Abstract
Inhibitors of the phosphatidylinositol 3-kinase (PI3 kinase)–FKBP-rapamycin-associated protein (FRAP) pathway, such as rapamycin and wortmannin, induce dephosphorylation and activation of the suppressor of cap-dependent translation, 4E-BP1. Encephalomyocarditis virus (EMCV) infection leads to activation of 4E-BP1 at the time of host translation shutoff. Consistent with these data, rapamycin mildly enhances the synthesis of viral proteins and the shutoff of host cell protein synthesis after EMCV infection. In this study, two defective EMCV strains were generated by deleting portions of the 2A coding region of an infectious cDNA clone. These deletions dramatically decreased the efficiency of viral protein synthesis and abolished the virus-induced shutoff of host translation after infection of BHK-21 cells. Both translation and processing of the P1-2A capsid precursor polypeptide are impaired by the deletions in 2A. The translation and yield of mutant viruses were increased significantly by the presence of rapamycin and wortmannin during infection. Thus, inhibition of the PI3 kinase-FRAP signaling pathway partly complements mutations in 2A protein and reverses a slow-virus phenotype.
The genome of picornaviruses, of which encephalomyocarditis virus (EMCV) is a member, is a single-stranded, positive-sense RNA of about 7,500 to 8,300 nucleotides (2). Picornavirus RNA is functionally monocistronic and, upon infection, is translated into a single polyprotein that is processed to yield structural and nonstructural virus proteins (49). EMCV polyprotein processing is performed solely by the 3C protease (3Cpro), except for the first cotranslational autoproteolytic cleavage at the 2A/2B junction (20, 38).
Infection with most picornaviruses is characterized by a strong inhibition of host cell protein synthesis at a time when virus-specific proteins are efficiently produced (reviewed in reference 9). Enteroviruses and rhinoviruses inhibit host translation, at least partially, by inactivation of eukaryotic translation initiation factor 4F (eIF4F), which binds to the cap structure of cellular mRNAs. eIF4F is composed of three polypeptides: eIF4E, eIF4A, and eIF4G (formerly p220). eIF4E is the cap-binding subunit (51). eIF4A possesses RNA-dependent ATPase activity and, in association with eIF4B, exhibits bidirectional RNA helicase activity (47, 48). eIF4G serves as a scaffold to bring together eIF4E, eIF4A, and eIF3 and bridges the mRNA and the ribosome (22). Picornavirus RNAs are naturally uncapped and translate by a cap- and eIF4E-independent mechanism, by which the ribosomes bind to an IRES (internal ribosome entry site) (1, 2, 24). Enteroviruses and rhinoviruses disrupt eIF4F by cleavage of the eIF4G subunit by 2Apro. This cleavage has been reported to be direct (18, 28) or indirect (60). eIF4G cleavage does not preclude but, rather, stimulates cap-independent initiation of viral protein synthesis, since the cap-binding subunit, eIF4E, remains associated with the N-terminal cleavage product (5, 28). The C-terminal cleavage fragment of eIF4G interacts with eIF4A and eIF3 to support IRES-dependent, but not cap-dependent, translation initiation (5, 37, 46). In contrast to enteroviruses and rhinoviruses, no cleavage of eIF4G occurs following infection of cells with cardioviruses, such as EMCV (36). Also, the 2A protein of EMCV is not similar to the enterovirus and rhinovirus 2Apro and does not possess protease consensus motifs or detectable proteolytic activity (31). It has long been assumed that the shutoff of host cell protein synthesis after EMCV infection results from the ability of viral RNA to efficiently compete with capped cellular mRNAs for some limiting component of the translational machinery (27, 53). Recently, it was suggested that EMCV causes the shutoff of host translation by dephosphorylation and activation of a suppressor of cap-dependent translation, 4E-BP1 (eIF4E-binding protein 1) (14). 4E-BP1 in its underphosphorylated form binds to eIF4E and inhibits its association with eIF4G (17, 32). 4E-BP1 does not inhibit cap-independent translation, such as that of picornaviruses, since this translation is independent of eIF4E (42). Another possible mechanism, which is not mutually exclusive, is the dephosphorylation of eIF4E (25).
Phosphorylation of 4E-BP1 is decreased by rapamycin and wortmannin, which inhibit the phosphatidylinositol 3-kinase (PI3 kinase)–FKBP-rapamycin-associated protein (FRAP) signal transduction pathway (3, 29, 57). PI3 kinase is activated by growth factors and hormones to deliver cell proliferation and survival signals. Upon activation, PI3 kinase phosphorylates the D3 position of PIs, which then act as second messengers to effect the different functions of PI3 kinase (reviewed in reference 12). Wortmannin inhibits PI3 kinase by binding irreversibly to its catalytic subunit (56). The immunosuppressive drug rapamycin is a potent inhibitor of FRAP (mTOR/RAFT), a member of the phosphatidylinositol kinase-related family, which is thought to be a downstream target of PI3 kinase (reviewed in reference 7).
Rapamycin augments the shutoff of host cell protein synthesis and the rate of synthesis of viral proteins after infection with poliovirus and EMCV (4), presumably because it inhibits cap-dependent translation, and thus confers an advantage to the viral mRNA. However, the observed effect of rapamycin is modest, probably because both EMCV and poliovirus replicate rapidly. To further explore this phenomenon, we wished to study the effect of rapamycin and wortmannin on the replication of a debilitated EMCV strain. We used EMCV mutants harboring deletions in the 2A coding region. These mutants were generated originally in an effort to determine whether 2A is required for virus replication. The deletions in 2A did not affect virus viability but greatly reduced the growth of the virus in BHK-21 cells. Translation of the mutant virus was inefficient, and the shutoff of host cell protein synthesis was significantly mitigated. Translation of viral mRNA was restored to its wild-type level and the shutoff of host cell protein synthesis was dramatically enhanced by rapamycin and wortmannin. Thus, inhibition of the PI3 kinase-FRAP pathway could be a useful tool in studying the replication of slow-growing and defective picornaviruses.
MATERIALS AND METHODS
Materials.
Rapamycin (100 μg/ml in ethanol [Calbiochem]) and wortmannin (1 mM in dimethyl sulfoxide [Sigma]) were kept in the dark at −20°C. Micrococcal nuclease-treated rabbit reticulocyte lysate was purchased from Promega. [35S]Methionine (>1,000 Ci/mmol) of translation or cell-labeling grade was from New England Nuclear. Recombinant mengovirus 3Cpro was obtained from D. J. Hall. The protease was expressed in Escherichia coli and purified as described previously (21).
Cells and viruses.
Baby hamster kidney cells (BHK-21) cells were grown in Dulbecco’s minimal essential medium (DMEM) (GIBCO) supplemented with 10% fetal bovine serum. Krebs-2 ascites tumor cells were grown in BALB/c mice for 7 to 8 days. EMCV with a short poly(C) tract that was derived from the infectious plasmid pE-C9 (19) was used as a wild-type virus. The 2A deletion mutants of EMCV, Δ2A and Δ2A*, were obtained following transfection of Krebs-2 cells with RNA transcribed from plasmids pE-C9-Δ2A and pE-C9-Δ2A*, respectively (see below). Virus titers were determined with eight replicates by using a 50% tissue culture infective dose (TCID50) assay on BHK-21 cells. We used this method rather than a plaque assay to determine the virus titer, because no discernible plaques were detected in BHK-21 cells after infection with the mutant viruses. Tenfold virus dilutions in 96-well microtiter plates were used (50). Cytopathic effects were evaluated 4 days after infection.
Plasmids.
Plasmid pE-C9 was described previously (19). Plasmids pE-C9-Δ2A and pE-C9-Δ2A* contain deletions of 174 (bases 3654 to 3827) and 360 (bases 3552 to 3911) nucleotides, respectively, in the 2A coding region. To generate these plasmids, pEss, a subclone containing the EMCV P2 coding region, was used. pEss was created by replacing the SpeI-SacII fragment of pBS-SK+ (Stratagene) with the corresponding fragment of EMCV. A 174-base deletion within pEss-Δ2A was engineered by amplifying by PCR a fragment from bases 3829 to 4386, digesting this fragment with EagI, and using it to replace the BbrPI-EagI fragment of pEss. A 360-base deletion within pEss-Δ2A* was engineered in a two-step PCR. In the first step, two separate reactions were performed, amplifying fragments N-terminal and C-terminal to the deletion. The negative-sense primer for the N-terminal fragment, N−, and the positive-sense primer for the C-terminal fragment, C+, are complementary to the template for 18 bases and have 18- or 19-base noncomplementary “tails.” The two are completely complementary to each other. In the second step, the products of the first step were purified and combined in a roughly equimolar ratio to serve as a template. These molecules can anneal through their short complementary sequence encoded by the N− and C+ primers, and extension with DNA polymerase creates a fusion product. The outer primers N+ and C− were also added for further amplification. The final product was digested with Bsu36I and XcmI and used to replace the corresponding segment of pEss. Full-length cDNA versions of the above deletion mutants were made by transferring the SpeI-SacII fragment of the relevant plasmids into pE-C9 (19). The full-length versions were named pE-C9-Δ2A and pE-C9-Δ2A*, accordingly.
In vitro transcription and transfection.
Plasmids were linearized with SalI and transcribed with T7 RNA polymerase (Promega) for 3 h at 37°C as recommended by the manufacturer. RNA integrity was examined by electrophoresis on formaldehyde-agarose gels. RNA was transfected into Krebs-2 cells by the method of Chumakov (8). Briefly, 108 washed cells were suspended in 1 ml of PSM buffer (0.15 M NaCl, 10 mM sodium phosphate [pH 7.3], 1 mM magnesium acetate). DEAE-dextran (Pharmacia) solution (100 mg/ml) was added to a final concentration of 2.5 mg/ml. After 3 min, 10 μg of RNA in 0.5 ml of STE buffer (0.1 M NaCl, 0.01 M Tris-HCl [pH 7.5], 0.1 mM EDTA) was added to 1 ml of cell suspension. RNA was left to adsorb under shaking for 60 min at room temperature. The cells were pelleted by centrifugation and suspended in 10 ml of Eagle’s medium (GIBCO) supplemented with 0.1% glucose, 0.15% sodium bicarbonate, and 50 U each of penicillin and streptomycin per ml. The cells were maintained in suspension (107 cells/ml) at 37°C for 24 h. After incubation, the cells were subjected to three cycles of freezing and thawing. The virus was passaged two more times in Krebs-2 cells, using a multiplicity of infection (MOI) of 0.1 TCID50 per cell, aliquoted, and stored at −70°C. To obtain Δ2A or Δ2A* EMCV for the purpose of viral RNA isolation, the infection was performed in the presence of 50 ng of rapamycin per ml and 1 μM wortmannin.
Metabolic labeling.
BHK-21 cells at 90 to 100% confluency in 35-mm petri dishes were infected with wild-type or 2A mutant EMCV at MOI of about 10 TCID50/cell in 0.5 ml of serum-free DMEM. After 30 min of adsorption at room temperature, the virus inoculum was removed by aspiration. The cells were washed once with methionine-free DMEM and incubated in 1 ml of methionine-free DMEM in the absence or presence of 50 ng of rapamycin per ml, 1 μg of wortmannin per ml, or a combination of the two drugs. The cells were labeled with [35S]methionine (50 μCi/ml) at 37°C for 30 min for different periods. They were lysed by being suspended in 0.5 ml of sample buffer and heated at 95°C for 8 min. Lysates from equal numbers of cells were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (15% polyacrylamide). After electrophoresis, the gels were processed for fluorography with En3Hance (Dupont).
Isolation of EMCV RNA.
EMCV RNA was extracted from virus purified by the method of Chumakov (8) with some modifications. The crude EMCV suspension (200 ml) was supplemented with 0.01 volume of 10% Nonidet P-40 and clarified by low-speed centrifugation. Then 0.1 volume of protamine sulfate (10 mg/ml) was added to the suspension, and the precipitate was discarded. The virus was further purified and concentrated by centrifugation at 26,000 rpm for 4 h through a 5-ml cushion containing 30% sucrose in 1 M NaCl–0.02 M Tris-HCl (pH 7.5) in an SW27 rotor. The virus was suspended in 5 ml of STMS buffer (0.1 M NaCl, 50 mM Tris-HCl [pH 7.5], 14 mM β-mercaptoethanol, 1% SDS). Polyethylene glycol 6000 as a 30% solution was added to the virus suspension to a final concentration of 5%. The precipitated virus was pelleted at 10,000 × g for 10 min. RNA from the purified virus preparation was extracted with phenol-chloroform-isoamyl alcohol (25:24:1) and purified by sucrose density gradient centrifugation (52, 55).
Assays.
EMCV RNA was translated in a rabbit reticulocyte lysate (12.5 μl) (Promega) in the presence of [35S]methionine at 30°C as recommended by the manufacturer. Translation products were analyzed on SDS–15% polyacrylamide gels as described above. Western blot analysis of 4E-BP1 was performed as described previously (14).
RESULTS
Construction and characterization of EMCV 2A deletion mutants.
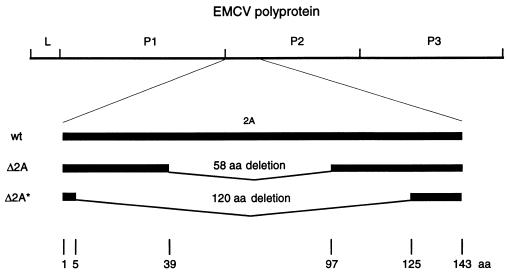
Two deletions were introduced into the 2A coding region of the infectious EMCV cDNA. One deletion, Δ2A (deletion of 58 of the 143 amino acids), eliminated an internal one-third of the 2A coding region, while another, Δ2A* (deletion of 120 amino acids), removed most of the 2A sequence (Fig. 1). Transfection of Krebs-2 cells with wild-type in vitro-transcribed RNA resulted in efficient virus production (titer of approximately 3 × 109 TCID50/ml) and complete cell lysis within 24 h. Transfection of cells with mutant RNAs generated infectious viruses but with a low yield (∼107 TCID50/ml), and most of the cells were alive after 24 h. Recovered mutant viruses also exhibited a small-plaque phenotype when assayed on HeLa cell monolayers (data not shown). In BHK-21 cells, the mutants also replicated more slowly and to lower titers than did wild-type EMCV (see below).
FIG. 1.
Schematic representation of EMCV wild type (wt) and 2A deletion mutants (Δ2A and Δ2A*). aa, amino acids.
Analysis of viral protein synthesis in vivo.
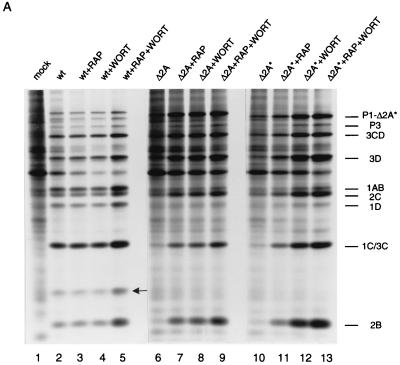
To determine which step of EMCV replication was affected by the mutations in 2A, protein synthesis in infected BHK-21 cells was examined. Cells were pulse-labeled with [35S]methionine 4.5 h postinfection, and extracts were analyzed by SDS-PAGE. Host cell protein synthesis was strongly inhibited, while the synthesis of viral proteins was efficient in wild-type-EMCV-infected cells (Fig. 2A, compare lane 2 to lane 1). However, infection with Δ2A or Δ2A* viruses had little effect on host cell protein synthesis (compare lanes 6 and 10 to lane 1). All proteins of mutant viruses were synthesized in much smaller amounts than those of wild-type virus (compare lanes 6 and 10 to lane 2). As anticipated, no 2A protein could be detected in either Δ2A or Δ2A* EMCV-infected cells.
FIG. 2.
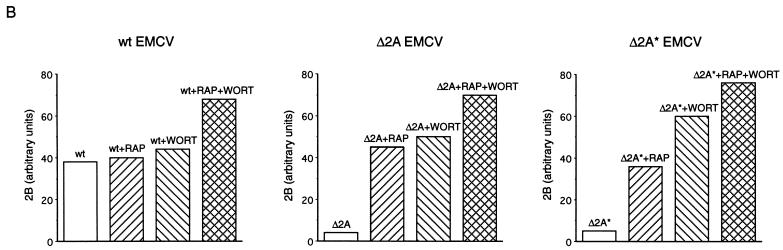
Rapamycin and wortmannin enhance EMCV protein synthesis. (A) Confluent BHK-21 cells were mock infected (lane 1) or infected with wild-type (wt) (lanes 2 to 5), Δ2A (lanes 6 to 9), or Δ2A* (lanes 10 to 13) EMCV at a MOI of 10 TCID50 per cell. The cells were incubated in methionine-free medium for 4.5 h and pulse-labeled for 30 min with [35S]methionine. Rapamycin (RAP) (50 ng/ml) and wortmannin (WORT) (1 μM), either alone or in combination, were present from the beginning of infection, where indicated. After labeling, cells were lysed in SDS-PAGE sample buffer and polypeptides were analyzed by electrophoresis. The positions of the major EMCV proteins are shown. The arrow indicates the position of 2A in wild-type-EMCV-infected cells. P1-Δ2A* and P1-Δ2A migrate slightly faster than P1-2A. (B) Quantification of 2B protein synthesis in wild-type-, Δ2A-, and Δ2A*-infected cells. The relative amount of radioactivity in the 2B bands (from panel A) was determined with BAS-2000 phosphorimager (Fuji Corp.).
We recently demonstrated that EMCV infection of Krebs-2 cells led to inhibition of phosphorylation of 4E-BP1 and that the decreased phosphorylation of 4E-BP1 coincided with the shutoff of host cell protein synthesis (14). Phosphorylation of 4E-BP1 is also inhibited by rapamycin and wortmannin, which block the PI3 kinase-FRAP signal transduction pathway (3, 29, 57). In EMCV-infected NIH 3T3 cells, the expression of viral proteins was augmented by rapamycin (4). It was therefore possible that rapamycin and wortmannin were able to rescue the replication of viruses defective in the shutoff of host protein synthesis. To test this, BHK-21 cells were infected with EMCV in the presence of rapamycin or wortmannin or both. With wild-type EMCV, the effects of rapamycin and wortmannin on viral protein synthesis were minimal, probably because the shutoff of host translation and the induction of viral protein synthesis were already evident at the time of the labeling—4.5 h postinfection (Fig. 2A, compare lanes 3 through 5 to lane 2). In contrast, for Δ2A and Δ2A* EMCV infections, both rapamycin and wortmannin strikingly stimulated the production of viral polypeptides (Δ2A, compare lanes 7 and 8 to lane 6; Δ2A*, compare lanes 11 and 12 to lane 10). The effect of the combination of the drugs was additive (lanes 9 and 13). As judged by the accumulation of the 2B polypeptide, which is generated by the first cleavage of the polyprotein (20, 40), the combination of rapamycin and wortmannin stimulated protein synthesis of Δ2A and Δ2A* EMCV by 21- and 16-fold, respectively, while the stimulation with wild-type virus was less than 2-fold (Fig. 2B).
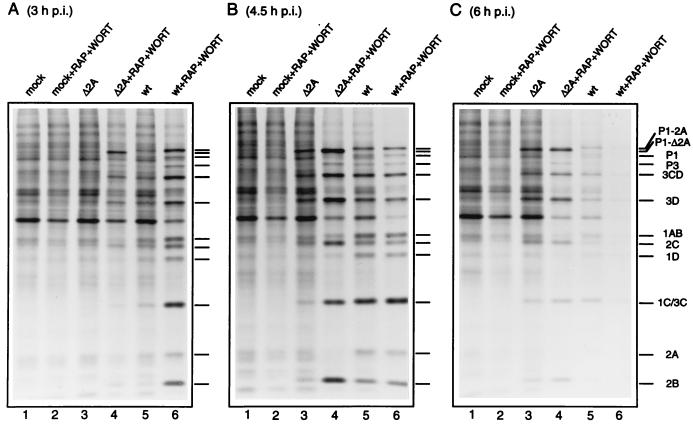
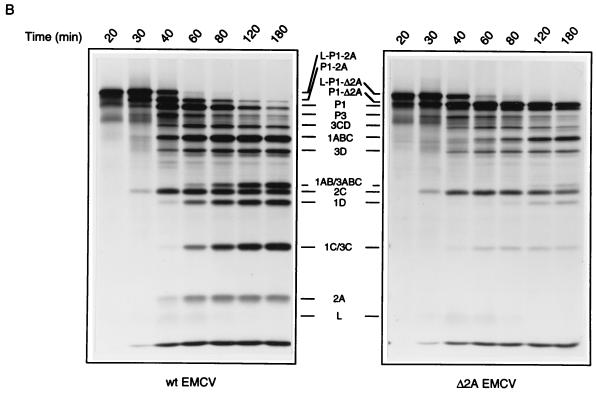
A time course of virus protein synthesis shows that Δ2A EMCV protein synthesis was enhanced by rapamycin and wortmannin to attain wild-type levels at all times of infection (Fig. 3). Also, the drugs partially restored the ability of the mutant virus to induce the shutoff of host translation. In contrast, in wild-type-virus-infected cells, viral protein synthesis was enhanced by rapamycin and wortmannin only early (3 h) after infection, when the shutoff of host protein synthesis and induction of viral protein synthesis were not fully manifested (Fig. 3A).
FIG. 3.
Effect of rapamycin and wortmannin on the time course of protein synthesis in cells infected with wild-type and Δ2A EMCV. Conditions for infections were as described in the legend to Fig. 2. Cells were mock infected or infected with Δ2A or wild-type EMCV for the indicated times. Rapamycin (50 ng/ml) and wortmannin (1 μM) were present where indicated. The positions of virus-specific polypeptides are indicated by bars. wt, wild type; p.i., postinfection.
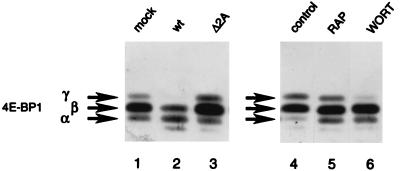
Next, we wished to determine whether dephosphorylation of 4E-BP1 correlates with the shutoff of host protein synthesis after infection with mutant EMCV; to do this, we used Western analysis. Three bands of 4E-BP1 were detected in mock-infected cells (Fig. 4, lane 1). Considering earlier reports, the bottom band (α) is hypophosphorylated compared to the top and middle bands (β and γ) (3, 14, 29). Mock-infected cells contained the hyperphosphorylated form of 4E-BP1 (form γ, lane 1), but infection with wild-type EMCV resulted in its disappearance due to its conversion to the less phosphorylated forms (forms α and β; compare lane 2 to lane 1). In contrast, Δ2A EMCV infection failed to change the ratio of the 4E-BP1 isoforms (compare lane 3 to lane 1). Similarly, the relative amounts of 4E-BP1 forms were not appreciably changed following Δ2A* EMCV infection (data not shown). These results provide additional evidence that the phosphorylation status of 4E-BP1 positively correlates with the level of host cell protein synthesis after EMCV infection. In a control experiment, rapamycin and wortmannin at the concentrations used in virus infections decreased 4E-BP1 phosphorylation as early as 30 min after exposure of cells to the drugs (compare lanes 5 and 6 to lane 4). The dephosphorylation was, however, not complete in either case.
FIG. 4.
Phosphorylation of 4E-BP1. BHK-21 cells were mock infected or infected for 4.5 h with wild-type or Δ2A EMCV. Where indicated, mock-infected cells were incubated for 30 min in the absence or presence of rapamycin (RAP, 50 ng/ml) or wortmannin (WORT, 1 μM). Heat-treated cell extracts were subjected to Western blotting and probed with a polyclonal anti-4E-BP1 antibody. The positions of 4E-BP1 isoforms α, β, and γ are indicated. wt, wild type.
Effects of rapamycin and wortmannin on virus growth.
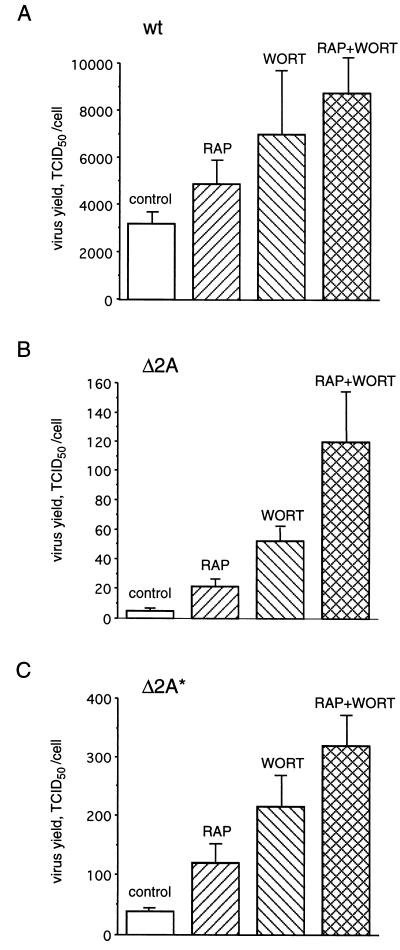
Considering the above results, it was pertinent to determine whether rapamycin and wortmannin could also rescue the growth of EMCV mutants. The effects of rapamycin and wortmannin on virus yield were determined by using a TCID50 assay. Rapamycin and wortmannin each had a small effect (∼1.5- to 2-fold) on the yield of wild-type virus (Fig. 5A). When used together, the drugs had an additive effect and increased the virus yield by threefold. The reason for this is not immediately clear, since rapamycin is known to inhibit only a subset of the phosphorylation events inhibited by wortmannin. We do not have an immediate explanation for the additive effect of the drugs, since both are supposed to act through the same pathway. Δ2A and Δ2A* EMCV yields were lower by 103- and 102-fold, respectively, than that of wild-type EMCV. Rapamycin and wortmannin increased mutant virus yield 3- to 10-fold, being especially effective with Δ2A EMCV. When the drugs were used in combination, their stimulatory effect was additive, yielding 21- and 9-fold more virus with Δ2A and Δ2A* EMCV, respectively. Thus, rapamycin and wortmannin preferentially stimulate the replication of mutant viruses over wild-type EMCV. However, the mutant virus production could not be fully restored by the drugs, and the titers of Δ2A and Δ2A* EMCV were considerably lower than those of the wild-type virus under all conditions examined. It is noteworthy that to induce a cytopathic effect, some slow mutant cardioviruses require more infecting viral particles than wild-type viruses do (61). Thus, the titers for 2A EMCV mutants shown in Fig. 5 could be underestimates.
FIG. 5.
Rapamycin and wortmannin increase EMCV yield. BHK-21 cells were infected with wild-type (wt) (A), Δ2A (B), or Δ2A* (C) EMCV in the absence (control) or presence of rapamycin and/or wortmannin, as indicated. Conditions for virus infections and drug treatments were essentially as described in the legend to Fig. 2. Virus yields (TCID50/cell) were determined 5 h postinfection as described in Materials and Methods. The mean values of three independent titer determinations and the error bars indicating the standard deviation from the mean are shown.
Δ2A and Δ2A* EMCV RNAs are deficient in reinitiating translation in vitro.
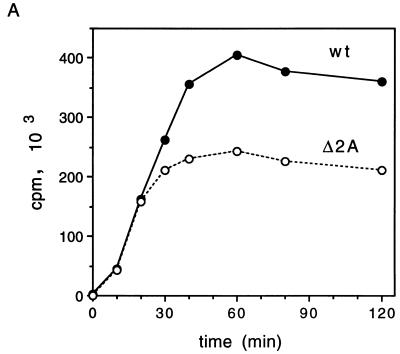
To address the question why EMCV protein synthesis is compromised by mutations in 2A, the translation of wild-type and mutant EMCV RNAs in a rabbit reticulocyte lysate were compared. EMCV RNA is translated efficiently in a rabbit reticulocyte lysate, and 3Cpro/3ABC-mediated polyprotein processing takes place in this system to generate an almost complete set of mature viral proteins (23, 39, 44). No difference in translation between wild-type and Δ2A EMCV RNA was observed after a short (up to 20-min) incubation (Fig. 6A). However, during a longer incubation, incorporation of [35S]methionine into protein, directed by wild-type EMCV RNA, was higher (up to 1.6-fold) than that directed by Δ2A EMCV RNA (Fig. 6A). At late times (e.g., 2 h), almost all virus-specific polypeptides accumulated in higher amounts in wild-type than in Δ2A EMCV RNA-programmed translations (Fig. 6B). A noticeable exception was P1-2A, which was less abundant than the P1-Δ2A counterpart. This probably reflects a more efficient cleavage of P1-2A than of P1-Δ2A (see below). A similar pattern was observed with Δ2A* EMCV RNA (Fig. 6C). Thus, both deletions within the 2A coding region impair viral translation in vitro.
FIG. 6.
Kinetics of protein synthesis in rabbit reticulocyte lysates programmed with wild-type, Δ2A or Δ2A* EMCV RNAs. (A and B) Rabbit reticulocyte lysate (0.1 ml) was incubated with 2 μg of wild-type (wt) or Δ2A EMCV RNA as specified in Materials and Methods. At the indicated times, aliquots were withdrawn from the reaction mixtures. One part of the aliquot (1 μl) was assayed for trichloroacetic acid-insoluble radioactivity, while another (5 μl) was processed for SDS-PAGE analysis. (A) Incorporation of [35S]methionine into trichloroacetic acid-insoluble material directed by wild-type or Δ2A EMCV RNAs. (B) Time course of accumulation of virus-specific polypeptides. Products of translation of wild-type and Δ2A EMCV RNAs are shown. (C) Rabbit reticulocyte lysate was programmed with wild-type (wt) or Δ2A* EMCV RNAs. Aliquots were withdrawn at the indicated times and subjected to SDS-PAGE as described for panels A and B. Products of the translation of wild-type EMCV RNA are shown.
Polyprotein processing is affected by deletions in 2A.
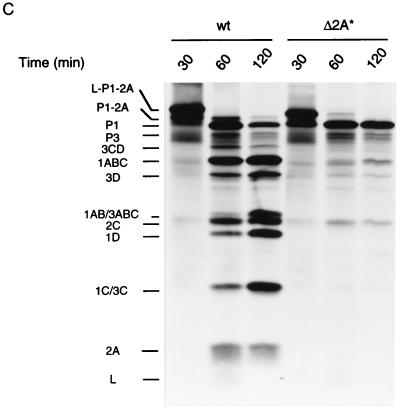
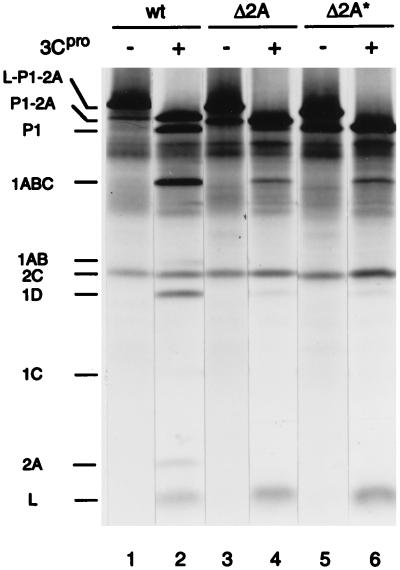
In addition to viral protein synthesis, processing of the capsid precursor polypeptide, P1-2A, is affected by the deletions in 2A. P1-Δ2A and P1-Δ2A* accumulated in higher amounts than did wild-type P1-2A (Fig. 2A, compare lanes 9 and 13 to lane 5; Fig. 3, compare lane 4 to lane 6). Concomitantly, mature virus proteins (1AB, 1D, and 1C) were generated more slowly for mutant EMCV than for wild-type EMCV. In the in vitro translation system, P1-Δ2A and P1-Δ2A* were also more stable than P1-2A (Fig. 6B and C). It is possible that P1-Δ2A and P1-Δ2A* are less susceptible to proteolysis than P1-2A. Alternatively, the failure of mutant capsid precursors to be efficiently processed could merely reflect low levels of 3Cpro. To distinguish between these two possibilities, wild-type, Δ2A and Δ2A* EMCV RNAs were translated in a rabbit reticulocyte lysate for a short period (25 min) in the absence or presence of purified 3Cpro from mengovirus. In the absence of 3Cpro, the polypeptides L-P1-2A and 2C were synthesized, but not 3Cpro or 3ABC, which are responsible for polyprotein processing (23, 39, 41, 54), or any other P3-derived polypeptides (Fig. 7). In the presence of exogenous 3Cpro (conditions which mimic those existing in virus-infected cells), the largest polypeptide synthesized was P1-2A (lane 2), which suggests that the leader peptide was rapidly cleaved off, apparently from the nascent polypeptide chain. Mutations in 2A affected neither this cotranslational cleavage of the leader peptide (compare lanes 2, 4, and 6) nor its cleavage from the presynthesized L-P1-2A precursor polypeptide (data not shown). However, subsequent processing of P1-2A, i.e., the cleavage at the P1/2A junction, was markedly diminished by the mutations. While the wild-type P1-2A was cleaved to a significant extent by 3Cpro into P1, 1ABC, 1AB, 1D, 1C, and 2A (lane 2), only a minute fraction of P1-Δ2A or P1-Δ2A* was converted to characteristic cleavage products within the same period (lanes 4 and 6). Thus, P1-Δ2A and P1-Δ2A* are less susceptible to cleavage by cardiovirus 3Cpro than is wild-type P1-2A.
FIG. 7.
Cleavage of wild-type, Δ2A, and Δ2A* EMCV capsid precursor polypeptides. Wild-type (wt), Δ2A, and Δ2A* EMCV RNAs were translated at 20 μg/ml in a rabbit reticulocyte lysate for 25 min in the absence or presence of purified mengovirus 3Cpro (20 μg/ml) as indicated. The positions of wild-type-EMCV-specific proteins are shown.
DISCUSSION
eIF4G serves as an adapter between mRNA and ribosomes and functions in both cap-dependent and cap-independent translations (22, 43, 45). Recently, a new homolog of eIF4G (termed eIF4GII) was identified which functions in a manner similar to the original isoform (termed eIF4GI) (16). eIF4G recruitment to capped mRNA is facilitated by its interaction with the cap-binding protein eIF4E. This interaction is regulated by a group of suppressor proteins, the 4E-BPs (30, 42). When bound to mRNA, eIF4G facilitates the binding of the 43S preinitiation complex to the mRNA, most probably through protein-protein interactions with the ribosome-associated eIF3 (26, 37). eIF4G also interacts with eIF4A, which, in conjunction with eIF4B, is thought to unwind the 5′ mRNA secondary structure.
To switch from translation of cellular mRNAs to efficient production of viral proteins, picornaviruses have evolved strategies to usurp eIF4G. Enteroviruses, rhinoviruses, and aphthoviruses encode proteases that cleave eIF4G to generate two fragments. The C-terminal fragment retains the capacity to interact with IRES elements, as well as with eIF3 and eIF4A, and is sufficient to promote cap-independent translation (5, 6, 37, 46). However, it lacks the eIF4E-binding site and is unable to support cap-dependent translation. Cardioviruses, as exemplified by EMCV, appear to affect eIF4G function in a different manner, namely, by inducing dephosphorylation and activation of 4E-BP1 (14). The dephosphorylated form of 4E-BP1 sequesters eIF4E into an inactive eIF4E–4E-BP1 complex (17) and thus inhibits the eIF4E-eIF4G interaction.
While protein synthesis of Δ2A EMCV in vivo is very inefficient relative to that of wild-type virus (Fig. 2), the translation of Δ2A virus mRNA in vitro is only ∼40% less efficient than that of the wild-type virus (Fig. 6A). This raises the possibility that the major effect of 2A deletion is not on translation but on some other steps of virus replication, such as viral RNA synthesis. An alternative interpretation, which we favor, is that the effect of 2A deletion is to attenuate virus mRNA translation in the cell, because rapamycin and wortmannin, which rescue viral protein synthesis, are known to affect the activity of translation initiation factors (3, 51, 57). The effect of 2A deletion could be either a cis effect, which would incapacitate the template, or a trans effect, whereby 2A protein would be required for virus mRNA translation. Since translation in vitro is not dramatically affected by the deletion of 2A, it is unlikely that the effect is in cis. It is thus conceivable that 2A is required for efficient translation of EMCV RNA in vivo to counteract the competition from cellular mRNAs. In contrast, in a nuclease-treated rabbit reticulocyte lysate, there is no competition from cellular mRNAs, and therefore translation of Δ2A EMCV RNA is less compromised. Addition of rapamycin and wortmannin, which inhibit capped-mRNA translation, would mitigate the competition and thus rescue Δ2A EMCV RNA translation.
The state of 4E-BP1 phosphorylation correlates with the efficiency of viral protein synthesis after EMCV infection (Fig. 4). The deletions in 2A, which abolished virus-induced 4E-BP1 dephosphorylation and inhibition of host cell protein synthesis, were also detrimental for the synthesis of viral proteins and resulted in a low virus yield (Fig. 2, 3, and 5). Rapamycin and wortmannin, which induce 4E-BP1 dephosphorylation, enhanced the replication of a defective virus. Thus, infection by a defective cardiovirus was significantly augmented by drugs that decrease 4E-BP1 phosphorylation. It should be noted, however, that other translation targets of the PI3 kinase pathway do exist (e.g., p70 S6 kinase and eIF2B) and could play a significant role in the phenomenon seen here (12, 59). In addition, the concentration of wortmannin used here (1 μM) is known to affect the mitogen-activated protein kinase pathway (10), which affects eIF4E phosphorylation (11, 13, 58). Inhibition of eIF4E phosphorylation leads to a decrease in cellular mRNA translation.
The reason why Δ2A EMCV is deficient in inducing 4E-BP1 dephosphorylation is not known. Perhaps 2A, either directly or indirectly, inhibits signaling through the PI3 kinase-FRAP pathway or somehow activates phosphatases targeting 4E-BP1. Alternatively, the lack of 4E-BP dephosphorylation following Δ2A EMCV infection could be a secondary effect resulting from inefficient virus replication and limited production of a protein other than 2A.
What is the nature of the impediments to efficient EMCV replication imposed by the deletions in 2A? Clearly, 2A disruption inhibits processing of the P1-2A precursor polypeptide. The 2A deletion mutants exhibited higher accumulation of the uncleaved P1-2A and less efficient formation of the mature capsid proteins than the wild-type EMCV (Fig. 2A and 3). Although the primary cotranslational cleavage at the 2A/2B junction (which results from an inherent instability of the corresponding peptide chain [20]) remained unaffected, the P1/2A cleavage by 3Cpro was significantly slowed both in vitro and in vivo (Fig. 2A, 3, 6B and C, and 7). It is not immediately clear why an intact amino acid sequence of 2A is important for efficient cleavage. The slow processing of the P1-2A junction in 2A mutants would leave some of the 2A fragments associated with VP1 and might hinder the proper assembly of capsids. This processing defect could account for the failure of rapamycin and wortmannin to fully restore mutant virus production, despite the potent activity of these drugs in rescuing virus-specific translation.
2A might also function as a positive regulatory factor in virus-specific translation and/or RNA replication. With respect to translation control, a minimal set of factors required for 48S initiation complex formation with EMCV RNA has recently been defined by using a reconstituted ribosome-binding system (45). No viral proteins are absolutely required for the activity of the EMCV IRES, since it functions efficiently in vivo with heterologous reporter sequences (6, 35). EMCV RNA is also translated early in infection and before any appreciable accumulation of viral products. However, although 2A is not critical for the IRES activity, it might facilitate its function. For example, 2A could bind to the IRES and stabilize an active conformation. Consistent with this proposal is the finding that EMCV 2A is basic and binds RNA (15). Moreover, a fraction of 2A is associated with ribosomes in virus-infected cells (33). However, evidence that 2A functions as a virus-specific translation factor is clearly lacking for EMCV. In addition, IRES activity and inhibition of host cell protein synthesis could be regulated by other viral proteins, for example, by the leader peptide (L), as was suggested for mengovirus (61). However, since the coding region of L is positioned very close to the IRES, it remains to be demonstrated that the contribution of the L sequences to efficient viral replication resides within the protein rather than within the RNA. Surprisingly, the 2A protein of Theiler’s murine encephalomyelitis virus, another cardiovirus, has been reported to be dispensable for RNA replication or virus production in BHK-21 cells (34). However, the 2A protein of Theiler’s murine encephalomyelitis virus diverges remarkably from that of EMCV and therefore could be involved in some other processes of the virus life cycle.
Finally, our results suggest that rapamycin and other immunosuppressive drugs with potential clinical applicability should be evaluated with respect to their ability to target 4E-BP1 and activate latent infections caused by viruses that use an internal ribosome entry mechanism.
ACKNOWLEDGMENTS
We thank B. Raught for critical reading of the manuscript.
This work was supported by grants from the National Institute of Canada to N.S. and by National Institutes of Health grant AI-17331 to A.C.P. and training grant GM-07215 to H.H. N.S. is a Medical Research Council Distinguished Scientist and a Howard Hughes International Scholar. A.-C.G. is a recipient of an NSERC 67 studentship.
REFERENCES
- 1.Agol V I. The 5′ untranslated region of picornaviral genomes. Adv Virus Res. 1991;40:103–180. doi: 10.1016/S0065-3527(08)60278-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belsham G J, Sonenberg N. RNA-protein interactions in regulation of picornavirus RNA translational. Microbiol Rev. 1996;60:499–511. doi: 10.1128/mr.60.3.499-511.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beretta L, Gingras A C, Svitkin Y V, Hall M N, Sonenberg N. Rapamycin blocks the phosphorylation of 4E-BP1 and inhibits cap-dependent initiation of translation. EMBO J. 1996;15:658–664. [PMC free article] [PubMed] [Google Scholar]
- 4.Beretta L, Svitkin Y V, Sonenberg N. Rapamycin stimulates viral protein synthesis and augments the shutoff of host protein synthesis upon picornavirus infection. J Virol. 1996;70:8993–8996. doi: 10.1128/jvi.70.12.8993-8996.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borman A M, Kirchweger R, Ziegler E, Rhoads R E, Skern T, Kean K M. elF4G and its proteolytic cleavage products: effect on initiation of protein synthesis from capped, uncapped, and IRES-containing mRNAs. RNA. 1997;3:186–196. [PMC free article] [PubMed] [Google Scholar]
- 6.Borman A M, Le Mercier P, Girard M, Kean K M. Comparison of picornaviral IRES-driven internal initiation of translation in cultured cells of different origins. Nucleic Acids Res. 1997;25:925–932. doi: 10.1093/nar/25.5.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown E J, Schreiber S L. A signaling pathway to translational control. Cell. 1996;86:517–520. doi: 10.1016/s0092-8674(00)80125-7. [DOI] [PubMed] [Google Scholar]
- 8.Chumakov K M. A highly efficient method of infection with encephalomyocarditis virus RNA. Acta Virol. 1980;24:225–231. [PubMed] [Google Scholar]
- 9.Ehrenfeld E. Initiation of translation by picornavirus RNAs. In: Hershey J W B, Mathews M B, Sonenberg N, editors. Translational control. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 549–573. [Google Scholar]
- 10.Ferby I M, Waga I, Hoshino M, Kume K, Shimizu T. Wortmannin inhibits mitogen-activated protein kinase activation by platelet-activating factor through a mechanism independent of p85/p110-type phosphatidylinositol 3-kinase. J Biol Chem. 1996;271:11684–11688. doi: 10.1074/jbc.271.20.11684. [DOI] [PubMed] [Google Scholar]
- 11.Flynn A, Proud G. Insulin-stimulated phosphorylation of initiation factor 4E is mediated by the MAP kinase pathway. FEBS Lett. 1996;389:162–166. doi: 10.1016/0014-5793(96)00564-9. [DOI] [PubMed] [Google Scholar]
- 12.Franke T F, Kaplan D R, Cantley L C. PI3K: downstream AKTion blocks apoptosis. Cell. 1997;88:435–437. doi: 10.1016/s0092-8674(00)81883-8. [DOI] [PubMed] [Google Scholar]
- 13.Fukunaga R, Hunter T. MNK1, a new MAP-kinase-activated protein kinase, isolated by a novel expression screening method for identifying protein kinase substrates. EMBO J. 1997;16:1921–1933. doi: 10.1093/emboj/16.8.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gingras A C, Svitkin Y, Belsham G J, Pause A, Sonenberg N. Activation of the translational suppressor 4E-BP1 following infection with encephalomyocarditis virus and poliovirus. Proc Natl Acad Sci USA. 1996;93:5578–5583. doi: 10.1073/pnas.93.11.5578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gorbalenya A E, Chumakov K M, Agol V I. RNA-binding properties of nonstructural polypeptide G of encephalomyocarditis virus. Virology. 1978;88:183–185. doi: 10.1016/0042-6822(78)90122-8. [DOI] [PubMed] [Google Scholar]
- 16.Gradi A, Imataka H, Svitkin Y V, Rom E, Raught B, Morino S, Sonenberg N. A novel functional human eukaryotic translation initiation factor 4G. Mol Cell Biol. 1998;18:334–342. doi: 10.1128/mcb.18.1.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haghighat A, Mader S, Pause A, Sonenberg N. Repression of cap-dependent translation by 4E-binding protein 1: competition with p220 for binding to eukaryotic initiation factor-4E. EMBO J. 1995;14:5701–5709. doi: 10.1002/j.1460-2075.1995.tb00257.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haghighat A, Svitkin Y, Novoa I, Kuechler E, Skern T, Sonenberg N. The eIF4G-eIF4E complex is the target for direct cleavage by the rhinovirus 2A proteinase. J Virol. 1996;70:8444–8450. doi: 10.1128/jvi.70.12.8444-8450.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hahn H, Palmenberg A C. Encephalomyocarditis viruses with short poly(C) tracts are more virulent than their mengovirus counterparts. J Virol. 1995;69:2697–2699. doi: 10.1128/jvi.69.4.2697-2699.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hahn H, Palmenberg A C. Mutational analysis of the encephalomyocarditis virus primary cleavage. J Virol. 1996;70:6870–6875. doi: 10.1128/jvi.70.10.6870-6875.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hall D J, Palmenberg A C. Mengo virus 3C proteinase: recombinant expression, intergenus substrate cleavage and localization in vivo. Virus Genes. 1996;13:99–110. doi: 10.1007/BF00568903. [DOI] [PubMed] [Google Scholar]
- 22.Hentze M W. eIF4G: a multipurpose ribosome adapter? Science. 1997;275:500–501. doi: 10.1126/science.275.5299.500. [DOI] [PubMed] [Google Scholar]
- 23.Jackson R J. A detailed kinetic analysis of the in vitro synthesis and processing of encephalomyocarditis virus products. Virology. 1986;149:114–127. doi: 10.1016/0042-6822(86)90092-9. [DOI] [PubMed] [Google Scholar]
- 24.Jackson R J, Kaminski A. Internal initiation of translation in eukaryotes: the picornavirus paradigm and beyond. RNA. 1995;1:985–1000. [PMC free article] [PubMed] [Google Scholar]
- 25.Kleijn M, Vrins C L, Voorma H O, Thomas A A. Phosphorylation state of the cap-binding protein eIF4E during viral infection. Virology. 1996;217:486–494. doi: 10.1006/viro.1996.0143. [DOI] [PubMed] [Google Scholar]
- 26.Lamphear B J, Kirchweger R, Skern T, Rhoads R E. Mapping of functional domains in eukaryotic protein synthesis initiation factor 4G (eIF4G) with picornaviral proteases. Implications for cap-dependent and cap-independent translational initiation. J Biol Chem. 1995;270:21975–21983. doi: 10.1074/jbc.270.37.21975. [DOI] [PubMed] [Google Scholar]
- 27.Lawrence C, Thach R E. Encephalomyocarditis virus infection of mouse plasmacytoma cells. I. Inhibition of cellular protein synthesis. J Virol. 1974;14:598–610. doi: 10.1128/jvi.14.3.598-610.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liebig H-D, Ziegler E, Yan R, Hartmuth K, Klump H, Kowalski H, Blaas D, Sommergruber W, Frasel L, Lamphear B, Rhoads R, Kuechler E, Skern T. Purification of two picornaviral 2A proteinases: interaction with eIF-4γ and influence on in vitro translation. Biochemistry. 1993;32:7581–7588. doi: 10.1021/bi00080a033. [DOI] [PubMed] [Google Scholar]
- 29.Lin T A, Kong X, Saltiel A R, Blackshear P J, Lawrence J C. Control of PHAS-I by insulin in 3T3-L1 adipocytes. Synthesis, degradation, and phosphorylation by a rapamycin-sensitive and mitogen-activated protein kinase-independent pathway. J Biol Chem. 1995;270:18531–18538. doi: 10.1074/jbc.270.31.18531. [DOI] [PubMed] [Google Scholar]
- 30.Lin T A, Kong X M, Haystead T, Pause A, Belsham G, Sonenberg N, Lawrence J C. PHAS-I as a link between mitogen-activated protein kinase and translation initiation. Science. 1994;266:653–656. doi: 10.1126/science.7939721. [DOI] [PubMed] [Google Scholar]
- 31.Lloyd R E, Grubman M J, Ehrenfeld E. Relationship of p220 cleavage during picornavirus infection to 2A proteinase sequencing. J Virol. 1988;62:4216–4223. doi: 10.1128/jvi.62.11.4216-4223.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mader S, Lee H, Pause A, Sonenberg N. The translation initiation factor eIF-4E binds to a common motif shared by the translation factor eIF-4γ and the translational repressors 4E-binding proteins. Mol Cell Biol. 1995;15:4990–4997. doi: 10.1128/mcb.15.9.4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Medvedkina O A, Scarlat I V, Kalinina N O, Agol V I. Virus-specific proteins associated with ribosomes of Krebs-II cells infected with encephalomyocarditis virus. FEBS Lett. 1974;39:4–8. doi: 10.1016/0014-5793(74)80003-7. [DOI] [PubMed] [Google Scholar]
- 34.Michiels T, Dejong V, Rodrigus R, Shaw-Jackson C. Protein 2A is not required for Theiler’s virus replication. J Virol. 1997;71:9549–9556. doi: 10.1128/jvi.71.12.9549-9556.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Molla A, Jang S K, Paul A V, Reuer Q, Wimmer E. Cardioviral internal ribosomal entry site is functional in a genetically engineered dicistronic poliovirus. Nature. 1992;356:255–257. doi: 10.1038/356255a0. [DOI] [PubMed] [Google Scholar]
- 36.Mosenkis J, Daniels-McQueen S, Janovec S, Duncan R, Hershey J W, Grifo J A, Merrick W C, Thach R E. Shutoff of host translation by encephalomyocarditis virus infection does not involve cleavage of the eucaryotic initiation factor 4F polypeptide that accompanies poliovirus infection. J Virol. 1985;54:643–645. doi: 10.1128/jvi.54.2.643-645.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ohlmann T, Rau M, Pain V M, Morley S J. The C-terminal domain of eukaryotic protein synthesis initiation factor (eIF) 4G is sufficient to support cap-independent translation in the absence of eIF4E. EMBO J. 1996;15:1371–1382. [PMC free article] [PubMed] [Google Scholar]
- 38.Palmenberg A C. Proteolytic processing of picornaviral polyprotein. Annu Rev Microbiol. 1990;44:603–623. doi: 10.1146/annurev.mi.44.100190.003131. [DOI] [PubMed] [Google Scholar]
- 39.Palmenberg A C, Pallansch M A, Rueckert R R. Protease required for processing picornaviral coat protein resides in the viral replicase gene. J Virol. 1979;32:770–778. doi: 10.1128/jvi.32.3.770-778.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Palmenberg A C, Parks G D, Hall D J, Ingraham R H, Seng T W, Pallai P V. Proteolytic processing of the cardioviral P2 region: primary 2A/2B cleavage in clone-derived precursors. Virology. 1992;190:754–762. doi: 10.1016/0042-6822(92)90913-a. [DOI] [PubMed] [Google Scholar]
- 41.Parks G D, Baker J C, Palmenberg A C. Proteolytic cleavage of encephalomyocarditis viral capsid region substrates by precursors to the 3C enzyme. J Virol. 1989;63:1054–1058. doi: 10.1128/jvi.63.3.1054-1058.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pause A, Belsham G J, Gingras A C, Donzé O, Lin T A, Lawrence J C, Sonenberg N. Insulin-dependent stimulation of protein synthesis by phosphorylation of a regulator of 5′-cap function. Nature. 1994;371:762–767. doi: 10.1038/371762a0. [DOI] [PubMed] [Google Scholar]
- 43.Pause A, Méthot N, Svitkin Y V, Merrick W C, Sonenberg N. Dominant negative mutants of mammalian translation initiation factor eIF-4A define a critical role for eIF-4F in cap-dependent and cap-dependent initiation of translation. EMBO J. 1994;13:1205–1215. doi: 10.1002/j.1460-2075.1994.tb06370.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pelham H R B. Translation of encephalomyocarditis virus RNA in vitro yields an active proteolytic processing enzyme. Eur J Biochem. 1978;85:457–462. doi: 10.1111/j.1432-1033.1978.tb12260.x. [DOI] [PubMed] [Google Scholar]
- 45.Pestova T V, Hellen C U T, Shatsky I N. Canonical eukaryotic initiation factors determine initiation of translation by internal ribosomal entry. Mol Cell Biol. 1996;16:6859–6869. doi: 10.1128/mcb.16.12.6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pestova T V, Shatsky I N, Hellen C U T. Functional dissection of eukaryotic initiation factor 4F: the 4A subunit and the central domain of the 4G subunit are sufficient to mediate internal entry of 43S preinitiation complexes. Mol Cell Biol. 1996;16:6870–6878. doi: 10.1128/mcb.16.12.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ray B K, Lawson T G, Kramer J C, Cladaras M H, Grifo J A, Abramson R D, Merrick W C, Thach R E. ATP-dependent unwinding of messenger RNA structure by eukaryotic initiation factors. J Biol Chem. 1985;260:7651–7658. [PubMed] [Google Scholar]
- 48.Rozen F, Edery I, Meerovitch K, Dever T E, Merrick W C, Sonenberg N. Bidirectional RNA helicase activity of eucaryotic translation initiation factors 4A and 4F. Mol Cell Biol. 1990;10:1134–1144. doi: 10.1128/mcb.10.3.1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rueckert R R. Picornaviridae: the viruses and their replication. In: Fields B N, editor. Fields virology. Vol. 1. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 609–654. [Google Scholar]
- 50.Schmidt N J. Cell culture techniques for diagnostic virology. In: Lennette E H, Schmidt N J, editors. Diagnostic procedures for viral, rickettsial and chlamydial infections. Washington, D.C: American Public Health Association; 1979. pp. 65–139. [Google Scholar]
- 51.Sonenberg N. mRNA 5′ cap-binding protein eIF4E and control of cell growth. In: Hershey J W B, Mathews M B, Sonenberg N, editors. Translational control. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 245–269. [Google Scholar]
- 52.Svitkin Y V, Agol V I. Complete translation of encephalomyocarditis virus RNA and faithful cleavage of virus-specific proteins in a cell-free system from Krebs-2 cells. FEBS Lett. 1978;87:7–11. doi: 10.1016/0014-5793(78)80121-5. [DOI] [PubMed] [Google Scholar]
- 53.Svitkin Y V, Ginevskaya V A, Ugarova T Y, Agol V I. A cell-free model of the encephalomyocarditis virus-induced inhibition of host cell protein synthesis. Virology. 1978;87:199–203. doi: 10.1016/0042-6822(78)90172-1. [DOI] [PubMed] [Google Scholar]
- 54.Svitkin Y V, Gorbalenya A E, Kazachkov Y A, Agol V I. Encephalomyocarditis virus-specific polypeptide p22 possessing a proteolytic activity: preliminary mapping on the viral genome. FEBS Lett. 1979;108:6–9. doi: 10.1016/0014-5793(79)81165-5. [DOI] [PubMed] [Google Scholar]
- 55.Svitkin Y V, Maslova S V, Agol V I. The genomes of attenuated and virulent poliovirus strains differ in their in vitro translation efficiencies. Virology. 1985;147:243–252. doi: 10.1016/0042-6822(85)90127-8. [DOI] [PubMed] [Google Scholar]
- 56.Ui M, Okada T, Hazeki K, Hazeki O. Wortmannin as a unique probe for an intracellular signalling protein, phosphoinositide 3-kinase. Trends Biochem Sci. 1995;20:303–307. doi: 10.1016/s0968-0004(00)89056-8. [DOI] [PubMed] [Google Scholar]
- 57.von Manteuffel S R, Gingras A-C, Ming X-F, Sonenberg N, Thomas G. 4E-BP1 phosphorylation is mediated by the FRAP-p70s6k pathway and is independent of mitogen-activated protein kinase. Proc Natl Acad Sci USA. 1996;93:4076–4080. doi: 10.1073/pnas.93.9.4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Waskiewicz A J, Flynn A, Proud C G, Cooper J A. Mitogen-activated protein kinases activate the serine/threonine kinases Mnk1 and Mnk2. EMBO J. 1997;16:1909–1920. doi: 10.1093/emboj/16.8.1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Welsh G I, Stokes C M, Wang X, Sakaue H, Ogawa W, Kasuga M, Proud C G. Activation of translation initiation factor eIF2B by insulin requires phosphatidyl inositol 3-kinase. FEBS Lett. 1997;410:418–422. doi: 10.1016/s0014-5793(97)00579-6. [DOI] [PubMed] [Google Scholar]
- 60.Wyckoff E E, Lloyd R E, Ehrenfeld E. Relationship of eukaryotic initiation factor 3 to poliovirus-induced p220 cleavage activity. J Virol. 1992;66:2943–2951. doi: 10.1128/jvi.66.5.2943-2951.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zoll J, Galama J M, van Kuppeveld F J, Melchers W J. Mengovirus leader is involved in the inhibition of host cell protein synthesis. J Virol. 1996;70:4948–4952. doi: 10.1128/jvi.70.8.4948-4952.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]