Abstract
Tomato bushy stunt virus (TBSV) is a plus-sense RNA virus which encodes a 33-kDa protein in its 5′-most open reading frame (ORF). Readthrough of the amber stop codon of the p33 ORF results in the production of a 92-kDa fusion protein. Both of these products are expressed directly from the viral genome and are suspected to be involved in viral RNA replication. We have investigated further the roles of these proteins in the amplification of viral RNAs by using a complementation system in which p33 and p92 are expressed from different viral RNAs. Our results indicate that (i) both of these proteins are necessary for viral RNA amplification; (ii) translation of these proteins can be uncoupled while maintaining amplification of viral RNAs; (iii) if compatibility requirements exist between p33 and p92, they are not exceptionally strict; and (iv) the C-terminal ∼6% of p33 is necessary for its functional activity. Interestingly, no complementation was observed when a p33-encoding replicon containing a deletion of a 3′-located segment, region 3.5, was tested. However, when 5′-capped transcripts of the same replicon were analyzed, complementation allowing for RNA amplification was observed. This ability to compensate functionally for the absence of region 3.5 by the addition of a 5′ cap suggests that this RNA segment may act as a translational enhancer for the expression of virally encoded products.
Plant RNA viruses have various genome organizations and use a wide array of gene expression strategies (4, 14, 30). Despite such differences, these viruses are unified by the fact that they all encode components of an RNA-dependent RNA polymerase activity involved in the replication of their genomes. Conserved motifs have been identified within these viral proteins, which have been implicated in specific functions in genome replication such as polymerase, helicase, and/or RNA-capping activities (5, 12, 17). These activities may be combined on a single protein or divided among two or more products, which, in turn, may be encoded in different RNA segments in multipartite genomes (30).
A common strategy used by several RNA plant virus genera (including Tobamovirus, Furovirus, Tobravirus, Necrovirus, Tombusvirus, and Carmovirus) is to encode their two protein replication components in the 5′-most open reading frame (ORF) and in a readthrough product of that ORF (30). Both of these products are translated directly from the viral genome, but the amount of readthrough product is generally orders of magnitude lower than that of the prereadthrough product (13, 22, 29), due to relatively inefficient suppression of the termination codon (23, 24). For tobacco mosaic virus, the 183-kDa readthrough product alone is sufficient for viral RNA replication but the viral RNA accumulation levels are significantly reduced compared to when both the pre- and readthrough products are present (7). This is in contrast to the Carmovirus turnip crinkle virus (TCV), where both the p28 prereadthrough and p88 readthrough products are required (29). For members of the genus Tombusvirus (1, 6, 16, 18, 25), there is some evidence suggesting a similar requirement for both products (1, 22).
The genome of tomato bushy stunt virus (TBSV, the prototype of the genus Tombusvirus) is a monopartite, positive-sense RNA of 4.7 kb (6). It is not polyadenylated, and it contains five functional ORFs (Fig. 1) (6). The genome is not thought to be 5′ capped, since that of the closely related carnation Italian ringspot Tombusvirus has been shown not to contain such a structure (18). The 5′-most ORF in the TBSV genome encodes a 33-kDa protein, which has been proposed to function in viral RNA replication (22); however, its amino acid sequence does not contain any known motifs that would define it as such. Readthrough of the amber stop codon of the p33 ORF results in the translation of a 92-kDa fusion protein, the readthrough portion of which contains highly conserved motifs found in catalytic subunits of RNA-dependent RNA polymerases (6, 12). The coat protein, a 41-kDa product, is encoded by ORF3, and the p22 and p19 movement/symptom-related products are encoded by the two 3′-most ORFs, which overlap in different reading frames (20, 21). The products of ORF3 through ORF5 are translated from two subgenomic (sg) mRNAs, which are synthesized during infections (6).
FIG. 1.
Schematic representation of TBSV genome and various defective viral RNAs. The wt TBSV genome (T-100) is shown at the top as a thick horizontal line, with coding regions depicted as boxes which include the approximate molecular weights (in thousands) of the encoded proteins (6). The translation product p33 and its readthrough product p92 are indicated as arrows below the genome, and the approximate position of region 3.5 is shown as a small black box near the 3′ end of the genome. HS-175 is a mutant of the genome in which the p33 termination codon has been replaced by a tyrosine codon to allow the expression of p92 (arrow) but not p33 (22). Various smaller defective viral RNAs are depicted in which shaded boxes correspond to regions of the genome retained in these molecules whereas thin horizontal lines correspond to genomic segments which are absent. DI-82 and DI-83 both encode p33 (open box) and are identical, except that region 3.5 is deleted from DI-82 (28). DI-83ΔMfeI is structurally similar to DI-83 but encodes a C-terminally truncated form of p33 (open box), which contains a C-terminal extension of 18 aa from a different reading frame (adjoining black box). DI-72 and DI-73 are composed of four and three noncontiguous regions, respectively (i.e., regions I through IV and I through III/IV, respectively), encode no functional proteins, and are identical except that region 3.5 is absent in DI-72 (28).
The TBSV p33 and p92 products are translated directly from the genome, and the ratio of their accumulation levels in vivo has been estimated at 20:1 (22). The precise efficiency of readthrough of the amber stop codon of p33 in TBSV is unknown, but in another member of the family Tombusviridae (i.e., TCV), readthrough occurs in vivo with an efficiency of approximately 1% (29). It is unclear how efficient translation of the p33 product (and to a lesser extent p92) is accomplished, since the TBSV genome does not appear to contain structures known to enhance translation in eukaryotic cells [i.e., a 5′ cap structure and a 3′ poly(A) tail]. It is possible, therefore, that the genome contains other structures whose role is to enhance translation; however, such elements have not yet been identified. A nonconventional translational enhancer element has been described in barley yellow dwarf virus (BYDV-PAV) (26). BYDV-PAV is a Luteovirus (subgroup I) which contains a plus-strand RNA genome which lacks a 5′ cap structure and poly(A) tail. Similar to the other viral genomes discussed, BYDV-PAV encodes two RNA replication-related products at the 5′ end of its genome; however, its corresponding fusion product is generated via a −1 frameshift mechanism rather than by readthrough (15). Based on amino acid comparisons of virus-encoded polymerase components, tombusviruses and luteoviruses (subgroup I) are related and have been assigned to the same supergroup of viruses (supergroup II) (3). Wang et al. (26) have shown that for BYDV-PAV the expression of the 5′ ORF is stimulated by an RNA segment located near the 3′ terminus. This stimulatory activity was lost when mutations were introduced into this 3′-translational enhancer (3′TE), but efficient translation was recovered by the addition of a 5′ guanosine cap but not by the addition of a poly(A) tail (26). Although TBSV has similar structural and genetic elements to BYDV-PAV, the 3′TE, which has also been identified in the genomes of both the necroviruses and the dianthoviruses, is not present in the TBSV genome (26). The mechanism(s) by which the p33 and p92 products of TBSV are expressed efficiently in vivo from a genome which lacks traditional translational enhancement elements is therefore unknown.
From previous studies, roles for p33 and p92 in TBSV genome replication have been inferred (22); however the evidence was not unequivocal. In the present study, we have further investigated the function of p33 and p92 and provide more definitive evidence that both products are essential for viral RNA amplification. In addition, we have identified a 3′ RNA segment within the TBSV genome which has properties that are consistent with a potential role as a translational enhancer. Finally, our data provide novel information on important cis-acting sequences within viral RNAs as well as on functionally relevant regions of p33.
MATERIALS AND METHODS
Viral and DI RNA constructs.
Plasmid constructs T-100 and K2/M5, containing cDNAs corresponding to the full-length viral genomes of TBSV and cucumber necrosis virus (CNV), respectively, have been described previously (6, 16). HS-175 is a mutant of T-100 in which the amber stop codon for the p33 ORF has been mutated to a tyrosine codon via a single-base substitution (22). Defective interfering (DI) RNA clones DI-82 and DI-83, both of which encode p33, have been described previously, as have DI-73 and DI-72, which represent two small prototypical TBSV DI RNAs (10, 28).
83ΔMfeI was constructed by digesting DI-83 with MfeI (at position 262 of the TBSV genomic sequence [6]) followed by filling in of the 3′-recessed ends with T4 DNA polymerase and then self-ligation. The resulting construct, DI-83ΔMfeI, contained a 4-nucleotide insertion at the MfeI site, causing a frameshift in the p33 ORF. For construction of DI-83M1, DI-83 was digested with BspEI (position 993) followed by filling in of the 3′-recessed ends with T4 DNA polymerase and then self-ligation. The resulting construct, DI-83M1, contained a 4-nucleotide insertion at the BspEI site, causing a frameshift in the p33 ORF. To create DI-83M2, DI-83 was digested with BspEI and StuI (position 1059) and the 5′ protruding end generated by BspEI was made blunt by filling in with T4 DNA polymerase followed by self-ligation of the gel-purified larger fragment. This created a construct, DI-83M2, that contained a 64-nucleotide deletion of residues 997 to 1060, which resulted in a frameshift. To construct DI-83M3, DI-83 was digested with Tth111I (position 928) and StuI and the 5′ overhang of Tth111I was filled in with T4 DNA polymerase. The larger fragment was gel purified and then self-ligated. The resulting construct, DI-83M3, contained a somewhat larger than expected deletion of 177 nucleotides, corresponding to positions 899 to 1075 and resulting in a shift in the reading frame. DI-83CNV was constructed in the following manner. K2/M5 was digested with StuI (position 1045) (16) and SphI (within the vector, 3′ to the CNV cDNA insert), and the larger vector/5′-CNV sequence-containing fragment was gel purified and ligated to a gel-purified StuI-SphI fragment (position 1059 to a more 3′ position within the vector) derived from digestion of DI-83. This generated a CNV/DI-83 hybrid molecule which contained the 5′ end of the CNV genome (which encodes p33 of CNV) fused to the 3′ portion of DI-83.
In vitro transcription.
Viral transcripts were generated in vitro via transcription of SmaI-linearized template DNAs with the Ampliscribe T7 RNA polymerase transcription kit (Epicentre Technologies). Following the transcription reaction, DNA templates were removed by treatment with DNase I (Epicentre Technologies) and unincorporated nucleotides were removed via column chromatography with a Sephadex G-25 spin column (Pharmacia). Ammonium acetate was added to the flowthrough to a final concentration of 2 M, and the transcripts were extracted twice with equal volumes of phenol-chloroform-isoamyl alcohol and then precipitated with ethanol. Subsequently, the transcripts were quantified spectrophotometrically and an aliquot was analyzed by agarose gel electrophoresis to verify integrity. Capped transcripts were prepared with the Ampliscribe T7 RNA polymerase transcription kit (Epicentre Technologies) by using the recommended 10:1 ratio of cap analog [m7G(5′)ppp(5′)G; New England Biolabs] to GTP. Under these conditions, the manufacturer estimates an average capping efficiency of 50%.
Isolation and inoculation of protoplasts.
Protoplasts were prepared from 6- to 8-day-old cucumber cotyledons (var. Straight 8) as described previously (8). Briefly, the lower epidermis of the cotyledons was peeled off with forceps and the cotyledons were digested in 20 ml of an enzyme mix (0.25 g of cellulase [Calbiochem], 0.025 g of pectinase [ICN], 0.025 g of bovine serum albumin [ICN]) for 3 to 5 h with gentle shaking (40 rpm) in the dark. The protoplasts were then washed in 10% mannitol and purified by banding twice on a 20% sucrose cushion. Quantification was carried out by bright-field microscopy with a hemacytometer. Purified protoplasts (approximately 3 × 105) were inoculated as described previously (8) with viral RNA transcripts (1 μg for DI-72 or DI-73; 5 μg for the others) and were incubated in a growth chamber under fluorescent lighting at 22°C for 24 h.
Analysis of viral RNAs.
Total nucleic acid was harvested from protoplasts 24 h postinoculation by resuspension in 300 μl of a buffer containing 2× STE (28) and 1% sodium dodecyl sulfate. After two extractions with phenol-chloroform-isoamyl alcohol, 100 μl of 8 M ammonium acetate was added to the aqueous phase and the mixture was precipitated with ethanol. Aliquots (1/10) of the total nucleic acid preparation were separated in nondenaturing 1.4% agarose gels. Viral RNAs were detected by electrophoretic transfer to nylon (Hybond-N; Amersham) followed by Northern blot analysis with a 32P-end-labeled oligonucleotide (P9) probe complementary to the 3′-terminal 23 nucleotides of the TBSV genome.
RESULTS
Viral RNA amplification via complementation of p33 and p92.
It was suggested previously that the gene products of both ORF1 and ORF2 are required for replication of the TBSV genome (22). This conclusion was based on the observation that mutant genomes which allow for the expression of either p33 or p92 do not accumulate when tested individually. HS-175 is a mutant TBSV genome that encodes only p92 due to the substitution of a tyrosine codon for the amber stop codon of the p33 ORF. Previously, it was observed that HS-175 does not accumulate when inoculated into protoplasts, and it was suggested that this result was likely a consequence of a defect in replication resulting from the absence of p33 (22). There are, however, equally plausible alternative explanations for this observation. For example, the genome modification introducing the tyrosine codon could cause (i) the production of a nonfunctional p92, (ii) the destabilization of the genome, (iii) negative effects on translation, and/or (iv) the disruption of a promoter sequence.
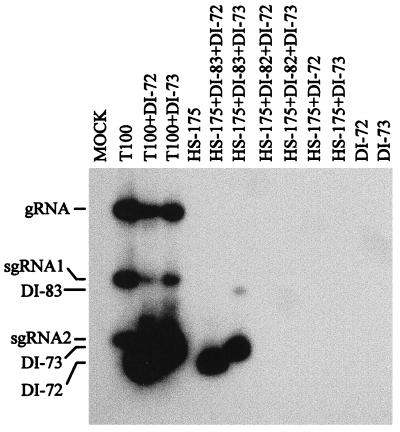
In order to determine more definitively whether both p33 and p92 are required for genome amplification, we tested the ability of mutant genomes, expressing either p33 or p92, to complement one another. Transcripts of the nonviable TBSV mutant HS-175 (Fig. 1), encoding p92, were coinoculated with either of the independently nonreplicable defective RNAs DI-82 and DI-83 (Fig. 1), both of which encode p33. Structurally, these two defective RNAs differ only in their 3′ regions, where a 167-nucleotide RNA segment, designated region 3.5, is present in DI-83 but is absent in DI-82. Protoplasts were coinoculated with various combinations of these viral transcripts, total nucleic acids were isolated 24 h postinoculation, and viral RNAs were detected by Northern blotting (Fig. 2). The inoculation with transcripts of wild-type (wt) TBSV genome (T-100) resulted in the accumulation of gRNA and its two sg mRNAs (Fig. 2). Individual inoculations of HS-175, DI-83, or DI-82 resulted in no detectable viral RNA accumulation. Coinoculation of either DI-82 or DI-83 with T-100 resulted in significant accumulation of defective RNA progeny, indicating that both of these molecules can be amplified efficiently in trans. When both DI-82 and DI-83 were coinoculated with T-100, DI-82 clearly dominated. The HS-175/DI-83 coinoculation, but not the HS-175/DI-82 coinoculation, led to significant accumulation of viral RNA (Fig. 2). In the HS-175/DI-83 coinoculation, only DI-83 accumulated to readily detectable levels; however, very small amounts of HS-175 were detectable after longer exposures of the blots (data not shown). Interestingly, DI-82 accumulation clearly dominated over that of DI-83 in the HS-175/DI-82/DI-83 coinoculation, similar to that observed in the T-100/DI-82/DI-83 coinoculation. It has been suggested previously that such differences in competitiveness are related primarily to replication competence (28). The observation that viral RNA amplification occurred only when the inocula contained both p33- and p92-encoding mutants is consistent with the concept that complementation occurred and that both products are necessary for the productive viral RNA amplification. Furthermore, these data indicate that expression of functional p33 and p92 can be uncoupled. Sequence analysis of reverse transcription-PCR products of the readthrough region, which were amplified from progeny HS-175 isolated from several independent coinfections, showed that the tyrosine codon was maintained in these molecules, thus indicating that reversion to wt had not occurred at significant levels (data not shown).
FIG. 2.
Northern blot analysis of progeny viral RNAs isolated from cucumber protoplasts inoculated with various combinations of viral RNA transcripts. The RNA transcripts used in the inoculations are indicated at the top, and the positions of the genome (gRNA), sg mRNAs (sgRNA1 and sgRNA2), and defective RNAs (DI-82 and DI-83) are shown at the left. Total nucleic acids were isolated from approximately 3 × 105 protoplasts after a 24-h incubation, separated in nondenaturing 1.4% agarose gels, transferred to a nylon membrane, and hybridized with a 32P-labeled oligonucleotide probe complementary to the 3′-terminal 23 nucleotides of the TBSV genome.
To verify further that clone HS-175 was incapable of producing any p33, either by proteolytic cleavage of p92 or by premature termination of translation, and that functional p33 was indeed being provided by DI-83, a mutant of DI-83 was constructed in which p33 was inactivated. The mutant, DI-83ΔMfeI, contains a frameshift early in the p33 ORF, which would severely truncate the product (Fig. 1). This mutant is predicted to encode only the N-terminal 33 amino acids (aa; plus an additional 18 non-p33 C-terminal residues) of the normally 296-aa p33. When DI-83ΔMfeI was coinoculated with HS-175, no viral RNA accumulation was detected (Fig. 3), even after longer exposures (data not shown). Coinoculation of HS-175/DI-83 demonstrated that the HS-175 transcripts used in HS-175/DI-83ΔMfeI coinoculations were biologically active and capable of complementation. Since 83ΔMfeI RNA accumulation was observed when 83ΔMfeI was coinoculated with T-100 (Fig. 3), it seems likely that the lack of amplification of DI-83ΔMfeI in coinfections with HS-175 is due to the absence of functional p33 rather than to a defect in a cis promoter element. This finding further supports the concept that p33 is required for viral RNA amplification and that in coinoculations with HS-175, functional p33 is being provided by DI-83.
FIG. 3.
Northern blot analysis of progeny viral RNAs isolated from cucumber protoplasts inoculated with various combinations of viral RNA transcripts. The RNA transcripts used in the inoculations are indicated at the top, and the positions of the genome (gRNA), sg mRNAs (sgRNA1 and sgRNA2), and defective RNAs (DI-83) are shown at the left. The position of DI-83ΔMfeI is the same as that indicated for DI-83. Total nucleic acids were isolated and analyzed as described in the legend to Fig. 2.
The activity induced by complementation can amplify DI-72 or DI-73.
To investigate further the properties of the amplification activity in HS-175/DI-83 coinoculations, we tested whether it could act on other amplifiable viral RNAs. DI-72 and DI-73 (Fig. 1) are two small DI viral RNAs which, when coinoculated with wt genomes, are amplified very efficiently (28). Assessment of the levels to which DI-72 and DI-73 accumulate in coinoculations provides a very sensitive means of monitoring replicase activity. Various combinations of viral RNA transcripts were inoculated into protoplasts, and the levels of viral RNA accumulation were determined. Either DI-72 or DI-73 accumulated very efficiently when coinoculated with T-100; however, both notably suppressed the accumulation of genomic and subgenomic RNAs (Fig. 4), as observed previously (28). These small DI RNAs also accumulated well when coinoculated with HS-175/DI-83 and showed a corresponding suppression of DI-83 accumulation (Fig. 4). The efficient accumulation of DI-72 and DI-73 indicates that the amplification activity induced by complementation is not only able to act on replicons which encode p33 (i.e., DI-82 and DI-83) but is also able to work efficiently in trans on other viral RNA templates. No accumulation of either DI-72 or DI-73 was observed when either was coinoculated with HS-175 or with HS-175/DI-82, further supporting the concept that HS-175, alone or in combination with DI-82, is incapable of directing viral RNA amplification.
FIG. 4.
Northern blot analysis of progeny viral RNAs isolated from cucumber protoplasts inoculated with various combinations of viral RNA transcripts. The RNA transcripts used in the inoculations are indicated at the top, and the positions of the genome (gRNA), sg mRNAs (sgRNA1 and sgRNA2), and various defective RNAs are shown at the left. Total nucleic acids were isolated and analyzed as described in the legend to Fig. 2.
Coinoculation of HS-175 and capped DI-82 allow for viral RNA amplification.
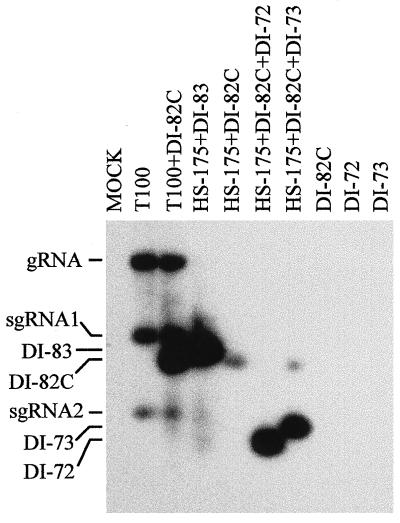
Our results indicated that DI-83, but not DI-82, was able to complement HS-175 for viral RNA amplification. DI-82 is identical to DI-83 except that it lacks a 3′ segment, region 3.5, which is present in DI-83. The apparent defect in DI-82 must therefore be related to the absence of this RNA segment. One possible role for region 3.5 in viral RNA amplification is to act as a translational enhancer for p33 production. To investigate this possibility, we synthesized 5′-capped transcripts of DI-82 and tested whether this modification (predicted to increase the translation efficiency of p33) would lead to productive viral RNA amplification. When capped DI-82 RNA transcripts (DI-82C) were coinoculated with HS-175, accumulation of DI-82C progeny was observed; however it was significantly lower than that observed for DI-83 in the HS-175/DI-83 coinoculation (Fig. 5). Efficient accumulation of DI-72 and DI-73 was also observed when they were coinoculated individually with HS-175/DI-82C (Fig. 5). These results show that the inability of DI-82 to complement HS-175 for viral RNA amplification can be partially compensated for by the presence of a 5′ cap structure. In addition, these data suggest that the p33 encoded in DI-82 is functional; therefore, the previously observed inability of uncapped DI-82 to complement HS-175 could not have been because it encoded a defective p33 ORF.
FIG. 5.
Northern blot analysis of progeny viral RNAs isolated from cucumber protoplasts inoculated with various combinations of viral RNA transcripts. The RNA transcripts used in the inoculations are indicated at the top (with capped transcripts indicated by the suffix C), and the positions of the genome (gRNA), sg mRNAs (sgRNA1 and sgRNA2), and various defective RNAs are shown at the left. Total nucleic acids were isolated and analyzed as described in the legend to Fig. 2.
Functional analysis of mutant forms of p33.
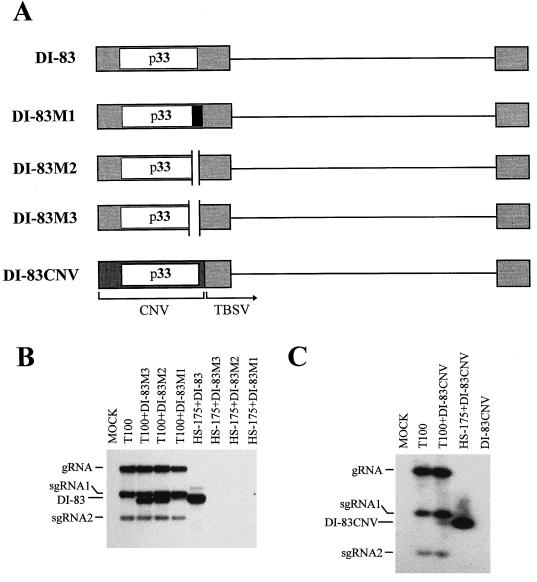
To determine the functional importance of the highly conserved C-terminal portion of p33 (6), several mutations were introduced into DI-83, which modified the encoded p33 ORF (Fig. 6A). The mutant transcripts were then coinoculated with either T-100 or HS-175 to determine if they were replicable and/or if they could complement to allow for viral RNA amplification, respectively. DI-83M1 has a 4-nucleotide insertion at position 997, which creates a frameshift, resulting in the replacement of the C-terminal 19 aa of the p33 ORF with 40 residues from an alternate reading frame. The mutant DI-83M2 contains a deletion of 64 nucleotides, corresponding to positions 997 to 1060, which truncates the C terminus of the p33 ORF by 19 aa and adds 1 residue (Ala) from an alternate reading frame. Mutant DI-83M3 has a 177-nucleotide deletion (positions 899 to 1075), which leads to removal of 52 aa from the C terminus of the p33 ORF and adds 3 residues (Leu-Gly-Leu) from another reading frame.
FIG. 6.
(A) Schematic representation of wt and mutant forms of DI-83. DI-83M1 has a 4-nucleotide insertion at position 997, which creates a frameshift, resulting in the replacement of the C-terminal 19 aa of p33 (open box) with 40 residues from an alternate reading frame (adjoining black box). DI-83M2 contains a deletion (indicated by a gap) of 64 nucleotides corresponding to positions 997 to 1060, which truncates the C terminus of p33 by 19 aa and adds 1 residue (Ala) from an alternate reading frame. DI-83M3 has a 177-nucleotide deletion (indicated by a gap; positions 899 to 1075), which leads to removal of 52 aa from the C terminus of p33 and adds 3 residues (Leu-Gly-Leu) from another reading frame. DI-83CNV has the 5′-terminal region of DI-83 replaced by the corresponding region from the CNV genome (darker shading) and encodes the CNV p33. (B) Northern blot analysis of mutant forms of DI-83. The RNA transcripts used in the inoculations are indicated at the top, and the positions of the genome (gRNA), sg mRNAs (sgRNA1 and sgRNA2), and defective RNAs (DI-83) are shown at the left. The positions of the mutant defective RNAs (M1, M2, and M3) are approximately the same as, or slightly lower than, that indicated for DI-83. (C) Northern blot analysis of DI-83CNV. The transcripts used in the inoculations are indicated at the top, and the positions of the genome (gRNA), sgmRNAs (sgRNA1 and sgRNA2), and defective RNAs (DI-83CNV) are shown at the left. For both panels B and C, total nucleic acids were isolated and analyzed as described in the legend to Fig. 2.
When transcripts corresponding to each of these three mutants were coinoculated individually with T-100, each accumulated to detectable levels, providing evidence that all were replicable (Fig. 6B). In coinoculations with T-100, DI-83M1 consistently accumulated to significantly lower levels than did either DI-82M2 or DI-83M3, suggesting that the modification to DI-83M1 did notably affect its viability. In coinoculations with HS-175, no viral RNA accumulation was detected for any of the mutants (Fig. 6B). Since these mutants were trans-amplifiable with wt helper (albeit DI-83M1 at reduced levels), the results suggest that the defect in amplification is related to the predicted C-terminal modifications in the encoded p33 ORF and that the C-terminal region of p33 is important for its functional activity.
To examine whether a high degree of compatibility is required between p33 and p92, a mutant of DI-83, DI-83CNV (Fig. 6A), was constructed in which the 5′ portion of DI-83 was replaced by the corresponding region from the genome of CNV (a closely related Tombusvirus). DI-83CNV thus encodes the CNV p33 homolog in place of the TBSV p33 (Fig. 6A). The p33 of CNV is 89.2% identical to the p33 of TBSV at the amino acid level (6). To determine whether these differences would affect viral RNA amplification, coinoculations of HS-175 and DI-83CNV were analyzed. These coinoculations resulted in the efficient accumulation of DI-83CNV progeny, indicating that the CNV p33 is compatible with the TBSV p92 (Fig. 6C).
DISCUSSION
In this study, we have investigated the roles of p33 and p92 in the accumulation of TBSV RNAs. Our results suggest that both of these proteins are essential for viral RNA accumulation, a finding consistent with their probable roles in viral RNA replication. The data also provide valuable information on important cis-acting sequences within viral RNAs as well as on functionally relevant regions of p33.
Both p33 and p92 are required for viral RNA amplification.
By first uncoupling the expression of functional p33 and p92 to generate independently nonviable RNAs and then using complementation to restore viability, we have shown that both of these proteins are likely required for amplification of the viral RNA. This conclusion is supported further by the observations that (i) the mutant DI-83ΔMfeI, containing an extensively truncated p33 ORF, could not complement HS-175 yet was able to replicate in trans with wt helper; (ii) no amplification was observed when HS-175 was coinoculated with the normally efficiently replicating DI-72 or DI-73; and (iii) reverse transcription-PCR analysis of the readthrough portion of HS-175 indicated that no high levels of reversion of the tyrosine codon had occurred in HS-175. These results indicate that the p92 expressed from HS-175 is functional and that p33 must be provided separately. The data thus support the concept that both products are required for viral RNA replication and that there is no strict prerequisite for their coupled expression.
The very low level of accumulation of HS-175 in HS-175/DI-83 coinoculations indicated that the nucleotide and/or codon substitution introduced into this RNA negatively impacts its viability. However, the efficient trans-amplification of DI-72 or DI-73 observed in coinoculations with HS-175 and DI-83 suggests that this low level is not due to the inability of the amplification activity to act in trans on templates other than DI-83. It therefore seems likely that this result may be due to altered properties of HS-175, which render it no longer able to accumulate efficiently (e.g., decreased stability and/or disruption of a cis-acting replication element). In addition, it is plausible that assembly of the functional replicase complex occurs preferentially on the RNA template which encodes p33 but that once formed, the active complex is able to dissociate subsequently and act on other templates (e.g., DI-72 and DI-73). Similar cis-preferential replication models, and the possible advantages thereof, have been discussed for turnip yellow mosaic virus and TCV (27, 29).
The presence of polymerase-specific motifs in the readthrough portion of p92 has led to the suggestion that it represents a component of the viral RNA replicase (6). Amino acid sequence analysis of the p33 products of tombusviruses has indicated no significant relationship to any proteins of known function (18, 25). Although the precise role of the TBSV p33 remains a mystery, it has been suggested that it may participate in genome replication (22). Results similar to those of this study were found when p28, the TCV homolog of p33, was analyzed (29). The p28 product is also required, along with its readthrough product, p88, for viral RNA amplification. The prereadthrough proteins from the Carmovirus and Tombusvirus genera therefore appear to share certain key properties and may have similar functions. In light of these results, it is conceivable that other members of the family Tombusviridae also require both prereadthrough and readthrough products for productive viral RNA amplification. It cannot, however, be entirely precluded that prereadthrough proteins may be functioning, alternatively or additionally, via a mechanism independent of replication (e.g., RNA stabilization).
Possible functional roles for region 3.5.
Region 3.5 could represent an important component of a promoter. If this were true, the absence of region 3.5 in DI-82 and other highly replicable molecules, such as DI-72, indicates that it would not be essential for trans-amplification of certain replicons. In fact, in various coinoculations, molecules lacking region 3.5 are more competitive than their counterparts containing it (Fig. 2) (28). It is possible that this region is important at an early stage in the replication process, such as the initial assembly of the replicase complex. Following assembly, region 3.5 may not be strictly required for the initiation or elongation steps of RNA synthesis on other RNA templates. Evidence contrary to this possibility comes from studies on the closely related Tombusvirus cymbidium ringspot virus (CyRSV) (11). Transgenic plants expressing only the CyRSV p33 and p92 were able to support the amplification of a CyRSV DI-72-like molecule (i.e., lacking its corresponding region 3.5). This result suggests that in the transgenic system, some assembly of functional replicase is possible in the absence of region 3.5; however, requirements under conditions of natural infections may differ.
It seems less likely that region 3.5 would represent a stability element, at least in the context of DI-82, since DI-82 is able to accumulate efficiently when coinoculated with T-100 and in other coinoculations. We have observed that capping DI-82 restored its ability to complement HS-175. This provides support for the concept that region 3.5 may act as a translational enhancer, similar to the 3′TE of BYDV-PAV (26). Interestingly, Scholthof and Jackson (19) found that modifications to the TBSV genome in a small ORF (encoding pX, a short polypeptide of unknown function), which resides entirely within region 3.5, led to nonviability in certain hosts. It was also determined that disruption of the RNA sequence, rather than of the pX ORF, was responsible for these defects. In vitro translation studies showed that these mutations did not appreciably affect the expression of p33 (or p92), but a possible role in translational regulation in vivo was not ruled out. Unlike the findings of the present study (Fig. 5), capping did not rescue the viability of transcripts containing modifications to the pX ORF (19). These differences in results may be related to variables between the systems used in the two studies (e.g., differences in hosts, viral RNAs, mutation types, and expression of p33 and p92). Studies are under way to delineate the sequences and/or structures in region 3.5 which confer its complementation activity and to determine whether this element represents a translational enhancer.
Functional and compatibility requirements of p33.
Amino acid comparisons of p33 from three different tombusviruses (TBSV, CNV, and CyRSV) showed that the level of identity was greatest in the C-terminal region (6). This high level of conservation suggested a somewhat strict requirement for maintenance of this region for functional activity. Three different mutations were introduced into the C-terminal region of p33, and all molecules harboring these modifications were unable to complement HS-175, yet all were amplified when coinoculated with T-100. These results suggest that the alterations to the p33 ORFs were responsible for the lack of viral RNA accumulation in the coinoculations. The smallest truncation in p33 that was not functional was one which removed 19 aa from the C terminus (DI-83M2), indicating an important role for these residues. Replacement of the missing 19 aa with 40 non-p33 residues (DI-83M1) did not restore the activity. These results suggest that the C-terminal ∼6% of p33 is critical for its functional activity.
The expression strategy of p33 and p92 from the TBSV genome requires that the N-terminal region of p92 be identical to p33. If both these proteins are involved in replication, it is possible that there is some compatibility requirement between the two products. For the bromoviruses, it has been shown that the 1a and 2a viral replication components interact directly (9) and that certain compatibility requirements exist (2). If a direct interaction between p33 and p92 is required for TBSV, it would be mediated by an association between either the two identical domains or p33 and the readthrough domain of p92. If the former were true, the overlapping nature of their ORFs would require that compatibility be maintained via a doubly coupled type of coevolution. If a compatibility requirement does exist between the TBSV p33 and p92, it is not exceptionally strict, since complementation was observed when the CNV p33 homolog was tested. This result also leaves open the possibility that p33 functions independently of direct interaction with p92.
ACKNOWLEDGMENTS
We thank Laurie Baggio and members of our laboratory for reviewing the manuscript. We are also grateful to Herman Scholthof for providing HS-175, D’Ann Rochon for providing K2/M5, and Lori Weisberg for constructing DI-83CNV.
This work was supported by grants to K.A.W. from the National Science and Engineering Research Council of Canada.
ADDENDUM IN PROOF
Following submission of the final draft of this article, evidence suggesting a requirement for both the pre- and readthrough products of artichoke mottled crinkle Tombusvirus for genome amplification was presented (P. Molinari, C. Marusic, A. Lucioli, R. Tavazza, and M. Tavazza, J. Gen. Virol. 79:639–647, 1998).
REFERENCES
- 1.Dalmay T, Rubino L, Burgyan J, Kollar A, Russo M. Functional analysis of cymbidium ringspot virus genome. Virology. 1993;194:697–704. doi: 10.1006/viro.1993.1310. [DOI] [PubMed] [Google Scholar]
- 2.Dinant S, Janda M, Kroner P A, Ahlquist P. Bromovirus RNA replication and transcription require compatibility between the polymerase- and helicase-like viral RNA synthesis proteins. J Virol. 1993;67:7181–7189. doi: 10.1128/jvi.67.12.7181-7189.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dolja V V, Carrington J C. Evolution of positive-strand RNA viruses. Semin Virol. 1992;3:315–326. [Google Scholar]
- 4.Futterer J, Hohn T. Translation in plants—rules and exceptions. Plant Mol Biol. 1996;32:159–189. doi: 10.1007/BF00039382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gorbalenya A E, Koonin E V, Donchenko A P, Blinov V M. A conserved NTP-motif in putative helicases. Nature. 1988;333:22. doi: 10.1038/333022a0. [DOI] [PubMed] [Google Scholar]
- 6.Hearne P Q, Knorr D A, Hillman B I, Morris T J. The complete genome structure and synthesis of infectious RNA from clones of tomato bushy stunt virus. Virology. 1990;177:141–151. doi: 10.1016/0042-6822(90)90468-7. [DOI] [PubMed] [Google Scholar]
- 7.Ishikawa M, Meshi T, Ohno T, Okada Y. Specific cessation of minus-strand RNA accumulation at an early stage of tobacco mosaic virus infection. J Virol. 1991;65:861–868. doi: 10.1128/jvi.65.2.861-868.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jones R W, Jackson A O, Morris T J. Defective-interfering RNAs and elevated temperatures inhibit replication of tomato bushy stunt virus in inoculated protoplasts. Virology. 1990;176:539–545. doi: 10.1016/0042-6822(90)90024-l. [DOI] [PubMed] [Google Scholar]
- 9.Kao C C, Ahlquist P. Identification of the domains required for direct interaction of the helicase-like and polymerase-like RNA replication proteins of brome mosaic virus. J Virol. 1992;66:7293–7302. doi: 10.1128/jvi.66.12.7293-7302.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Knorr D A, Morris T J. De novo generation of defective interfering RNAs of tomato bushy stunt virus by high multiplicity passage. Virology. 1991;181:193–202. doi: 10.1016/0042-6822(91)90484-S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kollar A, Burgyan J. Evidence that ORF 1 and 2 are the only virus-encoded replicase genes of cymbidium ringspot tombusvirus. Virology. 1994;201:169–172. doi: 10.1006/viro.1994.1280. [DOI] [PubMed] [Google Scholar]
- 12.Koonin E V. The phylogeny of RNA-dependent RNA polymerases of positive-strand RNA viruses. J Gen Virol. 1991;72:2197–2206. doi: 10.1099/0022-1317-72-9-2197. [DOI] [PubMed] [Google Scholar]
- 13.Lupo R, Rubino L, Russo M. Immunodetection of the 33K/92K polymerase proteins in cymbidium ringspot virus-infected and transgenic plant tissue extracts. Arch Virol. 1994;138:135–142. doi: 10.1007/BF01310044. [DOI] [PubMed] [Google Scholar]
- 14.Maia I G, Seron K, Haenni A, Bernardi F. Gene expression from viral RNA genomes. Plant Mol Biol. 1996;32:367–391. doi: 10.1007/BF00039391. [DOI] [PubMed] [Google Scholar]
- 15.Miller W A, Dinesh-Kumar S P, Paul C P. Luteovirus gene expression. Crit Rev Plant Sci. 1995;14:179–221. [Google Scholar]
- 16.Rochon D M, Johnston J C. Infectious transcripts from cloned cucumber necrosis virus cDNA: evidence for a bifunctional subgenomic promoter. Virology. 1991;181:656–665. doi: 10.1016/0042-6822(91)90899-m. [DOI] [PubMed] [Google Scholar]
- 17.Rozanov M N, Koonin E V, Gorbalenya A E. Conservation of the putative methyltransferase domain: a hallmark of the ‘Sindbis-like’ supergroup of positive-strand RNA viruses. J Gen Virol. 1992;73:2129–2134. doi: 10.1099/0022-1317-73-8-2129. [DOI] [PubMed] [Google Scholar]
- 18.Rubino L, Burgyan J, Russo M. Molecular cloning and complete nucleotide sequence of carnation Italian ringspot tombusvirus genomic and defective interfering RNAs. Arch Virol. 1995;140:2027–2039. doi: 10.1007/BF01322690. [DOI] [PubMed] [Google Scholar]
- 19.Scholthof H B, Jackson A O. The enigma of pX: a host-dependent cis-acting element with variable effects on tombusvirus RNA accumulation. Virology. 1997;237:56–65. doi: 10.1006/viro.1997.8754. [DOI] [PubMed] [Google Scholar]
- 20.Scholthof H B, Scholthof K B, Jackson A O. Identification of tomato bushy stunt virus host-specific symptom determinants by expression of individual genes from a potato virus X vector. Plant Cell. 1995;7:1157–1172. doi: 10.1105/tpc.7.8.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scholthof H B, Scholthof K B, Kikkert M, Jackson A O. Tomato bushy stunt virus spread is regulated by two nested genes that function in cell-to-cell movement and host-dependent systemic invasion. Virology. 1995;213:425–438. doi: 10.1006/viro.1995.0015. [DOI] [PubMed] [Google Scholar]
- 22.Scholthof K-B G, Scholthof H B, Jackson A O. The tomato bushy stunt virus replicase proteins are coordinately expressed and membrane associated. Virology. 1995;208:365–369. doi: 10.1006/viro.1995.1162. [DOI] [PubMed] [Google Scholar]
- 23.Skuzeski J M, Nichols L M, Gesteland R F. Analysis of leaky viral translation termination codons in vivo by transient expression of improved β-glucuronidase vectors. Plant Mol Biol. 1990;15:65–79. doi: 10.1007/BF00017725. [DOI] [PubMed] [Google Scholar]
- 24.Skuzeski J M, Nichols L M, Gesteland R F, Atkins J F. The signal for a leaky UAG stop codon in several plant viruses includes the two downstream codons. J Mol Biol. 1991;218:365–373. doi: 10.1016/0022-2836(91)90718-l. [DOI] [PubMed] [Google Scholar]
- 25.Tavazza M, Lucioli A, Calogero A, Pay A, Tavazza R. Nucleotide sequence, genomic organization and synthesis of infectious transcripts from a full-length clone of artichoke mottled crinkle virus. J Gen Virol. 1994;75:1515–1524. doi: 10.1099/0022-1317-75-7-1515. [DOI] [PubMed] [Google Scholar]
- 26.Wang S, Browning K S, Miller W A. A viral sequence in the 3′-untranslated region mimics a 5′ cap in facilitating translation of uncapped mRNA. EMBO J. 1997;16:4107–4116. doi: 10.1093/emboj/16.13.4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weiland J J, Dreher T W. Cis-preferential replication of the turnip yellow mosaic virus RNA genome. Proc Natl Acad Sci USA. 1993;90:6095–6099. doi: 10.1073/pnas.90.13.6095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.White K A, Morris T J. Nonhomologous RNA recombination in tombusviruses: generation and evolution of defective interfering RNAs by stepwise deletions. J Virol. 1994;68:14–24. doi: 10.1128/jvi.68.1.14-24.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.White K A, Skuzeski J M, Li W, Wei N, Morris T J. Immunodetection, expression strategy and complementation of turnip crinkle virus p28 and p88 replication components. Virology. 1995;211:525–534. doi: 10.1006/viro.1995.1434. [DOI] [PubMed] [Google Scholar]
- 30.Zaccomer B, Haenni A, Macaya G. The remarkable variety of plant RNA virus genomes. J Gen Virol. 1995;76:231–247. doi: 10.1099/0022-1317-76-2-231. [DOI] [PubMed] [Google Scholar]