Abstract
The fundamental event in prion disease is thought to be the posttranslational conversion of the cellular prion protein (PrPC) into a pathogenic isoform (PrPSc). The occurrence of PrPC on the cell surface and PrPSc in amyloid plaques in situ or in aggregates following purification complicates the study of the molecular events that underlie the disease process. Monoclonal antibodies are highly sensitive probes of protein conformation which can be used under these conditions. Here, we report the rescue of a diverse panel of 19 PrP-specific recombinant monoclonal antibodies from phage display libraries prepared from PrP deficient (Prnp0/0) mice immunized with infectious prions either in the form of rods or PrP 27-30 dispersed into liposomes. The antibodies recognize a number of distinct linear and discontinuous epitopes that are presented to a varying degree on different PrP preparations. The epitope reactivity of the recombinant PrP(90-231) molecule was almost indistinguishable from that of PrPC on the cell surface, validating the importance of detailed structural studies on the recombinant molecule. Only one epitope region at the C terminus of PrP was well presented on both PrPC and PrPSc, while epitopes associated with most of the antibodies in the panel were present on PrPC but absent from PrPSc.
Prion diseases are disorders of protein conformation that are characterized by a profound degeneration of the central nervous system (24, 25). The fundamental event in the pathogenesis of these diseases is the conversion of the cellular prion protein (PrPC) into the scrapie isoform (PrPSc). Evidence from modeling structural studies, including infrared spectroscopy, circular dichroism, and multidimensional heteronuclear solution nuclear magnetic resonance (NMR) argues that PrPSc formation involves an extensive conformational change in which the α-helical content of PrP diminishes and a large amount of β-sheet is acquired (3, 6, 11, 13, 19, 21, 28, 31, 35). Detailed structural studies of PrPSc have, however, been technically difficult to carry out. Limited proteinase K digestion employed during the purification of PrPSc yields PrP 27-30 which assembles into rod-shaped polymers with the ultrastructural and tinctorial properties of amyloid (18, 27).
Another approach to probing conformational transitions in prion proteins is to generate antibodies to diverse epitopes of PrPC and PrPSc. However, natural infection induces no humoral immune response to infectious scrapie particles (17), and immune tolerance to the highly conserved PrP amino acid sequence has restricted the generation of monoclonal antibodies in normal mice (2, 15, 30). To access a wider spectrum of PrP-specific monoclonal antibodies, we raised antisera recognizing mouse (Mo) and Syrian hamster (SHa) PrP in mice homozygous for PrP gene knockout (Prnp0/0) (4, 26) and prepared combinatorial phage antibody libraries from these animals as described previously (1, 5, 12, 34).
Antibody libraries were constructed from Prnp0/0 mice immunized either with prion rods containing MoPrP 27-30 or with disaggregated PrP 27-30 incorporated into liposomes (9, 10, 22). Mice immunized with prion rods received an immunization and three boosts. Animals immunized with PrP 27-30 in liposomes were divided into two groups and received either an immunization and two boosts (long immunization) or, in an attempt to increase the diversity of the antibody response, an immunization and a single boost (short immunization). For each mouse, PrP-specific reactivity in all four subclasses of serum immunoglobulin G (IgG) was determined by enzyme-linked immunosorbent assay (ELISA) against MoPrP 27-30 treated with the denaturant guanidium thiocyanate (GdnSCN). Mice immunized with prion rods generated PrP-specific serum antibody titers predominantly in the IgG1 and IgG2b subclasses, whereas mice immunized with PrP 27-30 liposomes produced a strong PrP-specific response in all IgG subclasses. Serum antibody reactivity has proven to be accurate in predicting the specificities rescued from the corresponding phage libraries (5, 33). We therefore prepared an IgG1κ and an IgG2bκ Fab library from a mouse immunized with prion rods. Additional IgG1κ, IgG2aκ, IgG2bκ, and IgG3κ Fab libraries were individually constructed from each of the two groups of mice given long and short immunizations with PrP liposomes. All of the libraries were prepared with total RNA extracted from spleen, bone marrow, and lymph node tissue, and all contained over 107 members.
The phage libraries were individually selected against denaturant-treated PrP 27-30, recombinant PrP(90-231) and detergent dispersed PrP 27-30 as previously described (1, 22). Phage recovered from the fourth or fifth round of panning were converted to express soluble Fab (1) and tested for specific PrP reactivity in ELISA against denaturant treated PrP 27-30 and SHaPrP(90-231). The heavy chain amino acid sequences were determined for antigen-reactive Fab clones, and this information allowed the clones to be sorted into distinct families, as illustrated in Table 1.
TABLE 1.
Partial heavy chain amino acid sequences of selected recombinant Fabs recovered from immunized mice
| Clonea | FR3 region sequence | CDR3 region sequence | FR4 region sequence | Epitope reactivityb |
|---|---|---|---|---|
| Immunization with MoPrP 27-30 rods | ||||
| PrP28 | KATLTADKSSSTAYLDLRSLTSEDSAVYFCAR | HDGYPFAY | WGQGTLVTVSA | DC |
| PrP3recPrP | KATLTADKTSSTAHIQLSSLTSEDSAVYFCAR | GFYYGSRYGPMDY | WGQGTSVIVSS | DC |
| PrP28DLPC | KATLTADKSSSTAYMDLRSLTSEDSAAYFCAR | VPISVY | WGQGTTLTVSS | DC |
| PrP1blocked | KATLTVDKSSSTAYIQPSSLTSEDSAVYYCAR | WGPFFYYGSRPSYYAMDS | WGQGGSVTVFS | DC |
| PrP34blocked | RATLTADKSSTTAHLQLFSLSSEDSAVYFCSR | SRSTNYFDY | WGQGTTLAVSS | DC |
| Immunization with dispersed SHaPrP 27-30 incorporated into liposomes | ||||
| R1 | KATLTVDTSSSTAYVDLSSLTSEDSAVYYCAR | EGHFPPDY | WGQGTTLTVSS | III |
| R2 | KATLTVDKSSSTAYIQLSRLTSEDSAVYYCAR | EGDAYPFGH | WVQGTLVTVSS | III |
| R5 | KATLTVDTSSSTAYVDLNSLTSEDSAVYYCTR | EDSSYPFAY | WGQGTTLTVSS | III |
| R10 | KATITADTSSNTVYLQLRSLTSEDTAIYYCGR | FDGNGWYFDV | WGAGTTVTVSS | I |
| R23 | KATLTVDKSSSTAYMQLSSLTSEDSAVYYCAR | GGYYGAMDY | WGQGTSVTVSS | DC |
| R25 | RATLTADTSSSTAYMQLSSLTSEDSTVYFCAR | RRLITTLVDSWSFDV | WGQGTTVTVSS | DC |
| R40 | KATLTADKSSSTAYMELRSLTSEDSAVYFCAR | DYVKGYFDV | WGTGTTVTVSS | DC |
| R72 | EATLTVDKSSSTAYMELRSLTSEDTAVYYCVR | RGIYHYAMDY | WGQGTSVTVSS | II |
| D2 | KATLTVDKSSSTAYMQLSRLTSEDSAVYYCAR | EGDYYPFGH | WGQGTLVTVSS | III |
| D4 | KATITADTSSNTVYLQLRSLTSEDTAIYYCGR | FDGNGWYLDV | WGAGTTVTVSS | I |
| D7 | RFAFSLETSASTAYLQINNLQNEDTATYFCVS | RGGDYGSSAFDY | WGQGTTLTVSS | III |
| D13 | RFTISRDNAKNTLYLQMSSLKSDDTAMYYCGR | LGGDYGGSYLDY | WGQGTTLTVSS | I |
| D14 | KATLTVDKSSSTAYMELRSLTSEDSAVYYCAA | YFYAMDY | WGQGTSVTVSS | DC |
| D18 | KATLTVDKSSSTAYMELRSLTSEDSAVYYCAG | FYYGMDY | WGQGTSVTVSS | DC |
Clones R1, R2, R5, R10, R23, R25, R40, and R72 were panned against recombinant SHaPrP(90-231) and clones D2, D4, D7, D13, D14, and D18 were panned against dispersed SHaPrP 27-30.
DC, discontinuous. I, II, and III are designations of linear epitope regions as described in the text.
Libraries constructed from mice immunized with Mo prions yielded five novel antibodies, designated Fabs PrP28, PrP1blocked, PrP34blocked, PrP3recPrP, and PrP28DLPC (34). Libraries constructed from mice immunized with PrP liposomes initially yielded a large number of closely related sequences, of which Fabs R1, R2, R5, and R10 are examples. Fabs D2, D4, D5, D7, D13, D14, and D18 were recovered by panning against dispersed SHaPrP 27-30. On a number of occasions, Fabs with similar sequences were recovered by panning against both SHaPrP(90-231) and SHaPrP 27-30 (e.g., R2 and D2, respectively). To generate greater diversity, the PrP antigens were masked with Fabs rescued from the first panning experiments, then re-presented to the libraries (7). Amino acid sequences of ELISA reactive Fab clones taken from these experiments contained several additional Fabs, R23, R25, R40, and R72, with novel heavy chain amino acid sequences.
The antigen-binding profiles of the novel recombinant Fabs were assessed against various PrP preparations as shown in Table 2. Reactivity of Fabs with cell surface MoPrPC was assessed by flow cytometry as described previously (34), using the mouse neuroblastoma line N2a (16) and a transfected Chinese hamster ovary (CHO) cell line expressing SHaPrPC. Antibody recognition of PrPC and PrPSc in situ was examined by immunostaining blotted cryostat sections of brains taken from normal uninoculated CD-1 mice and SHa, and from clinically ill CD-1 mice and SHa inoculated with Mo(RML) prions and Sc237 prions, respectively, as described previously (32). The Fabs were also assessed for their ability to immunoprecipitate SHaPrPC from transfected CHO cells (14, 22) and SHaPrP 27-30 from liposomes (22).
TABLE 2.
Reactivity of recombinant monoclonal Fabs against different PrP preparationsa
| Fab | Epitope reactivity | Cell-surface PrPC | REC PrP (90-231) | PrP 27-30 in ELISA (GdnSCN treated) | PrP 27-30 in ELISA (untreated) | PrP 27-30 immunoprecipitation | PrPC immunoprecipitation | PrP in situ (GdnSCN treated) |
|---|---|---|---|---|---|---|---|---|
| PrP28 | DC | + | + | + | − | nd | nd | + |
| PrP28DLPC | DC | + | + | + | − | nd | nd | + |
| PrP3recPrP | DC | ± | + | + | − | nd | nd | + |
| PrP1blocked | DC | + | + | + | − | nd | nd | + |
| PrP34blocked | DC | + | + | + | − | nd | nd | + |
| R10 | I | + | + | + | − | − | + | + |
| R72 | II | − | + | + | ± | ± | + | + |
| R1 | III | + | + | + | + | + | + | + |
| R23 | DC | + | + | + | − | − | + | + |
| R25 | DC | + | + | + | − | − | + | + |
| R40 | DC | + | + | + | − | − | + | + |
| D14 | DC | + | + | + | − | − | + | + |
| D18 | DC | + | + | + | − | − | + | + |
For an explanation of epitope reactivity data, see Table 1, footnote b. +, strong reactivity; ±, weak reactivity; −, no reactivity; nd, not done.
All of the antibodies reacted well in ELISA with recombinant SHaPrP(90-231) and with PrP 27-30 rods following incubation with 3 M GdnSCN. All of the antibodies also detected PrPSc in situ following treatment with denaturant and, with the exception of Fab R72, also efficiently immunoprecipitated SHaPrPC from transfected CHO cells. PrPC in its native state on the surface of the mouse neuroblastoma line N2a (16) and a transfected CHO cell line expressing SHaPrPC were recognized well by all Fabs, with the exceptions of PrP3rPrP, which bound weakly, and R23 and R72, which did not bind at all.
We next sought to identify the binding epitopes recognized by the recombinant antibodies by using both peptide-based ELISA studies and PrP fragment libraries displayed on filamentous phage. Initially, we studied antibody reactivity against a series of synthetic peptides representing residues 90 to 231 of SHaPrP. Twenty-seven peptides that were 15 residues in length and that overlapped by 5 residues at the N terminus were prepared and individually applied to ELISA plates to determine the reactivity of each antibody. In addition, protein fragment libraries of mouse PrP were prepared for display on the surface of M13 phage via fusion with coat protein III by using the phagemid vector pFRAG in a variation of the method of Petersen et al. (23). pFRAG was constructed by placing a 39-base-pair insert containing two distinct BglII sites into the XhoI and SfiI sites of the phage display vector pComb3 (1). The fragment libraries were panned individually over each recombinant Fab bound to ELISA wells. Following specific enrichment over sequential rounds of panning, the encoded PrP fragments of a representative population of phagemid clones were determined by DNA sequencing. Alignment of these sequences permitted the identification of a core sequence common to each clone, which likely approximates to the epitope of the antibody, as shown in Fig. 1.
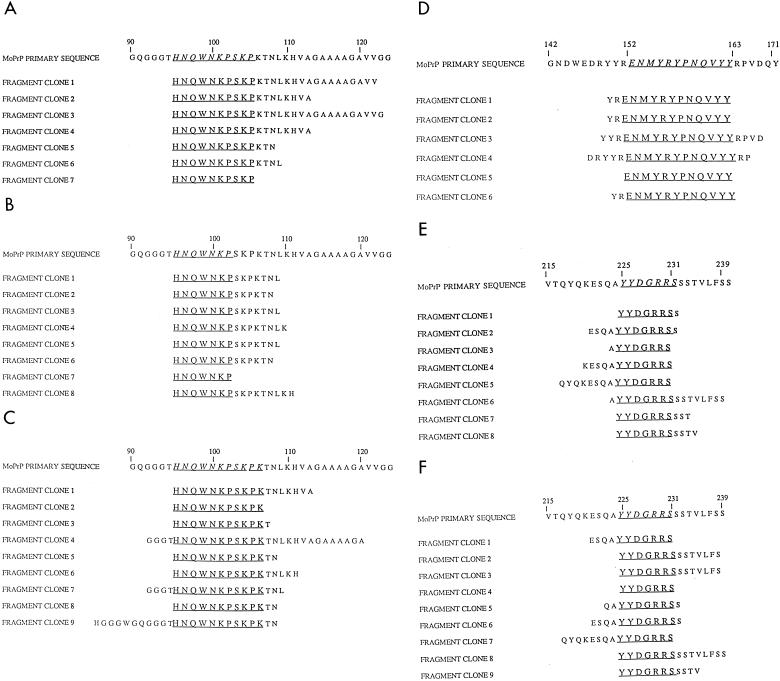
FIG. 1.
Identification of linear epitopes from PrP protein fragment phage display libraries. Three linear epitopes, designated I, II, and III, were identified by panning the PrP fragment libraries against recombinant Fab fragments applied to ELISA wells. Sequence alignments of clones taken following two or three rounds of panning are shown below the corresponding mouse PrP amino acid sequence. Regions of commonality are underlined. (A) Fab R10; (B) Fab D4; (C) Fab D13; (D) Fab R72; (E) Fab R1; (F) Fab R2.
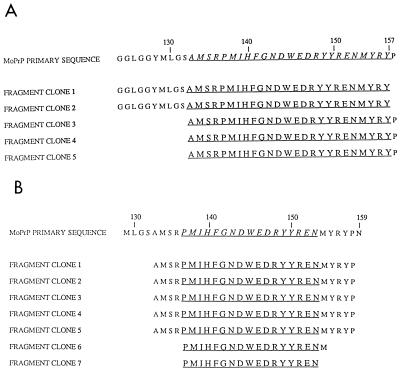
Data collected by both these approaches were highly consistent and indicated that a subset of recombinant antibodies, recovered following immunization with PrP liposomes, recognized three linear epitope regions, designated I, II, and III. Epitope region I was recognized by three antibodies (R10, D4, and D13) and lies within residues 96 to 104, a region of the protein shown in solution NMR studies to be largely disordered (8, 13, 29). Epitope region II was localized to residues 153 to 161 and was bound exclusively by Fab R72. This antibody was recovered when libraries prepared from mice immunized with PrP liposomes were panned against recombinant SHaPrP(90-231). Its epitope contains the final four residues of the first helical region of PrP(90-231) and extends to the beginning of a short β-strand (S2) (13). Epitope region III was assigned to residues 225 to 231 at the very C-terminal end of PrP, adjacent to the glycosylphosphatidylinositol anchor. We presume this region to be immunodominant in mice immunized with PrP liposomes since the majority of Fab-phage recovered from panning experiments against SHaPrP(90-231) and dispersed SHaPrP 27-30 reacted with this epitope. Of the recombinant Fabs that did not react with short peptides, only R40 and D14 specifically enriched phage from the PrP fragment library (Fig. 2). Fab R40 isolated phage that contained the amino acid sequence between residues 138 and 155, with a minimum consensus sequence of 17 amino acids between residues 137 and 153. Fab D18 enriched for phage bearing PrP sequence containing residues 133 through 157.
FIG. 2.
Identification of nonlinear epitopes from PrP fragment display libraries. Of the recombinant Fabs that did not recognize overlapping 15-mer PrP peptides only (A) D18 and (B) R40 specifically selected for phage bearing related PrP sequence. The sequences shown were obtained following five rounds of panning. Regions of commonality are underlined.
To determine whether any of the Fabs would specifically bind to PrP determinants in solution, we employed a competition ELISA technique in which antibody was preincubated with a range of concentrations of competing peptides, typically 50 μM to 50 pM, before being applied to PrP antigen. Sigmoidal binding curves were obtained for each antibody competition with Graphpac (ISI Software). The concentration of each peptide required to inhibit 50% of the recombinant Fab binding to the control polypeptide applied to the plate was then determined. The results are given in Table 3. Fabs R10 and D13, possessing divergent heavy chain amino acid sequences and both binding similar if not identical epitopes between residues 96 to 104, were competed effectively with synthetic peptides corresponding to residues 90 to 104 and 95 to 109. Fabs R1, R2, and D7 recognized epitope region III (residues 225 to 231) and were efficiently competed by peptides containing amino acids 220 to 231 and 225 to 231. Interestingly, Fab R72 was efficiently competed with a peptide containing residues 152 to 163 (the region identified as the binding epitope by the fragment libraries) but did not bind at all to recombinant SHaPrP(90-231) in solution. This epitope was, however, bound tightly when SHaPrP(90-231) was applied directly to ELISA wells, indicating that the epitope is normally either partially or completely inaccessible but becomes exposed when PrP is applied to ELISA plates.
TABLE 3.
Inhibition of Fab binding by various PrP peptides
| Fab | Concn (μM) required to inhibit 50% of Fab binding by:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| PrP(90-231) | PrP(90-104) | PrP(96-105) | PrP(147-167) | PrP(152-163) | PrP(154-163) | PrP(155-163) | PrP(220-231) | PrP(225-231) | |
| R10 | 3.0 | 0.8 | 0.4 | ||||||
| D13 | 0.06 | 0.1 | 0.08 | ||||||
| R72 | >50 | 1.2 | 1.9 | 7.8 | 18.6 | ||||
| R1 | 2.9 | 0.2 | 1.4 | ||||||
| R2 | 0.7 | 1.2 | 0.3 | ||||||
| D7 | 0.3 | 0.01 | 0.01 | ||||||
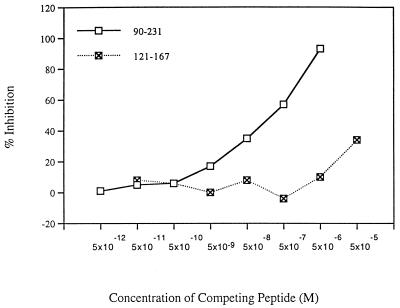
We reasoned that the antibodies recognizing discontinuous epitopes of PrP may bind longer synthetic peptides which may be able to adopt secondary structure arrangements found in the full-length protein (13, 29). We therefore synthesized a series of longer peptides corresponding to SHaPrP sequence between amino acids 90 and 145, 121 and 167, 147 and 167, 141 and 178, 159 and 201, 178 and 231 (containing protected cysteine side chains and therefore unable to form the disulfide bridge normally found between cysteine 179 and cysteine 214 in intact PrP), and 174 and 231. However, none of the recombinant Fabs was able to bind well to any of these peptides in a direct binding or competition ELISA, although recombinant SHaPrP(90-231) and SHaPrP(29-231) in α-helical states (8, 13) were bound tightly (data not shown). Similarly, in a competitive ELISA, with the exception of Fab R40, none of the Fabs was competed by the longer peptides. Fab R40 was partially competed with a peptide containing residues 127 to 167, which includes the region of sequence identified by this Fab from the protein fragment libraries (Fig. 3). We conclude that the discontinuous epitopes of PrP recognized by the antibodies may be fully formed only in the intact PrP(90-231) molecule.
FIG. 3.
Dose response of the competing antigens recombinant SHaPrP(90-231) and synthetic peptide SHaPrP(127-147) with recombinant Fab R40. Absorbance values were converted into percentages of inhibition.
To examine species cross-reactivity, the recombinant antibodies were reacted in ELISA with SHa-, Mo-, bovine, and human PrP (Table 4). Fabs binding epitope region I reacted very strongly with SHa- and MoPrP but had only very weak reactivity with bovine and human PrP. When amino acid sequences from the different species were examined in the region of epitope I, the only variation occurred at position 97, which is an asparagine residue in SHa- and MoPrP but is a serine residue in human PrP and a glycine residue in bovine PrP. The results suggest that the amino acid at position 97 makes direct contact with the group I antibodies. Epitope region II, recognized by Fab R72, is invariant across the species examined here, and predictably this antibody bound very strongly to all the PrP samples in ELISA. In contrast, residues 225 to 231 that compose epitope region III exhibit considerable diversity across different species. Fabs recognizing this region of PrP predictably bound to SHa- and MoPrP, which contain identical sequences between residues 225 to 231, but not to PrP from the other species tested, which contain markedly different sequences in this region.
TABLE 4.
Binding of selected recombinant antibodies and the hybridoma-derived antibody 3F4 against cellular PrPa
| Antibody | Reactivity of antibody against:
|
|||
|---|---|---|---|---|
| MoPrP | SHaPrP | Human PrP | Bovine PrP | |
| R10 | +++ | +++ | + | + |
| 3F4 | − | +++ | +++ | − |
| R72 | +++ | +++ | +++ | +++ |
| R2 | +++ | +++ | − | − |
| R23 | − | +++ | +++ | − |
| R40 | +++ | +++ | +++ | − |
| D14 | +++ | +++ | +++ | ++ |
| D18 | +++ | +++ | ++ | + |
| PrP28 | +++ | +++ | ++ | nd |
| PrP28DLPC | +++ | +++ | ++ | nd |
Number of plus signs (+) indicates degree of reactivity. −, no reaction; nd, not done.
In summary, we have generated a diverse panel of PrP-specific antibodies from immunized mice. These antibodies have been characterized in terms of their amino acid sequences, the binding epitopes recognized, and their reactivity with a number of PrP-antigenic presentations. Surprisingly, given that we immunized mice with infectious PrP 27-30 preparations, none of the rescued antibodies exclusively recognized this form of the protein, whereas all but one antibody clone reacted well with PrPC as it occurs on the cell surface. Significantly, the epitope reactivity of recombinant PrP was almost identical to that of the cell surface molecule. This finding provides direct evidence that the conformations adopted by the recombinant preparations used in structural studies of PrP closely approximate to that of PrPC in its native state.
Only Fabs binding to epitope region III recognized PrP 27-30 prior to treatment with denaturant (22). In contrast, although available in PrPC, epitope I was not reactive in PrP 27-30 prior to treatment with and removal of denaturant. This same pattern was observed for the antibody 3F4 which binds in the region of residues 109 to 112 (22, 30). These findings suggest that the C-terminal portion of PrPC, which contains a highly ordered structural core composed of helices B and C, remains relatively unaltered as PrPC is converted to PrPSc, whereas the N-terminal portion of the molecule undergoes extensive conformational rearrangement in which epitopes in the N terminus are either altered or buried in PrPSc. This conclusion is supported by protein engineering studies showing that this region of PrP is essential for PrPSc formation (20), by spectrophotometric studies which illustrate conformational plasticity in synthetic peptides corresponding to residues 90-145 (35), and by NMR studies which indicate that the N-terminal portion of PrP between residues 29 and 124 is highly flexible (8, 29).
Recombinant Fabs which did not recognize short linear amino acid sequences exhibited largely similar PrP reactivities, binding to PrPC on the cell surface, recombinant PrP(90-231), and PrP 27-30 rods following incubation with denaturing agents. These data imply similar epitope presentation between native PrPC, recombinant PrP(90-231), and denaturant-treated PrPSc. Hence, following denaturation in GdnSCN, presumably to a random coil state, PrP does not refold into the infective form but rather into a PrPC-like conformation. Although the epitopes of these antibodies have yet to be identified, this study does indicate that their binding sites are highly conformationally sensitive and are probably formed from secondary and possibly tertiary structural elements of PrP.
Acknowledgments
This work was supported by National Institutes of Health grants NS14069, AG02132, NS22786, and AG10770.
REFERENCES
- 1.Barbas C F, Lerner R A. Combinatorial immunoglobulin libraries on the surface of phage (Phabs): rapid selection of antigen-specific Fabs. In: Lerner R A, Burton D R, editors. Methods: a companion to methods in enzymology. Orlando, Fla: Academic Press; 1991. pp. 119–124. [Google Scholar]
- 2.Barry R A, Prusiner S B. Monoclonal antibodies to the cellular and scrapie prion proteins. J Infect Dis. 1986;154:518–521. doi: 10.1093/infdis/154.3.518. [DOI] [PubMed] [Google Scholar]
- 3.Borchelt D R, Scott M, Taraboulos A, Stahl N, Prusiner S B. Scrapie and cellular prion proteins differ in their kinetics of synthesis and topology in cultured cells. J Cell Biol. 1990;110:743–752. doi: 10.1083/jcb.110.3.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Büeler H, Fischer M, Lang Y, Bluethmann H, Lipp H P, DeArmond S J, Prusiner S B, Aguet M, Weissmann C. Normal development and behaviour of mice lacking the neuronal cell-surface PrP protein. Nature. 1992;356:577–582. doi: 10.1038/356577a0. [DOI] [PubMed] [Google Scholar]
- 5.Burton D R, Barbas C F. Human antibodies from combinatorial libraries. Adv Immunol. 1994;57:191–280. doi: 10.1016/s0065-2776(08)60674-4. [DOI] [PubMed] [Google Scholar]
- 6.Caughey B W, Raymond G J. The scrapie-associated form of PrP is made from a cell surface precursor that is both protease- and phospholipase-sensitive. J Biol Chem. 1991;266:18217–18223. [PubMed] [Google Scholar]
- 7.Ditzel H J, Binley J M, Moore J P, Sodroski J, Sullivan N, Sawyer L S W, Hendry R M, Yang W-P, Barbas C F, Burton D R. Neutralizing recombinant human antibodies to a conformational V2- and CD4-binding site-sensitive epitope of HIV-1 gp120 isolated by using an epitope-masking procedure. J Immunol. 1995;154:893–906. [PubMed] [Google Scholar]
- 8.Donne D G, Viles J H, Groth D, Mehlhorn I, James T L, Cohen F E, Prusiner S B, Wright P E, Dyson H J. Structure of the recombinant ful-length hamster prion protein PrP(29-231): the N-terminus is highly flexible. Proc Natl Acad Sci USA. 1997;94:13452–13457. doi: 10.1073/pnas.94.25.13452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gabizon R, McKinley M P, Prusiner S B. Purified prion proteins and scrapie infectivity copartition into liposomes. Proc Natl Acad Sci USA. 1987;84:4017–4021. doi: 10.1073/pnas.84.12.4017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gabizon R, Prusiner S B. Prion liposomes. Biochem J. 1990;266:1–14. doi: 10.1042/bj2660001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gasset M, Baldwin M A, Fletterick R J, Prusiner S B. Perturbation of the secondary structure of the scrapie prion protein under conditions that alter infectivity. Proc Natl Acad Sci USA. 1993;90:1–5. doi: 10.1073/pnas.90.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huse W D, Sastry L, Iverson S A, Kang A S, Alting-Mees M, Burton D R, Benkovic S J, Lerner R A. Generation of a large combinatorial library of the immunoglobulin repertoire in phage lambda. Science. 1989;246:1275–1281. doi: 10.1126/science.2531466. [DOI] [PubMed] [Google Scholar]
- 13.James T L, Liu H, Ulyanov N B, Farr-Jones S, Zhang H, Donne D, Kaneko K, Groth D, Mehlhorn I, Cohen F E, Prusiner S B. Solution structure of a 142-residue recombinant prion protein corresponding to the infectious fragment of the scrapie isoform. Proc Natl Acad Sci USA. 1997;94:10086–10091. doi: 10.1073/pnas.94.19.10086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaneko K, Peretz D, Pan K-M, Blochberger T C, Wille H, Gabizon R, Griffith O H, Cohen F E, Baldwin M A, Prusiner S B. Prion protein (PrP) synthetic peptides induce cellular PrP to acquire properties of the scrapie isoform. Proc Natl Acad Sci USA. 1995;92:11160–11164. doi: 10.1073/pnas.92.24.11160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kascsak R J, Rubenstein R, Merz P A, Tonna-DeMasi M, Fersko R, Carp R I, Wisniewski H M, Diringer H. Mouse polyclonal and monoclonal antibody to scrapie-associated fibril proteins. J Virol. 1987;61:3688–3693. doi: 10.1128/jvi.61.12.3688-3693.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klebe R J, Ruddle F H. Neuroblastoma: cell culture analysis of a differentiating stem cell system. J Cell Biol. 1969;43:69a. [Google Scholar]
- 17.Masters C L, Gajdusek D C, Gibbs C J., Jr Creutzfeldt-Jakob disease virus isolations from the Gerstmann-Straussler syndrome with an analysis of the various forms of amyloid plaque deposition in the virus-induced spongiform encephalopathies. Brain. 1981;104:559–588. doi: 10.1093/brain/104.3.559. [DOI] [PubMed] [Google Scholar]
- 18.McKinley M P, Meyer R K, Kenaga L, Rahbar F, Cotter R, Serban A, Prusiner S B. Scrapie prion rod formation in vitro requires both detergent extraction and limited proteolysis. J Virol. 1991;65:1340–1351. doi: 10.1128/jvi.65.3.1340-1351.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mehlhorn I, Groth D, Stockel J, Moffat B, Reilly D, Yansura D, Willet W S, Baldwin M, Fletterick R, Cohen F E, Vandlen R, Henner D, Prusiner S B. High-level expression and characterization of a purified 142-residue polypeptide of the prion protein. Biochemistry. 1996;35:5528–5537. doi: 10.1021/bi952965e. [DOI] [PubMed] [Google Scholar]
- 20.Muramoto T, Scott M, Cohen F E, Prusiner S B. Recombinant scrapie-like prion protein of 106 amino acids is soluble. Proc Natl Acad Sci USA. 1997;93:15457–15462. doi: 10.1073/pnas.93.26.15457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pan K-M, Baldwin M, Nguyen J, Gasset M, Serban A, Groth D, Mehlhorn I, Huang Z, Fletterick R J, Cohen F E, Prusiner S B. Conversion of α-helices into β-sheets features in the formation of the scrapie prion protein. Proc Natl Acad Sci USA. 1993;90:10926–10966. doi: 10.1073/pnas.90.23.10962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peretz D, Williamson R A, Matsunaga Y, Serban H, Pinilla C, Bastidas R, Rozenshteyn R, Papahadjopoulos D P, James T J, Houghten R A, Cohen F E, Prusiner S B, Burton D R. A conformational transition at the N-terminus of the prion protein features in formation of the scrapie isoform. J Mol Biol. 1997;273:614–622. doi: 10.1006/jmbi.1997.1328. [DOI] [PubMed] [Google Scholar]
- 23.Petersen G, Song D, Hugle-Dorr B, Oldenburg I, Bautz E K. Mapping of linear epitopes recognized by monoclonal antibodies with gene-fragment phage display libraries. Mol Gen Genet. 1995;249:425–431. doi: 10.1007/BF00287104. [DOI] [PubMed] [Google Scholar]
- 24.Prusiner S B. Transgenetics of prion diseases. Curr Top Microbiol Immunol. 1996;206:275–304. doi: 10.1007/978-3-642-85208-4_14. [DOI] [PubMed] [Google Scholar]
- 25.Prusiner S B. Molecular biology and pathogenesis of prion diseases. Trends Biochem Sci. 1996;21:482–487. doi: 10.1016/s0968-0004(96)10063-3. [DOI] [PubMed] [Google Scholar]
- 26.Prusiner S B, Groth D, Serban A, Koehler R, Foster D, Torchia M, Burton D R, Yang S-L, DeArmond S J. Ablation of the prion protein (PrP) gene in mice prevents scrapie and facilitates production of anti-PrP antibodies. Proc Natl Acad Sci USA. 1993;90:10608–10612. doi: 10.1073/pnas.90.22.10608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Prusiner S B, McKinley M P, Bowman K A, Bolton D C, Bendheim P E, Groth D F, Glenner G G. Scrapie prions aggregate to form amyloid-like birefringent rods. Cell. 1983;35:349–358. doi: 10.1016/0092-8674(83)90168-x. [DOI] [PubMed] [Google Scholar]
- 28.Riek R, Hornemann S, Wider G, Billeter M, Glockshuber R, Wuthrich K. NMR structure of the mouse prion protein domain PrP (121-231) Nature. 1996;382:180–182. doi: 10.1038/382180a0. [DOI] [PubMed] [Google Scholar]
- 29.Riek R, Hornemann S, Wider G, Glockshuber R, Wuthrich K. NMR characterization of the full-length recombinant murine prion protein, mPrP(23-231) FEBS Lett. 1997;413:282–288. doi: 10.1016/s0014-5793(97)00920-4. [DOI] [PubMed] [Google Scholar]
- 30.Rogers M, Serban D, Gyuris T, Scott M, Torchia T, Prusiner S B. Epitope mapping of the syrian hamster prion protein utilizing chimeric and mutant genes in a vaccinia virus expression system. J Immunol. 1991;147:3568–3574. [PubMed] [Google Scholar]
- 31.Safar J, Roller P P, Gajdusek D C, Gibbs C J., Jr Conformational transitions, dissociation and unfolding of scrapie amyloid (prion) protein. J Biol Chem. 1993;268:20276–20284. [PubMed] [Google Scholar]
- 32.Taraboulos A, Jendroska K, Serban D, Yang S-L, DeArmond S J, Prusiner S B. Regional mapping of prion proteins in brain. Proc Natl Acad Sci USA. 1992;89:7620–7624. doi: 10.1073/pnas.89.16.7620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Williamson R A, Burioni R, Sanna P P, Partridge L J, Barbas C F, Burton D R. Human monoclonal antibodies against a plethora of viral pathogens from single combinatorial libraries. Proc Natl Acad Sci USA. 1993;90:4141–4145. doi: 10.1073/pnas.90.9.4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Williamson R A, Peretz D, Smorodinsky N, Bastidas R, Serban A, Mehlhorn I, DeArmond S, Prusiner S B, Burton D R. Circumventing tolerance in order to generate autologous monoclonal antibodies to the prion protein. Proc Natl Acad Sci USA. 1996;93:7279–7282. doi: 10.1073/pnas.93.14.7279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang H, Kaneko K, Nguyen J T, Livshits T L, Baldwin M A, Cohen F E, James T L, Prusiner S B. Conformational transitions in peptides containing two putative α-helices of the prion protein. J Mol Biol. 1995;250:514–526. doi: 10.1006/jmbi.1995.0395. [DOI] [PubMed] [Google Scholar]