Abstract
This study introduces an innovative contour detection algorithm, PeakCET, designed for rapid and efficient analysis of natural product image fingerprints using comprehensive two-dimensional gas chromatogram (GC × GC). This method innovatively combines contour edge tracking with affinity propagation (AP) clustering for peak detection in GC × GC fingerprints, the first in this field. Contour edge tracking significantly reduces false positives caused by “burr” signals, while AP clustering enhances detection accuracy in the face of false negatives. The efficacy of this approach is demonstrated using three medicinal products derived from Curcuma wenyujin. PeakCET not only performs contour detection but also employs inter-group peak matching and peak-volume percentage calculations to assess the compositional similarities and differences among various samples. Furthermore, this algorithm compares the GC × GC fingerprints of Radix/Rhizoma Curcumae Wenyujin with those of products from different botanical origins. The findings reveal that genetic and geographical factors influence the accumulation of secondary metabolites in various plant tissues. Each sample exhibits unique characteristic components alongside common ones, and variations in content may influence their therapeutic effectiveness. This research establishes a foundational data-set for the quality assessment of Curcuma products and paves the way for the application of computer vision techniques in two-dimensional (2D) fingerprint analysis of GC × GC data.
Keywords: GC × GC, Image fingerprints, Contour detection, Clustering of mass spectra, Curcuma products
Graphical abstract
Highlights
-
•
Image fingerprints of GC × GC are analyzed by a novel contour detection algorithm.
-
•
Affinity propagation clustering optimizes the detection in the presence of false negatives.
-
•
The component percentage is deduced by the peak volume calculated by the pixels.
-
•
GC × GC fingerprints of three Curcuma wenyujin products are compared by PeakCET.
-
•
GC × GC fingerprints of dozens of Curcuma samples are differentiated by PeakCET.
1. Introduction
Natural products, rich in essential oils, play a significant role in the cosmetics industry [1]. These products also serve as medicinal materials, offering prevention and treatment for various diseases [2,3]. In this context, hundreds of phytochemicals deliver therapeutic effects in specific proportions [4]. However, the accumulation of secondary metabolites is heavily influenced by genetics and climate, echoing the well-known “Nanju Beizhi” phenomenon where environmental changes affect oranges [5]. Additionally, ethnic medicine reveals intricate patterns of “multiple medicines from a single plant” and “one medicine from multiple botanical sources” [6,7]. Consequently, finding a “Rosetta stone” to improve the quality of medicinal materials is a challenging task, especially with the increasing demand for efficacy and safety [[8], [9], [10]].
Over the past two decades, the fingerprinting strategy has emerged as a practical approach for assessing the quality of medicinal materials [[11], [12], [13], [14], [15], [16]]. Nonetheless, relying solely on one-dimensional (1D) separation techniques for natural products leads to issues such as co-elution of similar compounds and challenges in detecting trace compounds. Comprehensive two-dimensional gas chromatography (GC × GC) has advantages such as high peak capacity, and has found extensive applications in pharmacy, herbal medicine, and food-omics [[17], [18], [19], [20]]. The detectors in conjunction with GC × GC, such as the time of flight mass spectrometry (TOF MS), quadrupole mass spectrometry (qMS), quadrupole time of flight mass spectrometry (qTOF MS), and flame ionization detection (FID), have seen increasing use in medicinal material analysis. This includes work on Artemisia annua, vetiver, Cyperus rotundus, among others [[21], [22], [23], [24]].
Data mining from two-dimensional (2D) fingerprints is a key area of study for chemometric scholars [[25], [26], [27], [28]]. The core challenge lies in quickly and accurately detecting 2D peaks and extracting relevant information. Traditional methods, such as the drain algorithm, often lead to over-segmentation due to noise artifacts [29]. For instance, Peters et al. [30] adapted 1D chromatography algorithms for modulated peak detection, employing a decision tree for clustering and merging into 2D peaks. Vivó-Truyols [31] also used a conventional 1D peak detection algorithm, followed by Bayesian posterior probability calculations for optimal peak arrangement combinations. Kim et al. [32] introduced a normal-exponential Bernoulli (NEB) model for defining peak regions, peak selection, and subsequent merging based on mass spectral similarities. However, these existing algorithms, primarily based on 1D peak detection methods, struggle with noise sensitivity and difficulty in detecting overlapping peaks, presenting significant challenges in GC × GC analysis of complex samples like essential oils.
Advancements in computer vision, a branch of artificial intelligence that interprets information from visual inputs, have led to remarkable progress in image analysis [33], the food industry [34], remote sensing image registration [35], and scientific instrument “fingerprint” data. Yeap et al. [36] combined computer vision with gas chromatography/differential mobility spectrometry (GC/DMS) data for chemical signature alignment across datasets. The use of speeded up robust features (SURF) detection and fast library for approximate nearest neighbors (FLANN) matching algorithm has been effective in evaluating the similarity of GC × GC fingerprints of Chinese patent medicines [37]. Recently computer vision based on pattern recognition algorithm offer enhanced tracking of untargeted and targeted fingerprints [38]. However, challenges remain in reducing false positives/negatives and detecting overlapping peaks. Fully exploiting MS data, such as using subwindow factor analysis and mass spectral information for peak alignment, is another hurdle [39,40]. Hierarchical clustering has been beneficial in natural product discovery [41,42] and microbial classification [43,44].
This research introduces a new algorithm, PeakCET, for 2D peak (depicted in vertical view as contour) detection assisted by mass spectra clustering. Following a specific flowchart (Fig. 1), PeakCET effectively addresses over-segmentation and overlapped peak (contour) detection. It is applied to GC × GC fingerprints of three medicinal products derived from Curcuma wenyujin Y. H. Chen et C. Ling. PeakCET also facilitates comparative analysis of GC × GC data between Radix/Rhizoma Curcumae Wenyujin and other Radix/Rhizoma Curcumae samples from different botanical origins.
Fig. 1.
The parsing process of comprehensive two-dimensional gas chromatogram (GC × GC) fingerprint based on PeakCET algorithm. (A) Data pre-processing. (B) Contour detection (contour edge tracking and mass spectra clustering). 2D: two-dimensional; I: the scanning points in each modulation period; J: the mass-to-charge (m/z) range for each scan; K: the number of modulations. TIC: total ion chromatogram.
2. Material and methods
2.1. Materials
Three medicinal products (Rhizoma Wenyujin Concisum, Rhizoma Curcumae Wenyujin, and Radix Curcumae Wenyujin) were collected from the same plant (Curcuma wenyujin Y. H. Chen et C. Ling) in Taoshan Town, Rui'an, China. These samples were identified by Zhuzhou Institute for Food and Drug Inspection, China. Other samples of Radix Curcumae and Rhizoma Curcumae were obtained from Zhejiang Province, Sichuan Province, and Guangxi Zhuang Autonomous Region, respectively. Among them, Radix (Rhizoma) Curcumae phaeocaulis were also microscopically identified. The alkane standard solution (C7-30) was purchased from Fluka Chemika (Buchs, Switzerland).
2.2. Extraction of essential oil
50 g samples of Radix Curcumae or Rhizoma Curcumae was precisely measured and mixed with 400 mL of distilled water each. After being left at room temperature for 6 h, the samples underwent hydrodistillation for 3 h following the standard extraction method outlined in the Chinese Pharmacopoeia 2020 edition. The resulting essential oils were stored in sealed vials at 4 °C for immediate analysis.
2.3. GC × GC-TOF MS determination for different essential oils
A combination of the 7890B GC (Agilent Corporation, Santa Clara, CA, USA), electron ionization (EI)-0610 TOF MS (Hexin Instrument Co., Ltd., Guangzhou, China), and SSM1810E solid-state thermal modulator (J&X Technologies, Shanghai, China), were used to analyze essential oils from curcuma samples. The 1D and 2D columns used were DB-WAX (30 m × 0.25 mm × 0.25 μm) and DB-17 MS (1.1 m × 0.15 mm × 0.15 μm), respectively, while the modulation column was HV (C5–C30+) type (1.2 m × 0.25 mm) from J&X Technologies. The modulator period was set to 4 s. The injection temperature was set at 250 °C, with the initial temperature starting at 40 °C for 5 min, then increasing to 260 °C at a rate of 4 °C/min, and held for 1 min. The carrier gas used was helium with a purity of 99.999%, and a constant flow rate of 1.0 mL/min was maintained.
For the EI-TOF MS, the ionization temperature was set at 220 °C, the temperature of the MS transmission line was set to 240 °C, and the ionization energy was set at 70 eV. The scanning mass-to-charge (m/z) range was from 40 to 400 amu, and an acquisition frequency of 100 Hz was used. The data processing was done using the Canvas and GC Image software.
2.4. PeakCET algorithm for GC × GC-TOF MS fingerprint processing
2.4.1. Data preprocessing
As shown in Fig. 1A, data preprocessing is delineated in three distinct steps:
The initial raw signals are treated as a three-dimensional (3D) array X (I, J, K), where I (1, 2, …, i) indicates the scanning points in each modulation period, J (1, 2, …, j) represents the m/z range for each scan, and K (1, 2, …, k) denotes the number of modulations. In this dataset, the modulation period remains constant, and the m/z range is identical across all scanning points. The researchers first use MATLAB to process the raw data (in .CDF format), transforming it into a column-wise augmented data matrix X (I × K, J).
When visualizing the GC × GC contour matrix C (I, K) as a 2D image, signal size is indicated through contour lines or varying colors, with each component's corresponding area depicted as a 2D “spot”. In a 3D representation, each compound appears as a peak with a conical structure, where its volume signifies signal intensity. Hence, each peak within the GC × GC contour matrix is characterized by both its position and peak volume.
The contour detection function in OpenCV only recognizes grayscale images with pixel values ranging from 0 to 255. Thus, the signal intensity in the GC × GC contour matrix must be mapped onto pixels. This involves normalizing the intensity within the range of positive integers [0, 255]. The conversion of the contour matrix C (I, K) into the GC × GC pixel matrix C∗ (I, K) is executed using Eq. 1, as shown below:
| (1) |
In this equation, Amin and Amax symbolize the minimum and maximum intensity values in the GC × GC contour matrix, respectively. The variable “I” corresponds to the original intensity of each point in the matrix, while “Inew” signifies each point's normalized value. This normalization is crucial for allowing the direct input of the normalized GC × GC contour matrix for contour edge tracking, sidestepping the conventional method of image input [45] and subsequent detection.
2.4.2. Affinity propagation (AP) clustering-assisted contour detection
As observed in Fig. 1B, the contour edge tracking [46] process begins by applying OpenCV to the pixel matrix data C∗. This involves using the binary threshold method on C∗ for edge tracking. The primary objective is to establish an appropriate threshold such that pixel values below this threshold become 0, and those above it are set to 1. This binary division effectively separates the 2D peak clusters from the background, as outlined in Eq. 2.
| (2) |
Using the “cv2.findContours” function in OpenCV, a number of contours are identified, each composed of various points representing a peak cluster. The point with the highest response intensity within each contour is then identified and designated as the standard point for that contour.
Given the potential for overlapping edges, there is a risk of missing some 2D peaks during contour edge tracking. To address this, additional detection is performed to identify false negatives. This involves analyzing the mass spectra vectors of local maximum points within the contour using AP clustering [47], which optimizes the detection results. In this process, each data point is treated as a potential class center, and AP is utilized to identify the standard points of peaks that might have been missed within the contour. The AP algorithm procedure includes:
Input the dataset, , which contains mass spectrum vectors of length k for the n maximum points within the contour.
Step 1: Initialize the algorithm and calculate the initial similarity matrix using Euclidean distance , which is calculated by Eq. 3.
| (3) |
Step 2: Set the initial reference P and the number of iterations M, and is calculated by Eq. 4.
| (4) |
, is called the reference degree of xi, which refers to the reliability of using xi as the clustering center. n is the number of vectors, and it represents the number of local maximum points within the contour. Generally, the reference degree is set as the median of the similarity value, which means .
Step 3, compute the attraction index between data points as Eq. 5.
| (5) |
Step 4, determine the attribution ratio aij between data points using Eq. 6, with self-attribution akk being the sum of attractions from other points.
| (6) |
Step 5, to avoid oscillation, when AP algorithm is updated, the decay coefficient λ is introduced, followed the attraction index rij and the attribution ratio aij are updated iteratively by Eq. 7.
| (7) |
Among these variables, the convergence coefficient, represented by , is primarily used to regulate the convergence speed of the algorithm and the stability of the iteration process. The variable “old” denotes the original value. Eq. 7 shows that each data point is set to the sum of times the updated value from the previous iteration and times the original value.
Step 6, the process concludes if the number of actual iterations m exceeds M or if the cluster center remains unchanged after several iterations, assigning data points to their respective classes. If not, the loop restarts at Step 3.
The AP algorithm's key advantage is its independence from the need to predefine the number of clusters, allowing researchers to identify all standard points for contours with unknown compound quantities and minimize false negatives following contour edge tracking.
2.4.3. Inter-group peak matching and peak-list filling
To generate a peak list that includes inter-group information from various sets of fingerprints, an effective peak matching algorithm is essential. This algorithm ensures that a specific peak in one fingerprint corresponds accurately to the same peak in another. The procedure encompasses four key steps, detailed in Section S1 of the Supplementary data. Following the completion of these steps, the peak-list data can undergo further statistical analysis. One such method is the partial least squares-discriminant analysis (PLS-DA) models, which can be accessed through the libPLS platform available at www.libpls.net. This analysis allows for a more in-depth understanding and interpretation of the data, facilitating the identification of patterns and correlations within the peak lists derived from the fingerprint sets.
2.4.4. Visualize PeakCET as a software
Using the Pycharm 2022 compiler and the Python language, one can develop the PeakCET program. This program is adept at automatically creating 2D images for GC × GC data and conducting contour detection. It efficiently extracts both the position and intensity data from the identified 2D peaks. For access, PeakCET is available for free download upon contacting the corresponding author.
3. Results and discussion
3.1. Methodology of PeakCET
Data 1, 2, and 3 were derived from the aroma components of three mango cultivars using MEGA-WAX Plus (1D column) and DB-17 MS (2D column) in a GC × GC-TOF MS system. Data 4, characterized by “burr” signals, was obtained from Citrus reticulata Blanco using GC × GC-TOF MS system. Data 5 was acquired from morpankhi oil through an Rtx-5Sil MS (1D column) and BPX-50 (2D column) in a GC × GC-qMS system.
Initially, data 1, 2, and 3, which displayed excellent separation performance, were analyzed using PeakCET. The “minval” parameter plays a critical role in contour edge tracking as it sets the minimum threshold value for detection. Taking “Xiaotai mango” data 1 (Fig. S1) as an example, four different thresholds were applied for contour detection (Fig. S2A). The findings indicated that a higher threshold leads to reduced interference but increases the likelihood of false negatives in the PeakCET algorithm. Before comparing PeakCET with commercial software (Fig. S2B), theoretical lists for the contour count in the three mango fingerprints were compiled based on two principles: 1) Only spots within the intensity range of 5×104−12×106 were recognized as contours suitable for qualitative and quantitative analysis with practical significance; 2) true positives and true negatives were identified through manual comparison of contours in the images against the results detected by PeakCET or commercial software.
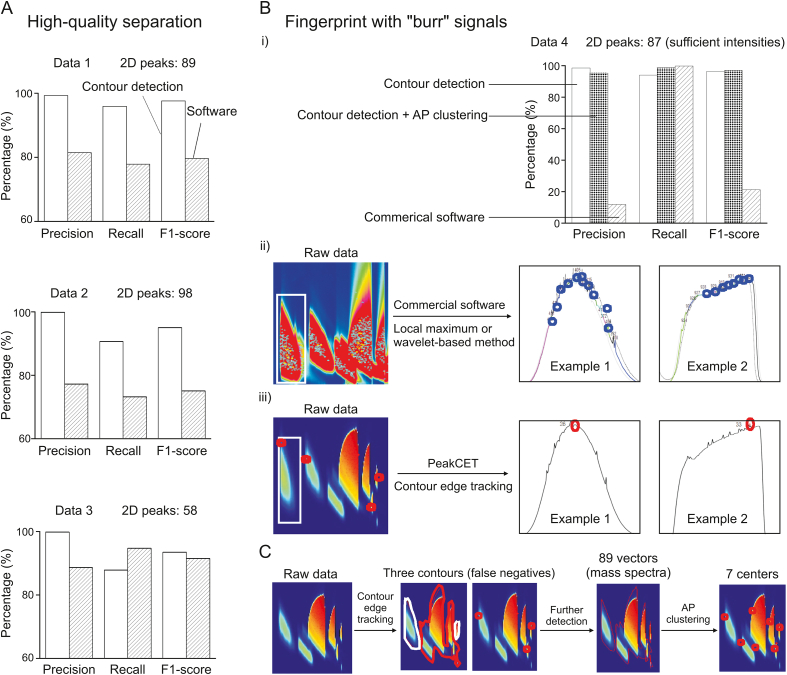
Subsequently, data 1, 2, and 3 were analyzed using PeakCET with thresholds of 2 or 5 (Fig. S3). The number of true positives, false positives, and false negatives were tallied for the test fingerprints by comparing them to the theoretical lists. Using Eqs. (8), (9), (10), the detection results were evaluated based on the accuracy rate (P), recall rate (R), and F1-score. A comprehensive evaluation of the PeakCET algorithm was then conducted against commercial software. As shown in Fig. 2A, both methods yielded accurate detection results, with F1-scores over 90% and 75%, respectively. The key difference was that PeakCET detection resulted in more false negatives, while the commercial software, relying on the local maximum principle, produced more false positives. In this context, AP clustering effectively improved false negatives due to “overlap” in PeakCET detection, e.g., 1 peak (“Xiaotai mango” data 1), 3 peaks (“Jinhuang mango” data 2), and 1 peak (“Qingpi mango” data 3). For the commercial software, increasing the threshold augmented the false negative rate, while decreasing it significantly raised the false positive rate, including “column bleeding” or other errors. This led to numerical discrepancies in precision, recall, and F1-score between the two methods.
| (8) |
| (9) |
| (10) |
Fig. 2.
An evaluation of the PeakCET algorithm was conducted against commercial software. (A) The detection results of two methods for three “high-quality” fingerprints. (B) The detection results of two methods for fingerprint with “burr” signals (i); the influence of “burr” on peak detection of commercial software (ii); and the influence of “burr” on peak detection of PeakCET (iii). (C) The PeakCET solution for the “overlap” contours. 2D: two-dimensional; AP: affinity propagation.
The researchers also analyzed data 4 (which comprised 87 peaks of sufficient intensity), as shown in Fig. S4 (supplementary data). Fig. 2Bi reveals that PeakCET successfully detected a reasonable number of peaks, whereas commercial software demonstrated reduced precision and F1-score due to its sensitivity to “burr” signals. In GC × GC detection, such “burr” signals are often the result of electrical disturbances, such as unstable power supply, instrument performance issues, or other interferences, which can adversely affect peak detection. As depicted in Fig. 2Bii, the commercial software erroneously interprets each “burr” signal as a peak, leading to a high number of false positives. However, as illustrated in Fig. 2Biii, the PeakCET algorithm, employing the contour edge tracking method, successfully avoids this error.
Additionally, the phenomenon of “overlap” sometimes occurs in GC × GC fingerprints, particularly when the physicochemical properties of analytes are very similar. Fig. 2C shows that several peaks, impacted by co-elution (indicated by the red area), are indistinguishable using contour edge tracking. To address the resultant false negatives, 89 MS vectors from all maximum points in the overlapped contours were analyzed. The researchers then applied the proposed AP clustering method to enhance contour detection, identifying seven clustering centers. This method, when applied to the complex data 4, yielded satisfactory results. As per Fig. 2Bi, contour edge tracking effectively reduced false positives caused by “burr” signals, achieving an F1-score of 96.47%, significantly higher than the 21.48% achieved using traditional software. Further refinement through AP clustering resulted in an improved F1-score of 97.18%, demonstrating its effectiveness in minimizing false negatives due to co-elution. While there is a potential for false positives, the overall F1-score has shown improvement, and almost all components were detected. In comparison to commercial software results, the PeakCET algorithm is particularly effective for analyzing complex fingerprints with “burr” signals. These findings suggest that PeakCET is adept at detecting well-separated fingerprints produced by reverse-phase column systems, with the added benefit of low false positives. Lowering the detection threshold and incorporating AP clustering can effectively mitigate the issue of false negatives.
To illustrate the algorithm's generalization, fingerprint 5, derived from the GC × GC-qMS system equipped with a normal-phase column, was analyzed (as shown in Fig. S5). At a threshold level of 10, PeakCET identified 117 peaks (contours), albeit with a few false negatives. However, reducing the threshold to 5 or 2 resulted in the detection of numerous false positive peaks. These examples demonstrate that PeakCET can yield effective results, particularly when the contours in the image fingerprint are well-separated. For comparison, commercial software also produced positive outcomes. In summary, PeakCET exhibits high accuracy with a low false positive rate. It is noteworthy, however, that PeakCET tends to be insensitive to contours of very low intensity and is less effective for image fingerprints that are densely packed (or circuitous) in the 2D space. This specificity underscores the need for careful threshold setting and consideration of the unique characteristics of each fingerprint dataset when using PeakCET for analysis.
3.2. Application of PeakCET in GC × GC fingerprints from Curcuma wenyujin products
3.2.1. Peak detection and inter-group peak matching of three fingerprints
Curcuma wenyujin Y. H. Chen et C. Ling, a member of the Curcuma genus within the ginger family, is a versatile plant from which three distinct types of medicinal materials are derived, a concept often referred to as “one plant, three medicines” in China [48] (as illustrated in Fig. S6). These medicinal materials are Radix Curcumae Wenyujin (known as Wen Yujin in Mandarin), Rhizoma Curcumae Wenyujin (Wen Ezhu in Mandarin), and Rhizoma Wenyujin Concisum (Pian Jianghuang in Mandarin). Each of these materials is rich in essential oils, and the specific type or concentration of their constituents contributes to their unique therapeutic properties.
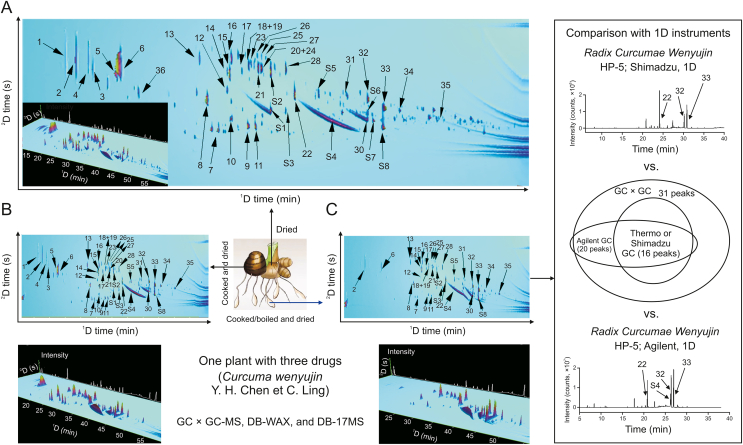
As depicted in Fig. 3, the researchers employed GC × GC-TOF MS to analyze the essential oils extracted from these three medicinal products. The GC × GC system, which combines the DB-WAX and DB-17 MS columns, is capable of effectively separating complex samples. From Fig. 3A (Rhizoma Wenyujin Concisum) and Fig. 3B (Rhizoma Curcumae Wenyujin), it can be seen that this system helps to avoid the issue of “co-eluted” peaks and enhances the detection of trace components. In the analysis of Radix Curcumae Wenyujin samples, it was observed that the number of peaks detected using GC × GC-TOF MS was significantly higher than those identified by three separate GC-MS instruments (Thermo, Shimadzu, and Agilent), as shown in Fig. 3C. Detailed information regarding the number of compounds, their names, and retention indices can be found in Table S1. This comprehensive analysis underscores the efficacy of GC × GC-TOF MS in providing detailed and nuanced insights into the chemical composition of these valuable medicinal materials.
Fig. 3.
Comprehensive two-dimensional gas chromatogram-time of flight mass spectrometry (GC × GC-TOF MS) determination of three medical products from Curcuma wenyujin: (A) Rhizoma Wenyujin Concisum, (B) Rhizoma Curcumae Wenyujin, and (C) Radix Curcumae Wenyujin. 1D: one-dimensional.
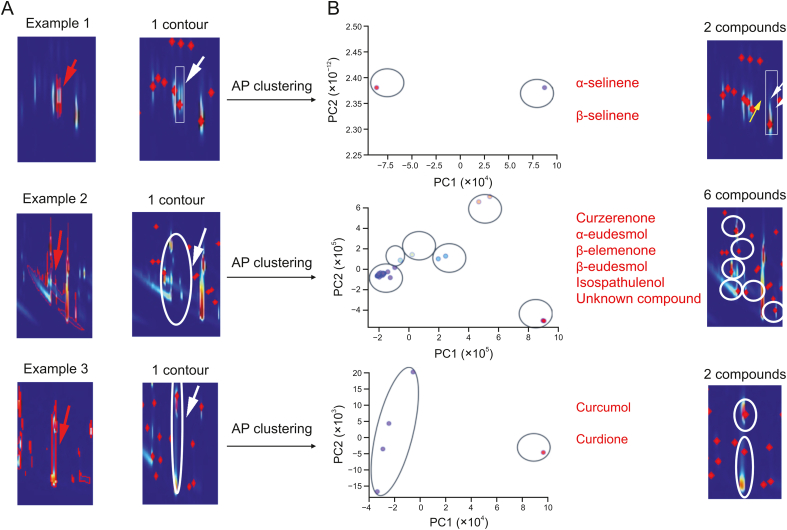
The 2D peaks were analyzed to discern the similarities and differences in the GC × GC fingerprints of three medicinal products. As expected, the PeakCET algorithm successfully tracked most of the 2D peaks. The researchers calculated the F1-scores to compare the detection outcomes of different samples using both individual and AP clustering-assisted contour tracking methods. According to Fig. S7A, AP clustering significantly enhanced the results of contour tracking, with the F1-scores for most detections exceeding 90%. However, it's important to note that the signal strength directly influences the tracking outcomes. For instance, samples 1, 2, 3, 4, and 5, with normal signal responses, achieved F1-scores over 95% in peak detection, whereas samples 7 and 8, with lower signal responses, showed a decrease in F1-scores. Fig. S7B demonstrates that AP clustering effectively reduces the number of false negatives in individual contour tracking, thereby optimizing detection. The researchers visualized the clustering effects within the missed-detection contours using the principal components analysis (PCA) method for clearer observation. Fig. 4A presents some false negatives in single contour tracking detection. On one side, the missed-detection range (red contour) is indicated by a red arrow; on the other, a red dot, pointed out by a white arrow, shows an area tracked by only one contour. AP clustering was applied to this suspected region to differentiate the mass spectra of the compounds present. For instance, in example 1 of Fig. 4B, the proximity of two peaks led to a missed detection in the 2D profile, resulting in data for only one component data. By applying AP clustering to the mass spectra at the contour's maximum point, data for two components, α-selinene and β-selinene, were obtained, compensating for the limitations of individual contour tracking detection. In the missed-detection contour of example 2, six overlapping compound data were identified using the AP clustering method. However, this process may also produce a small number of false-positive peaks. As detailed in Table S2, the F1-scores for PeakCET detection of the three medicinal products are approximately 95%, demonstrating the efficacy of the PeakCET algorithm in providing accurate and comprehensive analyses of GC × GC fingerprints in medicinal research.
Fig. 4.
Three cases of contour detection assisted by mass spectra clustering. (A) Some false negatives in single contour tracking detection. (B) Affinity propagation (AP) clustering was applied to these suspected regions to differentiate the mass spectra of the compounds present. PC: principal component.
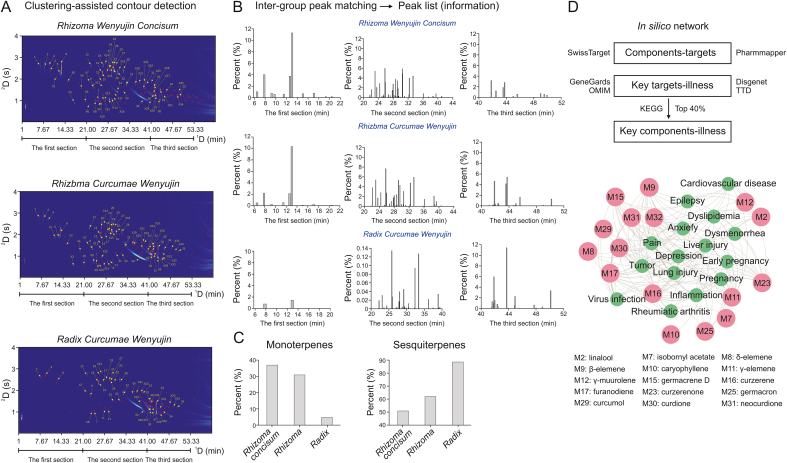
The analysis revealed that Rhizoma Wenyujin Concisum had the highest abundance of peaks, followed by Rhizoma Curcumae Wenyujin, and Radix Curcumae Wenyujin had the fewest. To further explore the relationships among these samples, the researchers conducted inter-group peak matching. This process revealed 52 common components between Rhizoma Wenyujin Concisum and Rhizoma Curcumae Wenyujin, 38 shared components between Rhizoma Curcumae Wenyujin and Radix Curcumae Wenyujin, and 35 common components between Rhizoma Wenyujin Concisum and Radix Curcumae Wenyujin. Detailed information on this process is available in Section S2 of the Supplementary data.
3.2.2. Quantitative comparisons of three fingerprints
After contour edge tracking, the researchers quantified the contour area by counting pixels and characterized the components using peak volume. For quantitative analysis of the GC × GC fingerprints, a normalization method was applied to calculate the component percentages, as shown in Eq. 11:
| (11) |
Among them, Pa represents the area of peak, and Ipeak represents the signal intensity at the peak standard-point.
As depicted in Fig. 5A, the PeakCET algorithm successfully detected the contours in the GC × GC fingerprints of the three medicinal products. The identified compounds and their respective percentages are detailed in Table S1. Based on their elution order, the components were categorized into three segments: 1–21 min (low boiling point components), 21–41 min (moderately high boiling point components), and 41–61 min (high boiling point components). The component percentages in each segment were calculated. Fig. 5B illustrates that the volatile profiles of Rhizoma Wenyujin Concisum and Rhizoma Curcumae Wenyujin were similar, especially in the first stage, whereas Radix Curcumae Wenyujin exhibited fewer volatiles in this stage, with more high boiling-point components in the second and third stages. This variance could be attributed to differences in processing methods: Rhizoma Wenyujin Concisum is sun-dried when freshly sliced, whereas Radix Curcumae Wenyujin undergoes high-temperature steaming before drying. The primary components were classified into five groups: monoterpenes, sesquiterpenes, alcohols, ketones, and esters. Fig. 5C shows that Radix Curcumae Wenyujin had the highest sesquiterpenoid content, followed by Rhizoma Curcumae Wenyujin, with Rhizoma Wenyujin Concisum having the lowest. Conversely, the monoterpene content was highest in Rhizoma Wenyujin Concisum. These findings suggest that while the three herbs share similar components, leading to comparable pharmacological activities, their varying concentrations contribute to their distinct efficacies. To elucidate the significance of these components in Curcumae Wenyujin, platforms like Swisstargetprediction and Pharmmapper were used for compound-target interaction prediction. Disease-related targets were collated from GeneCards, Online Mendelian Inheritance in Man (OMIM), Disgenet, and Therapeutic Target Database (TTD), taking the intersection of targets from both sources. The David platform was then employed for Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment. Ultimately, networks of key targets-components (top 10%), key targets-diseases (top 10%), and key components-diseases (top 40%), as seen in Fig. 5D, were depicted. This approach facilitates cross-cultural understanding between the global East and West. Detailed information on the specific components and their pharmacological distinctions can be found in Section S3 of the Supplementary data and Table S1.
Fig. 5.
Quantitative comparisons of three medical products from Curcumae Wenyujin. (A) The comprehensive two-dimensional gas chromatogram (GC × GC) fingerprints were detected by PeakCET algorithm. (B) The comparative analysis for the component-peaks in the three segments. (C) The comparative analysis for the monoterpenes and sesquiterpenes. (D) An in silico network of key components-illness. OMIM: Online Mendelian Inheritance in Man; TTD: Therapeutic Target Database; KEGG: Kyoto Encyclopedia of Genes and Genomes.
3.3. Application of PeakCET in Radix/Rhizoma Curcumae Wenyujin and other products
Radix Curcumae (Yujin in Mandarin Chinese) and Rhizoma Curcumae (Ezhu in Mandarin Chinese) are medicinal herbs with multiple sources. The former includes Radix Curcumae Wenyujin (Wen Yujin in Mandarin Chinese), Radix Curcumae kwangsiensis (Gui Yujin in Mandarin Chinese), Radix Curcumae phaeocaulis (Lvsi Yujin in Mandarin Chinese), and Radix Curcumae longae (Huangsi Yujin in Mandarin Chinese). Experts in traditional Chinese medicine generally believe that Radix (Rhizoma) Curcumae Wenyujin are widely planted in Rui'an, China; while Radix (Rhizoma) Curcumae phaeocaulis are widely planted in Chengdu, China and Radix/Rhizoma Curcumae kwangsiensis are planted in Guangxi Zhuang Autonomous Region, China. Accordingly, three types of Radix/Rhizoma Curcumae were collected from Taoshan Town, Rui'an, China (Fig. S6), Songqiao Village, Chongzhou, China (Fig. S8), and Luwu Town, Lingshan County, Guangxi Zhuang Autonomous Region, China (Fig. S9). These samples, representing different botanical origins, were analyzed using GC × GC technology. The GC × GC fingerprints revealed notable differences between samples from different origins, while variations within samples from the same origin were minimal (Fig. S10). PeakCET was employed for peak detection in different Radix Curcuma fingerprints. As shown in Table S3, Radix Curcumae wenyujin yielded better F1-scores compared to Radix Curcuma (Guangxi Zhuang Autonomous Region) and Radix Curcumae phaeocaulis when using contour detection alone, primarily due to the more complex composition and overlapping contours of Radix Curcumae phaeocaulis. However, after applying AP clustering, the recall performance of PeakCET improved significantly. It's important to note that the intensity of GC × GC fingerprints greatly influences peak detection, necessitating careful parameter setting. Fig. 6A shows the three sources for two products. Radix/Rhizoma Curcumae Wenyujin predominantly contains β-elemene, curzerene, furanodiene, β-elemenone, germacron, curcumol, and curdione. Radix/Rhizoma Curcumae phaeocaulis is characterized by β-elemene, epi-curzerenone, germacron, β-elemenone, curcumenol, furanodienone, and isofuranodienone. The Radix/Rhizoma Curcumae sample from Guangxi Zhuang Autonomous Region features β-elemene, curzerene, furanodiene, germacron, β-elemenone, β-eudesmol, and aromaticane B. The (semi)-volatile components of the three Curcuma species are listed in Table S4. Clearly, species or genetic differences significantly influence the content of each component. This variability raises concerns about the efficacy and safety of Radix/Rhizoma Curcumae medication and underscores the potential of characteristic components to serve as quality markers for different types of medicinal materials [49,50].
Fig. 6.
PeakCET was applied to comprehensive two-dimensional gas chromatogram (GC × GC) fingerprints from Radix/Rhizoma Curcumae Wenyujin and other products of different origins. (A) A heat map is depicted for three kinds of Radix (Rhizoma) Curcumae products. (B) The classification of Radix Curcumae Wenyujin and other Radix Curcumae samples with different botanical origins. PLS: partial least squares.
In addition to the previously mentioned data, researchers analyzed different GC × GC fingerprints from 18 commercial samples of Radix Curcumae. Initially, the PeakCET algorithm was employed for contour edge tracking, and AP clustering of mass spectra vectors was used to enhance the detection results. Subsequently, a peak matching algorithm was implemented to ensure that identical component-peaks in one fingerprint corresponded to the same component-peaks in another. This process, including the steps involved, is detailed in Section S1 of the Supplementary data. A notable advantage of the peak matching algorithm is its consideration of displacement factors, coupled with further validation through the calculation of MS similarity. Upon the completion of peak matching, a comprehensive peak list from the 18 Radix Curcumae fingerprints was generated. This data was then analyzed using PLS-DA to distinguish between the varieties and to uncover underlying patterns in the complex dataset [51]. The results, as shown in Fig. 6B, effectively differentiated the samples of Radix Curcumae Wenyujin from other Radix Curcumae varieties.
These findings demonstrate that both genetic factors and climatic conditions significantly influence the accumulation of secondary metabolites in these plants. However, given that more than ten Curcuma species are cultivated across various provinces and cities in China [52], there are notable differences in the compositions of Radix Curcumae (Yujin) and Rhizoma Curcumae (Ezhu). It is well-acknowledged that the quality of medicinal materials profoundly affects their therapeutic efficacy and safety. Consequently, the implementation and rigorous enforcement of good agricultural practices and quality traceability systems are essential in China to ensure the highest standards in traditional medicinal products.
4. Conclusion
Compared to 1D-GC, GC × GC offers superior sensitivity and resolution, enhancing the separation of complex essential oils from Curcuma wenyujin and other samples. However, efficiently extracting and analyzing the vast amount of information from GC × GC fingerprints is crucial for inter-group comparison and quality assessment. In this study, the PeakCET algorithm, based on mass spectra clustering-assisted contour detection, is developed to address these challenges. Firstly, PeakCET's capability to detect contour edges in GC × GC fingerprints helps avoid false positives typically caused by burrs. Additionally, to counter the issue of co-elution, AP clustering is applied to the MS vectors of local maximum points within a contour, thereby optimizing 2D peak detection results. Secondly, PeakCET is adept at calculating the 2D peak area using the pixels within a contour, facilitating quantitative analysis by deducing the peak volume and percentage. The algorithm was applied to analyze three fingerprints from Curcuma wenyujin (Rhizoma Wenyujin Concisum, Rhizoma Curcumae Wenyujin, and Radix Curcumae Wenyujin), achieving an average accuracy rate of over 95%. This enabled the researchers to conduct a comprehensive inter-group peak matching, effectively comparing the component similarities and differences among the three medicinal products. Furthermore, the researchers successfully differentiated Radix (Rhizoma) Curcumae Wenyujin from other samples of varying botanical origins. Despite these successes, it is important to recognize the limitations of the PeakCET algorithm. For instance, automating the removal of false negatives through a user-friendly interface remains a challenge. In practical applications, incorporating auxiliary functions such as peak matching, deconvolution, and pattern recognition is essential. In conclusion, the PeakCET algorithm offers a novel approach for analyzing complex GC × GC fingerprints. It holds potential not only for researchers analyzing chemical constituents in food and medicinal materials but also for advancing the standardization of 2D chromatographic fingerprints. However, continuous improvements and integration of additional functionalities are necessary to fully harness its capabilities in various practical scenarios.
CRediT author statement
Xinyue Yang: Data curation, Formal analysis, Software, Writing - Original draft preparation; Yingyu Sima: Data curation, Validation; Xuhuai Luo: Validation; Yaping Li: Investigation; Min He: Conceptualization, Methodology, Funding acquisition, Resources, Supervision, Writing - Reviewing and Editing.
Declaration of competing interest
The authors declare that there are no conflicts of interest.
Acknowledgments
We appreciate Mr. Dou Huang from Guangzhou Ingenious Laboratory Technology Co., Ltd., Guangzhou, China for his assistance in the provision of GC × GC fingerprints from “Mango and Citrus reticulata Blanco”. The same gratitude is also given to Dr. Zhijun Zhao from J&X Technologies, Shanghai, China and Mr. Sifan Luo from Guangdong MS Institute of Scientific Instrument Innovation, Guangzhou, China for their scientific assistance in GC × GC-TOF MS explanation.
This work is financially supported by Hunan 2011 Collaborative Innovation Center of Chemical Engineering & Technology with Environmental Benignity and Effective Resource Utilization, Hunan Province Natural Science Fund, China (Grant Nos.: 2020JJ4569 and 2023JJ60378) and Hunan Province College Students' Innovation and Entrepreneurship Training Program, China (Grant Nos.: S202110530044 and S202210530048).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jpha.2024.01.004.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Kant R., Kumar A. Review on essential oil extraction from aromatic and medicinal plants: Techniques, performance and economic analysis. Sustain. Chem. Pharm. 2022;30 [Google Scholar]
- 2.Bouyahya A., Abrini J., Dakka N., et al. Essential oils of Origanum compactum increase membrane permeability, disturb cell membrane integrity, and suppress quorum-sensing phenotype in bacteria. J. Pharm. Anal. 2019;9:301–311. doi: 10.1016/j.jpha.2019.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gu X., Hao D., Xiao P. Research progress of Chinese herbal medicine compounds and their bioactivities: Fruitful 2020. Chin. Herb. Med. 2022;14:171–186. doi: 10.1016/j.chmed.2022.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhao L., Zhang H., Li N., et al. Network pharmacology, a promising approach to reveal the pharmacology mechanism of Chinese medicine formula. J. Ethnopharmacol. 2023;309 doi: 10.1016/j.jep.2023.116306. [DOI] [PubMed] [Google Scholar]
- 5.Cao P., Wang G., Wei X., et al. How to improve CHMs quality: Enlighten from CHMs ecological cultivation. Chin. Herb. Med. 2021;13:301–312. doi: 10.1016/j.chmed.2021.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li Y., Wu Y., Li Y., et al. Review of the traditional uses, phytochemistry, and pharmacology of Curcuma wenyujin. Y. H. Chen et C. Ling, J. Ethnopharmacol. 2021;269 doi: 10.1016/j.jep.2020.113689. [DOI] [PubMed] [Google Scholar]
- 7.Zhang J., Wang C., Wu W., et al. Authentication of herbal medicines from multiple botanical origins with cross-validation mebabolomics, absolute quantification and support vector machine model, a case study of Rhizoma Alismatis. Arab. J. Chem. 2022;15 [Google Scholar]
- 8.Back to beginning: Searching for Rosetta Stone of enhancing herbal medicine quality. Chin. Herb. Med. 2021;13:299–300. doi: 10.1016/j.chmed.2021.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang D., Ding J., Feng X., et al. Identification of Q-Markers from Hedan Tablet by employing “spider-web” mode and taking compounds’ hepatotoxicity into account. Chin. Herb. Med. 2022;14:612–621. doi: 10.1016/j.chmed.2021.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guo S., Hu S., Jiang L., et al. Quantitative determination of multi-class bioactive constituents for quality control of Yiqi Jiangzhi Granules. Chin. Herb. Med. 2022;14:324–331. doi: 10.1016/j.chmed.2022.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sha M., Li X., Liu Y., et al. Comparative chemical characters of Pseudostellaria heterophylla from geographical origins of China. Chin. Herb. Med. 2023;15:439–446. doi: 10.1016/j.chmed.2022.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He M., Zhou Y. How to identify “Material basis-Quality markers” more accurately in Chinese herbal medicines from modern chromatography-mass spectrometry data-sets: Opportunities and challenges of chemometric tools. Chin. Herb. Med. 2021;13:2–16. doi: 10.1016/j.chmed.2020.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yan M., Zhang Z., Liu Y. Difference analysis of different parts of chicory based on HPLC fingerprint and multi-component content determination. Chin. Herb. Med. 2022;14:317–323. doi: 10.1016/j.chmed.2022.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kan H., Zhang D., Chen W., et al. Identification of anti-inflammatory components in Panax ginseng of Sijunzi Decoction based on spectrum-effect relationship. Chin. Herb. Med. 2023;15:123–131. doi: 10.1016/j.chmed.2022.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Han J., Xu K., Yan Q., et al. Qualitative and quantitative evaluation of Flos Puerariae by using chemical fingerprint in combination with chemometrics method. J. Pharm. Anal. 2022;12:489–499. doi: 10.1016/j.jpha.2021.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khan S., Rai A.K., Singh A., et al. Rapid metabolic fingerprinting with the aid of chemometric models to identify authenticity of natural medicines: Turmeric, Ocimum, and Withania somnifera study. J. Pharm. Anal. 2023;13:1041–1057. doi: 10.1016/j.jpha.2023.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He M., Yang Z., Yang T., et al. Chemometrics-enhanced one-dimensional/comprehensive two-dimensional gas chromatographic analysis for bioactive terpenoids and phthalides in Chaihu Shugan San essential oils. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2017;1052:158–168. doi: 10.1016/j.jchromb.2017.03.029. [DOI] [PubMed] [Google Scholar]
- 18.He M., Zeng J., Peng G., et al. Herbal Component Correlation and Matrix-based Resolution in Comprehensive two-dimensional Gas Chromatography - Mass Spectrometry data via Intelligent Clustering of Modulation Peaks. J. Pharm. Biomed. Anal. 2021;194 doi: 10.1016/j.jpba.2020.113800. [DOI] [PubMed] [Google Scholar]
- 19.Stilo F., Bicchi C., Robbat A., Jr., et al. Untargeted approaches in food-omics: The potential of comprehensive two-dimensional gas chromatography/mass spectrometry. Trac Trends Anal. Chem. 2021;135 [Google Scholar]
- 20.Zou M., Tang H., Chen X., et al. Insights into volatile flavor compound variations and characteristic fingerprints in Longpai soy sauce moromi fermentation via HS-GC-IMS and HS-SPME-GC × GC-ToF-MS. LWT. 2023;176 [Google Scholar]
- 21.Ma C., Wang H., Lu X., et al. Analysis of Artemisia annua L. volatile oil by comprehensive two-dimensional gas chromatography time-of-flight mass spectrometry. J. Chromatogr. A. 2007;1150:50–53. doi: 10.1016/j.chroma.2006.08.080. [DOI] [PubMed] [Google Scholar]
- 22.Filippi J.J., Belhassen E., Baldovini N., et al. Qualitative and quantitative analysis of vetiver essential oils by comprehensive two-dimensional gas chromatography and comprehensive two-dimensional gas chromatography/mass spectrometry. J. Chromatogr. A. 2013;1288:127–148. doi: 10.1016/j.chroma.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 23.He M., Yan P., Yang Z., et al. Multi-analytical strategy for unassigned peaks using physical/mathematical separation, fragmental rules and retention index prediction: An example of sesquiterpene metabolites characterization in Cyperus rotundus. J. Pharm. Biomed. Anal. 2018;154:476–485. doi: 10.1016/j.jpba.2018.03.042. [DOI] [PubMed] [Google Scholar]
- 24.Yang X., Zeng P., Wen J., et al. Gain deeper insights into traditional Chinese medicines using multidimensional chromatography combined with chemometric approaches. Chin. Herb. Med. 2024;16:27–41. doi: 10.1016/j.chmed.2023.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trinklein T.J., Cain C.N., Ochoa G.S., et al. Recent advances in GC × GC and chemometrics to address emerging challenges in nontargeted analysis. Anal. Chem. 2023;95:264–286. doi: 10.1021/acs.analchem.2c04235. [DOI] [PubMed] [Google Scholar]
- 26.Mikaliunaite L., Synovec R.E. Computational method for untargeted determination of cycling yeast metabolites using comprehensive two-dimensional gas chromatography time-of-flight mass spectrometry. Talanta. 2022;244 doi: 10.1016/j.talanta.2022.123396. [DOI] [PubMed] [Google Scholar]
- 27.Stilo F., Bicchi C., Jimenez-Carvelo A.M., et al. Chromatographic fingerprinting by comprehensive two-dimensional chromatography: Fundamentals and tools. Trac Trends Anal. Chem. 2021;134 [Google Scholar]
- 28.Nolvachai Y., Amaral M.S.S., Marriott P.J. Foods and contaminants analysis using multidimensional gas chromatography: An update of recent studies, technology, and applications. Anal. Chem. 2023;95:238–263. doi: 10.1021/acs.analchem.2c04680. [DOI] [PubMed] [Google Scholar]
- 29.Reichenbach S.E., Ni M., Kottapalli V., et al. Information technologies for comprehensive two-dimensional gas chromatography. Chemom. Intell. Lab. Syst. 2004;71:107–120. [Google Scholar]
- 30.Peters S., Vivó-Truyols G., Marriott P.J., et al. Development of an algorithm for peak detection in comprehensive two-dimensional chromatography. J. Chromatogr. A. 2007;1156:14–24. doi: 10.1016/j.chroma.2006.10.066. [DOI] [PubMed] [Google Scholar]
- 31.Vivó-Truyols G. Bayesian approach for peak detection in two-dimensional chromatography. Anal. Chem. 2012;84:2622–2630. doi: 10.1021/ac202124t. [DOI] [PubMed] [Google Scholar]
- 32.Kim S., Ouyang M., Jeong J., et al. A new method of peak detection for analysis of comprehensive two-dimensional gas chromatography mass spectrometry data. Ann. Appl. Stat. 2014;8:1209–1231. doi: 10.1214/14-aoas731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Victória Matias A., Atkinson Amorim J.G., Buschetto Macarini L.A., et al. What is the state of the art of computer vision-assisted cytology? A Systematic Literature Review. Comput. Med. Imaging Graph. 2021;91 doi: 10.1016/j.compmedimag.2021.101934. [DOI] [PubMed] [Google Scholar]
- 34.Kakani V., Nguyen V.H., Kumar B.P., et al. A critical review on computer vision and artificial intelligence in food industry. J. Agric. Food Res. 2020;2 [Google Scholar]
- 35.Wu Y., Ma W., Zhang J., et al. Point-matching algorithm based on local neighborhood information for remote sensing image registration. JARS. 2018;12 [Google Scholar]
- 36.Yeap D., McCartney M.M., Rajapakse M.Y., et al. Peak detection and random forests classification software for gas chromatography/differential mobility spectrometry (GC/DMS) data. Chemometr. Intell. Lab. Syst. 2020;203 doi: 10.1016/j.chemolab.2020.104085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.He M., Yang X., Li Y., et al. Development of image similarity strategy based on targeted filtration for non-targeted HS-SPME/GC × GC fingerprints of volatile oils from Chinese patent medicines: A case of Chaihu Shugan Wan. Microchem. J. 2023;191 [Google Scholar]
- 38.Caratti A., Squara S., Bicchi C., et al. Augmented visualization by computer vision and chromatographic fingerprinting on comprehensive two-dimensional gas chromatographic patterns: Unraveling diagnostic signatures in food volatilome. J. Chromatogr. A. 2023;1699 doi: 10.1016/j.chroma.2023.464010. [DOI] [PubMed] [Google Scholar]
- 39.Yang T., Yan P., He M., et al. Application of Subwindow Factor Analysis and Mass Spectral information for accurate alignment of non-targeted metabolic profiling. J. Chromatogr. A. 2018;1563:162–170. doi: 10.1016/j.chroma.2018.05.071. [DOI] [PubMed] [Google Scholar]
- 40.Zeng J., He M., Wu H., et al. Peak alignment for herbal fingerprints from liquid chromatography-high resolution mass spectrometry via diffusion model and bi-directional eigenvalues. Microchem. J. 2021;167 [Google Scholar]
- 41.Rawlinson C., Jones D., Rakshit S., et al. Hierarchical clustering of MS/MS spectra from the firefly metabolome identifies new lucibufagin compounds. Sci. Rep. 2020;10:6043. doi: 10.1038/s41598-020-63036-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chanana S., Thomas C.S., Zhang F., et al. hcapca: Automated hierarchical clustering and principal component analysis of large metabolomic datasets in R. Metabolites. 2020;10:297. doi: 10.3390/metabo10070297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Imanishi S.Y., Nakayama T., Asukabe H., et al. Application of MALDI biotyper to cyanobacterial profiling. Rapid Commun. Mass Spectrom. 2017;31:325–332. doi: 10.1002/rcm.7793. [DOI] [PubMed] [Google Scholar]
- 44.Teramoto K., Sato H., Sun L., et al. Phylogenetic classification of Pseudomonas putida strains by MALDI-MS using ribosomal subunit proteins as biomarkers. Anal. Chem. 2007;79:8712–8719. doi: 10.1021/ac701905r. [DOI] [PubMed] [Google Scholar]
- 45.Ferreira V.H.C., Hantao L.W., Poppi R.J. Use of color based chromatographic images obtained from comprehensive two-dimensional gas chromatography in authentication analyses. Talanta. 2021;234 doi: 10.1016/j.talanta.2021.122616. [DOI] [PubMed] [Google Scholar]
- 46.Suzuki S., Abe K. Topological structural analysis of digitized binary images by border following. Comput. Vis. Graph. Image Process. 1985;29 [Google Scholar]
- 47.Frey B.J., Dueck D. Clustering by passing messages between data points. Science. 2007;315:972–976. doi: 10.1126/science.1136800. [DOI] [PubMed] [Google Scholar]
- 48.Li Z., Hao E., Cao R., et al. Analysis on internal mechanism of zedoary turmeric in treatment of liver cancer based on pharmacodynamic substances and pharmacodynamic groups. Chin. Herb. Med. 2022;14:479–493. doi: 10.1016/j.chmed.2022.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lu X., Jin Y., Wang Y., et al. Multimodal integrated strategy for the discovery and identification of quality markers in traditional Chinese medicine. J. Pharm. Anal. 2022;12:701–710. doi: 10.1016/j.jpha.2022.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang Z., Li S. Network pharmacology in quality control of traditional Chinese medicines. Chin. Herb. Med. 2022;14:477–478. doi: 10.1016/j.chmed.2022.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li H., Xu Q., Liang Y. libPLS: An integrated library for partial least squares regression and linear discriminant analysis. Chemom. Intell. Lab. Syst. 2018;176:34–43. [Google Scholar]
- 52.Zhang L., Yang Z., Wei J., et al. Contrastive analysis of chemical composition of essential oil from twelve Curcuma species distributed in China. Ind. Crops Prod. 2017;108:17–25. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.