Abstract
The human adenovirus 5 E1B 55-kDa protein is required for efficient nucleocytoplasmic transport of late viral mRNAs. This protein is shown to have RNA-binding activity which maps to a region of the protein with homology to a family of RNA-binding proteins and which has been shown previously to be essential for functionality of the protein in vivo.
The expression of human adenovirus (Ad) genes during lytic infection is carefully regulated from the transcription of the earliest mRNAs right through to the final formation of progeny virions. Regulation occurs at all levels of the gene expression pathway, but posttranscriptional regulation is of particular importance since most Ad primary transcripts are differentially spliced to produce an array of distinct mRNA molecules (reviewed in references 16 and 21). Products of early genes E1B and E4 have been ascribed roles in regulating Ad late protein synthesis by modulating mRNA transport. Mutant viruses lacking either the E1B 55-kilodalton (55K) or the E4 Orf6 protein are deficient in transport of their late mRNAs from the nucleus to the cytoplasm, resulting in poor late protein synthesis and virus production (2, 12, 28, 36). The phenotypes of E1B 55K-E4 Orf6 double mutant viruses (5), as well as biochemical evidence (31), demonstrate that the 55K and Orf6 proteins interact and that complex formation is vital for efficient late viral mRNA transport. Deletion of a third protein, E4 Orf3, further exacerbates this defect (4, 15), but the exact role that the Orf3 protein plays in mRNA transport is still unclear (20). In addition to their role in mRNA transport, both the E1B 55K and E4 Orf6 proteins promote cellular transformation by binding to and inactivating the tumor suppressor protein p53 (8, 39).
Not all Ad mRNAs require 55K/Orf6 for export. Early RNAs are independent of 55K/Orf6 function, and the level of dependence of the late RNAs varies considerably, correlating with the number of unused splice sites or other potential intron sequences which they retain (22). Eukaryotic cells are thought to possess mechanisms for retaining RNA within the nucleus until processing is complete; this may be achieved by association with proteins, such as hnRNP-C1, which carry specific nuclear retention sequences (26). It has been suggested that 55K/Orf6 may act to block nuclear retention of susceptible late viral mRNAs and to divert them into an export pathway (6, 22), fulfilling a role in Ad infection similar to that of the Rev and Rex proteins in the life cycles of human immunodeficiency virus and human T-cell lymphotropic virus, respectively (reviewed in reference 13). Some evidence that 55K/Orf6 functions similarly to human immunodeficiency virus Rev has emerged from studies of Ad variants deficient in E1B 55K but producing instead the Rev protein and containing a specific Rev-binding site (RRE) within their major late transcription unit. The Rev/RRE system rescued, albeit to a modest extent, the cytoplasmic accumulation of viral mRNA (37). More recently, it was demonstrated that the 55K/Orf6 complex could shuttle between the nucleus and the cytoplasm (7). The Orf6 protein also inhibited Rev-mediated export of RRE-containing RNA, which implies that the 55K/Orf6 complex utilizes at least part of the same nuclear export pathway used by the retrovirus protein.
Similarity between the mechanism of action of the Ad E1B 55K/E4 Orf6 complex and those of the retroviral Rev and Rex proteins would suggest that the Ad proteins might perform specific interactions with their target mRNAs. No specific RRE-like sequences have been identified in Ad mRNAs, and there is no sequence that is common to all 55K/Orf6-dependent mRNAs; however, it is possible that the complex interacts with RNA via some feature of the unused splice sites or intron sequences or via a secondary-structure element. To begin to test for such interactions, the E1B 55K protein was examined for RNA-binding activity by using a series of RNA probes.
Preparation and purification of proteins.
E1B 55K was expressed in Escherichia coli as a glutathione S-transferase (GST) fusion protein from a plasmid kindly donated by Thomas Dobner, University of Regensberg, Regensberg, Germany (30); GST was expressed from pGEX2T (Pharmacia). Since the fusion protein was found to be very unstable, a range of strains was tested to find conditions which minimized degradation. Experiments described here used the GST-E1B 55K (GST-E1B) fusion protein from either XL1-Blue, TOPP 3, TOPP 5, or TOPP 6 cells (Stratagene), although TOPP 3 produced the highest-quality protein. GST was expressed in BL21 (Novagen) or XL1-Blue. Protein expression was induced in 1.0-liter cultures grown at 37°C to an optical density at 600 nm of 0.5 by the addition of IPTG (isopropyl-β-d-thiogalactopyranoside) at a final concentration of 0.1 mM, and incubation was continued at 30°C for a further 1 to 2 h (GST-E1B) or 4 h (GST). Cells were washed in 50 ml of ice-cold phosphate-buffered saline (PBS; 140 mM NaCl, 2.7 mM KCl, 8.1 mM Na2HPO4, 1.5 mM KH2PO4 [pH 7.2]) containing inhibitors (PBS/I; PBS, 500 μM dithiothreitol, 500 μM phenylmethylsulfonyl fluoride, 10 μM leupeptin, 2 μM pepstatin A, 10 mM EDTA and then resuspended in 10 ml of PBS/I and snap-frozen at −70°C. Cells were lysed with a French press at 1,010 lb/in2 (American Instruments Company), and cellular debris was removed by centrifugation at 32,500 × g for 60 min at 4°C. The lysate was diluted to 100 ml in PBS–1% Triton X-100 and incubated with 2 ml of a 50% suspension of glutathione-Sepharose 4B beads (Pharmacia) for 1 h at room temperature. The beads were washed four times in PBS, and protein was eluted with 50 mM Tris (pH 8)–10 mM reduced glutathione. Protein concentrations in fractions were determined by using the Bio-Rad protein assay, and samples were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) with either Coomassie blue staining or Western blotting. Fractions containing the highest concentrations of protein (equivalent also to those with the most full-length protein) were pooled and stored at −70°C in 50-μl aliquots. Since the proportion of full-length protein in GST-E1B preparations was small, care was taken to analyze each preparation by SDS-PAGE and Coomassie blue staining and to use preparations with similar full-length protein contents for all experiments.
Selection and synthesis of RNA probes.
RNA probes P1 to P8 represent adenovirus type 5 (Ad5) genomic sequences (5′-3′) of positions 35617 to 35357 (P1), 35357 to 35617 (P2), 35617 to 35469 (P3), 35357 to 35464 (P4), 35356 to 35094 (P5), 34935 to 34635 (P6), 12039 to 12357 (P7), and 16369 to 16593 (P8). These were selected from Ad E4, L1, and L2 mRNAs that are known to be highly dependent on 55K/Orf6 for efficient transport in late Ad-infected cells (6, 22) and were designed to contain splice sites or intron sequences that might act as targets for the action of the 55K/Orf6 complex in vivo. E4 mRNA A, which relies on the E1B 55K protein for export (6), encodes the Orf1 protein and differs from the other late E4 RNAs in that the 5′ intron is retained as coding sequence and the splice donor site D1 and acceptor sites A1a and A1b, etc., remain unused; probes P1, P3, P5, and P6 together cover the majority of this region, while P2 and P4 are antisense transcripts. Probes P7 and P8 are sections of major late transcription unit mRNAs for L1 52/55K and L2 III/pVII, respectively, each containing an unused splice acceptor site and part of the sequence to either side. T7 or SP6 RNA polymerase (Gibco BRL) was used in accordance with the manufacturer’s instructions to transcribe linearized cloned Ad5 genomic DNA in pGEM vectors (Promega) in the presence of 4 mCi of [α-32P]CTP (Amersham; 800 Ci/mmol) per ml, 500 μM each unlabelled GTP, ATP, and UTP, and 100 μM CTP. Unincorporated nucleotides were removed by gel filtration, and radioactivity incorporation was quantified by acid precipitation and scintillation counting. The length and quality of probes were verified by electrophoresis on 5% acrylamide–urea gels.
The GST-E1B fusion protein has RNA-binding activity.
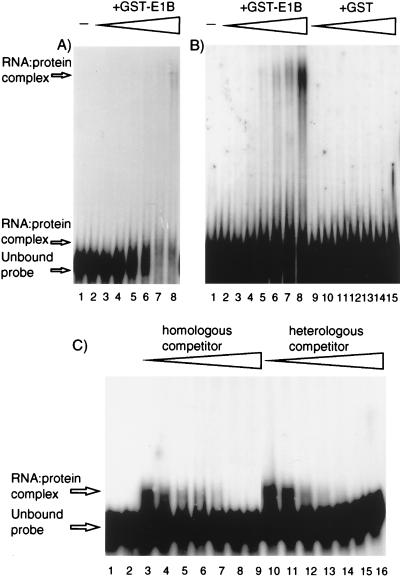
The RNA-binding activity of GST-E1B fusion protein was initially detected by gel mobility shift assays with probe P1. RNA was denatured by heating to 85°C for 10 min and then cooled on ice for 15 to 30 min before being mixed with purified GST-E1B or GST in binding buffer (10 mM HEPES [pH 7.6], 20 mM NaCl, 150 mM KCl, 0.5 mM EGTA, 10% glycerol, 1 mM dithiothreitol, 7.5 μg of bovine serum albumin per ml, 100 U of human placental RNase inhibitor per ml [adapted from reference 23]). Typically, 200 ng to 8 μg of protein was incubated with 2.5 fmol of 32P-RNA in a reaction volume of 25 μl for approximately 20 min on ice and protein-RNA complexes were resolved by electrophoresis through 4% polyacrylamide gels (acrylamide-to-bisacrylamide ratio, 79:1) in 0.5× Tris-borate-EDTA buffer at 4°C and visualized by autoradiography. GST-E1B caused a marked reduction in the level of unbound probe with increasing protein concentration, which was not observed for the equivalent GST samples, although protein-RNA complexes could not be visualized as specific shifted bands (data not shown). The possibility that the loss of the unshifted probe was due to RNA degradation was discounted by recovering RNA from binding reaction mixtures and analyzing it by denaturing gel electrophoresis. The presence of labelled probe retained in the wells with high inputs of GST-E1B suggested that complexes were being formed that were too large to enter the gel. Since GST is known to dimerize (17), protein-protein interactions via the GST domain probably contributed to this phenomenon. However, binding of multiple protein molecules to the RNA would also affect complex size. To minimize such effects and produce complexes better able to enter the gel, a shorter probe, P3, which consisted of the 5′-end 180 nucleotides (nt) of P1 (full length, 325 nt), was tested in similar assays with GST-E1B and GST. A short-exposure autoradiograph of the gel (Fig. 1A) showed the same reduction in the amount of unbound probe with increasing amounts of GST-E1B, as was seen for P1. In addition, small protein-RNA complexes were detected just above the unbound probe band. Upon longer exposure of the gel (Fig. 1B), these small complexes were obscured but larger GST-E1B-specific complexes were revealed migrating closer to the wells and, less clearly, as smears between these large complexes and the unbound probe. The various mobilities of the complexes observed might have been due to different fragments of GST-E1B since, despite optimization of expression conditions, GST-E1B preparations always contained mostly degraded protein (for example, see Fig. 4A). However, in comparisons of several independent preparations, the amount of full-length GST-E1B correlated positively with the observation of bands with decreased mobility of all types and not just with the observation of bands from the larger complexes (data not shown). An alternative explanation for the presence of different mobility-retarded bands is that they are due to protein binding to a different position(s) on the RNA to create complexes of differing shape and/or to the binding of multiple protein molecules to an RNA.
FIG. 1.
GST-E1B RNA binding and competition assays. (A) A short exposure of the gel shown in panel B, lanes 1 to 8. (B) Assays comprising 32P-probe RNA and various amounts of either GST-E1B or GST protein were analyzed on nondenaturing 4% polyacrylamide gels. The input protein is indicated above the lanes; −, no protein. Protein inputs were 0.1 (lanes 2 and 9), 0.2 (lanes 3 and 10), 0.5 (lanes 4 and 11), 1.0 (lanes 5 and 12), 1.5 (lanes 6 and 13), 2.0 (lanes 7 and 14), or 3.0 (lanes 8 and 15) μg. (C) Assays using 5 μg of GST-E1B protein and a constant amount of 32P-probe 3 together with either homologous (probe 3, unlabelled) or heterologous (probe 4, unlabelled) competitor RNA were analyzed on nondenaturing 4% polyacrylamide gels. Competitor-to-probe molar ratios were 0.0 (lanes 3 and 10), 1.0 (lanes 4 and 11), 2.0 (lanes 5 and 12), 5.0 (lanes 6 and 13), 10 (lanes 7 and 14), 100 (lanes 8 and 15), or 500 (lanes 9 and 16). Lanes 1 and 2 show probe 3 plus homologous and heterologous competitor RNAs, respectively, at molar ratios of 500 in the absence of GST-E1B protein.
FIG. 4.
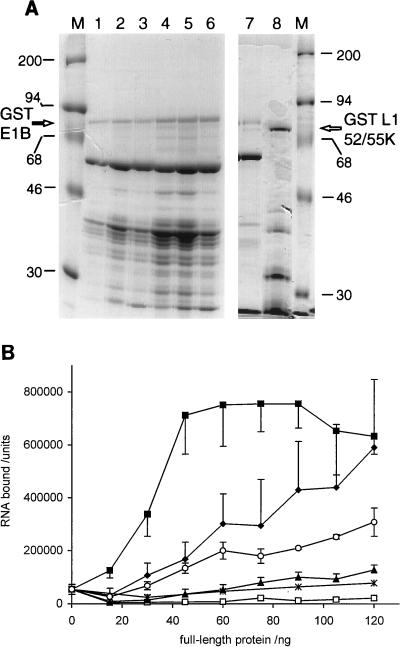
(A) GST-fusion proteins analyzed by SDS-PAGE and visualized by Coomassie blue staining. Lane 1, GST-E1B A284S (9.6 μg); lane 2, GST-E1B F285L-del287 (20.8 μg); lane 3, GST-E1B W289F (16.8 μg); lane 4, GST-E1B C288A (27.6 μg); lanes 5 to 7, independent wild-type GST-E1B preparations (34.4, 26.4, and 20.0 μg); lane 8, GST-L1 52/55K (20.0 μg). (B) Binding of wild-type and mutated forms of GST-E1B to RNA probe 3. Binding assay mixtures comprised various amounts of wild-type or mutated GST-E1B protein (panel A, lanes 1 to 6) or GST protein with 32P-labelled RNA probe 3. RNA-protein complexes were collected on nitrocellulose filters and detected by phosphorimaging. The amounts of radioactivity bound were quantified by using ImageQuant software and are indicated in arbitrary units as the mean of three determinations (two wild-type protein preparations were each analyzed in triplicate) ± standard deviations. Protein inputs for GST-E1B preparations are indicated as the amounts of full-length protein added. Actual protein inputs were significantly higher, due to the presence of degraded fragments (see panel A). To set the protein inputs for the GST control, the mean total GST-E1B (wild-type or mutated) protein input used to provide a given amount of full-length protein was calculated and this series of GST inputs was used for the experiment. Symbols: ⧫, wild-type GST-E1B; ✕⃒, GST-E1B A284S; ▴, GST-E1B F285L-del287; ■, GST-E1B W289F; ○, GST-E1B C288A; □, GST.
Since GST-E1B is a much larger protein than the GST control, it was possible that it might display different aggregation properties because of its size. An unrelated GST fusion protein of similar size to GST-E1B, carrying the L1 52/55K sequence of Ad5 (kindly provided by A. Arslanoglu), was therefore tested in a mobility shift assay with probes P3 and P4. No alteration to the mobility of either probe was observed in this experiment (data not shown), further confirming that the complexes formed by GST-E1B with RNA were specifically due to the presence of E1B protein sequences. The quality of the protein preparation used in this experiment is shown below (see Fig. 4A, lane 8) together with an example of a GST-E1B preparation which showed RNA-binding activity (see Fig. 4A, lane 7); these preparations show comparable levels of fragmentation. Thus, the RNA-binding activity of the GST-E1B protein preparation is unlikely to be a nonspecific property of partially folded protein fragments.
Sequence specificity of GST-E1B binding.
To determine whether the binding of GST-E1B to P1 and P3 represented a sequence-specific interaction, gel mobility shift assays were conducted with the antisense mRNA probe P2 (326 nt) and its shorter derivative, P4 (159 nt). Although transcribed from Ad DNA, these probes do not correspond to true Ad mRNA sequences. In analyses similar to those shown in Fig. 1A, GST-E1B bound to P2 and P4, causing probe shifts comparable to those of P1 and P3 at similar protein inputs while GST alone showed no binding (data not shown), suggesting that GST-E1B was binding RNA nonspecifically. In reciprocal competitor binding assays, unlabelled versions of P3 and P4 (C3 and C4), produced by using the Ampliscribe kit (Epicentre Technologies), were preincubated with GST-E1B for 20 min and then either the homologous or the heterologous probe was added to determine whether the protein displayed any binding preference for either of the two RNA sequences. As shown in Fig. 1C, C3 and C4 competed equally with P3 for GST-E1B binding, indicating that the RNA-binding activity of GST-E1B was not specific for the E4 mRNA sequence represented in P3. In the equivalent experiment, C3 and C4 also competed equally well for binding to P4 (data not shown).
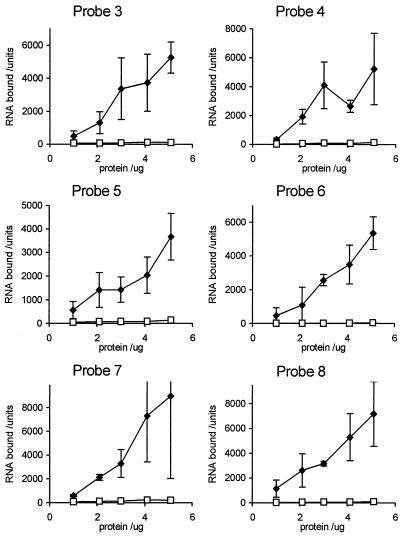
Although there was no specific affinity of GST-E1B for P3, it was possible that a high-affinity specific binding site for the protein existed elsewhere in E4 mRNA A or in another Ad transcript. To assess this, a filter binding assay which measured the total RNA bound by protein in each reaction was used to compare the binding levels of GST-E1B to a number of RNA probes within a single experiment. The binding conditions were similar to those used for mobility shift assays, but the incubation times were slightly increased. Nitrocellulose filters (Hybond-C; Amersham) were equilibrated in wash buffer (10 mM HEPES [pH 7.6], 20 mM NaCl, 150 mM KCl, 0.5 mM EGTA), and products of binding reactions were collected under gentle vacuum pressure by using a 96-well dot blot manifold. Filters were washed three times in wash buffer and air dried, and bound probe was visualized by phosphorimaging (Molecular Dynamics Corp.). Computer analysis of data was performed by using ImageQuant software. By using this technique, the binding of GST-E1B to probes P3 to P8 was tested (Fig. 2). The molar concentration of probe used in each case was the same. RNA-binding activity extended to all of the probes chosen, and the binding titration curves were similar for all cases. These data confirm the finding that GST-E1B shows non-sequence-specific binding to RNA of complex sequence.
FIG. 2.
GST-E1B protein binding to a range of RNA probes. Binding assays comprised various amounts of GST-E1B protein or GST protein and one of the 32P-labelled RNA probes 3 to 8. RNA-protein complexes were collected on nitrocellulose filters and detected by phosphorimaging. The amounts of radioactivity bound were quantified by using ImageQuant software and corrected for differences in length and percentage of C residues. Results are indicated in arbitrary units as the mean of three determinations ± standard deviations. Closed symbols, GST-E1B protein; open symbols, GST protein.
The E1B 55K protein has homology with a large number of RNA-binding proteins.
RNA-binding proteins have been classified by the nature of their RNA-binding domain(s). One large family of RNA-binding proteins with diverse origins and functions is defined by possession of the ribonucleoprotein (RNP) motif; family members can display various degrees of specificity in their RNA binding (14, 18). The RNP motif is composed of 80 to 100 residues and is characterized by two highly conserved sequences, RNP 1 and RNP 2, and a number of other, mostly hydrophobic, residues interspersed throughout the domain (3, 10). Folding of the RNP domain creates a β-sheet with the RNP 1 and RNP 2 consensus sequences lying on its two central strands (25, 38). This sheet forms a shallow platform for nonspecific RNA binding, whereas other parts of the domain, particularly the loops between β-strands, act to stabilize the interaction and are responsible for specific sequence recognition (reviewed in references 9 and 24).
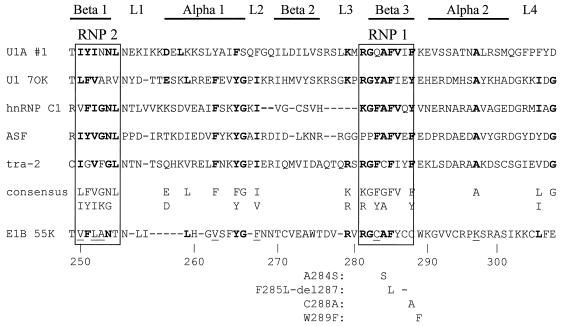
Analysis of the E1B 55K amino acid sequence revealed a region that had homology with the RNP consensus sequence. Stronger homology was evident when E1B 55K was compared with selected individual RNP family members (Fig. 3). Based on this alignment, a series of mutations was introduced into the potential RNP motif of E1B 55K. Three of the mutations were single amino acid substitutions, A284S and C288A within RNP 1 and W289F adjacent to RNP 1, and the fourth consisted of the substitution of one amino acid and the deletion of a second within the RNP 1 sequence, designated F285L-del287.
FIG. 3.
Protein sequence alignment between the Ad5 E1B 55K protein and selected members of the RNP family of proteins. The RNP 1 and RNP 2 consensus elements are boxed and the secondary-structure elements of the RNP domain are indicated above the alignment. The consensus sequence is taken from the work of Bandziulis et al. (3). Residues matching the consensus sequence are shown in boldface type; nonconsensus residues in consensus positions of E1B 55K which match those found at that position in at least one member of the RNP family are underlined. The mutations in E1B 55K described in this paper are indicated below the alignment at their respective positions by the amino acid substitution or deletion (−) in each case. Mutations were named by using the single-letter amino acid code and the residue positions in the 496-residue E1B 55K protein, the wild-type residue being listed first. The sequences for the following proteins are from the references indicated: U1A #1 (33), U1 70K (29), hnRNP C1 (34), ASF (11), tra-2 (1).
The RNP motif of GST-E1B is responsible for binding activity in vitro.
Mutated E1B sequences were expressed as GST fusions to assess the effects of the mutations on RNA-binding activity. As previously discussed in relation to Fig. 1, wild-type GST-E1B protein was very unstable when expressed in bacteria. Each mutated GST-E1B protein also showed great instability during purification; however, as shown in Fig. 4A, lanes 1 to 6, this pattern of degradation was both qualitatively and quantitatively similar for all four mutant protein preparations and the two wild-type protein preparations analyzed in parallel. Since, as already discussed, the RNA-binding activity of GST-E1B wild-type preparations correlated with the amount of full-length protein present, stained bands of full-length fusion protein were quantified by scanning densitometry in comparison with known amounts of a standard protein to provide estimates of the full-length protein concentration in each preparation. These data were then used to standardize protein inputs to binding assays.
Figure 4B shows the results of a filter binding assay comparing the abilities of wild-type GST-E1B and the four mutant proteins shown in Fig. 4A, lanes 1 to 6, to bind to probe P3. The two mutations affecting the central part of RNP 1 (A284S and F285L-del287) had the most severe effect on RNA binding activity, reducing binding to a level only marginally higher than the GST baseline binding. In contrast, mutating the last residue in the RNP 1 sequence (C288A) caused only a slight reduction in binding as compared with that of the wild-type protein and mutating the first residue outside the RNP 1 sequence (W289F) caused an increase in binding at low protein concentrations, while at high concentrations the binding activity was similar to that of wild-type GST-E1B. The fact that a single amino acid substitution in the E1B portion of GST-E1B could essentially destroy its RNA-binding activity suggested strongly that this protein, rather than any background contaminant present specifically in GST-E1B preparations, was responsible for the activity. However, since full-length protein comprised only a small part of the total protein input in these assays, it was important to consider whether the standardization of inputs based on concentrations of full-length protein might bias the results by ignoring the contribution of truncated proteins. An alternative representation of the data from Fig. 4B based on the total protein input to each assay did not change the relative positions of the curves significantly and did not affect any of the conclusions drawn (data not shown). Thus, these results demonstrate that the integrity of the RNP 1 consensus within the candidate RNP motif of the GST-E1B fusion protein is essential for this protein to display RNA-binding activity in vitro, although this does not mean that the domain necessarily adopts the RNP fold.
The region of the E1B 55K protein sequence implicated here in RNA binding, which includes residues 284, 285, and 287, has been shown previously to be crucial for normal virus growth (40). However, the lack of apparent specificity in this RNA-binding activity is at odds with the highly specific nature of the RNA export function which the protein provides in vivo. Many RNA-binding proteins, including members of the RNP family, display low-affinity nonspecific binding in addition to higher-affinity binding to a particular RNA sequence or secondary-structure element, so it is quite possible that such high-affinity, specific binding sites exist for E1B 55K in RNA sequences not tested in this study. Alternatively, it is possible that conditions within the infected cell nucleus may promote more specific interactions with RNA than those used in these experiments. First, the protein used in these assays, being expressed in bacteria, lacks the posttranslational modifications such as phosphorylation (32), which the E1B 55K protein normally sustains, and this may have affected its activity. However, the major sites of phosphorylation in E1B 55K map to its N- and C-terminal domains, away from the region implicated here in RNA binding (35). Second, the structure of RNA can be crucial for its correct recognition and might be an important factor in determining the specificity of interaction between E1B 55K and mRNA. This structure will in turn be affected by the association of RNA with protein to form RNP complexes, the natural form for all RNA in vivo and hence the true substrate for interaction with E1B 55K. Third, the E1B 55K protein is known to interact with both viral and cellular proteins, at least one of which, E4 Orf6, is also involved in the RNA export function, and some or all of these interactions may be required to impart specificity to RNA recognition. Alternative experimental approaches in which the E1B 55K protein is derived from eukaryotic expression systems, perhaps in complex with its in vivo partner, E4 Orf6, could be used to address these possibilities.
The secondary structure of RNA is difficult to control experimentally. For the binding assays described here, probe RNAs were heat denatured prior to incubation with the E1B 55K protein, and although cooling of the RNA before binding may have allowed some secondary structure to reform, this reformed structure cannot be assumed to be equivalent to the normal structure of the viral mRNAs from which the probes were derived. Furthermore, RNA inside the cell is always associated with protein; this association begins even before transcription is complete, acting to stabilize the nascent molecules and to direct processing and localization of the RNA within the nucleus (9). Such interactions alter RNA conformation and may affect binding of other proteins such as E1B 55K. Since all the probes used here were synthesized by in vitro transcription, these proteins were not present and this may have prevented the formation of the RNA secondary structure necessary for specific recognition by E1B 55K. In support of the relevance of RNA secondary structure to 55K protein-RNA interaction, we observed that the ability of tRNA to compete with labelled probe RNA for binding to E1B 55K varied between experiments (data not shown). Unlike the other probes used, tRNA has a defined and stable secondary structure to which it can refold after denaturation (19). Although all probes and competitor RNAs were heat denatured prior to their use in binding reactions, cooling of the tRNA before binding might have allowed the secondary structure to reform and this might have variably inhibited its binding to GST-E1B. While the secondary structure of tRNA may have inhibited binding to E1B 55K, the secondary structure of other types of RNA may of course promote binding.
The lack of specificity of E1B 55K RNA binding may indicate that the formation of the E1B 55K/E4 Orf6 complex is necessary for specific binding to occur. It is known that both proteins are necessary for efficient regulation of RNA transport and for the proteins to shuttle between nucleus and cytoplasm, so it follows that the Orf6 protein may be an important factor in E1B 55K-RNA interactions. Orf6 may be needed either to alter the conformation of E1B 55K to promote specific recognition of RNA or to make direct interactions with RNA itself. In preliminary experiments, Orf6 also displays RNA-binding activity (unpublished data). However, this could be related either to the RNA transport function of the protein (12) or to its role as a splicing factor (27). It will be important therefore to test whether formation of the 55K/Orf6 complex can cause selective targeting of mRNA in vitro that would correlate with the properties of these proteins in vivo. Finally, it is possible that other proteins, possibly host cell RNA-binding proteins or viral proteins such as the E4 Orf3 protein, might be important in generating the expected specific recognition of late viral RNA. The finding that E1B 55K has RNA-binding activity fits with the idea that it functions to facilitate RNA export by accompanying RNA out of the nucleus and suggests several lines of experiment which should lead to a full definition of the RNA-protein interactions involved in this mechanism.
Acknowledgments
We gratefully acknowledge the gift of a plasmid expressing the GST-E1B 55K fusion protein from Thomas Dobner, University of Regensberg, that was crucial to this work and for his advice on protein expression from this plasmid. We are also grateful to Alper Arslanoglu for the gift of negative-control GST L1 fusion protein and to Elizabeth Harfst for helpful discussions. The mutations in the E1B 55K protein tested here were originally isolated by Laurie Eden. J.J.H. was supported by a research studentship from the Medical Research Council.
REFERENCES
- 1.Amrein H, Gorman M, Nothinger R. The sex-determining gene tra2 of Drosophila encodes a putative RNA binding protein. Cell. 1988;55:1025–1035. doi: 10.1016/0092-8674(88)90247-4. [DOI] [PubMed] [Google Scholar]
- 2.Babiss L E, Ginsberg H S, Darnell J E. Adenovirus E1b proteins are required for accumulation of late viral mRNA and for effects on cellular mRNA translation and transport. Mol Cell Biol. 1985;5:2552–2558. doi: 10.1128/mcb.5.10.2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bandziulis R J, Swanson M S, Dreyfuss G. RNA-binding proteins as developmental regulators. Genes Dev. 1989;3:431–437. doi: 10.1101/gad.3.4.431. [DOI] [PubMed] [Google Scholar]
- 4.Bridge E, Ketner G. Redundant control of adenovirus late gene expression by early region 4. J Virol. 1989;63:631–638. doi: 10.1128/jvi.63.2.631-638.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cutt J R, Shenk T, Hearing P. Analysis of adenovirus early region 4-encoded polypeptides synthesized in productively infected cells. J Virol. 1987;61:543–552. doi: 10.1128/jvi.61.2.543-552.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dix I, Leppard K N. Regulated splicing of adenovirus type 5 E4 transcripts and regulated cytoplasmic accumulation of E4 mRNA. J Virol. 1993;67:3226–3231. doi: 10.1128/jvi.67.6.3226-3231.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dobbelstein M, Roth J, Kimberly W T, Levine A J, Shenk T. Nuclear export of the E1B 55-kDa and E4 34-kDa adenoviral oncoproteins mediated by a rev-like signal sequence. EMBO J. 1997;16:4276–4284. doi: 10.1093/emboj/16.14.4276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dobner T, Horikoshi N, Rubenwolf S, Shenk T. Blockage by adenovirus E4orf6 of transcriptional activation by the p53 tumor-suppressor. Science. 1996;272:1470–1473. doi: 10.1126/science.272.5267.1470. [DOI] [PubMed] [Google Scholar]
- 9.Dreyfuss G, Matunis M J, Pinol-Roma S, Burd C G. hnRNP proteins and the biogenesis of mRNA. Annu Rev Biochem. 1993;62:289–321. doi: 10.1146/annurev.bi.62.070193.001445. [DOI] [PubMed] [Google Scholar]
- 10.Dreyfuss G, Swanson M S, Pinol-Roma S. Heterogeneous nuclear ribonucleoprotein particles and the pathway of mRNA formation. Trends Biochem Sci. 1988;13:86–91. doi: 10.1016/0968-0004(88)90046-1. [DOI] [PubMed] [Google Scholar]
- 11.Ge H, Zuo P, Manley J L. Primary structure of the human splicing factor ASF reveals similarities with Drosophila regulators. Cell. 1991;66:373–382. doi: 10.1016/0092-8674(91)90626-a. [DOI] [PubMed] [Google Scholar]
- 12.Halbert D N, Cutt J R, Shenk T. Adenovirus early region 4 encodes functions required for efficient DNA replication, late gene expression, and host cell shutoff. J Virol. 1985;56:250–257. doi: 10.1128/jvi.56.1.250-257.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hammarskjold M L. Regulation of retroviral RNA export. Semin Cell Dev Biol. 1997;8:83–90. doi: 10.1006/scdb.1996.0127. [DOI] [PubMed] [Google Scholar]
- 14.Haynes S R. The RNP motif protein family. New Biol. 1992;4:421–429. [PubMed] [Google Scholar]
- 15.Huang M-M, Hearing P. Adenovirus early region 4 encodes two gene products with redundant effects in lytic infection. J Virol. 1989;63:2605–2615. doi: 10.1128/jvi.63.6.2605-2615.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Imperiale M J, Akusjarvi G, Leppard K N. Post-transcriptional control of adenovirus gene expression. Curr Top Microbiol Immunol. 1995;199(II):139–187. doi: 10.1007/978-3-642-79499-5_6. [DOI] [PubMed] [Google Scholar]
- 17.Ji X, Zhang P, Armstrong R N, Gilliland G L. The three-dimensional structure of a glutathione S-transferase from the Mu gene class. Structural analysis of the binary complex of isoenzyme 3-3 and glutathione at 2.2A resolution. Biochemistry. 1992;31:10169–10184. doi: 10.1021/bi00157a004. [DOI] [PubMed] [Google Scholar]
- 18.Kenan D J, Query C C, Keene J D. RNA recognition: towards identifying determinants of specificity. Trends Biochem Sci. 1991;16:214–220. doi: 10.1016/0968-0004(91)90088-d. [DOI] [PubMed] [Google Scholar]
- 19.Khorana H G. Transfer RNA: discovery, early work, and total synthesis of a tRNA gene. In: Soll D, RajBhandary U L, editors. tRNA: structure, biosynthesis, and function. Washington, D.C: ASM Press; 1995. pp. 5–16. [Google Scholar]
- 20.Leppard K N. E4 gene function in adenovirus, adenovirus vector and adeno-associated virus infections. J Gen Virol. 1997;78:2131–2138. doi: 10.1099/0022-1317-78-9-2131. [DOI] [PubMed] [Google Scholar]
- 21.Leppard K N. Regulated RNA processing and RNA transport during adenovirus infection. Semin Virol. 1998;8:301–307. [Google Scholar]
- 22.Leppard K N. Selective effects on adenovirus late gene expression of deleting the E1b 55K protein. J Gen Virol. 1993;74:575–582. doi: 10.1099/0022-1317-74-4-575. [DOI] [PubMed] [Google Scholar]
- 23.Malim M H, Tiley L S, McCarn D F, Rusche J R, Hauber J, Cullen B R. HIV-1 structural gene expression requires binding of the Rev trans-activator to its RNA target sequence. Cell. 1990;60:675–683. doi: 10.1016/0092-8674(90)90670-a. [DOI] [PubMed] [Google Scholar]
- 24.Moras D, Poterszman A. RNA-protein interactions: diverse modes of recognition. Curr Biol. 1995;5:249–251. doi: 10.1016/s0960-9822(95)00051-0. [DOI] [PubMed] [Google Scholar]
- 25.Nagai K, Oubridge C, Jessen T H, Li J, Evans P R. Crystal structure of the RNA-binding domain of the U1 small nuclear ribonucleoprotein A. Nature. 1990;348:515–520. doi: 10.1038/348515a0. [DOI] [PubMed] [Google Scholar]
- 26.Nakielny S, Dreyfuss G. The hnRNP C proteins contain a nuclear retention sequence that can override nuclear export signals. J Cell Biol. 1996;134:1365–1373. doi: 10.1083/jcb.134.6.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nordqvist K, Ohman K, Akusjarvi G. Human adenovirus encodes two proteins which have opposite effects on accumulation of alternatively spliced mRNAs. Mol Cell Biol. 1994;14:437–445. doi: 10.1128/mcb.14.1.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pilder S, Moore M, Logan J, Shenk T. The adenovirus E1b-55K transforming polypeptide modulates transport or cytoplasmic stabilization of viral and host-cell mRNAs. Mol Cell Biol. 1986;6:470–476. doi: 10.1128/mcb.6.2.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Query C C, Bentley R C, Keene J D. A common RNA recognition motif identified within a defined U1 RNA binding domain of the 70K U1 snRNP protein. Cell. 1989;57:89–101. doi: 10.1016/0092-8674(89)90175-x. [DOI] [PubMed] [Google Scholar]
- 30.Rubenwolf S, Schutt H, Nevels M, Wolf H, Dobner T. Structural analysis of the adenovirus type 5 E1B 55-kilodalton-E4orf6 protein complex. J Virol. 1997;71:1115–1123. doi: 10.1128/jvi.71.2.1115-1123.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sarnow P, Hearing P, Anderson C W, Halbert D N, Shenk T, Levine A J. Adenovirus early region 1B 58,000-dalton tumor antigen is physically associated with an early region 4 25,000-dalton protein in productively infected cells. J Virol. 1984;49:692–700. doi: 10.1128/jvi.49.3.692-700.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sarnow P, Sullivan C A, Levine A J. A monoclonal antibody detecting the Ad5 E1b 58K tumor antigen: characterization of the E1b 58K tumor antigen in adenovirus-infected and -transformed cells. Virology. 1982;120:510–517. doi: 10.1016/0042-6822(82)90054-x. [DOI] [PubMed] [Google Scholar]
- 33.Sillekens P T G, Habets W J, Beijer R P, van Venrooij W J. cDNA cloning of the human U1 snRNA-associated A protein: extensive homology between U1 and U2 snRNP-specific proteins. EMBO J. 1987;6:3841–3848. doi: 10.1002/j.1460-2075.1987.tb02721.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Swanson M S, Nakagawa T Y, LeVan K, Dreyfuss G. Primary structure of human nuclear ribonucleoprotein particle C proteins: conservation of sequence and domain structures in heterogeneous nuclear RNA, mRNA, and pre-rRNA-binding proteins. Mol Cell Biol. 1987;7:1731–1739. doi: 10.1128/mcb.7.5.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takayesu D, Teodoro J G, Whalen S G, Branton P E. Characterization of the 55K adenovirus type 5 E1B product and related proteins. J Gen Virol. 1994;75:789–798. doi: 10.1099/0022-1317-75-4-789. [DOI] [PubMed] [Google Scholar]
- 36.Weinberg D H, Ketner G. Adenoviral early region 4 is required for efficient viral DNA replication and for late gene expression. J Virol. 1986;57:833–838. doi: 10.1128/jvi.57.3.833-838.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Williams R D, Leppard K N. Human immunodeficiency virus type 1 Rev-dependent effects on the late gene expression of recombinant human adenovirus. Virus Genes. 1996;13:111–120. doi: 10.1007/BF00568904. [DOI] [PubMed] [Google Scholar]
- 38.Wittekind M, Gorlach M, Friedrichs M, Dreyfuss G, Mueller L. 1-H, 13-C and 15-N NMR assignments and global folding pattern of the RNA binding domain of the human hnRNP C proteins. Biochemistry. 1992;31:6254–6265. doi: 10.1021/bi00142a013. [DOI] [PubMed] [Google Scholar]
- 39.Yew P R, Burk A J. Inhibition of p53 transactivation required for transformation by adenovirus early 1B protein. Nature. 1992;357:82–85. doi: 10.1038/357082a0. [DOI] [PubMed] [Google Scholar]
- 40.Yew P R, Kao C C, Berk A J. Dissection of functional domains in the adenovirus 2 early 1b-55K polypeptide by suppressor linker insertional mutagenesis. Virology. 1990;179:795–805. doi: 10.1016/0042-6822(90)90147-j. [DOI] [PubMed] [Google Scholar]