produced OLs with more ramified myelin processes. As a medium originally developed for the culture of primary neurons, the most prominent feature that separates NBM from both DMEM and DMEM/F12 is its low osmolarity of approximately 235 mOsm when combined with the B27 supplement, compared with the osmolarity of DMEM or DMEM/F12, which is approximately 335 mOsm [44]. A previous study showed that increasing the osmolarity of NBM with NaCl or mannitol boosts its capacity for OPC proliferation to levels comparable to those of DMEM [45]. Prior research exploring the effect of extracellular matrix (ECM) stiffness and mechanosensing on the proliferation and differentiation of OPCs is also present, which may be a possible mechanism behind the effect of the low osmolarity of NBM, considering that changes in membrane tension caused by differing osmolarity levels influence the activation of the mechanoreceptor Piezo1/2 [[46], [47], [48]]. The results of our study suggest that the low osmolarity of NBM also contributes to a better induction of OL maturation. The dual effect of osmolarity on OL dynamics may not be limited to in vitro systems and may also play a role in vivo especially in the context of developmental myelination or remyelination after injury, warranting further research.
Another noteworthy feature of OLs that we discovered was the death of cells during differentiation, caused by the overly close distribution of OPCs. The formation of densely crowded OPC colonies during proliferation led to the development of entangled OLs in the colony center, many of which displayed markers of apoptosis. An inverse correlation was observed between the average space between OPCs and cell death during the induction of differentiation. The formation of overcrowded OPC colonies hints at their stemness, which is more obvious in the formation of “oligospheres” from neural progenitors observed in vitro [43,[49], [50], [51]]. Although the degree of OPC clustering shown by the E3 method or the generation of oligospheres may be a phenomenon limited to in vitro circumstances, there are numerous studies on the reactive proliferation of OPCs after injury and their insufficient differentiation and remyelination [[52], [53], [54]]. We speculate that the overcrowding of OPCs may cause insufficient differentiation and cell death. Additionally, the association between OPC clustering and cell death may provide insights for future research on OPC transplantation to promote remyelination after injury. Numerous studies on OPC transplantation have been conducted, and they have been shown to have a meaningful effect [[55], [56], [57], [58], [59], [60]]. However, in many cases, an excessively large number of OPCs are grafted to the site, which may recreate the OPC crowding observed in our study. The findings obtained during the development of the E3 method may provide information on the appropriate density of OPCs for successful transplantation.
During the development of the E3 method, we compared the use of growth factors commonly used in primary OL cultures. Growth factors such as PDGF and FGF have been widely used for OPC proliferation and insulin and T3 for OL differentiation. Additionally, we evaluated the effect of factors less known and used compared to the factors listed above, which were EGF for the proliferation of OPCs and CNTF for differentiation into mature OLs. The use of EGF during the proliferation of OPCs first isolated from the rat brain enhanced the proliferation of OPCs, surpassing media compositions without EGF in WST-8 proliferation assays. There was no difference in the proportion of OPCs positive for the proliferation markers EdU or Ki67 after 4.5 days of proliferation. The WST-8 proliferation assay is a colorimetric assay that reflects the total number of viable cells in a sample, while the proliferation markers EdU or Ki67 mark cells that are undergoing cell division at the time of analysis. The main difference between EdU and Ki67 is that while EdU is taken up by cells during the S phase of the cell cycle [61], Ki67 can be detected in cells at all stages of cell division (G1, S, G2, M phase) [62,63]. The most likely reason that differences between groups in the extent of proliferation were only observed in the WST-8 assay was that the difference took place at a different time point than that of analysis for EdU and Ki67. Another interesting aspect of EGF noted was that when applied to passaged OPCs for 2 days, it caused OPCs initially seeded randomly to rearrange into a clustered pattern. Taking into account the fact that the cell number per area was not different in conditions with EGF compared to those without (data not shown), a plausible explanation for this clustered arrangement is that OPCs acquired a heightened tendency to cluster together in EGF-included environments. Previous studies on EGF and oligodendrogenesis proposed that EGF promotes both the proliferation and differentiation of OPCs and causes them to transit into glial progenitor cells (GPCs) [25,26,64]. Although our study did not confirm the presence of GPCs, the proliferation and cluster-promoting properties of EGF in our proliferation media strongly imply that EGF potentiates the stemness of OPCs. With the use of CNTF, we did not observe a significant enhancement in OL maturation.
In conclusion, we introduced the E3 method of primary OL culture, a simple physical method for the isolation of OPCs from the neonatal rodent brain, along with the optimization of media composition for subsequent proliferation and differentiation, to ultimately generate OLs capable of myelination. The E3 method improves the clarity of the procedure and is an undemanding method for obtaining high-yield, high-quality primary OL cultures. In addition to its value as a practical tool for primary OL culture, the unique in vitro characteristics of the OL lineage revealed during the development of the E3 method offer insights for advancing research on oligodendrocyte physiology and pathophysiology.
Ethics statement
All animal experiments were reviewed and approved by the Institutional Animal Care and Use Committee of Ajou University School of Medicine (IACUC number; 2023-0021) and complied with the National Institutes of Health (NIH) Guide for the Care and Use of Laboratory Animals.
Funding information
This research was supported by a grant of the M.D.-Ph.D./Medical Scientist Training Program through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (to H. K.). This work was also supported by National Research Foundation of Korea (NRF) grants funded by the Korean government (MSIT; Ministry of Science and ICT) (NRF2019R1A5A2026045 and NRF-2021R1F1A1061819), a grant from the Korean Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (HR21C1003), and new faculty research fund of Ajou University School of Medicine (to J.Y.C.).
Data availability statement
The data and analyses in this study are available within the main text and the Supporting Information section. The R code used for spatial analysis of Poisson point processes is available at https://github.com/hanki313/Spatial-statistics-for-E3-methods/tree/main. Additional data related to the article will be made available upon reasonable request.
CRediT authorship contribution statement
Hanki Kim: Writing – review & editing, Writing – original draft, Visualization, Validation, Project administration, Methodology, Investigation, Funding acquisition, Formal analysis, Data curation, Conceptualization. Bum Jun Kim: Validation, Methodology, Investigation, Formal analysis. Seungyon Koh: Writing – review & editing, Methodology, Data curation. Hyo Jin Cho: Validation, Formal analysis, Data curation. Xuelian Jin: Writing – review & editing, Formal analysis, Data curation. Byung Gon Kim: Writing – review & editing, Supervision, Methodology. Jun Young Choi: Writing – review & editing, Writing – original draft, Validation, Supervision, Resources, Project administration, Methodology, Investigation, Funding acquisition, Conceptualization.
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: Hanki Kim reports financial support was provided by Korea Health Industry Development Institute. Jun Young Choi reports financial support was provided by Korea Health Industry Development Institute. Jun Young Choi reports financial support was provided by National Research Foundation of Korea. Jun Young Choi has patent pending to Ajou University Industry-Academic cooperation Foundation. If there are other authors, they declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Fig. 1, Fig. 4, Fig. 6c were created with Biorender.com. We thank Jonah R Chan from the University of California, San Francisco, for advice on designing the in vitro nanofiber myelination experiments.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e29359.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
figs1.
Fig. S1. Absence of astrocytes/microglia in the isolated primary OPC-enriched culture. a Representative images of isolated cells (after 5 days of proliferation), immunofluorescence for GFAP. P: PDGF, F: FGF, E: EGF. b Representative image of positive control (GFAP+ cell from media composition 2 from Table 1a) and quantification of GFAP+ / DAPI+ cells (%) according to media compositions corresponding to Table 1a (n=3 biologically independent samples/experiments, 6 groups per sample, 5 images per group, approximately 300 DAPI+ cells per image. One-way ANOVA. Data presented as mean values ± SEM.). c Representative images of isolated cells (after 5 days of proliferation), immunofluorescence for Iba1. P: PDGF, F: FGF, E: EGF. d Representative image of positive controls (isolated cells from the first centrifuge process in 12% OptiprepTM in Fig. 1a, cultured in OPC proliferation media for 5 days).
figs2.
Fig. S2. Reduction of neuron population after passaging of proliferated OPCs. a Representative images of passaged OPCs. Immunostaining for NG2, Tuj1. b Representative image of a Tuj1-positive neuron. c Quantification of Tuj1+/DAPI+ cells (%) according to media compositions corresponding to Table 1a (n=3 biologically independent samples/experiments, 6 groups per sample, 10 images per group, approximately 200 DAPI+ cells per image. One-way ANOVA. Data presented as mean values ± SEM.).
figs3.
Fig. S3. Oligodendrocyte stages during differentiation. a-b Representative images of differentiated OLs (after 2, 4 days of differentiation). Immunostaining for NG2, BCAS1, and MBP.
References
- 1.Bradl M., Lassmann H. Oligodendrocytes: biology and pathology. Acta Neuropathol. 2010;119:37–53. doi: 10.1007/s00401-009-0601-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Philips T., Rothstein J.D. Oligodendroglia: metabolic supporters of neurons. J. Clin. Invest. 2017;127:3271–3280. doi: 10.1172/JCI90610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Irvine K.A., Blakemore W.F. Remyelination protects axons from demyelination-associated axon degeneration. Brain. 2008;131:1464–1477. doi: 10.1093/brain/awn080. [DOI] [PubMed] [Google Scholar]
- 4.Simkins T.J., Duncan G.J., Bourdette D. Chronic demyelination and axonal degeneration in multiple sclerosis: pathogenesis and therapeutic implications. Curr. Neurol. Neurosci. Rep. 2021;21:26. doi: 10.1007/s11910-021-01110-5. [DOI] [PubMed] [Google Scholar]
- 5.Duncan I.D., Radcliff A.B. Inherited and acquired disorders of myelin: the underlying myelin pathology. Exp. Neurol. 2016;283:452–475. doi: 10.1016/j.expneurol.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deshmukh V.A., Tardif V., Lyssiotis C.A., Green C.C., Kerman B., Kim H.J., Padmanabhan K., Swoboda J.G., Ahmad I., Kondo T., Gage F.H., Theofilopoulos A.N., Lawson B.R., Schultz P.G., Lairson L.L. A regenerative approach to the treatment of multiple sclerosis. Nature. 2013;502:327–332. doi: 10.1038/nature12647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buntinx M., Vanderlocht J., Hellings N., Vandenabeele F., Lambrichts I., Raus J., Ameloot M., Stinissen P., Steels P. Characterization of three human oligodendroglial cell lines as a model to study oligodendrocyte injury: morphology and oligodendrocyte-specific gene expression. J. Neurocytol. 2003;32:25–38. doi: 10.1023/a:1027324230923. [DOI] [PubMed] [Google Scholar]
- 8.Pereira G.B., Dobretsova A., Hamdan H., Wight P.A. Expression of myelin genes: comparative analysis of Oli-neu and N20.1 oligodendroglial cell lines. J. Neurosci. Res. 2011;89:1070–1078. doi: 10.1002/jnr.22625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McCarthy K.D., de Vellis J. Preparation of separate astroglial and oligodendroglial cell cultures from rat cerebral tissue. J. Cell Biol. 1980;85:890–902. doi: 10.1083/jcb.85.3.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O’Meara R.W., Ryan S.D., Colognato H., Kothary R. Derivation of enriched oligodendrocyte cultures and oligodendrocyte/neuron myelinating co-cultures from post-natal murine tissues. J. Vis. Exp. 2011;54:e3324. doi: 10.3791/3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dugas J.C., Emery B. Purification and culture of oligodendrocyte lineage cells. Cold Spring Harb. Protoc. 2013;2013:810–814. doi: 10.1101/pdb.top074898. [DOI] [PubMed] [Google Scholar]
- 12.Emery B., Dugas J.C. Purification of oligodendrocyte lineage cells from mouse cortices by immunopanning. Cold Spring Harb. Protoc. 2013;2013:854–868. doi: 10.1101/pdb.prot073973. [DOI] [PubMed] [Google Scholar]
- 13.Dugas J.C., Emery B. Purification of oligodendrocyte precursor cells from rat cortices by immunopanning. Cold Spring Harb. Protoc. 2013;2013:745–758. doi: 10.1101/pdb.prot070862. [DOI] [PubMed] [Google Scholar]
- 14.Weil M.T., Schulz-Eberlin G., Mukherjee C., Kuo-Elsner W.P., Schafer I., Muller C., Simons M. Isolation and culture of oligodendrocytes. Methods Mol. Biol. 2019;1936:79–95. doi: 10.1007/978-1-4939-9072-6_5. [DOI] [PubMed] [Google Scholar]
- 15.Yoshida A., Takashima K., Shimonaga T., Kadokura M., Nagase S., Koda S. Establishment of a simple one-step method for oligodendrocyte progenitor cell preparation from rodent brains. J. Neurosci. Methods. 2020;342 doi: 10.1016/j.jneumeth.2020.108798. [DOI] [PubMed] [Google Scholar]
- 16.Nihonmatsu-Kikuchi N., Yu X.J., Matsuda Y., Ozawa N., Ito T., Satou K., Kaname T., Iwasaki Y., Akagi A., Yoshida M., Toru S., Hirokawa K., Takashima A., Hasegawa M., Uchihara T., Tatebayashi Y. Essential roles of plexin-B3(+) oligodendrocyte precursor cells in the pathogenesis of Alzheimer's disease. Commun. Biol. 2021;4:870. doi: 10.1038/s42003-021-02404-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Claude A. Fractionation of mammalian liver cells by differential centrifugation; experimental procedures and results. J. Exp. Med. 1946;84:61–89. [PubMed] [Google Scholar]
- 18.Livshits M.A., Khomyakova E., Evtushenko E.G., Lazarev V.N., Kulemin N.A., Semina S.E., Generozov E.V., Govorun V.M. Corrigendum: isolation of exosomes by differential centrifugation: theoretical analysis of a commonly used protocol. Sci. Rep. 2016;6 doi: 10.1038/srep21447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Swire M., Ffrench-Constant C. Oligodendrocyte-neuron myelinating coculture. Methods Mol. Biol. 2019;1936:111–128. doi: 10.1007/978-1-4939-9072-6_7. [DOI] [PubMed] [Google Scholar]
- 20.McKinnon R.D., Matsui T., Dubois-Dalcq M., Aaronson S.A. FGF modulates the PDGF-driven pathway of oligodendrocyte development. Neuron. 1990;5:603–614. doi: 10.1016/0896-6273(90)90215-2. [DOI] [PubMed] [Google Scholar]
- 21.Collarini E.J., Pringle N., Mudhar H., Stevens G., Kuhn R., Monuki E.S., Lemke G., Richardson W.D. Growth factors and transcription factors in oligodendrocyte development. J. Cell Sci. Suppl. 1991;15:117–123. doi: 10.1242/jcs.1991.supplement_15.16. [DOI] [PubMed] [Google Scholar]
- 22.Baron W., Metz B., Bansal R., Hoekstra D., de Vries H. PDGF and FGF-2 signaling in oligodendrocyte progenitor cells: regulation of proliferation and differentiation by multiple intracellular signaling pathways. Mol. Cell. Neurosci. 2000;15:314–329. doi: 10.1006/mcne.1999.0827. [DOI] [PubMed] [Google Scholar]
- 23.McKinnon R.D., Matsui T., Aranda M., Dubois-Dalcq M. A role for fibroblast growth factor in oligodendrocyte development. Ann. N. Y. Acad. Sci. 1991;638:378–386. doi: 10.1111/j.1749-6632.1991.tb49048.x. [DOI] [PubMed] [Google Scholar]
- 24.Hart I.K., Richardson W.D., Heldin C.H., Westermark B., Raff M.C. PDGF receptors on cells of the oligodendrocyte-type-2 astrocyte (O-2A) cell lineage. Development. 1989;105:595–603. doi: 10.1242/dev.105.3.595. [DOI] [PubMed] [Google Scholar]
- 25.Yang J., Cheng X., Qi J., Xie B., Zhao X., Zheng K., Zhang Z., Qiu M. EGF enhances oligodendrogenesis from glial progenitor cells. Front. Mol. Neurosci. 2017;10:106. doi: 10.3389/fnmol.2017.00106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang J., Cheng X., Shen J., Xie B., Zhao X., Zhang Z., Cao Q., Shen Y., Qiu M. A novel approach for amplification and purification of mouse oligodendrocyte progenitor cells. Front. Cell. Neurosci. 2016;10:203. doi: 10.3389/fncel.2016.00203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barres B.A., Hart I.K., Coles H.S., Burne J.F., Voyvodic J.T., Richardson W.D., Raff M.C. Cell death and control of cell survival in the oligodendrocyte lineage. Cell. 1992;70:31–46. doi: 10.1016/0092-8674(92)90531-g. [DOI] [PubMed] [Google Scholar]
- 28.Ahlgren S.C., Wallace H., Bishop J., Neophytou C., Raff M.C. Effects of thyroid hormone on embryonic oligodendrocyte precursor cell development in vivo and in vitro. Mol. Cell. Neurosci. 1997;9:420–432. doi: 10.1006/mcne.1997.0631. [DOI] [PubMed] [Google Scholar]
- 29.Billon N., Jolicoeur C., Tokumoto Y., Vennstrom B., Raff M. Normal timing of oligodendrocyte development depends on thyroid hormone receptor alpha 1 (TRalpha1) EMBO J. 2002;21:6452–6460. doi: 10.1093/emboj/cdf662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barres B.A., Lazar M.A., Raff M.C. A novel role for thyroid hormone, glucocorticoids and retinoic acid in timing oligodendrocyte development. Development. 1994;120:1097–1108. doi: 10.1242/dev.120.5.1097. [DOI] [PubMed] [Google Scholar]
- 31.Rodriguez-Pena A. Oligodendrocyte development and thyroid hormone. J. Neurobiol. 1999;40:497–512. doi: 10.1002/(sici)1097-4695(19990915)40:4<497::aid-neu7>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 32.Carre J.L., Demerens C., Rodriguez-Pena A., Floch H.H., Vincendon G., Sarlieve L.L. Thyroid hormone receptor isoforms are sequentially expressed in oligodendrocyte lineage cells during rat cerebral development. J. Neurosci. Res. 1998;54:584–594. doi: 10.1002/(SICI)1097-4547(19981201)54:5<584::AID-JNR3>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 33.Stankoff B., Aigrot M.S., Noel F., Wattilliaux A., Zalc B., Lubetzki C. Ciliary neurotrophic factor (CNTF) enhances myelin formation: a novel role for CNTF and CNTF-related molecules. J. Neurosci. 2002;22:9221–9227. doi: 10.1523/JNEUROSCI.22-21-09221.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Talbott J.F., Cao Q., Bertram J., Nkansah M., Benton R.L., Lavik E., Whittemore S.R. CNTF promotes the survival and differentiation of adult spinal cord-derived oligodendrocyte precursor cells in vitro but fails to promote remyelination in vivo. Exp. Neurol. 2007;204:485–489. doi: 10.1016/j.expneurol.2006.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bechler M.E. A neuron-free microfiber assay to assess myelin sheath formation. Methods Mol. Biol. 2019;1936:97–110. doi: 10.1007/978-1-4939-9072-6_6. [DOI] [PubMed] [Google Scholar]
- 36.Choi J.Y., Jin X., Kim H., Koh S., Cho H.J., Kim B.G. High mobility group box 1 as an autocrine chemoattractant for oligodendrocyte lineage cells in white matter stroke. Stroke. 2023;54:575–586. doi: 10.1161/STROKEAHA.122.041414. [DOI] [PubMed] [Google Scholar]
- 37.Choi J.Y., Cui Y., Chowdhury S.T., Kim B.G. High-mobility group box-1 as an autocrine trophic factor in white matter stroke. Proc. Natl. Acad. Sci. U. S. A. 2017;114:E4987–E4995. doi: 10.1073/pnas.1702035114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang Z., Watanabe M., Nishiyama A. Optimization of oligodendrocyte progenitor cell culture method for enhanced survival. J. Neurosci. Methods. 2005;149:50–56. doi: 10.1016/j.jneumeth.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 39.Zhu B., Zhao C., Young F.I., Franklin R.J., Song B. Isolation and long-term expansion of functional, myelinating oligodendrocyte progenitor cells from neonatal rat brain. Curr. Protoc. Stem Cell Biol. 2014;31(2D):17 11–15. doi: 10.1002/9780470151808.sc02d17s31. [DOI] [PubMed] [Google Scholar]
- 40.Mayer-Proschel M. Isolation and generation of oligodendrocytes by immunopanning. Curr. Protoc. Neurosci. 2001;3:3–13. doi: 10.1002/0471142301.ns0313s00. Chapter. [DOI] [PubMed] [Google Scholar]
- 41.Dincman T.A., Beare J.E., Ohri S.S., Whittemore S.R. Isolation of cortical mouse oligodendrocyte precursor cells. J. Neurosci. Methods. 2012;209:219–226. doi: 10.1016/j.jneumeth.2012.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brewer G.J., Torricelli J.R. Isolation and culture of adult neurons and neurospheres. Nat. Protoc. 2007;2:1490–1498. doi: 10.1038/nprot.2007.207. [DOI] [PubMed] [Google Scholar]
- 43.Chen Y., Balasubramaniyan V., Peng J., Hurlock E.C., Tallquist M., Li J., Lu Q.R. Isolation and culture of rat and mouse oligodendrocyte precursor cells. Nat. Protoc. 2007;2:1044–1051. doi: 10.1038/nprot.2007.149. [DOI] [PubMed] [Google Scholar]
- 44.Brewer G.J., Torricelli J.R., Evege E.K., Price P.J. Optimized survival of hippocampal neurons in B27-supplemented Neurobasal, a new serum-free medium combination. J. Neurosci. Res. 1993;35:567–576. doi: 10.1002/jnr.490350513. [DOI] [PubMed] [Google Scholar]
- 45.Kleinsimlinghaus K., Marx R., Serdar M., Bendix I., Dietzel I.D. Strategies for repair of white matter: influence of osmolarity and microglia on proliferation and apoptosis of oligodendrocyte precursor cells in different basal culture media. Front. Cell. Neurosci. 2013;7:277. doi: 10.3389/fncel.2013.00277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Domingues H.S., Cruz A., Chan J.R., Relvas J.B., Rubinstein B., Pinto I.M. Mechanical plasticity during oligodendrocyte differentiation and myelination. Glia. 2018;66:5–14. doi: 10.1002/glia.23206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Segel M., Neumann B., Hill M.F.E., Weber I.P., Viscomi C., Zhao C., Young A., Agley C.C., Thompson A.J., Gonzalez G.A., Sharma A., Holmqvist S., Rowitch D.H., Franze K., Franklin R.J.M., Chalut K.J. Niche stiffness underlies the ageing of central nervous system progenitor cells. Nature. 2019;573:130–134. doi: 10.1038/s41586-019-1484-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Savadipour A., Nims R.J., Rashidi N., Garcia-Castorena J.M., Tang R., Marushack G.K., Oswald S.J., Liedtke W.B., Guilak F. Membrane stretch as the mechanism of activation of PIEZO1 ion channels in chondrocytes. Proc. Natl. Acad. Sci. U. S. A. 2023;120 doi: 10.1073/pnas.2221958120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Avellana-Adalid V., Nait-Oumesmar B., Lachapelle F., Baron-Van Evercooren A. Expansion of rat oligodendrocyte progenitors into proliferative "oligospheres" that retain differentiation potential. J. Neurosci. Res. 1996;45:558–570. doi: 10.1002/(SICI)1097-4547(19960901)45:5<558::AID-JNR6>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 50.Vitry S., Avellana-Adalid V., Hardy R., Lachapelle F., Baron-Van Evercooren A. Mouse oligospheres: from pre-progenitors to functional oligodendrocytes. J. Neurosci. Res. 1999;58:735–751. [PubMed] [Google Scholar]
- 51.Zhang S.C., Lipsitz D., Duncan I.D. Self-renewing canine oligodendroglial progenitor expanded as oligospheres. J. Neurosci. Res. 1998;54:181–190. doi: 10.1002/(SICI)1097-4547(19981015)54:2<181::AID-JNR6>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 52.Back S.A., Han B.H., Luo N.L., Chricton C.A., Xanthoudakis S., Tam J., Arvin K.L., Holtzman D.M. Selective vulnerability of late oligodendrocyte progenitors to hypoxia-ischemia. J. Neurosci. 2002;22:455–463. doi: 10.1523/JNEUROSCI.22-02-00455.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Susarla B.T., Villapol S., Yi J.H., Geller H.M., Symes A.J. Temporal patterns of cortical proliferation of glial cell populations after traumatic brain injury in mice. ASN Neuro. 2014;6:159–170. doi: 10.1042/AN20130034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Back S.A. White matter injury in the preterm infant: pathology and mechanisms. Acta Neuropathol. 2017;134:331–349. doi: 10.1007/s00401-017-1718-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li W., He T., Shi R., Song Y., Wang L., Zhang Z., Tang Y., Yang G.Y., Wang Y. Oligodendrocyte precursor cells transplantation improves stroke recovery via oligodendrogenesis, neurite growth and synaptogenesis. Aging Dis. 2021;12:2096–2112. doi: 10.14336/AD.2021.0416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang L., Geng J., Qu M., Yuan F., Wang Y., Pan J., Li Y., Ma Y., Zhou P., Zhang Z., Yang G.Y. Oligodendrocyte precursor cells transplantation protects blood-brain barrier in a mouse model of brain ischemia via Wnt/beta-catenin signaling. Cell Death Dis. 2020;11:9. doi: 10.1038/s41419-019-2206-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xu L., Ryu J., Hiel H., Menon A., Aggarwal A., Rha E., Mahairaki V., Cummings B.J., Koliatsos V.E. Transplantation of human oligodendrocyte progenitor cells in an animal model of diffuse traumatic axonal injury: survival and differentiation. Stem Cell Res. Ther. 2015;6:93. doi: 10.1186/s13287-015-0087-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sun Y., Xu C.C., Li J., Guan X.Y., Gao L., Ma L.X., Li R.X., Peng Y.W., Zhu G.P. Transplantation of oligodendrocyte precursor cells improves locomotion deficits in rats with spinal cord irradiation injury. PLoS One. 2013;8 doi: 10.1371/journal.pone.0057534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wu B., Sun L., Li P., Tian M., Luo Y., Ren X. Transplantation of oligodendrocyte precursor cells improves myelination and promotes functional recovery after spinal cord injury. Injury. 2012;43:794–801. doi: 10.1016/j.injury.2011.09.013. [DOI] [PubMed] [Google Scholar]
- 60.Wang X., Zang J., Yang Y., Lu S., Guan Q., Ye D., Wang Z., Zhou H., Li K., Wang Q., Wu Y., Luan Z. Transplanted human oligodendrocyte progenitor cells restore neurobehavioral deficits in a rat model of preterm white matter injury. Front. Neurol. 2021;12 doi: 10.3389/fneur.2021.749244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Harris L., Zalucki O., Piper M. BrdU/EdU dual labeling to determine the cell-cycle dynamics of defined cellular subpopulations. J. Mol. Histol. 2018;49:229–234. doi: 10.1007/s10735-018-9761-8. [DOI] [PubMed] [Google Scholar]
- 62.Gerdes J., Lemke H., Baisch H., Wacker H.H., Schwab U., Stein H. Cell cycle analysis of a cell proliferation-associated human nuclear antigen defined by the monoclonal antibody Ki-67. J. Immunol. 1984;133:1710–1715. [PubMed] [Google Scholar]
- 63.Prayson R.A. The utility of MIB-1/Ki-67 immunostaining in the evaluation of central nervous system neoplasms. Adv. Anat. Pathol. 2005;12:144–148. doi: 10.1097/01.pap.0000163957.21409.52. [DOI] [PubMed] [Google Scholar]
- 64.Gonzalez-Perez O., Alvarez-Buylla A. Oligodendrogenesis in the subventricular zone and the role of epidermal growth factor. Brain Res. Rev. 2011;67:147–156. doi: 10.1016/j.brainresrev.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data and analyses in this study are available within the main text and the Supporting Information section. The R code used for spatial analysis of Poisson point processes is available at https://github.com/hanki313/Spatial-statistics-for-E3-methods/tree/main. Additional data related to the article will be made available upon reasonable request.