Abstract
Objective
Perianal Crohn’s disease (pCD) occurs in up to 40% of patients with CD and is associated with poor quality of life, limited treatment responses and poorly understood aetiology. We performed a genetic association study comparing CD subjects with and without perianal disease and subsequently performed functional follow-up studies for a pCD associated SNP in Complement Factor B (CFB).
Design
Immunochip-based meta-analysis on 4056 pCD and 11 088 patients with CD from three independent cohorts was performed. Serological and clinical variables were analysed by regression analyses. Risk allele of rs4151651 was introduced into human CFB plasmid by site-directed mutagenesis. Binding of recombinant G252 or S252 CFB to C3b and its cleavage was determined in cell-free assays. Macrophage phagocytosis in presence of recombinant CFB or serum from CFB risk, or protective CD or healthy subjects was assessed by flow cytometry.
Results
Perianal complications were associated with colonic involvement, OmpC and ASCA serology, and serology quartile sum score. We identified a genetic association for pCD (rs4151651), a non-synonymous SNP (G252S) in CFB, in all three cohorts. Recombinant S252 CFB had reduced binding to C3b, its cleavage was impaired, and complement-driven phagocytosis and cytokine secretion were reduced compared with G252 CFB. Serine 252 generates a de novo glycosylation site in CFB. Serum from homozygous risk patients displayed significantly decreased macrophage phagocytosis compared with non-risk serum.
Conclusion
pCD-associated rs4151651 in CFB is a loss-of-function mutation that impairs its cleavage, activation of alternative complement pathway, and pathogen phagocytosis thus implicating the alternative complement pathway and CFB in pCD aetiology.
INTRODUCTION
The inflammatory bowel diseases (IBD), Crohn’s disease (CD) and ulcerative colitis (UC), are chronic idiopathic disorders causing inflammation of the gastrointestinal tract and are significant causes of morbidity. CD is a clinically heterogeneous disease defined in part by transmural inflammation, which can lead to fistula and abscess formation around the anus and other locations. Perianal fistula are reported to originate as an extension from deep penetrating ulcers of the rectum or an infection of anal glands leading to local sepsis and tract formation.1 Perianal CD (pCD) is, arguably, the most debilitating manifestation of CD associated with significant morbidity, poor response to current therapies, need for surgery (often recurrent), and is therefore an area of significant unmet medical need.2–4 A total of 17%–50% of patients with CD develop perianal fistula and about one third of these patients suffer from recurring fistula formation.5 The development of perianal fistulae has genetic, microbiological and immunological components.2 5–8 Over the past decade, genomewide association studies (GWAS) have implicated over 240 loci in IBD,9–11 yet there is paucity of data understanding the functional effects of these variants and the underlying biology.
In this study, we evaluated clinical and serological variables associated with perianal status in 15 144 CD subjects from three independent cohorts and performed a within-case meta-analysis comparing 4056 patients with CD with perianal involvement (pCD+) with 11088 patients with CD without perianal disease (pCD-) to investigate genetic associations with pCD. We identified a missense variant (rs4151651) in complement factor B (CFB) to be associated with pCD and performed functional studies to interrogate the consequences of this non-synonymous polymorphism.
METHODS
Study population
Subjects included in final analyses were of European ancestry from three independent cohorts as detailed in online supplemental table 1, online supplemental methods: (1) Cedars-Sinai Medical Center (CSMC) MIRIAD IBD Research Repository; (2) International IBD Genetics Consortium (IIBDGC)10 12 and (3) the Sinai-Helmsley Alliance for Research Excellence (SHARE) consortium.13 Cases were confirmed as CD in accordance with the National Institute of Diabetes and Digestive and Kidney Diseases IBDGC phenotyping manual.14 All patients provided written informed consent approved by the local institutional review board.
Clinical and serological phenotyping
Demographic and clinical data for CSMC MIRIAD subjects were collected routinely by attending physicians and by chart review. Disease location and behaviour were characterised according to the Montreal Classification.15 For CSMC and SHARE, pCD+ was defined as the presence of perianal abscesses, perianal fistulae or recto-vaginal fistulae. Haemorrhoids, skin tags and anal fissures were not considered as perianal manifestations for this study. The IIBDGC applied the IBD Montreal Classification system for diagnosis and characterising disease location and disease behaviour including perianal disease modifier.12 16 All cohorts follow the Montreal classification standards of accumulated perianal disease diagnosis over time.15 Sera from the CSMC subjects only were analysed as previously described (online supplemental methods).17
Genotyping and quality control (QC)
Genotyping for CSMC, IIBDGC and SHARE was performed using the Illumina Immunochip array.2 10 13 18 We performed sample and SNP quality control (QC) measures consistently across all three cohorts (PLINK V.1.9)19 (online supplemental methods). Pre-QC and post-QC sample and SNP numbers are shown in online supplemental table 1. Admixture and principal component (PC) analyses were utilised to confirm European ancestry (online supplemental methods).
Statistical analyses
Clinical, demographic and serological parameters
Clinical and demographic variables were analysed by regression analyses to evaluate association with pCD, with Bonferroni corrected significance at p<0.0018 and nominal significance at p<0.05. Serological data were available for 756 pCD+ and 1620 pCD− CSMC subjects and included categorical (seropositivity) or continuous (antibody level, online supplemental methods) variables and were analysed by regression, with adjustment for disease behaviour, location and duration. Serology Quartile Sum Score (QSS) was calculated as previously described20 (online supplemental methods) and regression analyses with adjustment for disease behaviour, location and duration on 1139 subjects with available QSS was performed to identify association with perianal prevalence. All analyses were performed in R.21
Genetic analysis
Within-case association analyses were performed for CSMC, IIBDGC and SHARE using logistic regression correcting for population substructure with 5 PCs (PLINK V.1.9) and meta-analysis using METAL22 was performed for a total sample size of 4056 pCD+and 11 088 pCD-. Test statistics showed negligible genomic inflation (online supplemental methods). Gene-based analysis was performed for genomic regions with ≥3 SNPs associated at Pmeta<0.001 (VEGAS2).23 Plots were generated using R.21 Variant annotation was performed as described in online supplemental methods.
Gene enrichment analyses and prediction of post-translational modifications
Gene enrichment analyses uses lists of genes generated from association studies as input for computing enrichment with existing lists created from prior knowledge organised into gene-set libraries.24 We utilised the Genomic Regions Enrichment of Annotations Tool25 to annotate SNPs pmeta≤0.01 to nearby genes. This gene list was then evaluated in Enrichr, a gene set enrichment analysis tool, to identify pathways associated with pCD.24 Analyses were performed with and without inclusion of the MHC region. We queried AWESOME database to determine any post-translational modification (PTM) predictions associated with SNP of interest.26
Detailed methodologies of all in vitro experiments are described in online supplemental methods.
RESULTS
Demographic and clinical characteristics of study cohorts
CSMC, IIBDGC and SHARE had a total of 2315, 10 738 and 2091 subjects with CD, respectively. Perianal involvement across these cohorts ranged from ~17% to 31% of the study population. Most subjects were European ancestry and non-Jewish, with the exception of CSMC with 47% Jewish participants. Approximately 80% of the IIBDGC and SHARE participants had age at diagnosis ≥17 years, while CSMC had a larger percentage (35.7%) of paediatric onset patients. CSMC also had the lowest percentage of patients with non-stricturing, non-penetrating disease behaviour. All baseline demographic and clinical characteristics for study populations are shown in table 1.
Table 1.
Baseline demographic and clinical characteristics of all independent cohorts
| Clinical variable | N (%)- CSMC Total N=2315 | N (%)- IIBDGC Total N=10 738 | N (%)- SHARE Total N=2091 |
|---|---|---|---|
|
| |||
| Gender | |||
|
| |||
| Female | 1077 (46.5) | 5916 (55.1) | 1166 (55.8) |
|
| |||
| Ancestry | |||
|
| |||
| European | 2208 (95.4) | 10248 (95.44) | 2019 (96.6) |
| African American | 4 (0.2) | 2 (0.02) | 3 (0.1) |
| Asian | 21 (0.9) | 11 (0.10) | 25 (1.2) |
| Hispanic | 16 (0.7) | – | – |
| Middle Eastern | 18 (0.8) | 477 (4.44) | – |
| Missing and other | 48 (2.0) | – | 44 (2.1) |
|
| |||
| Ethnicity | |||
|
| |||
| Jewish | 1087 (47.0) | 283 (2.6) | 157 (7.5) |
| Non-Jewish | 1228 (53.0) | 7779 (72.4) | 1934 (92.5) |
| Unknown | – | 136 (1.3) | – |
| Missing | – | 2540 (23.7) | – |
|
| |||
| Median disease duration in years (range) | 15 (2.0–68.0) | – | – |
|
| |||
| Age at diagnosis | |||
|
| |||
| A1 (<17 years) | 829 (35.8) | 1737 (16.2) | 431 (20.6) |
| A2 (17–40 years) | 1179 (51.0) | 6570 (61.2) | 1309 (62.6) |
| A3 (>40 years) | 276 (11.9) | 1940 (18.1) | 351 (16.8) |
| Missing | 31 (1.3) | 491 (4.5) | – |
|
| |||
| Disease location | |||
|
| |||
| L1 (Ileal) | 368 (15.9) | 3348 (31.2) | 533 (25.5) |
| L2 (Colonic) | 394 (17.0) | 2567 (23.9) | 422 (20.2) |
| L3 (Ileocolonic) | 1515 (65.4) | 4359 (40.6) | 1129 (54.0) |
| Missing | 38 (1.6) | 464 (4.3) | 7 (0.3) |
| L4 (upper GI) | 390 (16.8) | 966 (9.0) | 104 (5.0) |
| Missing (L4) | 29 (1.3) | 2111 (19.7) | – |
|
| |||
| Disease behaviour | |||
|
| |||
| B1 (non- stricturing/ non-penetrating) B2 (stricturing) B3 (penetrating) Missing |
995 (43.0) | 5160 (48.1) | 1022 (48.9) |
| 577 (24.9) | 2653 (24.7) | 592 (28.3) | |
| 703 (30.4) | 2303 (21.4) | 477 (22.8) | |
| 40 (1.7) | 622 (5.8) | -- | |
|
| |||
| Perianal CD | 719 (31.1) | 2974 (27.7) | 363 (17.4) |
|
| |||
| Median time to first fibrotic surgery in years (range) | 8.07 (0.01–59.1) | – | – |
|
| |||
| Family history of IBD | |||
|
| |||
| Yes | 564 (24.4) | 2415 (22.5) | 324 (15.5) |
| Missing | 160 (6.9) | 1004 (9.3) | 152 (7.3) |
|
| |||
| Cigarette smoking (at diagnosis) | |||
|
| |||
| Current smoker | 455 (19.6) | 2658 (24.8) | 213 (10.2) |
| Former smoker | 36 (1.6) | 1326 (12.3) | 529 (25.3) |
| Never | 1432 (61.9) | 4788 (44.6) | 1338 (64.0) |
| Missing | 392 (16.9) | 1966 (18.3) | 11 (0.5) |
CD, Crohn’s disease; CSMC, Cedars- Sinai Medical Center; GI, gastrointestinal; IBD, inflammatory bowel disease; IIBDGC, International IBD Genetics Consortium; SHARE, Sinai- Helmsley Alliance for Research Excellence.
Clinical associations with pCD
Any CD colonic involvement was strongly associated with an increased risk for pCD across all cohorts, in keeping with perianal fistula arising at sites of inflammation in the colon (table 2).1 Compared with stricturing disease behaviour (B2), patients with internal penetrating (B3) disease were at significant increased risk of developing perianal complications across all cohorts. Similarly, patients with either stricturing or internal penetrating disease were at significant increased risk of pCD compared with those with inflammatory-only (B1) disease (table 2). Perianal disease was associated with lower age at diagnosis across all cohorts (table 2). Specifically, we observed a protective effect for developing perianal complications in subjects who developed CD older than 40 years of age (A3) when compared with <17 (A1) or 17–40 years (A2) (table 2). A family history of IBD was protective in SHARE yet a risk factor for pCD in IIBDGC (table 2). Cigarette smoking at the time of diagnosis was associated with increased risk of perianal complications in SHARE but was observed to be protective in CSMC (table 2). No association was observed in IIBDGC.
Table 2.
Associations between variables and presence of perianal Crohn’s disease in CSMC, IIBDGC and SHARE*
| CSMC |
IIBDGC |
SHARE |
||||
|---|---|---|---|---|---|---|
| Clinical variable | P value | OR (95% CI) | P value | OR (95% CI) | P value | OR (95% CI) |
|
| ||||||
| Gender (female) | 1.63E-01 | 0.88 (0.74 to 1.05) | 3.95E- 03 | 0.88 (0.81 to 0.96) | 5.29E-01 | 0.93 (0.74 to 1.17) |
|
| ||||||
| Ethnicity (Jewish) | 3.37E- 02 | 0.83 (0.69 to 0.99) | 1.81E-01 | 0.83 (0.63 to 1.09) | 1.72E-01 | 0.72 (0.45 to 1.15) |
|
| ||||||
| Age at diagnosis | ||||||
|
| ||||||
| A2 vs A1 | 7.46E-01 | 0.97 (0.80 to 1.17) | 5.99E-02 | 0.90 (0.80 to 1.00) | 2.52E-01 | 0.85 (0.65 to 1.12) |
| A3 vs A2 | 3.73E- 04 | 0.57 (0.42 to 0.78) | <2.00E- 16 | 0.49 (0.43 to 0.56) | 2.11E- 02 | 0.67 (0.47 to 0.94) |
| A3 vs A1 | 2.71E- 04 | 0.55 (0.40 to 0.76) | <2.00E- 16 | 0.44 (0.38 to 0.51) | 4.71E- 03 | 0.57 (0.38 to 0.84) |
| A2A3 vs A1 | 1.68E-01 | 0.88 (0.73 to 1.06) | 9.69E- 06 | 0.78 (0.70 to 0.87) | 8.27E-02 | 0.79 (0.60 to 1.03) |
|
| ||||||
| Age at diagnosis† | 4.86E- 04 | 0.99 (0.98 to 0.99) | <2.00E- 16 | 0.98 (0.975 to 0.98) | 1.57E- 03 | 0.99 (0.98 to 0.99) |
|
| ||||||
| Disease location | ||||||
|
| ||||||
| Ileal vs Colonic (L1 vs L2) | 7.32E- 07 | 0.43 (0.30 to 0.60) | <2.00E- 16 | 0.57 (0.51 to 0.65) | 3.41E- 12 | 0.26 (0.18 to 0.38) |
| Upper gastrointestinal (L4) | 4.69E-01 | 0.91 (0.72 to 1.16) | 6.35E-02 | 1.15 (0.99 to 1.33) | 4.35E-01 | 1.22 (0.74 to 1.99) |
| Any colonic involvement (L2L3 vs L1) | 4.63E- 08 | 2.19 (1.65 to 2.90) | <2.00E- 16 | 1.90 (1.72 to 2.09) | 2.50E- 10 | 2.95 (2.11 to 4.13) |
| Any SB Involvement (L1L3 vs L2) | 6.96E-02 | 0.81 (0.64 to 1.02) | 5.74E- 03 | 0.87 (0.79 to 0.96) | 1.61E- 06 | 0.53 (0.41 to 0.69) |
|
| ||||||
| Disease behaviour | ||||||
|
| ||||||
| B2 vs B1 | 3.37E- 02 | 1.28 (1.02 to 1.61) | 4.98E- 04 | 1.21 (1.09 to 1.35) | 5.86E-01 | 0.92 (0.69 to 1.23) |
| B3 vs B1 | 1.42E- 11 | 2.05 (1.67 to 2.53) | <2.00E- 16 | 2.26 (2.03 to 2.51) | 4.22E- 06 | 1.87 (1.43 to 2.45) |
| B2B3 vs B1 | 4.14E- 08 | 1.67 (1.39 to 2.01) | <2.00E- 16 | 1.64 (1.51 to 1.79) | 1.86E- 02 | 1.32 (1.05 to 1.65) |
| B3 vs B2 | 8.16E- 05 | 1.60 (1.27 to 2.02) | <2.00E- 16 | 1.87 (1.66 to 2.30) | 7.23E- 06 | 2.03 (1.49 to 2.76) |
|
| ||||||
| Family history of IBD | 9.25E-01 | 1.01 (0.82 to 1.25) | 2.22E- 04 | 1.21 (1.09 to 1.34) | 1.39E- 02 | 0.65 (0.46 to 0.92) |
|
| ||||||
| Cigarette smoking (at diagnosis) | ||||||
|
| ||||||
| Current versus never | 1.40E- 02 | 0.75 (0.59 to 0.94) | 3.69E-01 | 1.05 (0.94 to 1.17) | 1.31E- 02 | 1.55 (1.10 to 2.19) |
| Current versus never/former | 1.85E- 02 | 0.76 (0.60 to 0.95) | 7.50E-02 | 1.10 (0.99 to 1.21) | 7.79E- 03 | 1.58 (1.13 to 2.22) |
|
| ||||||
| Extraintestinal manifestations | ||||||
|
| ||||||
| Ankylosing spondylitis | 8.58E-01 | 1.03 (0.75 to 1.42) | 4.77E-01 | 0.94 (0.79 to 1.12) | 8.92E-01 | 1.14 (0.17 to 7.40) |
| Associated Arthalgias | 1.38E-01 | 1.22 (0.94 to 1.60) | – | – | 4.97E-01 | 0.87 (0.59 to 1.29) |
| Psoriasis | 6.98E-01 | 1.08 (0.74 to 1.56) | – | – | 2.94E-01 | 0.76 (0.31 to 1.42) |
| Oral ulcers | 9.72E-01 | 0.99 (0.46 to 2.11) | – | – | – | – |
| Erythema Nodosum | 4.42E- 04 | 2.08 (1.38 to 3.12) | – | – | 2.68E-01 | 1.61 (0.69 to 3.72) |
| Pyoderma gangrenosum | 8.80E-01 | 1.05 (0.55 to 2.00) | – | – | 7.72E-01 | 1.17 (0.41 to 3.36) |
| Uveitis- iritis | 5.03E-01 | 0.82 (0.47 to 1.46) | – | – | 1.64E-01 | 0.49 (0.18 to 1.34) |
| Primary sclerosing cholangitis | 4.78E-01 | 0.83 (0.50 to 1.38) | 5.84E-01 | 0.88 (0.55 to 1.41) | 1.64E-01 | 0.36 (0.08 to 1.52) |
| Deep vein thrombosis | 2.05E- 02 | 1.94 (1.11 to 3.40) | – | – | – | – |
| Kidney stone | 1.57E-01 | 1.34 (0.89 to 2.00) | – | – | – | – |
Variables associated p<0.05 are shown in bold; Bonferroni significance threshold p<0.0018. †Age at diagnosis evaluated as continuous variable.
CI, Confidence Interval; CSMC, Cedars- Sinai Medical Center; IBD, inflammatory bowel disease; IIBDGC, International IBD Genetics Consortium; OR, Odds Ratio; SHARE, Sinai- Helmsley Alliance for Research Excellence.
Genetic association with pCD
We performed a within-case meta-analysis comparing European ancestry patients with CD from the three independent cohorts with a total of 4056 pCD+ and 11 088 pCD-. No association achieved genome-wide threshold of significance. We identified putative novel associations with pCD for variants in loci previously unreported in IBD genetic association studies, including GABRB1, LRRC7, MAST4 and SDK1 (table 3; online supplemental table 2; meta-p<0.0005). We also observed associations with pCD and variants in known IBD susceptibility loci such as MHC (HLA-DRA, CFB and BTNL2), TGFBR3 and KIAA1109.9 Our most significant association was an intronic variant in HLA-DRA (p=4.68E-07, OR=1.24). Our second strongest association was with an exonic, non-synonymous mutation (A>G) in the CFB gene (rs4151651, p=9.35E-06 , OR=1.32), also located within the MHC (table 3; online supplemental table 2). We observe nominal associations in KIAA1109 which is located adjacent to known IBD locus containing IL2 and IL21.9 10 We performed a gene-based analyses on all variants with pmeta<0.001 and observed significance for CFB (nSNPs=33, p=2.01E-04), LRRC7 (nSNPs=14, p=1.13E-03), PITPNM2 (nSNPs=128, p=2.48E-03), MPHOSPH9 (nSNPs=149, p=3.58E-03) and GABRB1 (nSNPs=101, p=3.81E-03). The pathway enrichment analyses for genes associated with genomic regions annotated to SNPs pmeta≤0.01 demonstrated significant association with Systemic Lupus Erythematosus, IBD and activation of C3 and C5. When excluding MHC from enrichment analysis, we observed associations with JAK-ST AT, NF-KB and IL-2 signalling pathways (table 4).
Table 3.
Top genetic associations with pCD
| Meta- analysis | ||||||||
|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||
| CHR | SNP | Gene | Location | Novel (Y/N) | Allele | P value | OR | 95% CI |
|
| ||||||||
| 6 | rs17496549 | HLA-DRA | intronic | N | A | 4.68E-07 | 1.24 | 1.14 to 1.35 |
|
| ||||||||
| 6 | rs4151651 | CFB | exonic | N | A | 9.35E-06 | 1.32 | 1.17 to 1.49 |
|
| ||||||||
| 6 | rs2294884 | BTNL2 | intronic | N | A | 1.79E-05 | 0.86 | 0.80 to 0.92 |
|
| ||||||||
| 4 | rs13127214 | GABRB1 | intronic | Y | C | 5.04E-05 | 1.2 | 1.10 to 1.31 |
|
| ||||||||
| 1 | rs17325887 | LRRC7 | intronic | Y | A | 8.57E-05 | 0.73 | 0.63 to 0.86 |
|
| ||||||||
| 8 | rs6470362 | TRIB1 | intergenic | N | A | 9.55E-05 | 0.89 | 0.84 to 0.94 |
|
| ||||||||
| 19 | rs1325156 | LILRB4, LILRP2 | intergenic | N | A | 1.16E-04 | 1.15 | 1.07 to 1.23 |
|
| ||||||||
| 11 | rs1790353 | DHCR7 | intergenic | Y | A | 1.44E-04 | 0.88 | 0.83 to 0.94 |
|
| ||||||||
| 21 | rs7275203 | TSPEAR | intronic | N | A | 1.66E-04 | 0.89 | 0.84 to 0.95 |
|
| ||||||||
| 4 | rs227375 | MANBA | intronic | N | A | 1.91E-04 | 0.9 | 0.86 to 0.95 |
|
| ||||||||
| 1 | rs4542263 | LOC729987, SNX7 | intergenic | Y | A | 2.50E-04 | 1.19 | 1.08 to 1.31 |
|
| ||||||||
| 4 | rs6851362 | KIAA1109 | intronic | N | A | 2.58E-04 | 0.89 | 0.84 to 0.95 |
|
| ||||||||
| 12 | rs34484751 | MPHOSPH9 | intronic | Y | A | 2.67E-04 | 0.76 | 0.65 to 0.88 |
|
| ||||||||
| 4 | rs6848868 | KIAA1109 | exonic | N | A | 2.74E-04 | 0.85 | 0.78 to 0.93 |
|
| ||||||||
| 21 | rs2850146 | CBS, U2AF1 | intergenic | Y | C | 2.94E-04 | 0.84 | 0.77 to 0.92 |
|
| ||||||||
| 11 | rs10736469 | DRD2, TMPRSS5 | intergenic | Y | A | 3.04E-04 | 0.91 | 0.86 to 0.96 |
|
| ||||||||
| 6 | rs3763313 | BTNL2, HLA- DRA | intergenic | N | A | 3.06E-04 | 0.89 | 0.83 to 0.95 |
|
| ||||||||
| 4 | rs975403 | IL2, IL21 | intergenic | N | A | 3.39E-04 | 1.10 | 1.05 to 1.16 |
|
| ||||||||
| 2 | rs34228697 | GPR35, AQP12B | intergenic | N | A | 3.88E-04 | 1.14 | 1.06 to 1.22 |
|
| ||||||||
| 1 | rs4409689 | DOCK7, ATG4C | intergenic | N | A | 4.34E-04 | 1.11 | 1.05 to 1.17 |
|
| ||||||||
| 5 | rs468938 | MAST4 | intronic | Y | A | 4.36E-04 | 0.88 | 0.81 to 0.94 |
|
| ||||||||
| 1 | rs913059 | TGFBR3 | intronic | N | A | 4.69E-04 | 1.11 | 1.05 to 1.18 |
|
| ||||||||
| 7 | rs17133379 | SDK1 | intronic | Y | A | 4.83E-04 | 0.87 | 0.81 to 0.94 |
|
| ||||||||
| 6 | rs13211186 | LINC01526, IBTK | intergenic | Y | A | 4.94E-04 | 1.1 | 1.04 to 1.16 |
eta- analysis results are shown.
SNPs in the same genomic region are r2<0.5; Variants annotated as potentially novel fell outside of 1 MB of previously reported variants in an IBD-associated genomic region. 9 10 Allele, effect allele.
Study statistics per individual cohorts (CSMC, IIBDGC, and SHARE) are detailed in online supplemental table 2.
CHR, chromosome; CI, Confidence Interval; CSMC, Cedars-Sinai Medical Center; IBD, inflammatory bowel disease; IIBDGC, International IBD Genetics Consortium; OR, Odds Ratio; pCD, perianal Crohn’s disease; SHARE, Sinai- Helmsley Alliance for Research Excellence; SNP, single nucleotide polymorphism.
Table 4.
Pathway associations with pCD
| Including the MHC region | |||
|---|---|---|---|
|
| |||
| Pathway | Adjusted p value | OR | Genes |
|
| |||
| Systemic lupus | 3.83E-11 | 8.16 | HIST1H2BN; HIST1H2BL; erythematosus HIST1H2AJ; TNF; C4A; HLA- DRA; HIST1H3H; HIST1H3I; HIST1H4J; HIST1H2AA; HIST1H2BA; HLA- DQA2; HLA- DQA1; HLA- DRB1 |
|
| |||
| Inflammatory bowel | 2.86E-07 | 9.54 | IL21; MAF; IL5; IL12B; HLA- DRA; disease NOD2; TNF; HLA- DQA2; HLA- DQA1; IL2; NFKB1; HLA- DRB1 |
|
| |||
| Activation of C3 and C5 pathway | 2.72E-05 | 11.2 | C4A; CFB |
| Excluding the MHC region | |||
| JAK-ST AT signalling pathway | 9.16E-04 | 3.62 | IL21; CSF2; SOCS1; IL5; IL2RA; BCL2; IL12B; IL7R; IL2 |
|
| |||
| NF-κB signalling pathway | 5.86E-03 | 2.74 | PRKCB; BCL2; BCL10; NFKB1 |
|
| |||
| IL- 2 signalling pathway mediated with STAT5 | 4.07E-03 | 4.41 | IL2RA; BCL2; IL2 |
MHC, major histocompatibility complex; pCD, perianal Crohn’s disease.
Serological association with pCD
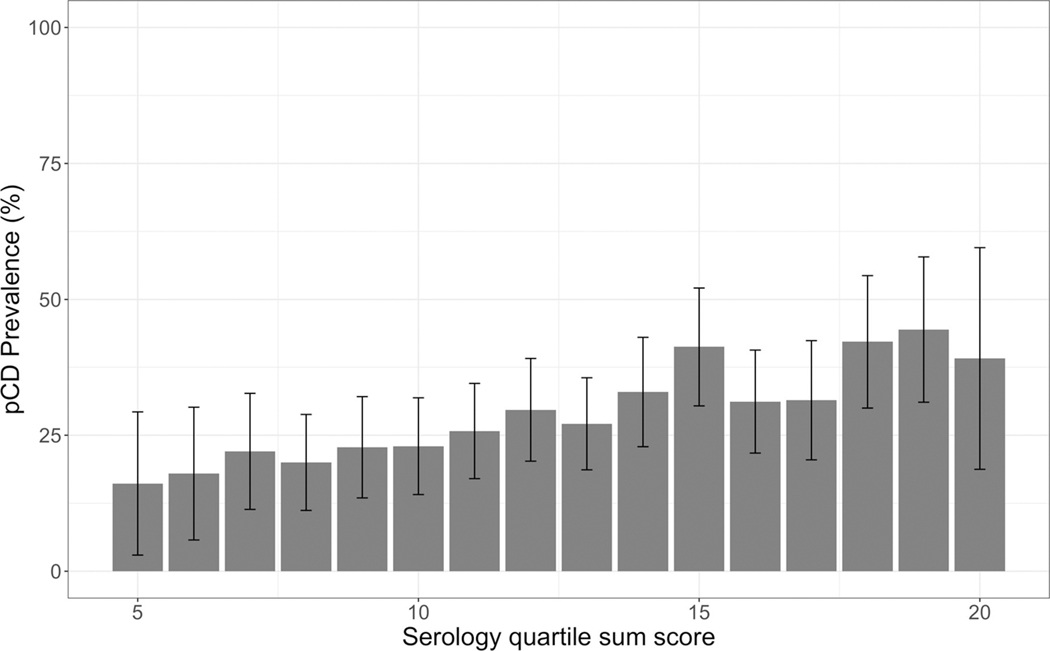
Serology data were available for 2376 patients with CD from the CSMC cohort. When compared with patients without perianal involvement pCD is associated with antibody positivity and level for both ASCA as well as OmpC (table 5). We observed a higher pCD prevalence rate with increasing serology QSS (p=1.39E-04 , OR 1.07, 95% CI 1.04 to 1.11) (figure 1).
Table 5.
Logistic regression analysis comparing perianal CD association with serology in CSMC cohort
| Positivity | Levels | |||
|---|---|---|---|---|
|
|
|
|||
| P value | OR (95% CI) | P value | OR (95% CI) | |
|
| ||||
| ANCA | 6.97E-10 | 1.05 (0.83 to 1.33) | 7.49E-01 | 1.00 (1.00 to 1.00) |
|
| ||||
| CBir1 | 1.93E-01 | 1.15 (0.93 to 1.42) | 1.98E-01 | 1.00 (1.00 to 1.00) |
|
| ||||
| I2 | 6.30E-01 | 0.94 (0.75 to 1.19) | 3.06E-01 | 1.00 (1.00 to 1.00) |
|
| ||||
| OmpC | 6.22E-03 | 1.39 (1.10 to 1.76) | 2.78E-02 | 1.01 (1.00 to 1.01) |
|
| ||||
| ASCA IgA | 2.78E-02 | 1.29 (1.03 to 1.63) | 1.48E-04 | 1.01 (1.00 to 1.01) |
|
| ||||
| ASCA IgG | 4.30E-02 | 1.26 (1.01 to 1.57) | 9.00E-05 | 1.01 (1.00 to 1.01) |
|
| ||||
| ASCA (any) | 1.15E-02 | 1.32 (1.06 to 1.65) | -- | -- |
CD, Crohn’s disease; CSMC, Cedars- Sinai Medical Center.
Figure 1.
Serology Quartile Sum Score (QSS) was strongly associated with pCD. Higher perianal CD prevalence was observed in CSMC patients with increasing serology quartile sum score (p=1.39×10–4; OR=1.07 (95% CI 1.04 to 1.11)). Error bars represent 95% CIs centred at the mean. CD, Crohn’s disease; CSMC, Cedars-Sinai Medical Center; pCD, perianal CD.
S252 CFB significantly decreases binding to C3b
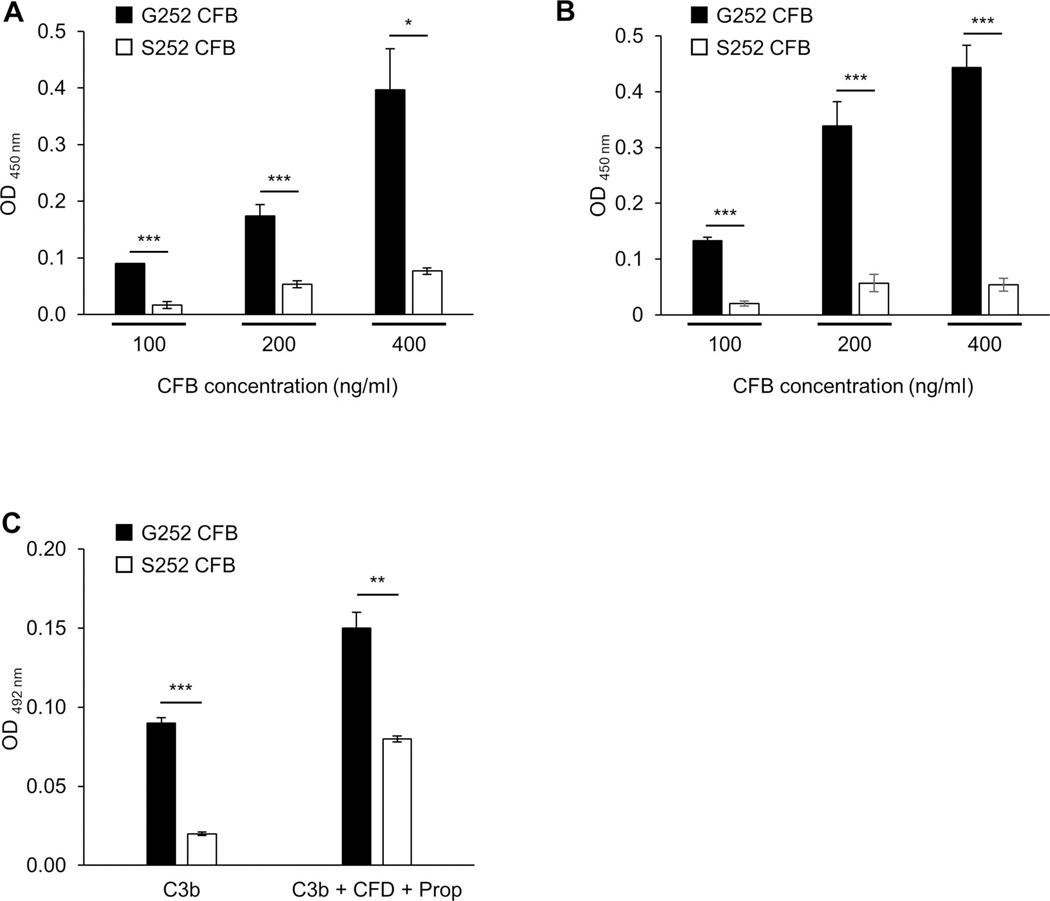
Given the genetic association for pCD and missense CFB variant, we aimed to characterise the functional consequences of rs4151651. We introduced the G252S mutation into human WT CFB using site-directed mutagenesis and overexpressed both G252 and S252 CFB in HEK293 cells. Using a cell-free binding assay and an anti-human CFB antibody we observed significantly reduced binding of S252 CFB to complement factor 3b (C3b) compared with G252 CFB at any of the concentrations tested (figure 2A). Binding of G252 or S252 CFB to C3b binding was dependent on Mg2+ cation and abolished in the presence of EDTA (data not shown). Addition of complement factor D (CFD) and properdin increased binding of G252 CFB to C3b but not S252 CFB (figure 2B). To rule out any differences in the affinity of the anti-CFB antibody for G252 compared with S252 CFB, we used plated-bound G252 or S252 CFB and detected the binding to C3b using an anti-C3 antibody. Consistent with our initial findings we observed significantly reduced binding of S252 CFB to C3b compared with G252 CFB (figure 2C). Similarly, in the presence of CFD and properdin binding of S252 CFB to C3b was significantly reduced compared with G252 CFB (figure 2C).
Figure 2.
S252 CFB impairs binding to C3b. (A, B) Plate-bound C3b was incubated with G252 or S252 CFB at the indicated concentration in the presence of C3b (A) or C3b + CFD + properdin (B) for 2 hours at 37°C. Binding of G252 or S252 CFB to C3b was detected using anti-human CFB antibody. (C) Plate-bound G252 or S252 were incubated with C3b or C3b + CFD + properdin for 2 h at 37°C. Binding of C3b to G252 or S252 CFB was detected using anti-human C3 antibody. Data are presented as means±SD. Data are representative of three independent experiments. *p<0.05, **p<0.01, ***p<0.001. CFB, complement factor B;
S252 CFB significantly impairs CFD-dependent CFB cleavage
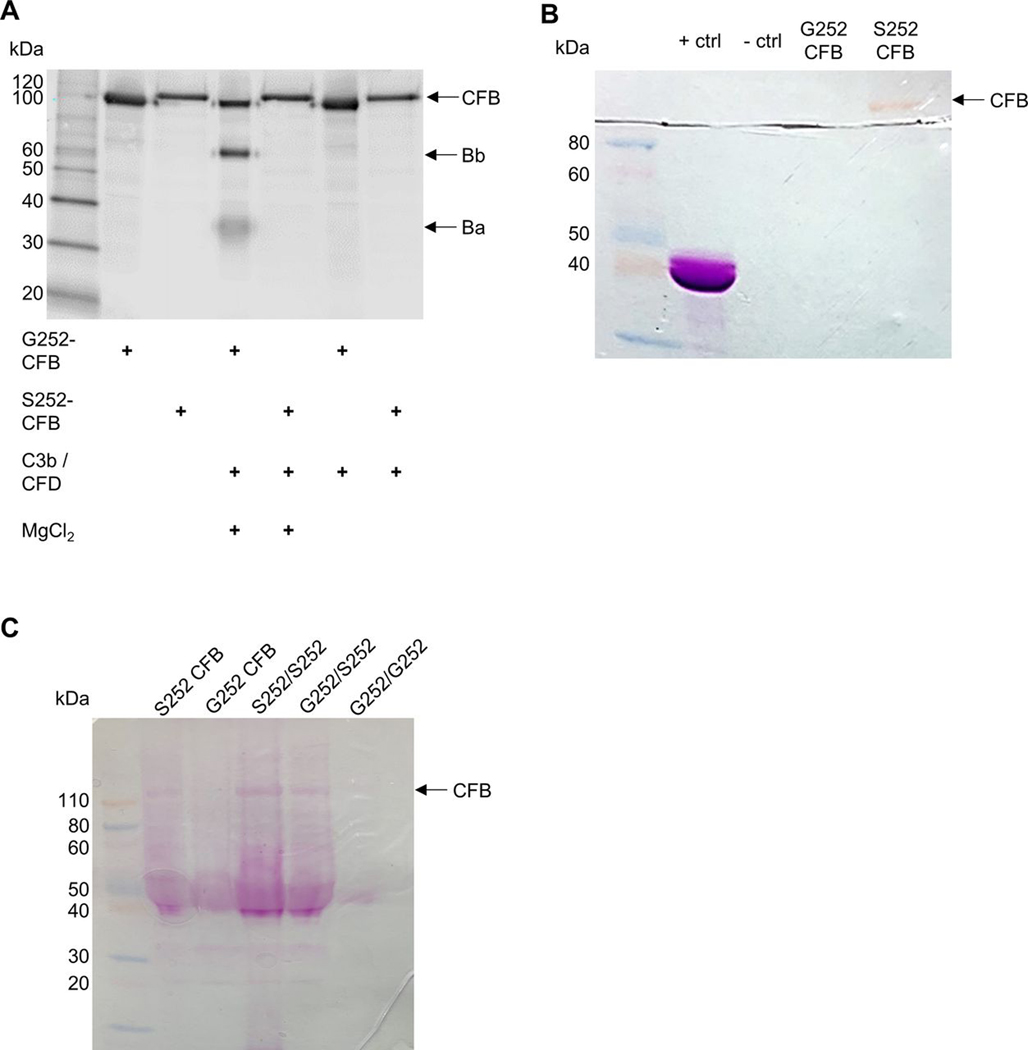
Next, we analysed the CFD-dependent cleavage of G252 or S252 CFB into its catalytic and non-catalytic subunits. Using a cell-free assay, G252 CFB was cleaved into its two subunits Bb (60 kDa), and Ba (33 kDa) in the presence of C3b and CFD (figure 3A). However, we only observed full length S252 CFB (90 kDa) suggesting that S252 CFB is impaired in CFD-dependent cleavage. Cleavage of G252 and S252 CFB was dependent on the presence of MgCl2. Our results demonstrate that S252 CFB abolished CFD-dependent cleavage into its two subunits.
Figure 3.
Serine 252 substitution in CFB impairs C3b-dependent CFB cleavage and results in a de novo glycosylation site. (A) G252 or S252 CFB were incubated with C3b and CFD for 30 min at 37°C. CFB cleavage products were detected with anti-human CFB antibody. (B) G252 or S252 CFB were separated by SDS-PAGE and stained with periodic acid-Schiff (PAS) reagent. (C) Recombinant G252 or S252 CFB (2 μg each), or pooled patient serum (S252/S252, G252/S252, G252/G252) (40 μg/genotype) were incubated with anti-human CFB antibody followed by immunoprecipitation. Samples were separated by SDS-PAGE and stained with PAS reagent. One representative experiment out of three independent experiments is shown (A, B). CFB, complement factor B; SDS-PAGE, sodium dodecyl-sulfate polyacrylamide gel electrophoresis.
Serine substitution at 252 generates a de novo glycosylation site in CFB
Surprisingly, we observed that uncleaved S252 CFB runs at a higher molecular weight compared with G252 CFB suggesting potential PTM of S252 CFB (figure 3A). Our PTM database query for rs4151651 SNP indicated a predicted gain of glycosylation (O-N-acetylgalactosamine) at this SNP with the glycine to serine substitution. To further investigate this PTM we separated G252 or S252 CFB by SDS-P AGE and stained with periodic acid-Schiff (P AS) reagent. We observed a distinct band for S252 CFB that was not present for G252 CFB demonstrating that serine 252 generates a de novo glycosylation site in CFB (figure 3B). To confirm glycosylation of S252 CFB in patient serum, we performed immunoprecipitation of CD serum from S252/S252, G252/S252 or G252/G252 carriers using anti-CFB antibody followed by SDS-PAGE separation and staining with PAS reagent. Due to the limited availability of patient serum from carriers of the risk alleles, we pooled patient serum with identical genotypes. We observed a distinct band of glycosylated S252 CFB from homozygous risk (S252/S252) and heterozygous (G252/S252) serum. However, we could not detect glycosylated CFB for homozygous protective (G252/G252) serum (figure 3C). Recombinant G252 and S252 CFB were used as controls. To understand the significance of glycosylated CFB at S252 within the linker region, we used molecular modelling. The putative glycosylation at the linker region forms a flexible loop structure. The region is also surface exposed and thus glycosylation does not interfere with CFB structure and should remain stable (online supplemental figure 1A). The linker region in CFB is also surface exposed when it forms a complex with C3b (online supplemental figure 1B,C) suggesting the glycosylation of CFB at this site is unlikely to have significant structural implication.
S252 CFB impairs phagocytosis of zymosan particles
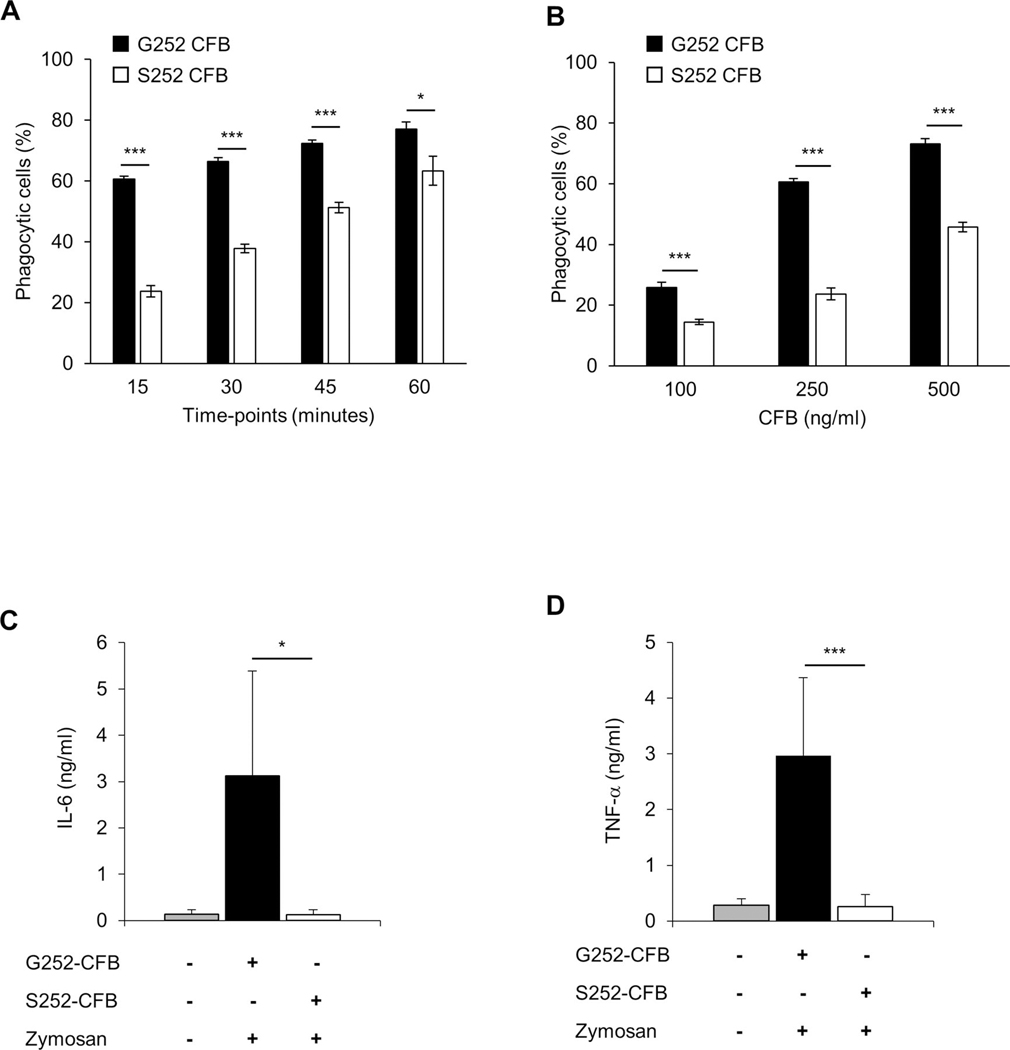
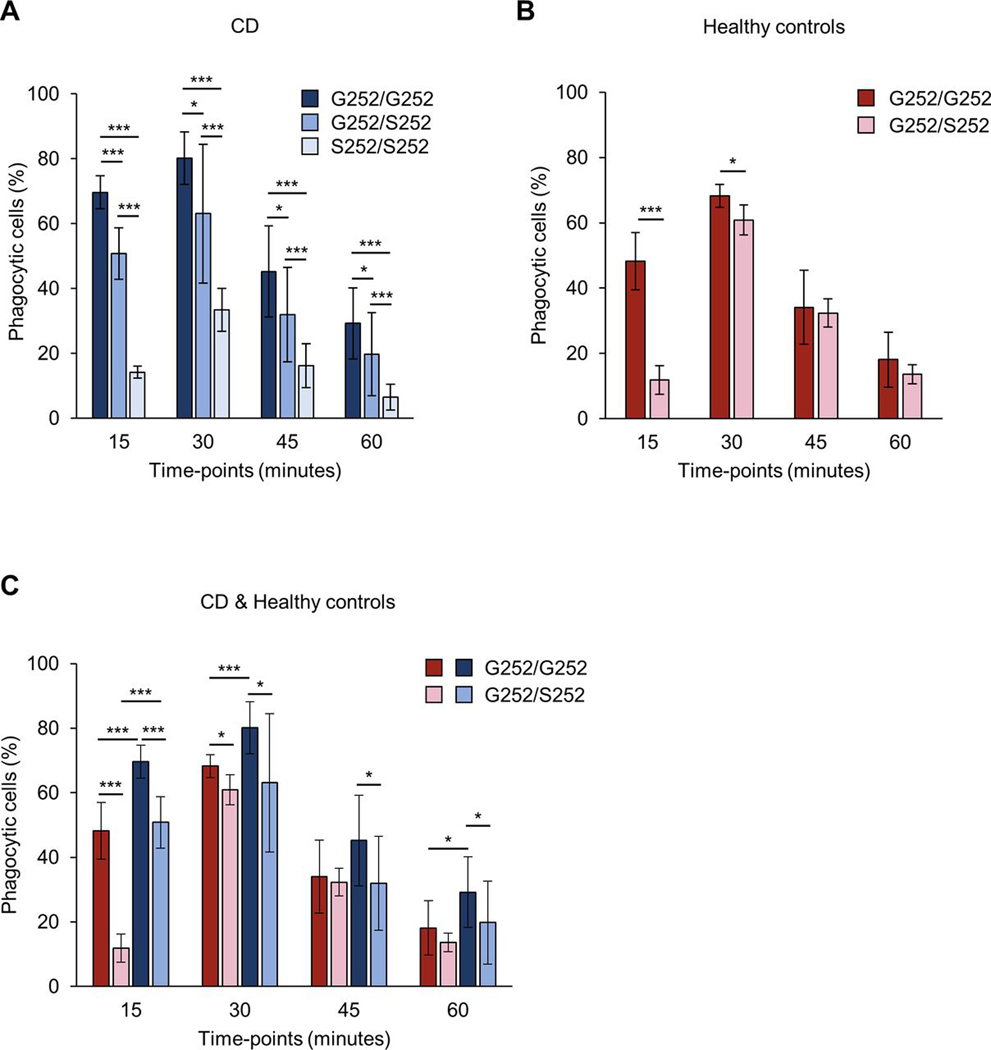
We hypothesised that the non-synonymous CFB SNP rs4151651 would decrease the CFB-dependent phagocytic ability of macrophages. To examine the impact of the CFB variants we used primary human PBMC-derived macrophages and incubated them with recombinant G252 or S252 CFB. We observed significantly reduced phagocytosis of zymosan particles with S252 CFB compared with G252 CFB at all time points (figure 4A). We observed significantly impaired phagocytosis for S252 CFB at all concentrations tested (figure 4B). Next, we determined cytokine secretion in human macrophages in response to CFB-mediated pathogen stimulation for G252 and S252 CFB. Zymosan stimulation in the presence of S252 CFB resulted in significantly reduced IL-6 and TNF-α secretion compared with G252 CFB (figure 4C,D). Reduced cytokine secretion was not due to differences in cell viability between G252 and S252 CFB (online supplemental figure 2). Next, we used serum from patients with CD carrying the S252/S252, G252/S252 or G252/G252 genotypes. We observed a significant decrease in phagocytosis of macrophages in homozygous risk (S252/S252) compared with homozygous protective (G252/G252) serum at all time points (15–60 min) (figure 5A). Furthermore, phagocytosis was also significantly decreased in G252/S252 heterozygous compared with S252/S252 risk serum at all time points, suggesting a gene-dose effect (figure 5A). To assess whether the observed difference is variant or disease specific, we used serum from non-IBD controls carrying the G252/S252 or G252/G252 genotype. Due to low minor allele frequency for rs4151651 (3% in European population), non-IBD controls with homozygous risk (S252/S252) genotype were not available. We observed a significant decrease in phagocytosis in G252/S252 compared with G252/G252 serum from non-IBD healthy controls at 15 and 30 min (figure 5B). Next, we compared phagocytosis in the presence of serum from non-IBD controls or patients with CD with G252/S252 or G252/G252 variants. We observed significant decrease in phagocytosis in both non-IBD controls and patients with CD in heterozygous (G252/S252) compared with homozygous protective (G252/G252) serum at 15 min (figure 5C). Although there was an overall slower kinetic for phagocytosis in the presence of non-IBD control serum, variant specific differences were preserved in non-IBD controls and CD sera. To determine whether phagocytosis of zymosan particles is specifically driven by CFB, we performed phagocytosis experiments in the presence of blocking anti-human CFB antiserum or control serum. We observed a dose-dependent inhibition of phagocytosis in the presence of healthy serum, while control serum had no effect on phagocytosis (data not shown). Furthermore, anti-CFB treatment resulted in a 90.4 and 85.1% inhibition of phagocytosis for CD serum from G252/G252 or S252/S252 patients, respectively (online supplemental figure 3). Our data suggest that the CFB risk variant leads to a significant decrease in phagocytosis of zymosan particles by macrophages.
Figure 4.
Recombinant S252 CFB decreases macrophage phagocytosis and cytokine secretion. (A, B) PBMC-derived macrophages from healthy donors were incubated with fluorescently labelled zymosan particles in the presence of recombinant G252 or S252 CFB. Percentage of phagocytic cells was determined by flow cytometry. (A) Time course of phagocytosis for G252 or S252 CFB (250 ng/mL). (B) Dose response of G252 or S252 CFB (100, 250 and 500 ng/mL) analysed at 15 min. (C, D) PBMC-derived Macrophages were incubated with G252 or S252 CFB (250 ng/mL) in the presence of zymosan (1 ng/mL) for 6 hours. IL-6 (C) and TNF-α (D) concentrations in supernatants were measured by ELISA. Data are presented as means±SD. Data are representative of three (A, B) or five (C, D) independent experiments. *p<0.05, ***p<0.005. CFB, complement factor B; PBMC, peripheral blood mononuclear cells.
Figure 5.
Serum from risk subjects (S252/S252, G252/S252) decreases macrophage phagocytosis. PBMC-derived macrophages from healthy donors were incubated with fluorescently labelled zymosan particles for the indicated time points in the presence of 10% serum from patients with CD (S252/S252 n=6, G252/S252 n=15, G252/G252 n=15) (A) or non-IBD controls (G252/S252 n=5, G252/G252 n=5) (B). Phagocytosis was measured by flow cytometry and the percentage of phagocytic cells is shown. Each serum sample was analysed in triplicates. (C) Comparison between G252/S252, G252/G252 non-IBD controls and G252/S252, G252/G252 patients with CD. Data are presented as means±SD. *p<0.05, ***p<0.005. CD, Crohn’s disease; IBD, inflammatory bowel disease; PBMC, peripheral blood mononuclear cells.
DISCUSSION
GWAS have been extremely successful in identifying genetic variants associated with risk for complex diseases. However, few studies have bridged the gap between genetic associations and an understanding of the altered biology consequent on the genetic variation. This study is the largest genetic study, to date, specifically studying pCD. Here, we performed within-case analyses comparing patients with CD with and without perianal complications in three independent cohorts totaling 4056 pCD+and 11 088 pCD-patients. Across all cohorts, patients with CD with colonic disease location and also with internal penetrating disease were at significantly higher risk of perianal manifestations. In the CSMC cohort alone, we observed reduced pCD prevalence in Jewish ancestry and other cohorts may have been underpowered to detect this association. Association of pCD with smoking at time of diagnosis was inconsistent between CSMC and SHARE, with no association observed for IIBDGC. The observed discrepancy may potentially be due, in part, to high percentage of missing smoking-related data in both CSMC and IIBDGC cohorts. Further work is needed to define the relationship between smoking and perianal disease, and studies determining smoking status at time of perianal disease development are necessary to assess whether a true relationship exists. Serological profiles are associated with CD prognosis,27 28 and in our study, we identified significant associations with OmpC and ASCA and increasing serology QSS. These profiles have previously been associated with complicated CD and small bowel disease location20 and our findings, after accounting for disease behaviour, location and duration, suggest these profiles are also associated with more complicated colonic disease. Serological associations may indicate aberrant immune responses to luminal microbes and in pCD this may be, in part, due to impaired bacterial phagocytosis and elimination.
Our most robust genetic associations were observed with variants within the MHC, a well-established IBD locus which has also been implicated in IBD disease location, behaviour and severity.12 29 We also identified associations with putative novel loci including MAST4 and LRRC7. While MAST4, a microtubule-associated serine/threonine protein kinase family member, has not been previously shown to be associated with IBD, another MAST family member MAST3 has been implicated and shown to modulate Toll-Like Receptor (TLR)-4-dependent NF-KB activity.30 MAST4 has also been reported to be a downstream target of IBD-associated microRNA-138 which is differentially expressed in inflamed versus non-inflamed colonic mucosa of UC patients.31 Our pCD-associated MAST4 variant rs468938 falls in a region demonstrating histone modifications consistent with enhancer elements in small and large intestine and is also an eQTL for MAST4-AS1 lncRNA in adipose tissue, cultured fibroblasts and sigmoid colon. Leucine Rich Repeat Containing 7 (LRRC7) locus has been implicated in psoriasis32 and associated with anti-TNFα responsiveness in paediatric patients with IBD.33 Pathway analyses confirmed our previously published association with JAK-STAT signalling pathway and pCD.2 JAK inhibitors are effective therapies in CD34 35 and our observations further support that patients with pCD might benefit from treatment with JAK inhibitors.
While NOD2 genetic variants are among the strongest risk factors associated with ileal CD,12 the role of NOD2 and perianal penetrating disease is less clear. Genetic variation in NOD2 and perianal disease have been shown as a ‘clinicogenetic variable’ to be strongly associated with increased risk of surgery in CD.36 NOD2 genetic variation has also been implicated in antibiotic treatment outcome of perianal fistulising CD37 and an association with NOD2/rs72796353 and the development of perianal fistulas has been reported, notably in the absence of key CD-associated NOD2 polymorphisms (rs2066844, rs2066845 and rs2066847).38 While our gene enrichment analysis identified NOD2 among a collection of genes involved in IBD to be associated with pCD, we were unable to replicate association with rs72796353, as this variant was not present on Immunochip array used in our study, and we did not observe associations (p<0.001) with pCD and any NOD2 variants including the frameshift mutation.
Our genetic associations highlighted CFB as a promising candidate for functional analyses. Here, we replicate across all three study populations an association for non-synonymous SNP rs4151651 with pCD. This variant has previously been strongly associated with CD colonic disease location, complicated CD disease behaviour, UC colectomy and UC age of diagnosis.12 Additionally, CFB mRNA and protein are abundantly expressed in human colonic epithelial cells and expression is upregulated in biopsies of active CD and UC.39 Previous GWAS have identified associations of CFB with multiple inflammatory diseases in addition to IBD including age-related macular degeneration, anterior uveitis and systemic lupus erythematosus.40–43 However, the functional consequences of CFB rs4151651 were not previously elucidated.
G252S is in exon 5 in the Ba subunit of CFB within the linker region connecting the CCP3 and Von Willebrand factor A domains.44 G252S is adjacent to the R259-K260 (sometimes referred to as R234-K235) scissile bond, the site of CFD-dependent cleavage into non-catalytic Ba and catalytic Bb subunits. First, we analysed the binding of S252 CFB to C3b. We observed significantly decreased binding of S252 CFB to C3b compared with G252 CFB. Additionally, G252 CFB protein underwent CFD-dependent cleavage into the Ba and Bb subunits, while there was no corresponding cleavage with S252 CFB with only full length S252 CFB protein detected. Our data suggest that glycine at position 252 is pivotal for its binding to C3b and its enzymatic activity and that a single amino acid exchange to serine is sufficient to impair C3b-dependent, CFD-mediated cleavage. Surprisingly, we observed that S252 CFB undergoes glycosylation at S252. Based on our molecular modelling we predicted that glycosylation at S252 should form a stable structure. Although the molecular modelling also predicted that the glycosylation of CFB at S252 is unlikely to have significant structural implication on the binding to C3b, a longer or bulkier sugar residues might induce conformational changes in CFB that might influence its binding to C3b. Indeed, we observed significant reduction in the binding of CFB to C3b in vitro. Our in vitro assays were performed in the presence of CFD and properdin. CFD directly binds to CFB leading to a conformational change in CFB and exposure of the linker region containing the sissle bond to the catalytic site within CFD.45 Furthermore, the downstream activation of the alternative complement pathway is impaired and leads to decreased phagocytosis of opsonised zymosan particles and decreased cytokine secretion. We observed, at all time points, a significant decrease in phagocytosis by macrophages in the presence of sera from CD subjects carrying risk (S252/S252) compared with non-risk (G252/G252) genotype. This phagocytic difference was observed in serum from both healthy controls and patients with CD, thus suggesting a variant-specific rather than a disease-specific effect (online supplemental figure 4). CFB expression is significantly up regulated in colonic biopsies of patients with active CD and UC.39 Recently, an increased abundance of several bacterial strains in fistula biopsies of patients with pCD including Corynebacterium has been reported.46 Corynebacterium can directly activate the alternative complement pathway in macrophages.47 48 Our study provides a possible mechanism by which high abundance of bacteria such as Corynebacterium in fistulae of patients with pCD could be the result of impaired CFB-mediated macrophage phagocytosis in patients carrying the CFB risk allele.
Despite being the largest genetic study of pCD to date, we did not observe genome-wide significant associations. This could be due, at least in part, to the heterogeneity of pCD. While all CSMC and SHARE cases are fistulising in nature, the perianal manifestation in the IIBDGC cohort likely contains subjects with perianal disease that is characterised by skin tags, strictures or fissures in the absence of fistula, although epidemiological studies suggest that the vast majority of patients with CD with perianal disease have fistula.49 Our study was limited with the availability of serological data in only a subset of subjects (CSMC cohort only). While we observed significant associations for pCD and increasing serology QSS, OmpC and ASCA, further investigations are warranted to ascertain the functional relationships of IBD-associated serologies and individual genetic variants. It is possible that the various serological markers define a subset of subjects characterised by a microbiome ‘profile’ and these microbiome changes may be a consequent of impaired phagocytosis. However, large-scale studies examining such connections between IBD-associated serologies and microbiome have not yet been performed. We further acknowledge the limitation of the current study with its observational nature. First, much of the clinical information were retrieved based on retrospective chart review. The potential information bias, although independent of genotypes, can potentially attenuate the power of our study. Second, patients enrolled in the current study are predominantly from referral centres, which may present a selection bias with regard to more severe disease. In this sense, further studies are warranted, preferably with more general patient populations, to validate the findings in the current study. Third, we used principal components to control for potential bias from population substructure. However, there remains a potential for other confounders that cannot be controlled in the current study, for example, different treatment strategies or lifestyles. Prospective studies using patients carrying specific genetic variants of interest can better address those potential confounding effects. Our study cohort was limited to subjects of European ancestry. Given the increased prevalence of pCD in African American and Hispanic patients with CD, genetic studies of perianal complications in non-Europeans may uncover additional findings. 50 51 Nevertheless, our results highlighted novel loci for further investigation and identified key associations within MHC, with a novel role for CFB in pCD.
In conclusion, we identified clinical characteristics, serological markers and novel genetic signals associated with pCD. Our functional follow-up studies defined the consequences of a non-synonymous SNP in CFB associated with pCD and has been associated with several other disease manifestations both in IBD and other immune-mediated diseases. We demonstrated that S252 CFB impairs binding to C3b, and CFD-mediated CFB cleavage and subsequent phagocytosis of complement-opsonised particles by macrophages. Our findings define a critical function of the alternative complement pathway and CFB in pCD by regulating microbial phagocytosis.
Supplementary Material
WHAT IS ALREADY KNOWN ON THIS TOPIC
Variant rs4151651 in complement factor B (CFB) has been associated with Crohn’s disease (CD) and ulcerative colitis (UC).
CFB mRNA and protein are expressed in human colonic epithelial cells and upregulated in biopsies of active CD and UC.
Genome-wide association studies have implicated a role for CFB in multiple inflammatory diseases.
WHAT THIS STUDY ADDS
Perianal CD-associated variant rs4151651 leads to glycine to serine amino acid substitution in CFB (G252S CFB) and confers significantly impaired binding to complement factor 3b and subsequent cleavage of CFB into Ba and Bb subunits. Serine substitution at 252 generates a de novo glycosylation site in CFB.
Serum from homozygous risk subjects (S252/S252) confers significantly decreased macrophage phagocytosis of zymosan particles compared with homozygous protective subjects (G252/G252).
HOW THIS STUDY MIGHT AFFECT RESEARCH, PRACTICE OR POLICY
This study demonstrates an important role for the alternative complement pathway and CFB in the development of perianal CD. This study suggests that targeting the alternative complement pathway may be a novel therapeutic approach for treating this disabling manifestation of CD.
Funding
This work was supported by grants from the National Institute of Health (U01DK062413, P01 DK046763 to DM), the F. Widjaja Foundation (SRT, KSM), the Leona M. & Harry B. Helmsley Charitable Trust (DM) and the Fred L. Hartley Family Foundation (DM). This study was supported by the Cedars-Sinai MIRIAD IBD Biobank. The MIRIAD IBD Biobank is supported by the Widjaja Foundation Inflammatory Bowel and Immunobiology Research Institute, National Institute of Diabetes and Digestive and Kidney Disease Grants P01DK046763 and U01DK062413 and The Leona M. and Harry B. Helmsley Charitable Trust. We are grateful to all patients and control subjects that volunteered to join this study. The authors would like to thank the Cedars-Sinai Medical Center Flow Cytometry Core.
Competing interests
Cedars-Sinai has financial interests in Prometheus Biosciences, a company which has access to the data and specimens in Cedars-Sinai’s MIRIAD Biobank (including the data and specimens used in this study) and seeks to develop commercial products. DM and SRT own stock in Prometheus Biosciences. DL, SRT and DM are consultants for Prometheus Biosciences. DM has consulted for Takeda, Sandoz Immunology, Gilead, Pfizer, Boehringer Ingelheim, Qu Biologics and Bridge Biotherapeutics. CH is on the Advisory Board or consultant for Abbvie, Bristol Myers Squibb, Ferring, Genentech, InDex Pharmaceuticals, Janssen, Pfizer, Takeda, received research support from Abbvie, Genentech, Eli Lilly, Pfizer, and educational grant funding from Pfizer. PF is a consultant for Takeda. GM is a consultant for Abbvie, Arena, Boehringer-Ingelheim, Bristol-Myers Squibb, Janssen, Medtronic, Techlab, Takeda, Pfizer, Samsung Bioepis, Entasis, Ferring, Shionogi and received research funding from Pfizer.
Collaborators
International IBD Genetics Consortium, Members of the International Inflammatory Bowel Disease Genetics Consortium: Clara Abraham, Jean-P aul Achkar, Tariq Ahmad, Leila Amininejad, Ashwin N Ananthakrishnan, Vibeke Andersen, Carl A Anderson, Jane M Andrews, Vito Annese, Guy Aumais, Leonard Baidoo, Robert N Baldassano, Peter A Bampton, Murray Barclay, Jeffrey C Barrett, Theodore M Bayless, Johannes Bethge, Alain Bitton, Gabrielle Boucher, Stephan Brand, Berenice Brandt, Steven R Brant, Carsten Büning, Angela Chew, Judy H Cho, Isabelle Cleynen, Ariella Cohain, Anthony Croft, Mark J Daly, Mauro D’Amato, Silvio Danese, Dirk De Jong, Martine De Vos, Goda Denapiene, Lee A Denson, Kathy L Devane, Olivier Dewit, Renata D’Inca, Marla Dubinsky, Richard H Duerr, Cathryn Edwards, David Ellinghaus, Jonah Essers, Lynnette R Ferguson, Eleonora A Festen, Philip Fleshner, Tim Florin, Denis Franchimont, Andre Franke, Karin Fransen, Richard Gearry, Michel Georges, Christian Gieger, Jürgen Glas, Philippe Goyette, Todd Green, Anne M Griffiths, Stephen L Guthery, Hakon Hakonarson, Jonas Halfvarson, Katherine Hanigan, Talin Haritunians, Ailsa Hart, Chris Hawkey, Nicholas K Hayward, Matija Hedl, Paul Henderson, Xinli Hu, Hailiang Huang, Jean-Pierre Hugot, Ken Y Hui, Marcin Imielinski, Andrew Ippoliti, Laimas Jonaitis, Luke Jostins, Tom H Karlsen, Nicholas A Kennedy, Mohammed Azam Khan, Gediminas Kiudelis, Krupa Krishnaprasad, Subra Kugathasan, Limas Kupcinskas, Anna Latiano, Debby Laukens, Ian C Lawrance, James C Lee, Charlie W Lees, Marcis Leja, Johan Van Limbergen, Paolo Lionetti, Jimmy Z Liu, Edouard Louis, Gillian Mahy, John Mansfield, Dunecan Massey, Christopher G Mathew, Dermot PB McGovern, Raquel Milgrom, Mitja Mitrovic, Grant W Montgomery, Craig Mowat, William Newman, Aylwin Ng, Siew C Ng, Sok Meng Evelyn Ng, Susanna Nikolaus, Kaida Ning, Markus Nöthen, Ioannis Oikonomou, Orazio Palmieri, Miles Parkes, Anne Phillips, Cyriel Y Ponsioen, Urõs Potocnik, Natalie J Prescott, Deborah D Proctor, Graham Radford-Smith, Jean-Francois Rahier, Soumya Raychaudhuri, Miguel Regueiro, Florian Rieder, John D Rioux, Stephan Ripke, Rebecca Roberts, Richard K Russell, Jeremy D Sanderson, Miquel Sans, Jack Satsangi, Eric E Schadt, Stefan Schreiber, Dominik Schulte, L Philip Schumm, Regan Scott, Mark Seielstad, Mark S Silverberg, Lisa A Simms, Jurgita Skieceviciene, Sarah L Spain, A. Hillary Steinhart, Joanne M Stempak, Laura Stronati, Jurgita Sventoraityte, Stephan R Targan, Kirstin M Taylor, Anje ter Velde, Emilie Theatre, Leif Torkvist, Mark Tremelling, Andrea van der Meulen, Suzanne van Sommeren, Eric Vasiliauskas, Severine Vermeire, Hein W Verspaget, Thomas Walters, Kai Wang, Ming-Hsi Wang, Rinse K Weersma, Zhi Wei, David Whiteman, Cisca Wijmenga, David C Wilson, Juliane Winkelmann, Ramnik J Xavier, Bin Zhang, Clarence K Zhang, Hu Zhang, Wei Zhang, Hongyu Zhao, Zhen Z Zhao.
Footnotes
Patient and public involvement Patients and/or the public were not involved in the design, or conduct, or reporting, or dissemination plans of this research.
Patient consent for publication Not applicable.
Ethics approval This study involves human participants and was approved by Subjects were included in this study after consent and with local IRB approval. Blood samples from healthy de-identified subjects were acquired under Cedars-Sinai IRB #3358. Participants gave informed consent to participate in the study before taking part.
Provenance and peer review Not commissioned; externally peer reviewed.
Supplemental material This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
Data are available on reasonable request. Meta-analysis summary statistics will be made available on publication through the NIDDK IBD Genetics Consortium (IBDGC) Data Commons portal currently under development.
REFERENCES
- 1.Safar B, Sands D. Perianal Crohn’s disease. Clin Colon Rectal Surg 2007;20:282–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kaur M, Panikkath D, Yan X, et al. Perianal Crohn’s disease is associated with distal colonic disease, stricturing disease behavior, IBD-associated serologies and genetic variation in the JAK-STAT pathway. Inflamm Bowel Dis 2016;22:862–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Panés J, Rimola J. Perianal Fistulizing Crohn’s disease: pathogenesis, diagnosis and therapy. Nat Rev Gastroenterol Hepatol 2017;14:652–64. [DOI] [PubMed] [Google Scholar]
- 4.Park SH, Aniwan S, Scott Harmsen W, et al. Update on the natural course of Fistulizing perianal Crohn’s disease in a population-based cohort. Inflamm Bowel Dis 2019;25:1054–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scharl M, Rogler G. Pathophysiology of fistula formation in Crohn’s disease. World J Gastrointest Pathophysiol 2014;5:205–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rieder F, Kessler S, Sans M, et al. Animal models of intestinal fibrosis: new tools for the understanding of pathogenesis and therapy of human disease. Am J Physiol Gastrointest Liver Physiol 2012;303:G786–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scharl M, Frei S, Pesch T, et al. Interleukin-13 and transforming growth factor β synergise in the pathogenesis of human intestinal fistulae. Gut 2013;62:63–72. [DOI] [PubMed] [Google Scholar]
- 8.Cleynen I, González JR, Figueroa C, et al. Genetic factors conferring an increased susceptibility to develop Crohn’s disease also influence disease phenotype: results from the ibdchip European project. Gut 2013;62:1556–65. [DOI] [PubMed] [Google Scholar]
- 9.de Lange KM, Moutsianas L, Lee JC, et al. Genome-Wide association study implicates immune activation of multiple integrin genes in inflammatory bowel disease. Nat Genet 2017;49:256–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jostins L, Ripke S, Weersma RK, et al. Host-Microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 2012;491:119–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu JZ, van Sommeren S, Huang H, et al. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat Genet 2015;47:979–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cleynen I, Boucher G, Jostins L, et al. Inherited determinants of crohn’s disease and ulcerative colitis phenotypes: a genetic association study. Lancet 2016;387:156–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peloquin JM, Goel G, Kong L, et al. Characterization of candidate genes in inflammatory bowel disease-associated risk loci. JCI Insight 2016;1:e87899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dassopoulos T, Nguyen GC, Bitton A, et al. Assessment of reliability and validity of IBD phenotyping within the National Institutes of diabetes and digestive and kidney diseases (NIDDK) IBD genetics Consortium (ibdgc). Inflamm Bowel Dis 2007;13:975–83. [DOI] [PubMed] [Google Scholar]
- 15.Satsangi J, Silverberg MS, Vermeire S, et al. The Montreal classification of inflammatory bowel disease: controversies, consensus, and implications. Gut 2006;55:749–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ananthakrishnan AN, Kwon J, Raffals L, et al. Variation in treatment of patients with inflammatory bowel diseases at major referral centers in the United States. Clin Gastroenterol Hepatol 2015;13:1197–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Landers CJ, Cohavy O, Misra R, et al. Selected loss of tolerance evidenced by Crohn’s disease-associated immune responses to auto-and microbial antigens. Gastroenterology 2002;123:689–99. [DOI] [PubMed] [Google Scholar]
- 18.Huang H, Fang M, Jostins L, et al. Fine-Mapping inflammatory bowel disease loci to single-variant resolution. Nature 2017;547:173–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007;81:559–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mow WS, Vasiliauskas EA, Lin Y-C, et al. Association of antibody responses to microbial antigens and complications of small bowel Crohn’s disease. Gastroenterology 2004;126:414–24. [DOI] [PubMed] [Google Scholar]
- 21.Team RDC. R: A language and environment for statistical computing [program]. Vienna, Austria: R Foundation for Statistical Computing, 2020. [Google Scholar]
- 22.Willer CJ, Li Y, Abecasis GR. Metal: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 2010;26:2190–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mishra A, Macgregor S. VEGAS2: software for more flexible gene-based testing. Twin Res Hum Genet 2015;18:86–91. [DOI] [PubMed] [Google Scholar]
- 24.Chen EY, Tan CM, Kou Y, et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 2013;14:128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McLean CY, Bristor D, Hiller M, et al. Great improves functional interpretation of cis-regulatory regions. Nat Biotechnol 2010;28:495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang Y, Peng X, Ying P, et al. Awesome: a database of SNPs that affect protein post-translational modifications. Nucleic Acids Res 2019;47:D874–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oliveira SB, Monteiro IM. Diagnosis and management of inflammatory bowel disease in children. BMJ 2017;357:j2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Amre DK, Lu S-E, Costea F, et al. Utility of serological markers in predicting the early occurrence of complications and surgery in pediatric Crohn’s disease patients. Am J Gastroenterol 2006;101:645–52. [DOI] [PubMed] [Google Scholar]
- 29.Goyette P, Boucher G, Mallon D, et al. High-Density mapping of the MHC identifies a shared role for HLA-DRB1*01:03 in inflammatory bowel diseases and heterozygous advantage in ulcerative colitis. Nat Genet 2015;47:172–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Labbé C, Goyette P, Lefebvre C, et al. MAST3: a novel IBD risk factor that modulates TLR4 signaling. Genes Immun 2008;9:602–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Valmiki S, Ahuja V, Paul J. Microrna exhibit altered expression in the inflamed colonic mucosa of ulcerative colitis patients. World J Gastroenterol 2017;23:5324–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsoi LC, Spain SL, Ellinghaus E, et al. Enhanced meta-analysis and replication studies identify five new psoriasis susceptibility loci. Nat Commun 2015;6:7001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dubinsky MC, Mei L, Friedman M, et al. Genome wide association (GWA) predictors of anti-TNFalpha therapeutic responsiveness in pediatric inflammatory bowel disease. Inflamm Bowel Dis 2010;16:1357–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sandborn WJ, Feagan BG, Loftus EV, et al. Efficacy and safety of upadacitinib in a randomized trial of patients with Crohn’s disease. Gastroenterology 2020;158:2123–38. [DOI] [PubMed] [Google Scholar]
- 35.Vermeire S, Schreiber S, Petryka R, et al. Clinical remission in patients with moderate-to-severe Crohn’s disease treated with filgotinib (the Fitzroy study): results from a phase 2, double-blind, randomised, placebo-controlled trial. Lancet 2017;389:266–75. [DOI] [PubMed] [Google Scholar]
- 36.Nasir BF, Griffiths L, Nasir A, et al. Perianal disease combined with NOD2 genotype predicts need for IBD-related surgery in Crohn’s disease patients from a population-based cohort. J Clin Gastroenterol 2013;47:242–5. [DOI] [PubMed] [Google Scholar]
- 37.Angelberger S, Reinisch W, Dejaco C, et al. Nod2/Card15 gene variants are linked to failure of antibiotic treatment in perianal fistulating Crohn’s disease. Am J Gastroenterol 2008;103:1197–202. [DOI] [PubMed] [Google Scholar]
- 38.Schnitzler, Friedrich M, Wolf C, et al. The NOD2 single nucleotide polymorphism rs72796353 (IVS4+10 a > C) is a predictor for perianal fistulas in patients with Cro’n’s disease in the absence of other NOD2 mutations. PLoS One 2015;10:e0116044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ostvik AE, vB Granlund A, Gustafsson BI, et al. Mucosal Toll-like receptor 3-dependent synthesis of complement factor B and systemic complement activation in inflammatory bowel disease. Inflamm Bowel Dis 2014;20:995–1003. [DOI] [PubMed] [Google Scholar]
- 40.Juyal G, Negi S, Sood A, et al. Genome-Wide association scan in North Indians reveals three novel HLA-independent risk loci for ulcerative colitis. Gut 2015;64:571–9. [DOI] [PubMed] [Google Scholar]
- 41.Thakkinstian A, McEvoy M, Chakravarthy U, et al. The association between complement component 2/complement factor B polymorphisms and age-related macular degeneration: a huge review and meta-analysis. Am J Epidemiol 2012;176:361–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang M, Lai TYY, Tam POS, et al. Association of C2 and CFB polymorphisms with anterior uveitis. Invest Ophthalmol Vis Sci 2012;53:4969–74. [DOI] [PubMed] [Google Scholar]
- 43.Yan Q, Ding Y, Liu Y, et al. Genome-Wide analysis of disease progression in age-related macular degeneration. Hum Mol Genet 2018;27:929–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Milder FJ, Gomes L, Schouten A, et al. Factor B structure provides insights into activation of the central protease of the complement system. Nat Struct Mol Biol 2007;14:224–8. [DOI] [PubMed] [Google Scholar]
- 45.Forneris F, Ricklin D, Wu J, et al. Structures of C3b in complex with factors B and D give insight into complement convertase formation. Science 2010;330:1816–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Haac BE, Palmateer NC, Seaton ME, et al. A distinct gut microbiota exists within Crohn’s disease-related perianal fistulae. J Surg Res 2019;242:118–28. [DOI] [PubMed] [Google Scholar]
- 47.McBride WH, Weir DM, Kay AB, et al. Activation of the classical and alternate pathways of complement by Corynebacterium parvum. Clin Exp Immunol 1975;19:143–7. [PMC free article] [PubMed] [Google Scholar]
- 48.Schorlemmer HU, Bitter-Suermann D, Allison AC. Complement activation by the alternative pathway and macrophage enzyme secretion in the pathogenesis of chronic inflammation. Immunology 1977;32:929–40. [PMC free article] [PubMed] [Google Scholar]
- 49.Peyrin-Biroulet L, Loftus EV Jr, Tremaine WJ, et al. Perianal Crohn’s disease findings other than fistulas in a population-based cohort. Inflamm Bowel Dis 2012;18:43–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Alli-Akintade L, Pruthvi P, Hadi N, et al. Race and Fistulizing perianal Crohn’s disease. J Clin Gastroenterol 2015;49:e21–3. [DOI] [PubMed] [Google Scholar]
- 51.Nguyen GC, Torres EA, Regueiro M, et al. Inflammatory bowel disease characteristics among African Americans, Hispanics, and non-Hispanic whites: characterization of a large North American cohort. Am J Gastroenterol 2006;101:1012–23. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are available on reasonable request. Meta-analysis summary statistics will be made available on publication through the NIDDK IBD Genetics Consortium (IBDGC) Data Commons portal currently under development.