Abstract
Nonstructural protein 5B (NS5B) of bovine viral diarrhea virus (BVDV) contains sequence motifs that are predictive of an RNA-dependent RNA polymerase activity. We describe the expression and purification of the BVDV NS5B protein derived from an infectious cDNA clone of BVDV (NADL strain). BVDV NS5B protein was active in an in vitro RNA polymerase assay using homopolymeric RNA or BVDV minigenomic RNA templates. The major product was a covalently linked double-stranded molecule generated by a “copy-back” mechanism from the input template RNA. In addition, a nucleotide-nonspecific and template-independent terminal nucleotidyl transferase activity was observed with the BVDV NS5B preparation.
Bovine viral diarrhea virus (BVDV) has been identified as the causative agent of viral diarrhea-mucosal disease in cattle (reviewed in reference 1) and represents an economically important disease in cattle. BVDV, classical swine fever virus, and ovine border disease virus are members of the Pestivirus genus in the family Flaviviridae (12–14, 20), which also contains the genera Flavivirus and the hepatitis C virus (HCV) group. Like other pestiviruses, BVDV is a small enveloped, positive-stranded RNA virus whose genome consists of a nonsegmented single-stranded RNA molecule of approximately 12.5 kb. BVDV genomic RNA encodes a single open reading frame (ORF) of approximately 3,900 amino acids (4, 9, 13, 16). The polyprotein translated from the ORF is subsequently processed by virally encoded or cellular proteases into 11 or 12 individual proteins (5, 6, 13, 16, 19, 22). All viral structural proteins (autoprotease Npro; capsid protein C; and the glycoproteins Erns, E1, and E2) are contained in the amino-terminal third of the polyprotein (10, 17, 18), while the nonstructural (NS) proteins are located in the carboxyl-terminal two-thirds of the polyprotein (see Fig. 2A) (reviewed in references 6 and 9). Nonstructural protein 5B (NS5B), or p75, is located at the carboxyl terminus of the polyprotein and contains the canonical amino acid motif, Gly-Asp-Asp (GDD), present in all positive-strand viral RNA polymerases (5). However, no direct biochemical evidence has yet established that the BVDV NS5B indeed contains RNA-dependent RNA polymerase (RdRp) activity. To address this issue, we obtained a full-length genomic cDNA clone of a cytopathic BVDV strain (NADL strain), pVVNADL, that yields RNA transcripts which generate infectious viral particles upon transfection into BVDV-susceptible cell lines (21), clearly indicating that all the viral proteins encoded by this genomic cDNA clone are fully functional.
FIG. 2.
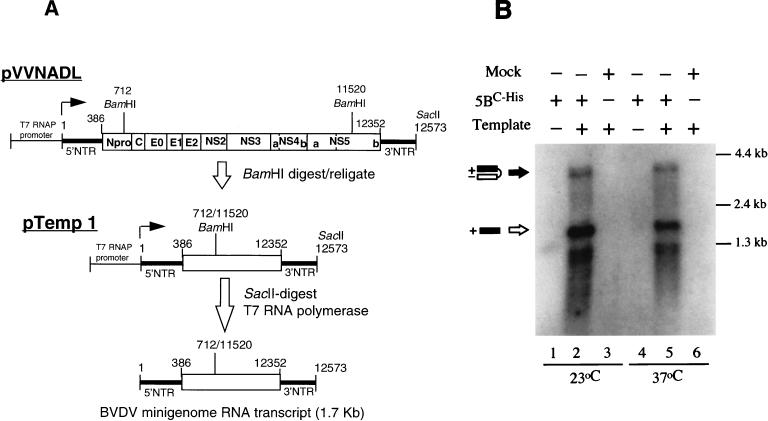
Detection of RdRp activity with purified BVDV NS5BC-His protein. (A) Schematic representation of the BVDV genome and minigenome RNA template. Individual BVDV proteins are shown as boxes; the T7 RNA polymerase (RNAP) promoter, the 5′ and 3′ NTR structures (thick lines) in pVVNADL, and the pTemp1 constructs are indicated with nucleotide positions relative to the BVDV genome (4). The solid arrows indicate the start of T7 RNA polymerase transcription at the first nucleotide of the BVDV genome (21). The structure of the pTemp1 minigenomic RNA template relative to the BVDV genomic RNA is shown with the nucleotide locations of the BamHI sites used to create the indicated internal deletion. The SacII restriction site at the end of the BVDV genome used to linearize template DNA before in vitro transcription with T7 RNA polymerase is also shown. (B) RdRp activity assay with purified NS5BC-His protein. Polymerase assays were performed at 23 (lanes 1 to 3) and 37°C (lanes 4 to 6) as indicated. Addition of pTemp1 RNA (Template), purified NS5BC-His protein (5BC-His), or negative control affinity eluates (Mock) made from wild-type AcNPV-infected Sf9 cells is indicated above the lanes. Products representing a putative monomer (open arrow) and a dimer hairpin (solid arrow) are indicated. Positions of RNA molecular size markers are indicated on the right.
As baculovirus has been employed previously to successfully express BVDV NS proteins (15), the coding region for NS5B was isolated from plasmid pVVNADL and cloned into baculovirus expression vector pVL1393. As the amino terminus of NS5B is generated normally by the proteolytic processing of the polyprotein precursor, the BVDV NS5B protein (NS5BC-His) was engineered with an initiating methionine codon at the amino terminus followed immediately by the entire NS5B coding sequence. Additionally, a hexahistidine affinity tag followed by a stop codon was added immediately after the last amino acid of NS5B. This design was based on previous work indicating that the addition of a hexahistidine tag at the C terminus of the HCV NS5B did not appear to adversely affect its RdRp activity (11). This vector, pVLBVDV-NS5BC-His, was used to generate recombinant baculovirus by standard methods.
Purification of the BVDV NS5B protein was achieved by using Ni-nitrilotriacetic acid affinity chromatography according to the protocol described by Lohmann et al. (11) with slight modifications. Briefly, 2.5 × 108 Sf9 cells infected with BVDV NS5B baculovirus were harvested 3 days postinfection, and the resulting cell pellet was resuspended in 5 ml of lysis buffer I (10 mM Tris-HCl [pH 7.5], 10 mM NaCl, 1.5 mM MgCl2, 10 mM 2-mercaptoethanol), incubated at 4°C for 30 min, and centrifuged for 10 min at 10,000 × g. The resultant supernatant was designated the S1 fraction. The pellet was resuspended in 5 ml of lysis buffer II (20 mM Tris-HCl [pH 7.5], 300 mM NaCl, 10 mM MgCl2, 0.5% Triton X-100, 20% glycerol, 10 mM 2-mercaptoethanol) and sonicated briefly. After a 10-min centrifugation at 10,000 × g, the supernatant (S2) was removed, and the pellet was resuspended in 5 ml of lysis buffer III (200 mM Tris-HCl [pH 7.5], 500 mM NaCl, 10 mM MgCl2, 2% Triton X-100, 10 mM imidazole, 50% glycerol). After further sonication and centrifugation steps, the supernatant (S3) was applied to a TALON column (Clontech). The bound NS5BC-His was eluted off the column with 400 μl of lysis buffer III containing 250 mM imidazole and 2 mM EGTA. Cell extracts from wild-type Autographa californica nuclear polyhedrosis virus (AcNPV)-infected Sf9 cells were similarly processed, and the final eluate was used in the assay as a negative control (mock).
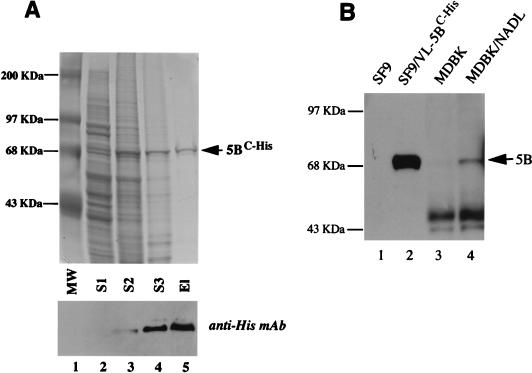
As shown in Fig. 1A, the expressed NS5BC-His protein, with a molecular mass between 70 and 75 kDa, was visible in the S2 fraction (lane 3) and became highly enriched in the S3 fraction (lane 4). In the final eluate, NS5BC-His protein was estimated to be approximately 90% pure (lane 5), although we cannot rule out the presence of contaminating proteins of identical size to BVDV NS5B. This material is referred to below as purified NS5BC-His. Western blot analysis demonstrated that the purified recombinant protein was immunoreactive with a mouse monoclonal antibody specific for a C-terminal hexahistidine tag (Invitrogen) (Fig. 1A, bottom, lanes 3 to 5).
FIG. 1.
Expression and purification of BVDV NS5BC-His. (A) Sf9 cells infected with NS5BC-His recombinant baculovirus were extracted with lysis buffers I, II, and III. A 10-μl aliquot of each supernatant fraction (S1, S2, and S3, lanes 2 to 4, respectively) and the final eluate (El) from the TALON column (lane 5) were analyzed by sodium dodecyl sulfate–8% polyacrylamide gel electrophoresis (SDS–8% PAGE) followed by Coomassie brilliant blue staining (top) or Western blotting (bottom) with a monoclonal antibody (MAb) directed against the hexahistidine affinity tag (Invitrogen). Lane 1 shows molecular weight (MW) markers (Gibco BRL). (B) Comparison of baculovirally expressed and native BVDV NS5B by Western blot analysis with BVDV NS5B-specific antiserum. Total protein samples from uninfected Sf9 cells (lane 1), BVDV NS5B recombinant baculovirus-infected Sf9 cells (lane 2), uninfected MDBK cells (lane 3), or BVDV(NADL)-infected MDBK cells (lane 4) were separated by SDS–8% PAGE, immunoblotted, and probed with a rabbit polyclonal antiserum raised against a peptide derived from BVDV NS5B.
To confirm the identity of the recombinant protein as BVDV NS5B, rabbit polyclonal antiserum was generated against a peptide derived from BVDV NS5B (amino acids 3389 to 3404 of the BVDV NADL strain) and used to compare the recombinant BVDV NS5B with native NS5B expressed during BVDV infection of Madin-Darby bovine kidney (MDBK) cells. Western blot analysis revealed that this antisera specifically reacted with a 75-kDa protein present in BVDV (NADL)-infected MBDK cells (Fig. 1B, lane 4) but absent in either uninfected MDBK cells (lane 3) or uninfected Sf9 cells (lane 1). This antisera also reacted with the recombinantly expressed protein, confirming its identity as BVDV NS5B (Fig. 1B, lane 2). Furthermore, the recombinantly expressed BVDV NS5B migrated with the same mobility as authentic BVDV NS5B produced during infection of MDBK cells with BVDV (NADL) (Fig. 1B, lanes 2 and 4).
RNA polymerase assays were performed to analyze whether the purified NS5BC-His protein had RdRp activity, as measured by incorporation of [α-32P]-labeled ribonucleotides with an RNA template. A BVDV “minigenome,” which contained both the 5′ and 3′ nontranslated regions (NTRs) of BVDV genomic RNA as well as a portion of the coding region, was used as the RNA template in the assay. This BVDV minigenome construct, pTemp1, was made by digesting plasmid pVVNADL (21) with BamHI followed by religation (Fig. 2A). We chose this template based on the assumption that the 5′ and 3′ NTRs would contain the cis-acting signals necessary for the initiation of minus-strand RNA synthesis. The highly ordered secondary structure at the 3′ NTR region (9) could be used for “copy-back” (cis) priming as in the case of HCV NS5B (3, 7, 11). The BVDV minigenomic RNA was transcribed in vitro by T7 RNA polymerase (Promega) by using SacII-linearized pTemp1 DNA as template and was gel purified (Fig. 2A). Linearization by SacII produces a 3′ end identical to that of infectious BVDV RNA transcripts (20). To assess RdRp activity, 0.5 μg of purified NS5BC-His protein was incubated in a 25-μl volume of standard reaction mixture (20 mM Tris-HCl [pH 7.5], 5 mM MgCl2, 1 mM dithiothreitol, 25 mM KCl, 1 mM EDTA, 40 U of RNasin (Promega), 50 μg of actinomycin D per ml, 10 μCi of [α-32P]ATP [Amersham], 500 μM [each] GTP, CTP, and UTP, and 20 μM ATP) in the presence or absence of pTemp1 RNA (0.3 μg per reaction) for 2 h. The reaction was carried out initially at both 23 and 37°C, and RNA was extracted following proteinase K treatment and resolved on a 1.2% agarose gel containing 2.2% formaldehyde. As shown in Fig. 2B, labeled products were observed in reactions containing both the exogenous RNA template and NS5BC-His protein (lanes 2 and 5). No products were observed in the absence of NS5BC-His protein, in the presence of mock-infected extracts (lanes 3 and 6), or in the absence of RNA template (lanes 1 and 4), indicating that the purification scheme had reduced potential endogenous RNA templates to undetectable levels. Interestingly, the reaction products were synthesized at comparable levels at both 23 and 37°C. This finding is somewhat different from the published HCV NS5B data in which HCV NS5B was found to work much more efficiently at the lower temperature (3, 11). As 37°C is closer to the natural replication temperature for BVDV, all following polymerase assays were performed at this temperature.
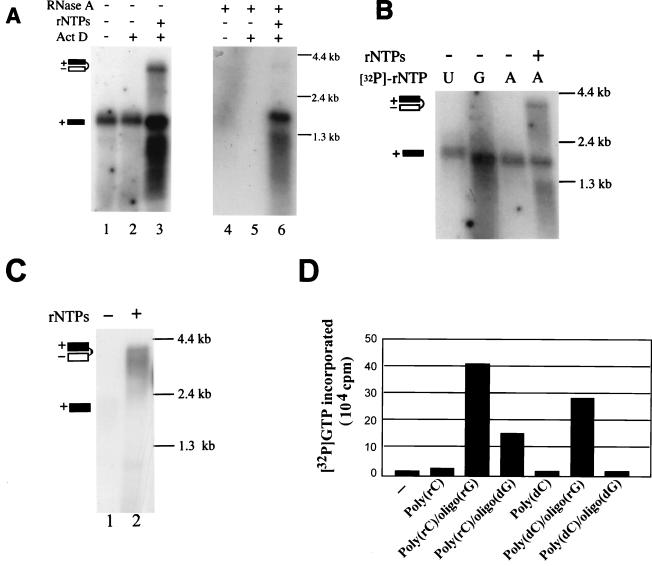
Three major product bands were observed in NS5BC-His polymerase reactions (Fig. 2B, lanes 2 and 5). One corresponded to the size of input template RNA (1.7 kb) while the other migrated much slower and was approximately twice the size of the input RNA. A third band migrating at 1.3 kb was observed only in reactions which contained NS5BC-His and all four ribonucleotide triphosphates (rNTPs). This band may represent prematurely terminated or fragmented RdRp product arising from a fragmented input template or from internal initiation by the RdRp. Together these results are similar to previously published results obtained with HCV NS5B in which a terminal nucleotidyl transferase activity (TNTase) was responsible for labeling the template-size product and an RdRp activity was responsible for synthesizing the dimer-sized product by a copy-back mechanism (3, 7, 11). To confirm these activities for BVDV NS5B, we performed polymerase assays in the absence or presence of either actinomycin D or unlabeled rNTPs. RNA extracted from the reactions was also treated with RNase A under conditions (40 μg of RNase A/ml for 30 min at room temperature in buffer containing 8% formamide, 10 mM HEPES [pH 7.5], 5 mM EDTA, and 350 mM NaCl) in which only single-stranded molecules would be degraded (2). As shown in Fig. 3A, in the absence of actinomycin D and in the presence of [α-32P]ATP alone, NS5BC-His was able to label the template RNA (lane 1). This activity was unaffected by the addition of actinomycin D (50 μg/ml) to the reaction mixture (lane 2). When treated with RNase A, the labeled products were no longer detected (lanes 4 and 5), indicating that they were either degraded or the end label had been removed. This result suggests that the NS5BC-His preparation contains a TNTase activity that incorporated labeled ATP into the template RNA. It is not clear whether this TNTase activity is intrinsic to NS5BC-His or is due to a cellular protein which copurified with the BVDV NS5BC-His, as suggested for HCV NS5B (11).
FIG. 3.
(A) Effects of RNase A and actinomycin D on NS5BC-His RdRp products. Addition of either actinomycin D (Act D), nonradioactive NTP mixture (500 μM [each] CTP, GTP, and UTP and 20 μM ATP), or postreaction digestion by RNase A is indicated above the lanes. Products were extracted and separated on a formaldehyde-agarose gel as described in the text. Products representing a putative monomer and a dimer hairpin are indicated schematically. The positions of RNA molecular size markers are shown on the right. (B) TNTase activity of NS5BC-His. Polymerase reactions were performed using [α-32P]UTP, [α-32P]GTP, or [α-32P]ATP as indicated above the lanes. The absence (−) or presence (+) of the three remaining rNTPs in the reaction is also indicated above the lanes. Other details are as described for panel A. (C) Northern blot analysis of RdRp products. Polymerase reactions were performed using pTemp1 RNA in the presence (+) or absence (−) of all four unlabeled rNTPs. Reaction products were analyzed by Northern blot analysis using a [32P]-labeled riboprobe specific for the negative strand. Other details are as described for panel A. (D) NS5BC-His activity on nonviral templates. Homopolymeric templates [poly(rC) or poly(dC)] or annealed primer-template pairs [poly(rC)-oligo(rG), poly(rC)-oligo(dG), poly(dC)-oligo(rG), or poly(dC)-oligo(dG)] were used in standard RdRp reactions. Reaction products were quantified by scintillation counting of acid-insoluble radioactivity. Activity without a template (−) was used to determine background radioactivity.
When unlabeled GTP, UTP, and CTP were included in the reaction, a dimer-sized product and a 1.3-kb RNA band were observed in addition to the monomer-sized product (Fig. 3A, lane 3). RNase A treatment caused the disappearance of the dimer-sized band, the appearance of a band whose size is consistent with that of monomer, and left the 1.3 kb RNA intact (lane 6). This result suggests that the dimer-sized product was generated by a copy-back mechanism; the plus-strand template and minus-strand product formed a covalently linked double-stranded molecule joined by an RNase-sensitive, single-stranded loop. With RNase treatment, this double-stranded molecule was no longer covalently linked and thus was separated into single-stranded monomeric molecules under denaturing gel conditions. Direct analysis of the purified individual products is needed to confirm this. The resistance of the 1.3-kb RNA band to RNase A digestion indicates that it is double stranded in nature and may represent prematurely terminated polymerase products. Additional experiments are needed to fully characterize the reaction products.
To determine if the TNTase activity in the NS5BC-His preparation required a specific rNTP as substrate, suggestive of template specificity, NS5BC-His was incubated with one of three different [α-32P]-labeled NTPs (UTP, GTP, and ATP) along with pTemp1 RNA. As shown in Fig. 3B, all three labeled nucleotides were incorporated into the template RNA (lanes 1 to 3), indicating the presence of template-independent transferase activity. In addition, this TNTase activity was not specific for BVDV RNA, as it was able to label an RNA molecular weight ladder with similar efficiency (data not shown). Furthermore, we cannot distinguish between the possibilities of either a 3′ TNTase or a 5′ ligase activity. We failed to detect this TNTase activity in the affinity elute of the mock-infected Sf9 cells (Fig. 2B, lanes 3 and 6), suggesting that either this TNTase activity is intrinsic to BVDV NS5B or it originates from a tightly associated cellular factor. To confirm that the RNA products generated truly represented negative-strand synthesis, polymerase reactions were carried out in which only pTemp1 RNA template with or without all four unlabelled rNTPs was added. RNA was extracted from these reactions and analyzed by Northern blotting using a [32P]-labeled negative-strand-specific riboprobe corresponding to the 3′ NTR and carboxy-terminal region of NS5B, which had been generated by in vitro transcription. This probe failed to hybridize when only template RNA was added to the reaction (Fig. 3C, lane 1). However, this probe hybridized to RNA corresponding to dimer-sized products (∼3.4 kb) (Fig. 3C, lane 2), suggesting that the dimer-sized RNAs contain genuine negative-strand product. The probe also hybridized to RNAs somewhat smaller than full-length dimer product. These could represent either incomplete synthesis products or partially degraded dimer product.
Lastly, we tested the in vitro template specificity of BVDV NS5B. As shown above, BVDV NS5B was able to use a minigenomic RNA as template. To see if BVDV NS5B could utilize RNA or DNA homopolymeric templates, BVDV NS5BC-His incorporation activity was assayed with either poly(C) or poly(dC) templates with either oligo(rG) or oligo(dG) as a primer. Four hundred nanograms of either poly(C) or poly(dC) alone or with 4 pmol of either oligo(rG) or oligo(dG) primer was heated for 2 min at 95°C followed by a 5-min incubation at 37°C to allow annealing. Templates were then added to the standard RdRp reaction, and the mixture was incubated at 37°C for 2 h. Reactions were terminated by the addition of 100 μg of calf thymus DNA and 1 ml of a solution containing 10% trichloroacetic acid (TCA) with 5% tetrasodium pyrophosphate (PPi). After a 30-min incubation at 4°C, the samples were filtered through GF/C glass microfiber filters (Whatman) and washed extensively with a solution containing 5% TCA with 1% PPi, and the radioactivity retained on the filters was counted with a scintillation counter. As shown in Fig. 3D, NS5BC-His incorporation of radioactivity into the primer-RNA template pair poly(rC)-oligo(rG) was approximately 20-fold higher than that observed with either poly(C) template alone, poly(dC) template alone, or the poly(dC)-oligo(dG) primer-template pair, suggesting a ribonucleotide primer dependency for RdRp activity on homopolymeric templates and an inability to use a DNA-DNA primer-template pair. The BVDV NS5B was able to utilize either an oligodeoxyribonucleotide primer on an RNA template, as has been similarly described for HCV NS5B (3, 7, 11), or an oligoribonucleotide primer on a DNA template, indicating that at least one strand must be RNA in order to detect activity. Furthermore, the inability of the BVDV NS5BC-His to utilize a DNA template [poly(dC)] in the presence of a deoxyribonucleotide primer confirms its RNA polymerase activity as being dependent upon an RNA template.
In this study, we successfully expressed the BVDV NS5B protein derived from an infectious cDNA clone by using a recombinant baculovirus. The inclusion of a hexahistidine affinity tag at the carboxyl terminus of the expressed protein allowed for affinity chromatography purification to approximately 90% homogeneity. When the BVDV NS5B protein was tested in a standard polymerase assay (3, 7, 11) with a minigenomic BVDV RNA as template, two types of activity were associated with the protein. The RdRp activity resulted in the synthesis of a double-stranded covalently linked molecule from the input template RNA via a copy-back mechanism. TNTase activity was also identified in the purified BVDV NS5BC-His but not in similarly prepared extracts from wild-type AcNPV-infected Sf9 cells, suggesting that it may be intrinsic to BVDV NS5B. However, it is still possible that a cellular TNTase activity is tightly associated with BVDV NS5B and copurified in the final affinity elution. The BVDV NS5B protein displays many characteristics which are similar to those of the HCV NS5B protein (3, 7, 11).
Purification of active viral polymerase protein is the first step towards understanding and elucidating the mechanisms underlying BVDV RNA replication. Unlike poliovirus, an in vitro replication system for members of the Flaviviridae family is not yet available. Such a system would provide a useful means for identifying viral or cellular factors that are required for viral RNA replication; however, a large number of difficult issues will need to be addressed before this is feasible. As a complete replication cycle for a positive-strand RNA virus is defined by the generation of positive-sense progeny RNA molecules, the activity described here has clearly not reached such capability, presumably because essential viral and cellular factors are not present. Future work will focus on identifying those factors. Finally, unlike BVDV, a robust culture system for the propagation of HCV in culture is currently unavailable. However, because of its similarity to HCV, BVDV has the potential of being used as surrogate system for HCV antiviral studies, particularly for antiviral agents directed against components of the viral RNA replicase. The initial characterization of the BVDV NS5B RdRp activity described here would support this idea.
Acknowledgments
We thank Ruben O. Donis and Ventzislav B. Vassilev for their most generous gift of the infectious BVDV cDNA clone, pVVNADL. We thank Christine Dabrowski, Susan Dillon, Klaus Esser, Baohua Gu, and Robert Sarisky for critically reading the manuscript.
REFERENCES
- 1.Baker J C. Bovine viral diarrhea virus: a review. J Am Vet Med Assoc. 1987;11:1449–1458. [PubMed] [Google Scholar]
- 2.Bartenschlager R, Junker-Niepmann M, Schaller H. The P gene product of hepatitis B virus is required as a structural component for genomic RNA encapsidation. J Virol. 1990;64:5324–5332. doi: 10.1128/jvi.64.11.5324-5332.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Behrens S-E, Tomei L, De Francesco R. Identification and properties of the RNA-dependent RNA polymerase of hepatitis C virus. EMBO J. 1996;15:12–22. [PMC free article] [PubMed] [Google Scholar]
- 4.Collett M S, Larson R, Gold C, Strick D, Anderson D K, Purchio A F. Molecular cloning and nucleotide sequence of the pestivirus bovine viral diarrhea virus. Virology. 1988;165:191–199. doi: 10.1016/0042-6822(88)90672-1. [DOI] [PubMed] [Google Scholar]
- 5.Collett M S, Larson R, Belzer S, Retzel E. Proteins encoded by bovine viral diarrhea virus: the genome organization of a pestivirus. Virology. 1988;165:200–208. doi: 10.1016/0042-6822(88)90673-3. [DOI] [PubMed] [Google Scholar]
- 6.Collett M S. Molecular genetics of pestiviruses. Comp Immun Microbiol Infect Dis. 1992;15:145–154. doi: 10.1016/0147-9571(92)90087-8. [DOI] [PubMed] [Google Scholar]
- 7.De Francesco R, Behrens S-E, Tomei L, Altamura S, Jiricny J. RNA-dependent RNA polymerase of hepatitis C virus. Methods Enzymol. 1996;275:58–67. doi: 10.1016/s0076-6879(96)75006-1. [DOI] [PubMed] [Google Scholar]
- 8.Deng R, Brock K V. 5′ and 3′ untranslated regions of pestivirus genome: primary and secondary structure analysis. Nucleic Acids Res. 1993;21:1949–1957. doi: 10.1093/nar/21.8.1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Donis R O. Molecular biology of bovine viral diarrhea virus and its interactions with the host. Vet Clin N Am Food Anim Pract. 1995;11:393–423. doi: 10.1016/S0749-0720(15)30459-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elbers K, Tautz N, Becher P, Rumenapf T, Thiel H-J. Processing in the pestivirus E2-NS2 region: identification of the nonstructural proteins p7 and E2p7. J Virol. 1996;70:4131–4135. doi: 10.1128/jvi.70.6.4131-4135.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lohmann V, Korner F, Herian U, Bartenschlager R. Biochemical properties of hepatitis C virus NS5B RNA-dependent RNA polymerase and identification of amino acid sequence motif essential for enzymatic activity. J Virol. 1997;71:8416–8428. doi: 10.1128/jvi.71.11.8416-8428.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meyers G, Rumenapf T, Thiel H-J. Molecular cloning and nucleotide sequence of the genome of hog cholera virus. Virology. 1989;171:555–567. doi: 10.1016/0042-6822(89)90625-9. [DOI] [PubMed] [Google Scholar]
- 13.Meyers G, Thiel H-J. Molecular characterization of pestiviruses. Adv Virus Res. 1996;47:53–118. doi: 10.1016/s0065-3527(08)60734-4. [DOI] [PubMed] [Google Scholar]
- 14.Moennig V, Plagemann P G. The pestiviruses. Adv Virus Res. 1992;41:53–98. doi: 10.1016/s0065-3527(08)60035-4. [DOI] [PubMed] [Google Scholar]
- 15.Petric M, Yolken R H, Dubovi E J, Wiskerchen M A, Collett M S. Baculovirus expression of pestivirus non-structural proteins. J Gen Virol. 1992;73:1867–1871. doi: 10.1099/0022-1317-73-7-1867. [DOI] [PubMed] [Google Scholar]
- 16.Rice C M. Flaviviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Vol. 1. Philadelphia, Pa: Lippincott-Raven Press; 1996. pp. 931–959. [Google Scholar]
- 17.Rumenapf T, Unger G, Strauss J H, Thiel H-J. Processing of the envelope glycoproteins of pestiviruses. J Virol. 1993;67:3288–3295. doi: 10.1128/jvi.67.6.3288-3294.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stark R, Meyers G, Rumenapf T, Thiel H-J. Processing of pestivirus polyprotein: cleavage site between autoprotease and nucleocapsid protein of classical swine fever virus. J Virol. 1993;67:7088–7095. doi: 10.1128/jvi.67.12.7088-7095.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tautz N, Elbers K, Stoll D, Meyers G, Thiel H-J. Serine protease of pestiviruses: determination of cleavage sites. J Virol. 1997;71:5415–5422. doi: 10.1128/jvi.71.7.5415-5422.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thiel H J, Plagemann G W, Moennig V. Pestiviruses. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Vol. 1. Philadelphia, Pa: Lippincott-Raven Press; 1996. pp. 1059–1073. [Google Scholar]
- 21.Vassilev V B, Collett M S, Donis R O. Authentic and chimeric full-length genomic cDNA clones of bovine viral diarrhea virus that yield infectious transcripts. J Virol. 1997;71:471–478. doi: 10.1128/jvi.71.1.471-478.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu J, Mendez E, Caron P R, Lin C, Murcko M A, Collett M C, Rice C M. Bovine viral diarrhea virus: polyprotein cleavage sites, cofactor requirements, and molecular model of an enzyme essential for pestivirus replication. J Virol. 1997;71:5312–5322. doi: 10.1128/jvi.71.7.5312-5322.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]