ABSTRACT
Anaerostipes hadrus (A. hadrus) is a dominant species in the human gut microbiota and considered a beneficial bacterium for producing probiotic butyrate. However, recent studies have suggested that A. hadrus may negatively affect the host through synthesizing fatty acid and metabolizing the anticancer drug 5-fluorouracil, indicating that the impact of A. hadrus is complex and unclear. Therefore, comprehensive genomic studies on A. hadrus need to be performed. We integrated 527 high-quality public A. hadrus genomes and five distinct metagenomic cohorts. We analyzed these data using the approaches of comparative genomics, metagenomics, and protein structure prediction. We also performed validations with culture-based in vitro assays. We constructed the first large-scale pan-genome of A. hadrus (n = 527) and identified 5-fluorouracil metabolism genes as ubiquitous in A. hadrus genomes as butyrate-producing genes. Metagenomic analysis revealed the wide and stable distribution of A. hadrus in healthy individuals, patients with inflammatory bowel disease, and patients with colorectal cancer, with healthy individuals carrying more A. hadrus. The predicted high-quality protein structure indicated that A. hadrus might metabolize 5-fluorouracil by producing bacterial dihydropyrimidine dehydrogenase (encoded by the preTA operon). Through in vitro assays, we validated the short-chain fatty acid production and 5-fluorouracil metabolism abilities of A. hadrus. We observed for the first time that A. hadrus can convert 5-fluorouracil to α-fluoro-β-ureidopropionic acid, which may result from the combined action of the preTA operon and adjacent hydA (encoding bacterial dihydropyrimidinase). Our results offer novel understandings of A. hadrus, exceptionally functional features, and potential applications.
IMPORTANCE
This work provides new insights into the evolutionary relationships, functional characteristics, prevalence, and potential applications of Anaerostipes hadrus.
KEYWORDS: Anaerostipes hadrus, butyrate, 5-fluorouracil, pan-genomics, metagenomics
INTRODUCTION
Trillions of microorganisms in the human gut are diverse and functionally rich. Species belonging to Anaerostipes, Eubacterium, Faecalibacterium, Clostridium, and other genera can produce butyrate through gastrointestinal bacterial fermentation (1). Butyrate is essential for maintaining intestinal epithelial cell barrier function, regulating the immune response of intestinal mucosa, and preventing cancer (2). The decrease in butyrate-producing species is closely associated with various diseases such as ulcerative colitis (UC) (3), Crohn’s disease (CD) (4), intestinal lymphoma (5), and type 2 diabetes (6). However, not all butyrate-producing bacteria have positive effects on the host. In addition to toxigenic strains of Clostridium butyricum that can cause botulism and necrotizing enterocolitis (7), Anaerostipes hadrus (A. hadrus) is also noteworthy as the first butyrate-producing species shown harmful effects on host health under certain disease-inducing conditions (8).
A. hadrus is a representative commensal bacterium with a relative abundance of 2%–7% in human intestines (9, 10). Previous studies have confirmed that A. hadrus can produce high levels of butyrate from sugars or acetate and lactate metabolized by other bacteria (11, 12). In addition, A. hadrus can also metabolize fructooligosaccharides to support the growth of other bacteria, such as Lacticaseibacillus rhamnosus (13). Furthermore, researchers identified that A. hadrus possesses biotin synthesis genes that regulate immunity and inflammation (14). Therefore, some scholars consider A. hadrus a beneficial bacterium (15). Consuming either milk (14) or galactooligosaccharides (16) can increase the abundance of A. hadrus in the gut microbiota. Nevertheless, a recent multi-omics study indicated that A. hadrus-mediated fatty acid biosynthesis influenced the availability of long-chain free fatty acids in the portal circulation and enhanced hepatic fibrosis (17). Another study mentioned that A. hadrus could inactivate the anticancer drug 5-fluorouracil (5-FU) (18), but the metabolic mechanism and pathway underlying this process still need to be better understood. The evidence above indicates that the precise role of A. hadrus in maintaining human health remains unclear. Thus, further culture independent and -dependent on A. hadrus is necessary.
With the advancement in microbiome research, metagenomics and culturomics have gradually become essential tools for studying microbes (19, 20). In this study, we completed the first large-scale population analysis of A. hadrus by integrating public genomic data. Moreover, we first characterized the ability of A. hadrus to convert 5-FU into α-fluoro-β-ureidopropionic acid (FUPA) as a dead-end metabolite using both in silico identifications of target genes and culture-based biotransformation assays. This work provides new insights into the evolutionary relationships, functional characteristics, prevalence, and potential applications of A. hadrus.
MATERIALS AND METHODS
Acquisition and quality control of public A. hadrus genomes
The overall analysis workflow is described in Fig. S1. First, we downloaded all A. hadrus genomes from GenBank (21) (https://ftp.ncbi.nlm.nih.gov/genomes/genbank/bacteria/Anaerostipes_hadrus/) and the Unified Human Gastrointestinal Genome [UHGG (22), http://ftp.ebi.ac.uk/pub/databases/metagenomics/mgnify_genomes/human-gut/v1.0/] collection, including isolate genomes and metagenome-assembled genomes (MAGs). Second, we used Kraken (23) (version 2.1.2) and Bracken (24) (version 2.6.1) to ensure the taxonomic classification. Then, we also used QUAST (25) (version 5.2.0) and BUSCO (26) (version 5.4.3) to assess the genome quality and core gene content, respectively. Finally, we applied CheckM (27) (version 1.2.2) to determine the genome completeness and contamination. Genomes analyzed in this study were required to have >90% completeness (CheckM), <5% contamination (CheckM), and >90% core genes (BUSCO). In addition, a size between 2.8 and 3.4 Mbp was required for isolate genomes. For MAGs, it should be between 2.5 and 3.4 Mbp. All software used default parameters during the analysis. More information about A. hadrus genomes can be found in Tables S1 and S2.
Pan-genome construction
We performed the gene annotation of screened A. hadrus genomes with Prokka (28) (version 1.14.6). Based on the annotation result (.gff file), we constructed the A. hadrus pan-genome with Roary (29) (version 3.10.2). Both Prokka and Roary used default parameters.
Phylogenetic and functional annotation analysis
According to the core gene alignment result generated by Roary, we constructed the phylogenetic tree of A. hadrus genomes using FastTree (30) (version 2.1.10) with the following parameters: "-nt -gtr". The average nucleotide identity (ANI) between A. hadrus genomes was calculated by Pyani (31) (version 0.2.12) with the parameter: "-m ANIm." We also constructed the multispecies phylogenetic tree of the genus Anaerostipes based on PhyloPhlAn (32) (version 3.0.67) and RAxML (33) (version 8.2.12). PhyloPhlAn used the parameters "--diversity low --fast -d phylophlan" and RAxML used the parameters "-f a -x 12,345 p 12345 -# 1000 m PROTGAMMAAUTO." The phylogenetic tree was visualized using ggtree (34) (version 3.2.1). Functional differences of representative genomes from different A. hadrus evolutionary clades were analyzed with the KEGG Automatic Annotation Server [KAAS (35), https://www.genome.jp/tools/kaas/]. Basic information of representative genomes for constructing the phylogenetic tree of the genus Anaerostipes is detailed in Table S3.
Analysis of butyrate-producing genes and 5-FU metabolism genes
We first selected reference protein sequences of butyrate-producing genes and 5-FU metabolism genes from Swiss-Prot as queries to perform tblastn (36) (version 2.13.0+) alignment with A. hadrus genomes (screening parameters: identity ≥30%, e-value <1e−10, query coverage ≥90%). Next, we verified alignment results with the non-redundant protein sequence database (NR) using blastx (36) (version 2.13.0+) alignment (screening parameters: identity ≥90%, query coverage ≥90%, subject coverage ≥90%, e-value <1e−10) and finally determined the sequences of target genes in A. hadrus genomes. Furthermore, we identified the upstream and downstream genes of butyrate-producing genes and 5-FU metabolism genes based on genome annotation files generated by Prokka. These gene sequences annotated by Prokka were also verified through the blastp [DIAMOND (37) version 2.0.15.153] alignment (screening parameters: identity ≥99%, e-value <1e−10) against NR. Finally, we used MEME Suite (38) (https://meme-suite.org/meme/meme_5.5.0/) to predict possible motifs lying upstream of 5-FU metabolism genes. The protein structure of A. hadrus PreT-PreA heterodimer was predicted by ColabFold (39) (https://colab.research.google.com/github/sokrypton/ColabFold/blob/v1.3.0/AlphaFold2.ipynb) with the following parameters: use_templates, true; use_amber, true; msa_mode, MMseqs2 (UniRef +Environmental); model_type, AlphaFold2-multimer-v2; num_models, 5; num_recycles, 6; pair_mode: unpaired +paired. The known protein structure was downloaded from the protein data bank (PDB). The alignment and visualization of protein structures were performed using PyMOL (http://www.pymol.org/pymol) (version 2.6.0a0).
Antibiotic resistance genes and virulence factor identification
To identify antibiotic resistance genes from CARD (version 3.2.6) in A. hadrus genomes, we analyzed the amino acid sequences of A. hadrus genomes using the Resistance Gene Identifier [RGI (40), version 4.0.3]. Genes identified by the Perfect algorithm in RGI were curated antibiotic resistance genes in CARD, while genes identified by RGI using the Strict algorithm were considered potential antibiotic resistance genes and required validation through comparison to NR. To investigate virulence factors in A. hadrus genomes, we performed a blastp (DIAMOND version 2.0.15.153) alignment between amino acid sequences of A. hadrus genomes and VFDB with filtering parameters of identity ≥60% and subject coverage ≥80%.
Determination of short-chain fatty acids
The concentrations of short-chain fatty acids (SCFAs, including acetate, propionate, butyrate, valerate, isobutyrate, and isovalerate) were determined using gas chromatography-mass spectrometry (GC-MS) as described in our previous research (9). In brief, the Anaerostipes hadrus CGMCC 1.32965 was incubated at 37°C anaerobically in modified mGAM broth for 72 h. Then, 1 mL of cell culture was extracted with 1 mL of ethyl acetate, and the supernatant was prepared for GC-MS analysis performed on a GCMS-QP2010 Ultra with an auto-sampler (SHIMADZU, Japan) and the DB-wax capillary column (30 m, 0.25 mm i.d., 0.25-µm film thickness, SHIMADZU, Japan). Standard curves of SCFAs were achieved by pure chemical agents of corresponding chemicals, purchased from Aladdin (Shanghai, China). The temperature of the oven was programmed from 35°C to 130°C at 5°C/min gradients, to 230°C at 30°C/min gradients, with a 16-min hold. Injection of 2 µL of samples was performed at 230°C. The carrier gas, helium, flowed at 1.0 mL/min. Ion source and interface temperature were both set at 230°C. The electronic impact was recorded at 70 eV.
Determination of 5-fluorouracil and its metabolites
To determine the degradation of 5-fluorouracil (5-FU) or production of α-fluoro-β-ureidopropionic acid (FUPA) by Anaerostipes hadrus cells in vitro, the Anaerostipes hadrus was incubated in modified MMGMB media for over 24 h until the microbes reached the stationary phase. The cells were harvested by centrifugation and washed with PBS buffer in anerobic chamber. After cell counting under microscopy, proper volume of resuspended cell solution was added to a 10-mL reaction system containing 5 mM of 5-FU at a final concentration of 109 cells/mL. The reaction system was incubated at 37°C under anerobic condition and at time points 0, 0.5, 1, 2, 3, and 6 h. A 1-mL reaction solution was sampled and centrifugated. The supernatant was used to analyze the concentration of 5-FU and FUPA with an Agilent Accurate-Mass-Q-TOF LC/MS 6520B instrument (Agilent, Germany) as described below: A Shim-pack GIST C18-AQ column (250 mm × 4.6 mm i.d.; 5 µm; SHIMADZU, Japan) was used at 35°C with a flow rate of 0.8 mL/min for liquid chromatography separation. The injection volume was 2 µL. The mobile phase A consisted of H2O with 0.1% formic acid, and the mobile phase B consisted of methanol. The gradient flow was set at 1% (vol/vol) B for 7 min, linearly increased to 95% B in the next 0.1 min and maintained for 5 min, then linearly decreased to 1% B in 0.1 min, and finally maintained at this composition for an additional 7.8 min. The ESI source of TOF mass spectrometry detection was negative ion mode, spray voltage was 3 kV, and the capillary temperature was set to 300°C. The sheath gas and auxiliary gas were both nitrogens, the flow rates were 30 and 10 (arbitrary units), and the scan range set to 60 to 1,000 m/z. The pure 5-FU (CAS Number: 51–21-8) and FUPA (CAS Number: 5006–64-4) were purchased from Aladdin (Shanghai, China). The standard curves of 5-FU and FUPA were constructed by HPLC-based quantification of the peak area under a series concentration of 0.1 0.25, 0.5, 1, and 2 mM.
Relative abundance calculation of A. hadrus and target genes
To investigate the relative abundance of A. hadrus and target genes, i.e., butyrate-producing genes and 5-FU metabolism genes, we downloaded the raw data of five cohorts from the Sequence Read Archive (SRA, https://www.ncbi.nlm.nih.gov/sra/), including a cohort of healthy males (41), an inflammatory bowel disease (IBD) cohort (42), a cohort of colorectal cancer (CRC) patients treated with FOLFOX (consisting of oxaliplatin and 5-FU) (43), a Chinese CRC cohort (44), and an Austrian CRC cohort (45). The metadata of five cohorts is detailed in Table S4. Then, we used Trim Galore (https://github.com/FelixKrueger/TrimGalore, version 0.6.7) and MultiQC (46) (version 1.13.dev0) to ensure the data quality. Finally, we applied Bowtie (47) (version 2.4.5) to remove potential host contamination (using the human genome sequence hg19 to build the index), resulting in clean data for subsequent analysis. All of the above software used default parameters. We calculated the relative abundance of A. hadrus and other species in five cohorts with MetaPhlAn (48) (version 3.0.14). Statistical analysis was performed using the ggpubr (49) package. Relative abundance on the species level was displayed using ggplot2 (50). We also used BWA (51) (version 0.7.17-r1188) to calculate the relative abundance of target genes with the following steps. First, butyrate-producing genes and 5-FU metabolism genes were extracted from the A. hadrus reference genome (GCF_000210695.1) to construct an index with BWA. Then, the BWA-MEM algorithm was chosen to align metagenomic data to the index. The number of mapped reads was calculated from the alignment results using Samtools (52) (version 1.6), and an R (53) (version 4.1.1) script was used to calculate TPM (transcripts per million) values. The specific calculation process of the R script is referenced from the study by Zhao et al. (54). Finally, TPM values were displayed using the pheatmap (55) package in R software.
RESULTS
Comparative genomic analysis uncovers an open pan-genome of A. hadrus
We screened and obtained 527 high-quality A. hadrus genomes from GenBank and UHGG, including 60 isolate genomes and 467 MAGs. The isolate genomes had an average size of 3.1 Mbp (range 2.8–3.4 Mbp), an average contig number of 114 (range 1–442), an average GC content of 37.0% (range 36.6%–37.5%), and an average N50 of 194.6 kbp (range 60.6 kbp–3.2 Mbp). The MAGs had an average size of 2.7 Mbp (range 2.5–3.3 Mbp), an average contig number of 168 (range 45–549), an average GC content of 37.2% (range 36.4%–38.1%), and an average N50 of 33.8 kbp (range 6.2–95.6 kbp). Each genome encoded an average of 2,555 predicted proteins (range 2,243–3,297).
We constructed the pan-genome of A. hadrus based on 527 genomes from 21 countries across four continents. The A. hadrus pan-genome contained 44,292 gene families, of which 1,196 were identified as core genes (present in more than 90% of 527 genomes), and the other 43,096 were identified as dispensable genes (present in less than 90% of 527 genomes). The average core gene content per A. hadrus genome was 46.8%. According to the Fig. S2A, the A. hadrus pan-genome is open, as the pan-genome size continuously increased with the addition of analyzed genomes. Moreover, the number of newly emerged gene families in the A. hadrus pan-genome decreased with the increase in analyzed genomes and eventually reached a plateau (Fig. S2B). When the number of analyzed genomes exceeds 500, adding each new genome resulted in an average of 46 new gene families. Thus, the A. hadrus pan-genome size expanded accordingly.
Phylogenetic analysis reveals three clades in A. hadrus genomes
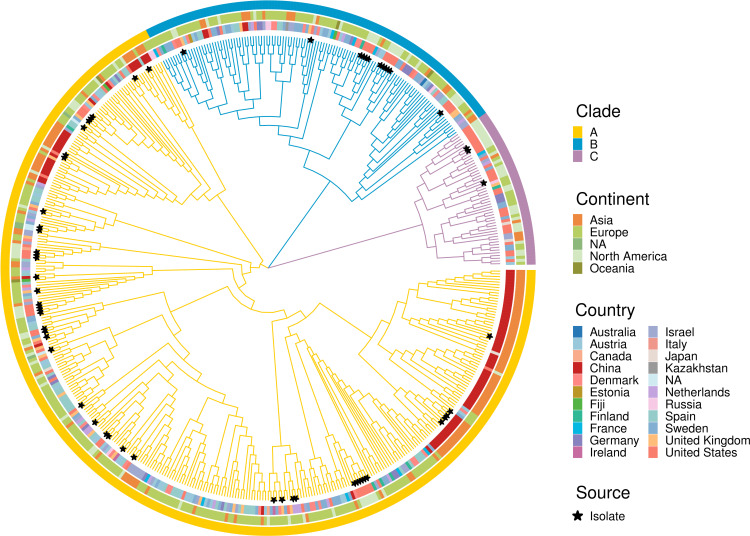
To investigate the evolutionary relationships of A. hadrus, we generated a maximum likelihood phylogenetic tree using 527 genomes. We observed three distinct evolutionary clades, A, B, and C, as shown in Fig. 1. Clade A comprised 356 genomes, including 45 isolate genomes. Clades B and C contained 120 and 51 genomes, respectively, with 12 and 3 isolate genomes each. These clades were detected in Asia, Europe, and North America, except clade C, which was not detected in Oceania. All three clades were observed in 10 countries, including the United States, Germany, France, Austria, and others. Clades A and B were found in China, Spain, Denmark, and Fiji. Clades A and C were both present in Japan. Clade A was detected only in Ireland, Italy, and Canada. Clade B was detected exclusively in Kazakhstan, Australia, and Estonia.
Fig 1.
Phylogenetic tree of 527 A. hadrus genomes. The colors of the outer ring and branches represent different evolutionary clades. The middle and inner ring colors represent different continents and countries, respectively. The black stars represent isolate genomes.
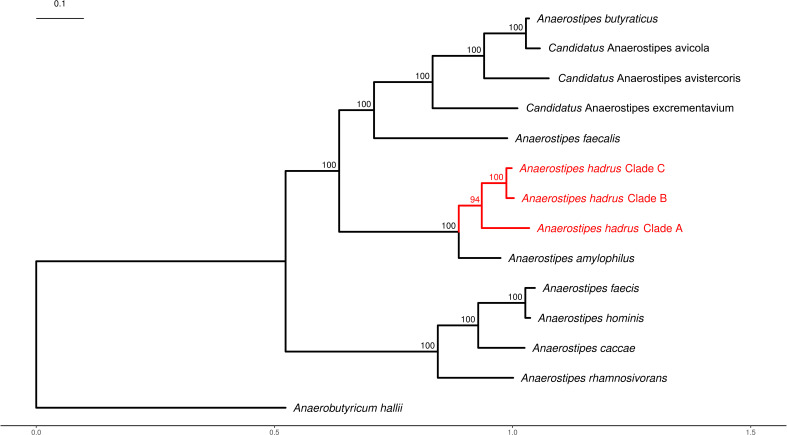
The genus Anaerostipes was first reported in 2002 (56). According to the NCBI Taxonomy database (http://www.ncbi.nlm.nih.gov/taxonomy) records, there are currently 11 species in the genus Anaerostipes, including eight confirmed and three candidatus species. We constructed a maximum likelihood phylogenetic tree of Anaerostipes species to study the evolutionary relationships between A. hadrus and other species (Fig. 2). The Anaerostipes species could be divided into two evolutionary clades. A. faecis, A. hominis, A. caccae, and A. rhamnosivorans existed on the smaller branch. While A. butyraticus, A. faecalis, A. hadrus, A. amylophilus, and three candidatus species were present on the bigger branch. A. amylophilus was the closest related species to A. hadrus. Compared to the minor clades B and C of A. hadrus, clade A was closer to A. amylophilus. The results of calculating the average nucleotide identity (ANI) between the three clades of A. hadrus also reflected this conclusion (Fig. S3). The ANI between 527 A. hadrus genomes was greater than 97%, and the average ANIs between clade A and clade B or clade C was 98.7%, while the mean ANI between clades B and C was 98.9%.
Fig 2.
Phylogenetic tree of the genus Anaerostipes. This tree reflects the evolutionary relationships between A. hadrus and other species within the genus Anaerostipes. We designated Anaerobutyricum hallii as an outgroup. The scale bar represents 0.1 substitutions per nucleotide. Bootstrap values are presented as a percentage of 1,000 replications. Detailed accession numbers can be found in Table S3.
The three clades of A. hadrus differed in genome content. We used a Venn diagram to illustrate the dissimilarities in gene families (Fig. S4A). The most significant number of gene families, 13,652, was shared between clades A and B, while clades B and C shared the least number of gene families (8,949). Overall, there are 8,602 gene families shared among all three clades. Additionally, it was found that the larger the clade, the more specific genes it contained, with the most specific genes in clade A (n = 20,737) and the least specific genes in clade C (n = 2,677). Although the three clades of A. hadrus differed significantly in gene family members, functional annotation analysis suggested that the main functions of different clades were the same (Fig. S4B). Sorted by the number of annotated genes, the main KEGG pathways of the representative genomes from different clades involved ribosome, ABC transporters, pyruvate metabolism, glycolysis/gluconeogenesis, and purine metabolism.
Widespread butyrate-producing genes drive the probiotic properties of A. hadrus
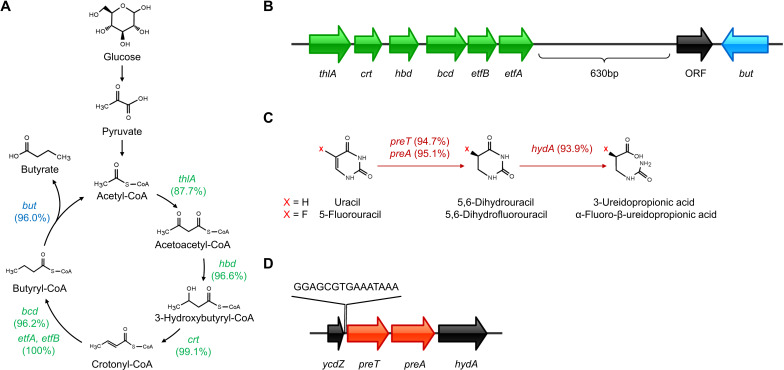
Butyrate is produced by the condensation of two acetyl-CoA molecules (57). Seven genes in A. hadrus are involved in this process (58). We then calculated the frequency of all genes involved in the microbial synthesis of butyrate from A. hadrus genomes (Fig. 3A), with the lowest frequency of 87.7% (462/527) for thlA and the highest frequency of 100% (527/527) for etfA and etfB. There were 84.3% (444/527) genomes carrying all seven butyrate-producing genes. In general, butyrate-producing genes were arranged in the order of catalyzed reactions in A. hadrus genomes, except for crt and hbd (Fig. 3B). The first six genes were arranged in the same direction and are less spaced apart (<100 bp). In contrast, but was farther from the first six genes (>1,600 bp). Between etfA and but, there existed an open reading frame (ORF) with an unknown function (>1,000 bp), which was aligned in the opposite direction to but. Additionally, we identified potentially harmful genes from the Comprehensive Antibiotic Resistance Database [CARD (40)] and the Virulence Factor Database [VFDB (59)] in A. hadrus genomes. Only 12.3% (65/527) of the A. hadrus genomes carried one to five antibiotic resistance genes (mainly related to antibiotics such as aminoglycosides, tetracyclines, and lincosamides). Concurrently, no virulence factors related to biological processes of invasion and exotoxin were identified in 527 A. hadrus genomes. To validate the probiotic properties of A. hadrus, we determined the production of butyrate and the other commonly found SCFAs by in vitro assays. The results revealed that the A. hadrus CGMCC 1.32965 were able to produce linear chain SCFAs as acetic, propanoic, butyric, and valeric acids during in vitro fermentation in modified mGAM media, other than branch chain ones represented by isobutyric and isovaleric acids (Fig. S5). The yields of C2–C4 SCFAs were 2.99, 22.16, 137.95, and 3.32 mg/L, respectively.
Fig 3.
Target pathways and gene structures in A. hadrus. (A) Butyrate production pathway of A. hadrus. The numbers in parentheses indicate the gene frequency of 527 A. hadrus genomes. The genes and their encoded proteins are as follows: bcd, butyryl-CoA dehydrogenase; but, butyryl-CoA:acetate CoA-transferase; crt, short-chain-enoyl-CoA hydratase; etfA, electron transfer flavoprotein subunit alpha; etfB, electron transfer flavoprotein subunit beta; hbd, 3-hydroxybutyryl-CoA dehydrogenase; thlA, acetyl-CoA acetyltransferase. (B) The structure of butyrate-producing genes. Green genes are involved in the reduction of acetyl-CoA to butyryl-CoA. The blue gene participates in the last step of butyrate production. (C) Reductive pyrimidine catabolic pathway and 5-FU metabolism pathway of A. hadrus. The numbers indicate the gene frequency in 527 A. hadrus genomes. The genes and their encoded proteins are as follows: hydA, bacterial dihydropyrimidinase; preA, NAD-dependent dihydropyrimidine dehydrogenase subunit PreA; preT, NAD-dependent dihydropyrimidine dehydrogenase subunit PreT. (D) The structure of 5-FU metabolism genes. The gene structure was displayed using IBS (60) software.
Ubiquitous 5-FU metabolism genes imply the complex role of A. hadrus
The preTA operon was first identified in Escherichia coli (E. coli) and encoded the bacterial dihydropyrimidine dehydrogenase (EcDPD) (61). EcDPD is not only involved in E. coli pyrimidine metabolism but also metabolizes 5-FU to the inactive dihydrofluorouracil (DHFU), functioning as human dihydropyrimidine dehydrogenase (DPD) (18). We identified the preTA operon in A. hadrus, suggesting that A. hadrus may have a similar reductive pyrimidine catabolic pathway as E. coli (Fig. 3C). In A. hadrus genomes, preT is located upstream of preA, and the two were adjacent to each other to form the preTA operon (Fig. 3D). ycdZ, the upstream gene of preT with unknown function, encoded a DUF1097 domain-containing protein with 38.4% amino acid identity to the intracellular membrane protein encoded by E. coli ycdZ. The downstream gene of preA, hydA, can encode the bacterial dihydropyrimidinase. However, there was only a 38.2% amino acid identity between the dihydropyrimidinase derived from A. hadrus (AhDHP) and human dihydropyrimidinase (DHP). DHP was reported to metabolize the catalytic product of DPD (62). We found that 93.2% (491/527) of A. hadrus genomes carried ycdZ, preTA operon, and hydA. Between ycdZ and preT, a potential motif (15 bp) that may be involved in regulating the preTA operon was predicted (Fig. 3D). Additionally, we noticed that both A. hadrus and E. coli carried the preTA operon but with different frequencies. We analyzed 2,565 complete E. coli genomes in GenBank and found that only 57.7% (1,480) of the genomes carried preT and preA. In contrast, 93.9% (495) of A. hadrus genomes carried preTA operon. Among 527 A. hadrus genomes analyzed in this study, the number of genomes carrying seven butyrate-producing genes and the preTA operon is 79.1% (417/527). This proportion reached 95% (57/60) in A. hadrus isolate genomes, indicating that the preTA operon metabolizing 5-FU was widely distributed within A. hadrus genomes as butyrate-producing genes.
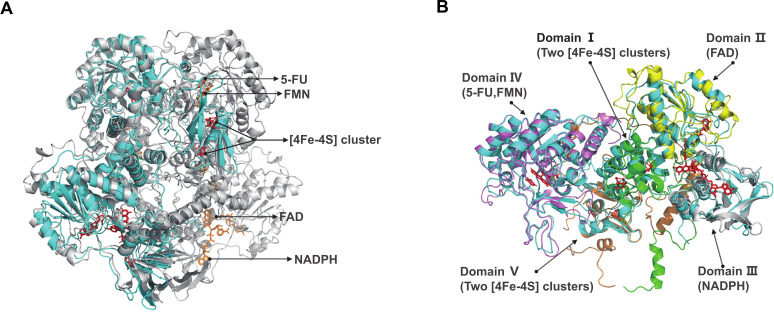
Mammalian DPD is a homodimer, whereas EcDPD is a heterotetramer consisting of two PreT and two PreA subunits (63). Moreover, the E. coli PreT-PreA heterodimer function is similar to one pig DPD monomer (63). Sequence alignment showed 58% amino acid identity between preT genes and 65% amino acid identity between preA genes encoded by A. hadrus and E. coli. Thus, the A. hadrus PreT-PreA heterodimer may also have similar functions with the pig DPD monomer. To demonstrate this, we used ColabFold to predict a high-quality protein structure of A. hadrus PreT-PreA heterodimer (predicted LDDT score = 94.6, predicted TM score = 0.915). Through comparing with the crystal structure of a ternary complex consisting of pig DPD, NADPH, and 5-FU (PDB ID: 1h7x), we found that the A. hadrus PreT-PreA heterodimer (820 AA) had a similar structure [root-mean-square deviation (RMSD) = 1.703 Å for 624 Cα atoms] to the pig DPD monomer (1,025 AA) (Fig. 4A). There are five functionally distinct domains (domains I–V) in the pig DPD monomer (11179210). We further performed a structure alignment between these five domains and predicted structure for a more in-depth study of the A. hadrus PreT-PreA heterodimer function (Fig. 4B). We found the similar domain I (RMSD = 0.786 Å for 85 Cα atoms), domain II (RMSD = 0.596 Å for 116 Cα atoms), domain III (RMSD = 2.284 Å for 85 Cα atoms), domain IV (RMSD = 0.619 Å for 255 Cα atoms), and domain V (RMSD = 1.023 Å for 79 Cα atoms) at corresponding positions of the predicted structure. In addition, structure-based amino acid sequence alignment showed that almost all sites involved in binding Fe–S clusters, FAD, NADPH, 5-FU, and FMN within pig DPD monomer were matched in the PreT-PreA heterodimer protein sequence of A. hadrus (Table S5). Therefore, A. hadrus DPD (AhDPD) encoded by the preTA operon is theoretically a heterotetramer, which has the same potential to metabolize 5-FU as EcDPD.
Fig 4.
Structural basis of 5-FU metabolism in A. hadrus. (A) Structure comparison between the predicted A. hadrus PreT-PreA heterodimer (cyan) and the pig DPD (PDB ID: 1h7x) (gray). Co-factors and substrates on the same monomer of the pig DPD are represented by the same color (red or orange). (B) Structure comparison between the predicted A. hadrus PreT-PreA heterodimer (cyan) and five distinct domains of the pig DPD (PDB ID: 1h7x) monomer. Domains are represented by green, yellow, gray, purple, and orange, respectively.
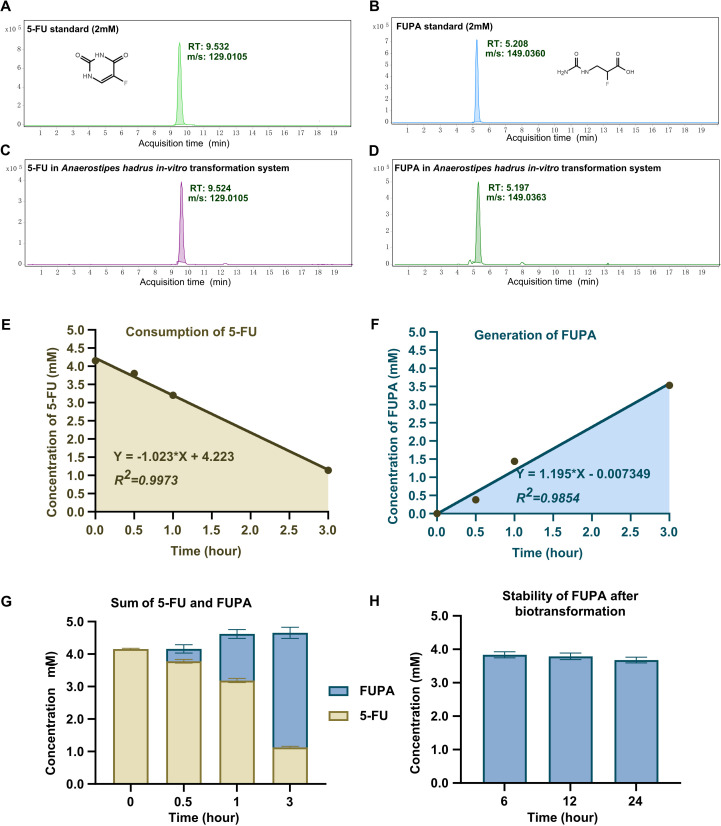
Considering the presence of hydA at the downstream of preTA operon, which encodes the bacterial dihydropyrimidinase, we deduced that the A. hadrus was able to transform 5-FU into α-fluoro-β-ureidopropionic acid (FUPA) as dead-end product other than DHFU as previously reported (18). Such deduction was then verified by in vitro biotransformation assay as described in the Materials and Methods section. We observed that the 5-FU was consumed in the presence of A. hadrus CGMCC 1.32965 in the system followed by the gradual generation of FUPA (Fig. 5A through G). Further regression analysis revealed that every 109 cells of A. hadrus CGMCC 1.32965 transform 5-FU into FUPA at an average velocity of 2.43 ± 1.66 mM/h. The level of the generated FUPA remained stable after an 18-h additional biotransformation (Fig. 5H), which indicated that the FUPA was a dead-end product that could not be transformed any further by A. hadrus CGMCC 1.32965.
Fig 5.
A. hadrus-mediated biotransformation of α-fluoro-β-alanine into α-fluoro-β-ureidopropionic acid in vitro. (A–D) The extracted iron chromographs of 5-FU standard (A), FUPA standard (B), and the remaining 5-FU (C) and generated FUPA (D) after 1 h of biotransformation by A. hadrus; RT, retention time; m/z, mass-to-charge ratio under negative source (−H). (E–F) The consumption of 5-FU (E) and generation of FUPA (F) by A. hadrus. The equation shown in the panel was calculated by simple linear regression analyzed by GraphPad Prism 9.0. (G) The total molar concentration of 5-FU and FUPA in one system after biotransformation by A. hadrus at different times.
High prevalence of A. hadrus and preTA orthologues across diverse cohorts
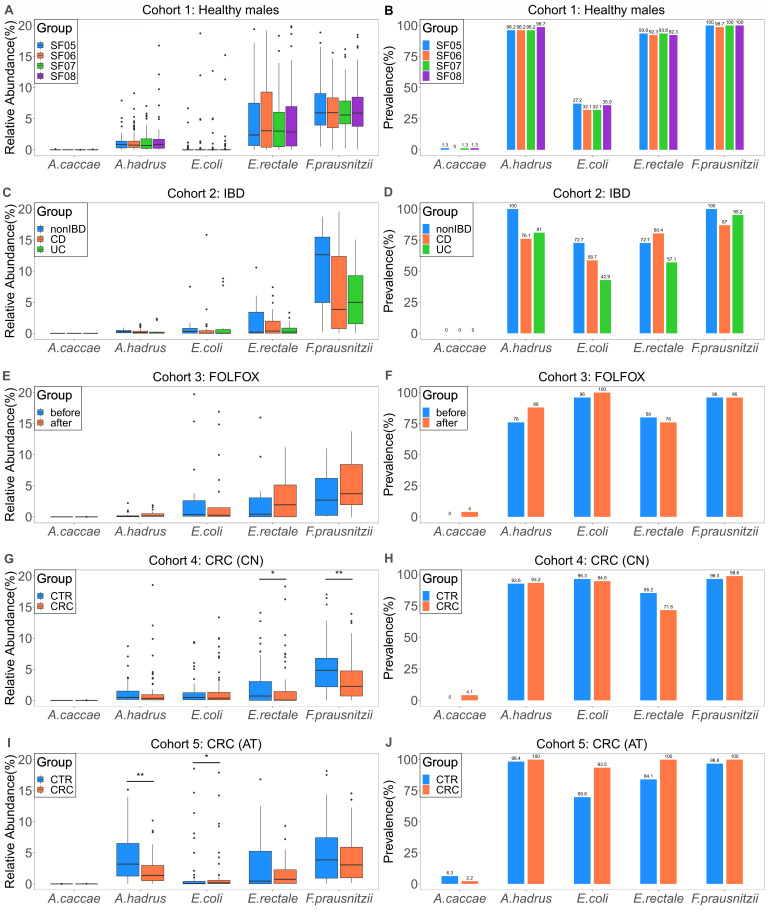
To further investigate the distribution of A. hadrus, we calculated its relative abundance in five cohorts. To better illustrate the relative abundance of A. hadrus, we compared A. hadrus with four other bacteria (Fig. 6), including Anaerostipes caccae (A. caccae), E. coli, Escherichia rectale (E. rectale), and Faecalibacterium prausnitzii (F. prausnitzii). A. caccae is the type species of the genus Anaerostipes and can produce butyrate (56). E. coli is a common conditional pathogen in the human intestinal tract and carries the preTA operon as A. hadrus. E. rectale and F. prausnitzii are high-abundance butyrate-producing bacteria in the human colon with essential healthy effects (10). In cohort 1, we found that the relative abundance and prevalence of A. hadrus in healthy men remained stable at different time points and were higher than that of E. coli (Fig. 6A and B). In cohort 2, compared with non-IBD individuals, the prevalence of A. hadrus in IBD patients decreased to 76.1%, still higher than that of E. coli (Fig. 6D), but its relative abundance did not change significantly (Fig. 6C). In cohort 3, after receiving FOLFOX treatment, A. hadrus prevalence in CRC patients increased (Fig. 6F). In cohorts 4 and 5, we found that almost all samples from CRC patients and healthy individuals carried A. hadrus (Fig. 6H and J). In these five metagenomic cohorts, only A. hadrus and A. caccae were detected within the genus Anaerostipes, with the relative abundance and prevalence of A. hadrus being much higher than that of A. caccae. Overall, in most healthy individuals, IBD patients, and CRC patients, the relative abundance of A. hadrus remained stable at less than 5%. Nevertheless, in some healthy individuals and CRC patients, the relative abundance of A. hadrus could reach around 15%. In addition, the prevalence rate of A. hadrus is similar to that of E. rectale and F. prausnitzii, maintaining a high level.
Fig 6.
Relative abundance and prevalence of A. hadrus and other species. The relative abundance is presented in the left boxplots, while the prevalence is shown in the right barplots. (A, B) Cohort 1 comprises 78 healthy males who contributed four stool samples over 6 months, resulting in 312 metagenomic data sets. (C, D Cohort 2 includes 28 individuals, comprising 15 with CD, nine with UC, and four non-IBD controls, who provided multiple stool samples over a year, resulting in 78 metagenomic data sets. (E, F) Cohort 3 includes 25 CRC patients who provided one stool sample before and after taking FOLFOX, resulting in 50 metagenomic data sets. (G, H) Cohort 4 consists of 128 Chinese individuals, including 74 with CRC and 54 healthy controls, who provided one stool sample each, resulting in 128 metagenomic data sets. (I, J) Cohort 5 comprises 109 Austrian individuals, 46 with CRC and 63 healthy controls, who provided one stool sample each, resulting in 109 metagenomic data sets. Statistical analysis was performed by a Wilcoxon rank sum test (*P < 0.05, **P < 0.01).
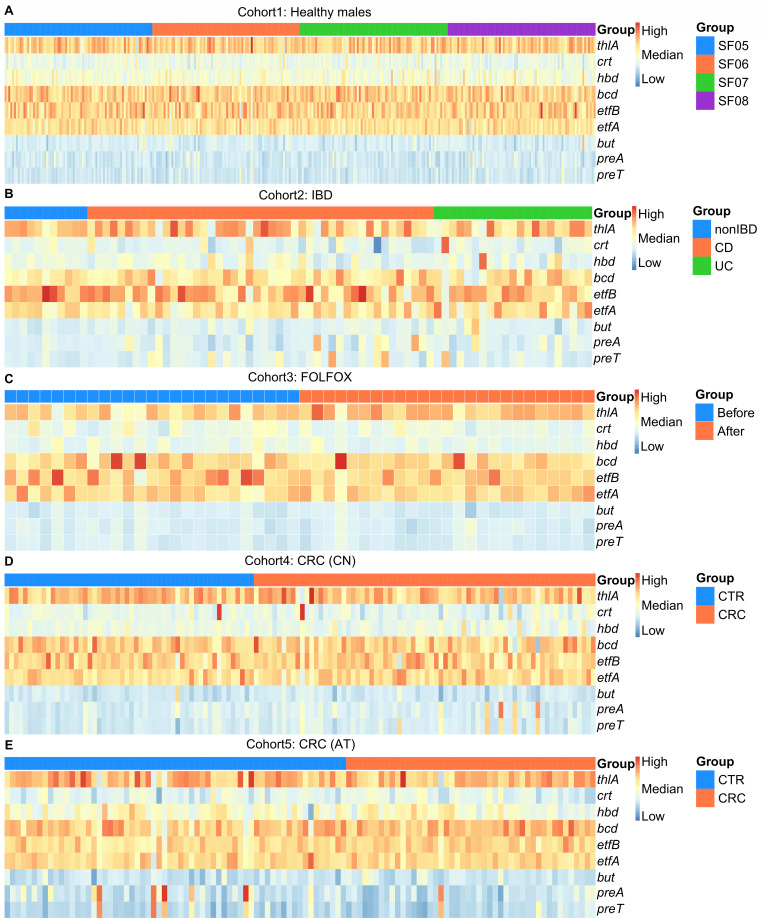
We further investigated the relative abundance of nine target genes, including butyrate-producing genes (thlA, crt, hbd, bcd, etfB, etfA, and but) and 5-FU metabolism genes (preT and preA), in five metagenomic cohorts. We found that these nine genes remained stable in relative abundance across cohorts and did not differ clearly between subgroups of the same cohort (Fig. 7). Overall, the relative abundance of thlA, bcd, etfB, and etfA was similar and at a higher level, while the relative abundance of but, preT, and preA was similar but lower. Additionally, in cohorts 4 and 5, we found that the relative abundance of preT and preA was significantly higher in a small number of samples from Chinese CRC patients, healthy Austrian individuals, and Austrian CRC patients.
Fig 7.
Relative abundance of butyrate-producing genes and 5-FU metabolism genes. The warmer color indicates a higher TPM value of the gene, corresponding to a higher relative abundance in one sample.
DISCUSSION
In this study, we conducted the first large-scale pan-genome analysis of A. hadrus (n = 527). We found that the proportion of core gene families in A. hadrus pan-genome (2.7%) is slightly lower than in other species (3%–84%) (64). Compared with other butyrate-producing bacteria, including F. prausnitzii [4.5% of core gene families in the pan-genome constructed by 84 strains (65)], Clostridium perfringens [3.8% of core gene families in the pan-genome constructed by 173 strains (66)], and Clostridium butyricum [9.9% of core gene families in the pan-genome constructed by 32 strains (67)], A. hadrus had a smaller core genome, suggesting the functional diversity and complexity. The phylogenetic and functional annotation analysis results showed no noticeable geographical distribution differences among different A. hadrus clades (Fig. 1), and their main functions were broadly consistent (Fig. S4B). However, we still need to pay attention to the impact of geographic factors on A. hadrus genomes. The latest study pointed out that the A. hadrus genome was prone to structural variations, and the core gene sequence identity cannot fully reflect functional similarity among A. hadrus genomes (68). Since a higher proportion of dispensable genes is in the A. hadrus pan-genome, the influence of strain isolation environment on dispensable genes should be fully considered when studying the function of a single A. hadrus strain.
5-FU is a first-line drug for chemotherapy in patients with CRC. However, host-derived DPD, DHP, and β-ureidopropionase from the reductive pyrimidine catabolic pathway (62) successively metabolize the majority of 5-FU entering the human body into non-anticancer DHFU, FUPA, and α-fluoro-β-alanine (69). In this study, we demonstrated that the A. hadrus genomes harbored homologs of human DPD and DHP encoded by the preTA operon and hydA, and observed for the first time that A. hadrus can convert 5-FU to FUPA. Concurrently, no homolog of β-ureidopropionase was found in any of the 527 A. hadrus genomes, which explains that the final product of 5-FU metabolism by A. hadrus is FUPA other than α-fluoro-β-alanine. Furthermore, we found that the location of hydA in E. coli genomes is far from the preTA operon, which may be the reason E. coli metabolizes 5-FU into DHFU (18), indicating that the conservation of the reductive pyrimidine catabolic pathway varies among different bacteria. Since humans and various microorganisms metabolize 5-FU into different final products, this character may help us distinguish different participants in 5-FU metabolism.
Although A. hadrus may interfere with the therapeutic effect of 5-FU due to the presence of the preTA operon, on the other hand, A. hadrus is expected to become probiotics for CRC patients suffering from DPD deficiency. It has been reported that 10%–30% of patients experience severe adverse reactions after receiving fluoropyrimidine treatment, and 30%–80% of them are due to the lack of DPD (70). In theory, A. hadrus can exert the same 5-FU rate-limiting effect as mammalian DPD and produce beneficial butyrate for the human body. Thus, A. hadrus has broad application prospects in helping CRC patients reduce 5-FU toxicity. Through metagenomic analysis, this study revealed that the distribution of A. hadrus was characterized by wide and stable features across different cohorts (Fig. 6). Notably, in the cohort consisting of non-IBD individuals and IBD patients, the prevalence of A. hadrus in stool samples from CD and UC patients was significantly lower, suggesting that butyrate-producing A. hadrus may be associated with the occurrence and development of IBD. Besides, in the cohort of CRC patients treated with FOLFOX, we found that this first-line chemotherapeutic agent increased the prevalence of A. hadrus in CRC patients' stool samples. Previous studies have indicated that the gut microbiome regulates the efficacy of FOLFOX (71, 72). Thus, it is worth exploring whether the increase in A. hadrus abundance will affect the subsequent therapeutic effect of FOLFOX. Our study also found that the relative abundance of preT and preA was close to that of but. In nature, most butyrate-producing bacteria rely on the butyryl-CoA:acetate CoA transferase encoded by but to complete the final step of butyrate production (73). Therefore, we should take seriously the potential impact of preT and preA from gut microbiota on fluoropyrimidine drugs. Additionally, we found that samples with a higher abundance of A. hadrus carried more preT and preA, which may support the idea that preTA operons in the population are mainly derived from Anaerostipes (18).
Despite exploratory analyses, there are still some limitations in our study. First, due to the strict culture conditions, the genome resources of A. hadrus strains that can be publicly obtained for analysis are limited. Therefore, we incorporated more MAGs to study the A. hadrus pan-genome comprehensively. However, MAGs can cause the loss of core genes (74), so the core genome size of the A. hadrus pan-genome we described may be slightly smaller than the actual situation. Second, although we calculated the relative abundance of butyrate-producing genes and 5-FU metabolism genes in five metagenomic cohorts, this only preliminary indicated that different populations carry a certain number of preT and preA. More research is needed on the level of gene expression. Third, our functional description of A. hadrus needs to be entirely adequate. Through protein structure prediction and amino acid sequence alignment, we speculated the binding sites of co-factors and substrates in AhDPD (Table S8). Nevertheless, these speculations have yet to be verified due to experimental limitations.
Conclusion
Through a large-scale A. hadrus population analysis, we systematically studied the evolutionary relationship of A. hadrus and found that butyrate-producing genes and genes involved in 5-FU metabolism (the preTA operon and hydA) are core genes. Through culture-based in vitro biotransformation assay, we then confirmed that the A. hadrus metabolizes 5-FU into FUPA as dead-end product for the first time. Based on the distribution of A. hadrus, preT, and preA in different metagenomic cohorts, we suggested that butyrate-producing A. hadrus may interfere with the efficacy of fluoropyrimidine drugs or reduce adverse reactions in CRC patients, which may depend on the level of human DPD. In conclusion, this study found that A. hadrus has the potential to exert beneficial or harmful effects on hosts, which expands our understanding of bacterial duality and inspires us to study the role of A. hadrus in the human body deeply, to better apply A. hadrus to clinical diagnosis and treatment of related diseases.
ACKNOWLEDGMENTS
We thank Dr. DeFeng Li (Institute of Microbiology, Chinese Academy of Sciences) for generously offering insightful suggestions and constructive comments during the preparation of this manuscript.
This work was supported by the National Key Research and Development Program of China (2022YFA1304103).
Conceptualization: Z.H., C.L. Methodology: Z.H., C.L., S.-J.L., S.H. Investigation: D.L., C.L., L.X., S.L., K.L., Y.Y., W.-W. Visualization: D.L., C.L. Supervision: Z.H., C.L. Writing—original draft: D.L., C.L. Writing—review and editing: Z.H., C.L., S.-J.L., S.H.
Contributor Information
Chang Liu, Email: liu.c@sdu.edu.cn.
Zilong He, Email: hezilong@buaa.edu.cn.
Hideyuki Tamaki, National Institute of Advanced Industrial Science and Technology, Tsukuba, Ibaraki, Japan.
DATA AVAILABILITY
The genomic data of A. hadrus are available in Table S1. The raw sequencing data can be found in the National Center for Biotechnology Information Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra/) under the following identifiers: PRJNA354235, PRJNA389280, PRJNA484031, PRJEB10878, PRJEB7774. Metadata for these samples are available in Table S6.
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/msphere.00816-23.
Figures S1 to S5.
Supplemental material legends.
Basic information of 527 publicly available A. hadrus genomes.
Quality assessment results of 527 A. hadrus genomes.
Basic information of representative genomes for constructing the phylogenetic tree of the genus Anaerostipes.
Basic information of the five cohorts used in this study.
Structure-based amino acid sequence alignment between predicted A. hadrus PreT-PreA heterodimer and five pig DPD domains.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Fu X, Liu Z, Zhu C, Mou H, Kong Q. 2019. Nondigestible carbohydrates, butyrate, and butyrate-producing bacteria. Crit Rev Food Sci Nutr 59:S130–S152. doi: 10.1080/10408398.2018.1542587 [DOI] [PubMed] [Google Scholar]
- 2. Liu H, Wang J, He T, Becker S, Zhang G, Li D, Ma X. 2018. Butyrate: a double-edged sword for health? Adv Nutr 9:21–29. doi: 10.1093/advances/nmx009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Machiels K, Joossens M, Sabino J, De Preter V, Arijs I, Eeckhaut V, Ballet V, Claes K, Van Immerseel F, Verbeke K, Ferrante M, Verhaegen J, Rutgeerts P, Vermeire S. 2014. A decrease of the butyrate-producing species Roseburia hominis and Faecalibacterium prausnitzii defines dysbiosis in patients with ulcerative colitis. Gut 63:1275–1283. doi: 10.1136/gutjnl-2013-304833 [DOI] [PubMed] [Google Scholar]
- 4. Quévrain E, Maubert MA, Michon C, Chain F, Marquant R, Tailhades J, Miquel S, Carlier L, Bermúdez-Humarán LG, Pigneur B, et al. 2016. Identification of an anti-inflammatory protein from Faecalibacterium prausnitzii, a commensal bacterium deficient in Crohn's disease. Gut 65:415–425. doi: 10.1136/gutjnl-2014-307649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lu H, Xu X, Fu D, Gu Y, Fan R, Yi H, He X, Wang C, Ouyang B, Zhao P, Wang L, Xu P, Cheng S, Wang Z, Zou D, Han L, Zhao W. 2022. Butyrate-producing Eubacterium rectale suppresses lymphomagenesis by alleviating the TNF-induced TLR4/MyD88/NF-κB axis. Cell Host Microbe 30:1139–1150. doi: 10.1016/j.chom.2022.07.003 [DOI] [PubMed] [Google Scholar]
- 6. Tilg H, Moschen AR. 2014. Microbiota and diabetes: an evolving relationship. Gut 63:1513–1521. doi: 10.1136/gutjnl-2014-306928 [DOI] [PubMed] [Google Scholar]
- 7. Cassir N, Benamar S, La Scola B. 2016. Clostridium butyricum: from beneficial to a new emerging pathogen. Clin Microbiol Infect 22:37–45. doi: 10.1016/j.cmi.2015.10.014 [DOI] [PubMed] [Google Scholar]
- 8. Zhang Q, Wu Y, Wang J, Wu G, Long W, Xue Z, Wang L, Zhang X, Pang X, Zhao Y, Zhao L, Zhang C. 2016. Accelerated dysbiosis of gut microbiota during aggravation of DSS-induced colitis by a butyrate-producing bacterium. Sci Rep 6:27572. doi: 10.1038/srep27572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Abdugheni R, Wang W, Wang Y, Du M, Liu F, Zhou N, Jiang C, Wang C, Wu L, Ma J, Liu C, Liu S. 2022. Metabolite profiling of human‐originated Lachnospiraceae at the strain level. iMeta 1. doi: 10.1002/imt2.58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Walker AW, Duncan SH, Louis P, Flint HJ. 2014. Phylogeny, culturing, and metagenomics of the human gut microbiota. Trends Microbiol 22:267–274. doi: 10.1016/j.tim.2014.03.001 [DOI] [PubMed] [Google Scholar]
- 11. Louis P, Duncan SH, McCrae SI, Millar J, Jackson MS, Flint HJ. 2004. Restricted distribution of the butyrate kinase pathway among butyrate-producing bacteria from the human colon. J Bacteriol 186:2099–2106. doi: 10.1128/JB.186.7.2099-2106.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Allen-Vercoe E, Daigneault M, White A, Panaccione R, Duncan SH, Flint HJ, O’Neal L, Lawson PA. 2012. Anaerostipes hadrus comb. nov., a dominant species within the human colonic microbiota; reclassification of Eubacterium hadrum Moore et al. 1976. Anaerobe 18:523–529. doi: 10.1016/j.anaerobe.2012.09.002 [DOI] [PubMed] [Google Scholar]
- 13. Endo A, Tanno H, Kadowaki R, Fujii T, Tochio T. 2022. Extracellular fructooligosaccharide degradation in Anaerostipes hadrus for co-metabolism with non-fructooligosaccharide utilizers. Biochem Biophys Res Commun 613:81–86. doi: 10.1016/j.bbrc.2022.04.134 [DOI] [PubMed] [Google Scholar]
- 14. Tian B, Yao JH, Lin X, Lv WQ, Jiang LD, Wang ZQ, Shen J, Xiao HM, Xu H, Xu LL, Cheng X, Shen H, Qiu C, Luo Z, Zhao LJ, Yan Q, Deng HW, Zhang LS. 2022. Metagenomic study of the gut microbiota associated with cow milk consumption in Chinese peri-/postmenopausal women. Front Microbiol 13:957885. doi: 10.3389/fmicb.2022.957885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hu X, Li H, Zhao X, Zhou R, Liu H, Sun Y, Fan Y, Shi Y, Qiao S, Liu S, Liu H, Zhang S. 2021. Multi-omics study reveals that statin therapy is associated with restoration of gut microbiota homeostasis and improvement in outcomes in patients with acute coronary syndrome. Theranostics 11:5778–5793. doi: 10.7150/thno.55946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Schoemaker MH, Hageman JHJ, Ten Haaf D, Hartog A, Scholtens P, Boekhorst J, Nauta A, Bos R. 2022. Prebiotic galacto-oligosaccharides impact stool frequency and fecal microbiota in self-reported constipated adults: a randomized clinical trial. Nutrients 14:309. doi: 10.3390/nu14020309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ali RO, Quinn GM, Umarova R, Haddad JA, Zhang GY, Townsend EC, Scheuing L, Hill KL, Gewirtz M, Rampertaap S, Rosenzweig SD, Remaley AT, Han JM, Periwal V, Cai H, Walter PJ, Koh C, Levy EB, Kleiner DE, Etzion O, Heller T. 2023. Longitudinal multi-omics analyses of the gut-liver axis reveals metabolic dysregulation in hepatitis C infection and cirrhosis. Nat Microbiol 8:12–27. doi: 10.1038/s41564-022-01273-y [DOI] [PubMed] [Google Scholar]
- 18. Spanogiannopoulos P, Kyaw TS, Guthrie BGH, Bradley PH, Lee JV, Melamed J, Malig YNA, Lam KN, Gempis D, Sandy M, Kidder W, Van Blarigan EL, Atreya CE, Venook A, Gerona RR, Goga A, Pollard KS, Turnbaugh PJ. 2022. Host and gut bacteria share metabolic pathways for anti-cancer drug metabolism. Nat Microbiol 7:1605–1620. doi: 10.1038/s41564-022-01226-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhang X, Li L, Butcher J, Stintzi A, Figeys D. 2019. Advancing functional and translational microbiome research using meta-omics approaches. Microbiome 7:154. doi: 10.1186/s40168-019-0767-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lagier JC, Dubourg G, Million M, Cadoret F, Bilen M, Fenollar F, Levasseur A, Rolain JM, Fournier PE, Raoult D. 2018. Culturing the human microbiota and culturomics. Nat Rev Microbiol 16:540–550. doi: 10.1038/s41579-018-0041-0 [DOI] [PubMed] [Google Scholar]
- 21. Benson DA, Cavanaugh M, Clark K, Karsch-Mizrachi I, Ostell J, Pruitt KD, Sayers EW. 2018. GenBank. Nucleic Acids Res 46:D41–D47. doi: 10.1093/nar/gkx1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Almeida A, Nayfach S, Boland M, Strozzi F, Beracochea M, Shi ZJ, Pollard KS, Sakharova E, Parks DH, Hugenholtz P, Segata N, Kyrpides NC, Finn RD. 2021. A unified catalog of 204,938 reference genomes from the human gut microbiome. Nat Biotechnol 39:105–114. doi: 10.1038/s41587-020-0603-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wood DE, Salzberg SL. 2014. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol 15:R46. doi: 10.1186/gb-2014-15-3-r46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lu J, Breitwieser FP, Thielen P, Salzberg SL. 2017. Bracken: estimating species abundance in metagenomics data. PeerJ Comput Sci 3:e104. doi: 10.7717/peerj-cs.104 [DOI] [Google Scholar]
- 25. Gurevich A, Saveliev V, Vyahhi N, Tesler G. 2013. QUAST: quality assessment tool for genome assemblies. Bioinformatics 29:1072–1075. doi: 10.1093/bioinformatics/btt086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Seppey M, Manni M, Zdobnov EM. 2019. BUSCO: assessing genome assembly and annotation completeness. Methods Mol Biol 1962:227–245. doi: 10.1007/978-1-4939-9173-0_14 [DOI] [PubMed] [Google Scholar]
- 27. Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. 2015. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res 25:1043–1055. doi: 10.1101/gr.186072.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153 [DOI] [PubMed] [Google Scholar]
- 29. Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, Holden MTG, Fookes M, Falush D, Keane JA, Parkhill J. 2015. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31:3691–3693. doi: 10.1093/bioinformatics/btv421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Price MN, Dehal PS, Arkin AP. 2010. FastTree 2--approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pritchard L, Glover RH, Humphris S, Elphinstone JG, Toth IK. 2016. Genomics and taxonomy in diagnostics for food security: soft-rotting enterobacterial plant pathogens. Anal Methods 8:12–24. doi: 10.1039/C5AY02550H [DOI] [Google Scholar]
- 32. Asnicar F, Thomas AM, Beghini F, Mengoni C, Manara S, Manghi P, Zhu Q, Bolzan M, Cumbo F, May U, Sanders JG, Zolfo M, Kopylova E, Pasolli E, Knight R, Mirarab S, Huttenhower C, Segata N. 2020. Precise phylogenetic analysis of microbial isolates and genomes from metagenomes using PhyloPhlAn 3.0. Nat Commun 11:2500. doi: 10.1038/s41467-020-16366-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. doi: 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yu G. 2020. Using ggtree to visualize data on tree-like structures. Curr Protoc Bioinformatics 69:e96. doi: 10.1002/cpbi.96 [DOI] [PubMed] [Google Scholar]
- 35. Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M. 2007. KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res 35:W182–W185. doi: 10.1093/nar/gkm321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Johnson M, Zaretskaya I, Raytselis Y, Merezhuk Y, McGinnis S, Madden TL. 2008. NCBI BLAST: a better web interface. Nucleic Acids Res 36:W5–W9. doi: 10.1093/nar/gkn201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Buchfink B, Xie C, Huson DH. 2015. Fast and sensitive protein alignment using DIAMOND. Nat Methods 12:59–60. doi: 10.1038/nmeth.3176 [DOI] [PubMed] [Google Scholar]
- 38. Bailey TL, Johnson J, Grant CE, Noble WS. 2015. The MEME suite. Nucleic Acids Res 43:W39–W49. doi: 10.1093/nar/gkv416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mirdita M, Schütze K, Moriwaki Y, Heo L, Ovchinnikov S, Steinegger M. 2022. ColabFold: making protein folding accessible to all. Nat Methods 19:679–682. doi: 10.1038/s41592-022-01488-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Alcock BP, Huynh W, Chalil R, Smith KW, Raphenya AR, Wlodarski MA, Edalatmand A, Petkau A, Syed SA, Tsang KK, et al. 2023. CARD 2023: expanded curation, support for machine learning, and resistome prediction at the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res 51:D690–D699. doi: 10.1093/nar/gkac920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nguyen LH, Ma W, Wang DD, Cao Y, Mallick H, Gerbaba TK, Lloyd-Price J, Abu-Ali G, Hall AB, Sikavi D, Drew DA, Mehta RS, Arze C, Joshi AD, Yan Y, Branck T, DuLong C, Ivey KL, Ogino S, Rimm EB, Song M, Garrett WS, Izard J, Huttenhower C, Chan AT. 2020. Association between sulfur-metabolizing bacterial communities in stool and risk of distal colorectal cancer in men. Gastroenterology 158:1313–1325. doi: 10.1053/j.gastro.2019.12.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Schirmer M, Franzosa EA, Lloyd-Price J, McIver LJ, Schwager R, Poon TW, Ananthakrishnan AN, Andrews E, Barron G, Lake K, Prasad M, Sauk J, Stevens B, Wilson RG, Braun J, Denson LA, Kugathasan S, McGovern DPB, Vlamakis H, Xavier RJ, Huttenhower C. 2018. Dynamics of metatranscription in the inflammatory bowel disease gut microbiome. Nat Microbiol 3:337–346. doi: 10.1038/s41564-017-0089-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Li J, Li J, Lyu N, Ma Y, Liu F, Feng Y, Yao L, Hou Z, Song X, Zhao H, Li X, Wang Y, Xiao C, Zhu B. 2020. Composition of fecal microbiota in low-set rectal cancer patients treated with FOLFOX. Ther Adv Chronic Dis 11:2040622320904293. doi: 10.1177/2040622320904293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yu J, Feng Q, Wong SH, Zhang D, Liang QY, Qin Y, Tang L, Zhao H, Stenvang J, Li Y, et al. 2017. Metagenomic analysis of faecal microbiome as a tool towards targeted non-invasive biomarkers for colorectal cancer. Gut 66:70–78. doi: 10.1136/gutjnl-2015-309800 [DOI] [PubMed] [Google Scholar]
- 45. Feng Q, Liang S, Jia H, Stadlmayr A, Tang L, Lan Z, Zhang D, Xia H, Xu X, Jie Z, et al. 2015. Gut microbiome development along the colorectal adenoma-carcinoma sequence. Nat Commun 6:6528. doi: 10.1038/ncomms7528 [DOI] [PubMed] [Google Scholar]
- 46. Ewels P, Magnusson M, Lundin S, Käller M. 2016. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32:3047–3048. doi: 10.1093/bioinformatics/btw354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Beghini F, McIver LJ, Blanco-Míguez A, Dubois L, Asnicar F, Maharjan S, Mailyan A, Manghi P, Scholz M, Thomas AM, Valles-Colomer M, Weingart G, Zhang Y, Zolfo M, Huttenhower C, Franzosa EA, Segata N. 2021. Integrating taxonomic, functional, and strain-level profiling of diverse microbial communities with bioBakery 3. Elife 10:e65088. doi: 10.7554/eLife.65088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kassambara A. 2023. ggpubr: 'ggplot2' based publication ready plots
- 50. Ginestet C. 2011. ggplot2: elegant graphics for data analysis. J R Stat Soc Ser A Stat Soc 174:245–246. doi: 10.1111/j.1467-985X.2010.00676_9.x [DOI] [Google Scholar]
- 51. Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv:1303.3997. doi: 10.48550/arXiv.1303.3997 [DOI]
- 52. Danecek P, Bonfield JK, Liddle J, Marshall J, Ohan V, Pollard MO, Whitwham A, Keane T, McCarthy SA, Davies RM, Li H. 2021. Twelve years of SAMtools and BCFtools. Gigascience 10:giab008. doi: 10.1093/gigascience/giab008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. R Core Team . 2021. R: a language and environment for statistical computing. Available from: https://www.R-project.org
- 54. Zhao Y, Li M-C, Konaté MM, Chen L, Das B, Karlovich C, Williams PM, Evrard YA, Doroshow JH, McShane LM. 2021. TPM, FPKM, or normalized counts? A comparative study of quantification measures for the analysis of RNA-seq data from the NCI patient-derived models repository. J Transl Med 19:269. doi: 10.1186/s12967-021-02936-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kolde R. 2019. pheatmap: pretty heatmaps
- 56. Schwiertz A, Hold GL, Duncan SH, Gruhl B, Collins MD, Lawson PA, Flint HJ, Blaut M. 2002. Anaerostipes caccae gen. nov., sp. nov., a new saccharolytic, acetate-utilising, butyrate-producing bacterium from human faeces. Syst Appl Microbiol 25:46–51. doi: 10.1078/0723-2020-00096 [DOI] [PubMed] [Google Scholar]
- 57. Miller TL, Wolin MJ. 1996. Pathways of acetate, propionate, and butyrate formation by the human fecal microbial flora. Appl Environ Microbiol 62:1589–1592. doi: 10.1128/aem.62.5.1589-1592.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Louis P, Flint HJ. 2009. Diversity, metabolism and microbial ecology of butyrate-producing bacteria from the human large intestine. FEMS Microbiol Lett 294:1–8. doi: 10.1111/j.1574-6968.2009.01514.x [DOI] [PubMed] [Google Scholar]
- 59. Liu B, Zheng D, Zhou S, Chen L, Yang J. 2022. VFDB 2022: a general classification scheme for bacterial virulence factors. Nucleic Acids Res 50:D912–D917. doi: 10.1093/nar/gkab1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Liu W, Xie Y, Ma J, Luo X, Nie P, Zuo Z, Lahrmann U, Zhao Q, Zheng Y, Zhao Y, Xue Y, Ren J. 2015. IBS: an illustrator for the presentation and visualization of biological sequences. Bioinformatics 31:3359–3361. doi: 10.1093/bioinformatics/btv362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Mihara H, Hidese R, Yamane M, Kurihara T, Esaki N. 2008. The iscS gene deficiency affects the expression of pyrimidine metabolism genes. Biochem Biophys Res Commun 372:407–411. doi: 10.1016/j.bbrc.2008.05.019 [DOI] [PubMed] [Google Scholar]
- 62. Hidese R, Mihara H, Kurihara T, Esaki N. 2011. Escherichia coli dihydropyrimidine dehydrogenase is a novel NAD-dependent heterotetramer essential for the production of 5,6-dihydrouracil. J Bacteriol 193:989–993. doi: 10.1128/JB.01178-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Yoshioka H, Ishida T, Mihara H. 2021. Overexpression and characterization of Escherichia coli dihydropyrimidine dehydrogenase: a four iron-sulphur cluster containing flavoprotein. J Biochem 170:511–520. doi: 10.1093/jb/mvab067 [DOI] [PubMed] [Google Scholar]
- 64. McInerney JO, McNally A, O’Connell MJ. 2017. Why prokaryotes have pangenomes. Nat Microbiol 2:17040. doi: 10.1038/nmicrobiol.2017.40 [DOI] [PubMed] [Google Scholar]
- 65. Bai Z, Zhang N, Jin Y, Chen L, Mao Y, Sun L, Fang F, Liu Y, Han M, Li G. 2022. Comprehensive analysis of 84 Faecalibacterium prausnitzii strains uncovers their genetic diversity, functional characteristics, and potential risks. Front Cell Infect Microbiol 12:919701. doi: 10.3389/fcimb.2022.919701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Feng Y, Fan X, Zhu L, Yang X, Liu Y, Gao S, Jin X, Liu D, Ding J, Guo Y, Hu Y. 2020. Phylogenetic and genomic analysis reveals high genomic openness and genetic diversity of Clostridium perfringens. Microb Genom 6:mgen000441. doi: 10.1099/mgen.0.000441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Benamar S, Cassir N, Merhej V, Jardot P, Robert C, Raoult D, La Scola B. 2017. Multi-spacer typing as an effective method to distinguish the clonal lineage of Clostridium butyricum strains isolated from stool samples during a series of necrotizing enterocolitis cases. J Hosp Infect 95:300–305. doi: 10.1016/j.jhin.2016.10.026 [DOI] [PubMed] [Google Scholar]
- 68. Kogawa M, Nishikawa Y, Saeki T, Yoda T, Arikawa K, Takeyama H, Hosokawa M. 2023. Revealing within-species diversity in uncultured human gut bacteria with single-cell long-read sequencing. Front Microbiol 14:1133917. doi: 10.3389/fmicb.2023.1133917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Vodenkova S, Buchler T, Cervena K, Veskrnova V, Vodicka P, Vymetalkova V. 2020. 5-fluorouracil and other fluoropyrimidines in colorectal cancer: past, present and future. Pharmacol Ther 206:107447. doi: 10.1016/j.pharmthera.2019.107447 [DOI] [PubMed] [Google Scholar]
- 70. Laures N, Konecki C, Brugel M, Giffard A-L, Abdelli N, Botsen D, Carlier C, Gozalo C, Feliu C, Slimano F, Djerada Z, Bouché O. 2022. Impact of guidelines regarding dihydropyrimidine dehydrogenase (DPD) deficiency screening using uracil-based phenotyping on the reduction of severe side effect of 5-fluorouracil-based chemotherapy: a propension score analysis. Pharmaceutics 14:2119. doi: 10.3390/pharmaceutics14102119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Iida N, Dzutsev A, Stewart CA, Smith L, Bouladoux N, Weingarten RA, Molina DA, Salcedo R, Back T, Cramer S, Dai RM, Kiu H, Cardone M, Naik S, Patri AK, Wang E, Marincola FM, Frank KM, Belkaid Y, Trinchieri G, Goldszmid RS. 2013. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science 342:967–970. doi: 10.1126/science.1240527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Scott TA, Quintaneiro LM, Norvaisas P, Lui PP, Wilson MP, Leung KY, Herrera-Dominguez L, Sudiwala S, Pessia A, Clayton PT, Bryson K, Velagapudi V, Mills PB, Typas A, Greene NDE, Cabreiro F. 2017. Host-microbe co-metabolism dictates cancer drug efficacy in C. elegans. Cell 169:442–456. doi: 10.1016/j.cell.2017.03.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Vital M, Karch A, Pieper DH. 2017. Colonic butyrate-producing communities in humans: an overview using omics data. mSystems 2:e00130-17. doi: 10.1128/mSystems.00130-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Li T, Yin Y. 2022. Critical assessment of pan-genomic analysis of metagenome-assembled genomes. Brief Bioinform 23:bbac413. doi: 10.1093/bib/bbac413 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figures S1 to S5.
Supplemental material legends.
Basic information of 527 publicly available A. hadrus genomes.
Quality assessment results of 527 A. hadrus genomes.
Basic information of representative genomes for constructing the phylogenetic tree of the genus Anaerostipes.
Basic information of the five cohorts used in this study.
Structure-based amino acid sequence alignment between predicted A. hadrus PreT-PreA heterodimer and five pig DPD domains.
Data Availability Statement
The genomic data of A. hadrus are available in Table S1. The raw sequencing data can be found in the National Center for Biotechnology Information Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra/) under the following identifiers: PRJNA354235, PRJNA389280, PRJNA484031, PRJEB10878, PRJEB7774. Metadata for these samples are available in Table S6.