Abstract
Akv is an endogenous, ecotropic murine leukemia virus (MuLV) of the AKR strain. It has served as a prototype nonpathogenic or weakly pathogenic reference virus for studies of closely related potent lymphomagenic viruses such as the T-lymphomagenic SL3-3. We here report that Akv and an Akv mutant (Akv1-99) with only one copy of the 99-bp transcriptional enhancer induce malignant lymphomas with nearly 100% incidence and mean latency periods of 12 months after injection into newborn NMRI mice. Molecular analysis of tumor DNA showed that the majority of the tumors were of the B-cell type. Sequence analysis of proviral transcriptional enhancers in DNA of B-cell lymphomas revealed conservation of the enhancer sequence, as well as a lack of sequence duplications of the Akv1-99 variant, while the repeat copy number in Akv was subject to fluctuations. In support of a B-cell specificity of the Akv enhancer, a murine plasmacytoma cell line was found to sustain three- to fivefold-higher transient transcriptional activity upon the Akv and Akv1-99 enhancers than upon the enhancer of the T-lymphomagenic SL3-3 MuLV. Thus, the overall picture is that Akv MuLV possesses a B- lymphomagenic potential and that the second copy of the 99-bp sequence seems to be of minor importance for this potential. However, in one animal the lymphomas induced by Akv1-99 were of the T-cell type. Among the 24 tumors analyzed only this one harbored a clonal proviral integration in the c-myc locus. This provirus had undergone a duplication of a 113-bp sequence of the enhancer region, partly overlapping with the 99-bp repeat of Akv, as well as a few single nucleotide alterations within and outside the repeats. Taken together with previous studies, our results suggest that T- versus B-lymphomagenic specificity of the enhancer is governed by more than one nucleotide difference and that alterations in binding sites for transcription factors of the AML1 and nuclear-factor-1 families may contribute to this specificity.
Ecotropic murine leukemia viruses (MuLVs) are transmitted through the germ line of many inbred mouse strains (61), where in several instances they contribute to mouse strain characteristic patterns of disease (38). These viruses, which are carried at loci emv-1 through emv-18 (48, 61), are all closely related, and a prototype of the family is Akv (12, 23), which is found at one or more loci (emv-11, emv-12, emv-13, and emv-14) in the inbred AKR strain (61). Akv is an essential genetic determinant of the high incidence of thymic lymphoma development in AKR; however, the virology in AKR animals is complex, and recombinants between Akv and nonecotropic endogenous viruses play important roles in the disease process (62).
A number of commonly used laboratory strains of exogenous ecotropic MuLVs have been isolated after selecting for the potency and specificity of disease induction. Within this group of viruses major determinants for the induction of specific tumors of the hematopoietic compartment have been mapped to the enhancer region of U3, a DNA region characterized by repeat elements that interact with the transcriptional machinery of the host in a cell-type-specific manner (8, 10, 13, 25, 29, 30, 58). Of the exogenous MuLVs that have been subject to detailed investigations of viral disease determinants and virus-host interactions, the highly T-lymphomagenic virus SL3-3 is more closely related to Akv than are the T-lymphomagenic Moloney MuLV and the erythroleukemic Friend MuLV (45, 66). SL3-3 induces thymic lymphomas with high incidence within 2 to 4 months when injected into newborn mice of a number of strains, while Akv has been found to be nonpathogenic for observation periods of less than 1 year (22, 29, 45). By using SL3-3/Akv chimeric proviruses, the U3 region of the SL3-3 long terminal repeat (LTR) was identified as a major determinant of disease induction (29). The U3 regions of Akv and SL3-3 differ in the organization of the tandem repeats of the enhancer, where Akv has two copies of a 99-bp sequence and SL3-3 has 2.5 copies of a 72-bp sequence. Moreover, the two U3 regions show three point differences within the tandem repeat region and one point difference in the upstream U3 region (29, 36, 37).
Relative to Akv, the SL3-3 MuLV U3 shows a transcriptional cell type preference for T-lymphoid cells as analyzed by transient expression studies of cell cultures (5, 7, 43, 54). The tandem repeat region of SL3-3 confers high levels of T-cell transcription, and within the repeat sequences the enhancer core 5′-TGTGGTTAA-3′ (here termed the SL3-3 core), which serves as a binding site for a family of transcription factors variously called CBF, PEBP2, AML1/CBFβ, and SEF1, is a major determinant of T-cell tropism (9, 63, 69). The corresponding sequence of Akv 5′-TGTGGTCAA-3′ (here termed the Akv core) differs from the SL3-3 core by one pyrimidine transition. It has been found that an SL3-3 mutant with a replacement of the SL3-3 core sequence by the Akv sequence causes the appearance of T lymphomas with a longer latency period than does the wild-type SL3-3 (37). Moreover, alterations from the Akv core sequence to the SL3-3 core or to the Soule core version 5′-TGCGGTCAA-3′ found in the T-lymphomagenic Soule virus (6) were associated with the pathogenic process in a majority of cases (37). These results show that the Akv core sequence is less potent than the corresponding SL3-3 sequence in terms of T lymphomagenesis when analyzed in the context of SL3-3 U3 but do not address the function of this sequence in its natural Akv context.
The CWD strain carries emv-1, whose U3 sequence differs from that of Akv by the lack of the second copy of the 99-bp repeat and by a single nucleotide alteration outside the repeat region (35). The nucleotide sequence of the core site in recombinant proviruses of spontaneous T- and B-cell lymphomas in mice of this strain was found to frequently deviate from the Akv (emv-1) version. While the Akv core site was found predominantly in B-cell tumors, the SL3-3 core version of this site was also, somewhat unexpectedly, recovered from B-cell tumors, suggesting that the SL3-3 core site alone does not confer T-cell tropism (35). Additionally, a virus, AKL-E1, which harbors the emv-1 LTR in an Akv background, was found to induce B-cell lymphomas in NIH Swiss mice with a mean latency period of 18 months (28).
The sequence of the Akv enhancer conforms to the general pattern of organization of MuLV enhancers and includes a conserved cluster of the sequence motif termed the enhancer framework (18). The Akv enhancer has been analyzed in NIH fibroblasts, where it is stronger than that of SL3-3. Major cis determinants of Akv enhancer function in these cells were localized to sequences known to interact with the nuclear factor 1 (NF-1) family of proteins (32, 42). NF-1 sites are also found in SL3-3, where they are neutral or have a negative effect on SL3-3 expression in T-lymphoid cells (9).
While Akv has frequently been used as a negative control in pathogenicity experiments and Akv and related endogenous viruses are identified as disease determinants in inbred or recombinant inbred strains, little work has focused on the ability of Akv to induce tumors and on the function of its transcriptional enhancer in mouse tissues. We report here on the pathogenic properties of Akv and of a deletion mutant, Akv1-99, harboring only one copy of the 99-bp enhancer sequence. We find that Akv and Akv1-99 induce predominantly B-cell lymphomas and that no recurring pattern of alteration is found in the B- lymphoma-associated viruses. However, a provirus located at the c-myc locus in the T-lymphoma DNA of one mouse injected with Akv1-99 showed alterations reminiscent of those found in other T-lymphomagenic viruses. Thus, our results may contribute to an understanding of determinants of T- and B-cell specificity of MuLVs.
MATERIALS AND METHODS
Cell culture.
NIH 3T3 fibroblast cells, L-691 T-lymphoma cells, and the murine plasmacytoma B-cell line, MPC11, were grown in Dulbecco’s modified Eagle medium containing Glutamax-1 (GIBCO BRL and Life Technologies) supplemented with 10% newborn calf serum, 100 U of penicillin per ml, and 100 μg of streptomycin per ml.
Plasmids.
Expression vectors pAkv6-cat and pAkv1-99-cat were as described previously (32). pSL3-3-cat was constructed by exchanging a U3-R fragment (PstI-KpnI) from pAkv6-cat with the corresponding SL3-3 fragment (43). Plasmid pAKR-59 consists of the complete Akv without LTR duplication derived from λ623 (33) and cloned as a PstI-PstI fragment into pBR322 (12). The Akv1-99 provirus was generated by replacement of the U3 region of Akv with the U3 region of pAkv1-99-cat by a multistep restriction enzyme cleavage and ligation procedure.
Generation of virus stocks.
To generate Akv and Akv1-99 virus stocks, DNA of the proviral plasmids was digested with PstI, ligated to concatamers, and transfected into NIH 3T3 cells (10). Productive infection of the cultures was monitored by measuring reverse transcriptase activity and the number of infectious virus particles in cell supernatants (14, 26, 53).
Animal experiments.
Newborn NMRI/Nhg mice (less than 36 h old) were injected intraperitoneally with 0.1 ml of virus grown on NIH 3T3 cells. The titers were 105 to 106 infectious particles per ml as determined by endpoint dilution on NIH 3T3 cells and immunohistochemical analysis as described previously (53). Control mice were mock injected with complete medium. The mice were checked for tumor development on 5 days of the week. Mice were killed, X rayed, and autopsied when they showed illness or tumor development or at the end of the 2-year observation period. Tumors were diagnosed on the basis of gross appearance of lymphoid organs as previously described (51) and according to cytologic and anatomic criteria as described by Pattengale (44). Skeletal lesions were diagnosed as described previously (52).
Southern blotting and hybridization.
DNA was extracted from frozen tumor tissues and analyzed by Southern blotting and hybridization as described previously (56).
DNA probes.
The ecotropic-virus-specific probe was a SmaI fragment from the Akv MuLV (positions 6240 to 6570 [66]). The immunoglobulin heavy-chain probe was a BamHI-EcoRI fragment from the J region of the murine immunoglobulin heavy-chain (IgH) gene (denoted J11 [34]). The c-myc and T-cell receptor β (TCRβ) probes J1 and J2 were prepared by PCR as described previously (1, 56). The immunoglobulin kappa light-chain (Igκ) probe was generated by PCR with the primers described below. The PCR amplification products were electrophoresed in a 1.5% low-melting-point agarose (SeaKem; FMC Corp.) gel; fragments were excised from the gel and purified by phenol-chloroform extractions or with the Wizard PCR Preps DNA Purification System (Promega).
Immunohistochemistry.
Cryostat sections (6 μm thick) of representative tumors were prepared from unfixed frozen tissue, placed on silane-coated slides, and fixed with methanol-acetone (1:1) at −20°C for 10 min. The sections were preincubated (1 h, 37°C) with 5% bovine serum albumin in phosphate-buffered saline (PBS), followed by incubations with the following monoclonal and polyclonal antibodies and reagents: rat anti-mouse Ly-5-Biotin (Medac, Hamburg, Federal Republic of Germany); rat anti-mouse κ light chain-Biotin (Amersham); rat anti-mouse IgM-Biotin (Dianova, Hamburg, Federal Republic of Germany), rat anti-mouse CD3, rat anti-mouse CD4, and rat anti-mouse CD8 (kindly provided by S. Thierfelder, GSF, Munich, Germany); rabbit anti-mouse Thy-1 (Cedarlane Laboratories, Hornby, Ontario, Canada); and goat anti-rat IgG-Biotin (Dianova) and peroxidase-coupled goat anti-rabbit IgG as a secondary antibody. Purified IgG fractions of the antibodies (200 μg/ml) or complete antisera (dilutions of 1:20 and 1:1,000, respectively) in PBS containing 1% bovine serum albumin were applied to the sections and incubated for 30 min at room temperature in a moist chamber. Slides were washed three times, reacted with peroxidase-coupled streptavidin (1:2,000) for 10 min, and stained with amino-ethyl-carbazole in 50 mM Tris buffer (pH 5.5) containing 0.05% (vol/vol) H2O2 for 15 min. Control sections included tissues from healthy thymus, spleen, lymph node, and kidney of young ad ult mice; control reactions included incubations with normal rat serum, normal rabbit serum, PBS, and substrate to rule out endogenous peroxidase activity.
Transfection and reporter assays.
NIH 3T3 and MPC11 cells were transfected by calcium phosphate-mediated precipitation (32) with 3 μg of the various chloramphenicol acetyltransferase (CAT) constructs and 1 μg of the pCH110 (SV40-lacZ) (32) internal control plasmid. L-691 cells were transfected by the DEAE-dextran method as described previously (44). CAT assays were performed by the method of Gorman et al. (19) and Lovmand et al. (32). β-Galactosidase activity was measured by using an o-nitrophenyl-β-d-galactopyranoside (ONPG) assay (32). All transfections were done in duplicate or triplicate and repeated two to six times.
PCR and DNA sequencing analysis.
PCR amplifications, fragment analysis, and sequence determinations were done as described previously (56, 57).
Primer sequences.
The c-myc primers (primers a, b, and c [see Fig. 4]) used to generate the probe for the Southern hybridizations and to verify integration of provirus were as described previously (1, 56). The TCRβ primers used to generate the J1 and J2 probes for the Southern hybridizations were also as described previously (1). The primers used to generate the Igκ probe had the sequences 5′-GGTCTGACTGCAGGTAGCGTGGTCTTCT- 3′ and 5′-CGTTCCTACAGAGTCTCTCATTTTGACAT-3′, corresponding to nucleotide positions 2361 to 2388 and 2848 to 2820, respectively (nucleotide accession number g51657). The LTR primers used correspond to the following positions of the Akv LTR (66) (1 = first base in Akv U3): 377 to 360 (primer d [see Fig. 4]) (57), 360 to 377 (primer e) (56), 354 to 319 (56), 550 to 578 (56), 14 to 41 (5′-TTCATAAGGCTTAGCCAGCTAACTGCAG-3′), 68 to 91 (5′-AATACCAGAGCTGATGTTCTCAGA-3′), 83 to 110 (5′-GTTCTCAGAAAAACAAGAACAAGGAAGT-3′), and 129 to 101 (5′-TACTTTCCAGCCTCTCTGTACTTCCTTGT-3′).
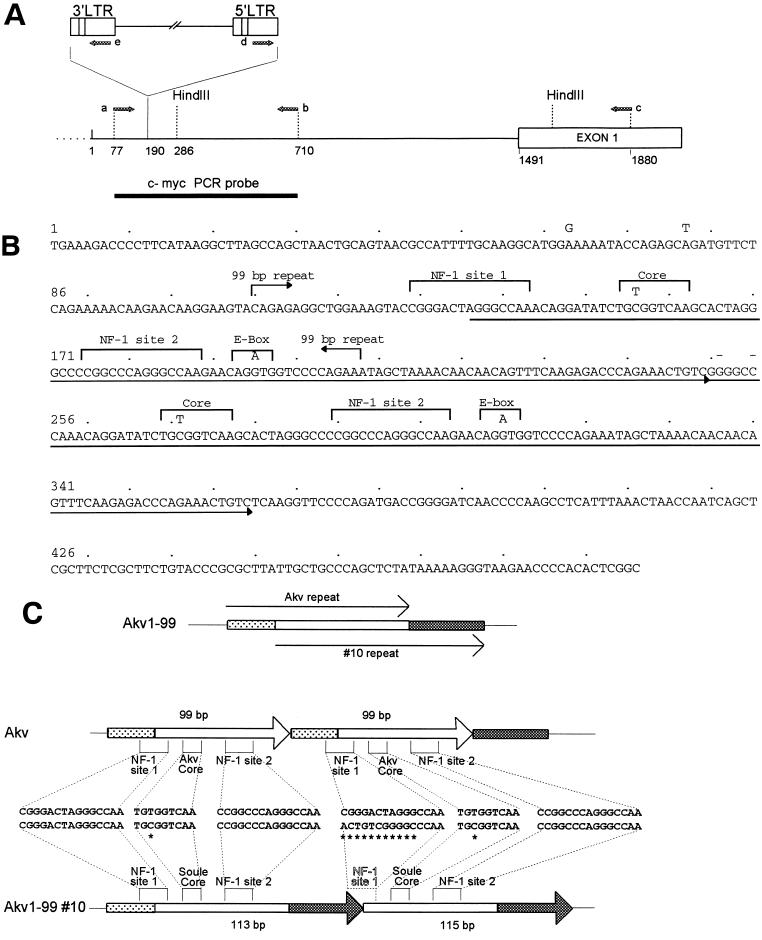
FIG. 4.
Analysis of a provirus integrated at the c-myc locus in Akv1- 99 tumor 10. (A) Integration site and orientation of provirus at c-myc. Indicated are the c-myc probe used for Southern hybridization, the c-myc-specific primers a, b, and c used for PCR analysis, and the provirus-specific primers d and e. The proviral site of integration and orientation was determined by sequence analysis of PCR products with the primers described in Materials and Methods. The position numbering corresponds to a c-myc promoter sequence from GenBank and EMBL (nucleotide sequence accession no. M12345). (B) Nucleotide sequence of the complete U3 region of the c-myc integrated provirus. The nucleotide sequence was determined from PCR amplification products of provirus–c-myc junction fragments with LTR-specific primers as described in Materials and Methods. Marked above the sequence are nucleotide differences in Akv1- 99 and the location of the 99-bp tandem repeat of Akv. The arrows underlining the sequence mark an imperfect tandem repeat of 113(115) bp. Two nucleotide insertions in the second repeat are marked with (−). (C) U3 repeat organization of Akv and the c-myc integrated provirus of Akv1- 99 10. Differences in core and NF-1 sites are shown.
RESULTS
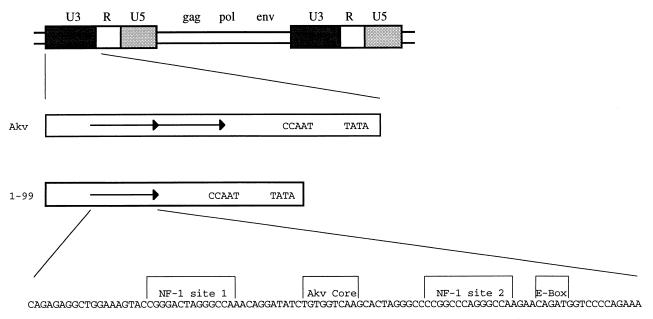
The standard source of infectious Akv has been the molecular clone λ623 harboring an integrated provirus derived from productively infected mouse cells (33). The U3 enhancer of the Akv virus in λ623 carries a 99-bp tandem repeat, whereas several closely related endogenous viruses lack this repeat (35, 47). Since enhancer repeats of MuLVs have been found to influence the pathogenicity of the viruses (10, 24, 29, 30, 65), it was of interest to also analyze an Akv mutant in which one copy of the tandem repeat had been deleted. The genetic organization of Akv and its U3 region is depicted in Fig. 1. The mutant Akv1- 99 was generated from Akv by restriction enzyme cleavage-mediated deletion of one copy of the 99-bp sequence (32).
FIG. 1.
Proviral structures of Akv and Akv1- 99. Akv contains a 99-bp perfect tandem repeat in the U3 enhancer region, while Akv1- 99 contains only one copy of the 99-bp sequence. Shown is the 99-bp sequence with demarcation of NF-1 sites 1 and 2 and the Akv core and E-box sequences.
Pathogenicity of Akv and Akv1- 99.
Viral stocks were generated by transfection of the molecular clones of Akv and Akv1- 99 into NIH 3T3 cells and subsequent spread of the viruses. A total of 104 to 105 infectious viruses were injected into newborn mice of the NMRI strain, and the animals were monitored for development of disease. As shown in Table 1, both viruses induced lymphomas at high incidences and with similar latency periods of about 12 months. The primary manifestations of the lymphomas were seen in the spleens and in lymph nodes and partly also in liver and kidneys. The mean sizes of the thymuses in the Akv1- 99-infected group and in the Akv-infected group were 5 to 6 mm, which was very similar to that found in the control mice. These data suggest that Akv1- 99 and Akv induced primarily nonthymic lymphomas. However, in addition to having greatly enlarged spleen and lymph nodes, one mouse (mouse 10 of the Akv1- 99 series) showed enlargement of the thymus, indicating the development of a thymic lymphoma. Among the 22 mock-infected control mice there was one mouse with a follicular center cell lymphoma, which was detected at the end of the observation period of 721 days.
TABLE 1.
Pathogenicity of Akv and Akv1- 99
| Virus | No. of micea | % Lymphoma incidence | Latency period (days ± SD) |
|---|---|---|---|
| Akv | 16 | 94 (15/16)b | 357 ± 116 |
| Akv1- 99 | 15 | 93 (14/15)c | 349 ± 150 |
| Control | 22 | 4 (1/22)d |
All animals were NMRI mice.
One mouse with mammary carcinoma was sacrificed at day 273.
One mouse with a stomach tumor was sacrificed at day 373.
One mouse developed lymphoma at day 721.
Both viruses also induced skeletal lesions. Akv induced osteopetrosis in 3 of 16 mice and osteomas in 5 of 16 mice, whereas Akv1- 99 did not induce osteopetrosis but did induce osteomas in 3 of 15 mice. Osteopetrosis or osteomas were not detected in mock-infected control mice.
Akv- and Akv1- 99-induced lymphomas were mainly of the B-cell type as indicated by molecular characterization.
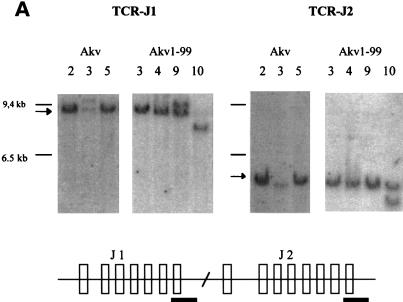
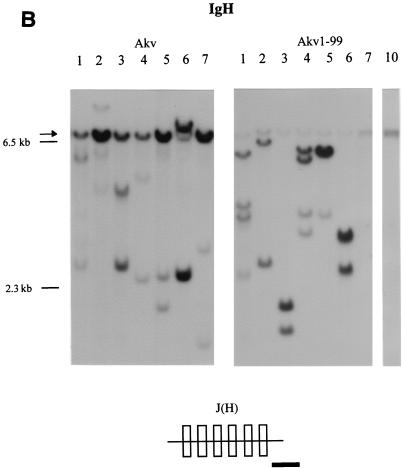
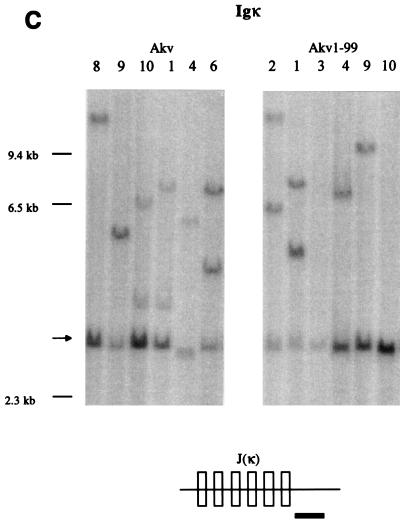
Lymphomas induced by Akv and Akv1- 99 were further characterized by Southern blotting analysis with probes derived from the TCRβ-chain locus, the IgH locus and the Igκ locus. DNA samples from 11 tumors of the Akv series and 13 tumors of the Akv1- 99 series were analyzed as summarized in Table 2. Analysis with the TCRβ probes J1 and J2 (Fig. 2A) with HindIII- cleaved tumor DNA showed rearrangements in only a few samples. Rearrangements were detected with the J1 or J2 probe in three of the Akv1- 99 tumors (tumors 2, 9, and 10) and in one of the Akv tumors (tumor 3) (Table 2). In contrast, most of the tumors showed rearrangements of the IgH locus (Fig. 2B). Rearrangements were seen in all 11 Akv lymphomas and in 11 of 13 Akv1- 99 lymphomas (Table 2). Similarly, Southern blotting analysis with the Igκ probe revealed rearrangements in all 11 Akv lymphomas and in 10 of 13 Akv1-99 lymphomas (Fig. 3C and Table 2). The low number of rearranged bands and their intensities indicate that the tumors are monoclonal or nearly monoclonal with respect to IgH as well as Igκ rearrangements. Notably, the IgH rearrangements differ from the specific pattern seen in Ly1+ lymphomas (21, 49), a pattern which may also be found in spontaneous B lymphomas of old NMRI mice (55). DNA samples from two tumor tissues of the same animal revealed the same animal-specific pattern of rearrangement, a finding supporting the absence of complex patterns of polyclonality in the five tumors Akv 6 and Akv1- 99 2, 5, 6, and 8 (Table 2).
TABLE 2.
Molecular characterization of lymphomas
| Virus and animal no. | Tumor tissuea | Latency (days) | DNA rearrange- ments
|
Tumor typeb | Eco- tropicc | c-mycd | |||
|---|---|---|---|---|---|---|---|---|---|
| J1 | J2 | IgH | Igκ | ||||||
| Akv | |||||||||
| 1 | L | 273 | − | − | + | + | B | + | − |
| 2 | S | 366 | − | − | + | + | B | + | − |
| 3 | S | 366 | + | + | + | + | T, Be | + | − |
| 4 | L | 429 | − | − | + | + | B | + | − |
| 5 | L | 491 | − | − | + | + | B | + | − |
| 6 | S, L | 509 | − | − | + | + | B | + | − |
| 7 | S | 593 | − | − | + | + | B | + | − |
| 8 | L | 211 | − | − | + | + | B | + | − |
| 9 | L | 243 | − | − | + | + | B | + | − |
| 10 | L | 245 | − | − | + | + | B | + | − |
| 11 | S | 260 | − | − | + | + | B | + | − |
| Akv1- 99 | |||||||||
| 1 | L | 183 | − | − | + | + | B | + | − |
| 2 | S, L | 197 | + | − | + | + | T, Bf | + | − |
| 3 | L | 204 | − | − | + | − | preB | + | − |
| 4 | L | 252 | − | − | + | + | B | + | − |
| 5 | Sc, S | 293 | − | − | + | + | B | + | − |
| 6 | S, L | 306 | − | − | + | + | B | + | − |
| 7 | L | 310 | − | − | − | − | Nullf | + | − |
| 8 | S, L | 310 | − | − | + | + | B | + | − |
| 9 | L | 349 | + | − | + | + | T, Bg | + | − |
| 10 | L | 495 | + | + | − | − | T | − | + |
| 11 | Sc | 359 | − | − | + | + | B | + | − |
| 12 | S | 359 | − | − | + | + | B | − | − |
| 13 | S | 730 | − | − | + | + | B | + | − |
L, Lymphoma; S, spleen; Sc, subcutaneous tumor.
Tumor types as determined by hybridization analysis.
Southern blot detection of hybridization to an ecotropic-specific probe.
Southern blot hybridization of c-myc promoter rearrangement.
Mixed T- and B-cell lymphoma as determined by histopathology.
Follicular center cell lymphoma as determined by histopathology.
Large-cell lymphoblastic lymphoma as determined by histopathology.
FIG. 2.
Tumor classification by Southern hybridizations. (A) DNA from Akv-induced tumors 2, 3, and 5 and Akv1- 99-induced tumors 3, 4, 9, and 10 was cleaved by HindIII and analyzed by using the TCRβ J1 and TCRβ J2 probes. (B) DNA from Akv-induced tumors 1, 2, 3, 4, 5, 6, and 7 and Akv1- 99-induced tumors 1, 2, 3, 4, 5, 6, 7, and 10 was cleaved by EcoRI and analyzed by using IgH probe. The significance of the tendency towards lower intensity of the IgH germ line band for the Akv1- 99 tumors than for the Akv tumors is not clear. (C) DNA from Akv1- 99-induced tumors 2, 1, 3, 4, 9, and 10 and Akv-induced tumors 8, 9, 10, 1, 4, and 6 was cleaved by HindIII and analyzed by using the Igκ probe. Arrows mark the position of the unrearranged genomic band. Size markers of 9.4, 6.5, and 2.3 kb are indicated. The location of the probes in relation to the joining regions is indicated by a horizontal black bar in the diagrams below each panel.
FIG. 3.
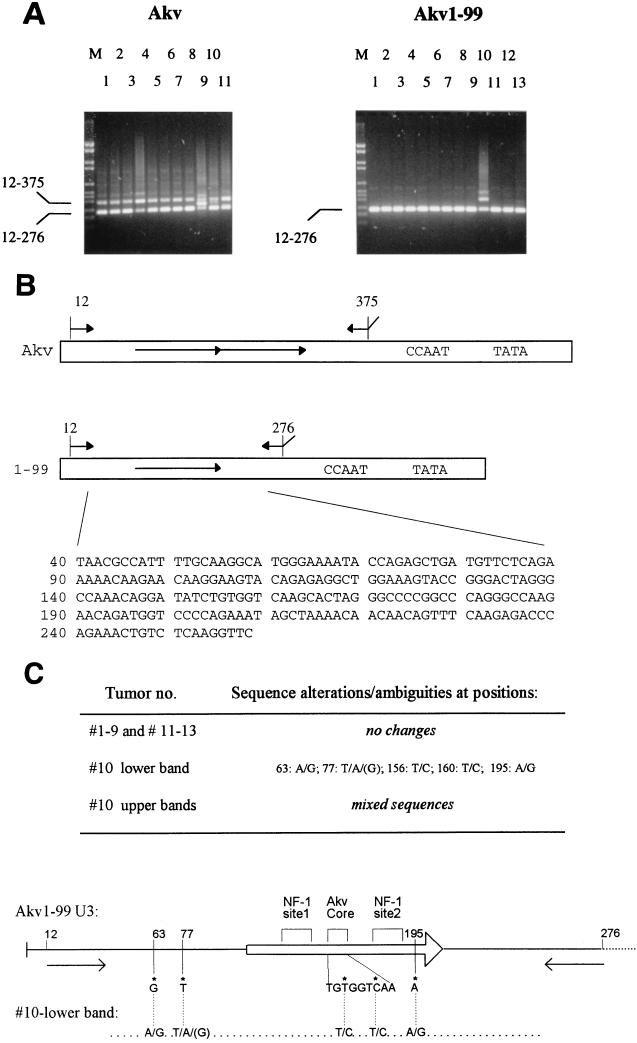
Analysis of proviral U3 regions in tumor DNAs. (A) PCR amplification of U3 enhancer sequences of DNA of Akv- and Akv1- 99-induced tumors; the numbers refer to the animal numbers presented in Table 2. M, DNA size markers. The positions of amplified fragments resulting from Akv (positions 12 to 375) and Akv1- 99 (positions 12 to 276) are shown. (B) Akv and Akv1- 99 U3 structures showing the PCR primers used in panel A. Below is shown the input Akv1- 99 sequence (positions 40 to 258). (C) All nucleotide sequences of PCR fragments of Akv1- 99-induced tumors were determined from positions corresponding to 40 to 258 of the input U3 sequence shown in panel A. Only tumor 10 showed changes as shown in the lower band.
Based upon rearrangements in IgH and Igκ loci but not in the TCRβ locus, we classified most of the tumors as B-cell lymphomas; this diagnosis was consistent in all cases with the morphological diagnoses and immunohistochemical analyses. One tumor (Akv1- 99 3) showed rearrangements only in IgH and was classified as pre-B-cell type. One tumor (Akv1- 99 7) that lacked rearrangements in all three loci was diagnosed as a follicular center cell lymphoma, i.e., a mature B-cell tumor. Among the three tumors that show rearrangements at all three loci, Akv 3 represented a mixed T- and B-cell lymphoma, whereas the two others (Akv1- 99 2 and Akv1- 99 9) were classified as follicular center cell lymphoma and large-cell lymphoblastic lymphoma, respectively. Among the 24 tumors analyzed, only 1 tumor (Akv1- 99 10) was classified as a T-cell lymphoma. It showed TCRβ rearrangement and lacked immunoglobulin rearrangements. This mouse also showed a considerably enlarged thymus.
Morphological and immunohistochemical analysis of early and late lymphomas.
For further characterization, two tumors of each group with different latency periods and represented by cervical or mesenterial lymph nodes were investigated in more detail. The early Akv-induced tumor (tumor 8) was a large-cell pleomorphic lymphoma. In agreement with the IgH chain rearrangement, many cells expressed Ly-5 (panB) and IgM. The late Akv-induced tumor (tumor 4), which also showed IgH rearrangements, was a follicular center cell lymphoma. Consistent with these findings, many tumor cells expressed Ly-5 (panB) and IgM. Numerous admixed mature lymphocytes expressed CD4 or CD8.
The early Akv1- 99 tumor (tumor 1) was a lymphoblastic lymphoma with numerous mitotic figures. In agreement with the IgH rearrangement detected by Southern blotting, many tumor cells showed expression of Ly-5 (panB) and IgM. The late tumor (tumor 10) was a lymphoblastic lymphoma; it showed diffuse infiltration of the significantly enlarged thymus. In agreement with the TCRβ rearrangements, the majority of the tumor cells expressed Thy-1. Single cells in the tumor also expressed CD3, CD4, and CD8.
Integrated ecotropic proviruses in most tumor DNAs.
The tumors were analyzed for clonality or oligoclonality with respect to integration of ecotropic proviruses (Table 2). The 24 DNA samples used for tumor classification were analyzed by Southern blotting with HindIII-cleaved DNA and hybridized with an ecotropic-specific probe. In 22 of the samples, clonal or oligoclonal integrations of hybridizing proviruses could be detected (data not shown). The number of hybridizing bands ranged from one to five. In two tumors induced by Akv1- 99 (tumors 10 and 12) no bands were detected, indicating a lack of clonally integrated ecotropic proviruses. In all five cases analyzed, the hybridization patterns of the two lymphoma samples taken from the same animal were identical (Table 2).
Organization of Akv and Akv1- 99 U3 regions in tumor DNAs.
The above analysis suggests that the pathogenic properties of Akv and the deletion mutant Akv1- 99 are largely similar. Since it is known that fluctuation in the number of tandem repeats in the enhancers of MuLV is commonly observed in tumors, it was of interest to analyze the enhancer dynamics of the two Akv viruses, with or without the tandem repeat, and to assess its relation to the disease process. The enhancer structure was analyzed by bulk PCR amplification from tumor DNA of a U3 fragment (Fig. 3) by using two primers located upstream and downstream of the 99-bp repeat of Akv. The resulting fragments were then analyzed by agarose gel electrophoresis as shown in Fig. 3A. The use of DNA from mock-infected NMRI mice as a template for PCR amplification did not lead to detectable bands, while DNA from tumors of virus-infected animals in all cases gave rise to PCR bands. As seen in the left panel, PCR amplification with DNA of Akv-induced tumors as a template resulted in most cases in two bands of sizes compatible with the 364- and 265-bp fragments predicted to result from the Akv enhancer with one or both copies of the 99-bp tandem repeat (Akv tumors 1, 2, 3, 5, 6, 7, 8, and 11). The appearance of more than one band did not just reflect an artifact of the PCR amplification, as was demonstrated by the fact that the band intensity differences among the tumors were reproducible in independent PCRs. Moreover, fluctuations in U3 repeat numbers have previously been found for tumors induced by other MuLVs as confirmed by Southern blotting analysis (9, 10, 37). The analysis revealed three cases (tumors Akv 4, 9, and 10) of clear bands with sizes that were distinct from the input and the 99-bp-deletion sizes. Akv tumor 4 contains a band of increased size relative to Akv, while further sequence analysis of the fragments amplified from Akv tumors 9 and 10 showed that both of the additional fragments harbor repetitions of sequences upstream the 99-bp repeat sequence (data not shown).
In Akv1- 99-induced tumors, on the other hand, PCR analysis revealed no detectable alterations in 12 of the 13 tumor DNAs analyzed. One tumor, Akv1- 99 10, indicated a more complex pattern in which at least two larger bands were present in addition to the band corresponding to the input enhancer structure. Interestingly, this was the only tumor that was classified as a T-cell tumor by pathological examination, immunohistochemistry, and DNA analysis.
Conservation of proviral enhancer sequences in tumor DNA.
In a number of cases minor nucleotide alterations in MuLV enhancers have been found to affect enhancer specificity and viral pathogenicity. Moreover, it is known that selection for single nucleotide alterations that appear during viral replication may contribute to the pathogenic process. The above analysis indicated that the Akv1- 99 enhancer had not undergone major alterations during tumorigenesis, with the exception of that of tumor Akv1- 99 10.
The sequences of PCR products of Akv1- 99-induced tumors were analyzed. In Akv1- 99 tumors 1 through 9 and 11 through 13 all nucleotides from 40 to 258 were unambiguously determined to harbor nucleotides identical to those of the input Akv1- 99 virus (Fig. 3). The PCR amplification products resulting from Akv1- 99 tumor 10 were also subject to nucleotide sequence analysis. The lower band of a size similar to that of the 265- bp Akv1- 99 band was found to deviate from Akv1- 99 at only five positions by the presence of a mixture of bases (Fig. 3). Analysis of the larger amplification product appearing from Akv1- 99 tumor 10 did not lead to a coherent nucleotide sequence, although sequences of Akv1- 99 origin were clearly present. Notably, the nucleotide sequence of the lower band of Akv1- 99 tumor 10 differs from the input U3 at two positions within the Akv core (Fig. 3C).
Viral enhancer strength in a B-lymphoid cell line.
The recovery of input enhancers in B lymphomas induced by Akv and Akv1- 99 raised the possibility that the enhancer carries specific determinants of B lymphomagenicity. Among previous studies that have assessed the enhancer strengths of MuLV enhancers in cell lines, one has also analyzed the Akv enhancer in the M12 B-cell line (15). In this cell line, the Akv enhancer was in fact found to be stronger than those of Moloney, SL3-3, and Friend MuLVs. This may support the notion that the Akv enhancer may be particularly strong in B-lineage cells and that this specificity may contribute to its B-lymphomagenic properties. To analyze these properties of the enhancer further, we chose the mouse plasmacytoma cell line MPC11. Transient expression driven by the U3s of Akv, Akv1- 99, and SL3-3 (Table 3) showed that Akv directed stronger expression than the T-cell tropic SL3-3. The transcriptional activity of Akv1- 99 was not significantly different from Akv in MPC11 cells, indicating that the duplication is not critical for enhancer strength in this cell line. Transient expression values of the three constructs in the T-cell line L-691 showed that SL3-3 was much stronger and Akv1- 99 was somewhat stronger than Akv. In NIH 3T3 fibroblast cells, Akv was somewhat stronger than SL3-3 and Akv1- 99. The similarity in transient expression values of Akv and Akv1- 99 in the B-cell line parallels the similar pathogenicities of the two viruses and is thus compatible with a direct role of the Akv enhancer in B-cell lymphomagenicity.
TABLE 3.
Transient expression of CAT reporter constructs
| Cell line | CAT activity (SD)a
|
||
|---|---|---|---|
| pAkv6-cat | pAkv1- 99cat | pSL3-3-cat | |
| MPC11 | 100 | 82 (19) | 20 (7) |
| L-691 | 100 | 171 (12) | 1,107 (176) |
| NIH 3T3 | 100 | 40 (9) | 18 (4) |
Transient-transfection assays with CAT reporter constructs. Transfections were done in series with each construct included three to six times. Within each series, the mean of the values for each construct was normalized to the wild-type level, which was arbitrarily set at 100. Standard deviations of three to six independent transfection series are given in parentheses.
No clonal c-myc rearrangements in B-cell lymphomas.
The c-myc locus is a common target for insertional mutagenesis in MuLV-induced lymphomas. DNA samples of the 24 tumors were analyzed by Southern blotting analysis with a c-myc probe to detect possible rearrangements at this locus. In only 1 of the 24 tumors analyzed were rearrangements detected (Table 2). Interestingly, this tumor was Akv1- 99 10, which was the only one classified as a T-cell tumor (Table 2). We conclude that integration in c-myc is not a key mechanism in B-cell lymphoma induction by Akv.
An Akv1- 99 provirus at c-myc in T-cell lymphoma DNA shows enhancer duplication and point mutations.
To investigate if the rearrangement at the c-myc locus of Akv1- 99 tumor 10 was caused by provirus integration, combination PCR analysis was done with c-myc-specific and LTR-specific primers a, b, c, d, and e as shown in Fig. 4A. DNA fragments resulting from the PCRs appeared that were compatible with a provirus integration at the locus in a transcriptional orientation opposite to c-myc (data not shown). Sequence analysis of these proviral host junction fragments identified the integration site as nucleotide 190 in the c-myc promoter (Fig. 4A), which is within the normal region of provirus integration in T-cell lymphomas. The two proviral c-myc junction fragments were further sequenced by using the LTR primers specified in Materials and Methods. The resulting sequence of the complete U3 region of 496 bp, combined from analysis of the two LTRs, is shown in Fig. 4B. The U3 of the Akv1- 99 tumor 10 provirus at the c-myc locus differs from that of Akv1- 99 at a few nucleotide positions as well as by the presence of an imperfect tandem repeat of 113 (versus 115) bp (Fig. 4B).
The repeat organization of the Akv1- 99 tumor 10 U3 is compared to that of Akv in Fig. 4C. The 5′ and 3′ ends of the repeated sequence of Akv1- 99 10 are located 27 and 41 bp, respectively, downstream of the 5′ and 3′ ends of the 99-bp sequence repeated in Akv. The 5′ end is located between the two half-sites of the NF-1 binding site 1, causing a loss of this site from the second repeat copy, which moreover carries two nucleotide insertions in the remaining half-site (Fig. 4C). A T-to-C change relative to Akv1- 99 is found in both tandem repeat copies within the Akv core. Interestingly, the sequence found at this site of Akv1- 99 10 corresponds to that of the T-lymphomagenic Soule MuLV (Soule core). Moreover, this exact nucleotide transition from the Akv core to the Soule core was previously found to take place during T-cell lymphomagenesis by SL3-3 viruses mutated to carry the Akv core (37). A second nucleotide difference of both repeats of Akv1- 99 10 relative to Akv1- 99 is an A-to-G transition at nucleotide positions 195 and 310. We cannot ascribe any function to this change, but we note that the transition is located between the half-sites of a basic helix-loop-helix factor binding (41) and that it generates a perfect six-nucleotide match, 5′-GTGGTC-3′, to the Akv core. Of the two nucleotide differences between Akv1- 99 10 and Akv upstream of the repeat region, we note that the G-to-A transition at position 63 corresponds to a difference between the closely related viruses Akv and Gross passage A MuLVs and that the SL3-3 MuLV harbors a nucleotide insertion next to this position (29). We note, moreover, that the T-to-A mutation upstream of the repeats recreates at this upstream position the exact basic helix-loop-helix factor site (5′-CAGATG-3′) of the Akv repeat.
A U3 structure similar to that of the c-myc integrated provirus may also be present in bulk PCR-amplified DNA from Akv1- 99 tumor 10, since the size and nucleotide sequence are compatible with available information on the larger fragment (Fig. 3). Notably, the lower band of the bulk PCR-amplified fragments from Akv1- 99 tumor 10 has single-nucleotide alterations or mixtures at the same four positions (numbers 63, 77, 156, and 195 [Fig. 4]) as has the U3 of the c-myc integrated provirus. Moreover, the bulk PCR fragment has the C-to-T alteration characteristic of the SL3-3 core.
The altered U3 sequences of Akv1- 99 tumor 10 are not endogenous to NMRI mice.
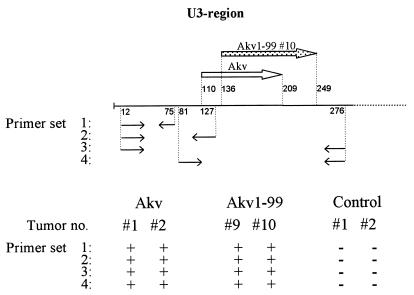
The finding that bulk PCR-amplified, as well as c-myc locus-derived, U3 sequences of Akv1- 99 tumor 10 harbored multiple alterations from the input Akv1- 99 U3 raised the question of the origin of these variant sequences. To understand their possible relation to the T-cell phenotype of tumor Akv1- 99 10, it was particularly important to determine whether they originated from recombination with a stretch of endogenous viral sequences of NMRI mice or whether they represent independent mutations of the Akv1- 99 viral genome. The possibility of recombination is highly relevant, since the DNA of tumor Akv1- 99 10 did not hybridize to the ecotropic-envelope-specific probe (Table 2). Hence, the provirus integrated at the c-myc locus must either be defective or carry a recombinant envelope gene. We note, however, that the U3 sequences derived from tumor Akv1- 99 10 has none of the characteristics of endogenous or recombinant nonecotropic proviruses (18). Moreover, since PCRs covering nucleotides 12 to 276 of Akv1- 99 U3 did not amplify fragments from the DNA of mock-infected NMRI mice, we can exclude the possibility that the complete variant U3 regions found in tumor Akv1- 99 10 were of endogenous origin. To further analyze if parts of the U3 sequences may be of endogenous origin, a series of PCRs were done on control NMRI DNA and on DNA samples of Akv tumors 1 and 2 and Akv1- 99 tumors 9 and 10 (Fig. 5). For all primer sets the amplification reaction was negative on control NMRI DNA, while fragments of the predicted sizes were amplified from the DNAs of the virus-induced tumors included (Fig. 5). The results obtained by using primer set 1 show that the stretch of nucleotides from 12 to 75, harboring the alteration at position 63, cannot be of pure endogenous origin. Analysis with primer set 2 excludes that the whole upstream part of U3 comprising the altered positions 63 and 77 of tumor Akv1- 99 10 can be detected in endogenous viruses. Analysis with primer set 4 further excludes that the variant U3 regions of tumor Akv1- 99 10 are endogenous sequences modified by a single recombination event in the upstream U3. Primer set 3, included again as a control, is identical to that used to amplify the fragments of Fig. 4.
FIG. 5.
Analysis of the presence of specific U3 sequences in DNA of virus-induced tumors and noninfected control tissues. The upper panel shows the U3 region, with the arrows indicating the repeated sequence in Akv and Akv1- 99 10. Also shown are the four primer sets used to verify the nonendogenous nature of the altered U3 region in the Akv1- 99-induced tumor 10. The lower panel shows the results of PCRs on DNA of the two Akv-induced tumors 1 and 2, the two Akv1- 99-induced tumors 9 and 10, and the tissues from two mock-injected control mice. +, Amplified fragments of the expected size were detected; −, no amplified bands were detected.
We may conclude based upon this analysis that if the variant U3 sequences of tumor 10 were derived by recombination with endogenous sequences, they would have to be composed of at least three blocks of sequences. Although our analysis does not exclude the possibility that endogenous sequences contribute to the final pattern, we infer that the variant U3 sequences result from a process involving multiple events. The finding of differences between U3 sequences from the same tumor and the appearance of the variant U3 as ecotropic provirus sequences with a few scattered point alterations may add support to this notion.
DISCUSSION
This study analyzed the lymphomagenic properties of Akv MuLV and one of its derivatives lacking the second copy of the 99-bp tandem repeat sequence of the transcriptional enhancer of U3. Both viruses induce lymphomas in NMRI mice, with incidences of more than 90% and mean latency periods of 12 months. Among the 24 lymphomas induced by Akv or Akv1-99, 22 were classified as being of B-cell origin.
It is well established that Akv is involved in leukemogenesis of the AKR mouse, where it contributes to formation of leukemogenic mink cell focus-forming viruses (62). Akv harboring loci also contribute to the spontaneous development of lymphomas in AKXD recombinant inbred strains, derived by crossing of AKR/J and DBA2/J strains (40). In some of these strains the predominant lymphoma type is the B-cell type. In the AKXD strains that develop primarily B-cell lymphomas, the somatically acquired proviruses in tumor DNA are mostly ecotropic, whereas strains developing primarily T-cell lymphomas show newly integrated proviruses of the ecotropic and MCF types (17, 39). Our finding that Akv induces mostly B-cell lymphomas and that most of the lymphomas harbor ecotropic proviruses may thus be reminiscent of the situation in these AKXD strains. B-cell lymphomagenesis associated with the somatic acquirement of ecotropic proviruses is also a characteristic feature of other inbred mouse lines, such as CWD/LeAg1 and SEA/GnJ (38). While several loci have been identified as frequent targets of proviral insertional mutagenesis in T-cell lymphomas, the number of loci that have been implicated in B-cell lymphomagenesis in inbred (2, 27) or oncogene-activated transgenic mouse strains is small (50, 67, 68). Hence, the model presented here may aid the search for additional target genes for insertional mutagenesis in B-cell lymphomagenesis. The tumors analyzed here were screened for proviral insertions at the c-myc promoter, which is a common target for provirus insertions in T-cell lymphomas (64). In none of the B-cell lymphomas did we find alterations at the c-myc promoter, which is in agreement with previous reports that proviral insertions at c-myc in B-cell lymphomas are infrequent.
Although a number of studies of more potent MuLVs have used Akv as a nonleukemogenic or weakly leukemogenic reference virus, studies on Akv pathogenesis are limited (28, 46, 59). Tandem repeat sequences of MuLV transcriptional enhancers are important determinants of transcriptional strength and cell-type specificity and have in some cases been demonstrated to be important for the leukemogenic potency of a virus. The 99-bp tandem repeat of U3 in the lambda clone 623 isolate of Akv has previously been found to increase the strength of the U3 promoter-enhancer in NIH 3T3 fibroblasts, and we have here presented data on the effect of removal of one copy of the repeat sequence on U3-directed transcription in a T-lymphoma cell line and a plasmacytoma cell line. We found that the Akv1- 99 enhancer with one copy of the 99-bp sequence deleted is stronger than the Akv enhancer in a T-lymphoma cell line and similar to Akv in a B-cell line.
Our pathogenicity studies show that the presence of one or two copies of this sequence make no significant difference in relation to the overall latency period and incidence of disease induction. The AKL-E1 virus studied by Lawrenz-Smith et al. (28) is very similar to Akv1- 99 and exhibits a similar pathogenicity phenotype in that it was found to induce B-cell lymphomas in NIH Swiss mice with a mean latency period of 18 months. Thus, the pathogenic properties of Akv-like viruses have now been demonstrated in two different, although probably closely related (60), mouse strains.
In agreement with earlier studies of MuLV U3 regions in tumors, we found a fluctuation in the number of direct repeats in tumors induced by Akv, where forms with two copies and only a single copy of the 99-bp sequence were present. In contrast to results of previous studies of the SL3-3 wild type and mutants (1, 10, 37) and a Moloney mutant (4), we observed mainly cases of alterations leading to a reduced size of the Akv enhancer. The organization of the Akv1- 99 enhancers was stable during B-cell lymphomagenesis, as was evident from an analysis of the bulk PCR products of U3. Presumably, the lack of a tandem repeat in the input virus makes alterations mediated by a template shift of reverse transcriptase less frequent. In case of the B-cell lymphomas, the general conservation of the Akv and Akv1- 99 enhancer sequences and the lack of duplications of Akv1- 99 sequence suggest that no single simple change, such as a point mutation or duplication, will make Akv and Akv1- 99 more potent in terms of B-cell lymphomagenesis. Specifically, there seems to be no strong selection for single nucleotide changes of the Akv core site in B-cell lymphomagenesis.
The only Akv1- 99 induced tumor, tumor 10, in which enhancer repeats were generated was a T-cell tumor harboring a clonal provirus integrated at the c-myc promoter. Moreover, this tumor carried no clonal ecotropic proviruses. The U3 region of the Akv1- 99 tumor 10 provirus at the c-myc locus differed from Akv1- 99 by an imperfect tandem repeat of 113 (versus 115) bp. In addition to two nucleotide insertions carried only in the second repeat copy, the Akv1- 99 tumor 10 diverged from Akv1- 99 at two nucleotide positions upstream of the repeat and two nucleotide positions within the repeated sequence. Although the provirus in question most likely was the result of recombination with endogenous nonecotropic viral sequences, we believe that the U3 of Akv1- 99 10 is derived from the input Akv1- 99 rather than from one consecutive stretch of endogenous sequences. Since PCRs covering various parts of the Akv1- 99 10 provirus were negative for control DNA of NMRI mice, mutations and duplication of the Akv1- 99 sequence most likely account for the derivation of the U3 of tumor 10. However, we cannot exclude the possibility that shorter stretches of sequence were the result of recombination with endogenous viruses as suggested by Massey et al. (35). A bulk-amplified PCR product from the same tumor was found to harbor nucleotide differences from Akv1- 99 at the same positions as the c-myc integrated provirus. Interestingly, this bulk-amplified PCR product also harbored a C-to-T alteration characteristic of the SL3-3 core.
An interesting and not unlikely possibility is that the U3 regions of Akv1- 99 tumor 10 was the result of stepwise alterations that gave a selective advantage for T-cell lymphomagenesis. The observation that the U3 carries multiple changes, while the U3 regions of Akv1- 99 proviruses in B-cell lymphomas were fully conserved, suggests a model of sequential alterations leading to more T-cell-lymphomagenic viruses. Whether env or U3 changes or other events might have been involved in the initiation of such a possible selectional pathway is a matter of speculation. We note a number of characteristic features of the U3 region of the c-myc integrated provirus suggestive of a role in altering the Akv1- 99 U3 towards a more T-cell-lymphomagenic structure.
Duplication of enhancer sequences is frequently associated with MuLV induction of T-cell lymphomas, which when compared with the absence of duplications in the proviruses of Akv1- 99 induced B-cell lymphomas may indicate a role of the 113(115)-bp tandem duplication in T-cell-lymphoma induction. The mutation in the Akv core site is reminiscent of the pattern of mutations found in c-myc integrated proviruses at the c-myc locus of tumors induced by SL3-3 viruses mutated at a single nucleotide of the SL3-3 core to provide a match to the Akv core 5′-TGTGGTCAA-3′ (37). Alterations in the enhancer of U3 of these were specifically located at the core sites, including true reversions to the SL3-3 core, as well as alterations to the Soule core version 5′-TGCGGTCAA-3′, the latter corresponding to the alteration observed in the c-myc integrated provirus of Akv1- 99 tumor 10. Like the SL3-3 core, the Soule core has higher affinity for AML1 proteins than has the Akv core (63). It therefore seems likely that the mutation of the Akv core to the Soule or SL3-3 core contributes to the T-cell-tropic properties of U3. While it is likely that the generation of the repeat as well as the Akv core mutation of Akv1- 99 10 contribute to T-cell lymphomagenicity, it seems unlikely that just one of the two changes will serve to change Akv1- 99 into a potent T-lymphomagenic virus. Had the change of Akv1- 99 to a T-lymphomagenic virus required just one nucleotide alteration in the Akv core site, as was the case for the single nucleotide Akv core mutants of SL3-3 (37), we would have expected to observe a higher frequency of Akv1- 99 induction of T-cell lymphomas, which generally develop faster than B-cell lymphomas. Thus, more than one change in the U3 seems required for a change from B- to T-cell specificity and vice versa (35, 37). That additional parts of the SL3-3 enhancer serve to make this virus T lymphomagenic is also supported by the induction of T-cell lymphomas by SL3-3 mutants harboring nonrevertable triple mutations of the SL3-3 core site (1, 10, 20). The finding in bulk PCR-amplified U3 of tumor Akv1- 99 10 of an additional alteration of the core site similar to the SL3-3 core suggests that multiple mutational pathways towards T-lymphoid enhancer strength may be followed in the same tumor, a pattern reminiscent of that seen by Morrison et al. (37).
We note that the 5′ border of the repeat of Akv1- 99 tumor 10 is located between the half sites of a binding site for NF-1 proteins and thus determines the loss of this site from the downstream repeat copy, which furthermore harbors two nucleotide insertions at the remaining half site. Although contributing positively to the T lymphomagenicity of Moloney MuLV, NF-1 sites have been found to be neutral or act negatively on the ability of SL3-3 to induce T-cell lymphomas (9–11). Moreover, we have found that an Akv1- 99 U3 mutant in which the NF-1 site 1 is impaired by four nucleotide substitutions has a twofold-stronger enhancer in T-cell line L691 than does Akv1- 99 (31). Hence, the loss of an NF-1 site at the junction of the repeats of Akv1- 99 tumor 10 may contribute to T-lymphomagenic properties.
A puzzling observation concerns two cases of loss and recurrence of hexameric sequence motifs in the U3 sequence of the c-myc integrated provirus of Akv1- 99 tumor 10. By the first of these changes a six-nucleotide match to the Akv core was generated by an A-to-G alteration at the basic helix-loop-helix factor bonding site (E-box) of the Akv enhancer (41). This alteration will still allow binding of basic helix-loop-helix proteins but may affect the affinity of the site and the nature of the complexes formed (3). While basic helix-loop-helix proteins are important for gene expression in lymphoid cells (70), roles of other factors interacting with this site in U3 are not unlikely (16). By the second pattern of loss and recurrence of motifs, the six-nucleotide core of the E-box sequence of the Akv enhancer was formed by the T-to-A change upstream of the repeat, which presumably leads to a new binding site for factors of the basic helix-loop-helix family.
Altogether, our results show that Akv MuLV has B-lymphomagenic properties and that these properties are retained after removal of one copy of the enhancer repeat in Akv1- 99 and suggests that occasional T-cell-lymphoma induction by Akv1- 99 may be an interesting model for studies of the lymphomagenic specificity of MuLVs toward B or T cells.
ACKNOWLEDGMENTS
The technical assistance of Angelika Appold, Anna Nickl, Elenore Samson, and Lone Højgaard is gratefully acknowledged. We also thank J. S. Diaz-Cano for discussions of the histopathology.
This project was supported by the Danish Cancer Society, the Karen Elise Jensen Foundation, the Danish Natural Sciences and Medical Research Councils, the Danish Biotechnology Programme, and the Leo Nielsen Foundation and by European Commission contracts CT-950100 (Biotechnology), CT-95067 (Biomed-2), and FI4P-CT95-0008 (Radiation protection).
REFERENCES
- 1.Amtoft H W, Sørensen A B, Bareil C, Schmidt J, Luz A, Pedersen F S. Stability of AML1 (core) site enhancer mutations in T lymphomas induced by attenuated SL3-3 murine leukemia virus mutants. J Virol. 1997;71:5080–5087. doi: 10.1128/jvi.71.7.5080-5087.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bergeron D, Poliquin L, Houde J, Barbeau B, Rassart E. Analysis of proviruses integrated in Fli-1 and Evi-1 regions in Cas-Br-E MuLV-induced non-T, non-B-cell leukemias. Virology. 1992;191:661–669. doi: 10.1016/0042-6822(92)90241-g. [DOI] [PubMed] [Google Scholar]
- 3.Bonven B J, Nielsen A L, Nørby P L, Pedersen F S, Jørgensen P. E-box variants direct formation of distinct complexes with the basic helix-loop-helix protein ALF1. J Mol Biol. 1995;249:564–575. doi: 10.1006/jmbi.1995.0319. [DOI] [PubMed] [Google Scholar]
- 4.Brightman B K, Farmer C, Fan H. Escape from in vivo restriction of Moloney mink cell focus-inducing viruses driven by the Mo-PyF101 long terminal repeat (LTR) by LTR alteration. J Virol. 1993;67:7140–7148. doi: 10.1128/jvi.67.12.7140-7148.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Celander D, Haseltine W A. Tissue-specific transcription preference as a determinant of cell tropism and leukaemogenic potential of murine retroviruses. Nature. 1984;312:159–162. doi: 10.1038/312159a0. [DOI] [PubMed] [Google Scholar]
- 6.Corcoran L M, Adams J, Dunn A R, Cory S. Murine T lymphomas in which the cellular myc oncogene has been activated by retroviral insertion. Cell. 1984;37:113–122. doi: 10.1016/0092-8674(84)90306-4. [DOI] [PubMed] [Google Scholar]
- 7.Dai H Y, Etzerodt M, Bækgaard A J, Lovmand S, Jørgensen P, Kjeldgaard N O, Pedersen F S. Multiple sequence elements in the U3 region of the leukemogemic murine retrovirus SL3-2 contribute to cell-dependent gene expression. Virology. 1990;175:581–585. doi: 10.1016/0042-6822(90)90445-w. [DOI] [PubMed] [Google Scholar]
- 8.DesGroseillers L, Jolicoeur P. The tandem direct repeats within the long terminal repeat of murine leukemia viruses are the primary determinant of their leukemogenic potential. J Virol. 1984;52:945–952. doi: 10.1128/jvi.52.3.945-952.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ethelberg S, Hallberg B, Lovmand J, Schmidt J, Luz A, Grundstrøm T, Pedersen F S. Second-site proviral enhancer alterations in lymphomas induced by enhancer mutants of SL3-3 murine leukemia virus: negative effect of nuclear factor 1 binding site. J Virol. 1997;71:1196–1206. doi: 10.1128/jvi.71.2.1196-1206.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ethelberg S, Lovmand J, Schmidt J, Luz A, Pedersen F S. Increased lymphomagenicity and restored disease specificity of AML1 site (core) mutant SL3-3 murine leukemia virus by a second-site enhancer variant evolved in vivo. J Virol. 1997;71:7273–7280. doi: 10.1128/jvi.71.10.7273-7280.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ethelberg S, Sørensen A B, Schmidt J, Luz A, Pedersen F S. An SL3-3 murine leukemia virus enhancer variant more pathogenic than wild type obtained by assisted molecular evolution in vivo. J Virol. 1997;71:9796–9799. doi: 10.1128/jvi.71.12.9796-9799.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Etzerodt M, Mikkelsen T, Pedersen F S, Kjeldgaard N O, Jørgensen P. The nucleotide sequence of the Akv murine leukemia virus genome. Virology. 1984;134:196–207. doi: 10.1016/0042-6822(84)90285-x. [DOI] [PubMed] [Google Scholar]
- 13.Fan H. Influences of the long terminal repeats on retrovirus pathogenicity. Semin Virol. 1990;1:165–174. [Google Scholar]
- 14.Gallagher R E, Gallo R C. Type C RNA tumor virus isolated from cultured human acute myelogenous leukemia cells. Science. 1975;187:350–353. doi: 10.1126/science.46123. [DOI] [PubMed] [Google Scholar]
- 15.Gama Sosa M A, Rosas D H, DeGasperi R, Morita E, Hutchison M R, Ruprecht R M. Negative regulation of the 5′ long terminal repeat (LTR) by the 3′ LTR in the murine proviral genome. J Virol. 1994;68:2662–2670. doi: 10.1128/jvi.68.4.2662-2670.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Genetta T, Ruezinsky D, Kadesch T. Displacement of an E-box-binding repressor by basic helix-loop-helix proteins: implications for B-cell specificity of the immunoglobulin heavy-chain enhancer. Mol Cell Biol. 1994;14:6153–6163. doi: 10.1128/mcb.14.9.6153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gilbert D J, Neumann P E, Taylor B A, Jenkins N A, Copeland N G. Susceptibility of AKXD recombinant inbred mouse strains to lymphomas. J Virol. 1993;67:2083–2090. doi: 10.1128/jvi.67.4.2083-2090.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Golemis E A, Speck N A, Hopkins N. Alignment of U3 region sequences of mammalian type C viruses: identification of highly conserved motifs and implications for enhancer design. J Virol. 1990;64:534–542. doi: 10.1128/jvi.64.2.534-542.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gorman C, Padhmanabhan R, Howard B H. High efficiency DNA-mediated transformation of primate cells. Science. 1983;221:551–553. doi: 10.1126/science.6306768. [DOI] [PubMed] [Google Scholar]
- 20.Hallberg B, Schmidt J, Luz A, Pedersen F S, Grundström T. SL3-3 enhancer factor 1 transcriptional activators are required for tumor formation by SL3-3 murine leukemia virus. J Virol. 1991;65:4177–4181. doi: 10.1128/jvi.65.8.4177-4181.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haran-Ghera N, Peled A, Brightman B K, Fan H. Lymphomagenesis in AKR Fv-1b congenic mice. Cancer Res. 1993;53:3433–3438. [PubMed] [Google Scholar]
- 22.Hays E F, Levy J A. Differences in lymphomagenic properties of AKR mouse retroviruses. Virology. 1984;138:49–57. doi: 10.1016/0042-6822(84)90146-6. [DOI] [PubMed] [Google Scholar]
- 23.Herr W. Nucleotide sequence of AKV murine leukemia virus. J Virol. 1984;49:471–478. doi: 10.1128/jvi.49.2.471-478.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Holland C A, Thomas C Y, Chattopadhyay S K, Koehne C, O’Donnell P V. Influence of enhancer sequences on thymotropism and leukemogenicity of mink cell focus-forming viruses. J Virol. 1989;63:1284–1292. doi: 10.1128/jvi.63.3.1284-1292.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ishimoto A, Takimoto M, Adachi A, Kakuyama M, Kato S, Kakimi K, Fukuoka K, Ogiu T, Matsuyama M. Sequences responsible for erythroid and lymphoid leukemia in the long terminal repeats of Friend-mink cell focus-forming and Moloney murine leukemia viruses. J Virol. 1987;61:1861–1866. doi: 10.1128/jvi.61.6.1861-1866.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jørgensen E C, Kjeldgaard N O, Pedersen F S, Jørgensen P. A nucleotide substitution in the gag N terminus of the endogenous DBA/2 virus prevents Pr65-gag myristylation and virus replication. J Virol. 1988;62:3217–3223. doi: 10.1128/jvi.62.9.3217-3223.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Justice M J, Morse III H C, Jenkins N A, Copeland N G. Identification of Evi-3, a novel common site of retroviral integration in mouse AKXD B-cell lymphomas. J Virol. 1994;68:1293–1300. doi: 10.1128/jvi.68.3.1293-1300.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lawrenz-Smith S C, Massey A C, Innes D J, Thomas C Y. Pathogenic determinants in the U3 region of recombinant murine leukemia viruses isolated from CWD and HRS/J mice. J Virol. 1994;68:5174–5183. doi: 10.1128/jvi.68.8.5174-5183.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lenz J, Celander D, Crowther R L, Patarca R, Perkins D W, Haseltine W A. Determination of the leukaemogenicity of a murine retrovirus by sequences within the long terminal repeats. Nature. 1984;308:467–470. doi: 10.1038/308467a0. [DOI] [PubMed] [Google Scholar]
- 30.Li Y, Golemis E, Hartley J W, Hopkins N. Disease specificity of nondefective Friend and Moloney murine leukemia viruses is controlled by a small number of nucleotides. J Virol. 1987;61:693–700. doi: 10.1128/jvi.61.3.693-700.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lovmand, J., and F. S. Pedersen. Unpublished results.
- 32.Lovmand S, Kjeldgaard N O, Jørgensen P, Pedersen F S. Enhancer functions in U3 of Akv virus: a role for cooperativity of a tandem repeat unit and its flanking DNA sequences. J Virol. 1990;64:3185–3191. doi: 10.1128/jvi.64.7.3185-3191.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lowy D R, Rands E, Chattopadhyay S K, Garon C F, Hager G L. Molecular cloning of infectious integrated murine leukemia virus DNA from infected mouse cells. Proc Natl Acad Sci USA. 1980;77:614–618. doi: 10.1073/pnas.77.1.614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marcu K B, Banerji J, Penncavage N A, Lang R, Arnheim N. 5′ Flanking region of immunoglobulin heavy chain constant region genes displays length heterogeneity in germlines of inbread mouse strains. Cell. 1980;22:187–196. doi: 10.1016/0092-8674(80)90167-1. [DOI] [PubMed] [Google Scholar]
- 35.Massey A C, Lawrenz-Smith S C, Innes D J, Thomas C Y. Origins of enhancer sequences of recombinant murine leukemia viruses from spontaneous B- and T-cell lymphomas of CWD mice. J Virol. 1994;68:3773–3783. doi: 10.1128/jvi.68.6.3773-3783.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morrison H L, Dai H Y, Pedersen F S, Lenz J. Analysis of the significance of two single-base-pair differences in the SL3-3 and Akv long terminal repeats. J Virol. 1991;65:1019–1022. doi: 10.1128/jvi.65.2.1019-1022.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morrison H L, Soni B, Lenz J. Long terminal repeat enhancer core sequences in proviruses adjacent to c-myc in T-cell lymphomas induced by a murine retrovirus. J Virol. 1995;69:446–455. doi: 10.1128/jvi.69.1.446-455.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mucenski M L, Bedigian H G, Shull M M, Copeland N G, Jenkins N A. Comparative molecular genetic analysis of lymphomas from six inbred mouse strains. J Virol. 1988;62:839–846. doi: 10.1128/jvi.62.3.839-846.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mucenski M L, Taylor B A, Copeland N G, Hopkins N A. Characterization of somatically acquired ecotropic and mink cell focus-forming viruses in lymphomas of AKXD recombinant inbred mice. J Virol. 1987;61:2929–2933. doi: 10.1128/jvi.61.9.2929-2933.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mucenski M L, Taylor B A, Jenkins N A, Copeland N G. AKXD recombinant inbred strains: models for studying the molecular genetic basis of murine lymphomas. Mol Cell Biol. 1986;6:4236–4243. doi: 10.1128/mcb.6.12.4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nielsen A L, Nørby P L, Pedersen F S, Jørgensen P. Various modes of basic helix-loop-helix protein-mediated regulation of murine leukemia virus transcription in lymphoid cell lines. J Virol. 1996;70:5893–5901. doi: 10.1128/jvi.70.9.5893-5901.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Olsen H S, Lovmand S, Lovmand J, Jørgensen P, Kjeldgaard N O, Pedersen F S. Involvement of nuclear factor I-binding sites in control of Akv virus gene expression. J Virol. 1990;64:4152–4161. doi: 10.1128/jvi.64.9.4152-4161.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Paludan K, Dai H Y, Duch M, Jørgensen P, Kjeldgaard N O, Pedersen F S. Different relative expression from two murine leukemia virus long terminal repeats in unintegrated transfected DNA and in integrated retroviral vector proviruses. J Virol. 1989;63:5201–5207. doi: 10.1128/jvi.63.12.5201-5207.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pattengale P K. Neoplastic lesions of the mouse lymphoid system. In: Bannasch P, Goessner W, editors. Pathology of neoplasia and preneoplasia in rodents. Stuttgart, Germany: Schattauer; 1994. pp. 168–176. [Google Scholar]
- 45.Pedersen F S, Crowther R L, Tenney D Y, Reimold A M, Haseltine W A. Novel leukaemogenic retroviruses isolated from cell line derived from spontaneous AKR tumor. Nature. 1981;292:167–170. doi: 10.1038/292167a0. [DOI] [PubMed] [Google Scholar]
- 46.Pedersen F S, Etzerodt M, Lovmand S, Dai H Y, Bækgaard A J, Sørensen J, Jørgensen P, Kjeldgaard N O, Schmidt J, Leib-Mösch C, Luz A, Erfle V. Transcriptional control and oncogenicity of murine leukemia viruses. In: Kjeldgaard N O, Forchhammer J, editors. Viral carcinogenesis. Alfred Benzon Symposium 24. Copenhagen, Denmark: Munksgaard; 1987. pp. 17–35. [Google Scholar]
- 47.Pedersen L, Strauss P G, Schmidt J, Luz A, Erfle V, Jørgensen P, Kjeldgaard N O, Pedersen F S. Pathogenicity of endogenous N-tropic BALB/c viruses. Virology. 1990;179:931–935. doi: 10.1016/0042-6822(90)90171-m. [DOI] [PubMed] [Google Scholar]
- 48.Robson I B, Mowat M, Bernstein A. Tissue-specific expression of the newly acquired ecotropic Emv-18 provirus in Fv-2 congenic mice. J Virol. 1985;55:54–59. doi: 10.1128/jvi.55.1.54-59.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rosner A, Peled A, Haran-Ghera N, Canaani E. Analysis of Ly-1 B-cell populations and IgH rearrangements in “normal” spleens and in lymphomas of AKR/J and AKR Fv-1 mice. Cancer Res. 1993;53:2147–2153. [PubMed] [Google Scholar]
- 50.Scheijen B, Jonkers J, Acton D, Berns A. Characterization of pal-1, a common proviral insertion site in murine leukemia virus-induced lymphomas of c-myc and Pim-1 transgenic mice. J Virol. 1997;71:9–16. doi: 10.1128/jvi.71.1.9-16.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schmidt J, Erfle V, Pedersen F S, Rohmer H, Schetters H, Marquart K-H, Luz A. Oncogenic retrovirus from spontaneous murine osteomas. I. Isolation and biological characterization. J Gen Virol. 1984;65:2237–2248. doi: 10.1099/0022-1317-65-12-2237. [DOI] [PubMed] [Google Scholar]
- 52.Schmidt J, Krump-Konvalinkova V, Luz A, Goralczyk R, Snell G, Wendel S, Dorn S, Pedersen L, Strauss P G, Erfle V. Akv murine leukemia virus enhances bone tumorigenesis in hMT-c-fos-LTR transgenic mice. Virology. 1995;206:85–92. doi: 10.1016/s0042-6822(95)80022-0. [DOI] [PubMed] [Google Scholar]
- 53.Schmidt J, Luz A, Erfle V. Endogenous murine leukemia viruses: frequency of radiation-activation and novel pathogenic effects of viral isolates. Leukocyte Res. 1988;12:393–403. doi: 10.1016/0145-2126(88)90058-6. [DOI] [PubMed] [Google Scholar]
- 54.Short M K, Okenquist S A, Lenz J. Correlation of leukemogenic potential of murine retroviruses with transcriptional tissue preference of the viral long terminal repeats. J Virol. 1987;61:1067–1072. doi: 10.1128/jvi.61.4.1067-1072.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sørensen, A. B., J. Schmidt, and F. S. Pedersen. Unpublished results.
- 56.Sørensen A B, Duch M, Amtoft H W, Jørgensen P, Pedersen F S. Sequence tags of provirus integration sites in DNAs of tumors induced by the murine retrovirus SL3-3. J Virol. 1996;70:4063–4070. doi: 10.1128/jvi.70.6.4063-4070.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sørensen A B, Duch M, Jørgensen P, Pedersen F S. Amplification and sequence analysis of DNA flanking integrated proviruses by a simple two-step polymerase chain reaction method. J Virol. 1993;67:7118–7124. doi: 10.1128/jvi.67.12.7118-7124.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Speck N A, Renjifo B, Golemis E, Fredrickson T N, Hartley J W, Hopkins N. Mutation of the core or adjacent Lvb elements of the Moloney murine leukemia virus enhancer alters disease specificity. Genes Dev. 1990;4:233–242. doi: 10.1101/gad.4.2.233. [DOI] [PubMed] [Google Scholar]
- 59.Speth C, Luz A, Strauss P G, Wendel S, Zeidler R, Dorn S, Erfle V, Brem G, Lipp M, Schmidt J. Akv murine leukemia virus enhances lymphomagenesis in myc-kappa transgenic and in wild-type mice. Virology. 1995;206:93–99. doi: 10.1016/s0042-6822(95)80023-9. [DOI] [PubMed] [Google Scholar]
- 60.Staats J. Standardized nomenclature for inbred strains of mice: eighth listing. Cancer Res. 1985;45:945–977. [PubMed] [Google Scholar]
- 61.Stoye J, Coffin J M. Endogenous retroviruses. In: Weiss R, Teich N, Varmus H, Coffin J, editors. RNA tumor viruses. 2nd ed., supplements and appendices. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1985. pp. 357–404. [Google Scholar]
- 62.Stoye J P, Moroni C, Coffin J M. Virological events leading to spontaneous AKR thymomas. J Virol. 1991;65:1273–1285. doi: 10.1128/jvi.65.3.1273-1285.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thornell A, Hallberg B, Grundström T. Binding of SL3-3 enhancer factor 1 transcriptional activators to viral and chromosomal enhancer sequences. J Virol. 1991;65:42–50. doi: 10.1128/jvi.65.1.42-50.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tsichlis P N, Lazo P A. Virus-host interactions and the pathogenesis of murine and human oncogenic retroviruses. Curr Top Microbiol Immunol. 1991;171:95–171. doi: 10.1007/978-3-642-76524-7_5. [DOI] [PubMed] [Google Scholar]
- 65.Tupper J C, Chen H, Hays E F, Bristol G C, Yoshimura F K. Contributions to transcriptional activity and to viral leukemogenicity made by sequences within and downstream of the MCF13 murine leukemia virus enhancer. J Virol. 1992;66:7080–7088. doi: 10.1128/jvi.66.12.7080-7088.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Van Beveren C. In: RNA tumor viruses. 2nd ed., supplements and appendices. Weiss R, Teich N, Varmus H, Coffin J, editors. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1985. [Google Scholar]
- 67.van Lohuizen M, Verbeek S, Scheijen B, Weintjens E, van der Gulden H, Berns A. Identification of cooperating oncogenes in Eμ-myc transgenic mice by provirus tagging. Cell. 1991;65:737–752. doi: 10.1016/0092-8674(91)90382-9. [DOI] [PubMed] [Google Scholar]
- 68.Verbeek S, van Lohuizen M, van der Valk M, Domen J, Kraal G, Berns A. Mice bearing the Eμ-myc and Eμ-pim-1 transgenes develop pre-B-cell leukemia prenatally. Mol Cell Biol. 1991;11:1176–1179. doi: 10.1128/mcb.11.2.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zaiman A L, Lewis A F, Crute B E, Speck N A, Lenz J. Transcriptional activity of core binding factor α (AML1) and β subunits on murine leukemia virus enhancer cores. J Virol. 1995;69:2898–2906. doi: 10.1128/jvi.69.5.2898-2906.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhuang Y, Cheng P, Weintraub H. B-lymphocyte development is regulated by the combined dosage of three basic helix-loop-helix genes, E2A, E2-2, and HEB. Mol Cell Biol. 1996;16:2898–2905. doi: 10.1128/mcb.16.6.2898. [DOI] [PMC free article] [PubMed] [Google Scholar]