Summary
Plants establish symbiotic associations with arbuscular mycorrhizal fungi (AMF) to facilitate nutrient uptake, particularly in nutrient-limited conditions. This partnership is rooted in the plant’s ability to recognize fungal signaling molecules, such as chitooligosaccharides (chitin) and lipo-chitooligosaccharides. In the legume Medicago truncatula, chitooligosaccharides trigger both symbiotic and immune responses via the same lysin-motif-receptor-like kinases (LysM-RLKs), notably CERK1 and LYR4. The nature of plant-fungal engagement is opposite according to the outcomes of immunity or symbiosis signaling, and as such, discrimination is necessary, which is challenged by the dual roles of CERK1/LYR4 in both processes. Here, we describe a LysM-RLK, LYK8, that is functionally redundant with CERK1 for mycorrhizal colonization but is not involved in chitooligosaccharides-induced immunity. Genetic mutation of both LYK8 and CERK1 blocks chitooligosaccharides-triggered symbiosis signaling, as well as mycorrhizal colonization, but shows no further impact on immunity signaling triggered by chitooligosaccharides, compared with the mutation of CERK1 alone. LYK8 interacts with CERK1 and forms a receptor complex that appears essential for chitooligosaccharides activation of symbiosis signaling, with the lyk8/cerk1 double mutant recapitulating the impact of mutations in the symbiosis signaling pathway. We conclude that this novel receptor complex allows chitooligosaccharides activation specifically of symbiosis signaling and helps the plant to differentiate between activation of these opposing signaling processes.
Keywords: Medicago truncatula, arbuscular mycorrhizal fungi, chitooligosaccharides, lysin motif receptor-like kinase, plant immunity, plant symbiosis
Graphical abstract
Highlights
-
•
Medicago LYK8 is functionally redundant with CERK1 for mycorrhizal fungi symbiosis
-
•
LYK8 cannot bind to chitin but is involved in chitin-mediated symbiosis signaling
-
•
LYK8 is not required for chitin-induced plant immunity
-
•
LYK8 forms a receptor complex with CERK1 and DMI2 to activate symbiosis signaling
The establishment of arbuscular mycorrhizal fungi (AMF) symbiosis necessitates the recognition of fungal signals by plant receptors. Zhang et al. discover a LysM receptor that is functionally redundant with CERK1 for perceiving chitin produced by AMF, thereby specifically activating symbiosis signaling and enhancing AMF colonization.
Introduction
The majority of terrestrial plant species can establish a symbiotic relationship with arbuscular mycorrhizal fungi (AMF) to enhance nutrient uptake.1,2 This symbiosis boosts plant growth and plays a pivotal role in the regulation of the global terrestrial biogeochemical cycles.3 For this relationship to be established, plants must recognize AMF through the activation of a conserved symbiosis signaling pathway.1,4 This activation occurs through the perception of fungal signaling molecules, known as “Myc Factors.” 5 Myc factors comprise chitooligosaccharides (Myc-COs) from fungal cell walls,6,7 which are oligomers of N-acetyl glucosamine, and lipo-chitooligosaccharides (Myc-LCOs) secreted by AMF.8 Myc-LCO is distinguished by substitutions on the nonreducing end of the molecule of N-acyl moieties and by additional decorations onto this basic backbone.8 These LCOs are structurally similar to the nodulation factors (NFs) generated by symbiotic nitrogen-fixing rhizobium bacteria.8 Upon CO/LCO perception, symbiosis signaling is activated through nuclear calcium oscillations and resultant symbiotic gene expression to facilitate AMF infection.4 Additionally, co-inoculation of CO and AMF promotes strigolactone biosynthesis, stimulating intracellular accommodation and arbuscule development of AMF in plant roots.9
Studies in various plant species have demonstrated that plant lysin-motif receptor-like kinases (LysM-RLKs) play a central role in recognizing CO and LCO molecules.10,11,12,13,14 In the legume Medicago truncatula (M. truncatula), CO oligomers ranging from 4 to 8 units can activate symbiosis signaling.6,7,15 Receptors required for this process include two LysM-RLKs, MtCERK1 (hereafter designated as CERK1) and MtLYR4 (hereafter LYR4).7,16 Both receptors function in the CO-activation of nuclear calcium oscillations and resultant transcriptional upregulation of symbiotic gene expression, upon treatments with either CO4 (N-acetyl chitotetraose) or CO8 (N-acetyl chitooctaose).7 cerk1 displays a significant reduction in AMF colonization, highlighting a role for CO signaling in arbuscular mycorrhizal symbiosis (AMS).7,16 This has been similarly observed in other species where CERK1 orthologs have been mutated, such as rice (Oryza sativa), pea (Pisum sativum), banana (Musa spp.), and Parasponia andersonii (P. andersonii).17,18,19,20,21,22,23 Besides CO, AMF also produce LCO,8 and several LysM-RLKs have been shown to bind LCO in legumes24,25,26,27,28,29,30 and act as pivotal components for LCO detection in the root nodule symbiosis (RNS), notably MtNFP/LjNFR5 and MtLYK3/LjNFR1 in legumes M. truncatula and Lotus japonicus.31,32,33,34,35,36,37 These LCO receptors, which are essential for the RNS, also contribute to the AMS, as evidenced by a quantitative reduction in AMF colonization in the cerk1/nfp double mutant, as compared with cerk1 alone.7 Furthermore, NFP orthologs in tomato (Solanum lycopersicum), Petunia hybrida, and barley (Hordeum vulgare) contribute to AMF colonization,38,39,40,41 although, at least in barley, the function of NFP homologs is not limited to LCO signaling alone, as they also contribute to CO signaling.41 Unlike the case for the RNS, for AMF infection, no mutation in a single LysM-RLK, in any species, leads to almost complete abolishment of AMF colonization,7,29,42,43 suggesting the presence of additional unknown receptors involved in AMF induction of symbiosis signaling.
Besides their role as symbiotic signals, COs also act as microbe-associated molecular patterns (MAMPs), triggering plant immune responses, such as the generation of reactive oxygen species (ROS), mitogen-activated protein kinase (MAPK) phosphorylation, and initiation of defense gene expression.2,14,44,45 Recognition of CO8, and to a lesser extent CO4, activates defense responses in various plant species, and this requires CERK1 and LYR4 in M. truncatula.7,45 Activation of both immunity and symbiosis signaling through the same receptors7,16,18,19,20,21 suggests that plants might not be able to differentiate AMF from pathogenic fungi, solely through CO recognition.7 To establish a successful infection in plant roots, AMF can suppress plant immunity through LCO, short-chain CO, or secreted effectors and proteins.7,40,46,47,48,49 However, how plants discriminate between the activation of immunity and symbiosis signaling remains unknown.
CERK1 is classified within the LYK subfamily of LysM-RLKs with potentially active kinase domains.11 This LYK family has 11 members (MtLYK1–MtLYK11; CERK1 is MtLYK9)11 in M. truncatula, which is greater than the equivalent in the non-symbiotic plant species Arabidopsis thaliana (A. thaliana). This expansion may suggest an evolutionary adaptation for engaging in symbiotic microbe associations. In M. truncatula, CERK1 is the major receptor mediating AMF recognition and symbiosis, but its mutant phenotype suggests functional redundancy with other receptors that still allow some level of AMF colonization.7,16 In this study, we identified a further LysM-RLK in the CERK1 family, MtLYK8 (LYK8 hereafter), which exhibits functional redundancy with CERK1 in the AMS but plays no role in CO-activated plant immunity. We demonstrate the dominance of CO signaling in AMS and show how LYK8 differentiates between CO-activation of symbiosis and immunity signaling.
Results
LYK8 is functionally redundant with CERK1 for AMF colonization
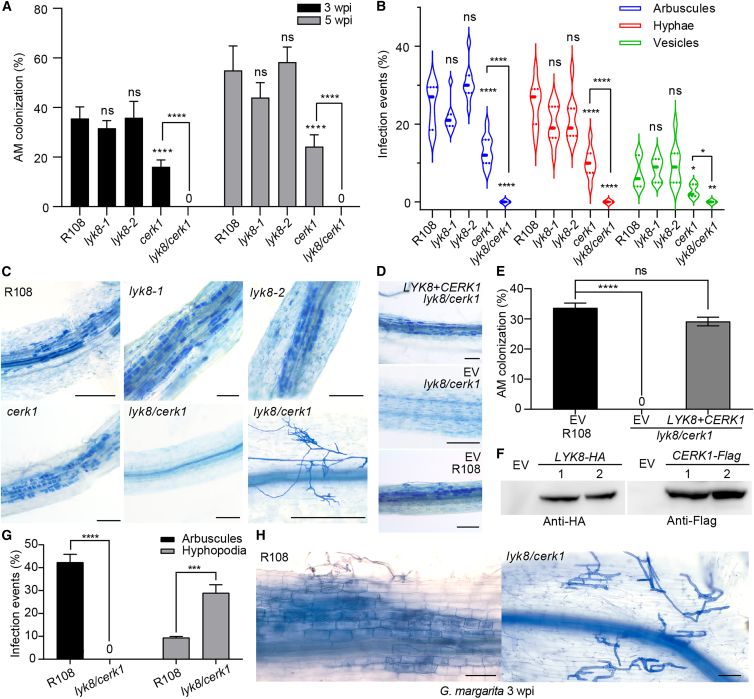
To uncover LysM-RLKs in the LYK family that might participate in AMS, we obtained Tnt1 insertion mutant lines for several LysM-RLK genes,11,50 including MtLYK6, MtLYK7, MtLYK8, and MtLYK10 in M. truncatula. The homozygous mutant alleles were validated for the absence of detectable transcripts using semi-quantitative reverse transcription polymerase chain reaction, and they were designated as lyk6, lyk7, lyk8 (lyk8-1 and lyk8-2; two mutant alleles with insertions in different domains of LYK8), and lyk10, respectively (Figures S1A–S1C). Subsequently, wild-type and mutant plant roots were inoculated with Rhizophagus irregularis (R. irregularis) spores to assess fungal colonization. As a control, the cerk1 mutant displayed significantly reduced fungal infection and colonization, consistent with prior observations7,16 (Figures 1A, 1B, S1D, and S1E). The mutations of LYK6, LYK7, LYK8, and LYK10, individually, appeared to have no effect on total AMF colonization and fungal infection (Figures 1A, 1B, S1D, and S1E). To investigate whether these LysM-RLKs function redundantly with CERK1 for AMS, we genetically crossed cerk1 with the new receptor mutants, producing several double-mutant lines. Surprisingly, when inoculated with a low concentration of spores, lyk8-1/cerk1 (lyk8/cerk1 hereafter) roots exhibited a complete loss of arbuscules, intercellular hyphae, and vesicles, while infection events in lyk6/cerk1, lyk7/cerk1, and lyk10/cerk1 were comparable to those in cerk1 alone (Figures 1A–1C, S1D, and S1E). Higher doses of AMF inoculum led to higher levels of hyphopodium development on the root surface of lyk8/cerk1 than on the root surface of wild-type plants, but the fungal hyphae did not penetrate the rhizodermis to form arbuscules in the mutant (Figures 1C and S2A). This deficiency in fungal colonization persisted, even extending to 7 weeks post inoculation (Figure S2C). Additionally, co-cultivation of wild-type nurse plants with lyk8/cerk1 increased the number of hyphopodia development in the mutant but failed to restore AMF infection and arbuscule development (Figures S2A and 2B). Altogether, this demonstrates the persistent lack of fungal infection, even with increased inoculum strength and extended inoculation time.
Figure 1.
lyk8/cerk1 double mutant exhibited no AMF colonization
(A) Colonization of R. irregularis is represented as the percentage of root length colonization, performed at 3 and 5 weeks post inoculation (wpi).
(B) Violin plot illustrates infection events quantified at 3 wpi. This experiment was repeated three times with similar results.
(C) Images of R. irregulars inoculated roots stained with ink. The hyphopodia (bottom right image) was observed in the roots of 5 wpi of high concentration of fungal spores. Scale bars, 150 μm.
(D and E) AMF colonization and corresponding images from a complementation experiment in lyk8/cerk1 double mutant by hairy root transformation. The empty vector (EV) was transferred as a control. Scale bars, 150 μm.
(F) The expression levels of LYK8 and CERK1 in the complementation roots were detected by western blot. Results from two individual transgenic roots are shown.
(G) Arbuscules and hyphopodia were quantified in plants inoculated with G. margarita at 3 wpi. Statistically significant differences were determined using Student’s t test (mean ± SEM, n = 10).
(H) Images show the ink-stained colonization of G. margarita in plant roots. Scale bars, 100 μm.
(A, B, and E) Asterisks denote statistical significance as calculated by one-way ANOVA and Tukey’s multiple comparison (mean ± SEM, n = 10).
See also Figures S1–S3.
To confirm that the mutation of LYK8 in the cerk1 background hinders fungal colonization, we undertook a genetic complementation of the lyk8/cerk1 mutant using the coding sequences of LYK8 and CERK1. We introduced both genes in a single construct, driven by their native promoters, into lyk8/cerk1 roots via Agrobacterium rhizogenes-mediated hairy root transformation. Successful transformation resulted in complete restoration of fungal infection and colonization (Figures 1D and 1E), accompanied by 100% complementary events in ten individual transgenic roots in which the expression of LYK8 and CERK1 proteins were detected in the roots (Figure 1F). These data reveal that LYK8 is redundant with CERK1 in AMS. In the single lyk8 mutant, the signaling function is fulfilled by the major receptor CERK1, resulting in fungal colonization similar to that of the wild-type plant (Figures 1A–1C).
To determine whether the lyk8/cerk1 mutant is specifically deficient in symbiosis with R. irregularis or impacts root symbiosis with other AMF as well, we inoculated wild-type and lyk8/cerk1 mutant plants with Gigaspora margarita (G. margarita), which is phylogenetically distant from the model R. irregularis. 3 weeks post inoculation, only hyphopodia were observed, with a complete absence of arbuscules in the cortical cells of the lyk8/cerk1 mutant (Figures 1G and 1H). This finding further supports the critical role of LYK8 and CERK1 in facilitating symbiotic relationships between plant roots and various AMF species.
AMS and RNS share the common symbiosis signaling pathway.4 To examine whether LYK8 and CERK1 play a role in the recognition of rhizobia, we inoculated Ensifer meliloti Em1021 on wild-type and mutant roots. We observed that mutation neither in LYK8 alone nor in conjunction with CERK1 impaired nodulation, suggesting their specific relevance in the AMS (Figure S2D).
Studies have shown that AMF can induce lateral root primordia formation and increase lateral root densities in a CERK1-dependent manner in diverse plant species, including M. truncatula.51 To investigate whether LYK8 also plays a role in root development in response to AMF, we assessed the root architecture response of wild-type and receptor mutant plants. Interestingly, both lyk8 and cerk1 plants exhibited an increase in first-order lateral root numbers in the absence of AMF inoculation, suggesting their potential involvement in lateral root development (Figure S2G). However, LYK8 appears to have no role in the regulation of primary root growth (Figure S2H). Notably, the promotion of lateral root number by AMF was absent in lyk8 plants, similar to the observations in cerk1 and lyk8/cerk1 mutants (Figure S2G), highlighting that LYK8 and CERK1 are both essential for AMF-induced root responses and do not play redundant roles. This indicates that these two receptors might form a receptor complex for the perception of AMF-produced signal molecules, thereby regulating root development.
To understand how CERK1 and LYK8 occupy overlapping signaling functions in the AMS, we examined their promoter activity during AMF infection. Both LYK8 and CERK1 exhibited similar expression patterns in root tissues, as evidenced by promoter β-glucuronidase (GUS) assays and wheat germ agglutinin (WGA) staining (Figures S3A–S3H). Without R. irregularis inoculation, the promoters of both LYK8 and CERK1 were expressed throughout various root cell layers (Figures S3A and S3B). These expression patterns were maintained during the early stages of AMF infection, including hyphopodium formation (Figures S3C and S3D) and the extension of intraradical hyphae into the root cells (Figures S3E and S3F). Interestingly, fungal colonization slightly increased receptor expression in arbuscule-containing cells (Figures S3G and S3H), consistent with the observations found in the Medicago gene expression database,47 suggesting a modality of positive feedback colonization in root cells and receptor expression. However, neither LYK8 nor CERK1 alone are required for arbuscule development7 (Figure S1F). Quantitative real-time polymerase chain reaction (real-time qPCR) revealed an induction of LYK8 expression post fungal root colonization (Figure S4A) that is likely the result of LYK8 transcriptional induction by CO and LCO (Figure S4B). Taken together, these findings demonstrate that LYK8 functions synergistically with CERK1 in AMF colonization and that these two receptors show very similar expression patterns.
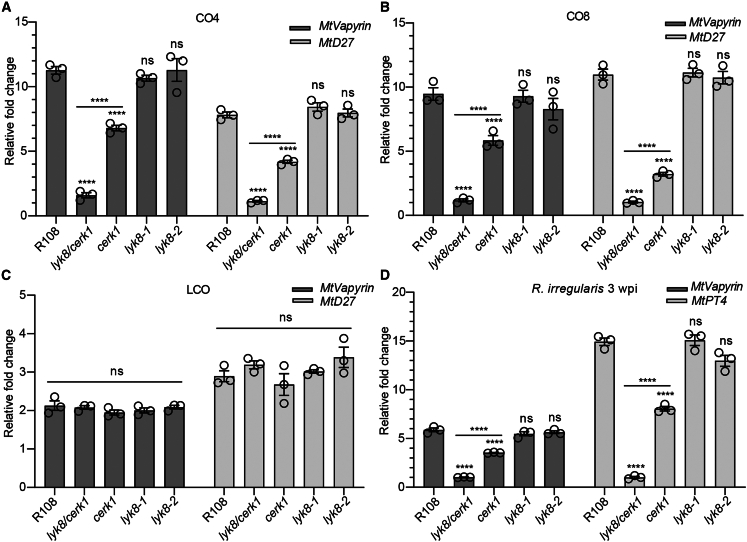
LYK8 cannot bind to CO but is involved in CO-medicated symbiosis signaling
The essential role of LYK8 and CERK1 in fungal infection implies that similar to CERK1, LYK8 may participate in early symbiosis signaling initiated by symbiotic elicitors. To test this, we evaluated CO- and LCO-triggered gene expression of symbiotic marker genes7 in lyk8-1, lyk8-2, cerk1, and lyk8/cerk1 mutants. We found that wild-type plants exhibited CO4- and CO8-driven induction of symbiotic genes, with this induction being partially dependent on CERK1, but independent of LYK8 (Figures 2A, 2B, S4C, and S4D). However, the lyk8/cerk1 showed no detectable induction of the tested genes by CO, while maintaining their induction by LCO (Figures 2A–2C, S4C, and S4D). During R. irregularis inoculation, the induction of MtVapyrin, MtPT4, and MtLYK10 was disrupted in lyk8/cerk1 roots after 3 and 5 weeks of inoculation, in line with the colonization of this mutant (Figures 2D, S4E, and S4F). Our previous studies highlighted the combined importance of CO and LCO signaling for AMS, noting a significant reduction of AMF colonization in cerk1/nfp roots compared with cerk1 alone.7 Interestingly, AMF colonization and infection patterns showed no defects in lyk8/nfp roots, indicating the dominant role of CERK1 in CO signaling that might compensate for the deficiency of lyk8/nfp in AMF colonization (Figures S2E and S2F). Collectively, our findings demonstrate that LYK8 and CERK1 have overlapping roles in modulating CO-triggered symbiotic gene expression.
Figure 2.
lyk8/cerk1 completely interrupted the expression of symbiotic gene induced by CO and AMF
(A–C) Real-time qPCR analysis assessed the expression of symbiotic marker genes in the roots of both the wild type and receptor mutants following treatments with H2O, 10−8 M CO4 (A), 10−8 M CO8 (B), and 10−8 M LCO (C) for 6 h. Relative fold change compared with individual water treatments is shown.
(D) The relative fold change represents the expression of MtVapyrin and MtPT4 detected in the plant roots 3 wpi with R. irregularis, compared with the expression in non-inoculated roots.
(A–D) Asterisks denote statistical significance as calculated by one-way ANOVA and Tukey’s multiple comparison (mean ± SEM, n = 8). These results have three independent biological replications.
See also Figure S4.
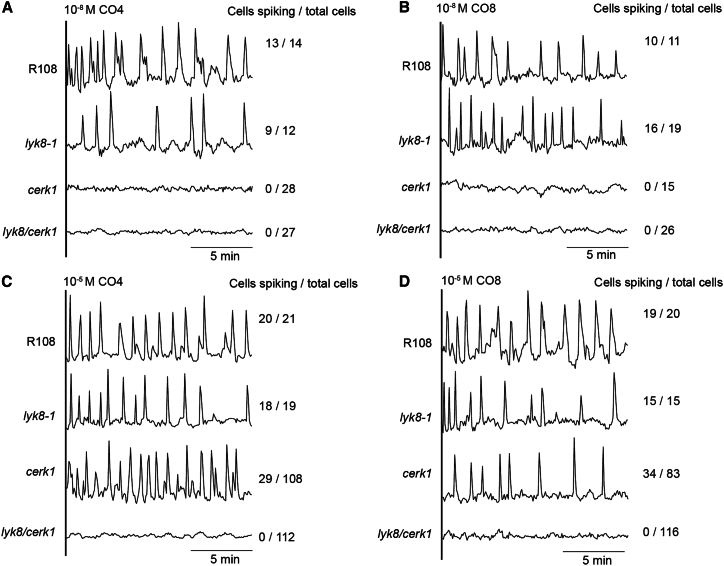
Periodic calcium oscillations in the nucleus serve as an indicator of symbiosis signaling, activated by both CO and LCO.15 To delve deeper into the role of LYK8 in CO-triggered symbiosis signaling, we analyzed calcium oscillations in the roots of wild type and receptor mutants using the calcium reporter, Yellow Cameleon YC3.6.7 In line with prior research, the cerk1 single mutant displayed a disruption in calcium oscillations in atrichioblast cells when exposed to low concentrations of CO4 and CO87 (10−8 M; Figures 3A and 3B). However, cerk1 still showed a response when exposed to high concentrations of these molecules (10−5 M; Figures 3C and 3D), suggesting the involvement of additional receptor(s) at these concentrations of CO. In contrast to CERK1, the mutation of LYK8 alone exhibited no defect in CO4- and CO8-induced nuclear calcium oscillations (Figures 3A–3D). In the cerk1/lyk8 double mutant, we observed a complete loss of CO-induced calcium response, even at exceptionally high concentrations of CO4 and CO8 (10−4 M; Figures 3C, 3D, and S5). This observation underscores the redundant role of LYK8 and CERK1 in CO signaling. The LCO-triggered calcium response in lyk8 and lyk8/cerk1 mutants mirrored that of wild-type plants (Figure S5). These data, combined with the gene expression and root nodulation findings (Figures 2C and S2D), indicate that LYK8 plays a specific role in CO, but not LCO, signaling in M. truncatula.
Figure 3.
CO-triggered calcium oscillations are abolished in lyk8/cerk1
(A–D) Representative calcium traces of M. truncatula atrichoblasts from lateral roots were recorded in wild type, lyk8-1, cerk1, and lyk8/cerk1 in response to both a low concentration (10−8 M) of CO4 (A) and CO8 (B), as well as a high concentration (10−5 M) of CO4 (C) and CO8 (D). The traces denote the ratio of yellow fluorescent protein (YFP) to cyan fluorescent protein (CFP) in arbitrary units. The numbers marked on the right of the traces indicate the number of cells responding compared with the total number of cells analyzed.
See also Figure S5.
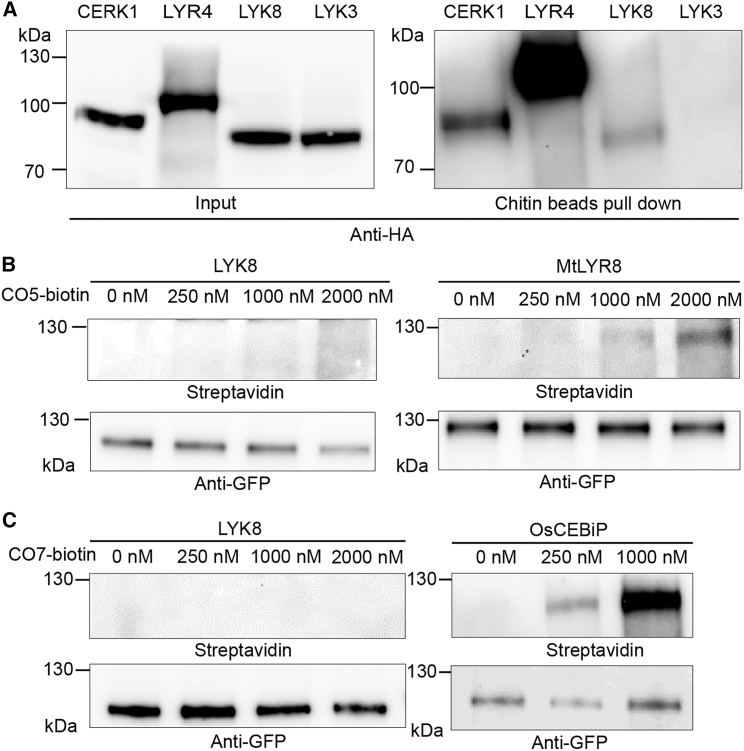
To determine whether LYK8 functions as a CO receptor, we transiently expressed LYK8 in Nicotiana benthamiana (N. benthamiana) leaves and assessed its ability to bind to chitin resin beads through a pull-down assay. The previously reported CO receptors, CERK1 and LYR4, served as positive controls, while the LCO receptor LYK3 served as the negative control.33,36 The result demonstrated that both LYR4 and CERK1 possess a strong affinity for chitin beads, consistent with previous studies7 (Figure 4A). LYK8 displayed much weaker chitin binding relative to CERK1 and LYR4, while no binding was evident for LYK3 (Figure 4A). However, the weak binding observed was unable to withstand the stringent washes following the chitin bead pull-down assay (data not shown). Additionally, we employed cross-linkable biotinylated versions of CO5 (CO5-biotin) and CO7 (CO7-biotin) to evaluate LYK8’s binding affinity to COs. Consistent with the results from the chitin beads, neither CO5-biotin nor CO7-biotin was able to bind to LYK8 at any concentration (Figures 4B and 4C). In contrast, as positive controls, the M. truncatula MtLYR8 protein demonstrated binding ability to CO5-biotin,52 and the rice OsCEBiP exhibited high-affinity binding ability to CO7-biotin53 (Figures 4B and 4C). These data suggest that LYK8 might not be sufficient to bind with CO and is likely to function as a co-receptor for LYK9 and LYR4 in the perception of CO molecules. Nevertheless, we cannot exclude the possibility of a weak binding affinity of LYK8 for COs, undetectable in the assays that we performed.
Figure 4.
LYK8 protein weakly binds to chitin beads but not to CO5 and CO7
(A) The LYK8 protein expressed in N. benthamiana leaves exhibits weak binding to chitin resin beads. CERK1 and LYR4 are positive controls. LYK3, the LCO receptor, is the negative control.
(B and C) Detecting the binding affinity of the LYK8 protein with CO5 and CO7. Chimeric YFP-tagged proteins were expressed in N. benthamiana leaves followed by incubation with CO5-biotin or CO7-biotin. The proteins were enriched using anti-GFP beads, and their presence was confirmed via western blotting with anti-GFP antibody and streptavidin. MtLYR8 served as the positive control for CO5 binding (B), and OsCEBiP was the positive control for CO7 binding (C).
LYK8 appears to have no role in CO-triggered plant immunity
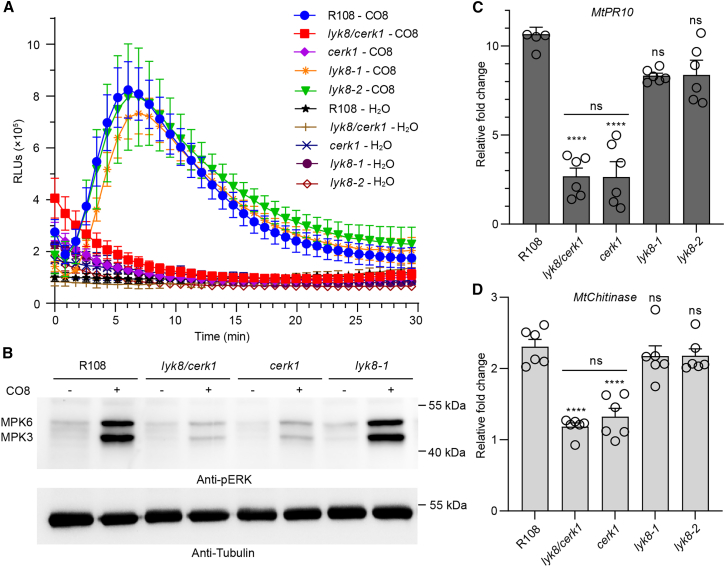
CERK1 is implicated in both CO8-induced plant immunity and symbiosis.7 This prompted us to postulate a potential role for LYK8 in plant immunity. To this end, we assessed early immune responses, specifically ROS production and MAPK activation, in the roots of wild type and receptor mutants following CO8 treatment. Notably, lyk8 roots exhibited ROS response and MPK3/6 phosphorylation at levels comparable to those of the wild type (Figures 5A, 5B, S6A, and S6B). Although cerk1 plants exhibited a modest activation of MPKs, this activation was LYK8 independent, as shown by the similar levels of activation between lyk8/cerk1 and cerk1 (Figures 5B and S6B). This suggests no functional overlap between LYK8 and CERK1 in CO8-triggered MAPK activation. Furthermore, through real-time qPCR analysis, we examined CO8-induced expression of defense genes such as MtPR10, MtChitinase, and MtRbohA.7 LYK8 is dispensable for the induction of these genes upon CO8 exposure and does not have a redundant role with CERK1 in defense regulation (Figures 4C, 4D, and S6C). Consequently, these data indicate that LYK8 does not act as a redundant receptor alongside CERK1 in CO8-trigged immune signaling.
Figure 5.
LYK8 is not involved in CO8-triggered immune signaling
(A) ROS production was measured in the roots of M. truncatula in response to both water and 10−6 M CO8 (mean ± SEM, n = 6).
(B) The phosphorylation of MPK3 and MPK6 in plant roots was detected by the anti-pERK antibody. The Tubulin protein served as a control to ensure equal loading.
(C and D) The real-time qPCR analysis of defense marker genes MtPR10 (C) and MtChitinase (D) induced by 10−6 M CO8 in wild type and receptor mutants. The relative fold change compared with water treatment is shown. This experiment was independently replicated three times. The asterisks denote statistical significance as calculated by one-way ANOVA and Tukey’s multiple comparison (mean ± SEM, n = 8).
See also Figure S6.
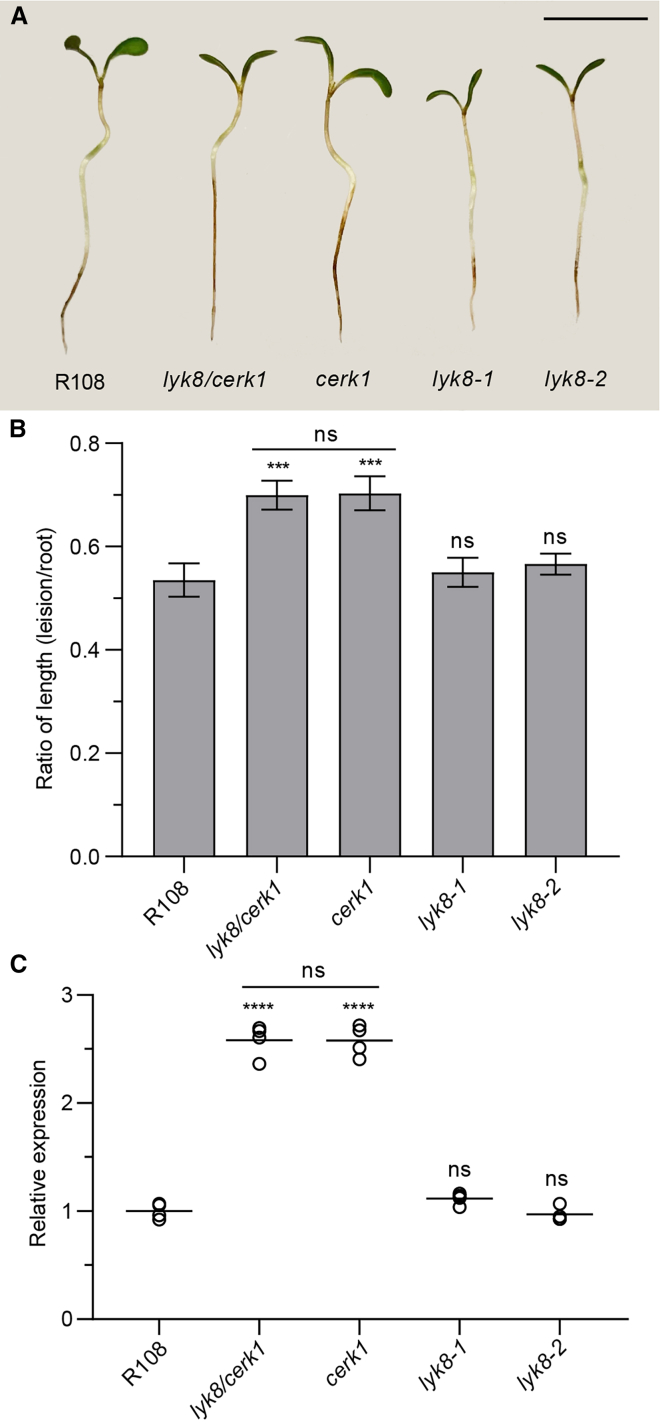
To investigate the role of LYK8 in regulating plant disease resistance, we evaluated the susceptibility to the root fungal pathogen, Fusarium oxysporum (F. oxysporum), in both the wild type and receptor mutants. By measuring the root lesion size of the infected samples, we found that roots of cerk1 were more susceptible to F. oxysporum (Figures 6A and 6B), while lyk8 showed susceptibility similar to the wild type, and lyk8/cerk1 exhibited susceptibility comparable to cerk1 (Figures 6A and 6B). This observation is corroborated by real-time qPCR analyses quantifying F. oxysporum biomass, which reflected the fungal infection trends observed in the measurement of root lesion sizes (Figure 6C). Collectively, these findings indicate that LYK8 is not involved in plant immunity.
Figure 6.
LYK8 is dispensable for M. truncatula resistance against a fungal pathogen
(A–C) 5-day-old M. truncatula seedling roots were inoculated with F. oxysporum spores.
(A) Root disease symptoms (scale bars, 1 cm).
(B) Quantification of the ratio of lesion size to the total length of the roots.
(C) Assessment of F. oxysporum biomass measured using the fungal gene FOW1, relative to the M. truncatula gene Ubiquitin.
(B and C) These experiments were independently replicated twice. Asterisks denote statistical significance as calculated by one-way ANOVA and Tukey’s multiple comparison (mean ± SEM, n = 15).
LYK8 forms a receptor complex with CERK1 for the activation of symbiosis signaling
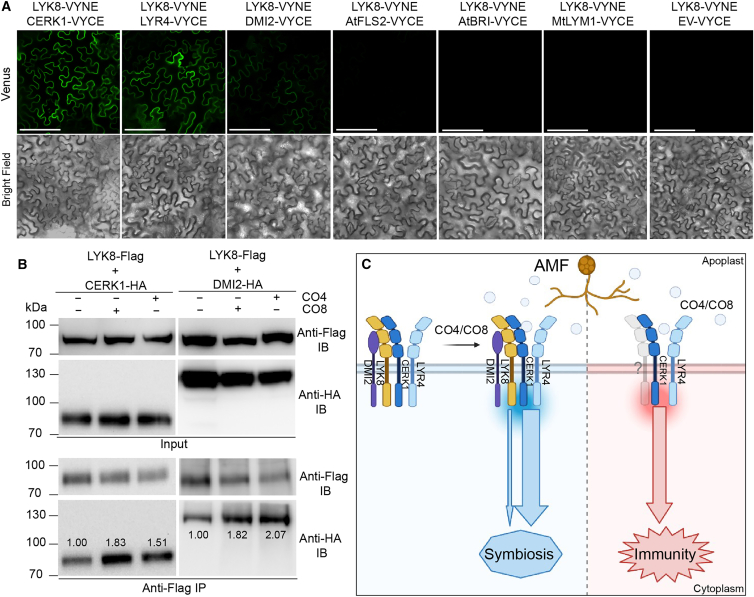
We hypothesized that LYK8 might partner with the identified CO receptors to form a receptor complex pivotal for the regulation of symbiosis signaling. To test this hypothesis, we employed a bimolecular fluorescence complementation (BiFC) assay, fusing LYK8 to the N-terminal part of yellow fluorescent protein Venus and the other receptors to the C-terminal part of Venus, subsequently expressing them in N. benthamiana. The results showed that LYK8 associates with CERK1 and LYR4 at the plasma membrane. However, we observed no interactions between LYK8 and the leucine-rich repeat receptor-like kinases AtFLS254 and AtBRI155 from A. thaliana or between LYK8 and the LysM receptor-like protein MtLYM156 from M. truncatula (Figure 7A). Additionally, while LYK8 can interact with DMI2 on the membrane, the fluorescence appears weak due to low expression of DMI2 in N. benthamiana (Figure 7A). Extending our investigation using a co-immunoprecipitation assay, we confirmed interactions between LYK8 and other receptors, namely CERK1, LYR4, and DMI2 (Figure S7A). Upon exposure to CO4 and CO8, the interactions of LYK8 with CERK1 and DMI2 were notably enhanced, while the interactions of LYK8 with LYR4 or LYK8 themselves remained unchanged (Figures 7B and S7B). This implies that the CERK1-LYK8-DMI2 receptor complex might play a pivotal role in mediating CO-triggered symbiosis signaling.
Figure 7.
LYK8 interacts with CERK1 and DMI2, and these interactions are enhanced by CO4 and CO8
(A) LYK8 interacts with CERK1, LYR4, and DMI2, but not the plasma-membrane localized protein AtFLS2, AtBRI1, and MtLYM1 in N. benthamiana leaves, evidenced by split-Venus analysis. Scale bars, 150 μm.
(B) CoIP validation of the interactions between LYK8 and CERK1/DMI2. The indicated proteins were transiently expressed in N. benthamiana leaves with or without treatment with 10−6 M of CO4 and CO8 for 10 min before protein extraction. The intensity of the bands for IP samples was quantified by ImageJ.
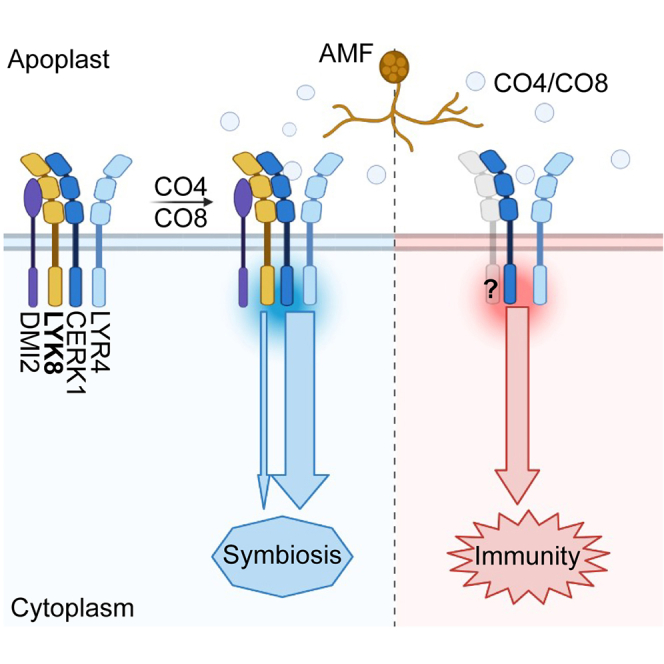
(C) Proposed model of LYK8- and CERK1-mediated CO perception for activation of symbiosis and immunity. LYK8 is associated with CERK1/DMI2/LYR4 in the pre-symbiotic status. AMF-produced CO molecules, such as CO4 and CO8, stimulate LYK8 expression, strengthening its interaction with CERK1/DMI2 to initiate symbiosis signaling. CERK1, a key CO receptor, may partner with another LysM-RLK for immune signaling.
See also Figure S7.
Discussion
AMF generate a range of signaling molecules, notably CO and LCO.6,8 These molecules activate symbiosis signaling and eventually enable host infection. This study reveals that CO-mediated symbiosis signaling is pivotal and sufficient to explain AMF root colonization, with LYK8 working alongside CERK1 for CO recognition (Figure 7C). The expression of LYK8 is upregulated by CO, AMF, and nutrient deprivation41 (Figures S4A and S4B), implying a role of LYK8 in reinforcing fungal perception, alongside constitutive CERK1. However, CERK1’s dominant role in CO signaling has overshadowed LYK8’s function, making it challenging to pinpoint mutants of LysM-RLKs that completely lack AMF colonization. The enhanced expression of LYK8 and CERK1 in arbuscule-containing cells suggests that CO recognition also occurs in the late stage of fungal infection. This is consistent with the observation of calcium oscillations in root cortical cells coincident with the fungal colonization of the root cortex,57 as well as the observation that application of CO can promote arbuscule development.9 This recognition likely promotes either the interactions between LYK8/CERK1 and CKL (CYCLIN-DEPENDENT-KINASE-LIKE) proteins or the phosphorylation of CKLs by the receptors, potentially enhancing lipid provisioning to the fungus.58 It seems that the role of LYK8 in the AMS is evolutionarily conserved in dicots. For instance, in P. andersonii, concurrent mutations of the potential LYK8-orthologous gene, PanLYK1, and the CERK1-orthologous gene, PanLYK3 (Figure S1G), lead to an absence of mature arbuscules in the cortical cells and a drastic reduction in fungal colonization.20 Similarly, suppressing the LYK8-ortholog SlLYK12 in tomatoes severely inhibits the AMS.59 However, unlike M. truncatula, mutants in these receptors still partially sustain fungal colonization, implying the presence of other receptors or signaling pathways for the AMS in these species.
Beyond CO signaling, we previously showed that LCO signaling plays a role in the AMS,7 albeit to a lesser extent than CO signaling. AMF might leverage LCO molecules to amplify CO-mediated symbiosis signaling under specific scenarios. Supporting this, rice and barley roots respond to LCO only under nutrient-deprived conditions.15,41 Recent work indicates that LCO is not exclusive to AMF but is also synthesized by other beneficial and pathogenic fungi.60 This implies that the function of LCO extends beyond simply activating symbiosis, possibly acting as a general signal for fungi.60 This signal could either regulate the metabolic profile of microbes in the soil to compete across microbial kingdoms61 or influence various host responses, like enhancing lateral root growth and suppressing plant immunity.7,8,15,46,48,51,62,63,64,65,66,67 Importantly, our work demonstrates that abolishing CO signaling alone is sufficient to block AMF colonization, analogous to the situation observed in mutants of the symbiosis signaling pathway.5 As such, we conclude that CO recognition acts as the principal factor for recognizing AMF and is sufficient to explain the activation of symbiosis signaling by AMF. Consistent with our conclusion, the application of CO in plant roots has a long-term effect through the development of AMS signaling.9 We proposed that LCO must play much more subtle roles in AMF colonization, with their negative impact on immunity signaling7,46,48,62,64 being one modality of action.
The differential responses of LYK8 and CERK1 in mediating immunity are intriguing. CERK1 functions in both immunity and symbiosis, while LYK8 is dedicated to CO-mediated symbiosis signaling. It remains possible that LYK8 might also contribute to immunity, potentially masked by the overlapping functions of other LysM-RLKs in M. truncatula. Current findings align with the observations on SlLYK12 and PanLYK1, where both receptors appear to be important for AMF colonization, without direct roles in plant immunity.20,59 Additionally, the L. japonicus receptor EPR3a, which can bind β-1,3/β-1,6-glucans from fungi, appears to be involved only in symbiosis and is not required for β-glucan-triggered plant immunity.68 A recent study demonstrated that nanobody induction of the formation of a receptor complex can specifically activate symbiosis signaling.69 Interestingly, CO treatments intensify interactions of LYK8 with CERK1 and DMI2, but not LYR4 (Figures 7B and S7B), hinting that receptors might form specialized complexes in the presence of CO to differentiate between immunity and symbiosis (Figure 7C). The underlying mechanisms of these interactions remain an avenue for further study. Taken together, this suggests that plants might recognize identical molecules but employ different signaling routes for symbiosis and immunity, potentially due to the combination of receptors and their downstream associates.
LysM-RLKs, responsible for detecting CO and LCO, predominantly pair an active kinase from the LYK subfamily with an inactive one from the LYR subfamily.11 While LYR4 is required for CO-induced symbiosis and immunity signaling, unlike CERK1, its mutation does not influence AMF colonization.7 This leads us to propose that other LYR receptors might be functionally interchangeable with LYR4 for the AMS, much in the way LYK8 is for CERK1. Notably, cerk1 and lyk8/cerk1 roots still maintain a similarly low level of MAPK activation in response to CO8 (Figure 5B). Considering LYK8 is not involved in immunity signaling, this suggests that another LYK receptor might have functional redundancy with CERK1, specifically regulating immune signaling (Figure 7C). The expansion of the LysM-RLK family in legumes highlights the adaptive evolution of these receptors to engage in microbial associations. Future studies could focus on deciphering the functional diversity of LysM-RLKs and their downstream components in regulating plant immunity and symbiosis.
Our study extends the understanding of the molecular mechanisms underpinning the AMS. The newly discovered role of LYK8, in particular its synergy with CERK1, serves as an illuminating addition to the broader puzzle of how plants recognize AMF to engage in symbiosis. This knowledge could be pivotal in future efforts to optimize and harness AMS for agricultural advantage. In addition, our findings contribute to a deeper understanding of the complex interplay between CO-mediated symbiosis signaling and immunity, revealing new insights into the strategies plants employ to dissect the two different signaling pathways to interact with beneficial microbes while maintaining defense mechanisms.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Phospho-p44/42 MAPK (Erk1/2) (Thr202/Tyr204) Rabbit mAb | Cell Signaling Technology | Cat#4370; RRID: AB_2315112 |
| Mouse monoclonal anti-Tubulin (clone B-5-1-2) | Sigma-Aldrich | Cat#T5168; RRID: AB_477579 |
| Mouse monoclonal ANTI-FLAG M2-Peroxidase antibody | Sigma-Aldrich | Cat#A8592-2MG; RRID: AB_439702 |
| Mouse monoclonal anti-GFP (clones 7.1 and 13.1) | Roche | Cat# 11814460001; RRID: AB_390913 |
| Rat monoclonal Anti-HA-Peroxidase antibody | Roche | Cat# 12013819001; RRID: AB_390917 |
| Goat anti-mouse IgG (H+L)-HRP Conjugate | Biorad | Cat# 1706516; RRID: AB_2921252 |
| Goat anti-rabbit IgG-HRP Conjugate | Sigma-Aldrich | Cat# A0545-1ML; RRID: AB_257896 |
| Bacterial and fungal strains | ||
| Ensifer meliloti Em1021 | Lab stock | N/A |
| Agrobacterium tumefaciens GV3101 | Lab stock | N/A |
| Agrobacterium rhizogenes AR1193 | Lab stock | N/A |
| Fusarium oxysporum | From Stephen Marek | N/A |
| Rhizophagus irregularis | Premier Tech | N/A |
| Gigaspora margarita | From Paola Bonfante | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| X-Gluc | Goldbio | B735 |
| WGA-Alexa Fluor 488 | Thermo Fisher | W11261 |
| Aminoethoxyvinylglycine (AVG) | Sigma-Aldrich | A6685 |
| CO4 | Megazyme | O-CHI4 |
| CO8 | From Sébastien Fort | N/A |
| LCOs | From Sébastien Fort | N/A |
| CO5-biotin | Cullimore et al.29 | N/A |
| CO7-biotin | Cullimore et al.29 | N/A |
| Peroxidase from horseradish | Sigma-Aldrich | P6782 |
| L-012 | Wako chemicals | 120-04891 |
| cOmplete EDTA-free Protease Inhibitor Cocktail | Roche | 11873580001 |
| PhosSTOP | Roche | 4906837001 |
| Chitin Resin | New England Biolabs | S6651 |
| ANTI-FLAG M2 Affinity Agarose Gel | Sigma-Aldrich | A2220 |
| ChromoTek GFP-Trap® Magnetic Agarose | Proteintech | gtma |
| Streptavidin, HRP conjugate | Thermo Fisher | S911 |
| RQ1 RNase-Free DNase | Promega | M6101 |
| Typan Blue | Sigma-Aldrich | T6146 |
| Critical commercial assays | ||
| NEBridge® Ligase Master Mix | New England Biolabs | M1100S |
| BsaI-HF®v2 | New England Biolabs | R3733 |
| BbsI-HF | New England Biolabs | R3539 |
| LunaScript RT SuperMix Kit | New England Biolabs | E3130 |
| Luna Universal qPCR Master Mix | New England Biolabs | M3003 |
| OneTaq Quick-Load 2X Master Mix | New England Biolabs | M0486 |
| Spectrum™ Plant Total RNA Kit | Sigma-Aldrich | STRN250 |
| Turface MVP | American Plant Products & Services | N/A |
| Vermiculite | American Plant Products & Services | N/A |
| Experimental models: Organisms/strains | ||
| M. truncatula R108 | Lab stock | N/A |
| M. truncatula R108 YC3.6 | Feng et al.7 | N/A |
| M. truncatula cerk1 | Feng et al.7 | NF16753 |
| M. truncatula nfp-3 | Feng et al.7 | NF7796 |
| M. truncatula lyk6 | Oklahoma State University | NF14155 |
| M. truncatula lyk7 | Oklahoma State University | NF8175 |
| M. truncatula lyk8-1 | Oklahoma State University | NF9395 |
| M. truncatula lyk8-2 | Oklahoma State University | NF11260 |
| M. truncatula lyk10 | Oklahoma State University | NF20763 |
| M. truncatula lyk8/cerk1 | This study | N/A |
| M. truncatula lyk8/nfp | This study | N/A |
| M. truncatula cerk1 YC3.6 | This study | N/A |
| M. truncatula lyk8-1 YC3.6 | This study | N/A |
| M. truncatula lyk8/cerk1 YC3.6 | This study | N/A |
| M. truncatula lyk6/cerk1 | This study | N/A |
| M. truncatula lyk7/cerk1 | This study | N/A |
| M. truncatula lyk10/cerk1 | This study | N/A |
| N. Benthamiana | Lab stock | N/A |
| Oligonucleotides | ||
| Primers | Table S1 | N/A |
| Recombinant DNA | ||
| pL2B-tYFPNLS-CERK1-3×FLAG-LYK8-3×HA | This study | N/A |
| pL2B-LAP1-proLYK8-GUS | This study | N/A |
| pL2B-LAP1-proCERK1-GUS | This study | N/A |
| pL1M-R2-pro35S-LYK8-3×HA-T35S | This study | N/A |
| pL1M-R2-pro35S-LYK8-3×FLAG-T35S | This study | N/A |
| pL1M-R2-pro35S-LYR4-3×HA-T35S | This study | N/A |
| pL1M-R2-pro35S-LYK3-3×HA-T35S | This study | N/A |
| pL1M-R2-pro35S-CERK1-3×HA-T35S | This study | N/A |
| pL1M-R1-pro35S-LYK8-VYNE-T35S | This study | N/A |
| pL1M-R2-pro35S-CERK1-VYCE-TOcs | This study | N/A |
| pL1M-R2-proLjUBI1-DMI2-VYCE-TOcs | This study | N/A |
| pL1M-R2-pro35S-LYR4-VYCE-TOcs | This study | N/A |
| pL1M-R2-pro35S-AtFLS2-VYCE-TOcs | This study | N/A |
| pL1M-R2-pro35S-AtBRI1-VYCE-TOcs | This study | N/A |
| pL1M-R2-pro35S-MtLYM1-VYCE-TOcs | This study | N/A |
| pL1M-R2-pro35S-VYCE-TOcs | This study | N/A |
| pL1M-R3-proLjUBI1-DMI2-3×HA-TNOS | This study | N/A |
| pL1M-R2-pro35S-AtFLS2-3×HA-T35S | This study | N/A |
| pL1M-R3-proLjUBI1-AtBRI1-3×HA-TNOS | This study | N/A |
| pL1M-R2-pro35S-MtLYM1-3×HA-T35S | This study | N/A |
| pCAMBIA-pro35S-LYK8ect-NFPtmic-YFP | This study | N/A |
| pCAMBIA-pro35S-MtLYR8ect-NFPtmic-YFP | Ding et al.52 | N/A |
| pCAMBIA-pro35S-OsCEBiP8ect-NFPtmic-YFP | Cullimore et al.29 | N/A |
| Software and algorithms | ||
| ImageJ | National Institute of Health | Version 1.54f |
| ZEN Blue Version | Zeiss | Version 3.8 |
| Prism | GraphPad Software | Version 9.5.0 |
| MEGA | www.megasoftware.net/ | Version 11 |
| iTOL | itol.embl.de/ | Version 6.8.1 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Feng Feng (feng.feng@okstate.edu).
Materials availability
The plasmids and plant materials generated in this study can be provided upon request and following the requisite material transfer agreement (MTA) with the originating institution(s) responsible for generating the mutant line(s).
Data and code availability
-
•
No sequencing data or accession numbers were produced as part of this study. Original data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Experimental model and subject details
Plant Materials and growth conditions
M. truncatula cv. R108 was used as the wild-type, cerk1 (NF16753) and nfp-3 (NF7796) mutants were reported previously.7 The Tnt1 transposon insertion mutants lyk6 (NF14155), lyk7 (NF8175), lyk10 (NF20763) lyk8-1 (NF9395) and lyk8-2 (NF11260) were obtained from the Oklahoma State University. The calcium reporter Yellow Cameleon (YC) 3.6 was introduced into the above-mentioned mutant lines by crossing them with an existing stable YC3.6 transgenic line. M. truncatula seeds were lightly scarified using sandpaper, sterilized in 10% sodium hypochlorite for 1.5 minutes, and then rinsed five times with sterilized water. The seeds were subsequently transferred to 1.5% water agar plates and stored in the dark at 4°C for three days before being germinated at room temperature. Seedlings were then either cultivated on responsive medium or grown in a mixture of soil substrates, depending on the requirements of the experiments (a 3:3:1 mixture of Turface MVP, play sand, and LC1 grower mix for mycorrhizal inoculation; a 1:1 mixture of vermiculite and Turface MVP for rhizobial inoculation). Five-week-old Nicotiana benthamiana leaves were used for protein expression. The plants were grown in a plant growth room at 22°C under a 16:8 photoperiod. Primers used for mutant identification are listed in Table S1.
Bacterial and Fungal Strains
Ensifer meliloti strain Em1021 was cultivated at 28°C on TY medium supplemented with appropriate antibiotics. Agrobacterium tumefaciens strain GV3101 and Agrobacterium rhizogenes strain AR1193 were incubated at 28°C on LB medium supplemented with antibiotics. Rhizophagus irregularis powder was purchased from Premier Tech, Canada or maintained in a carrot root organ culture as described previously.51 Gigaspora margarita BEG-34 was kindly provided by Professor Paola Bonfante and grown in co-culture with Tagetes patula (French marigold) in a sterile substrate of sand-terragreen mix in a glasshouse for approximately 4 months until maximum mycorrhizal colonization was achieved. The pots were then dried, plant material removed, and the substrate sieved to produce a crude inoculum containing spores and infective propagules. The inoculum was stored at 4°C under low (< 7 %) relative humidity. Fusarium oxysporum was originally isolated from alfalfa roots and cultivated on Synthetischer Nährstoffarmer Agar (SNA)70 for at least 7 days before collection of spores.
METHOD DETAILS
Mycorrhizal inoculation
M. truncatula wild-type and mutant plants were cultivated in 48-well trays containing a mixture of soil substrates (3:3:1 mixture of Turface MVP, play sand, and LC1 grower mix). They were inoculated with Rhizophagus irregularis at a rate of 100 spores per plant for low-concentration inoculation and 300 spores per plant for high-concentration inoculation. For Gigaspora margarita inoculation, an inoculation strength of 5 % (v/v) per plant was used. For the co-cultivation of lyk8/cerk1 plants with nurse plants, one wild-type M. truncatula R108 was grown alongside one lyk8/cerk1 plant. Roots colonized by mycorrhizae were harvested at specified time points and subjected to staining with ink or 0.05% Trypan Blue. To assess the extent of mycorrhizal colonization, we used the gridline intersect method.71 Briefly, root segments were cut into smaller pieces and randomly distributed across a square Petri dish, which featured a 1 cm × 1 cm grid on its base. Using a Leica EZ4 stereomicroscope, 100 grid intersections were examined for each root sample to evaluate the AMF colonization and infection events. Representative images were taken using a Keyence VHX-5000 Digital Microscope (Keyence, UK).
Root architecture measurement
To quantify root development, root samples were harvested three weeks post-inoculation with R. irregularis, rinsed with tap water, and subsequently preserved in 50% (v/v) ethanol as described previously.51 The length of the primary root was determined using a ruler, and the first-order of lateral roots were counted using a Leica EZ4 stereomicroscope.
Nodulation test
Wild-type R108 and receptor mutant seedlings were grown in a 1:1 mixture of vermiculite and Turface MVP. The seedlings were watered twice weekly with B&D liquid medium and received additional tap water once per week. After 7 days of growth, the plant roots were inoculated with E. meliloti 1021 at an OD600 of 0.03. Following inoculation, the plants were continuously watered with B&D medium and tap water for three weeks. The roots were then harvested, rinsed with tap water, and the number of pink and white nodules on each root was quantified.
Molecular cloning and plant complementation
In the promoter-GUS assay, a 2.3 kb sequence upstream of either the LYK8 or CERK1 coding region was synthesized by Twist Bioscience, USA. This upstream sequence, serving as the native promoter, was then fused to the GUS reporter module to generate the construct by Golden Gate cloning,72 which was subsequently introduced into the A. rhizogenes AR1193 for hairy root transformation. To complement the lyk8/cerk1 double mutant, we utilized Golden Gate cloning to create a single construct comprising both the LYK8 and CERK1 genes, as well as a nucleus localized triple YFP fluorescence marker (tYFP-NLS) for the selection of transformed plant roots. In this construct, the synthesized coding sequences of LYK8 and CERK1 were individually fused with a 3×HA and a 3×Flag peptide, respectively, each under the control of its own native promoter. We then introduced this construct into A. rhizogenes and transferred it into lyk8/cerk1 plants through hairy root transformation. Roots with YFP signal and transformed with either an empty vector or with the LYK8 and CERK1 construct were then inoculated with R. irregularis spores. To assess the protein expression levels of LYK8 and CERK1, an equal amount of transformed roots was harvested three weeks post-AMF inoculation for protein extraction. Western blotting, employing anti-HA and anti-FLAG antibodies, was then conducted to verify the expression of LYK8 and CERK1 proteins in the complementation plants.
To generate constructs for protein expression in N. benthamiana, synthesized coding sequences of various genes, including LYK8, CERK1, DMI2, LYR4, LYK3, AtFLS2, AtBRI1, and MtLYM1, were fused with either a 3×HA or 3×FLAG tag. This resulted in the generation of constructs such as LYK8-HA, CERK1-HA, DMI2-HA, LYR4-HA, LYK3-HA, AtFLS2-HA, AtBRI1-HA, and MtLYM1-HA, as well as LYK8-FLAG. For the CO5/CO7 binding assay, we amplified the sequence representing the extracellular domain of LYK8 (LYK8ect, amino acids 1-223) from M. truncatula cDNA using PCR (primer details in Table S1). This sequence was then fused in-frame with the coding regions for the transmembrane and intracellular domains of MtNFP (NFPtmic), followed by a YFP tag, all under the control of CaMV 35S promoter. This fusion construct of LYK8 was used to maintain consistency with the constructs of two positive controls, MtLYR8ect-NFPtmic-YFP and OsCEBIPect-NFPtmic-YFP, which were created similarly.29,52,73 For the bimolecular fluorescence complementation (BiFC) assay, the LYK8 coding sequence was fused with the N-terminal fragment of the yellow fluorescent protein Venus (VYNE). The coding sequences for CERK1, LYR4, DMI2, AtFLS2, AtBRI1, and MtLYM1 were fused with the C-terminal fragment of Venus (VYCE). All these constructs were subsequently transformed into Agrobacterium GV3101 for transient expression in the leaves of N. benthamiana.
GUS and WGA staining
M. truncatula wild-type roots harboring a promoter-GUS construct, introduced through hairy root transformation, were cultivated in a soil mixture inoculated with mycorrhizal spores. These plant roots were harvested at intervals of one week, two weeks, and three weeks after inoculation. Subsequently, the roots were rinsed and subjected to GUS staining using 1 mg/mL X-Gluc staining buffer, which consists of 100 mM NaP buffer, 10 mM EDTA, and 1 mM potassium ferricyanide. The samples were incubated at 37°C for 6 hours and then washed with 70% ethanol and cleared in a 20% KOH solution. A final staining step was carried out using WGA-Alexa Fluor 488 dye (Thermo Fisher), dissolved in PBS buffer at a concentration of 0.5 μg/mL, and the samples were incubated for an additional 6 hours. Images of the stained samples were captured using a fluorescence microscope (BX51, Olympus, Japan).
Gene expression analysis
M. truncatula wild-type and mutant plants were cultured on Buffered Nodulation Media (BNM) agar plates for seven days. Afterward, the plants were moved to liquid BNM with or without adding elicitors such as 10-7 M CO4 (Megazyme), 10-7 M CO8 (produced by Sébastien Fort) and 10-8 M LCO (LCO from E. meliloti 1021, produced by Sébastien Fort). To induce symbiotic gene expression, the plant roots were treated with CO or LCO for 6 hours. For the induction of immune marker genes, a 30-minute incubation of CO8 was utilized. We used eight roots for each treatment and conducted three biological replicates for each time point. Regarding gene expression triggered by AMF, roots infected for 3 and 5 weeks were washed and collected for RNA extraction. The root samples were frozen using liquid nitrogen, and total RNA was extracted using a plant RNA extraction kit (Sigma). Genomic DNA was eliminated by treating with RNase-free DNase (Sigma) according to the manufacturer’s instructions. The concentration of the produced total RNA was determined using the NanoDrop-1000 Spectrophotometer (Thermo Fisher).
For real-time qPCR analysis, 300 ng of total RNA from each sample was utilized for cDNA synthesis with a LunaScript RT SuperMix Kit (New England Biolabs). Gene expression was determined using a CFX96 touch real-time PCR system (Bio-Rad), with 10 ng of cDNA template amplified using a Luna-SYBR green PCR master mix (New England Biolabs). Expression data were analyzed using an M. truncatula endogenous Ubiquitin gene as a reference. The fold change in gene expression was calculated for samples treated with elicitors or AMF in comparison to those subjected to water treatment or no AMF inoculation. For the Semi-quantitative RT-PCR analysis in Figures S1B and S1C, 10 ng of synthesized cDNA was added into PCR reaction. The primers for detecting the expression of symbiotic genes (MtVapyrin, MtLYK10, MtD27, MtPT4), defense genes (MtPR10, MtChitinase, MtRbohA), and the receptors (LYK8, MtLYK6, MtLYK7) can be found in Table S1.
Nuclear calcium oscillation
To assess periodic calcium oscillation, seedlings of M. truncatula wild-type and different receptor mutants containing the YC3.6 reporter were cultured on BNM agar plates supplemented with 100 nM AVG until lateral roots emerged. Only plant roots exhibiting strong YFP fluorescence were selected to assess the calcium response upon exposure to CO and LCO at specified doses. Calcium spiking was measured using an inverted epifluorescence microscope (TE2000, Nikon, Japan). Analysis of the fluorescent signal and the calcium spike curve followed the methodology outlined in a previous study.15
Reactive oxygen species production
M. truncatula wild-type and mutant plants were grown on BNM agar plates supplemented with 100 nM AVG for four days. Following this, the primary roots, which were about 4 cm in length, were segmented into 0.5 cm strips and placed in a 96-well microplate containing 200 μL of water in each well. An overnight incubation was conducted to eliminate internal ROS generated during the process of root segmentation. After incubation, water was removed from each well and replaced with 100 μL of reaction buffer containing 10-6 M CO8, 10 μg/mL horseradish peroxidase and 0.5 mM L-012. Luminescence was recorded with a Synergy H1 microplate reader (BioTek, USA) at indicated time points with a 1000 ms integration time. For each genotype and treatment, at least six samples were used as biological replication.
MAPK activity assay
Roots of four-day-old M. truncatula plants, cultivated on BNM agar plates supplemented with 100 nM AVG, were cut into small segments and subsequently incubated in water for 4 hours. The root samples were then exposed to either 10-6 M CO8 for 10 minutes or were left untreated before being rapidly frozen in liquid nitrogen for protein extraction. Root proteins were then homogenized using an extraction buffer composed of 50 mM HEPES-KOH pH 7.5, 50 mM NaCl, 1 mM PMSF, 5% Glycerol, 1 mM NaF, 1 mM EDTA, 0.2% Triton X-100, 2 mM DTT, complete protease inhibitors and the PhoSTOP phosphatase inhibitor. For the assessment of MPK3 and MPK6 phosphorylation, an anti-pERK antibody was employed. A duplicate blot was used to detect M. truncatula Tubulin, serving as the endogenous reference for equal loading, utilizing an anti-tubulin antibody.
Pathogen infection
F. oxysporum was cultured on SNA solid medium for a minimum of seven days. Sterilized, prechilled water (between 5-10 mL) was added to each plate and allowed to stand for 1 hour to release spores. The spore concentration was quantified using a hemacytometer. M. truncatula wild -type and mutant seedlings were grown on 0.8 % agarose plates for 36 hours. The root tip regions were inoculated with F. oxysporum spores at a concentration of 1 × 106 spores/mL. For assessing F. oxysporum growth, 15 seedlings were collected at 48 hours post-inoculation to measure lesion size, which was then normalized against individual root length. The same root samples were then frozen in liquid nitrogen to extract DNA for quantitative PCR analysis, which utilized the F. oxysporum FOW174 gene in relation to the M. truncatula Ubiquitin gene.
Agrobacterium-mediated transient expression
The A. tumefaciens GV3101 carrying the specified binary vectors were cultivated in Luria-Bertani (LB) medium, supplemented with the appropriate antibiotics, at 28 °C overnight. Following this step, the bacterial cells were collected and resuspended in an infiltration buffer (10 mM MgCl2, 10 mM MES, pH 5.7, and 100 μM acetosyringone), followed by a 3-hour incubation at 28 °C. The optical density of the bacterial suspension was adjusted to a final concentration of OD600 = 0.5. This suspension was then combined with A. tumefaciens containing a P19 protein before being infiltrated into the fully expanded N. benthamiana leaves using a syringe without a needle. Subsequently, the plants were covered with a black plastic bag for 12 hours to facilitate A. tumefaciens infection. Infiltrated leaves were collected at either 24 or 36 hours, depending on the specific experimental requirements.
BiFC assay
The N. benthamiana leaves expressing split-Venus proteins were harvested 36 hours after A. tumefaciens infiltration. To visualize the fluorescent signal, leaf tissue near the infiltration site was employed for microscopic analysis under a laser scanning confocal microscope (LSM 980, Zeiss, Germany), utilizing YFP filters: 514 nm for excitation and 540 nm for emission.
Chitin beads and CO-biotin binding assays
Agrobacterium GV3101 transformants carrying the LYK8-HA, CERK1-HA, LYR4-HA, and LYK3-HA constructs were infiltrated into N. benthamiana leaves and left for 36 hours to induce protein expression. Subsequently, the infiltrated leaves were ground into fine powder using liquid nitrogen, and the total powder was mixed with 1 mL of cold lysis buffer (50 mM HEPES-KOH pH 7.5, 50 mM NaCl, 1 mM PMSF, 5% Glycerol, 1 mM NaF, 1 mM EDTA, 0.2% Triton X-100, 2 mM DTT, complete protease inhibitors, and 2% PVPP) for extracting receptor proteins. The protein lysate was then subjected to centrifugation, and the resulting supernatant was transferred to a new tube for the chitin binding assay. To initiate the binding process, 20 μL of chitin resin beads (New England Biolabs) were washed three times by adding 1 mL of lysis buffer (without PVPP) and subsequently incubated with the extracted proteins for 3 hours in a cold room. After incubation, the beads were centrifuged and washed once with 1 mL of wash buffer I (50 mM HEPES-KOH pH 7.5, 400 mM NaCl, 1 mM EDTA, 0.2% Triton X-100, and 2 mM DTT), followed by three washes with buffer II (buffer I without NaCl), and finally, a last wash with buffer III (buffer I without NaCl and Triton X-100). The bound protein was recovered by boiling beads with SDS loading buffer.
To assess the binding affinity of receptors with CO5 and CO7, constructs encoding LYK8ect-NFPtmic-YFP, MtLYR8ect-NFPtmic-YFP, and OsCEBIPect-NFPtmic-YFP were expressed in N. benthamiana leaves, and the membrane fractions were isolated as previously described.39 The synthesis of cross-linkable and biotinylated versions of CO5-biotin and CO7-biotin has been detailed in earlier work.29 Binding assays with different concentrations of CO-biotin were conducted using membrane fractions in a buffer (25 mM NaCacodylate pH 6.0, 1 mM MgCl2, 1 mM CaCl2, 250 mM Saccharose and protease inhibitors) for 1 hour on ice. After incubation, samples were centrifuged at 31,000 g for 30 minutes at 4 °C, and the pellets were resuspended in IP buffer (25 mM Tris-HCl pH 7.5, 150 mM NaCl, 10 % glycerol, supplemented with protease inhibitors and the phosphatase inhibitor). The proteins were then solubilized in the IP buffer containing 0.2 % DDM detergent for 1 hour at 4 °C and immunoprecipitated using GFP-trap magnetic agarose beads (ChromoTek). Following washing in IP buffer, proteins were eluted with Laemmli buffer. Western blot analysis utilized anti-HA, anti-GFP and Streptavidin to verify the presence of the receptor proteins and their binding with chitin and CO-biotin, respectively.
Co-IP assay
N. benthamiana leaves were infiltrated with Agrobacterium GV3101 transformants carrying designated constructs and incubated for 36 hours to facilitate protein expression. Afterward, the leaves were infiltrated with 1 μM of CO4, CO8, or water for 10 minutes, followed by protein extraction. One gram of Agrobacterium-infected leaf tissue was homogenized into a fine powder in liquid nitrogen using a mortar and pestle. Protein extraction was performed by adding cold lysis buffer (50 mM HEPES-KOH pH 7.5, 150 mM NaCl, 1 mM PMSF, 5% Glycerol, 1 mM NaF, 1 mM EDTA, 0.5 % Triton X-100, 2 mM DTT, complete protease inhibitors, and 2% PVPP) to the samples, followed by a 10-minute incubation on ice. Subsequently, the protein lysate underwent centrifugation, and the resulting supernatants were filtered through a 0.45 μm filter. Ten microliters of cell extracts were used as input to demonstrate the expression of each protein. The remaining cell extracts were combined with 25 μL of pre-washed anti-FLAG M2 agarose resin beads (Sigma) and incubated for 4 hours at 4 °C. Afterward, the agarose beads were subjected to centrifugation and washed with washing buffers as detailed in the chitin binding section. The protein complexes associated with the beads were recovered by boiling them with SDS loading buffer for 5 minutes. The presence of HA- and FLAG-tagged proteins was subsequently assessed using anti-HA and anti-FLAG antibodies in a western blot analysis.
Phylogenetic analysis
Protein sequences of the LYK-type LysM-RLK family across several species, including M. truncatula, L. japonicus, S. lycopersicum, O.sativa, Brachypodium distachyon, A. thaliana and P. andersonii were obtained from National Center for Biotechnology Information (details listed in Table S2). These sequences were aligned using MUSCLE. Subsequently, a neighbor-joining phylogenetic tree was constructed using MEGA software and modified by iTOL website.
Quantification and statistical analysis
Statistical analysis was performed using GraphPad Prism 9.5.0 software. This analysis involved employing one-way or two-way analysis of variance (ANOVA) followed by Tukey’s multiple comparisons test or multiple Student’s t-tests. The error bars represent the standard error of the mean (S.E.M.). Statistically significant differences were denoted by asterisks (ns, p > 0.05; ∗, p < 0.05; ∗∗, p < 0.01; ∗∗∗, p < 0.001, ∗∗∗∗, p < 0.0001) to indicate samples with significant distinctions. The real-time qPCR data were shown using dot plots to reflect the data distribution, and the normality of samples was confirmed by the Shapiro-Wilk test. Detailed sample size and statistical differences are available in the figure legends.
Acknowledgments
We thank Stephen Marek for providing F. oxysporum isolate and Doris Albinsky, Eirini Vlachaki, and Mara Sgroi for amplifying and providing G. margarita inoculum. This work was supported by the OCAST Plant Science Research Program PS21-008 to F.F. and the NIFA grant 2022-67014-38607 as part of the NSF/NIFA Plant Biotic Interactions Program to F.F. G.E.D.O. was funded by the Biotechnology and Biological Sciences as BB/X011933/1 and by the Bill and Melinda Gates Foundation and the Foreign, Commonwealth and Development Office as Enabling Nutrient Symbioses in Agriculture (OPP1172165). S.F. acknowledges NanoBio ICMG (UAR 2607) for providing access to mass spectrometry and NMR facilities and the French National Research Agency-ANR through LABEX ARCANE/EUR CBH-EUR-GS (ANR-17-EURE-0003), Glyc@Alps (ANR-15-IDEX-02), and Carnot Polynat (CARN-025-01) for partial financial support. M. truncatula Tnt1 seeds were developed and maintained with funding from NSF (DBI-0703285; IOS-1127155; IOS-1733470; DBI-2233714).
Author contributions
J.Z., J.S., C.H.C., F.F., and G.E.D.O. conceived and designed the experiments. J.Z., J.S., C.H.C., and K.L., performed the study. D.L. and B.L. provided biotinylated-CO5/CO7 and conducted the binding assay. J.W. and K.S.M. provided the M. truncatula Tnt1 mutant seeds. S.F. produced CO and LCO molecules. J.Z., F.F., and G.E.D.O. wrote the manuscript with comments from J.S. and C.H.C.
Declaration of interests
The authors declare no competing interests.
Published: April 3, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.cub.2024.03.015.
Contributor Information
Giles E.D. Oldroyd, Email: gedo2@cam.ac.uk.
Feng Feng, Email: feng.feng@okstate.edu.
Supplemental information
References
- 1.Harrison M.J. Signaling in the arbuscular mycorrhizal symbiosis. Annu. Rev. Microbiol. 2005;59:19–42. doi: 10.1146/annurev.micro.58.030603.123749. [DOI] [PubMed] [Google Scholar]
- 2.Zipfel C., Oldroyd G.E.D. Plant signaling in symbiosis and immunity. Nature. 2017;543:328–336. doi: 10.1038/nature22009. [DOI] [PubMed] [Google Scholar]
- 3.Hawkins H.J., Cargill R.I.M., Van Nuland M.E., Hagen S.C., Field K.J., Sheldrake M., Soudzilovskaia N.A., Kiers E.T. Mycorrhizal mycelium as a global carbon pool. Curr. Biol. 2023;33:R560–R573. doi: 10.1016/j.cub.2023.02.027. [DOI] [PubMed] [Google Scholar]
- 4.Oldroyd G.E.D. Speak, friend, and enter: signalling systems that promote beneficial symbiotic associations in plants. Nat. Rev. Microbiol. 2013;11:252–263. doi: 10.1038/nrmicro2990. [DOI] [PubMed] [Google Scholar]
- 5.Catoira R., Galera C., de Billy F., Penmetsa R.V., Journet E.P., Maillet F., Rosenberg C., Cook D., Gough C., Dénarié J. Four genes of Medicago truncatula controlling components of a nod factor transduction pathway. Plant Cell. 2000;12:1647–1665. doi: 10.1105/tpc.12.9.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Genre A., Chabaud M., Balzergue C., Puech-Pagès V., Novero M., Rey T., Fournier J., Rochange S., Bécard G., Bonfante P., Barker D.G. Short-chain chitin oligomers from arbuscular mycorrhizal fungi trigger nuclear Ca2+ spiking in Medicago truncatula roots and their production is enhanced by strigolactone. New Phytol. 2013;198:190–202. doi: 10.1111/nph.12146. [DOI] [PubMed] [Google Scholar]
- 7.Feng F., Sun J., Radhakrishnan G.V., Lee T., Bozsóki Z., Fort S., Gavrin A., Gysel K., Thygesen M.B., Andersen K.R., et al. A combination of chitooligosaccharide and lipochitooligosaccharide recognition promotes arbuscular mycorrhizal associations in Medicago truncatula. Nat. Commun. 2019;10:5047. doi: 10.1038/s41467-019-12999-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maillet F., Poinsot V., André O., Puech-Pagès V., Haouy A., Gueunier M., Cromer L., Giraudet D., Formey D., Niebel A., et al. Fungal lipochitooligosaccharide symbiotic signals in arbuscular mycorrhiza. Nature. 2011;469:58–63. doi: 10.1038/nature09622. [DOI] [PubMed] [Google Scholar]
- 9.Volpe V., Chialva M., Mazzarella T., Crosino A., Capitanio S., Costamagna L., Kohlen W., Genre A. Long-lasting impact of chitooligosaccharide application on strigolactone biosynthesis and fungal accommodation promotes arbuscular mycorrhiza in Medicago truncatula. New Phytol. 2023;237:2316–2331. doi: 10.1111/nph.18697. [DOI] [PubMed] [Google Scholar]
- 10.Kelly S., Radutoiu S., Stougaard J. Legume LysM receptors mediate symbiotic and pathogenic signalling. Curr. Opin. Plant Biol. 2017;39:152–158. doi: 10.1016/j.pbi.2017.06.013. [DOI] [PubMed] [Google Scholar]
- 11.Buendia L., Girardin A., Wang T., Cottret L., Lefebvre B. LysM receptor-like kinase and LysM receptor-like protein families: an update on phylogeny and functional characterization. Front. Plant Sci. 2018;9:1531. doi: 10.3389/fpls.2018.01531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khokhani D., Carrera Carriel C., Vayla S., Irving T.B., Stonoha-Arther C., Keller N.P., Ané J.M. Deciphering the chitin code in plant symbiosis, defense, and microbial networks. Annu. Rev. Microbiol. 2021;75:583–607. doi: 10.1146/annurev-micro-051921-114809. [DOI] [PubMed] [Google Scholar]
- 13.Gough C., Cullimore J. Lipo-chitooligosaccharide signaling in endosymbiotic plant-microbe interactions. Mol. Plant Microbe Interact. 2011;24:867–878. doi: 10.1094/MPMI-01-11-0019. [DOI] [PubMed] [Google Scholar]
- 14.Chiu C.H., Paszkowski U. Receptor-like kinases sustain symbiotic scrutiny. Plant Physiol. 2020;182:1597–1612. doi: 10.1104/pp.19.01341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun J., Miller J.B., Granqvist E., Wiley-Kalil A., Gobbato E., Maillet F., Cottaz S., Samain E., Venkateshwaran M., Fort S., et al. Activation of symbiosis signaling by arbuscular mycorrhizal fungi in legumes and rice. Plant Cell. 2015;27:823–838. doi: 10.1105/tpc.114.131326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gibelin-Viala C., Amblard E., Puech-Pages V., Bonhomme M., Garcia M., Bascaules-Bedin A., Fliegmann J., Wen J., Mysore K.S., le Signor C., et al. The Medicago truncatula LysM receptor-like kinase LYK9 plays a dual role in immunity and the arbuscular mycorrhizal symbiosis. New Phytol. 2019;223:1516–1529. doi: 10.1111/nph.15891. [DOI] [PubMed] [Google Scholar]
- 17.Miyata K., Kozaki T., Kouzai Y., Ozawa K., Ishii K., Asamizu E., Okabe Y., Umehara Y., Miyamoto A., Kobae Y., et al. The bifunctional plant receptor, OsCERK1, regulates both chitin-triggered immunity and arbuscular mycorrhizal symbiosis in rice. Plant Cell Physiol. 2014;55:1864–1872. doi: 10.1093/pcp/pcu129. [DOI] [PubMed] [Google Scholar]
- 18.Zhang X., Dong W., Sun J., Feng F., Deng Y., He Z., Oldroyd G.E.D., Wang E. The receptor kinase CERK1 has dual functions in symbiosis and immunity signaling. Plant J. 2015;81:258–267. doi: 10.1111/tpj.12723. [DOI] [PubMed] [Google Scholar]
- 19.Zhang L., Yuan L., Staehelin C., Li Y., Ruan J., Liang Z., Xie Z., Wang W., Xie J., Huang S. The LYSIN MOTIF-CONTAINING RECEPTOR-LIKE kinase 1 protein of banana is required for perception of pathogenic and symbiotic signals. New Phytol. 2019;223:1530–1546. doi: 10.1111/nph.15888. [DOI] [PubMed] [Google Scholar]
- 20.Rutten L., Miyata K., Roswanjaya Y.P., Huisman R., Bu F., Hartog M., Linders S., van Velzen R., van Zeijl A., Bisseling T., et al. Duplication of symbiotic lysin motif receptors predates the evolution of nitrogen-fixing nodule symbiosis. Plant Physiol. 2020;184:1004–1023. doi: 10.1104/pp.19.01420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leppyanen I.V., Pavlova O.A., Vashurina M.A., Bovin A.D., Dolgikh A.V., Shtark O.Y., Sendersky I.V., Dolgikh V.V., Tikhonovich I.A., Dolgikh E.A. LysM receptor-like kinase LYK9 of Pisum Sativum L. may regulate plant responses to chitooligosaccharides differing in structure. Int. J. Mol. Sci. 2021;22:711. doi: 10.3390/ijms22020711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leppyanen I.V., Shakhnazarova V.Y., Shtark O.Y., Vishnevskaya N.A., Tikhonovich I.A., Dolgikh E.A. Receptor-like kinase LYK9 in Pisum sativum L. is the CERK1-Like receptor that controls both plant immunity and AM symbiosis development. Int. J. Mol. Sci. 2017;19:8. doi: 10.3390/ijms19010008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carotenuto G., Chabaud M., Miyata K., Capozzi M., Takeda N., Kaku H., Shibuya N., Nakagawa T., Barker D.G., Genre A. The rice LysM receptor-like kinase OsCERK1 is required for the perception of short-chain chitin oligomers in arbuscular mycorrhizal signaling. New Phytol. 2017;214:1440–1446. doi: 10.1111/nph.14539. [DOI] [PubMed] [Google Scholar]
- 24.Bouchiba Y., Esque J., Cottret L., Maréchaux M., Gaston M., Gasciolli V., Keller J., Nouwen N., Gully D., Arrighi J.F., et al. An integrated approach reveals how lipo-chitooligosaccharides interact with the lysin motif receptor-like kinase MtLYR3. Protein Sci. 2022;31 doi: 10.1002/pro.4327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Broghammer A., Krusell L., Blaise M., Sauer J., Sullivan J.T., Maolanon N., Vinther M., Lorentzen A., Madsen E.B., Jensen K.J., et al. Legume receptors perceive the rhizobial lipochitin oligosaccharide signal molecules by direct binding. Proc. Natl. Acad. Sci. USA. 2012;109:13859–13864. doi: 10.1073/pnas.1205171109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fliegmann J., Jauneau A., Pichereaux C., Rosenberg C., Gasciolli V., Timmers A.C.J., Burlet-Schiltz O., Cullimore J., Bono J.J. LYR3, a high-affinity LCO-binding protein of Medicago truncatula, interacts with LYK3, a key symbiotic receptor. FEBS Lett. 2016;590:1477–1487. doi: 10.1002/1873-3468.12191. [DOI] [PubMed] [Google Scholar]
- 27.Gysel K., Laursen M., Thygesen M.B., Lironi D., Bozsóki Z., Hjuler C.T., Maolanon N.N., Cheng J., Bjørk P.K., Vinther M., et al. Kinetic proofreading of lipochitooligosaccharides determines signal activation of symbiotic plant receptors. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2111031118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Malkov N., Fliegmann J., Rosenberg C., Gasciolli V., Timmers A.J., Nurisso A., Cullimore J., Bono J.J. Molecular basis of lipo-chitooligosaccharide recognition by the lysin motif receptor-like kinase LYR3 in legumes. Biochem. J. 2016;473:1369–1378. doi: 10.1042/BCJ20160073. [DOI] [PubMed] [Google Scholar]
- 29.Cullimore J., Fliegmann J., Gasciolli V., Gibelin-Viala C., Carles N., Luu T.B., Girardin A., Cumener M., Maillet F., Pradeau S., et al. Evolution of lipochitooligosaccharide binding to a LysM-RLK for nodulation in Medicago truncatula. Plant Cell Physiol. 2023;64:746–757. doi: 10.1093/pcp/pcad033. [DOI] [PubMed] [Google Scholar]
- 30.Murakami E., Cheng J., Gysel K., Bozsoki Z., Kawaharada Y., Hjuler C.T., Sørensen K.K., Tao K., Kelly S., Venice F., et al. Epidermal LysM receptor ensures robust symbiotic signalling in Lotus japonicus. eLife. 2018;7 doi: 10.7554/eLife.33506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Amor B.B., Shaw S.L., Oldroyd G.E., Maillet F., Penmetsa R.V., Cook D., Long S.R., Dénarié J., Gough C. The NFP locus of Medicago truncatula controls an early step of Nod factor signal transduction upstream of a rapid calcium flux and root hair deformation. Plant J. 2003;34:495–506. doi: 10.1046/j.1365-313x.2003.01743.x. [DOI] [PubMed] [Google Scholar]
- 32.Arrighi J.F., Barre A., Ben Amor B., Bersoult A., Soriano L.C., Mirabella R., de Carvalho-Niebel F., Journet E.P., Ghérardi M., Huguet T., et al. The Medicago truncatula lysin motif-receptor-like kinase gene family includes NFP and new nodule-expressed genes. Plant Physiol. 2006;142:265–279. doi: 10.1104/pp.106.084657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Limpens E., Franken C., Smit P., Willemse J., Bisseling T., Geurts R. LysM domain receptor kinases regulating rhizobial Nod factor-induced infection. Science. 2003;302:630–633. doi: 10.1126/science.1090074. [DOI] [PubMed] [Google Scholar]
- 34.Madsen E.B., Madsen L.H., Radutoiu S., Olbryt M., Rakwalska M., Szczyglowski K., Sato S., Kaneko T., Tabata S., Sandal N., Stougaard J. A receptor kinase gene of the LysM type is involved in legume perception of rhizobial signals. Nature. 2003;425:637–640. doi: 10.1038/nature02045. [DOI] [PubMed] [Google Scholar]
- 35.Radutoiu S., Madsen L.H., Madsen E.B., Felle H.H., Umehara Y., Grønlund M., Sato S., Nakamura Y., Tabata S., Sandal N., Stougaard J. Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature. 2003;425:585–592. doi: 10.1038/nature02039. [DOI] [PubMed] [Google Scholar]
- 36.Smit P., Limpens E., Geurts R., Fedorova E., Dolgikh E., Gough C., Bisseling T. Medicago LYK3, an entry receptor in rhizobial nodulation factor signaling. Plant Physiol. 2007;145:183–191. doi: 10.1104/pp.107.100495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moling S., Pietraszewska-Bogiel A., Postma M., Fedorova E., Hink M.A., Limpens E., Gadella T.W.J., Bisseling T. Nod factor receptors form heteromeric complexes and are essential for intracellular infection in medicago nodules. Plant Cell. 2014;26:4188–4199. doi: 10.1105/tpc.114.129502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Buendia L., Wang T., Girardin A., Lefebvre B. The LysM receptor-like kinase SlLYK10 regulates the arbuscular mycorrhizal symbiosis in tomato. New Phytol. 2016;210:184–195. doi: 10.1111/nph.13753. [DOI] [PubMed] [Google Scholar]
- 39.Girardin A., Wang T., Ding Y., Keller J., Buendia L., Gaston M., Ribeyre C., Gasciolli V., Auriac M.C., Vernié T., et al. LCO receptors involved in arbuscular mycorrhiza are functional for rhizobia perception in legumes. Curr. Biol. 2019;29:4249–4259.e5. doi: 10.1016/j.cub.2019.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang C., He J., Dai H., Wang G., Zhang X., Wang C., Shi J., Chen X., Wang D., Wang E. Discriminating symbiosis and immunity signals by receptor competition in rice. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2023738118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li X.R., Sun J., Albinsky D., Zarrabian D., Hull R., Lee T., Jarratt-Barnham E., Chiu C.H., Jacobsen A., Soumpourou E., et al. Nutrient regulation of lipochitooligosaccharide recognition in plants via NSP1 and NSP2. Nat. Commun. 2022;13:6421. doi: 10.1038/s41467-022-33908-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miyata K., Hasegawa S., Nakajima E., Nishizawa Y., Kamiya K., Yokogawa H., Shirasaka S., Maruyama S., Shibuya N., Kaku H. OsCERK2/OsRLK10, a homolog of OsCERK1, has a potential role for chitin-triggered immunity and arbuscular mycorrhizal symbiosis in rice. Plant Biotechnol. (Tokyo) 2022;39:119–128. doi: 10.5511/plantbiotechnology.21.1222a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rasmussen S.R., Füchtbauer W., Novero M., Volpe V., Malkov N., Genre A., Bonfante P., Stougaard J., Radutoiu S. Intraradical colonization by arbuscular mycorrhizal fungi triggers induction of a lipochitooligosaccharide receptor. Sci. Rep. 2016;6:29733. doi: 10.1038/srep29733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jones J.D.G., Dangl J.L. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 45.Cao Y., Halane M.K., Gassmann W., Stacey G. The role of plant innate immunity in the legume-rhizobium symbiosis. Annu. Rev. Plant Biol. 2017;68:535–561. doi: 10.1146/annurev-arplant-042916-041030. [DOI] [PubMed] [Google Scholar]
- 46.Rey T., André O., Nars A., Dumas B., Gough C., Bottin A., Jacquet C. Lipo-chitooligosaccharide signalling blocks a rapid pathogen-induced ROS burst without impeding immunity. New Phytol. 2019;221:743–749. doi: 10.1111/nph.15574. [DOI] [PubMed] [Google Scholar]
- 47.Zeng T., Rodriguez-Moreno L., Mansurkhodzaev A., Wang P., van den Berg W., Gasciolli V., Cottaz S., Fort S., Thomma B.P.H.J., Bono J.J., et al. A lysin motif effector subverts chitin-triggered immunity to facilitate arbuscular mycorrhizal symbiosis. New Phytol. 2020;225:448–460. doi: 10.1111/nph.16245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang T., Gasciolli V., Gaston M., Medioni L., Cumener M., Buendia L., Yang B., Bono J.J., He G., Lefebvre B. LysM receptor-like kinases involved in immunity perceive lipo-chitooligosaccharides in mycotrophic plants. Plant Physiol. 2023;192:1435–1448. doi: 10.1093/plphys/kiad059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yu H., Bai F., Ji C., Fan Z., Luo J., Ouyang B., Deng X., Xiao S., Bisseling T., Limpens E., Pan Z. Plant lysin motif extracellular proteins are required for arbuscular mycorrhizal symbiosis. Proc. Natl. Acad. Sci. USA. 2023;120 doi: 10.1073/pnas.2301884120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tadege M., Wen J., He J., Tu H., Kwak Y., Eschstruth A., Cayrel A., Endre G., Zhao P.X., Chabaud M., et al. Large-scale insertional mutagenesis using the Tnt1 retrotransposon in the model legume Medicago truncatula. Plant J. 2008;54:335–347. doi: 10.1111/j.1365-313X.2008.03418.x. [DOI] [PubMed] [Google Scholar]
- 51.Chiu C.H., Roszak P., Orvošová M., Paszkowski U. Arbuscular mycorrhizal fungi induce lateral root development in angiosperms via a conserved set of MAMP receptors. Curr. Biol. 2022;32:4428–4437.e3. doi: 10.1016/j.cub.2022.08.069. [DOI] [PubMed] [Google Scholar]
- 52.Ding, Y., Gasciolli, V., Medioni, L., Gaston, M., de-Regibus, A., Rem-blière, C., Bono, J.J., Cullimore, J., Dalmais, M., Saffray, C., et al. A new group of LysM-RLKs involved in symbiotic signal perception and arbuscular mycorrhiza establishment. Preprint at BioRxiv. 10.1101/2024.03.06.583654. [DOI]
- 53.Shinya T., Osada T., Desaki Y., Hatamoto M., Yamanaka Y., Hirano H., Takai R., Che F.S., Kaku H., Shibuya N. Characterization of receptor proteins using affinity cross-linking with biotinylated ligands. Plant Cell Physiol. 2010;51:262–270. doi: 10.1093/pcp/pcp185. [DOI] [PubMed] [Google Scholar]
- 54.Gómez-Gómez L., Boller T. FLS2: an LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol. Cell. 2000;5:1003–1011. doi: 10.1016/S1097-2765(00)80265-8. [DOI] [PubMed] [Google Scholar]
- 55.Li J., Chory J. A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell. 1997;90:929–938. doi: 10.1016/s0092-8674(00)80357-8. [DOI] [PubMed] [Google Scholar]
- 56.Fliegmann J., Uhlenbroich S., Shinya T., Martinez Y., Lefebvre B., Shibuya N., Bono J.J. Biochemical and phylogenetic analysis of CEBiP-like LysM domain-containing extracellular proteins in higher plants. Plant Physiol. Biochem. 2011;49:709–720. doi: 10.1016/j.plaphy.2011.04.004. [DOI] [PubMed] [Google Scholar]
- 57.Sieberer B.J., Chabaud M., Fournier J., Timmers A.C.J., Barker D.G. A switch in Ca2+ spiking signature is concomitant with endosymbiotic microbe entry into cortical root cells of Medicago truncatula. Plant J. 2012;69:822–830. doi: 10.1111/j.1365-313X.2011.04834.x. [DOI] [PubMed] [Google Scholar]
- 58.Ivanov S., Harrison M.J. Receptor-associated kinases control the lipid provisioning program in plant-fungal symbiosis. Science. 2024;383:443–448. doi: 10.1126/science.ade1124. [DOI] [PubMed] [Google Scholar]
- 59.Liao D., Sun X., Wang N., Song F., Liang Y. Tomato LysM receptor-like kinase SlLYK12 is involved in arbuscular mycorrhizal symbiosis. Front. Plant Sci. 2018;9:1004. doi: 10.3389/fpls.2018.01004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rush T.A., Puech-Pagès V., Bascaules A., Jargeat P., Maillet F., Haouy A., Maës A.Q., Carriel C.C., Khokhani D., Keller-Pearson M., et al. Lipo-chitooligosaccharides as regulatory signals of fungal growth and development. Nat. Commun. 2020;11:3897. doi: 10.1038/s41467-020-17615-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rush T.A., Tannous J., Lane M.J., Gopalakrishnan Meena M., Carrell A.A., Golan J.J., Drott M.T., Cottaz S., Fort S., Ané J.M., et al. Lipo-chitooligosaccharides induce specialized fungal metabolite profiles that modulate bacterial growth. mSystems. 2022;7 doi: 10.1128/msystems.01052-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shaw S.L., Long S.R. Nod factor inhibition of reactive oxygen efflux in a host legume. Plant Physiol. 2003;132:2196–2204. doi: 10.1104/pp.103.021113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Oláh B., Brière C., Bécard G., Dénarié J., Gough C. Nod factors and a diffusible factor from arbuscular mycorrhizal fungi stimulate lateral root formation in Medicago truncatula via the DMI1/DMI2 signalling pathway. Plant J. 2005;44:195–207. doi: 10.1111/j.1365-313X.2005.02522.x. [DOI] [PubMed] [Google Scholar]
- 64.Liang Y., Cao Y., Tanaka K., Thibivilliers S., Wan J., Choi J., Kang Ch., Qiu J., Stacey G. Nonlegumes respond to rhizobial Nod factors by suppressing the innate immune response. Science. 2013;341:1384–1387. doi: 10.1126/science.1242736. [DOI] [PubMed] [Google Scholar]
- 65.Tanaka K., Cho S.H., Lee H., Pham A.Q., Batek J.M., Cui S., Qiu J., Khan S.M., Joshi T., Zhang Z.J., et al. Effect of lipo-chitooligosaccharide on early growth of C4 grass seedlings. J. Exp. Bot. 2015;66:5727–5738. doi: 10.1093/jxb/erv260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Buendia L., Maillet F., O'Connor D., van de-Kerkhove Q., Danoun S., Gough C., Lefebvre B., Bensmihen S. Lipo-chitooligosaccharides promote lateral root formation and modify auxin homeostasis in Brachypodium distachyon. New Phytol. 2019;221:2190–2202. doi: 10.1111/nph.15551. [DOI] [PubMed] [Google Scholar]
- 67.Bonhomme M., Bensmihen S., André O., Amblard E., Garcia M., Maillet F., Puech-Pagès V., Gough C., Fort S., Cottaz S., et al. Distinct genetic basis for root responses to lipo-chitooligosaccharide signal molecules from different microbial origins. J. Exp. Bot. 2021;72:3821–3834. doi: 10.1093/jxb/erab096. [DOI] [PubMed] [Google Scholar]
- 68.Kelly S., Hansen S.B., Rübsam H., Saake P., Pedersen E.B., Gysel K., Madland E., Wu S., Wawra S., Reid D., et al. A glycan receptor kinase facilitates intracellular accommodation of arbuscular mycorrhiza and symbiotic rhizobia in the legume Lotus japonicus. PLoS Biol. 2023;21 doi: 10.1371/journal.pbio.3002127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rübsam H., Krönauer C., Abel N.B., Ji H., Lironi D., Hansen S.B., Nadzieja M., Kolte M.V., Abel D., de Jong N., et al. Nanobody-driven signaling reveals the core receptor complex in root nodule symbiosis. Science. 2023;379:272–277. doi: 10.1126/science.ade9204. [DOI] [PubMed] [Google Scholar]
- 70.Leslie J.F., Summerell B.A. John Wiley & Sons; 2008. The Fusarium Laboratory Manual. [Google Scholar]
- 71.Giovannetti M., B M., Mosse B. An evaluation of techniques for measuring vesicular arbuscular mycorrhizal infection in roots. New Phytol. 1980;84:489–500. doi: 10.1111/j.1469-8137.1980.tb04556.x. [DOI] [Google Scholar]
- 72.Weber E., Engler C., Gruetzner R., Werner S., Marillonnet S. A modular cloning system for standardized assembly of multigene constructs. PLoS One. 2011;6 doi: 10.1371/journal.pone.0016765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fliegmann J., Canova S., Lachaud C., Uhlenbroich S., Gasciolli V., Pichereaux C., Rossignol M., Rosenberg C., Cumener M., Pitorre D., et al. Lipo-chitooligosaccharidic symbiotic signals are recognized by LysM receptor-like kinase LYR3 in the legume Medicago truncatula. ACS Chem. Biol. 2013;8:1900–1906. doi: 10.1021/cb400369u. [DOI] [PubMed] [Google Scholar]
- 74.Li Y., Mao L., Yan D., Ma T., Shen J., Guo M., Wang Q., Ouyang C., Cao A. Quantification of Fusarium oxysporum in fumigated soils by a newly developed real-time PCR assay to assess the efficacy of fumigants for Fusarium wilt disease in strawberry plants. Pest Manag. Sci. 2014;70:1669–1675. doi: 10.1002/ps.3700. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
No sequencing data or accession numbers were produced as part of this study. Original data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.