Abstract
Background
While the size of chloroplast genomes (cpDNAs) is often influenced by the expansion and contraction of inverted repeat regions and the enrichment of repeats, it is the intergenic spacers (IGSs) that appear to play a pivotal role in determining the size of Pteridaceae cpDNAs. This provides an opportunity to delve into the evolution of chloroplast genomic structures of the Pteridaceae family. This study added five Pteridaceae species, comparing them with 36 published counterparts.
Results
Poor alignment in the non-coding regions of the Pteridaceae family was observed, and this was attributed to the widespread presence of overlong IGSs in Pteridaceae cpDNAs. These overlong IGSs were identified as a major factor influencing variations in cpDNA size. In comparison to non-expanded IGSs, overlong IGSs exhibited significantly higher GC content and were rich in repetitive sequences. Species divergence time estimations suggest that these overlong IGSs may have already existed during the early radiation of the Pteridaceae family.
Conclusions
This study reveals new insights into the genetic variation, evolutionary history, and dynamic changes in the cpDNA structure of the Pteridaceae family, providing a fundamental resource for further exploring its evolutionary research.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12864-024-10296-0.
Keywords: Pteridaceae, Chloroplast, Evolutionary genomics, Structural comparison, Divergence time
Introduction
Ferns are one of the oldest and most primitive vascular plant groups on Earth [1]. They are a group of vascular plants with independent gametophyte and sporophyte generations, mainly undergoing sexual reproduction through spores. Pteridaceae is the second most genera-rich fern family. According to the Pteridophyte Phylogeny Group I (PPG I) classification, Pteridaceae contain five subfamilies, 53 genera, with an estimated 1,211 species contributing to about 10% of extant leptosporangiate fern diversity [2, 3]. These species have multiple values. For example, the Pteris species can accumulate arsenic, which is of great significance for the remediation of heavy metals in soil [4]. Plenty of the Adiantum species can be used in medicine and are used in different parts of the world [5–8]. Pteridaceae species have a cosmopolitan distribution concentrated in wet tropical and arid regions, occupying various ecosystems such as terrestrial, epiphytic, rupestral, and even aquatic [9].
In plants, chloroplasts are the site of photosynthesis and play an important role in the synthesis of defense-related hormones, which sustain life on Earth [10, 11]. Chloroplasts also participate in some metabolic processes [11] and play important roles in plant adaptation to environmental stress [12–14]. Chloroplasts typically possess independent genes and mechanisms for gene expression [15]. The chloroplast genomes (cpDNAs) of land plants are typically 110–160 kb in size [16], usually divided into a large single-copy (LSC) region and a small single-copy (SSC) region by a pair of inverted repeats (IRa and IRb), forming a typical quadripartite structure [17]. The cpDNA is mostly inherited from one parent, its structure is conservative, recombination is less, and the substitution rate is much lower than that of the nuclear genome [18–20]. With the advancement of sequencing technology, cpDNA has become more accessible, therefore, it has become increasingly common to use chloroplasts to explore plant evolutionary events [21, 22].
Comparing complete cpDNAs contributes to the study of mechanisms underlying genome evolution, revealing evolutionary relationships and phylogenies among species. For instance, lycophytes share a similar chloroplast gene order with mosses, while displaying an inverted gene order compared to all other vascular plants, providing evidence for the ancient evolution of early vascular land plants [23]. Similarly, the expansion and contraction of the IR region often serve as evidence for interspecies phylogenetic relationships in chloroplast genome studies [24, 25]. In the context of evolution, the substantial loss of genes was initially linked to endosymbiotic events, but subsequent research indicates that gene loss independently occurred in different lineages [26]. This suggests that a series of complex evolutionary constraints, selection, and convergence led to the conservation of chloroplast genome structure and content. For instance, the evolution of parasitic angiosperms has resulted in the relaxation of evolutionary constraints associated with the maintenance of photosynthetic functionality. Therefore, in the early stages of parasitic evolution, some photosynthetic genes (such as ndh-) were lost, leading to significant changes in chloroplast genome content [27].
Research has shown that the size of cpDNA is often influenced by changes in the IR boundaries [28]. Furthermore, a high number of repetitive sequences in cpDNA has been recognized as a contributing factor to variations in genome size [29–31]. In this study, the variations in sizes of Pteridaceae cpDNAs were ascribed to alterations in the length of overlong intergenic spacers (IGSs), with these IGSs exhibiting species-specific differences. Through sequencing the cpDNAs of five Pteridaceae species and comparing their structures with 36 other reported cpDNAs in the Pteridaceae family, this study aimed to uncover the evolutionary dynamics, genetic variations, and evolutionary relationships of cpDNAs among different species within the Pteridaceae family.
Results
Basic characteristics of Pteridaceae
The sizes of the Pteridaceae cpDNAs in this study ranged from 145,327 bp to 165,631 bp, with a GC content varied from 36.7% to 45.3%. They all possessed typical quadripartite structures, of which the LSC region was 80,810 − 89,030 bp, the SSC region was 19,930 − 27,974 bp, and the IR regions were 42,054 − 61,842 bp (Table 1). The accuracy of gene annotations for these 41 Pteridaceae species was rechecked, using the reference sequences of Adiantum capillus-veneris and Adiantum shastense, and missing annotations were supplemented by conducting local BLAST to retrieve homologous sequences (Figure S1). The statistics of lost chloroplast genes showed that Paragymnopteris bipinnata var. bipinnata and Acrostichum speciosum had relatively more gene losses, with 7 (psbF, rpl2, rpl21, ycf2, ycf12, ycf94, and trnT-UGU) and 9 (psbF, rpl2, rps11, ycf1, ycf2, ycf12, ycf94, trnR-UCG, and trnT-UGU) missing genes, respectively. In addition, trnR-UCG and trnT-UGU were frequently absent in the 41 Pteridaceae cpDNAs, while ycf94 showed a phenomenon of universal loss.
Table 1.
CpDNA features of the 41 Pteridaceae species
| Subfamily | Name | Size (bp) | GC% | Length (bp) | Accession No. | ||
|---|---|---|---|---|---|---|---|
| LSC | IR | SSC | |||||
| Vittarioideae | Adiantum aleuticum | 157,519 | 45.3 | 82,785 | 53,138 | 21,596 | NC_040209.1 |
| Adiantum capillus-veneris | 150,568 | 42.0 | 82,282 | 46,894 | 21,392 | NC_004766.1 | |
| Adiantum flabellulatum | 152,063 | 43.3 | 83,384 | 47,230 | 21,449 | NC_064144.1 | |
| Adiantum malesianum | 154,671 | 42.6 | 89,030 | 44,154 | 21,487 | NC_063331.1 | |
| Adiantum nelumboides | 149,956 | 42.8 | 83,281 | 45,192 | 21,483 | NC_050350.1 | |
| Adiantum reniforme var. sinense | 150,102 | 42.8 | 83,267 | 45,376 | 21,459 | NC_062433.1 | |
| Adiantum shastense | 150,414 | 44.3 | 82,113 | 46,762 | 21,539 | NC_037478.1 | |
| Adiantum tricholepis | 150,470 | 42.5 | 82,606 | 46,403 | 21,461 | NC_040172.1 | |
| Antrophyum semicostatum | 150,274 | 40.1 | 87,392 | 42,054 | 20,828 | NC_040176.1 | |
| Haplopteris elongata | 156,002 | 40.1 | 80,810 | 54,376 | 20,816 | NC_040215.1 | |
| Scoliosorus ensiformis | 145,327 | 40.0 | 82,358 | 42,156 | 20,813 | NC_040218.1 | |
| Vaginularia trichoidea | 147,102 | 39.2 | 84,017 | 43,155 | 19,930 | NC_040175.1 | |
| Vittaria appalachiana | 149,531 | 40.1 | 84,330 | 44,370 | 20,831 | NC_040219.1 | |
| Vittaria graminifolia | 151,035 | 40.1 | 86,058 | 44,132 | 20,845 | NC_040217.1 | |
| Pteridoideae | Gastoniella chaerophylla | 148,099 | 40.3 | 81,915 | 44,646 | 21,538 | NC_040210.1 |
| Onychium japonicum | 150,156 | 41.2 | 82,290 | 46,838 | 21,028 | NC_040205.1 | |
| Pityrogramma trifoliata | 148,156 | 40.0 | 82,321 | 44,930 | 20,905 | NC_040207.1 | |
| Pteris arisanensis | 160,191 | 42.4 | 81,989 | 57,086 | 21,116 | NC_083994.1 | |
| Pteris ensiformis | 148,985 | 41.7 | 81,778 | 46,094 | 21,113 | NC_083995.1 | |
| Pteris multifida | 153,916 | 42.2 | 82,027 | 50,760 | 21,129 | NC_058883.1 | |
| Pteris semipinnata | 162,270 | 42.3 | 81,963 | 59,182 | 21,125 | NC_060734.1 | |
| Pteris vittata | 154,106 | 41.7 | 82,602 | 50,550 | 20,954 | MH173082.1 | |
| Taenitis blechnoides | 157,301 | 40.4 | 88,369 | 47,996 | 20,936 | NC_083996.1 | |
| Tryonia myriophylla | 156,327 | 40.0 | 87,296 | 48,224 | 20,807 | NC_040208.1 | |
| Parkerioideae | Acrostichum speciosum | 156,095 | 38.4 | 84,476 | 49,734 | 21,885 | NC_053768.1 |
| Ceratopteris cornuta | 149,424 | 36.7 | 83,623 | 44,574 | 21,227 | MH173068.1 | |
| Ceratopteris thalictroides | 149,399 | 36.7 | 83,580 | 44,577 | 21,241 | NC_062137.1 | |
| Cryptogrammoideae | Coniogramme intermedia | 153,561 | 45.0 | 82,817 | 49,508 | 21,236 | NC_057002.1 |
| Cryptogramma acrostichoides | 150,162 | 42.3 | 83,763 | 45,231 | 21,168 | NC_040211.1 | |
| Llavea cordifolia | 149,387 | 41.9 | 81,944 | 46,416 | 21,027 | NC_040216.1 | |
| Cheilanthoideae | Bommeria hispida | 156,749 | 42.6 | 82,491 | 46,284 | 27,974 | NC_040206.1 |
| Calciphilopteris ludens | 157,068 | 43.5 | 82,423 | 53,170 | 21,475 | NC_040214.1 | |
| Cheilanthes micropteris | 157,257 | 41.4 | 88,145 | 46,550 | 22,562 | NC_040174.1 | |
| Hemionitis subcordata | 165,631 | 42.8 | 82,607 | 61,842 | 21,182 | NC_040173.1 | |
| Myriopteris covillei | 155,548 | 42.7 | 83,079 | 51,148 | 21,321 | NC_039724.1 | |
| Myriopteris lindheimeri | 155,770 | 42.7 | 83,059 | 51,388 | 21,323 | NC_014592.1 | |
| Myriopteris scabra | 162,051 | 42.1 | 82,874 | 54,230 | 24,947 | NC_040213.1 | |
| Notholaena standleyi | 159,556 | 42.4 | 83,769 | 54,522 | 21,265 | NC_040203.1 | |
| Paragymnopteris bipinnata var. bipinnata | 150,736 | 42.5 | 82,926 | 46,516 | 21,294 | NC_061171.1 | |
| Pellaea truncata | 150,713 | 42.5 | 82,865 | 46,480 | 21,368 | NC_040202.1 | |
| Pentagramma triangularis | 153,445 | 41.8 | 85,668 | 46,763 | 21,014 | NC_040171.1 | |
LSC: Large-single copy, SSC: Small-single copy, IR: Inverted repeat
Sequence variation analysis
Multiple alignments of the 41 Pteridaceae cpDNAs revealed higher divergence in non-coding sequences than in coding regions (Figure S2). Particularly, IGS regions exhibited significant differentiation, while coding regions like matK, cemA, rpoC2 and ycf1 also showed variation. Overall, the IR region of the 41 Pteridaceae cpDNAs had the highest degree of conservation, while the single copy region had less conservation. Nucleotide diversity (Pi) values ranged from 0.006 to 0.376 for common genes and from 0 to 0.603 for common IGS regions. MatK, ndhF, ndhH - rps15, and trnL - ccsA showed notably higher Pi values, indicating substantial single nucleotide polymorphism (Figure S3). These markers could be utilized for distinguishing different species or populations, with matK already recognized as the core DNA barcode for ferns [32].
Repetitive sequence analyses
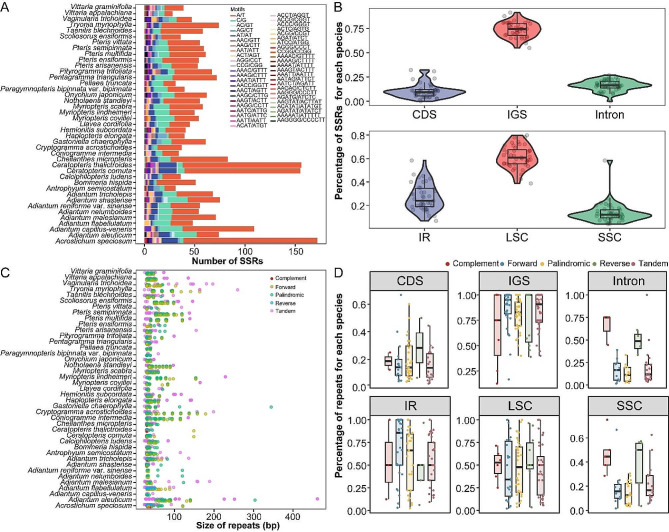
The number of simple sequence repeats (SSRs) in the 41 Pteridaceae cpDNAs ranged from 28 in Vittaria appalachiana to 172 in A. speciosum (Fig. 1A, Table S1). A/T motifs, especially in C. cornuta, A. speciosum, and C. thalictroides, dominated the SSR motifs. Hexanucleotide repeats were the least common, accounting for 0.64% (C. thalictroides) to 4.88% (H. subcordata). SSRs were predominantly located in the LSC region (median: 60.81%), followed by the IR regions (median: 24%) and SSC region (median: 12.05%) (Fig. 1B, Table S1). In comparison to the CDS regions (median: 9.26%) and intron regions (median: 16.36%), most SSRs were found in the IGS regions (median: 74.36%) (Fig. 1B, Table S1).
Fig. 1.
Comparison of repetitive sequences among the 41 Pteridaceae cpDNAs. (A) The number of SSRs among each species. (B) The percentage of SSRs located in different cpDNA regions and gene sequence regions. (C) The size distribution of dispersed repeats and tandem repeats among the 41 Pteridaceae cpDNAs. (D) The percentage of dispersed repeats and tandem repeats located in different cpDNA regions and gene sequence regions
Dispersed repeats, predominantly forward and palindromic, were found in the cpDNAs of all 41 Pteridaceae species, with complement and reverse repeats observed in a few species (Table S2). Tandem repeats were identified in all other species except Adiantum nelumboides and Adiantum reniforme var. sinense (Table S3). Most repeats were within 100 bp, with some exceeding 200 bp (Fig. 1C). The majority of repeats were in the LSC (median: 48.80%) and IR regions (median: 54.21%), compared to the SSC region (median: 7.84%) (Fig. 1D). Additionally, repeats are more prevalent in the IGS regions (median: 86.96%) compared to the CDS (median: 12.01%) and intron regions (median: 4.41%) (Fig. 1D).
Expansion and contraction of IR boundary analysis
The IR/SC boundary genes of the 41 Pteridaceae species had varying degrees of expansion and contraction. No similar patterns were found among the IR/SC boundaries in five different subfamilies (Fig. 2). The genes located at the IR/SC boundaries of these species, primarily included rpl23, trnI-CAU, trnT-UGU, trnR-ACG, ndhF, chlL, trnN-GUU, and ndhB. The IR/SSC boundary genes in these species were consistent, with only slight displacement near the boundary. The main reason for the differences was the inversion of the SSC region. In contrast, the LSC/IR boundary underwent much greater changes, such as trnI-CAU of Adiantum malesianum, Ceratopteris cornuta, Ceratopteris thalictroides and Vaginularia trichoidea all entering the IRb region; while the trnI-CAU of other species was located in the LSC region or on the LSC/IRb boundary. Another reason for differences in IR/LSC boundary genes was the absence of trnT-UGU in some species.
Fig. 2.
Comparison of IR/SC boundaries among the 41 Pteridaceae cpDNAs. The numbers above, below, or adjacent to genes represent gene length or the distances from the front or end of genes to the boundary sites. Figure features are not to scale
The relationship between overlong IGS and CpDNA size
Overlong IGSs seemed to be common in the Pteridaceae cpDNAs, but no reliable patterns of occurrence were found between subfamilies or genera (Fig. 2). In this study, within the same IGS across different species, lengths greater than the mean of these IGSs were defined as overlong IGS. By comparing the length of the positions with overlong IGS in the Pteridaceae cpDNAs (Table 2), it was found that they frequently occurred in the rpoB - trnD region of the LSC and the rps12 - rrn16 region of the IRs. Additionally, in some species, the trnD - trnY (LSC), ndhC- trnV (LSC), psbE - petL (LSC), and rps15 - ycf1 (SSC) IGSs also had longer lengths. Chloroplast genes were usually relatively conservative, so this random change of overlong IGSs was likely the main reason for the difference in cpDNA sizes of Pteridaceae species (Fig. 3A). For instance, Hemionitis subcordata, Myriopteris scabra, and Pteris semipinnata had larger cpDNAs, and their interiors contained overlong IGS regions. The lengths of the LSC, SSC, and IR regions of these species were separately calculated, and it was found that most of the factors contributing to the differences in these region lengths were largely due to the presence of these overlong IGSs (Fig. 3B). Moreover, the length of IGSs in each species was linearly related to their cpDNA sizes (Fig. 3C), and there was a highly significant positive correlation (r = 0.819, p = 5.798e-11 < 0.001).
Table 2.
Comparison of overlong IGSs in the 41 Pteridaceae cpDNAs
| Species | rpoB-trnD (bp) | trnD-trnY (bp) | ndhC-trnV (bp) | psbE-petL (bp) | rps12-rrn16 (bp) | rps15-ycf1 (bp) | cpDNA size |
|---|---|---|---|---|---|---|---|
| Adiantum aleuticum | 932 | 110 | 402 | 566 | 4785 | 305 | 157,519 |
| Adiantum capillus-veneris | 944 | 114 | 448 | 740 | 1600 | 290 | 150,568 |
| Adiantum flabellulatum | 935 | 114 | 380 | 749 | 1593 | 289 | 152,063 |
| Adiantum malesianum | 6877 | 116 | 447 | - | 1265 | 295 | 154,671 |
| Adiantum nelumboides | 833 | 124 | 433 | - | 1260 | 292 | 149,956 |
| Adiantum reniforme var. sinense | 669 | 124 | 433 | - | 1270 | 292 | 150,102 |
| Adiantum shastense | 929 | 119 | 402 | 568 | 1584 | 293 | 150,414 |
| Adiantum tricholepis | 878 | 110 | 405 | 765 | 1378 | 286 | 150,470 |
| Antrophyum semicostatum | 7061 | 92 | - | 664 | - | 279 | 150,274 |
| Haplopteris elongata | 944 | 87 | 419 | 797 | - | 232 | 156,002 |
| Scoliosorus ensiformis | 901 | 99 | - | 757 | - | 208 | 145,327 |
| Vaginularia trichoidea | 6880 | 90 | - | 537 | 1200 | 211 | 147,102 |
| Vittaria appalachiana | 3067 | 95 | - | 723 | - | 201 | 149,531 |
| Vittaria graminifolia | 4817 | 72 | - | 735 | - | 183 | 151,035 |
| Gastoniella chaerophylla | 826 | 99 | 360 | 751 | 1301 | 209 | 148,099 |
| Onychium japonicum | 820 | 106 | 379 | 807 | 3221 | 332 | 150,156 |
| Pityrogramma trifoliata | 831 | 108 | 394 | 751 | 1237 | 273 | 148,156 |
| Pteris arisanensis | 779 | 105 | 493 | 747 | 7391 | 320 | 160,191 |
| Pteris ensiformis | 778 | 105 | 461 | 700 | 1923 | 321 | 148,985 |
| Pteris multifida | 773 | 111 | 518 | 744 | 4076 | 325 | 153,916 |
| Pteris semipinnata | 783 | 105 | 498 | 873 | 8272 | 321 | 162,270 |
| Pteris vittata | 826 | 106 | 454 | 776 | 3549 | 243 | 154,106 |
| Taenitis blechnoides | 7346 | 113 | 530 | 766 | 2617 | 310 | 157,301 |
| Tryonia myriophylla | 584 | 6943 | 493 | 750 | 2564 | 294 | 156,327 |
| Acrostichum speciosum | 443 | 116 | 3953 | 503 | 3730 | - | 156,095 |
| Ceratopteris cornuta | 879 | 134 | 1984 | 492 | 1684 | 322 | 149,424 |
| Ceratopteris thalictroides | 886 | 133 | 1984 | 491 | 1683 | 322 | 149,399 |
| Coniogramme intermedia | 2593 | 105 | 393 | 725 | 1590 | 305 | 153,561 |
| Cryptogramma acrostichoides | 744 | 105 | 397 | 741 | 1241 | 306 | 150,162 |
| Llavea cordifolia | 818 | 103 | 416 | 721 | 1563 | 229 | 149,387 |
| Bommeria hispida | 793 | 118 | 447 | 765 | 1581 | 6872 | 156,749 |
| Calciphilopteris ludens | 846 | 119 | 416 | 752 | 4874 | 301 | 157,068 |
| Cheilanthes micropteris | 875 | 109 | 4760 | 722 | 1568 | 199 | 157,257 |
| Hemionitis subcordata | 851 | 119 | 422 | 758 | 9179 | 304 | 165,631 |
| Myriopteris covillei | 858 | 125 | 440 | 765 | 1570 | 306 | 155,548 |
| Myriopteris lindheimeri | 865 | 121 | 453 | 769 | 1567 | 306 | 155,770 |
| Myriopteris scabra | 849 | 122 | 438 | 761 | 5336 | 3877 | 162,051 |
| Notholaena standleyi | 863 | 113 | 445 | 751 | 6635 | 199 | 159,556 |
| Paragymnopteris bipinnata var. bipinnata | 862 | 133 | - | 771 | 1565 | 303 | 150,736 |
| Pellaea truncata | 862 | 124 | 461 | 694 | 1567 | 305 | 150,713 |
| Pentagramma triangularis | 908 | 114 | 443 | 3890 | 1558 | 194 | 153,445 |
trnD: trnD-GUC; trnY: trnY-GUA; trnV: trnV-UAC. The presence of “-” in the data indicates the presence of corresponding gene loss in cpDNAs. Bold values represents overlong IGS
Fig. 3.
Comparison of chloroplast genome features in the 41 Pteridaceae species. (A) Comparison of the cpDNA sizes with the overlong IGSs; even if the overlong IGS is in the IR regions, the figure only shows the length of one copy of the IGS. (B) Comparison of the lengths of the LSC, SSC, and IR regions; with * indicating regions containing overlong IGS. (C) The correlation between IGS length and cpDNA sizes
Characteristics and analysis of overlong IGSs
This study analyzed the GC content of these sequences and found that the overall GC content of cpDNAs was less affected by these overlong IGSs. However, specific IGSs encompassing both overlong and non-overlong situations exhibited a significant difference (p = 1.189e-11 < 0.001); overlong IGSs tended to exhibit higher GC content (Fig. 4A). These expanded IGSs exhibited collinearity across diverse intergenic regions in various species (Figure S4). Upon conducting homologous sequence alignment of these elongated IGSs in the NCBI database, it was found that the majority of these homologous sequences originated from fern cpDNAs. In certain overlong IGSs, such as rps12-rrn16, alignments were observed with sequences from mitochondrial genomes of Haplopteris ensiformis (Pteridaceae), suggesting that they may transfer within organelles through mechanisms such as gene transfer or horizontal gene transfer. In addition, repetitive sequences and transposable elements located within these IGS were screened. The results of the Mann-Whitney U test revealed that compared to non-overlong IGSs, there were significantly higher numbers of SSRs (p = 0.016), tandem repeats (p = 2.2e-16 < 0.001), and dispersed repeats (p = 2.2e-16 < 0.001) in overlong IGSs (Fig. 4B). Regarding the length relationship between repetitive sequences and IGSs, although not statistically significant, a strong positive correlation was observed between tandem repeats and dispersed repeats with the expansion of the IGSs (r = 0.77 and 0.72, respectively) (Fig. 4C). For transposable elements, relevant sequences could not be retrieved structurally, but similar short fragments of different types of transposable elements were identified in A. malesianum (Gypsy, 48 bp) and Pteris arisanensis (Copia, 65 bp) (Table 3).
Fig. 4.
Comparison of (A) GC content and (B) number of repeats between overlong and non-overlong IGSs. (C) Correlation among SSRs, tandem repeats, dispersed repeats, and the length of overlong IGSs. *p < 0.05; **p < 0.01; ***p < 0.001
Table 3.
The homologous fragments of transposable elements contained in the overlong IGSs
| Species | Overlong IGS | Start-End | Matching repeat | Repeat class/family | Start-End |
|---|---|---|---|---|---|
| Adiantum malesianum | rpoB - trnD | 28,584–28,631 | Gypsy-14_SB-I | LTR/Gypsy | 2773–2821 |
| Pteris arisanensis | rrn16 - rps12 | 93,518–93,582 | Copia-7_Mad-I | LTR/Copia | 6358−6295 |
| Pteris arisanensis | rps12 - rrn16 | 148,599–148,663 | Copia-7_Mad-I | LTR/Copia | 6295–6358 |
Phylogenetic relationship and divergence time estimate
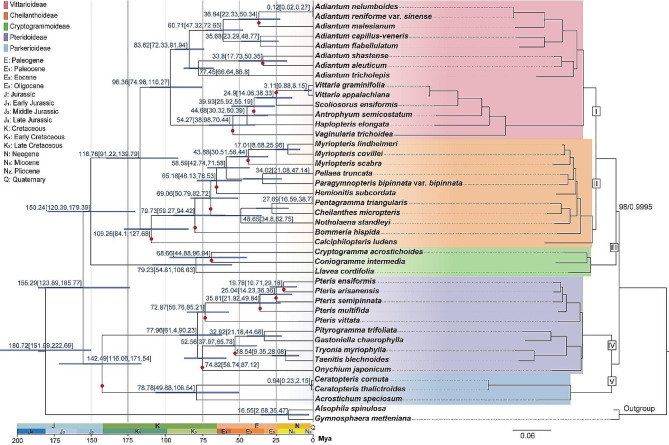
The BI tree and ML tree, constructed using the common protein-coding sequences of all species, were consistent (Fig. 5). Reconstructed phylogenetic relationships received high support, with the lowest node support being 98%. Here, Pteridaceae species were divided into five subfamily clades: clade I (Vittarioideae), clade II (Cheilanthoideae), clade III (Cryptogrammoideae), clade IV (Pteridoideae), and clade V (Parkerioideae). Their divergence from the outgroup could be traced back to the Jurassic period (∼ 180.72 Mya). Clades I, II, and III share a more recent common ancestor, indicating a closer phylogenetic relationship; this common ancestor diverged in the Late Jurassic period, approximately 155.29 Mya. The common ancestor of clades I and II further diverged around 150.24 million years ago in the same period. Additionally, clades I and II diverged during the Early Cretaceous period (∼ 116.76 Mya). Clades IV and V shared a common ancestor dating back to approximately 142.49 Mya, near the Jurassic-Cretaceous (J/K) boundary. The phylogenetic tree strongly supported Pteris and Adiantum as monophyletic clades, and both of their ancestral clades diverged during the Late Cretaceous period, during which most genera of the fern family began to rapidly differentiate. Overlong IGSs were present during the early divergence of the family, and as species rapidly diversified, this type of overlong IGSs gradually became more prevalent.
Fig. 5.
Phylogenetic relationship (right) and divergent time estimate (left) of the 41 Pteridaceae species. The mean divergence time of the nodes is shown next to the nodes while the blue bars correspond to the 95% highest posterior density (HPD). The red dots represent species within the branch that contain overlong IGS. Bootstrap value/posterior probabilities < 100%/1 are displayed on the branches
Discussion
This study sequenced the cpDNA structures of five Pteridaceae species and examined those of 41 species, covering all subfamilies. They exhibited typical quadripartite structures, with genome sizes ranging from 145,327 bp to 165,631 bp and GC contents between 36.7% and 45.3%. Upon re-aligning and completing missing gene annotations, higher gene losses were observed in P. bipinnata var. bipinnata (7 chloroplast genes lost) and A. speciosum (9 lost) among the Pteridaceae cpDNAs. Additionally, trnR-UCG and trnT-UGU were frequently lost among the 41 Pteridaceae cpDNAs, along with a common loss of ycf94. The alignment of all 41 Pteridaceae cpDNAs revealed poor alignment in non-coding regions, especially in the IGS regions. Four regions with significantly higher Pi values compared to other genes/IGSs were identified: matK, ndhF, ndhH - rps15, and trnL - ccsA. Among these, matK has been used as a core DNA barcode for ferns, and the other three markers may serve as candidate DNA barcodes for species within the Pteridaceae family.
Repetitive sequences can be dispersed widely or found in simple tandem arrays. SSRs, also known as microsatellites, consist of 1–6 nucleotide tandem repeat motifs and are distributed throughout the genome [33, 34]. SSRs are highly polymorphic and specific, making them valuable for studying molecular evolution, genetic diversity, and developing molecular markers [35, 36]. The diversity in repeat length, copy number, and distribution within species is attributed to slipped-strand mispairing during DNA replication on a single strand [37, 38]. Mononucleotide repeats, especially A/T motifs, were the most common in this study. A potential reason for the higher frequency of A/T repeats is that during chloroplast genome replication, the separation of AT strands is relatively easier compared to GC, which increases slip mismatching [39]. In the Pteridaceae cpDNAs, SSRs were mainly located in the LSC region (38.71–79.73%) and tended to occur in IGS (54.84–91.18%), possibly due to stronger constraints in coding regions [40]. Short repeat units can also be further extended into longer tandem repeats through slipped-strand mispairing or recombination [41–43], with the number of tandem repeats varying due to susceptibility to slippage events during DNA replication [44]. Dispersed repeats are often associated with and contribute significantly to the chloroplast genome rearrangement in plants [25, 45]. Here, all species except A. nelumboides and A. reniforme var. sinense exhibited tandem and dispersed repeats, primarily less than 100 bp in size, with forward and palindromic repeat motifs predominating. Furthermore, these repeats were more prevalent in the IGS regions.
The substitution rate in chloroplast IR region genes is significantly lower than that in the SC region, thus greater conservation in the IR region [46]. However, structural variations in the IR/SC boundary regions are still common [47–49]. Among the 41 Pteridaceae cpDNAs, varying degrees of IR/SC boundary expansion and contraction were observed, even within the same genus. The IR/SSC boundary genes remained consistent in the Pteridaceae cpDNAs, with differences primarily attributed to SSC region inversions. In contrast, the LSC/IR boundary varied more due to changes in the trnI-CAU position and the absence of trnT-UGU in some species. The variation in cpDNA size is often associated with changes in the IR/SC boundary [50–52] and the expansion of repetitive sequences [30, 31]. In this study, the movement in the IR/SC boundary genes of Pteridaceae cpDNA only led to minor differences in cpDNA size. For instance, the cpDNA size of H. subcordata was the largest, with a longer IR region due to the expansion of the rps12 - rrn16 IGS, rather than significant expansion of its IR/SC boundaries. A strong correlation between cpDNAs and IGSs was observed, and there was a common occurrence of overlong IGSs in species within this family. These overlong IGSs consistently aligned with the changes in the Pteridaceae cpDNA size, implying their primary influence on cpDNA size and their potential role in driving cpDNA structure evolution [53]. In cases like A. malesianum, overlong IGS amplifies cpDNA size and triggers sequential movement of LSC region genes, affecting IR/SC boundaries [48].
The overlong IGSs prevalent in the Pteridaceae family were found in various intergenic regions across different species and showed a degree of collinearity (Figure S4). Mobile elements are present in the fern cpDNAs and are often found near genome inversion sites [53]. In this study, only a few inversions occurred in the Pteridaceae cpDNAs, such as the ndhJ - psbE, the rrn5 - rrn16, and the SSC region. Some overlong IGSs were also found near inversion sites, such as rrn16 - rps12, which may have served as hotspots for IGS expansion. Additionally, the psbE - petL IGS of Pentagramma triangularis also underwent expansion. Within the same IGS, the GC content of overlong IGSs that underwent expansion was consistently higher, showing a significant difference compared to non-overlong IGSs. An important characteristic of GC base pairs is their higher thermal stability compared to AT base pairs [54]. These interactions appear to be crucial for the overall structural stability of DNA and RNA transcripts [55, 56]. Significant differences in GC content exist among different genomes and within different regions of genomes. Some studies suggest a correlation between GC content and the length of coding genes, where the length of exons often increases with higher GC content [57]. This is because stop codons are rich in AT, consequently resulting in a lower frequency of stop codon occurrence in GC-rich exons [58]. The increase in GC content may also be attributed to the presence of more GC-rich sequences within these overlong IGSs, such as repetitive sequences. In the Pteridaceae cpDNAs, the overlong IGSs contained significantly more repetitive sequences, especially tandem repeats and dispersed repeats; meanwhile, these repeats had a strong positive correlation with the expansion of IGSs (r = 0.77 and 0.72, respectively), although statistical significance was not achieved. This suggests that repetitive sequences may promote the occurrence of chloroplast genome structural variation (SV). For instance, the location of SV in the Carex cpDNAs is closely related to the location of long repeats [59]. The amplification of the Cyripedium cpDNAs is associated with a surge in AT-biased repeats [30]. In addition, similar fragments were observed in the mitochondrial genomes of H. ensiformis and detected transposable element-like fragments in a few species, suggesting that they may transfer among different organelles.
According to this study, the Pteridaceae family had clear boundaries in both subfamilies and genera. The Pteris and Adiantum were both monophyletic, consistent with previous research [2, 60–62]. Based on fossil evidence, ferns are believed to have originated in the Devonian period [63], and their dominance continued into the Paleozoic era [64]. Here, the MCMCTree model suggested that the divergence of the Pteridaceae family from the outgroup occurred during the Jurassic period (∼ 180.72 Mya). Fossil records from the Jurassic period indicate significant fern evolution [65, 66], with favorable climate and environmental conditions contributing to their survival and reproduction during this time. As a result, ferns occupied a crucial ecological niche on Earth during this period, evolving a wide range of morphological and ecological characteristics that had a significant impact on the evolution and diversity of terrestrial ecosystems [67]. The J/K boundary period represents a time of environmental upheaval, characterized by intense transgressive phases due to rapidly changing sea levels [68]. The subfamily of Parkerioideae represents an aquatic branch, with its species thriving in wet aquatic environments [69, 70], diverged around the J/K boundary period (∼ 142.49 Mya) and possibly underwent adaptive evolution. In addition, the divergence of the genus Acrostichum, within Parkerioideae, occurred around the Late Cretaceous (∼ 78.78 Mya), overlapping with the fossil record of this genus [71]. During the Late Cretaceous period, the genus of this family began to rapidly diverge. Overlong IGSs were present during the early divergence stages of the Pteridaceae family, indicating that this structural feature may have an ancient origin in related species. As species rapidly diversified, the prevalence of these overlong IGSs gradually increased.
Conclusion
This study offers comprehensive insights into Pteridaceae cpDNAs. Chloroplast gene numbers were mostly stable, except for P. bipinnata var. bipinnata and A. speciosum. Changes in LSC/IR boundaries resulted from trnI-CAU movement and trnT-UGU deletion. SSC/IR boundary shifts were mainly due to SSC region inversion. The Pteridaceae cpDNAs often had overlong IGSs, increasing non-coding region variability and affecting cpDNA size changes. These overlong IGSs had higher GC content and were rich in repetitive sequences. Divergence time analysis traced Pteridaceae separation to the Jurassic (∼ 180.72 Mya), with rapid diversification within the genera beginning in the Late Cretaceous period. Additionally, overlong IGSs may have already existed during the early differentiation stages of this family. This study provides further theoretical support for the classification of Pteridaceae species, genetic diversity, and the evolution of genomic structure.
Materials and methods
Plant materials, DNA extraction and De Novo sequencing
Obtaining a more complete chloroplast genome helps to understand their structural evolution. This study added cpDNAs of species Pteris ensiformis, P. arisanensis, Taenitis blechnoides, Adiantum flabellulatum and A. malesianum. Fresh leaves of the first three were sampled from the campus of Shenzhen Fairy Lake Botanical Garden [72]. Fresh leaves of the latter two were sampled from the campus of South China Agricultural University (SCAU) [48]. The plant materials used in the study were identified by Ting Wang and deposited in the Herbarium of SCAU with specimen numbers GXL20210901 (A. flabellulatum), GXL20210902 (A. malesianum), GXL20210903 (P. ensiformis), GXL20210904 (P. arisanensis), and GXL20210905 (T. blechnoides). DNA was extracted from the samples using a Tiangen Plant Genome DNA Kit (Tiangen Biotech Co., Ltd., Beijing, China) according to the manufacturer’s instructions. The Illumina NovaSeq6000 platform was used for sequencing.
Sequence assembly and annotation
The complete cpDNAs were assembled using GetOrganelle [73] and Novoplasty [74]. NUMER [75] was used to check their collinearity. The cpDNAs were annotated by GeSeq [76] with A. capillus-veneris as the reference, and manually corrected. The cpDNAs were submitted to NCBI (National Center for Biotechnology Information) under GenBank accession numbers NC_083994.1 (P. arisanensis), NC_083995.1 (P. ensiformis), NC_083996.1 (T. blechnoides), NC_064144.1 (A. flabellulatum), and NC_063331.1 (A. malesianum).
Comparative genome and boundary regions analysis
The complete cpDNAs of 36 Pteridaceae species were downloaded from GenBank (Table 1). Combining our five sequenced species, a total of 41 Pteridaceae species were examined, covering all subfamilies. The accuracy of gene annotation for these 41 Pteridaceae cpDNAs was rechecked, and local BLAST was used for homologous sequence retrieval to complete some missing annotation genes (Figure S1). The boundary region of the 41 Pteridaceae cpDNAs was rechecked using Geneious [77] and displayed using Adobe Illustrator 2020, to better observe the expansion/contraction of IR regions.
Repetitive sequence analyses
The Perl script MISA (http://pgrc.ipk-gatersleben.de/misa/) [78] was used with the filter thresholds set to detect SSRs. The following parameters were set: a minimal repeat number of 10 for mononucleotide repeats, 5 for di-, 4 for tri-, and 3 for tetra-, penta-, and hexanucleotide SSRs. Tandem repeats were found with the tandem repeats finder (TRF) using default parameters [79]. To identify complex repetitive sequences such as forward, reverse, complement and palindromic, REPuter online software [80] was used with a minimum repeat size of 30 bp and 90% sequence identity (Hamming distance of 3). The transposable elements were retrieved using RepeatMasker [81], with the rmblast database selected and the reference species aligned to “viridiplantae”, with all other parameters set to default.
Sequence divergence analysis
The comparative analysis was carried out by using the shuffle-LAGAN mode in mVISTA online tool [82] to analyze the cpDNA divergence of the 41 Pteridaceae species, with A. capillus-veneris (NC_004766.1) as a reference. Extracted the common genes and IGSs of these cpDNAs as independent datasets, aligned each dataset in MAFFT v7.475 using default parameters [83], and calculated nucleotide diversity in DnaSP v6.0 [84].
Phylogenetic analysis and divergence time estimates
Phylogenetic reconstruction of the above 41 Pteridaceae species with Gymnosphaera metteniana and Alsophila spinulosa as outgroups. 76 common but not repetitive protein-coding sequences of these species were retained, and MAFFT was used to perform sequence alignment, and remove 90% of the gaps in each multi-alignment sequence using trimAl [85]. PhyloSuite [86] was used to concatenate these sequences into a dataset for phylogenetic analysis. The ML tree was inferred using RAxML [87], GTRGAMMAI was selected as the nucleotide substitution model. The Bayesian inference (BI) tree was established by MrBayes [88] and was estimated by running 2,000,000 generations (Nst = 6, rates = invgamma).
In this study, the differentiation time estimated by TimeTree [89] is used to calibrate the time tree (A. aleuticum & Calciphilopteris ludens: 57.8–129.7 Mya; A. speciosum & A. spinulosa: 154.7–228.8 Mya; Pteris multifida & Pteris vittate: 51.3–89 Mya; A. nelumboides & Adiantum tricholepis: 34.8–88.4 Mya). Inferring the time tree of Pteridaceae using the MCMCTree software package of PAML [90], the model was set to GTR, and the MCMC procedures had a burn-in of 2,000 iterations and then ran for 20,000 iterations. MCMCTree analysis was performed twice, which generated similar results, confirming the robustness of the analysis. The final tree was visualized and edited in FigTree v.1.4.3 (http://tree.bio.ed.ac.uk/software/figtree/).
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The plant materials in this study are from cultivated plants, and leaf collection has been approved by Shenzhen Fairy Lake Botanical Garden and South China Agricultural University.
Author contributions
The study was conceptualized by X.G. and T.W. Data analyses, visualization and curation were conducted by X.G. Sample collection was conducted by X.G., X.Z. and L.L. Funding and supervision were contributed by Y.S. and T.W. X.G. wrote the manuscript together with Y.S. and T.W. All authors contributed to writing the manuscript.
Data availability
All data generated or analysed during this study are included in this published article and its supplementary information files.
Declarations
Appropriate permissions and/or licences for collection of plant or seed specimens
The plant materials in this study are from cultivated plants, and leaf collection has been approved by Shenzhen Fairy Lake Botanical Garden and South China Agricultural University.
Ethics approval and consent to participate
The authors declare that the collection of plant materials for this study complies with relevant institutional, national and international guidelines and legislation.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Yingjuan Su, Email: suyj@mail.sysu.edu.cn.
Ting Wang, Email: tingwang@scau.edu.cn.
References
- 1.Rastogi S, Pandey MM, Rawat A. Ethnopharmacological uses, phytochemistry and pharmacology of genus Adiantum: a comprehensive review. J ETHNOPHARMACOL. 2018;215:101–19. doi: 10.1016/j.jep.2017.12.034. [DOI] [PubMed] [Google Scholar]
- 2.Zhang L, Zhou XM, Lu NT, Zhang LB. Phylogeny of the fern subfamily Pteridoideae (Pteridaceae; Pteridophyta), with the description of a new genus: Gastoniella. MOL PHYLOGENET EVOL. 2017;109:59–72. doi: 10.1016/j.ympev.2016.12.037. [DOI] [PubMed] [Google Scholar]
- 3.Schuettpelz E, Schneider H, Smith AR, Hovenkamp P, Prado J, Rouhan G, Salino A, Sundue M. A community-derived classification for extant lycophytes and ferns. J SYST EVOL. 2016;54(6):563–603. doi: 10.1111/jse.12229. [DOI] [Google Scholar]
- 4.Kohda YH, Endo G, Kitajima N, Sugawara K, Chien MF, Inoue C, Miyauchi K. Arsenic uptake by Pteris vittata in a subarctic arsenic-contaminated agricultural field in Japan: an 8-year study. SCI TOTAL ENVIRON. 2022;831:154830. doi: 10.1016/j.scitotenv.2022.154830. [DOI] [PubMed] [Google Scholar]
- 5.Singh M, Singh N, Khare PB, Rawat AK. Antimicrobial activity of some important Adiantum species used traditionally in indigenous systems of medicine. J ETHNOPHARMACOL. 2008;115(2):327–9. doi: 10.1016/j.jep.2007.09.018. [DOI] [PubMed] [Google Scholar]
- 6.Kasabri V, Al-Hallaq EK, Bustanji YK, Abdul-Razzak KK, Abaza IF, Afifi FU. Antiobesity and antihyperglycaemic effects of Adiantum capillus-veneris extracts: in vitro and in vivo evaluations. PHARM BIOL. 2017;55(1):164–72. doi: 10.1080/13880209.2016.1233567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoseinifar SH, Jahazi MA, Mohseni R, Raeisi M, Bayani M, Mazandarani M, Yousefi M, Van Doan H, Torfi MM. Effects of dietary fern (Adiantum capillus-veneris) leaves powder on serum and mucus antioxidant defence, immunological responses, antimicrobial activity and growth performance of common carp (Cyprinus carpio) juveniles. FISH SHELLFISH IMMUNOL. 2020;106:959–66. doi: 10.1016/j.fsi.2020.09.001. [DOI] [PubMed] [Google Scholar]
- 8.Nonato FR, Nogueira TM, Barros TA, Lucchese AM, Oliveira CE, Santos RR, Soares MB, Villarreal CF. Antinociceptive and antiinflammatory activities of Adiantum latifolium Lam.: evidence for a role of IL-1beta inhibition. J ETHNOPHARMACOL. 2011;136(3):518–24. doi: 10.1016/j.jep.2010.05.065. [DOI] [PubMed] [Google Scholar]
- 9.Schuettpelz E, Schneider H, Huiet L, Windham MD, Pryer KM. A molecular phylogeny of the fern family Pteridaceae: assessing overall relationships and the affinities of previously unsampled genera. MOL PHYLOGENET EVOL. 2007;44(3):1172–85. doi: 10.1016/j.ympev.2007.04.011. [DOI] [PubMed] [Google Scholar]
- 10.Lu Y, Yao J. Chloroplasts at the crossroad of photosynthesis, pathogen infection and plant defense. INT J MOL SCI 2018, 19(12). [DOI] [PMC free article] [PubMed]
- 11.Daniell H, Lin CS, Yu M, Chang WJ. Chloroplast genomes: diversity, evolution, and applications in genetic engineering. GENOME BIOL. 2016;17(1):134. doi: 10.1186/s13059-016-1004-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khan M, Nawaz N, Ali I, Azam M, Rizwan M, Ahmad P, Ali S. Regulation of photosynthesis under metal stress. PHOTOSYNTHESIS PRODUCTIVITY Environ STRESS 2019:95–105.
- 13.Luo S, Kim C. Current understanding of temperature stress-responsive chloroplast FtsH metalloproteases. INT J MOL SCI. 2021;22(22):12106. doi: 10.3390/ijms222212106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao C, Haigh AM, Holford P, Chen ZH. Roles of chloroplast retrograde signals and ion transport in plant drought tolerance. INT J MOL SCI. 2018;19(4):963. doi: 10.3390/ijms19040963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Song Y, Feng L, Alyafei M, Jaleel A, Ren M. Function of chloroplasts in plant stress responses. INT J MOL SCI 2021, 22(24). [DOI] [PMC free article] [PubMed]
- 16.Wicke S, Schneeweiss GM, DePamphilis CW, Muller KF, Quandt D. The evolution of the plastid chromosome in land plants: gene content, gene order, gene function. PLANT MOL BIOL. 2011;76(3–5):273–97. doi: 10.1007/s11103-011-9762-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu XF, Zhu GF, Li DM, Wang XJ. Complete chloroplast genome sequence and phylogenetic analysis of Spathiphyllum ‘Parrish’. PLoS ONE. 2019;14(10):e224038. doi: 10.1371/journal.pone.0224038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith DR. Mutation rates in plastid genomes: they are lower than you might think. GENOME BIOL EVOL. 2015;7(5):1227–34. doi: 10.1093/gbe/evv069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wolfe KH, Li WH, Sharp PM. Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. PROC NATL ACAD SCI U S A. 1987;84(24):9054–8. doi: 10.1073/pnas.84.24.9054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Drouin G, Daoud H, Xia J. Relative rates of synonymous substitutions in the mitochondrial, chloroplast and nuclear genomes of seed plants. MOL PHYLOGENET EVOL. 2008;49(3):827–31. doi: 10.1016/j.ympev.2008.09.009. [DOI] [PubMed] [Google Scholar]
- 21.Dong W, Liu Y, Xu C, Gao Y, Yuan Q, Suo Z, Zhang Z, Sun J. Chloroplast phylogenomic insights into the evolution of Distylium (Hamamelidaceae) BMC Genomics. 2021;22(1):293. doi: 10.1186/s12864-021-07590-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu Q, Li X, Li M, Xu W, Schwarzacher T, Heslop-Harrison JS. Comparative chloroplast genome analyses of Avena: insights into evolutionary dynamics and phylogeny. BMC PLANT BIOL. 2020;20(1):406. doi: 10.1186/s12870-020-02621-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Raubeson LA, Jansen RK. Chloroplast DNA evidence on the ancient evolutionary split in vascular land plants. Science. 1992;255(5052):1697–9. doi: 10.1126/science.255.5052.1697. [DOI] [PubMed] [Google Scholar]
- 24.Wu CS, Wang YN, Liu SM, Chaw SM. Chloroplast genome (cpDNA) of Cycas taitungensis and 56 cp protein-coding genes of Gnetum parvifolium: insights into cpDNA evolution and phylogeny of extant seed plants. MOL BIOL EVOL. 2007;24(6):1366–79. doi: 10.1093/molbev/msm059. [DOI] [PubMed] [Google Scholar]
- 25.Wang RJ, Cheng CL, Chang CC, Wu CL, Su TM, Chaw SM. Dynamics and evolution of the inverted repeat-large single copy junctions in the chloroplast genomes of monocots. BMC EVOL BIOL. 2008;8:36. doi: 10.1186/1471-2148-8-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martin W, Stoebe B, Goremykin V, Hansmann S, Hasegawa M, Kowallik KV. Gene transfer to the nucleus and the evolution of chloroplasts. Nature. 1998;393:162–5. doi: 10.1038/30234. [DOI] [PubMed] [Google Scholar]
- 27.Bungard RA. Photosynthetic evolution in parasitic plants: insight from the chloroplast genome. BioEssays. 2004;26(3):235–47. doi: 10.1002/bies.10405. [DOI] [PubMed] [Google Scholar]
- 28.Goulding SE, Olmstead RG, Morden CW, Wolfe KH. Ebb and flow of the chloroplast inverted repeat. MOL GEN GENET. 1996;252(1–2):195–206. doi: 10.1007/BF02173220. [DOI] [PubMed] [Google Scholar]
- 29.Sawicki J, Bączkiewicz A, Buczkowska K, Górski P, Krawczyk K, Mizia P, Myszczyński K, Ślipiko M, Szczecińska M. The increase of simple sequence repeats during diversification of Marchantiidae, an early land plant lineage, leads to the first known expansion of inverted repeats in the evolutionarily-stable structure of liverwort plastomes. GENES-BASEL. 2020;11(3):299. doi: 10.3390/genes11030299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guo YY, Yang JX, Li HK, Zhao HS. Chloroplast genomes of two species of cypripedium: expanded genome size and proliferation of AT-biased repeat sequences. FRONT PLANT SCI. 2021;12:609729. doi: 10.3389/fpls.2021.609729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu S, Wang Z, Su Y, Wang T. Comparative genomic analysis of Polypodiaceae chloroplasts reveals fine structural features and dynamic insertion sequences. BMC PLANT BIOL. 2021;21(1):31. doi: 10.1186/s12870-020-02800-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuo LY, Li FW, Chiou WL, Wang CN. First insights into fern matK phylogeny. MOL PHYLOGENET EVOL. 2011;59(3):556–66. doi: 10.1016/j.ympev.2011.03.010. [DOI] [PubMed] [Google Scholar]
- 33.Bhattarai G, Shi A, Kandel DR, Solis-Gracia N, Da SJ, Avila CA. Genome-wide simple sequence repeats (SSR) markers discovered from whole-genome sequence comparisons of multiple spinach accessions. Sci Rep. 2021;11(1):9999. doi: 10.1038/s41598-021-89473-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li YC, Korol AB, Fahima T, Beiles A, Nevo E. Microsatellites: genomic distribution, putative functions and mutational mechanisms: a review. MOL ECOL. 2002;11(12):2453–65. doi: 10.1046/j.1365-294X.2002.01643.x. [DOI] [PubMed] [Google Scholar]
- 35.Thakur O, Randhawa GS. Identification and characterization of SSR, SNP and InDel molecular markers from RNA-Seq data of guar (Cyamopsis tetragonoloba, L. Taub.) Roots. BMC Genomics. 2018;19(1):951. doi: 10.1186/s12864-018-5205-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhu M, Feng P, Ping J, Li J, Su Y, Wang T. Phylogenetic significance of the characteristics of simple sequence repeats at the genus level based on the complete chloroplast genome sequences of Cyatheaceae. ECOL EVOL. 2021;11(20):14327–40. doi: 10.1002/ece3.8151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gandhi SG, Awasthi P, Bedi YS. Analysis of SSR dynamics in chloroplast genomes of Brassicaceae family. BIOINFORMATION. 2010;5(1):16–20. doi: 10.6026/97320630005016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ochoterena H. Homology in coding and non-coding DNA sequences: a parsimony perspective. PLANT SYST EVOL. 2009;282(3–4):151–68. doi: 10.1007/s00606-008-0095-y. [DOI] [Google Scholar]
- 39.George B, Bhatt BS, Awasthi M, George B, Singh AK. Comparative analysis of microsatellites in chloroplast genomes of lower and higher plants. CURR GENET. 2015;61(4):665–77. doi: 10.1007/s00294-015-0495-9. [DOI] [PubMed] [Google Scholar]
- 40.Kelchner SA, Wendel JF. Hairpins create minute inversions in non-coding regions of chloroplast DNA. CURR GENET. 1996;30(3):259–62. doi: 10.1007/s002940050130. [DOI] [PubMed] [Google Scholar]
- 41.Macas J, Koblížková A, Navrátilová A, Neumann P. Hypervariable 3’ UTR region of plant LTR-retrotransposons as a source of novel satellite repeats. Gene. 2009;448(2):198–206. doi: 10.1016/j.gene.2009.06.014. [DOI] [PubMed] [Google Scholar]
- 42.Ellegren H. Microsatellites: simple sequences with complex evolution. NAT REV GENET. 2004;5(6):435–45. doi: 10.1038/nrg1348. [DOI] [PubMed] [Google Scholar]
- 43.Levinson G, Gutman GA. Slipped-strand mispairing: a major mechanism for DNA sequence evolution. MOL BIOL EVOL. 1987;4(3):203–21. doi: 10.1093/oxfordjournals.molbev.a040442. [DOI] [PubMed] [Google Scholar]
- 44.Zhu L, Wu H, Li H, Tang H, Zhang L, Xu H, Jiao F, Wang N, Yang L. Short tandem repeats in plants: genomic distribution and function prediction. ELECTRON J BIOTECHN. 2021;50:37–44. doi: 10.1016/j.ejbt.2020.12.003. [DOI] [Google Scholar]
- 45.Maul JE, Lilly JW, Cui L, DePamphilis CW, Miller W, Harris EH, Stern DB. The Chlamydomonas reinhardtii Plastid chromosome: islands of genes in a sea of repeats. Plant Cell. 2002;14(11):2659–79. doi: 10.1105/tpc.006155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Perry AS, Wolfe KH. Nucleotide substitution rates in legume chloroplast DNA depend on the presence of the inverted repeat. J MOL EVOL. 2002;55(5):501–8. doi: 10.1007/s00239-002-2333-y. [DOI] [PubMed] [Google Scholar]
- 47.Guo YY, Yang JX, Bai MZ, Zhang GQ, Liu ZJ. The chloroplast genome evolution of Venus slipper (Paphiopedilum): IR expansion, SSC contraction, and highly rearranged SSC regions. BMC PLANT BIOL. 2021;21(1):248. doi: 10.1186/s12870-021-03053-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gu X, Zhu M, Su Y, Wang T. A large intergenic spacer leads to the increase in genome size and sequential gene movement around IR/SC boundaries in the chloroplast genome of Adiantum malesianum (Pteridaceae) INT J MOL SCI. 2022;23(24):15616. doi: 10.3390/ijms232415616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wei N, Perez-Escobar OA, Musili PM, Huang WC, Yang JB, Hu AQ, Hu GW, Grace OM, Wang QF. Plastome evolution in the hyperdiverse genus Euphorbia (Euphorbiaceae) using phylogenomic and comparative analyses: large-scale expansion and contraction of the inverted repeat region. FRONT PLANT SCI. 2021;12:712064. doi: 10.3389/fpls.2021.712064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chumley TW, Palmer JD, Mower JP, Fourcade HM, Calie PJ, Boore JL, Jansen RK. The complete chloroplast genome sequence of Pelargonium x hortorum: organization and evolution of the largest and most highly rearranged chloroplast genome of land plants. MOL BIOL EVOL. 2006;23(11):2175–90. doi: 10.1093/molbev/msl089. [DOI] [PubMed] [Google Scholar]
- 51.Guisinger MM, Kuehl JV, Boore JL, Jansen RK. Extreme reconfiguration of plastid genomes in the angiosperm family Geraniaceae: rearrangements, repeats, and codon usage. MOL BIOL EVOL. 2011;28(1):583–600. doi: 10.1093/molbev/msq229. [DOI] [PubMed] [Google Scholar]
- 52.Park S, An B, Park S. Reconfiguration of the plastid genome in Lamprocapnos spectabilis: IR boundary shifting, inversion, and intraspecific variation. SCI REP 2018, 8(1). [DOI] [PMC free article] [PubMed]
- 53.Robison TA, Grusz AL, Wolf PG, Mower JP, Fauskee BD, Sosa K, Schuettpelz E. Mobile elements shape plastome evolution in ferns. GENOME BIOL EVOL. 2018;10(10):2558–71. doi: 10.1093/gbe/evy189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yakovchuk P, Protozanova E, Frank-Kamenetskii MD. Base-stacking and base-pairing contributions into thermal stability of the DNA double helix. NUCLEIC ACIDS RES. 2006;34(2):564–74. doi: 10.1093/nar/gkj454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Correlation between nucleotide composition and folding energy of coding sequences with special attention to wobble bases. THEOR BIOL MED MODEL 2008, 5:14. [DOI] [PMC free article] [PubMed]
- 56.Šmarda P, Bures P. The variation of base composition in plant genomes. Springer Vienna; 2012.
- 57.Xia X, Xie Z, Li WH. Effects of GC content and mutational pressure on the lengths of exons and coding sequences. J MOL EVOL. 2003;56(3):362–70. doi: 10.1007/s00239-002-2406-1. [DOI] [PubMed] [Google Scholar]
- 58.Oliver JL, Marin A. A relationship between GC content and coding-sequence length. J MOL EVOL. 1996;43(3):216–23. doi: 10.1007/BF02338829. [DOI] [PubMed] [Google Scholar]
- 59.Xu S, Teng K, Zhang H, Gao K, Wu J, Duan L, Yue Y, Fan X. Chloroplast genomes of four Carex species: long repetitive sequences trigger dramatic changes in chloroplast genome structure. FRONT PLANT SCI. 2023;14:1100876. doi: 10.3389/fpls.2023.1100876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lu JM, Wen J, Lutz S, Wang YP, Li DZ. Phylogenetic relationships of Chinese Adiantum based on five plastid markers. J PLANT RES. 2012;125(2):237–49. doi: 10.1007/s10265-011-0441-y. [DOI] [PubMed] [Google Scholar]
- 61.Schuettpelz E, Davila A, Prado J, Hirai RY, Yatskievych G. Molecular phylogenetic and morphological affinities of Adiantum senae (Pteridaceae). Taxon. 2014;63(2):258–64.
- 62.Zhang L, Zhang LB. Phylogeny and systematics of the brake fern genus Pteris (Pteridaceae) based on molecular (plastid and nuclear) and morphological evidence. MOL PHYLOGENET EVOL. 2018;118:265–85. doi: 10.1016/j.ympev.2017.09.011. [DOI] [PubMed] [Google Scholar]
- 63.Kenrick P, Crane PR. The origin and early evolution of plants on land. Nature. 1997;389:33–9. doi: 10.1038/37918. [DOI] [Google Scholar]
- 64.Niklas KJ, Tiffney BH, Knoll AH. Patterns in vascular land plant diversification. Nature. 1983;303(5918):614–6. doi: 10.1038/303614a0. [DOI] [Google Scholar]
- 65.Kawai H, Kanegae T, Christensen S, Kiyosue T, Sato Y, Imaizumi T, Kadota A, Wada M. Responses of ferns to red light are mediated by an unconventional photoreceptor. Nature. 2003;421(6920):287–90. doi: 10.1038/nature01310. [DOI] [PubMed] [Google Scholar]
- 66.Taylor EL, Taylor TN. The biology and evolution of fossil plants. Englewood Cliffs, N.J: Prentice Hall; 1993. [Google Scholar]
- 67.Tidwell WD, Ash SR. A review of selected triassic to early cretaceous ferns. J PLANT RES 1994(107):417–42.
- 68.Tennant JP, Mannion PD, Upchurch P, Sutton MD, Price GD. Biotic and environmental dynamics through the late jurassic-early cretaceous transition: evidence for protracted faunal and ecological turnover. BIOL REV CAMB PHILOS SOC. 2017;92(2):776–814. doi: 10.1111/brv.12255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang Z, He Z, Xu S, Li X, Guo W, Yang Y, Zhong C, Zhou R, Shi S. Transcriptome analyses provide insights into the phylogeny and adaptive evolution of the mangrove fern genus Acrostichum. SCI REP. 2016;6:35634. doi: 10.1038/srep35634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lloyd RM. Systematics of the genus Ceratopteris (Parkeriaceae), I. sexual and vegetative reproduction in hawaiian Ceratopteris thalictroides. AM FERN J. 1973;69(1):12–8. doi: 10.2307/1546563. [DOI] [Google Scholar]
- 71.Bonde SD, Kumaran KPN. The oldest macrofossil record of the mangrove fern Acrostichum L. from the late cretaceous Deccan Intertrappean beds of India. Cretac RES. 2002;23(1):149–52. doi: 10.1006/cres.2001.0307. [DOI] [Google Scholar]
- 72.Gu X, Li L, Li S, Shi W, Zhong X, Su Y, Wang T. Adaptive evolution and co-evolution of chloroplast genomes in Pteridaceae species occupying different habitats: overlapping residues are always highly mutated. BMC PLANT BIOL. 2023;23(1):511. doi: 10.1186/s12870-023-04523-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jin JJ, Yu WB, Yang JB, Song Y, DePamphilis CW, Yi TS, Li DZ. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. GENOME BIOL. 2020;21(1):241. doi: 10.1186/s13059-020-02154-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dierckxsens N, Mardulyn P, Smits G. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. NUCLEIC ACIDS RES. 2017;45(4):e18. doi: 10.1093/nar/gkw955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Delcher AL, Phillippy A, Carlton J, Salzberg SL. Fast algorithms for large-scale genome alignment and comparison. NUCLEIC ACIDS RES. 2002;30(11):2478–83. doi: 10.1093/nar/30.11.2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. GeSeq - versatile and accurate annotation of organelle genomes. NUCLEIC ACIDS RES. 2017;45(W1):W6–11. doi: 10.1093/nar/gkx391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28(12):1647–9. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Beier S, Thiel T, Munch T, Scholz U, Mascher M. MISA-web: a web server for microsatellite prediction. Bioinformatics. 2017;33(16):2583–5. doi: 10.1093/bioinformatics/btx198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Benson G. Tandem repeats finder: a program to analyze DNA sequences. NUCLEIC ACIDS RES. 1999;27(2):573–80. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kurtz S, Choudhuri JV, Ohlebusch E, Schleiermacher C, Stoye J, Giegerich R. REPuter: the manifold applications of repeat analysis on a genomic scale. NUCLEIC ACIDS RES. 2001;29(22):4633–42. doi: 10.1093/nar/29.22.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bergman CM, Quesneville H. Discovering and detecting transposable elements in genome sequences. BRIEF BIOINFORM. 2007;8(6):382–92. doi: 10.1093/bib/bbm048. [DOI] [PubMed] [Google Scholar]
- 82.Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I. VISTA: computational tools for comparative genomics. NUCLEIC ACIDS RES 2004, 32(Web Server issue):W273–9. [DOI] [PMC free article] [PubMed]
- 83.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. MOL BIOL EVOL. 2013;30(4):772–80. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rozas J, Ferrer-Mata A, Sanchez-DelBarrio JC, Guirao-Rico S, Librado P, Ramos-Onsins SE, Sanchez-Gracia A. DnaSP 6: DNA sequence polymorphism analysis of large data sets. MOL BIOL EVOL. 2017;34(12):3299–302. doi: 10.1093/molbev/msx248. [DOI] [PubMed] [Google Scholar]
- 85.Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T. TrimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25(15):1972–3. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhang D, Gao F, Jakovlic I, Zou H, Zhang J, Li WX, Wang GT. PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. MOL ECOL RESOUR. 2020;20(1):348–55. doi: 10.1111/1755-0998.13096. [DOI] [PubMed] [Google Scholar]
- 87.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30(9):1312–3. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Hohna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. MrBayes 3.2: efficient bayesian phylogenetic inference and model choice across a large model space. SYST BIOL. 2012;61(3):539–42. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kumar S, Suleski M, Craig JM, Kasprowicz AE, Sanderford M, Li M, Stecher G, Hedges SB. TimeTree 5: an expanded resource for species divergence Times. MOL BIOL EVOL 2022, 39(8). [DOI] [PMC free article] [PubMed]
- 90.Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. MOL BIOL EVOL. 2007;24(8):1586–91. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analysed during this study are included in this published article and its supplementary information files.