Abstract
It has been suggested that the inability of the immune response to control human immunodeficiency virus type 1 (HIV-1) replication may be due, at least in part, to the capacity of this virus to escape from immune recognition through mutation. While there is increasing evidence for the importance of HIV-1-specific CD4+ T cells in containing HIV-1 spread in the infected individual, little is known about the consequences of HIV-1 mutation on virus-specific CD4+ T-cell function. The impact of HIV-1 sequence variation on CD4+ T-helper (Th)- cell function was assessed with a rhesus monkey model for immune recognition of the HIV-1 envelope (Env) glycoprotein. A series of HIV-1 Env(484-496) variant peptides were shown to retain the ability to bind to the appropriate rhesus monkey major histocompatibility complex class II DR molecule. Peptides bearing substitutions at position 490, however, failed to drive the proliferation or cytokine secretion of two well-characterized HXBc2 Env-specific rhesus monkey CD4+ Th-cell lines. Exogenous costimulation was ineffective in complementing the ability of the nonstimulatory peptides to induce [3H]thymidine incorporation by these cells. Finally, HIV-1 Env(484-496) variant peptides with substitutions at position 490 antagonized the HXBc2 Env peptide-induced proliferative response of the CD4+ Th-cell lines. Thus, HIV-1 variants appear to have the capacity to neutralize the function of virus-specific CD4+ T lymphocytes.
Human immunodeficiency virus type 1 (HIV-1) rapidly mutates in an infected individual because of its high replication rate and the infidelity of its reverse transcriptase (11, 12, 29). The greatest sequence variation in HIV-1 is seen in regions of the viral envelope (Env) glycoprotein. Most infected individuals mount a vigorous immune response against HIV-1. Nevertheless, in the absence of therapeutic interventions, high levels of viral replication persist and profound deterioration of the immune system usually occurs. It has been suggested that the inability of the immune response to contain this viral replication may be due to the capacity of the mutating virus to escape immune recognition.
There is increasing evidence for the importance of CD4+ T cells in containing HIV-1 spread in the infected individual (28). The extent to which HIV-1 can avoid recognition by virus-specific CD4+ T lymphocytes has not been well defined. Early reports suggested that human HIV-1 Env (410-429)-specific CD4+ T-helper (Th)-cell clones could recognize peptides corresponding to divergent viral isolates (3, 31). Similarly, studies with a large panel of HIV-1 Env (428-443) variant peptides bearing empirically introduced amino acid substitutions indicated that evasion of H-2Ek-restricted murine Th-cell recognition was an unlikely event (2). In contrast, the absence of recognition of HIV-1 Env (236-251) variant peptides by human CD4+ Th-cell clones suggested that significant differences exist between ubiquitous clade B viruses (20). More recent efforts have documented the inability of human CD4+ Th-cell clones to recognize third-hypervariable-loop peptides, HIV-1 Env (303- 338), corresponding to heterologous viral isolates (9, 10, 26). These reports suggest that HIV-1 may be able to escape from CD4+ Th-cell recognition. Little structural information, however, has been gathered to define the amino acid substitutions in HIV-1 peptides required to alter their binding to major histocompatibility complex (MHC) class II molecules or to T- cell receptors (TCR). Furthermore, the functional consequences of alterations in an HIV-1-specific CD4+ T-cell epitope for Th cells have not been thoroughly examined.
With the development of chimeric simian-human immunodeficiency viruses containing HIV-1 env on a simian immunodeficiency virus backbone, the role of HIV-1 Env in AIDS pathogenesis can be explored with rhesus monkeys (19, 30). We previously established HIV-1 Env-specific CD4+ Th-cell lines from plasmid DNA-vaccinated rhesus monkeys (18). Two of these CD4+ Th-cell lines were found to recognize the HIV-1 Env (484-496) epitope in an MHC class II DR-restricted fashion (17). In this study, we examined the effect of substitutions in this HIV-1-specific CD4+ Th-cell epitope on MHC class II molecule binding and CD4+ Th-cell recognition and function.
MATERIALS AND METHODS
Cell lines.
Rhesus monkey B lymphocytes were transformed with a supernatant from an immortalized B-cell line (S594) productively infected with the baboon herpesvirus Herpes papio (23). Transformed rhesus monkey B-lymphoblastoid-cell lines (B-LCL) were expanded and maintained in RPMI 1640 medium (GIBCO BRL, Gaithersburg, Md.) supplemented with HEPES (25 mM), l-glutamine (2 mM), penicillin (50 U/ml), streptomycin (40 μg/ml), gentamicin (50 μg/ml), and 10% fetal calf serum (FCS) (BioWhittaker, Walkersville, Md.).
HIV-1 Env-specific CD4+ Th-cell lines were established from peripheral blood lymphocytes of HIV-1 env DNA-vaccinated rhesus monkeys by stimulation with recombinant gp120 (Intracel, Cambridge, Mass.) followed by expansion with recombinant interleukin 2 (IL-2) (kindly provided by Hoffmann-La Roche, Nutley, N.J.) as previously described (18). Cells were maintained in cultures in 24-well plates containing RPMI 1640 medium supplemented with antibiotics and 10% FCS through repeated rounds of antigen restimulation at 7- to 14-day intervals. The HIV-1 Env-specific Th-cell lines were used in functional assays at least 7 days following restimulation.
Rhesus monkey MHC class II DR α- and β-chain cDNAs were cloned into pREP4 (Invitrogen, San Diego, Calif.) after insertion of the encephalomyocarditis virus internal ribosomal entry sequence (6). Plasmid DNA was electroporated into MHC class II-negative, human Epstein-Barr virus-transformed RM3 B-LCL (4). Stable RM3 transfectants expressing rhesus monkey MHC class II DR molecules were isolated through hygromycin B (Calbiochem Novabiochem Corp., La Jolla, Calif.) selection in RPMI 1640 medium supplemented with antibiotics and 10% FCS (17). Cell surface expression of MHC class II DR molecules was confirmed by staining with the phycoerythrin-conjugated, HLA-DR-specific monoclonal antibody L243 (Becton Dickinson Immunocytometry Systems, San Jose, Calif.) followed by flow cytometric analysis on an EPICS XL apparatus (Coulter Corp., Miami, Fla.).
Synthetic peptides.
The 20-amino-acid HIV-1 HXBc2 gp120 synthetic peptides used to map epitopes recognized by Env-specific CD4+ Th-cell lines were kindly provided by the Medical Research Council courtesy of the AIDS Reagent Project (Hertfordshire, United Kingdom). The sequences of the peptides used in this study are as follows: p15/Env (172-191), EYAFFYKLDIIPIDNDTTSY; p22/Env (242-261), VSTVQCTHGIRPVVSTQLLL; and p46/Env (482-501), ELYK Y K V V K I E PLGVAPTKA. The sequences of the 13-amino-acid HIV-1 Env (484-496) variant peptides are listed in Table 3. These peptides were obtained from Quality Controlled Biochemicals (Hopkinton, Mass.).
TABLE 3.
Effects of variation in an HIV-1 Env Th- cell epitope on the proliferative responses of MHC class II-restricted CD4+ Th- cell lines
| Isolate | Sequence | Peptidea | Proliferative responseb of Th-cell line:
|
|
|---|---|---|---|---|
| 233.00 | 431.78 | |||
| HXBc2 | YKYKVVKIEPLGV | Index | +++ | +++ |
| VE3 | -----I------- | V489I | +++ | ++ |
| RF | ------R------ | K490R | − | − |
| VE7 | ------Q------ | K490Q | − | − |
| MN | ------T------ | K490T | − | − |
| QZ4589 | ------E------ | K490E | − | − |
| 4995 | -------V----- | I491V | +++ | +++ |
| GUN | ----------I-- | L494I | +++ | +++ |
| SF2B13 | ------------I | V496I | +++ | +++ |
| 68A | ------------L | V496L | +++ | +++ |
HIV-1 Env variant peptides were named with the nomenclature X#Y, where X represents the index peptide amino acid residue, # represents the amino acid position within Env, and Y represents the amino acid substitution observed at position #.
Results are expressed as the concentration of peptide required to give 50% maximal proliferation of the Mamu-DR*W201-restricted rhesus monkey CD4+ Th- cell lines: +++, ≤0.5 μg/ml; ++, 0.5 to 5.0 μg/ml; +, 5.0 to 50 μg/ml; −, >50 μg/ml. In the presence of medium alone, both T-cell lines had a background of less than 100 cpm. Fifty percent maximal proliferation was defined as 2,000 cpm for cell line 233.00 and 11,900 cpm for cell line 431.78.
Mamu-DR*W201-binding assay.
The binding of HIV-1 Env (484-496) variant peptides to the Macaca mulatta leukocyte antigen (Mamu)-DR*W201 molecule expressed on the surface of RM3 transfectants was measured in a cellular competition assay similar to that used by others (3, 33). Briefly, the ability of HIV-1 Env (484-496) variant peptides to inhibit the proliferation of a Mamu-DR*W201-restricted, CD4+ Th-cell line specific for HIV-1 Env (172-191) was measured. X-irradiated (5,000 rads) Mamu-DR*W201-expressing RM3 cells were incubated in 96-well flat-bottom microtiter plates in the presence of 25 μg of the Env (484-496) peptide competitor per ml for 2 h prior to the addition of 0.5 μg of the Env (172-191) peptide per ml and an equivalent number of Env (172-191)-specific CD4+ Th cells. Cultures were pulsed with [3H]thymidine on day 2 and harvested on day 3. The incorporation of [3H]thymidine was counted by liquid scintillation, as described below.
Proliferation assay.
CD4+ Th-cell lines were cultured at 2 × 104 cells/well with an equivalent number of X-irradiated (5,000 rads) autologous B-LCL in the presence of 0 to 50 μg of peptide per ml in 96-well flat-bottom microtiter plates. After 2 days, 1 μCi of [3H]thymidine was added per well. The cultures were harvested onto glass fiber filter mats on day 3 with an automated plate harvester (Tomtec, Orange, Conn.). [3H]thymidine incorporation was determined with Optiphase Betamix scintillation fluid and a Microbeta 1450 liquid scintillation counter (Wallac, Gaithersburg, Md.).
To reconstitute the ability of HIV-1 Env (484-496) variant peptides bearing substitutions at position 490 to stimulate the proliferation of HXBc2 Env (484-496)-specific CD4+ Th cells, 40 ng of phorbol myristate acetate (PMA) per ml, 200 ng of ionomycin (ION) per ml, or 100 pg of human recombinant IL-1β per ml was added to cultures containing 5 μg of Env (484-496) variant peptides per ml. Both PMA and ION were obtained from Sigma Chemical Co. (St. Louis, Mo.); recombinant human IL-1β was purchased from Genzyme (Cambridge, Mass.). These studies were in all other respects identical to the proliferation assays already described.
Cytokine secretion.
To measure cytokine production by CD4+ Th-cell lines, 106 T cells were cultured with an equivalent number of X-irradiated (5,000 rads) autologous B-LCL in 24-well plates containing 1 ml of RPMI 1640 medium supplemented with antibiotics and 5% FCS in the presence or absence of 5 μg of peptide per ml. Supernatants were harvested after 48 h and frozen prior to analysis. Cytokine measurements were made according to manufacturers’ instructions with commercially available enzyme-linked immunosorbent assay (ELISA) kits. The human IL-4 kit, the human IL-10 kit, and the rhesus monkey gamma interferon kit were obtained from Biosource International (Camarillo, Calif.), while the human transforming growth factor β kit and the human tumor necrosis factor alpha kit were obtained from Genzyme. The human ELISA kits were selected on the basis of cross-reactivity with cytokines present in the supernatants of activated rhesus monkey peripheral blood lymphocytes.
Antagonism assay.
The ability of HIV-1 Env (484-496) variant peptides to antagonize the proliferative response of HXBc2-specific CD4+ Th-cell lines was examined with a prepulse assay to eliminate peptide competition for MHC class II occupancy (7). Autologous B-LCL were pulsed for 2 h with 0.5 μg of the HIV-1 HXBc2 Env (484-496) index peptide per ml. The B-LCL were then washed extensively and X-irradiated (5,000 rads) prior to a 2-h pulse with a 0.5 to 50 μg/ml amount of the index peptide, the Env (484-496) variant peptides, or the nonstimulatory, Mamu-DR*W201-binding Env (172-191) peptide p15. The B-LCL were again washed thoroughly and plated at 2 × 104 cells/well in 96-well flat-bottom microtiter plates with an equivalent number of HXBc2 Env (484-496) peptide-specific CD4+ Th cells. Cultures were pulsed with [3H]thymidine on day 2 and harvested on day 3. The incorporation of [3H]thymidine was counted by liquid scintillation, as described above.
RESULTS
Variation of HIV-1 clade B Env sequences in the region of a CD4+ Th-cell epitope is greatest at amino acid residues 490 and 496.
We previously identified the nonamer YKVVKIEPL (amino acids 486 to 494) as a minimal HIV-1 Env CD4+ Th- cell epitope by using long-lived cell lines isolated from HIV-1 env DNA-vaccinated rhesus monkeys (18). Although this CD4+ Th- cell epitope is located in the fifth conserved region of gp120, some sequence variation has been observed in this location, both across HIV-1 clades and among clade B isolates (Tables 1 and 2). In fact, when 92 clade B Env sequences were compared to the HIV-1 HXBc2 index sequence, two frequent conservative substitutions were seen: arginine for lysine at position 490 and isoleucine for valine at position 496 (21). These two positions appear to be relative hot spots for viral variation.
TABLE 1.
Intra-clade B variation in HIV-1 CD4+ Th- cell epitope Env (484-496) variant peptides
HIV-1 HXBc2 Env (484-496) index sequence. All HIV-1 Env sequences were obtained from the Human Retrovirus and AIDS database (21).
Subscripts indicate the number of times a given substitution was observed in 92 HIV-1 clade B Env sequences.
TABLE 2.
Cross-clade variation in HIV-1 CD4+ Th- cell epitope Env (484-496) variant peptides
| Clade | Sequencea |
|---|---|
| HXBc2 | YKYKVVKIEPLGV |
| A | ------------- |
| B | ------------- |
| C | ------E-K---- |
| D | ------R------ |
| E | ------Q-----I |
| F | ------E------ |
| G | --------K---- |
| H | No consensus |
| O | F-----RVK-FS- |
| U | ------------- |
HIV-1 Env consensus sequences were obtained from the Human Retrovirus and AIDS database (21). Dashes indicate amino acid identity.
HIV-1 Env (484-496) variant peptides retain the ability to bind to the MHC class II molecule Mamu-DR*W201.
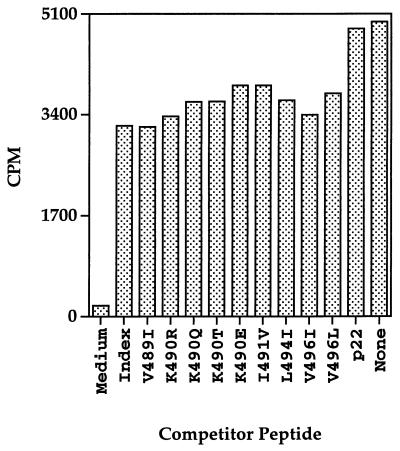
We also previously showed that recognition of both HIV-1 Env (482-501) and HIV-1 Env (172-191) peptides by CD4+ Th- cell lines is restricted by the same MHC class II DR molecule, Mamu-DR*W201 (17). It is conceivable that some Env (484-496) variant peptides may not bind to Mamu-DR*W201 and therefore may go unrecognized by HIV-1 HXBc2 Env (484-496)-specific, Mamu-DR*W201-restricted CD4+ Th cells. We used the Env (171-192)-specific, Mamu-DR*W201-restricted CD4+ Th- cell line 431.08 in a functional assay to determine whether any of the Env (484-496) variant peptides could compete with Env (171-192) peptide binding to a Mamu-DR*W201-expressing antigen-presenting cell. Specifically, 431.08 Th cells and an RM3 transfectant expressing Mamu-DR*W201 were incubated in the presence or absence of Env (484-496) variant peptides, as well as a suboptimal concentration of the Env (171-192) peptide. As expected, the HXBc2 Env (484-496) peptide, but not an irrelevant Env (241-262) peptide, inhibited the Env (171-192) peptide-driven proliferative response of 431.08 (Fig. 1). Interestingly, all of the Env (484-496) variant peptides tested inhibited Env (171-192) peptide-stimulated [3H]thymidine incorporation, suggesting that the corresponding amino acid substitutions within these peptides were not sufficient to abrogate peptide binding to Mamu-DR*W201.
FIG. 1.
Peptides corresponding to naturally occurring variants of the CD4+ Th- cell epitope Env (484-496) compete with the Env (171-192) peptide for binding to Mamu-DR*W201 molecules expressed by RM3-transfected antigen-presenting cells. Mamu-DR*W201-expressing RM3 cells were plated at 2 × 104 cells/ well in 96-well flat-bottom microtiter plates and incubated for 2 h with 25 μg of the Env (484-496) variant peptides or an irrelevant Env (241-262) peptide per ml. Approximately 2 × 104 431.08 CD4+ Th cells were then added to each well along with 0.5 μg of the Env (171-192) peptide per ml. Cultures were pulsed with 1 μCi of [3H]thymidine per well after 2 days and harvested on day 3.
HIV-1 Env (484-496) variant peptides with substitutions at position 490 fail to drive proliferation and cytokine secretion by two HXBc2-specific CD4+ Th- cell lines.
Proliferation assays were then done to determine whether the Env (484-496) variant peptides, which retained the ability to bind to the restricting MHC class II DR molecule, were able to stimulate HIV-1 HXBc2 Env (484-496)-specific CD4+ Th cells. As shown in Table 3, five of the nine Env (484-496) variant peptides tested stimulated the HXBc2-specific cell lines to proliferate. In contrast, the four Env (484-496) variant peptides bearing substitutions at position 490 did not drive [3H]thymidine incorporation above what was observed in the presence of medium alone. Moreover, only Env (484-496) variant peptides capable of stimulating a proliferative response were able to induce the expression of the high-affinity IL-2 receptor CD25 (data not shown).
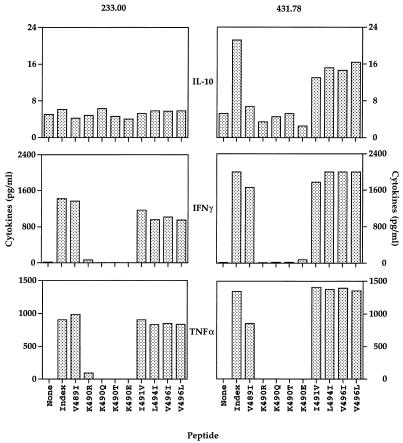
Since it has been shown in other systems that a peptide ligand can trigger cytokine secretion by a T-cell population in the absence of proliferation, the ability of the Env (484-496) variant peptides to induce cytokine production was measured (8). As shown in Fig. 2, only Env (484-496) variant peptides capable of stimulating [3H]thymidine incorporation were able to induce substantial gamma interferon and tumor necrosis factor alpha production. In addition, the CD4+ Th- cell line 431.78, which produces IL-10 in response to the HXBc2 index peptide, also did so in the presence of the I491V, L494I, V497I, and V496L peptides. This cell line, however, did not secrete IL-10 in the presence of Env (484-496) variant peptides bearing substitutions at position 489 or 490. Neither of the CD4+ Th- cell lines produced IL-4 or transforming growth factor β in response to either the HXBc2 index peptide or any of the nine Env (484-496) variant peptides examined (data not shown). Thus, variation at position 490 of the HIV-1 Env (484-496) epitope abolished rather than modulated cytokine secretion by both of the HXBc2-specific CD4+ Th- cell lines.
FIG. 2.
Peptides corresponding to naturally occurring variants of the CD4+ Th- cell epitope Env (484-496) bearing substitutions at amino acid position 490 fail to stimulate cytokine secretion by two CD4+ Th- cell lines. Approximately 106 CD4+ Th cells were cultured in the presence of an equivalent number of autologous B-LCL in 1 ml of medium in the presence or absence of Env (484-496) variant peptides. After 2 days, supernatants were harvested and frozen. Cytokine content was later measured with commercially available ELISA kits.
Exogenous costimulation fails to restore the ability of HIV-1 Env (484-496) variant peptides bearing substitutions at position 490 to stimulate HXBc2-specific CD4+ Th- cell lines.
It was formally possible that the K490R, K490Q, K490T, and K490E peptides, when bound to Mamu-DR*W201, partially induced Th- cell activation. To assess this possibility, the pharmacological agent PMA, the calcium ionophore ION, or the cytokine IL-1β was added to cultures in an attempt to complement any T-cell signaling ability of the Env (484-496) variant peptides bearing substitutions at amino acid 490. PMA, ION, and IL-1β, while able to enhance the proliferative responses of the two CD4+ Th- cell lines to stimulatory peptides, failed to induce substantial [3H]thymidine incorporation by these cell lines in response to the K490R, K490Q, K490T, and K490E peptides (data not shown). Thus, if the Env (484-496) variant peptides with substitutions at position 490 induce a partial activation signal, this signal is weak at best.
HIV-1 Env (484-496) variant peptides antagonize the HXBc2- specific proliferative responses of two CD4+ Th- cell lines.
It has been demonstrated in both murine and human model systems that altered peptide ligands can inhibit the ability of wild-type peptide ligands to activate CD4+ Th cells and CD8+ cytotoxic T lymphocytes (13, 15). A prepulse assay was used to determine whether the Env (484-496) variant peptides with substitutions at position 490 could antagonize the proliferative responses of the HXBc2-specific CD4+ Th cell lines driven by the wild-type peptide. Specifically, autologous B-LCL were prepulsed with a suboptimal concentration of the index peptide. The index peptide-pulsed antigen-presenting cells were then washed and pulsed with increasing concentrations of the Env (484-496) variant peptides for use in a [3H]thymidine incorporation assay with HXBc2-specific CD4+ Th cells. As demonstrated in Fig. 3, at high concentrations the K490R, K490Q, K490T, and K490E peptides antagonized the index peptide-driven proliferative responses of HXBc2-specific CD4+ Th cells. In contrast, another Env peptide whose recognition was restricted by Mamu-DR*W201 had no effect on the index peptide-specific proliferative responses.
FIG. 3.
HIV-1 Env (484-496) variant peptides with substitutions at position 490 antagonize the recognition of wild-type Env (484-496) as measured in a proliferation assay. Autologous B-LCL were prepulsed with 0.5 μg of the index HXBc2 Env (484-496) peptide per ml for 2 h to stimulate the CD4+ Th- cell lines to the levels indicated by the horizontal broken lines. Prepulsed B-LCL were then pulsed for an additional 2 h with the indicated concentrations of the index peptide, the Env (484-496) variant peptides, or the nonstimulatory, Mamu-DR*W201-binding Env (172-191) peptide p15. Results are expressed as [3H]thymidine incorporation in counts per minute. In the absence of peptides, the background proliferation of both Th- cell lines was less than 400 cpm.
DISCUSSION
Certain structural requirements for peptide binding to MHC class II molecules have been determined through the characterization of naturally processed peptides eluted from purified MHC class II complexes. As is the case for the MHC class II- associated invariant-chain peptide, the major anchor residues of the HLA-DR1, HLA-DR17, HLA-DR4, and H-2E ligands are located at relative positions 1, 4, 6, and 9 of the bound peptides (25, 32). This finding suggests that the amino acid residues of the HIV-1 Env nonamer YKVVKIEPL that are critical for binding to Mamu-DR*W201 correspond to Y486, V489, I491, and L494 (18). The only substitutions at these presumed anchor positions in naturally occurring HIV-1 isolates are conservative ones and have little or no effect on MHC class II binding and CD4+ Th- cell recognition in this system.
Position 5, on the other hand, is the primary TCR contact residue for two well-characterized H-2Ek-restricted CD4+ Th-cell epitopes (24). In fact, both a T-cell line and a T-cell hybridoma specific for a moth cytochrome c epitope having lysine at position 5 did not recognize any of the 19 possible amino acid substitutions introduced at this position into a moth cytochrome c peptide (27). HIV-1 Env peptides synthesized with each of the four naturally occurring amino acid substitutions for K490 at position 5 within the rhesus monkey CD4+ Th- cell epitope were unable to drive proliferation and cytokine secretion by two HIV-1 Env-specific Th- cell lines in the present study. The inability of peptides with unconservative substitutions (K490Q, K490T, and K490E) to drive T-cell proliferation and cytokine secretion was not surprising, particularly since these substitutions were only infrequently observed among HIV-1 clade B isolates. In contrast, the failure to tolerate the substitution of one basic residue for another, K490R, was remarkable. The K490R substitution has been observed in numerous HIV-1 clade B, clade D, and clade O viruses. The exquisite sensitivity of the two rhesus monkey CD4+ Th- cell lines to substitutions at residue 490 is consistent with the premise that lysine at position 5 is the primary TCR contact residue of the HIV-1 Env (484-496) epitope.
The induction of different patterns of cytokine secretion by CD4+ Th- cells following interactions with altered peptide ligands has been documented with both human multiple sclerosis and murine experimental autoimmune encephalomyelitis systems (22, 34). In light of the potential importance of the cytokine milieu in AIDS immunopathogenesis and the suggested harmful effects of Th2 cytokines in this setting, it was important to determine whether HIV-1 variation may cause a shift in Th- cell cytokine secretion from a Th1 to a Th2 profile (5). None of the HIV-1 Env (484-496) variant peptides tested, however, induced secretion of IL-4 or increased levels of IL-10 production by two HIV-1 HXBc2-specific rhesus monkey CD4+ Th1-like cell lines. Hence, no evidence of an altered peptide ligand-induced change in the CD4+ Th- cell cytokine secretion profile was found in this study.
The ability of empirically chosen altered peptide ligands to inhibit the activation of murine CD4+ T-cell populations specific for the model antigens hemoglobin β and moth cytochrome c has been convincingly demonstrated (24). The significance of these observations for cellular immune responses in virus-induced diseases, however, has been unclear. Interestingly, naturally occurring hepatitis B virus and HIV-1 variants which are capable of inhibiting the lysis of wild-type peptide-pulsed targets by CD8+ T-lymphocyte populations have been described (1, 16). In addition, wild-type peptide-induced activation of an HLA-DR1-restricted CD4+ Th- cell clone specific for influenza virus hemagglutinin can be hindered by several variant peptides (7). More recently, an altered peptide ligand corresponding to the sequence of an HIV-1 isolate obtained from an infected individual was shown to antagonize the cytolytic and proliferative responses of a vaccine-induced Env-specific human CD4+ Th2-like clone (14). The ease with which antagonist sequences were identified suggests that variant peptide antagonism of T-lymphocyte function may be a widespread phenomenon among virus-specific CD4+ Th cells as well as CD8+ cytotoxic T lymphocytes.
ACKNOWLEDGMENTS
We are grateful to R. E. Bachelder and M. E. Grigg for critically reading the manuscript. The HIV-1 Env (171-192), Env (241-262), and Env (481-502) peptides were provided by the AIDS Reagent Project of the Medical Research Council.
This work was supported by Public Health Service grants AI20729, AI35351, and CA50139.
REFERENCES
- 1.Bertoletti A, Sette A, Chisari F V, Penna A, Levrero M, DeCarli M, Fiaccadori F, Ferrari C. Natural variants of cytotoxic epitopes are T-cell receptor antagonists for antiviral cytotoxic T cells. Nature. 1994;369:407–410. doi: 10.1038/369407a0. [DOI] [PubMed] [Google Scholar]
- 2.Boehncke W-H, Takeshita T, Pendleton C D, Houghton R A, Sadegh-Nasseri S, Racioppi L, Berzofsky J A, Germain R N. The importance of dominant negative effects of amino acid side chain substitutions in peptide-MHC molecule interactions and T cell recognition. J Immunol. 1993;150:331–341. [PubMed] [Google Scholar]
- 3.Callahan K M, Fort M M, Obah E A, Reinherz E L, Siliciano R F. Genetic variability in HIV-1 gp120 affects interactions with HLA molecules and T cell receptor. J Immunol. 1990;144:3341–3346. [PubMed] [Google Scholar]
- 4.Calman A, Peterlin B. Mutant human B cell lines deficient in class II major histocompatibility complex transcription. J Immunol. 1987;139:2489–2495. [PubMed] [Google Scholar]
- 5.Clerici M, Shearer G M. A Th1-to-Th2 switch is a critical step in the etiology of HIV infection. Immunol Today. 1993;14:107–111. doi: 10.1016/0167-5699(93)90208-3. [DOI] [PubMed] [Google Scholar]
- 6.Davies M, Kaufman R. The sequence context of the initiation codon in the encephalomyocarditis virus leader modulates the efficiency of internal translation initiation. J Virol. 1992;66:1924–1932. doi: 10.1128/jvi.66.4.1924-1932.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DeMagistris M T, Alexander J, Coggeshall M, Altman A, Gaeta F C A, Grey H M, Sette A. Antigen analog-major histocompatibility complexes act as antagonists of the T cell receptor. Cell. 1992;68:625–634. doi: 10.1016/0092-8674(92)90139-4. [DOI] [PubMed] [Google Scholar]
- 8.Evagold B D, Allen P M. Separation of IL-4 production from Th cell proliferation by an altered T cell receptor ligand. Science. 1991;252:1308–1310. [PubMed] [Google Scholar]
- 9.Fernandez M H, Faith A, Higgins J A, Weber J, Rees A D M. The effect of a single amino acid substitution within the V3 loop of HIV-1 gp120 on HLA-DR1-restricted CD4 T-cell recognition. Immunology. 1995;85:176–183. [PMC free article] [PubMed] [Google Scholar]
- 10.Fernandez M H, Fidler S J, Pitman R J, Weber J N, Rees A D M. CD4+ T-cell recognition of diverse clade B HIV-1 isolates. AIDS. 1997;11:281–288. doi: 10.1097/00002030-199703110-00004. [DOI] [PubMed] [Google Scholar]
- 11.Fisher A G, Ensoli B, Looney D, Rose A, Gallo R C, Saag M S, Shaw G M, Hahn B H, Wong-Staal F. Biologically diverse molecular variants within a single HIV-1 isolate. Nature. 1988;334:444–447. doi: 10.1038/334444a0. [DOI] [PubMed] [Google Scholar]
- 12.Hahn B H, Shaw G M, Taylor M E, Redfield R R, Markham P D, Salahuddin S Z, Wong-Staal F, Gallo R C, Parks E S, Parks W P. Genetic variation in HTLV-III/LAV over time in patients with AIDS or at risk for AIDS. Science. 1986;232:1548–1553. doi: 10.1126/science.3012778. [DOI] [PubMed] [Google Scholar]
- 13.Jameson S C, Bevan M J. T cell receptor antagonists and partial agonists. Immunity. 1995;2:1–11. doi: 10.1016/1074-7613(95)90074-8. [DOI] [PubMed] [Google Scholar]
- 14.Kent S J, Greenberg P D, Hoffman M C, Akridge R E, McElrath M J. Antagonism of vaccine-induced HIV-1-specific CD4+ T cells by primary HIV-1 infection. J Immunol. 1997;158:807–815. [PubMed] [Google Scholar]
- 15.Kersh G J, Allen P M. Essential flexibility in the T-cell recognition of antigen. Nature. 1996;380:495–498. doi: 10.1038/380495a0. [DOI] [PubMed] [Google Scholar]
- 16.Klenerman P, Rowland-Jones S, McAdam S, Edwards J, Daenke S, Lalloo D, Koppe B, Rosenberg W, Boyd D, Edwards A, Giangrande P, Phillips R E, McMichael A J. Cytotoxic T-cell activity antagonized by naturally occurring HIV-1 Gag variants. Nature. 1994;369:403–407. doi: 10.1038/369403a0. [DOI] [PubMed] [Google Scholar]
- 17.Lekutis C, Letvin N L. HIV-1 envelope-specific CD4+ T helper cells from simian/human immunodeficiency virus-infected rhesus monkeys recognize epitopes restricted by MHC class II DRB1*0406 and DRB*W201 molecules. J Immunol. 1997;159:2049–2057. [PubMed] [Google Scholar]
- 18.Lekutis C, Shiver J W, Liu M A, Letvin N L. HIV-1 env DNA vaccine administered to rhesus monkeys elicits MHC class II- restricted CD4+ T helper cells that secrete IFNγ and TNFα. J Immunol. 1997;158:4471–4477. [PubMed] [Google Scholar]
- 19.Li J, Lord C I, Haseltine W, Letvin N L, Sodroski J. Infection of cynomolgus monkeys with a chimeric HIV-1/SIVmac virus that expresses the HIV-1 envelope glycoprotein. J Acquired Immune Defic Syndr. 1992;5:639–646. [PubMed] [Google Scholar]
- 20.Manca F, Habeshaw J A, Dalgleish A G, Fenoglio D, Pira G L, Sercarz E E. Role of flanking variable sequences in antigenicity of consensus regions of HIV gp120 for recognition by specific human T helper clones. Eur J Immunol. 1993;23:269–274. doi: 10.1002/eji.1830230142. [DOI] [PubMed] [Google Scholar]
- 21.Myers G, Korber B, Foley B, Jeang K-T, Mellors J W, Wain-Hobson S. Human retrovirus and AIDS. Los Alamos, N.Mex: Los Alamos National Laboratory; 1996. [Google Scholar]
- 22.Nicholson L B, Greer J M, Sobel R A, Lees M B, Kuchroo V K. An altered peptide ligand mediates immune deviation and prevents autoimmune encephalomylelitis. Immunity. 1995;3:397–405. doi: 10.1016/1074-7613(95)90169-8. [DOI] [PubMed] [Google Scholar]
- 23.Rabin H, Neubauer B H, Hopkins III R F, Dzhikidze E K, Shevtsova Z V, Lapin B A. Transforming activity and antigenicity of an Epstein-Barr-like virus from lymphoblastoid cell lines of baboons with lymphoid disease. Intervirology. 1977;8:240–249. doi: 10.1159/000148899. [DOI] [PubMed] [Google Scholar]
- 24.Rabinowitz J D, Beeson C, Wulfing C, Tate K, Allen P M, Davis M M, McConnell H M. Altered T cell receptor ligands trigger a subset of early T cell signals. Immunity. 1996;5:125–135. doi: 10.1016/s1074-7613(00)80489-6. [DOI] [PubMed] [Google Scholar]
- 25.Rammensee H-G, Friede T, Stevanovic S. MHC ligands and peptide motifs: first listing. Immunogenetics. 1995;41:178–228. doi: 10.1007/BF00172063. [DOI] [PubMed] [Google Scholar]
- 26.Ratto S, Sitz K V, Scherer A M, Loomis L D, Cox J H, Redfield R R, Birx D L. CD4+ T-lymphocyte lines developed from HIV-1-seropositive patients recognize different epitopes within the V3 loop. J Acquired Immune Defic Syndr. 1996;11:128–136. doi: 10.1097/00042560-199602010-00003. [DOI] [PubMed] [Google Scholar]
- 27.Reay P A, Kantor R M, Davis M M. Use of global amino acid replacements to define the requirements for MHC binding and T cell recognition of moth cytochrome c (93-103) J Immunol. 1994;152:3946–3957. [PubMed] [Google Scholar]
- 28.Rosenberg E S, Billingsley J M, Caliendo A M, Boswell S L, Sax P E, Kalams S A, Walker B D. Vigorous HIV-1-specific CD4+ T cell responses associated with control of viremia. Science. 1997;278:1447–1450. doi: 10.1126/science.278.5342.1447. [DOI] [PubMed] [Google Scholar]
- 29.Saag M S, Hahn B H, Gibbons J, Li Y X, Parks E S, Parks W P, Shaw G M. Extensive variation of human immunodeficiency virus type-1 in vivo. Nature. 1988;334:440–444. doi: 10.1038/334440a0. [DOI] [PubMed] [Google Scholar]
- 30.Sakuragi S, Shibata R, Mukai R, Komatsu T, Fukasawa M, Sakai H, Sakuragi J-I, Kawamura M, Ibuki K, Hayami M, Adachi A. Infection of macaque monkeys with a chimeric human and simian immunodeficiency virus. J Gen Virol. 1992;73:2983–2987. doi: 10.1099/0022-1317-73-11-2983. [DOI] [PubMed] [Google Scholar]
- 31.Siliciano R F, Lawton T, Knall C, Karr R W, Berman P, Gregory T, Reinherz E L. Analysis of host-virus interactions in AIDS with anti-gp120 T cell clones: effect of HIV sequence variation and a mechanism for CD4+ cell depletion. Cell. 1988;54:561–575. doi: 10.1016/0092-8674(88)90078-5. [DOI] [PubMed] [Google Scholar]
- 32.Sinigaglia F, Hammer J. Motifs and supermotifs for MHC class II binding peptides. J Exp Med. 1995;181:449–451. doi: 10.1084/jem.181.2.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsitoura D C, Verhoef A, Gelder C M, O’Hehir R E, Lamb J R. Altered T cell ligands derived from a major house dust mite allergen enhance IFNγ but not IL-4 production by human CD4+ T cells. J Immunol. 1996;157:2160–2165. [PubMed] [Google Scholar]
- 34.Windhagen A, Scholz C, Hollsberg P, Fukaura H, Sette A, Hafler D A. Modulation of cytokine patterns of human autoreactive T cell clones by a single amino acid substitution of their peptide ligand. Immunity. 1995;2:373–380. doi: 10.1016/1074-7613(95)90145-0. [DOI] [PubMed] [Google Scholar]