Abstract
Atopic dermatitis (AD) is a persistent and recurring inflammatory condition affecting the skin. An expanding corpus of evidence indicates the potential participation of transforming growth factor-β1 (TGF-β1) in the modulation of inflammation and tissue remodeling in AD. The primary objective of this study was to examine the aberrant modulation of TGF-β1/small mothers against decapentaplegic homolog 3 (SMAD3) signaling through a comprehensive analysis of their molecular and protein expression profiles. The study encompassed an aggregate of 37 participants, which included 25 AD patients and 12 controls. The assessment of mRNA and protein levels of TGF-β1 and SMAD3 was conducted utilizing quantitative real-time PCR and immunohistochemistry (IHC), whereas serum IgE and vitamin D levels were estimated by ELISA and chemiluminescence, respectively. Quantitative analysis demonstrated a 2.5-fold upregulation of TGF-β1 mRNA expression in the lesional AD skin (P < 0.0001). IHC also exhibited a comparable augmented pattern, characterized by moderate to strong staining intensities. In addition, TGF-β1 mRNA showed an association with vitamin D deficiency in serum (P < 0.02), and its protein expression was linked with the disease severity (P < 0.01) Furthermore, a significant decrease in the expression of the SMAD3 gene was observed in the affected skin (P = 0.0004). This finding was further confirmed by evaluating the protein expression and phosphorylation of SMAD3, both of which exhibited a decrease. These findings suggest that there is a dysregulation in the TGF-β1/SMAD3 signaling pathway in AD. Furthermore, the observed augmentation in mRNA and protein expression of TGF-β1, along with its correlation with the disease severity, holds considerable clinical significance and emphasizes its potential role in AD pathogenesis.
Keywords: atopic dermatitis, TGF-β1 mRNA expression, TGF-β1 protein, TGF-β1 signaling, SMAD3, TGF-β/SMAD signaling
The study highlights the upregulation of transforming growth factor-β1 (TGF-β1) mRNA and protein expression in the affected skin of patientswith atopic dermatitis (AD), suggesting a potential role for TGF-β1 in the modulation of inflammation and tissue remodeling in AD. The association of TGF-β1 mRNA with vitamin D deficiency in serum, and its protein expression with the severity of AD emphasizes the clinical significance of TGF-β1 in AD pathogenesis and its potential role as a marker for disease severity. A significant decrease in the expression of small mothers against decapentaplegic homolog 3 (SMAD3) in the affected skin of AD patients suggests a dysregulation in the TGF-β1/SMAD3 signaling pathway in AD.
Graphical Abstract
Graphical Abstract.
Introduction
Atopic dermatitis (AD) is a hypersensitive manifestation of the skin with severe pruritis and relapsing eczematous lesions as the predominant features, often followed by thickening of the affected skin due to the chronicity of the disease course. With a prevalence rate of 15–20% among children and up to 10% among adults, this condition ranks as the 15th most common nonfatal disease, and the skin disease with the greatest disease burden [1, 2]. The disorder tends to be linked to a familial predisposition to allergies [3] and may lead to the subsequent development of other atopic conditions, such as allergic rhinitis, allergic asthma, and food allergy [4].
AD is a multifaceted disorder that exhibits considerable variability in clinical manifestations and disease progression. AD pathogenesis involves the interplay among genetic factors, environmental agents, dysfunction of the skin barrier, immune system dysregulation, and microbial imbalance [3, 5, 6]. The initiation of inflammation is believed to occur due to impairment in the epidermal barrier integrity and subsequent induction of immune cells, which lead to the activation of various T helper cell pathways. Activated T cells secrete cytokines that promote pruritus, inflammation, and synthesis of antigen-specific IgE from activated B cells and plasma cells [2]. Numerous cytokines have been associated with the etiology of AD. Among them, transforming growth factor-β1 (TGF-β1) plays a pivotal role in sustaining and amplifying the inflammatory response, making it a significant player in the pathophysiology of allergic diseases, including AD. The implication of TGF-β1 in the pathogenesis of allergic disease was reported in a study of autosomal dominant Loeys–Dietz syndrome (LDS), characterized by mutations in the TGF-β1 signaling pathway and higher incidence of allergic rhinitis, asthma, food allergy, and eczema as compared to the general population [7]
TGF-β1 is a pleiotropic cytokine expressed in variety of cell types and serves a crucial function in regulating diverse cellular events including differentiation, proliferation, adhesion, and migration in various cells [8, 9]. This cytokine influences the differentiation of various T cell subsets, as well as the class switching of B cell immunoglobulins subsequent to the initial exposure to an allergen. It also mediates inflammation in various allergic diseases [10] and functions as a fibrogenic agent, capable of stimulating fibroblast differentiation and promoting the deposition of collagen and fibronectin in the extracellular matrix [11]. As a result, it leads to the remodeling of the affected tissue due to repeated cycles of injury and tissue repair during the chronic stages of allergic asthma and AD [11–13].
TGF-β1 is typically inactive due to its binding with latency-associated peptide (LAP). Once activated, it binds to its heterotetrameric receptors and initiates intracellular signaling resulting in the phosphorylation of small mothers against decapentaplegic homolog 2 (SMAD2) and SMAD3. These activated SMADs subsequently form oligomeric complexes with SMAD4 and translocate into the nucleus to regulate gene expression alongside other transcription factors [14, 15]. A mouse model of LDS showed that decreased TGF-β1 response upregulates its synthesis, which hyperactivates intracellular SMAD signaling [10]. Thus, it is unclear whether decreased TGF-β1 signaling at the receptor level or augmented signaling at the cellular stage enhances the allergic reaction. This study aimed to examine the aberrant modulation of TGF-β1/SMAD3 signaling through a comprehensive analysis of their molecular and protein expression profiles to gain insight into the potential mechanisms by which altered TGF-β1 signaling could influence the progression and severity of AD.
Materials and methods
Ethics and subject recruitment
The present case–control study was approved by the SKIMS ethics committee (IEC-SKIMS) via protocol ID #RP 62/2020, dated 24 January 2021. A cohort of 25 individuals diagnosed with AD in the Kashmir valley of J&K (North India) who sought medical consultation from March 2020 to October 2022 at the Allergy clinic of the Immunology and Molecular Medicine, Sher-i-Kashmir Institute of Medical Sciences (SKIMS, Srinagar) and Department of Dermatology, GMC Srinagar, were selected as cases. Prior to their participation in the study, all subjects signed the written informed consent. Patients treated with desensitization immunotherapy or systemic immunosuppressants within a period of 6 months, as well as those who had received local immunosuppressents within 1 week were excluded from the study. The control group comprised 12 normal skin samples obtained from healthy donors who underwent aesthetic or reconstructive surgery in the Department of Plastic and Reconstructive Surgery at SKIMS.
Sample collection
Punch biopsy:
A 4-mm punch biopsy obtained from the lesional skin of patients was divided into 2 bits of 2 mm each. One bit was formalin-fixed and paraffin-embedded (FFPE) for hematoxylin and eosin (H&E) staining and immunohistochemistry (IHC) and the other bit was collected in RNA later, stored at −80°C for mRNA expression analysis.
Peripheral blood sample:
Each subject provided a total of 2 ml of peripheral blood samples, 1 ml in EDTA vials and 1 ml in serum separating tube was centrifuged, and serum was stored at −80°C till further use.
Assessment of disease severity in patients with AD
The scoring atopic dermatitis (SCORAD) is a comprehensive score used to assess the severity of AD. According to this, AD is classified as mild if the score is below 25, moderate if it falls between 25 and 50, and severe if the score exceeds 50.
Skin prick test
The skin prick test (SPT) involving eczema panel was conducted by applying a series of allergen extracts (obtained from Greer Labs, USA; Supplemantary Table S1) on lateral aspect of the forearm. The skin was subjected to gentle pricking, and the resulting wheal diameters were assessed approximately 15–20 min later. A positive reaction was determined if the diameter of the wheal was 3 mm × 4 mm larger than that of the negative control.
Estimation of serum levels of total IgE and vitamin D
Serum levels of Total IgE were determined using the Human IgE total ELISA kit from Biogenix. Similarly, vitamin D levels were estimated using a chemiluminescence unicell DXI 800 analyzer (Beckman Coulter, USA).
RNA extraction and cDNA synthesis
The Trizol-Chloroform method (Sigma Aldrich) was employed to isolate total RNA from skin tissues. The integrity of the RNA was confirmed on 1% agarose gel electrophoresis, and its quantification was performed based on the A260/280 ratio. The RNA was then treated with a DNase I enzyme (GCC biotech) in order to eliminate any residual genomic DNA. The synthesis of complementary DNA (cDNA) was performed using the Maxima First Strand cDNA synthesis kit (Thermo Scientific; Cat no: #K1641) in accordance with the manual provided by the manufacturer.
TGF-β1 and SMAD3 mRNA quantification
The primers for TGF-β1, SMAD3, and GAPDH were designed using IDT software, specifically targeting exon–exon junctions (Table 1). The SYBR Green Master Mix was utilized for the qPCR reaction and each sample was analysed in triplicate to ensure accuracy and reproducibility. The temperature profile involved initial denaturation at 95°C for 5 min, followed by 40 cycles of 95°C for 15 s and 60°C for 30 s, 72°C for 20 s, and finally melt curve analysis consisting of 90 s of pre-melt conditioning on the first step and 5 s on subsequent steps. The results were analysed using the Delta Delta cycle threshold (ΔΔCT) method. The amplified qPCR products were checked on a 2% agarose gel electrophoresis in order to verify the adequacy of product amplification (Supplementary Fig. S1).
Table 1.
Primers used for qPCR amplification of TGF-β1, SMAD3, and GAPDH with their PCR amplicon size
| Gene | Primer sequence (5ʹ-3ʹ) | Amplicon size (bp) |
|---|---|---|
| TGF-β1 | F:5ʹ-CGTGGAGCTGTACCAGAAATAC-3ʹ R:5ʹ-CACAACTCCGGTGACATCAA-3ʹ |
112 |
| SMAD3 | F: 5ʹ-CCCAGAGCAATATTCCAGAGAC-3ʹ R: 5ʹ-GTCCATGCTGTGGTTCATCT-3ʹ |
89 |
| GAPDH | F: 5ʹ-AGAAGGCTGGGGCTCATTTG-3ʹ R:5ʹ-AGGGGCCATCCACAGTCTTC-3ʹ |
258 |
H&E staining
For elucidation of cellular and tissue structure details in AD, 25 skin biopsies from the lesional skin of AD patients and 8 normal skin tissue samples of healthy donors were fixed in formalin and embedded in paraffin blocks. A manual microtome (Leica Biosystems, India) was used to slice thin tissue sections of a 4.5-μm thickness from these paraffin blocks, mounted on glass slides and proceeded for routine H&E staining [16]. Briefly, the tissue sections were deparaffinized with xylene and serially rehydrated with 100%, 90%, 50% ethanol, and distilled water. All slides were stained with hematoxylin, differentiated by giving 1–2 dips in 1% HCl in 70% alcohol, and then thoroughly washed in water. The tissue sections were counterstained in eosin and dehydrated through graded alcohols. Sections were finally cleared in xylene twice and mounted with mounting media (DPX). Photomicrographs were obtained using Nikon DS-Ri1microscope (Nikon corporation Tokyo, Japan).
Protein expression and localization of TGF-β1 and phospho-SMAD3 via IHC
The protein expression of TGF-β1 and phospho-SMAD3 (pSMAD3) was conducted in the Department of Pathology, SKIMS. The FFPE tissues were sliced into 5-μm sections and settled on charged adhesive glass slides (Cancer Diagnostics, Inc. Durham, Lot# 20210125). After drying the slides at 37°C for 15 min, the sections were proceeded for deparaffinization with xylene, followed by rehydration using a series of graded alcohol solutions and final washing in phosphate-buffered saline (PBS). This was followed by antigen retrieval with citrate buffer (10 mM, pH 6.0) at 100°C for 20 min in the steamer. Following a gradual cooling process to room temperature, the endogenous peroxidase activity was inhibited by treatment with a 3% hydrogen peroxide solution and blockage of non-specific binding with 10% normal goat serum. The slide edges were dried with blotting tissue and a Pap pen (Abcam, Cambridge, MA, USA) was employed to delineate the boundaries of the tissue. Then, the sections were incubated with primary antibodies i.e. TGF-β1 Rabbit polyclonal antibody (Abclonal, Catalog no: A2124) and Phospho-SMAD3 (Ser423, Ser425) polyclonal antibody (Rockland, Catalog no: 600-401-919S), respectively at 4°C overnight. The sections were then washed three times with PBS followed by immunodetection using MACH 1 Universal horseradish peroxidase (HRP)-Polymer (Catalog no. #MRH538L10) anti-rabbit HRP conjugate secondary antibody (BioCare Medical) and visualized with 3,3ʹ-diaminobenzidine. The sections were counterstained, dehydrated, and finally mounted with DPX. Negative controls consisted of tissue sections incubated in PBS instead of primary antibody under the same experimental conditions. Photomicrographs were obtained using Nikon DS-Ri1microscope (Nikon corporation Tokyo, Japan). All the slides were assessed by two proficient pathologists in a manner that ensured their independence. A consensus-based semi-quantitative method (IR score; Remmele score) was used to obtain staining results.
Statistical analysis
The statistical analysis was conducted using SPSS 26.0 software (SPSS Inc., Chicago, Illinois) and Graph Pad Prism version 5 (La Jolia, California, USA). The difference in data groups was analysed by various parametric and non-parametric tests, wherever applicable. The P-value was obtained from two-sided testing and was considered significant when its value was <0.05.
Results
Patient characteristics
The baseline data of AD patients are presented in Supplementary Table S2. The cases comprised of 15 (60%) males and 10 (40%) females. The mean age was 13.0 ± 10.6 years and 11 patients were <5 years, 7 patients in the age group of 6–18, and 7 were >19 years of age. The control group had 8 (66.6%) males and 4 (33.3%) females with a mean age of 18.3 ± 12.11 years. There were no significant differences observed in the mean age and gender distributions between the cases and controls, indicating that there was appropriate frequency matching. Among all the cases, 18 (72%) lived in rural areas and 7(28%) lived in urban areas.
SCORAD score and severity of AD
Out of 25 AD cases, 24% (6/25) were mild cases with a SCORAD score of <25, 52% (13/25) were moderate in the range of 25–50, and 24% (6/25) were severe cases with a SCORAD score of >50 (Supplementary Table S2).
SPT results
Out of 25 AD cases, 84% (21/25) were positive for more than 1 species of dust mite and 76% (19/25) were positive for more than 1 foods that were tested in the study (Supplementary Table S2).
Serum IgE levels and vitamin D levels in AD
Total serum IgE levels were raised in 72% (18/25) of the patients with AD whereas only 28% (7/25) of patients had normal IgE levels. In addition, 52% (13/25) were vitamin D deficient, 32% (8/25) had insufficient levels, and 16% (4/25) had sufficient vitamin D levels (Supplementary Table S2).
TGF-β1 and SMAD3 mRNA expression in lesional AD skin
TGF-β1 and SMAD3 mRNA expression was quantified in 25 AD cases and 12 healthy controls using the 2−ΔΔCt method, as described previously [17]. The TGF-β1 mRNA in the lesional skin of AD cases showed a significant difference as compared to the controls. (Mean ± SD of 5.6 ± 1.7 in cases vs 2.3 ± 0.6 in controls; P < 0.0001; Fig. 1A). Overall, we observed an average fold change of 2.4, which signifies a greater than 2-fold increased TGF-β1 gene expression in the lesional AD skin. Similarly, an average fold change of 0.27 was detected in SMAD3 with a mean ± SD of 0.3 ± 0.2 in cases as compared to controls with a mean ± SD of 1.1 ± 0.8 (P < 0.0001; (Fig. 1B). This shows overall decreased expression of SMAD3 in the lesional skin of patients with AD.
Figure 1.
Box and Whisker plots showing the relative expression of TGF-β1 (A) and SMAD3 (B) in the lesional skin of AD patients in comparison to healthy controls.
Comparison of mRNA expression of TGF-β1 and SMAD3 with various clinicopathological and lab parameters
The mRNA expression of TGF-β1 and SMAD3 was evaluated with relation to various parameters, where we found that TGF-β1 mRNA in lesional skin was negatively associated serum vitamin D levels (P = 0.02; Supplementary Table S3; and Fig. S2). However, SMAD3 mRNA levels in lesional skin did not show any association with clinicopathological or laboratory parameters (P > 0.05).
Correlation analysis of TGF-β1 and SMAD3 mRNA expression with various clinical and laboratory parameters
The Pearson’s correlation analysis of TGF-β1 and SMAD3 mRNA with various parameters showed negative correlation of TGF-β1 mRNA with age (r = −0.437, P = 0.029) and serum vitamin D levels (r = −0.437, P = 0.018) as depicted in Supplementary Fig. S3. However, SMAD3 mRNA expression did not show any correlation with studied parameters (P > 0.05).
Histological features of AD skin
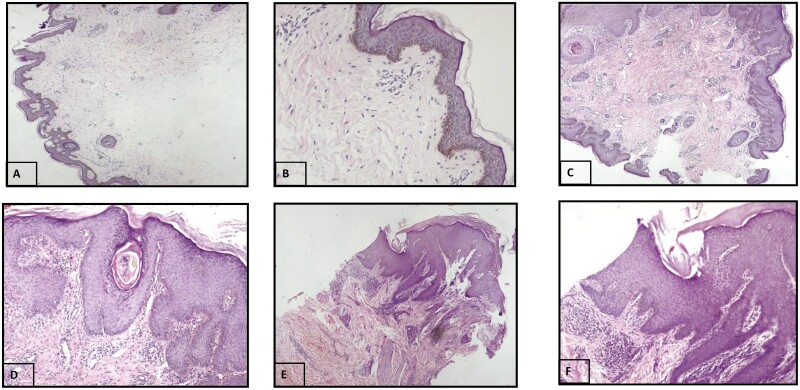
The tissue structure details were analysed in the lesional skin of AD cases by H&E staining and the slides were analysed by experienced pathologists. Out of 25 AD cases, 22 (88%) showed acanthosis, 16 (64%) showed parakeratosis, 13 (52%) showed hyperkeratosis, 10 (40%) showed spongiosis, and 16 (64%) showed inflammatory infiltrate (Supplementary Fig. S4; Fig. 2).
Figure 2.
Morphology assessments of AD skin by H&E stain. (A) Control skin with normal epidermis and stratum corneum (magnification 4×). (B) Control skin with normal epidermis and stratum corneum (magnification 10×). (C) AD skin section showing irregular acanthosis along with focal parakeratosis. Superficial dermis shows proliferation of capillaries along with a moderate degree of lymphohistiocytic infiltrate (magnification 4×). (D) Same as (C) (magnification 10×). (E) AD skin showing acanthosis, hyperkeratosis, and parakeratosis. The dermis shows dense perivascular chronic inflammatory infiltrate and dermal edema (magnification 4×). (F) Same as E (magnification 10×).
Protein expression and localization of TGF-β1 and pSMAD3 in lesional skin of patients with AD
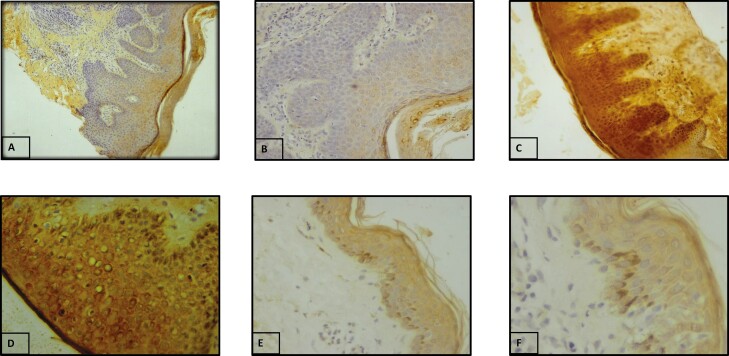
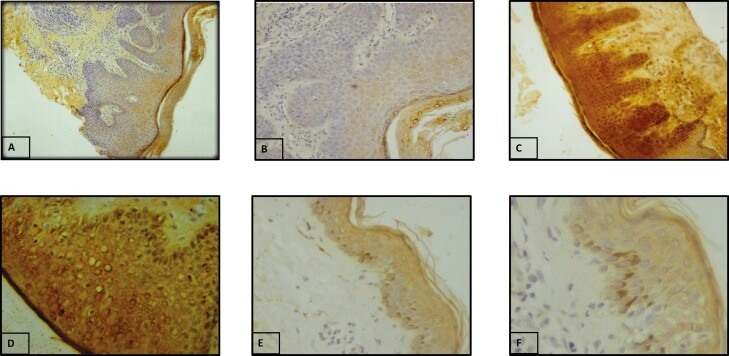
IHC was employed to determine the protein expression and localization of TGF-β1 and pSMAD3 in lesional AD skin. In case of TGF-β1, 6 cases (24%) showed membranous expression only, whereas 19 cases (76%) showed both cytoplasmic and membranous expression. The staining was mostly moderate to strong in AD cases, wherein 40% (10/25) cases showed moderate expression and 60% (15/25) showed strong expression of TGF-β1 protein as compared to normal tissues. The TGF-β1 expression was also evaluated with various clinicopathological features where we found a significant association of TGF-β1 protein levels with the severity of AD (P = 0.01; Supplementary Table S4). Figure 3 represents the staining pattern of TGF-β1 in AD lesional skin and controls.
Figure 3.
Protein expression of TGF-β1 by IHC. (A) Negative control (10×). (B) Negative control (20×). (C) Strong staining in AD lesional skin (magnification 20×). (D) Strong staining in AD lesional skin (magnification 40×). (E) Weak staining in normal skin tissue (magnification 20×). (F) Weak staining in normal skin tissue (magnification 40×).
In case of SMAD3, expression, phosphorylation, and localization were analysed. Out of 25 AD cases, 8% (2/25) showed weak expression, 52% (13/25) showed mild expression and 40% (10/25) showed strong expression of pSMAD3 whereas in controls 25% (3/12) showed weak expression and 75% (9/12) showed mild expression of pSMAD3. The localization of pSMAD3 was predominantly cytoplasmic in the majority of cases (76%), whereas in 24% of cases localization was both cytoplasmic and nuclear (Fig. 4). In controls, 66.7% showed nuclear expression whereas 33.3% showed both nuclear and cytoplasmic expressions. Overall, phosphorylation and the nuclear expression were considered a positive activation of signaling and cytoplasmic as negative. Stratification analysis of pSMAD3 expression with various clinicopathological features did not reveal any significant association with any clinical or laboratory parameter. Figure 4 represents the staining pattern of pSMAD3 in AD lesional skin and controls.
Figure 4.
Protein expression and localization of pSMAD3 by IHC. (A) Negative control (20×). (B) Negative control (40×). (C) Cytoplasmic staining in AD lesional skin (magnification 20×). (D) Cytoplasmic staining in AD lesional skin (magnification 40×). (E) Nuclear staining in normal skin tissue (magnification 20×). (F) Nuclear staining in normal skin tissue (magnification 40×).
Discussion
Transforming growth factor-β1
AD is a pruritic inflammatory skin disease characterized by intensely eczematous skin lesions that undergo relapsing and remitting cycles. This study was aimed to assess the molecular and protein-level expression of crucial signaling molecules, TGF-β1 and SMAD3, within skin lesions associated with AD to enhance our comprehension of the involvement of these molecules in the development and progression of AD. Our findings show that TGF-β1 mRNA and protein expression is increased in patients with AD. The TGF-β1 mRNA was increased by 2.4-fold in AD cases as compared to normal skin controls. The results of our study indicate a notable upregulation of TGF-β1 mRNA and protein expression in patients with AD. It was observed that the expression of TGF-β1 mRNA exhibited a significant increase of 2.4-fold when compared to the control group. The observed results align with our prior investigation, wherein we documented the upregulation of this specific marker in the peripheral blood and serum of patients with AD [17]. Taken together, our studies provide the first evidence of elevated TGF-β1 expression in peripheral blood, lesional skin, as well as serum of AD patients. The present study has identified comparable observations to those reported by Fedenko et al. [18], wherein an upregulation of TGF-β1 mRNA expression was observed in the cutaneous tissue of patients diagnosed with AD. In contrast, previous investigations have failed to observe any discernible disparity in TGF-β1 mRNA levels when comparing individuals diagnosed with AD and those with unaffected skin as control subjects [19, 20]. The observed discrepancies among various studies can be ascribed to variances in the sizes of the samples, the selection criteria for control groups, and the existence of heterogeneity stemming from the ethnic makeup of the population being examined. In order to understand the clinical significance of TGF-β1 in AD, we analysed its mRNA expression with various laboratory parameters where we found that its expression in lesional skin was negatively associated with serum vitamin D levels. This can be explicated by considering the fact that vitamin D functions in proliferation of keratinocytes and maintenance of epidermal barrier integrity [21], and in AD, its decreased serum levels may be one of the causes of skin barrier dysfunction. Furthermore, a study has found that vitamin D receptor (VDR) negatively regulates TGF-β/Smad signaling and suggested that decreased expression of VDR and its ligand can lead to an overactive TGF-β signaling pathway [22]. These findings are consistent with our study where we also found decreased serum vitamin D levels associated with increased TGF-β1 mRNA in lesional skin of AD. The augmented mRNA expression observed in our investigation was subsequently validated through protein expression analysis utilizing IHC techniques. Notably, the immunohistochemical analysis revealed robust staining intensities of TGF-β1 in AD cases that also showed a positive association with the severity of AD. The results presented here align with the research conducted by Toda et al. [13], which similarly reported heightened protein expression of TGF-β1 in both acute and chronic skin lesions associated with AD [13]. These results are further corroborated by a study that demonstrated the partial alleviation of AD symptoms through the blockade of TGF-βR1. This blockage effectively led to suppression of serum fibrosis markers, including TGF-β1 and LAP, that were previously induced by AD [23]. The upregulation of TGF-β1 can be ascribed to its involvement in wound healing and tissue remodeling, as during the progression, recurrent course of tissue damage and subsequent repair, often leads to tissue remodeling and fibrosis, leading to lichenification of skin lesions that characterizes the chronic course of the disease. In summary, our investigation has contributed to the advancement of knowledge regarding the role of TGF-β1 in AD and highlights its potential importance in the development and progression of AD.
Small mothers against decapentaplegic homolog 3
The SMAD3 protein serves as a major mediator of the TGF-β signaling, regulating the transcription of specific genes within the cell nucleus. It is located on chromosome 15q, where evidence for the AD linkage was found [24]. The implication of TGF-β/SMAD3 signaling in the development of allergic disorders stands limited. To date, there is a limited body of research on the role of SMAD3 in AD. We investigated the mRNA expression of SMAD3 and phosphorylation status of SMAD3 protein in lesional AD skin, in order to effectively address the current understanding of the role of the TGF-β/SMAD pathway in AD. Our findings suggest that the SMAD3 mRNA expression in lesional AD skin is markedly reduced as compared to normal skin. The results were confirmed further by analysing the protein expression and phosphorylation of SMAD3 by IHC, both of which were reduced in the lesional AD skin. The cytoplasmic expression of phospho-SMAD3 indicated the inactive SMAD signaling in lesional AD skin. Contrary to the outcomes of our previous inquiry, which involved the observation of elevated levels of SMAD3 expression in the peripheral blood of patients with AD [25], we have now identified a divergent pattern in the affected skin tissue during the current investigation. The results of the current investigation align with prior research that has shown a reduction in SMAD3 expression in the skin of individuals with AD [20], as well as in mouse models of AD [26, 27]. Conversely, a study on Smad3 knockout mice showed that upon exposure to high doses of ionizing irradiation, these mice exhibited a protective effect against the development of acanthosis, hyperkeratosis, and dermal inflammation. Furthermore, these mice also showed resistance to ovalbumin-induced peribronchial fibrosis in the lungs [28]. A separate study found that SMAD3−/− mice exhibited a notable decrease in dermis thickness and expression of proinflammatory cytokines. On the contrary, there was an increase in mast cells, eosinophils infiltrating the skin, and specific IgE levels [26]. This indicates that the overproduction of allergen-specific IgE, mast cells, and eosinophils, as observed in AD, has an inhibitory effect on the SMAD3 signaling in the lesional skin but not in the peripheral blood. Although there is no clear evidence that the inhibition of SMAD3 expression in lesional skin is a cause or consequence of AD, Ostuka et al. [27] speculated that the decreased expression of SMAD3 in the lesional skin of patients with AD and mouse models of AD may be attributed to the potential protective mechanism against skin inflammation-induced injury. The reduced expression of SMAD3 also leads to a decrease in the expression of FoxP3 resulting in altered Treg function and on the other hand, results in intensified Th17 differentiation that plays a role in the AD pathogenesis [29]. The decreased expression of SMAD3, besides upregulation of TGF-β1 in the lesional skin, suggests that TGF-β1 in the lesional skin of AD functions through different axis/pathway since it can exhibit its downstream effects through both canonical SMAD as well as Non-SMAD mediated pathway [30]. In conclusion, the findings of our study which show decreased expression of SMAD3 in lesional skin are in accordance with the results of the prior studies [20, 26, 27], However, it is necessary to establish the foundation of these findings, as SMAD3 plays a role in various processes ranging from the deposition of extracellular matrix to the development of Treg, Th17, and Th9 cells. Hence, the findings of the present study, along with previous research conducted on mouse models underscore the potential involvement of SMAD3 in the pathogenesis of AD. Nevertheless, it is important to consider the limited number of participants and the scarcity of existing literature in this field, specifically regarding studies conducted on human subjects. Therefore, additional research is necessary to gain a comprehensive understanding of the involvement of SMAD3 in AD.
Limitations of the study
One notable constraint inherent in our study pertained to the notably small sample size of the study cohort. Furthermore, it is worth noting that the potential weak point of this study lies in the possibility of selection or recall bias. However, it is important to acknowledge that the presence of only one interviewer in the data collection process mitigates this bias to some extent.
Conclusion
Based on our findings, it can be inferred that there exists an aberrant regulation of TGF-β1 and its subsequent signaling mediator, namely SMAD3, in the context of AD. The results of this study provide substantial evidence to underscore the significance of TGF-β1 in the development and progression of AD, particularly in relation to processes such as inflammation and tissue remodeling. Furthermore, these findings shed light on potential associations between vitamin D levels and the severity of the disease. Further investigation within this domain may yield the development of precise therapeutic interventions focused on the modulation of TGF-β1 signaling, thereby enhancing the overall efficacy of AD management in forthcoming times.
Supplementary Material
Glossary
Abbreviations
- AD
atopic dermatitis
- TGF-β1
transforming growth factor-β1
- SMAD3
small mothers against decapentaplegic homolog 3
- SPT
skin prick test
- SCORAD score
scoring atopic dermatitis
- LDS
Loeys–Dietz syndrome
Contributor Information
Tabasum Shafi, Department of Immunology and Molecular Medicine, SKIMS, Srinagar, India-, 190011.
Roohi Rasool Wani, Department of Immunology and Molecular Medicine, SKIMS, Srinagar, India-, 190011.
Showkat Hussain, Department of Immunology and Molecular Medicine, SKIMS, Srinagar, India-, 190011.
Imtiyaz A Bhat, Department of Immunology and Molecular Medicine, SKIMS, Srinagar, India-, 190011.
Rumana Makhdoomi, Department of Pathology, SKIMS, Srinagar, India-, 190011.
Sheikh Adil Bashir, Department of Plastic and Reconstructive Surgery, SKIMS, Srinagar, India-, 190011.
Iffat Hassan, Department of Dermatology, Venereology, and Leprosy, GMC, Srinagar, India-, 190010.
Zafar A Shah, Department of Immunology and Molecular Medicine, SKIMS, Srinagar, India-, 190011.
Funding
This work was supported by the Indian council of Medical Research (ICMR), New Delhi under the grant head 45/08/2022-IMM/BMS.
Conflict of Interests
The authors declare no conflict of interest.
Author Contributions
Tabasum Shafi: Conceptualization, Data curation, Methodology, Investigation, Visualization. Roohi Rasool: Project Administration, Funding acquisition, Resources. Showkat Hussain: Investigation, Visualization. Imtiyaz A Bhat: Formal analysis, Validation. Rumana Makhdoomi: Methodology and Supervision, Sheikh Adil Bashir: Resources, Iffat Hassan: Resources, Zafar A. Shah: Supervision.
Ethical Approval
The present study received approval from the Institutional ethics committee (IEC-SKIMS) via protocol ID #RP 62/2020, dated January 24, 2021.
Data Availability
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
References
- 1. Laughter MR, Maymone MBC, Mashayekhi S, Arents BWM, Karimkhani C, Langan SM, et al. The global burden of atopic dermatitis: lessons from the Global Burden of Disease Study 1990–2017. Br J Dermatol 2021, 184, 304–9. doi: 10.1111/bjd.19580 [DOI] [PubMed] [Google Scholar]
- 2. Ständer S. Atopic dermatitis. N Engl J Med 2021, 384, 1136–43. doi: 10.1056/NEJMra2023911 [DOI] [PubMed] [Google Scholar]
- 3. Langan SM, Irvine AD, Weidinger S.. Atopic dermatitis. Lancet 2020, 396, 345–60. doi: 10.1016/S0140-6736(20)31286-1 [DOI] [PubMed] [Google Scholar]
- 4. Silverberg JI. Comorbidities and the impact of atopic dermatitis. Ann Allergy Asthma Immunol 2019, 123, 144–51. doi: 10.1016/j.anai.2019.04.020 [DOI] [PubMed] [Google Scholar]
- 5. Narla S, Silverberg JI.. The role of environmental exposures in atopic dermatitis. Curr Allergy Asthma Rep 2020, 20, 74. doi: 10.1007/s11882-020-00971-z [DOI] [PubMed] [Google Scholar]
- 6. Patrick GJ, Archer NK, Miller LS.. Which way do we go? Complex interactions in atopic dermatitis pathogenesis. J Invest Dermatol 2021, 141, 274–84. doi: 10.1016/j.jid.2020.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Frischmeyer-Guerrerio PA, Guerrerio AL, Oswald G, Chichester K, Myers L, Halushka MK, et al. TGFβ receptor mutations impose a strong predisposition for human allergic disease. Sci Transl Med 2013, 5, 195ra94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nickel J, Ten Dijke P, Mueller TD.. TGF-β family co-receptor function and signaling. Acta Biochim Biophys Sin (Shanghai) 2018, 50, 12–36. doi: 10.1093/abbs/gmx126 [DOI] [PubMed] [Google Scholar]
- 9. Meng XM, Nikolic-Paterson DJ, Lan HY.. TGF-β: the master regulator of fibrosis. Nat Rev Nephrol 2016, 12, 325–38. doi: 10.1038/nrneph.2016.48 [DOI] [PubMed] [Google Scholar]
- 10. Weissler KA, Frischmeyer-Guerrerio PA.. Genetic evidence for the role of transforming growth factor-β in atopic phenotypes. Curr Opin Immunol 2019, 60, 54–62. doi: 10.1016/j.coi.2019.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Redington AE, Madden J, Frew AJ, Djukanovic R, Roche WR, Holgate ST, et al. Transforming growth factor-beta 1 in asthma measurement in bronchoalveolar lavage fluid. Am J Respir Crit Care Med 1997, 156, 642–7. doi: 10.1164/ajrccm.156.2.9605065 [DOI] [PubMed] [Google Scholar]
- 12. Vignola AM, Chanez P, Chiappara G, Merendino A, Pace E, Rizzo A, et al. Transforming growth factor-beta expression in mucosal biopsies in asthma and chronic bronchitis. Am J Respir Crit Care Med 1997, 156, 591–9. doi: 10.1164/ajrccm.156.2.9609066 [DOI] [PubMed] [Google Scholar]
- 13. Toda M, Leung DYM, Molet S, Boguniewicz M, Taha R, Christodoulopoulos P, et al. Polarized in vivo expression of IL-11 and IL-17 between acute and chronic skin lesions. J Allergy Clin Immunol 2003, 111, 875–81. doi: 10.1067/mai.2003.1414 [DOI] [PubMed] [Google Scholar]
- 14. Hata A, Chen YG.. TGF-β signaling from receptors to Smads. Cold Spring Harb Perspect Biol 2016, 8, a022061. doi: 10.1101/cshperspect.a022061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Massagué J, Seoane J, Wotton D.. Smad transcription factors. Genes Dev 2005, 19, 2783–810. doi: 10.1101/gad.1350705 [DOI] [PubMed] [Google Scholar]
- 16. Fischer AH, Jacobson KA, Rose J, Zeller R.. Hematoxylin and eosin staining of tissue and cell sections. CSH Protoc 2008, 2008, pdb.prot4986. doi: 10.1101/pdb.prot4986 [DOI] [PubMed] [Google Scholar]
- 17. Shafi T, Rasool R, Ayub S, Bhat IA, Shah IH, Hussain S, et al. Unveiling the TGF- β1 paradox: significant implication of TGF- β1 promoter variants and its mRNA and protein expression in atopic dermatitis. Mol Immunol 2023, 157, 214–24. doi: 10.1016/j.molimm.2023.04.006 [DOI] [PubMed] [Google Scholar]
- 18. Fedenko ES, Elisyutina OG, Filimonova TM, Boldyreva MN, Burmenskaya OV, Rebrova OY, et al. Cytokine gene expression in the skin and peripheral blood of atopic dermatitis patients and healthy individuals. Self/Nonself 2011,2,120-24. doi: 10.4161/self.2.2.16939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jeong CW, Ahn KS, Rho NK, Park YD, Lee DY, Lee JH, et al. Differential in vivo cytokine mRNA expression in lesional skin of intrinsic vs. extrinsic atopic dermatitis patients using semiquantitative RT-PCR. Clin Exp Allergy 2003,33:1717-24. doi: 10.1111/j.1365-2222.2003.01782.x [DOI] [PubMed] [Google Scholar]
- 20. Gambichler T, Tomi NS, Skrygan M, Altmeyer P, Kreuter A.. Alterations of TGF-β/Smad mRNA expression in atopic dermatitis following narrowband ultraviolet B phototherapy: Results of a pilot study. J Dermatol Sci 2006, 44, 56-8. doi: 10.1016/j.jdermsci.2006.06.004 [DOI] [PubMed] [Google Scholar]
- 21. Mesquita K de C, Igreja AC de SM, Costa IMC.. Atopic dermatitis and vitamin D: facts and controversies. An Bras Dermatol 2013, 88, 945–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zerr P, Vollath S, Palumbo-Zerr K, Tomcik M, Huang J, Distler A, et al. Vitamin D receptor regulates TGF-β signalling in systemic sclerosis. Ann Rheum Dis 2015, 74, e20. doi: 10.1136/annrheumdis-2013-204378 [DOI] [PubMed] [Google Scholar]
- 23. Alyoussef A. Blocking TGF-β type 1 receptor partially reversed skin tissue damage in experimentally induced atopic dermatitis in mice. Cytokine 2018, 106, 45–53. doi: 10.1016/j.cyto.2018.02.025 [DOI] [PubMed] [Google Scholar]
- 24. Enomoto H, Noguchi E, Iijima S, Takahashi T, Hayakawa K, Ito M, et al. Single nucleotide polymorphism-based genome-wide linkage analysis in Japanese atopic dermatitis families. BMC Dermatol 2007, 7, 5. doi: 10.1186/1471-5945-7-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Shafi T, Rasool R, Ayub S, Bhat IA, Gull A, Hussain S, et al. Analysis of intronic SNP (rs4147358) and expression of SMAD3 gene in atopic dermatitis: a case-control study. Immunobiology 2023, 228, 152390. doi: 10.1016/j.imbio.2023.152390 [DOI] [PubMed] [Google Scholar]
- 26. Anthoni M, Wang G, Deng C, Wolff HJ, Lauerma AI, Alenius HT.. Smad3 signal transducer regulates skin inflammation and specific IgE response in murine model of atopic dermatitis. J Invest Dermatol 2007, 127, 1923–9. doi: 10.1038/sj.jid.5700809 [DOI] [PubMed] [Google Scholar]
- 27. Otsuka K, Takeshita S, Enomoto H, Takahashi T, Hirota T, Tamari M, et al. SMAD3 as an atopic dermatitis susceptibility gene in the Japanese population. J Dermatol Sci 2009, 55, 200–2. doi: 10.1016/j.jdermsci.2009.06.007 [DOI] [PubMed] [Google Scholar]
- 28. Flanders KC, Sullivan CD, Fujii M, Sowers A, Anzano MA, Arabshahi A, et al. Mice Lacking Smad3 Are Protected Against Cutaneous Injury Induced by Ionizing Radiation. Am J Pathol 2002, 160, 1057–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Martinez GJ, Zhang Z, Chung Y, Reynolds JM, Lin X, Jetten AM, et al. Smad3 differentially regulates the induction of regulatory and inflammatory T cell differentiation. J Biol Chem 2009, 284, 35283–6. doi: 10.1074/jbc.C109.078238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Clayton SW, Ban GI, Liu C, Serra R.. Canonical and noncanonical TGF-β signaling regulate fibrous tissue differentiation in the axial skeleton. Sci Rep 2020, 10, 21364. doi: 10.1038/s41598-020-78206-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.