Abstract
A small number of cases of human immunodeficiency virus (HIV) infection have been reported in individuals with no identified risk factors for transmission. We report on the seroconversion of the 61-year-old mother and the subsequent finding of HIV seropositivity in the 66-year-old father of a 31-year-old AIDS patient. Extensive investigation failed to identify any risk factor for intrafamilial transmission. We conducted a genetic analysis and determined the amino acid signature patterns of the V3, V4, and V5 hypervariable domains and flanking regions in the HIV-1 gp120 env gene of 26 clones derived from proviral DNA in peripheral blood mononuclear cells of the members of the family. env sequences of the viruses isolated from the patients were compared with sequences of HIV-1 subtype B viruses from Europe and local field isolates. Phylogenetic analysis revealed that the sequences of the viruses isolated from the patients were genetically related and formed an intrafamilial cluster of HIV-1 distinct from other subtype B viruses. Interindividual nucleotide variability in the C2-V3 and V4-C4-V5 domains ranged between 1.2 and 5.0% and between 2.2 and 7.5%, respectively, whereas divergence between HIV strains from the patients and control viral strains ranged from 6.6 to 29.3%. The amino acid signature patterns of viral clones from the three patients were closely related. In the C2-V3 region, two minor clones derived from the son’s virus showed less nucleotide divergence (mean, 3.5 and 3.9%) than did the clones derived from the viruses of both parents or the seven other predominant clones derived from the virus from the son (mean, 5.4%). The top of the V3 loop of the last two clones and of all viral clones from the parents exhibited an unusual GPGG sequence. This is the first report of genotypic relatedness of HIV-1 in three adults of the same family in the absence of identified risk factor for transmission between the members of the family. Our findings suggest that atypical transmission of HIV may occur.
A small number of cases of human immunodeficiency virus (HIV) infection have been reported in adults and children with no identified risk factors for transmission or an unproven mode of transmission despite thorough investigation (5, 6, 14, 34, 45). Two reports of such cases have included detailed genetic analyses of variable env sequences of the viral envelope in five patients of a dentist infected with HIV-1 (34) and in a child possibly contaminated through unrecognized exposure to blood by an HIV-1-infected child living in the same home (14). Although epidemiologic evidence argues against casual transmission of HIV (4, 12, 17, 19, 42), these few cases raise the possibility that atypical transmission by means other than those usually evidenced does occur.
We report on a cluster of HIV-1 infection in three adults of the same family whom we found to be infected with genetically related viruses and in whom no risk factor for intrafamilial viral transmission was identified.
CASE REPORTS
Patient 1 was a 31-year-old man identified to be seropositive for HIV-1 in December 1994. The patient had used intravenous drugs between 1984 and 1989 and claimed to have been free of drugs since then. The patient lived in his parents’ home. He was admitted to our hospital on 4 December 1994 because of high fever, productive cough, hemoptysis, and respiratory distress. The chest X ray and thoracic computed tomogram showed extensive bilateral bullous lesions and a voluminous cavity in the apex of the right lung. Pneumocystis carinii pneumonia was diagnosed upon analysis of the bronchoalveolar lavage fluid. A diagnosis of tuberculosis was also suspected, although Ziehl’s staining of bronchoalveolar lavage fluid gave negative results. The patient was treated simultaneously with high-dose trimethoprim-sulfamethoxazole and a quadruple combination of antituberculous drugs. Cultures and PCR for Mycobacterium tuberculosis remained negative. At the time of admission, the CD4 T-cell count was 106/liter and the HIV RNA level in plasma was 209,700 equivalent (Eq) copies/ml as assessed retrospectively (Chiron, Emeryville, Calif.). Analysis of the pol 215 codon of proviral DNA from the patient’s peripheral blood mononuclear cells (PBMC) by selective PCR (26) revealed a mixed sensitive and resistant genotype to zidovudine with a predominance of sensitive variants. In the next 16 months, the patient developed a disseminated form of Kaposi’s sarcoma (KS). He died in April 1996.
Patient 2, the mother of patient 1, is a 61-year-old woman who had had no medical history until December 1994, except for mild hypertension and hypothyroidism. She was admitted to our hospital on 10 December 1994 with acute febrile meningitis. Cerebrospinal fluid analysis showed 30 lymphocytes/ mm3 and hyperalbuminorachia (1.34 g/liter). No bacteria, including M. tuberculosis and Treponema pallidum, and no viruses (herpes simplex virus, varicella-zoster virus, cytomegalovirus, Epstein-Barr virus, human herpesvirus 6) were found by PCR or isolated upon culture of cerebrospinal fluid and blood samples from this patient. However, because of her son’s recent history, the patient was given antituberculous drugs. Antimycobacterial therapy was discontinued after 18 weeks because PCR for M. tuberculosis and cultures for mycobacteria remained negative. A first HIV screening serologic test performed on 12 December was negative. However, p24 antigenemia was found to be positive (64 pg/ml). On 19 December, Western blot analysis revealed an incomplete HIV-1 pattern, with the presence of bands at 24, 41, and 55 kDa. One week later, the Western blot showed a pattern typical of HIV-1 infection. Therefore, we interpreted the patient’s meningitis as being associated with acute primary HIV infection. Analysis of the pol 215 codon in the patient’s PBMC revealed a similar genotype for zidovudine resistance to that observed in her son. Patient 2 recovered from her initial symptoms within 2 weeks. She received a combination of zidovudine and zalcitabine for 11 months; the therapy was then switched to a combination of zidovudine and didanosine. In April 1995, the HIV RNA level in plasma was below the threshold of detection (<10,000 Eq copies/ml) and the CD4 T-cell count was 600 × 106/liter.
Patient 3, the father of patient 1 and the husband of patient 2, is a 66-year-old baker with a medical history of hypertension for the last 20 years. In April 1995, patient 3 volunteered for an HIV test, which was found to be positive for HIV-1. The CD4 T-cell count was 220 × 106/liter. The patient had no history suggestive of an acute retroviral syndrome. His plasma viral load at the time of diagnosis of the HIV infection was 683,700 Eq copies/ml. Analysis of the pol 215 codon revealed a similar genotype for zidovudine resistance to that observed in patients 1 and 2. Patient 3 received a combination of zidovudine and zalcitabine for 13 months. He remains clinically asymptomatic with a stable CD4 T-cell count of 300 × 106/liter.
Epidemiological investigation.
Extensive discussions conducted by three independent physicians, not involved in the patients’ care, with both parents of patient 1 did not reveal any risk factor for HIV in patients 2 and 3, including sexual transmission, blood transfusion, usage of intravenous drugs, and identified contact with the blood of patient 1. Patient 1 lived at home, except for the periods when the disease required him to stay in hospital. While in hospital in the terminal phase of his illness, patient 1 received frequent visits from his parents. The patient died at hospital. The parents and their son had separate rooms and did not sleep together. They did not share toothbrushes or razors, but they did share eating utensils. The parents have not been involved at any time in the nursing care of their son. They claimed not to have been in contact with blood or other body fluids of their son, including stools, urine, vomitus, saliva, and nasal secretions. They did not use gloves at home. Patient 1 had no other skin lesions besides KS lesions. There was no bleeding associated with the KS lesions. There were no skin lesions on the hands of patients 2 and 3. Patient 1 had a poor dental condition and presented with oral lesions of KS. He did not present with nasal bleeding. He had had a severe cough and hemoptysis, while at home, prior to his first referral to hospital. Epidemiological investigation also did not evidence how transmission had occurred between the parents. Patient 2 works in her husband’s bakery, which is located on the first floor of their lodging. Patients 2 and 3 claimed not to have had sexual intercourse for more than 5 years. Patient 3 had consulted one of us in previous years for sexual dysfunction associated with α-methyldopa therapy.
MATERIALS AND METHODS
Blood samples were obtained on 2 January 1995 from patient 2, on 29 March 1995 from patient 1, and on 1 June 1995 from patient 3. Plasma and PBMC were frozen until use. We also analyzed PBMC obtained from all three patients 1 to 3 months after the initial sampling and plasma from two unrelated HIV-seropositive patients from the same ward, obtained at the same time as the 2 January 1995 sample from patient 2.
PBMC (2 × 106) were incubated overnight in lysis buffer containing proteinase K. DNA was extracted by the phenol-chloroform method. Amplification of proviral env DNA fragments in the hypervariable V3, V4, and V5 loops and their flanking interspersed constant (C) regions was performed by nested PCR with oligonucleotide primers derived from HIV-1 consensus env sequences, as described previously (10). The final amplified product of 667 bp was allowed to migrate on a low-melting-point agarose gel at 6.0%. Relevant bands were eluted, and DNA was extracted by the phenol-chloroform method. The amplified products were inserted into the pMOS blue T-vector plasmid (Amersham Life Science, Little Chalfont, United Kingdom). Clones carrying HIV-1 env sequences were identified by blue-white screening of recombinants and confirmed by direct-colony env PCR.
The nucleotide sequences were determined with the T7 sequencing kit from Amersham, using the dideoxynucleotide chain termination method. DNA sequences were aligned with CLUSTAL software (22) and corrected manually. Twenty-five env sequences of European isolates and 28 env sequences of local field isolates from Paris (8) were used as nucleotide and amino acid reference sequences for the HIV-1 subtype B C2-V3 region. The “local control” sequences that were used originated from patients in three hospitals in different areas of Paris from that where the family lives. The distance between those hospitals and the area where the family lives is less than 5 miles. We also used C2-V3 sequences from the previous studies of atypical HIV transmission reported by Ou et al. (GenBank accession numbers are as follows: M90847 for the dentist [US1.Dent], M90854 for patient A [US1.PtA], and M90964 [US1.LC9] and M90939 [US1.LC35] for two local controls) (34), and by Fitzgibbon et al. (GenBank accession numbers are as follows: L12752 for child 1 [US12.CHA], L19695 for child 2 [US12.CHB], and L12753 [US12.LC1] and L12754 [US12.LC2] for two local controls) (14). C2-V3 consensus sequences were generated from control European and Parisian HIV-1 sequences. Nine env sequences from Europe isolates were used as nucleotide and amino acid reference sequences for the V4-C4-V5 region. We also used V4-C4-V5 sequences from those previously published by Ou et al. (GenBank accession numbers are as follows: M91084 for the dentist [US1.Dent], M91090 for patient A [US1.PtA], and M91132 [US1.LC1] and M91133 [US1.LC2] for two local controls) (34). Sibling sequences (sets of viral sequences from the same patients) were not included in the reference sets. A V4-C4-V5 consensus sequence was generated from the control European HIV-1 sequences.
The genetic distances between the HIV-1 env sequences from the same patient and from those of one patient to those of another patient or set of reference sequences were defined as the average percentage of sequence divergence of all available pairs of C2-V3 and V4-C4-V5 nucleotide sequences. Only single-nucleotide differences were scored, and positions with gaps were excluded. The nonparametric U test of Mann-Whitney was used to evaluate the significance of the differences in the genetic variability between env sequences. The patients’ sequences were also compared with all sequences accessible in the GenEMBL database by using the FASTA search tool (35).
Phylogeny construction and evaluation were performed with the Phylip software package (11), the neighbor-joining algorithm (36), the matrix distance Fitch and Margoliash method (13), and the Fitch and Wagner maximum parsimony method.
The hypervariable env amino acid sequences deduced from the patients’ viruses were subjected to signature pattern analysis (14, 24, 34) with VESPA software (24). This method permits us to define particular sites in sequences at which residues are shared among certain groups of virus (24, 39). The amino acid C2-V3 or V4-C4-V5 sequences of patients’ viruses were then scanned for amino acids that occurred at homologous positions in less than 50% of the reference set, defined by all the previously selected C2-V3 (n = 48) and V4-C4-V5 (n = 9) sequences of HIV-1 subtype B isolates from Europe and from Paris.
Nucleotide sequence accession numbers.
The nucleotide sequences presented in this report have been deposited in the GenBank database under accession no. U87171 to U87221.
RESULTS
Nucleotide env sequences were obtained from 26 clones of PCR products amplified from the PBMC of the patients. After truncating and gap-stripping, the sequences spanned 243 bp in the C2-V3 region, comprising the V3 loop flanked at the 5′ and 3′ ends by 87 and 51 bp, respectively; the sequences spanned 265 bp in the V4-C4-V5 region. All sequences belonged to the European/North American group M, subtype B.
Mean intrapatient diversity ranged between 1.1 and 2.6% in the C2-V3 region and between 0.7 and 3.5% in the V4-C4-V5 region (Table 1). In the C2-V3 region, intrapatient diversity was higher in patient 1 than in patients 2 and 3 (P < 0.0003). Genetic diversity in the V4-C4-V5 region was higher in patient 1 than in patients 2 and 3 (P < 0.0004) and higher in patient 3 than in patient 2 (P < 0.0001). The mean nucleotide distances in C2-V3 between clones PA7 and PA12, and all clones from viruses of patients 2 and 3 were 3.9 and 3.5%, respectively; the distances were significantly smaller than the mean divergence calculated between clones PA1, PA4, PA11, PA14, PA15, PA12, and all clones derived from viruses of patients 2 and 3 (5.4%; P < 0.0001). Means of inter-patient diversity ranged between 1.2% and 5.0% in the C2-V3 region, and between 2.2% and 7.5% in the V4-C4-V5 region (Table 1). In contrast, means of genetic diversity between the patients’ cloned sequences and control sequences ranged between 16.8 and 17.2% for the C2-V3 regions and between 13.8 and 14.7% for the V4-C4-V5 regions. Means of genetic diversity in the C2-V3 region between patients’ clones and local field isolates ranged between 15.1 and 15.7%. The results suggest that the viruses from patients 1 through 3 are genetically related. Moreover, the genetic diversity in C2-V3 was higher between patients’ env sequences and European control sequences (mean, 17.0%; range, 10.0 to 29.3%) than between European sequences (mean, 13.2%; range, 3.2 to 30.1%) (P < 0.0001); similarly, genetic diversity in C2-V3 was higher between patients’ env sequences and sequences from Parisian viruses than between the Parisian env sequences themselves (P < 0.0001). Similar results were obtained upon analysis of the V4-C4-V5 region (data not shown).
TABLE 1.
Nucleotide diversity in the C2-V3 and V4-C4-V5 regions of the gp120 env gene of clones derived from patients’ proviruses
| Patient | C2-V3a
|
V4-C4-V5a
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| No. of clones | Intraperson variation (%) | Interindividual variation (%)
|
No. of clones | Intraperson variation (%) | Interindividual variation (%)
|
||||||
| To patient 3 | To patient 2 | To LC from Europeb | To LC from Parisc | To patient 3 | To patient 2 | To LC from Europed | |||||
| 1 | 9 | 2.6 (0.4–6.0) | 5.0 (3.8–9.2) | 4.7 (2.3–9.7) | 16.8 (13.1–28.4) | 15.1 (12.1–21.3) | 9 | 3.5 (0.0–7.3) | 7.2 (5.7–9.6) | 7.5 (6.8–10.2) | 14.7 (7.9–21.1) |
| 3 | 9 | 1.4 (0.0–6.0) | 1.2 (0.0–7.8) | 17.2 (13.2–29.3) | 15.7 (11.1–22.2) | 9 | 1.9 (0.7–4.1) | 2.2 (1.1–4.5) | 13.8 (6.6–21.8) | ||
| 2 | 8 | 1.1 (0.0–3.8) | 17.1 (10.0–26.9) | 15.6 (8.9–21.3) | 7 | 0.7 (0.0–1.9) | 14.0 (7.2–21.0) | ||||
Results are means and ranges of differences between sequences.
Local controls (LC) from Europe: 25 C2-V3 sequences of unrelated subtype B strains from Europe (32), including 5 sequences from Paris (8).
Local controls from Paris: 28 C2-V3 sequences of unrelated subtype B strains from Paris (8).
Local controls from Europe: 9 V4-C3-V5 sequences of unrelated subtype B strains from Europe (32).
Upon comparison of the patients’ data with the GenEMBL database by using the FASTA search tool, the sequences of the patients’ viruses did not appear related to previously published HIV-1 sequences; the less divergent sequences in C2-V3 (HIV-1 clone LC02; 10.6% divergence) as well as in V4-C4-V5 (HIV-1 clone RT1.21; 13.7% divergence) originated from North America.
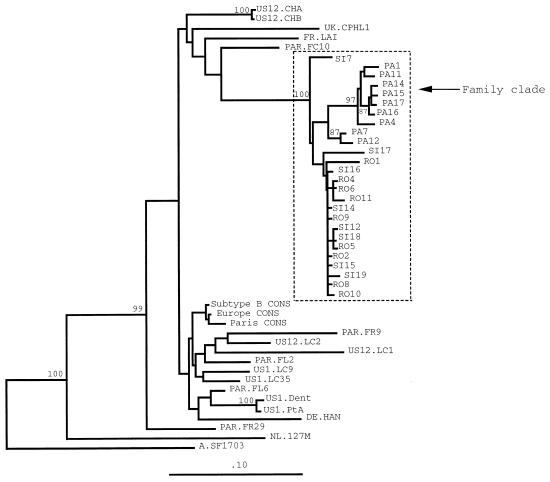
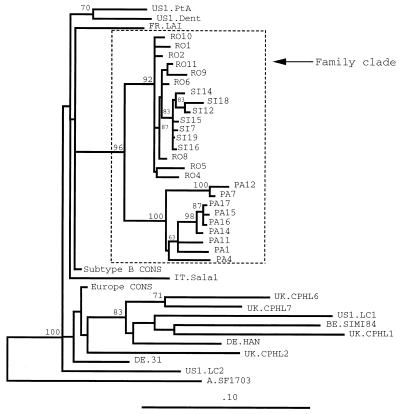
The genetic relatedness of the patients’ viruses was then confirmed by the analysis of phylogenetic trees. The sequences in C2-V3 (Fig. 1) and V4-C4-V5 (Fig. 2) of patients’ viruses clustered with each other and diverged from control sequences of strains from Europe and Paris, as well as from published sequences of viruses from patients with atypical HIV transmission. The branch dividing the familial cluster from the other groups of sequences was resolved in 100% of bootstrap replicates in the C2-V3 domain and in 96% in the V4-C4-V5 domain, supporting the monophyletic grouping of the sequences of viruses from patients 1 through 3.
FIG. 1.
Neighbor-joining phylogenetic tree of the coding sequences for the HIV-1 env C2-V3 region of 26 clones derived from proviruses of patients 1, 2, and 3 (isolates PA, SI, and RO, respectively), sequences of field isolates from Paris (PAR), published sequences from patients with atypical HIV transmission and from their local controls (US) (14, 34), and consensus sequences obtained from 25 unrelated controls from Europe (Europe CONS) (FR.LAI, IT.Sala1, IT.115, IT.193, UK.CPHL1, UK.CPHL2, UK.CPHL6, UK.CPHL7, UK.V77, UK.V87, CH.K11, CH.K16, DE.HAN, DE.31, BE.SIMI84, NL.H466, NL.B130, NL.127M, NL.114M, and NL.H36 [32], including 5 sequences from Paris [PAR.FC10, PAR.FL2, PAR.FL6, PAR.FR9, and PAR.FR29]), from 28 controls from Paris (Paris CONS), and the consensus sequence of HIV-1 subtype B (Subtype B CONS) (32). An HIV-1 subtype A consensus sequence (A.SF1703) was used as an outgroup. Vertical branches are for clarity only; the lengths of the horizontal branches are proportional to the single-base changes. Numbers at nodes represent the percentage of bootstrap samples for 100 replications, for which the corresponding cluster is depicted to the right. Only bootstrap values above 70% are indicated. Phylogenetic trees constructed by using the Fitch and Margoliash algorithm and by using the maximum-parsimony method resulted in similar branching patterns.
FIG. 2.
Phylogenetic tree analysis (obtained by using the Fitch and Margoliash algorithm) comparing the coding sequences for the HIV-1 env V4-C4-V5 region of the 26 clones derived from proviruses of patients 1, 2, and 3 (isolates PA, SI, and RO, respectively), published sequences of viruses of patients with atypical HIV transmission and their local controls (US) (34), the consensus sequence obtained from nine unrelated controls from Europe (Europe CONS) (FR.LAI, IT.Sala1, UK.CPHL1, UK.CPHL2, UK.CPHL6, UK.CPHL7, DE.HAN, DE.31, and BE.SIMI84), and the consensus sequence of HIV-1 subtype B (Subtype B CONS). Phylogenetic trees constructed by using the neighbor-joining method and by using the maximum-parsimony method resulted in similar branching patterns.
Phylogenetic analysis of the C2-V3 sequences showed two separate lineages (Fig. 1), one comprising all sequences of viruses from patient 1, including two predominant variants, among which sequences of clones PA7 and PA12 were minor variants, and the other comprising all sequences of viruses from patients 2 and 3. Although there was a distinctive branch favoring a separate lineage for the C2-V3 sequences of the viruses from patient 1 within the family cluster, it was not strongly supported by the bootstrap value (which was only 56% in the neighbor-joining tree). Within the subcluster of variants of viruses from patients 2 and 3, the C2-V3 sequences did not discriminate between the patients.
Phylogenetic analysis of the V4-C4-V5 sequences clearly differentiated two lineages: the first subcluster comprised all sequences of viruses from patient 1, with 100% of bootstrap replicates; the second subcluster comprised all sequences of viruses from patients 2 and 3, with 92% of bootstrap replicates (Fig. 2). Within the subcluster of viruses from patient 1, there was a strong grouping of sequences of clones PA7 and PA12, with 100% of bootstrap replicates. In addition, all sequences of viruses from patient 2 formed a distinct subgroup in the subcluster of sequences from patients 2 and 3.
The phylogenies inferred by using the neighbor-joining, the Fitch and Margoliash, and the maximum parsimony methods were highly congruent. When hypervariable sites in the V4-C4-V5 domain were removed, the branching patterns were reproduced (data not shown). Taken together, the data demonstrate a strong genetic linkage among env sequences of viruses from patients 1 through 3 and an additional subgrouping of env sequences for viruses from patients 2 and 3.
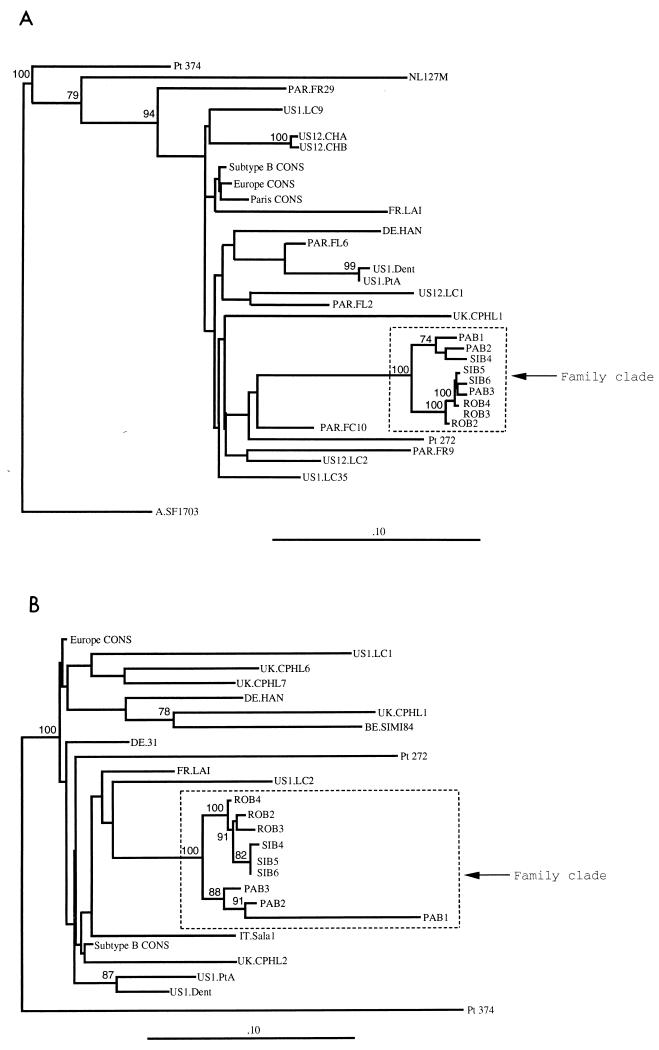
Cross-contamination between patients’ samples upon amplification was ruled out by the following lines of evidence. First, samples from patients 1, 2, and 3 were not amplified, cloned, and sequenced in the same week. Second, we have used a second blood sample obtained from each of the patients at 1 to 3 months after the initial sample, cloned the PCR product from the env gene, and sequenced three clones from the samples from each patient in a different laboratory from that in which the original sequencing work was performed. Minor changes from the initial sequences were observed (data not shown). The newly obtained sequences in C2-V3 and V4-C4-V5 fitted within the phylogenic subgroup corresponding to the family cluster (Fig. 3). The only difference in C2-V3 observed with the second samples from the patients, compared with the initial samples (Fig. 1), was that one clone from patient 2 belonged to the PA subcluster (clones obtained from patient 1) and vice versa (Fig. 3A). This difference, however, was not significant. Thus, the clones from the second samples behaved similarly to the clones from the initial samples upon phylogenetic analysis. The results of the phylogenetic analysis of the second samples in V4-C4-V5 (Fig. 3B) were similar to those of the analysis of the initial samples (Fig. 2), with distinct subgrouping according to the patients. The fact that similar features have been found in the initial samples and the second samples, which have been independently analyzed, rules out the possibility that our results correspond to cross-contamination between samples. Third, we further sequenced the amplification product of viruses from PBMC of two unrelated HIV-1-seropositive patients (patients 272 and 374) obtained at the same time as the first sample from patient 2. Unexpectedly, HIV-1 from patient 374 was found to be of subtype A. C2-V3 as well as V4-C4-V5 sequences from the viruses derived from patients 272 and 374 were clearly distinct from the family cluster (Fig. 3).
FIG. 3.
Phylogenetic tree analyses comparing the coding sequences for the HIV-1 env C2-V3 region as obtained by the neighbor-joining method (A) and the env V4-C4-V5 region as obtained by the Fitch-Margolias method (B) for the 9 clones derived from proviruses of patients 1 (clones PAB1, PAB2, and PAB3), 3 (clones ROB2, ROB3, and ROB4), and 2 (clones SIB4, SIB5, and SIB6) obtained at a second blood sampling; published sequences of viruses from patients with atypical HIV transmission and their local controls (US) (14, 29); the consensus C2-V3 sequence from 28 controls from Paris (Paris CONS); the consensus C2-V3 and V4-C4-V5 sequences obtained from unrelated controls from Europe (Europe CONS); and the consensus C2-V3 and V4-C4-V5 sequences of HIV-1 subtype B (Subtype B CONS) (32), as previously defined in the legends to Fig. 1 and 2, respectively. An HIV-1 subtype A consensus sequence (A.SF1703), and the V4-C4-V5 sequence of HIV-1 of patient 374 were used as outgroups for the phylogenetic analysis in C2-V3 and V4-C4-V5, respectively.
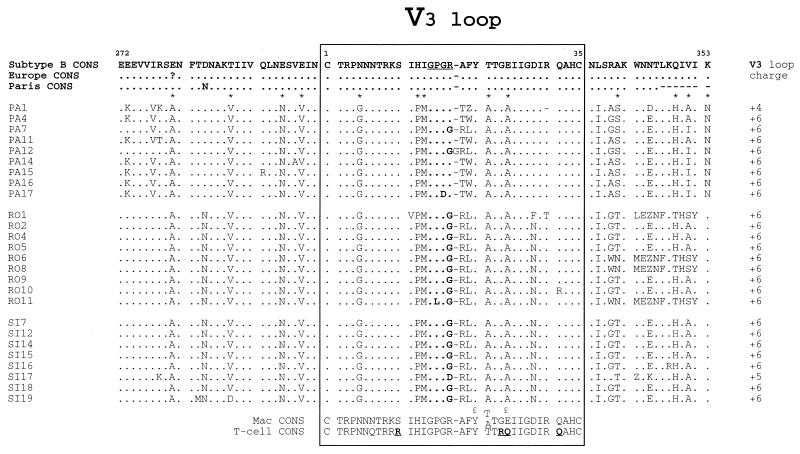
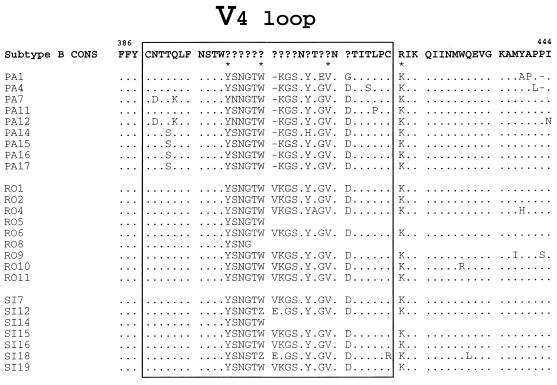
The relatedness of patients’ viruses was further assessed by analyzing the deduced amino acid sequences of the V3 and V4 hypervariable regions of gp120. The deduced amino acid sequences obtained from patients 1 through 3 were aligned (Fig. 4 and 5). The top of the V3 loop harbored the GPGR motif in five of nine clones derived from patient 1. GPGR is the most common motif in HIV-1 subtype B sequences (32). The other clones harbored the GPGG and GPDR motifs. Both clones with the GPGG motif belonged to the phylogenetic subcluster of clones PA7 and PA12 (Fig. 1). The GPGG motif also represented the amino acid sequence of the top of the V3 loop in most clones derived from the viruses from patients 2 and 3. As previously reported for the V4 region of HIV-1 (32, 40), multiple insertions of 3 nucleotides were observed in the V4 loop, thus maintaining the reading frame downstream. Two V4 motifs, YSNGTW and KGSNYTGVND, that are not present in HIV-1 subtype B, were frequently observed, indicating a high degree of relatedness among the V4 loop sequences of the viruses from these three patients. Of 13 residues that constituted the signature pattern of patient 1, 11 were found with high frequencies (0.96 to 1.0) in clones derived from patients 2 and 3 (Table 2). Only one amino acid residue in clones from patients 2 and 3 was present with a 100% frequency in the 13 amino acids of the reference set of unrelated isolates of HIV-1 subtype B at the homologous positions of the signature pattern of patient 1, supporting the conclusion that the viruses of patients 1 through 3 were placed together within the cluster. Only 3 of 10 positions of the common signature pattern of the three patients corresponded to residues in the same position in the signature pattern reported by Ou et al. (34), and only 2 positions corresponded to residues in the signature pattern reported by Fitzgibbon et al. (14). The amino acid patterns in the V4-C4-V5 region were similar for the three patients (Table 2).
FIG. 4.
Multiple alignments of the inferred amino acid sequence of the HIV-1 env C2-V3 region coding sequences of clones from patients 1, 2, and 3 (isolates PA, SI, and RO, respectively). The consensus C2-V3 amino acid sequence of European and North American HIV-1 subtype B (Subtype B CONS), that from 25 unrelated controls from Europe (Europe CONS), and that from 28 local controls from Paris (Paris CONS) are shown at the top. The macrophage (Mac CONS) and T-cell (T-cell CONS) consensus sequences of the V3 loop defined by Chesebro et al. (9) are shown at the bottom. Positions 11, 24, 25, and 32, in the consensus T-cell CONS defined as amino acid positions that may encode basic residues in T-cell-tropic isolates, are indicated in bold and underlined (15, 31). Positions 21 and 25, which has been identified as important in macrophage tropism by Westervelt et al. (44), are indicated by the symbol £. The V3 loop is boxed; its top is in bold. The V3 loop charge of each sequence is indicated on the right. Numbers within the box refer to amino acid position in the consensus sequence of the V3 loop of HIV-1 subtype B env gp120 (32), counting from the cysteine at the amino terminus. Amino acids are numbered according to their position in the HIV-1 LAI sequence (32). Dots indicate sequence homology, and dashes indicate gaps introduced for optimal alignment. Stop codons in the sequence are indicated by Z. Asterisks designate the noncontiguous amino acids of the signature pattern in the C2-V3 region for patient 1 (Table 2). Single-letter abbreviations for the amino acid residues are as follows: A, Ala; C, Cys; D, Asp; E, Glu; F, Phe; G, Gly; H, His; I, Ile; K, Lys; L, Leu; M, Met; N, Asn; P, Pro; Q, Gln; R, Arg; S, Ser; T, Thr; V, Val; W, Trp; Y, Tyr.
FIG. 5.
Multiple alignments of the inferred amino acid sequence of the HIV-1 env V4 loop coding sequences and its flanking regions of clones from patients 1, 2, and 3 (isolates PA, SI, and RO, respectively). The consensus V4-C3 amino acid sequence of European and North American HIV-1 subtype B (Subtype B CONS) is shown at the top. The V4 loop is boxed. Amino acids are numbered according to their position in the HIV-1 LAI sequence (32). The question marks indicate hypervariable sites at indicated positions. Asterisks designate the noncontiguous amino acids of the signature pattern in the V4-C4 region for patient 1 (Table 2). The amino acid region located after the frame shift in clones RO5, RO8, and SI14 was kept blank. Other symbols are defined in the legend to Fig. 4.
TABLE 2.
Amino acid signature patterns of viruses of patients 1 through 3 with reference to unrelated HIV-1 subtype B local controls
| Signature pattern and virus | Amino acid frequency or reference amino acida
|
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C2-V3 | V4-C4-V5 | |||||||||||||||||
| Patient 1 signature patternb | A | V | N | V | G | P | M | A | A | S | H | I | N | Y | W | V | K | |
| Patient 3 viruses | 1.0 | 0.96 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 0.0 | 0.56 | 0.0 | 0.0 | 1.0 | 1.0 | 1.0 | 1.0 | |
| Patient 2 viruses | 1.0 | 0.96 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 0.0 | 1.0 | 0.0 | 0.0 | 1.0 | 0.62 | 1.0 | 1.0 | |
| Unrelated local field isolatesc | 0.13 | 0.02 | 0.04 | 0.22 | 0.04 | 0.07 | 0.07 | 0.44 | 0.05 | 0.05 | 0.00 | 0.08 | 0.0 | 0.00 | 0.13 | 0.00 | 0.07 | |
| Unrelated local field isolates | E | T | E | E | N | H | I | T | E | A | Q | K | K | — | — | — | R | |
| Patient 1 viruses | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | ||||
| Patient 3 viruses | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | ||||
| Patient 2 viruses | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | ||||
| Common signature patterns of patients 1–3 | A | — | N | V | G | P | M | A | A | — | H | I | — | Y | W | V | K | |
| Unrelated local field isolates | 0.13 | 0.04 | 0.22 | 0.04 | 0.07 | 0.07 | 0.44 | 0.05 | 0.00 | 0.08 | 0.0 | 0.0 | 0.0 | 0.0 | ||||
| Reference 34d | A | — | A | — | — | — | — | A | — | — | — | — | — | M | W | E | R | |
| Reference 14e | E | — | — | — | W | — | — | — | — | — | — | — | — | NA | NA | NA | NA | |
Numbers depict the frequency at which the same amino acid is found at a given position within the signature pattern. —, absence of amino acid signature at that position. NA, not available. Top panel: frequencies of the amino acids defining the signature pattern of patient 1 in viruses of patients 2 and 3, as well as in the reference set of unrelated local field isolates of HIV-1 subtype B; middle panel: frequencies of the amino acids of the reference set of unrelated local field isolates of HIV-1 subtype B at the homologous positions of the signature pattern of patient 1, in viruses of patients 2 and 3; bottom panel: frequencies of the amino acids of the reference set of unrelated local field isolates of HIV-1 subtype B at positions of the common amino acid signature pattern of viruses of patients 1 to 3, and signature patterns previously reported by Ou et al. (34) and Fitzgibbon et al. (14).
The signature pattern corresponds to amino acids which differ by more than 50% of those present in the same positions in the reference set of C2-V3 and V4-C4 sequences of local HIV-1 subtype B isolates from Europe or Paris.
Forty-eight distinct sequences in the C2-V3 domain and nine sequences in the V4-C4-V5 domain of field isolates from Europe and Paris.
Amino acid signature pattern of the viruses of the dentist reported by Ou et al. (34).
Common amino acid signature pattern of the viruses of the children reported by Fitzgibbon et al. (14).
DISCUSSION
We report on a family in which the parents of an HIV-positive adult man became HIV positive with a virus that was phylogenetically linked to their son’s virus. We identified no risk factor for intrafamilial viral transmission and no unproven mode of transmission despite conducting a thorough investigation. Our attention was initially drawn by the unexpected seroconversion of the mother of a 31-year-old HIV-infected drug addict and the subsequent finding of HIV seropositivity in his father. We initiated a molecular analysis of cell-associated viruses of the three family members. A similar genotypic pattern of the pol 215 codon was observed, suggesting some similarity between the viral populations of the three patients. We then documented the genotypic relatedness between the viruses of the patients by three separate approaches, including assessment of genetic distance, phylogenetic analysis, and analysis of amino acid signature patterns within the hypervariable regions of the gp120 env gene. Analysis of the nucleotide sequences in the hypervariable env regions of viruses of the three family members showed an interpatient diversity of 1.2 to 5.0% and 2.2 to 7.5% in the C2-V3 and V4-C4-V5 regions, respectively. Interpatient diversities below 5.0% in C2-V3 region (3, 7, 14, 29, 30, 34, 46) and below 8.5% in the V4-C4-V5 region (3, 34) have been reported previously in the setting of epidemiologically linked sexual and parenteral HIV transmission. Interpatient diversity in the V4-C4-V5 domain was in a similar range to that estimated in the analysis of an epidemiologically linked cluster of infection reported by Ou et al. (34). In contrast, nucleotide sequences yielded more than 13.5% divergence between the C2-V3 and V4-C4-V5 sequences studied and sequences of unrelated local field isolates. Similar degrees of genetic divergence have been reported in nonepidemiologically linked HIV infections (2, 3, 14, 18, 34, 46, 47). These findings strongly suggested that the viruses from the three members of the family originated from a common source of infection.
The genetic relatedness of the viruses was further confirmed by the finding that all sequences clustered tightly within the same monophyletic group; the sequences diverged significantly from selected reference sequences, as well as from sequences from patients with atypical HIV transmission (14, 34). In addition, phylogenetic analysis demonstrated that within the familial HIV cluster, variants of the virus from patient 1 belong to a subcluster that is phylogenetically distinct from a subcluster comprising the variants of viruses from patients 2 and 3. In the subcluster of variants from patient 1, the C2-V3 and V4-C4-V5 sequences of two minor clones, PA7 and PA12, were strongly subgrouped. Obviously, one cannot totally exclude that the fact that PA7 and PA12 appear as minor strains is not due to a sampling bias. In the C2-V3 region, these clones showed less nucleotide divergence than did the clones derived from the viruses from both parents and the other predominant clones of the virus from the son. Analysis of the amino acid sequences of the latter two clones further revealed the presence of an unusual GPGG motif at the top of the V3 loop, reported to occur with a frequency of only 2.1% among the published HIV-1 sequences of North American and European isolates (32), that was shared with viruses from patients 2 and 3. Furthermore, the majority of the viruses from patients 1 to 3 showed several similar amino acid changes in the C2-V3 region compared with the HIV-1 B consensus sequence. Signature patterns were almost identical for the clones from the three patients but distinct from reference sequences, supporting the notion that the viruses from these patients harbored very similar quasispecies (20, 32), with marked similarities in highly functionally relevant domains of the env gp120 gene including the V3 loop (15, 27, 41). Indeed, the viruses from all three patients had the same predicted macrophage-tropic, non-syncytium-inducing (NSI) phenotype; i.e., all studied V3 sequences showed a serine at position 11 and an alanine at position 25, resulting in global electrostatic neutrality at these positions, compatible with an NSI phenotype (15) and highly characteristic of macrophage-tropic viruses (9, 44). Each of the patients’ variants exhibited more homology to the consensus V3 sequence of macrophage-tropic variants than to the consensus V3 sequence of T-cell-line-tropic variants (9) (Fig. 4). Only one of the variants encoded a basic amino acid at one of the four positions in the V3 loop that are associated with T-cell tropism (31).
The mode of cross-infection of HIV among members of the family remains unknown. The clinical features of HIV disease and the genetic analysis of viruses suggest a chronology of infection where patient 1 was infected before patient 3, who, in turn, was infected before patient 2. Sequences obtained from patient 2 demonstrated a limited nucleotide and amino acid genetic diversity, as previously documented in primary infections (1, 25, 32, 48, 49). The relatively high homogeneity of the viral populations in patient 3 is compatible with a recent infection (46). In contrast, the C2-V3 and C4-V4-V5 domain nucleotide sequences of the variants from patient 1, who had most probably been infected while using intravenous drugs, i.e., at least 5 years before sampling, were significantly more heterogeneous than those derived from patients 2 and 3. Analysis of the intraindividual variations in the V4-C4-V5 domain confirmed the higher divergence of viral strains from patient 1 than of those from patients 2 and 3 and further revealed that strains from patient 3 were significantly more divergent than those from patient 2. The results are consistent with patient 1 having been infected for a longer time than patients 2 and 3 and with patient 3 having been infected before patient 2. In addition, patient 3 already presented with a high HIV load in plasma and low CD4 cell counts 4 months after patient 2 had seroconverted. Since the majority of the variants from patients 2 and 3 were genetically related among themselves and were related to minor variants from patient 1, one may speculate that patient 1 first contaminated patient 3 and later contaminated patient 2; in the latter case, the same minor variants from patient 1 would have represented the common source of infection of patients 2 and 3, and therefore should have been selected twice during intrafamilial transmission. There is evidence that minor strains are commonly transmitted from HIV-positive donors to the recipients (49, 50). Alternatively, patient 1 may have infected patient 3, who, in turn, contaminated patient 2; in this case, minor variants of the virus from patient 1 should have been initially selected during transmission to patient 3; major variants of the virus from patient 3 would then have been the source of infection in patient 2.
Despite extensive epidemiological investigation, no risk factor for cross-infection was identified among family members. The risk of transmission of HIV in the household in the absence of sexual or percutaneous exposure has been extensively investigated (4, 12, 17, 19, 28, 33, 37, 38, 42, 43). It is known that viral transmission has not occurred after tens of thousands of days of sharing eating utensils, towels, combs, toilets, bathtubs, and beds and of hugging and kissing between family members (16). A meta-analysis of several studies in the United States and Europe has revealed no case of infection in the follow-up of more than 1,700 person-years after household contact with HIV-infected people (42). However, a small number of cases of infection has been reported in which no risk factor could be identified despite extensive investigation (5, 6, 14, 34, 45). The contact of cutaneous or mucous membranes with infected blood or body fluids has been considered to be the explanation for most such cases of HIV transmission (38, 43). The estimated risk of transmission after mucous and cutaneous exposure to HIV-infected blood in prospective studies of health care workers is below 0.1% (21, 23). Transmission of the virus from the son to one of his parents through casual contact as the parents were caring for their son is highly unlikely to have occurred on two independent occasions. These findings raise the issue of whether the virus exhibited distinct phenotypic properties or the recipients exhibited specific genetic susceptibility to infection. Taken together, our observations suggest that atypical transmission of HIV may occur.
ACKNOWLEDGMENTS
We are indebted to Xavier Jeunemaître, Françoise Ferchal, Catherine Letondal, Elisabeth Menu, Sylvie Corbet, and Marie-Charlotte Hallouin for assistance with nucleotide sequencing or analyses; Sentob Saragosti and Marie-Laure Chaix for providing sequences of field isolates in Paris; and Denis Beaumont and Eric Gressier (Laboratoire Cedric, CNAM, Paris, France) for the opportunity to use the “veryfastDNAml” software.
M.C.M.-T. is a recipient of grants from Agence Nationale de Recherches sur le SIDA (ANRS) and from Fondation pour la Recherche Médicale (SIDACTION), France. This work was supported by Institut National de la Santé et de la Recherche Médicale, ANES, and SIDACTION, France.
REFERENCES
- 1.Ait-Khaled M, Emery V C. Sequence variation within the human immunodeficiency virus V3 loop at seroconversion. J Med Virol. 1993;41:270–274. doi: 10.1002/jmv.1890410403. [DOI] [PubMed] [Google Scholar]
- 2.Arnold C, Balfe P, Clewley J P. Sequence distances between env genes of HIV-1 from individuals from the same source: implications for the investigation of possible transmission events. Virology. 1995;211:198–203. doi: 10.1006/viro.1995.1391. [DOI] [PubMed] [Google Scholar]
- 3.Balfe P, Simmonds P, Ludlam C A, Bishop J O, Leigh-Brown A J. Concurrent evolution of human immunodeficiency virus type 1 in patients infected from the same source: rate of sequence change and low frequency of inactivating mutations. J Virol. 1990;64:6221–6223. doi: 10.1128/jvi.64.12.6221-6233.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berthier A, Chamaret S, Fauchet R, Fonlupt J, Genetet N, Gueguen M, Pommereuil M, Ruffault A, Montagnier L. Transmissibility of human immunodeficiency virus in hemophilic and non-hemophilic children living in a private school in France. Lancet. 1986;ii:598–601. doi: 10.1016/s0140-6736(86)92427-x. [DOI] [PubMed] [Google Scholar]
- 5.Blank S, Simonds R J, Weisfuse I, Rudnick J, Chiasson M A, Thomas P. Possible nosocomial transmission of HIV. Lancet. 1994;344:512–514. doi: 10.1016/s0140-6736(94)91900-3. [DOI] [PubMed] [Google Scholar]
- 6.Browstein A, Fricke W. HIV transmission between two adolescent brothers with hemophilia. Morbid Mortal Weekly Rep. 1993;42:948–951. [PubMed] [Google Scholar]
- 7.Burger H, Weiser B, Flaherty K, Gulla J, Nguyen P N, Gibbs R A. Evolution of human immunodeficiency virus type 1 nucleotide sequences diversity among close contacts. Proc Natl Acad Sci USA. 1991;88:11236–11240. doi: 10.1073/pnas.88.24.11236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chaix M-L, Chappey C, Couillin I, Rozenbaum W, Levy J-P, Saragosti S. Diversity of the V3 region of HIV in Paris, France. AIDS. 1993;7:1199–1204. doi: 10.1097/00002030-199309000-00008. [DOI] [PubMed] [Google Scholar]
- 9.Chesebro B, Wherly K, Nishio J, Perryman S. Macrophage-tropic human immunodeficiency virus isolates from different patients exhibit unusual V3 envelope sequence homogeneity in comparison with T-cell-tropic isolates: definition of critical amino acids involved in cell tropism. J Virol. 1992;66:6547–6554. doi: 10.1128/jvi.66.11.6547-6554.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Delwart E L, Shaper E G, Louwagie J, McCutchan F E, Grez M, Rübsamen-Waigmann H, Mullins J I. Genetic relationships determined by a DNA heteroduplex mobility assay: analysis of HIV-1 env genes. Science. 1993;262:1257–1261. doi: 10.1126/science.8235655. [DOI] [PubMed] [Google Scholar]
- 11.Felsenstein J. PHYLIP—phylogeny inference package. Cladistics. 1989;5:164–166. [Google Scholar]
- 12.Fischl M A, Dickinson G M, Scott G B, Klimas N, Flechter M A, Parks W. Evaluation of heterosexual partners, children, and household contacts of adults with AIDS. JAMA. 1987;257:640–644. [PubMed] [Google Scholar]
- 13.Fitch W M, Margoliash E. Construction of phylogenetic trees. Science. 1967;155:279–284. doi: 10.1126/science.155.3760.279. [DOI] [PubMed] [Google Scholar]
- 14.Fitzgibbon J E, Gaur S, Frenkel L D, Laraque F, Edlin B R, Dubin D T. Transmission from one child to another of human immunodeficiency virus type 1 with a zidovudine-resistance mutation. N Engl J Med. 1993;329:1835–1841. doi: 10.1056/NEJM199312163292502. [DOI] [PubMed] [Google Scholar]
- 15.Fouchier R A M, Groenink M, Kootstra N A, Tersmette M, Huisman H G, Miedema F, Schuitemaker H. Phenotype-associated sequence variation in the third variable region of the human immunodeficiency virus type 1 gp120 molecule. J Virol. 1992;66:3183–3187. doi: 10.1128/jvi.66.5.3183-3187.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Friedland G, Kahl P, Saltzman B, Rogers M, Feiner C, Mayers M, Schable C, Klein R S. Additional evidence for lack of transmission of HIV infection by close interpersonal (casual) contact. AIDS. 1990;4:639–644. doi: 10.1097/00002030-199007000-00005. [DOI] [PubMed] [Google Scholar]
- 17.Friedland G H, Saltzman B R, Rogers M F, Kahl P A, Lesser M L, Mayers M M, Klein R S. Lack of transmission of HTLV-III/LAV infection to household contacts of patients with AIDS or AIDS-related complex with oral candidiasis. N Engl J Med. 1986;314:344–349. doi: 10.1056/NEJM198602063140604. [DOI] [PubMed] [Google Scholar]
- 18.Furuta Y, Bergström T, Norkrans G, Horal P. HIV type 1 V3 sequence diversity in contact-traced Swedish couples at the time of sexual transmission. AIDS Res Hum Retroviruses. 1994;10:1187–1189. doi: 10.1089/aid.1994.10.1187. [DOI] [PubMed] [Google Scholar]
- 19.Gershon R R, Vlahov D, Nelson K E. The risk of transmission of HIV-1 through non-percutaneous, non-sexual modes—a review. AIDS. 1990;4:645–650. doi: 10.1097/00002030-199007000-00006. [DOI] [PubMed] [Google Scholar]
- 20.Goodenow M, Huet T, Saurin W, Kwok S, Sninsky J J, Wain-Hobson S. HIV-1 isolates are rapidly evolving quasispecies: evidence for viral mixtures and preferred nucleotides substitutions. J Acquired Immune Defic Syndr. 1989;2:344–352. [PubMed] [Google Scholar]
- 21.Henderson D K, Fahey B J, Willy M, Schmitt J M, Carey K, Koziol D E, Lane H C, Fedio J, Saah A J. Risk for occupational transmission of human immunodeficiency virus type 1 (HIV-1) associated with clinical exposure: a prospective evaluation. Ann Intern Med. 1990;113:740–746.17. doi: 10.7326/0003-4819-113-10-740. [DOI] [PubMed] [Google Scholar]
- 22.Higgins D G, Bleasby A J, Fuchs R. CLUSTAL V: improved software for multiple sequence alignment. Comp Appl Biosci. 1992;8:189–191. doi: 10.1093/bioinformatics/8.2.189. [DOI] [PubMed] [Google Scholar]
- 23.Ippolito G, Puro V, De Carli G Italian Study Group on Occupational Risk of HIV Infection. The risk of occupational human immunodeficiency virus infection in health care workers: Italian Multicenter Study. Arch Intern Med. 1993;153:1451–1458. [PubMed] [Google Scholar]
- 24.Korber B, Myers G. Signature pattern analysis: a method for assessing viral sequence relatedness. AIDS Res Hum Retroviruses. 1992;8:1549–1560. doi: 10.1089/aid.1992.8.1549. [DOI] [PubMed] [Google Scholar]
- 25.Kuiken, C. L., and B. T. M. Korber. 1994. Epidemiological significance of intra- and inter-person variation of HIV-1. AIDS 8(Suppl. 1): S73–S83.
- 26.Larder B A, Kellam P, Kemp S D. Zidovudine resistance predicted by direct detection of mutation in DNA from HIV-infected lymphocytes. AIDS. 1991;5:137–144. doi: 10.1097/00002030-199102000-00002. [DOI] [PubMed] [Google Scholar]
- 27.Leigh Brown A J, Monaghan P. Evolution of the structural proteins of human immunodeficiency virus: selective constraints on nucleotide substitution. AIDS Res Hum Retroviruses. 1988;4:399–407. doi: 10.1089/aid.1988.4.399. [DOI] [PubMed] [Google Scholar]
- 28.Lifson A R. Do alternate modes for transmission of human immunodeficiency virus exist? JAMA. 1988;259:1353–1356. [PubMed] [Google Scholar]
- 29.McNearney T, Hornickova Z, Kostler B, Birdwell A, Storch G A, Polmar S H, Arens M, Ratner L. Evolution of sequence diversity among human immunodeficiency type 1 isolates derived from a blood donnor and a recipient. Pediatr Res. 1993;33:36–42. doi: 10.1203/00006450-199301000-00008. [DOI] [PubMed] [Google Scholar]
- 30.McNearney T, Westervelt P, Thielan B J, Trowbridge D B, Garcia J, Whittier R, Ratner L. Limited sequence heterogeneity among biologically distinct human immunodeficiency virus type 1 isolates from individuals involved in a clustered infectious outbreak. Proc Natl Acad Sci USA. 1990;87:1917–1921. doi: 10.1073/pnas.87.5.1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Milich L, Margolin B, Swanstrom R. V3 loop of the human immunodeficiency virus type 1 env protein: interpreting sequence variability. J Virol. 1993;67:5623–5634. doi: 10.1128/jvi.67.9.5623-5634.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Myers G, Korber B, Wain-Hobson S, Jeang K-T, Henderson L E, Pavlakis D N. Human retroviruses and AIDS: a compilation and analysis of nucleic acid and amino acid sequences. Los Alamos, N.M: Theoretical Biology and Biophysics Group, Los Alamos National Laboratory; 1995. [Google Scholar]
- 33.Operskaski E A, Mosley J W. Risk of HTLV-III/LAV transmission to household contacts. N Engl J Med. 1986;315:257. doi: 10.1056/NEJM198607243150413. [DOI] [PubMed] [Google Scholar]
- 34.Ou C-Y, Ciesielski C A, Myers G, Bandea C I, Luo C-C, Korber B T M, Mullins C I, Schochetman G, Berkelman R L, Economou A N, Witte J J, Furman L J, Satten G A, MacInnes K A, Curran J W, Jaffe H W. Molecular epidemiology of HIV transmission in a dental practice. Science. 1992;256:1165–1171. doi: 10.1126/science.256.5060.1165. [DOI] [PubMed] [Google Scholar]
- 35.Pearson W R, Lipman D J. Improved tools for biological sequence comparison. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 37.Sande M A. Transmission of AIDS. The case against casual contagion. N Engl J Med. 1986;314:380–382. doi: 10.1056/NEJM198602063140609. [DOI] [PubMed] [Google Scholar]
- 38.Sattar S A, Springthorpe V S, Conway B, Xu Y. Inactivation of the human immunodeficiency virus: an update. Rev Med Microbiol. 1994;5:139–150. [Google Scholar]
- 39.Sharp, P. M., D. L. Robertson, F. Gao, and B. H. Hahn. 1994. Origins and diversity of human immunodeficiency viruses. AIDS 8(Suppl. 1): S27–S42.
- 40.Simmonds P, Balfe P, Ludlam C A, Bishop J O, Brown A J. Analysis of sequence diversity of hypervariable regions of the external glycoprotein of human immunodeficiency virus type 1. J Virol. 1990;64:5840–5850. doi: 10.1128/jvi.64.12.5840-5850.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Simmonds P, Zhang L Q, McOmish F, Balfe P, Ludlam C A, Leigh Brown A J. Discontinuous sequence change of human immunodeficiency virus (HIV) type 1 env sequences in plasma viral and lymphocyte-associated proviral populations in vivo: implications for models of HIV pathogenesis. J Virol. 1991;65:6266–6276. doi: 10.1128/jvi.65.11.6266-6276.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Simonds R J, Chanock S. Medical issues related to caring for human immunodeficiency virus-infected children in and out of the home. Pediatr Infect Dis J. 1993;12:845–852. doi: 10.1097/00006454-199310000-00010. [DOI] [PubMed] [Google Scholar]
- 43.Simonds R J, Rogers M F. HIV prevention—bringing the message home. N Engl J Med. 1993;329:1883–1885. doi: 10.1056/NEJM199312163292511. [DOI] [PubMed] [Google Scholar]
- 44.Westervelt P, Trowbridge D B, Epstein L G, Blumberg B M, Li Y, Hahn B H, Shaw G M, Price R W, Ratner L. Macrophage tropism determinants of human immunodeficiency virus type 1 in vivo. J Virol. 1992;66:2577–2582. doi: 10.1128/jvi.66.4.2577-2582.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Whan V, Kramer H H, Voit T, Bruister H T, Scrampical B, Scheid A. Horizontal transmission of HIV between two siblings. Lancet. 1986;ii:694. doi: 10.1016/s0140-6736(86)90209-6. [DOI] [PubMed] [Google Scholar]
- 46.Wolfs T F, Zwart G, Bakker M, Goudsmit J. HIV-1 genomic RNA diversification following sexual and parenteral virus transmission. Virology. 1992;189:103–110. doi: 10.1016/0042-6822(92)90685-i. [DOI] [PubMed] [Google Scholar]
- 47.Wolinsky S M, Wike C M, Korber B T M, Hutto C, Parks W P, Rosenblum L L, Kunstman K J, Furtado M R, Munoz J L. Selective transmission of human immunodeficiency virus type 1 variants from mothers to infants. Science. 1992;255:1134–1137. doi: 10.1126/science.1546316. [DOI] [PubMed] [Google Scholar]
- 48.Zhang L Q, MacKenzie P, Cleland A, Holmes E C, Leigh Brown A J, Simmonds P. Selection of specific sequences in the external envelope protein of human immunodeficiency virus type 1 upon primary infection. J Virol. 1993;67:3345–3356. doi: 10.1128/jvi.67.6.3345-3356.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhu T, Mo H, Wang N, Nam D S, Cao Y, Koup R A, Ho D D. Genotypic and phenotypic characterization of HIV-1 in patients with primary infection. Science. 1993;261:1179–1181. doi: 10.1126/science.8356453. [DOI] [PubMed] [Google Scholar]
- 50.Zhu T, Wang N, Carr A, Nam D S, Moor-Jankowski R, Cooper D A, Ho D D. Genetic characterization of human immunodeficiency virus type 1 in blood and genital secretions: evidence for viral compartmentalization and selection during sexual transmission. J Virol. 1996;70:3098–3107. doi: 10.1128/jvi.70.5.3098-3107.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]