Abstract
Packaging of type C retrovirus genomic RNAs into budding virions requires a highly specific interaction between the viral Gag precursor and unique cis-acting packaging signals on the full-length RNA genome, allowing the selection of this RNA species from among a pool of spliced viral RNAs and similar cellular RNAs. This process is thought to involve RNA secondary and tertiary structural motifs since there is little conservation of the primary sequence of this region between retroviruses. To confirm RNA secondary structures, which we and others have predicted for this region, disruptive, compensatory, and deletion mutations were introduced into proviral constructs, which were then assayed in a permissive cell line. Disruption of either of two predicted stem-loops was found to greatly reduce RNA encapsidation and replication, whereas compensatory mutations restoring base pairing to these stem-loops had a wild-type phenotype. A GGNGR motif was identified in the loops of three hairpins in this region. Results were consistent with the hypothesis that the process of efficient RNA encapsidation is linked to dimerization. Replication and encapsidation were shown to occur at a reduced rate in the absence of the previously described kissing hairpin motif.
Retroviral 5′ untranslated leader sequences contain cis acting sites that are important for RNA encapsidation (1, 9, 21, 28, 35, 43, 54), dimerization (3, 12, 14, 31, 40, 60), and efficient gag translation (44). Retroviral RNA packaging is a highly specific process involving interactions between the viral Gag precursor and cis acting packaging signals in the 5′ leader sequence of their RNA genomes (5, 37, 58). The interaction is thought to involve RNA secondary structure since there is little conservation of primary sequences in the region. However, some conserved motifs have been identified. The GACG motif identified in several type C retroviruses (29) has been shown to be important for efficient encapsidation of avian spleen necrosis virus RNA (65) and murine leukemia virus RNA (45). A GAYC motif was found in the loop of a region 5′ to the gag initiation codon of Mason-Pfizer monkey virus and several other retroviruses (19). We (18) and others (3, 57) presented an RNA secondary-structure model for the human immunodeficiency virus type 1 (HIV-1) 5′ leader sequence region based on biochemical and enzymatic probing, comparison of the sequences of HIV-1 quasispecies, and free-energy minimization algorithms. Neither the GACG nor the GAYC motif is found in this region of HIV-1.
Electron microscopy has shown that retroviral RNAs under partially denaturing conditions are joined together in an apparent parallel orientation at a structure referred to as the dimer linkage site near the 5′ end of the genomic RNAs (4, 47). It is thought that a parallel orientation of the dimeric RNAs exists in HIV-1, and two recent publications have lent support to this theory (10, 22). Dimerization may modulate several steps of the virus life cycle, such as translation, encapsidation, recombination, and reverse transcription. Earlier work on RNA dimerization in HIV-1 using synthetic RNAs in vitro (2, 61) suggested that guanine tetrads (64) might be involved in dimer formation. This model has not been supported by in vitro studies of guanine or purine sequences in the region. Since then, stem-loop one (SL1), 5′ positions 240 to 280 (18) (now known as the kissing hairpin), has been proposed as the dimer initiation signal (7, 14, 16, 31–34, 40, 46, 49–51, 60). The present study has addressed the role of structure and sequence motifs in the HIV-1 leader in their effect on encapsidation. Putative dimer linkage sites have been included in the analysis. RNA encapsidation in nonpermissive cells (11, 38, 41, 42) has in some cases differed qualitatively and quantitatively from encapsidation in permissive cell lines (27, 34). In this study, we have examined the role of discrete RNA secondary structures in the HIV-1 core packaging signal region in encapsidation, in permissive cells, by introduction of disruptive, compensatory, and deletion mutations. RNA packaging was analyzed by using RNase protection assays (RPAs), which are both quantitative and qualitative.
MATERIALS AND METHODS
Cells and viruses.
The cell line Jurkat-tat (56) was grown in RPMI 1640 medium supplemented with 10% fetal calf serum, penicillin, and streptomycin. COS-1 cells were grown in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum, penicillin, and streptomycin. The infectious proviral clone HIV-1 HXB2 (13) was used in all experiments. In common with the majority of recent publications, we have numbered the viral sequence from the RNA cap site. Thus, the G of the splice donor is nucleotide (nt) 290, and the A of the gag initiation codon is nt 336.
Construction of mutants.
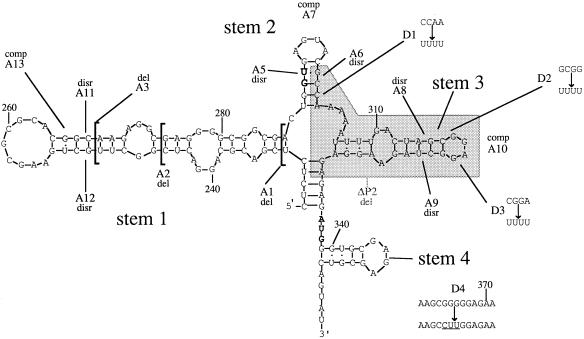
The BglII fragment of pSVC21 from base 21 to base 1644 (numbering of viral sequences as described above) was cloned into the BamHI site of the expression vector pBluescript KSII (Stratagene) to create a plasmid which we refer to as pKSBgl II. Mutagenesis was done essentially in accordance with the method of Kunkel et al. (30). The plasmid pKSBgl II was transformed into Escherichia coli CJ236, and a single-stranded DNA was rescued by using the manufacturer’s helper phage. Synthetic oligonucleotide primers were purchased from R&D Ltd. (Oxford, United Kingdom). The sequences of mutants are shown in Fig. 1, and their locations are shown in Fig. 2.
FIG. 1.
Nucleic acid sequences of mutants created by introducing mutations into the HIV-1 HXB2 provirus. Altered bases are underlined. The positions of deletions are indicated by vertical lines. The protein sequence of the D4 mutant is shown below the primary sequence, and the glycine-to-leucine change is underlined.
FIG. 2.
Positions of mutations within the RNA secondary structures of the HIV-1 HXB2 5′ leader sequence showing our original prediction for the structure of SL2 (stem 2), which differs from that of Sakaguchi et al. (57). The sequence deleted in the ΔP2 mutation is shaded. Abbreviations: del, deletion: disr, disruption: comp, compensatory mutation. Stems 1 to 4, SL1 to SL4.
Once the mutants had been identified by sequencing, the NarI-to-ApaI fragment (from bases 186 to 1558) was excised from the mutant pKSBgl II plasmids and the fragments were cloned into the unique NarI-to-ApaI sites in the HXB2 provirus which were then revalidated by sequencing, amplifying the region from bases 202 to 464 by using a biotinylated oligodeoxynucleotide primer, 5′-B-CTG AAA GCG AAA GGG AAA CC-3′, and a nonbiotinylated oligodeoxynucleotide primer, 5′-TTCTAGCTCCCTGCTTGCCC-3′. Single-stranded DNA was prepared by bonding the amplified fragment to Dynabeads (Dynal, Cheshire, United Kingdom) and by alkali treatment using the manufacturer’s recommended protocol. Single-stranded DNAs were sequenced to check that the designed mutation was present in each proviral clone.
Replication studies.
Equal numbers of virus particles, as judged by using reverse transcriptase (RT), from transient COS-1 transfections were used to infect 2 × 106 Jurkat-tat cells for 4 h. These cells were washed in serum-free RPMI 1640 medium, and fresh RPMI 1640 medium was added (with fetal calf serum, penicillin, and streptomycin). During replication studies, two 10-μl samples of the supernatant of each culture were taken out every fourth day and frozen at −70°C until 30 days postinfection, when the RT levels were assayed simultaneously. The cells were split every fourth day, with the cell numbers kept approximately equal in all cultures.
RT levels were determined by using the RT assay of Potts (53), incorporating [α-32P]dTTP (Amersham, Amersham, United Kingdom).
Protein studies.
For analysis of proteins, proviral mutants were transfected into Jurkat-tat cells by using DEAE dextran (59). Cells were cultured in RPMI 1640 medium supplemented with 10% fetal calf serum, penicillin, and streptomycin. At 72 h posttransfection, the cells were counted and then harvested by centrifugation at 2,000 rpm for 5 min in an MSE Falcon 6/300 and resuspended in 100 μl of phosphate-buffered saline with 200 μl of sample buffer (6.25 mM Tris-HCl [pH 6.8], 2% sodium dodecyl sulfate [SDS], 10% glycerol, 5% β-mercaptoethanol, 0.02% bromophenol blue). The supernatants were filtered through 0.45-μm-pore-size filters and incubated at 4°C overnight with 0.5 volume of 30% polyethylene glycol 8000 in 0.4 M NaCl. The precipitate was collected by centrifugation at 2,000 rpm for 45 min in an MSE Falcon 6/300, and the pellet was resuspended in 100 μl of phosphate-buffered saline and 200 μl of sample buffer. Following sonication, the protein equivalent of 650,000 cells was loaded per lane (equalized quantities of supernatants were also loaded) on an SDS-polyacrylamide gel electrophoresis gel with 12.5 or 10% polyacrylamide (probing for Gag or Env, respectively). Proteins were electrophoresed for approximately 3.5 h at 50 mA and then transferred to Hybond C-extra nitrocellulose (Amersham) overnight at 250 mA. The filters were probed by using monoclonal antibodies to p55/p24 (MRCADP 313) or gp120/160 (MRCADP 323), both supplied through the United Kingdom Medical Research Council AIDS Programme. The Gag probe was used at a concentration of 3 μg/ml, and the Env probe was used at a dilution of 1 in 1,000; both were incubated for 1 h at room temperature. Bands were visualized by sheep anti-mouse horseradish peroxidase-linked whole antibody and enhanced chemiluminescence Western detection reagents (Amersham) as described in the manufacturer’s instructions. Rainbow molecular weight markers (Amersham) were used.
RNA extraction and quantitation.
Cytoplasmic RNAs were extracted from Jurkat-tat cells by rapid lysis at 4°C in NP40 buffer (50 mM Tris HCl [pH 8.00], 100 mM NaCl, 5 mM MgCl2 0.5% [vol/vol] Nonidet P-40). Cell debris and nuclei were removed by a 1-min centrifugation at 13,000 rpm in a microcentrifuge. The supernatants were adjusted to 0.2% SDS and 125 μg of proteinase K per ml, incubated at 37°C for 15 min, and extracted twice with phenol-chloroform and once with chloroform. Virion RNAs were prepared from the supernatants of chronically infected Jurkat-tat cells. RT assays were carried out on two 10-μl aliquots from the supernatant of each culture. Based on these data, equivalent numbers of particles were collected from each supernatant (about 2 ml from the wild-type [WT] virus supernatant). HIV-1 particles were harvested as described above and then resuspended in 0.5 ml of TNE (10 mM Tris HCl, 150 mM NaCl, 1 mM EDTA [pH 7.5]). This suspension was layered over an equal volume of TNE containing 20% sucrose and centrifuged at 98,000 × g in a Beckman TLA45 rotor for 2 h. The pellets were resuspended in TNE and stored at 4°C while another RT assay was carried out on the suspension. Equivalent amounts of particles were then used for RNA extractions. The suspensions were made up to equal volumes with TNE; an equal volume of 2X proteinase K buffer (100 mM Tris HCl [pH 7.5], 200 mM NaCl, 20 mM EDTA, 2% SDS, 200 μg of proteinase K per ml, 200 μg of tRNA per ml) was then added and the mixture was incubated for 30 min at 37°C. Two phenol-chloroform extractions were carried out. RNAs from both cytoplasmic and virion samples were precipitated under ethanol and stored at −70°C.
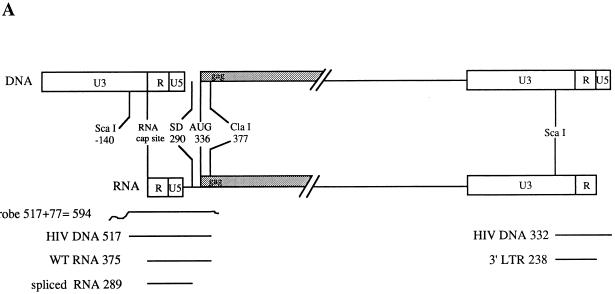
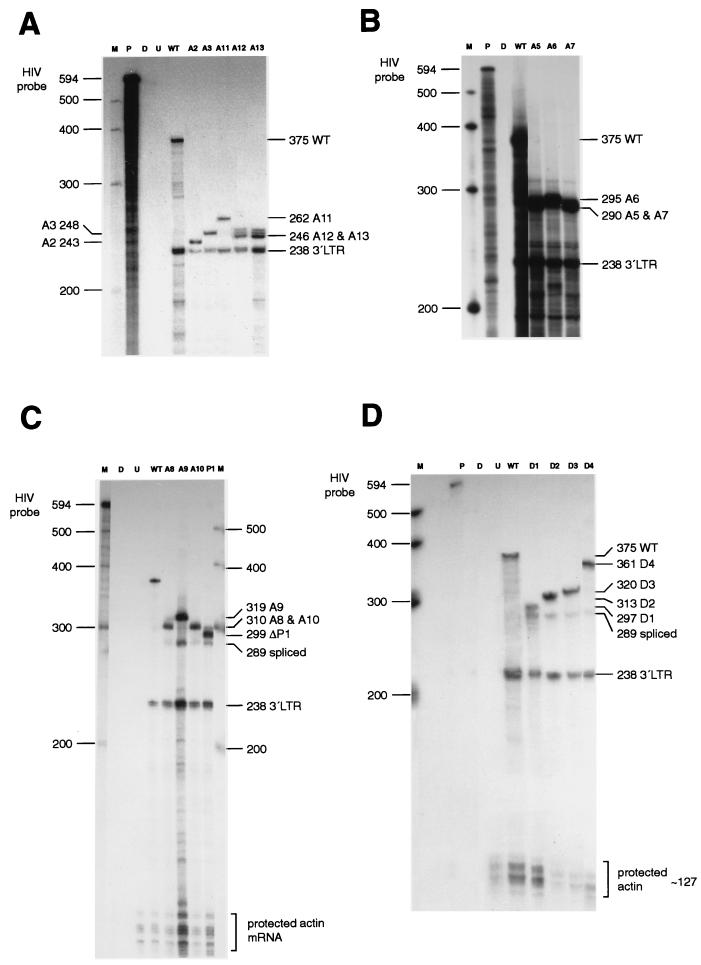
RPAs were carried out with the RPA II kit supplied by Ambion (Austin, Tex.). For expression of the HIV-1 antisense riboprobe, the plasmid pKSΨCS (26), which has the ScaI-to-ClaI fragment of pSVC21 (from 140 bases upstream of the RNA cap site to 377 bases downstream) cloned into the EcoRV and ClaI sites of pBluescript KSII, was used. The plasmid was linearized with XbaI, and RNA was transcribed from the T3 promoter by using an in vitro transcription kit (Promega) incorporating Redivue [α-32P]UTP (Amersham). The riboprobe contained 517 bases of WT HIV-1 sequence, with 77 bases from the vector making a total length of 594 nt (Fig. 3A).
FIG. 3.
(A) Predicted sizes (in bases) of the protected RNA fragments for the WT sequence of the HIV-1 HXB2 provirus after RNase protection assays. LTR, long terminal repeat. (B) Predicted sizes (in bases) of the protected RNA fragments which characterize mutant sequences after RNase protection assays using the WT riboprobe.
A riboprobe for the human actin mRNA, pTRI-B actin-125-human (Ambion), was used to determine the levels of actin mRNA in cytoplasmic RNA preparations and to determine whether any actin mRNA was encapsidated. The actin probe was 160 nt in length and protected RNAs of 127 nt. The HIV-1 riboprobe distinguished between spliced RNA (289 bases), unspliced RNA (WT, 375 bases), and 3′ long-terminal-repeat RNA (236 bases) (Fig. 3A). During the RPAs, the WT riboprobe was digested at the sites of mutations, resulting in protected fragments of characteristic length. The predicted sizes of the larger protected unspliced RNA fragments are detailed in Fig. 3B.
RNA markers were transcribed from the Century marker template (Ambion) with T7 polymerase incorporating Redivue [α-32P]UTP (Amersham). The intensities of protected HIV-1 RNAs on gels were determined with a laser densitometer (Molecular Dynamics model 300A) and were calculated as a percentage of that of WT. The results of at least three separate experiments were used to calculate the standard deviations.
Secondary-structure predictions.
In this paper, we refer to the stem-loop from 232U to 286A as SL1, structures in the region from 288U to 301A as SL2, the stem-loop from 306U to 330A as SL3, and the stem-loop from 339G to 352C as SL4. The nucleic acid folding program MFold (24, 66) adapted for GCG (Genetics Computer Group, University of Wisconsin, Madison) was used for free-energy minimization predictions with the graphical presentation of Squiggles (48) in the GCG program Plotfold.
RESULTS
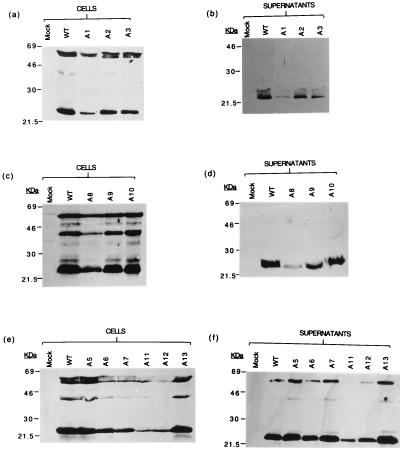
Figures 4 to 6 show Western blots of viral proteins produced by the mutants. None of the mutations abolish protein production as judged by analysis of cells and supernatants from transient transfections. Lower-than-WT levels of Gag and Env were detected on Western blots for mutants A11 (SL1) and A12 (SL1) as well as A8 (SL3) and ΔP1 (SL3). D4, which disrupts Gag coding, led to a decrease in Gag but not Env protein that was detectable in both cells and supernatants. The integrity of SL1 and SL3 thus has an influence on viral protein production.
FIG. 4.
Western blots of proteins extracted from cytoplasms (a, c, and e) and supernatants (b, d, and f) of Jurkat-tat cells transfected with the WT and mutant HIV-1 proviruses A1 to A3 and A5 to A13, probed with a monoclonal antibody to p55/24. Mock, nontransfected cells.
FIG. 6.
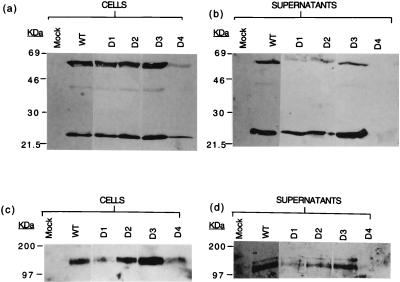
Western blots of proteins extracted from the cytoplasms (a and c) and supernatants (b and d) of Jurkat-tat cells transfected with the WT and mutant HIV-1 proviruses D1 to D4, probed with a monoclonal antibody to p55/24 (a and b) or to gp120/160 (c and d). Mock, nontransfected cells.
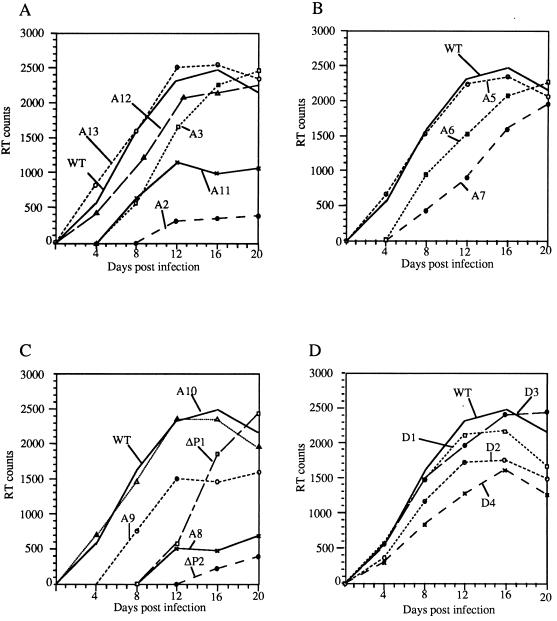
The A1 mutant was lethal, and no functional infective viruses were produced. Other than this mutant, sufficient viral particles were produced from infected Jurkat-tat cells to analyze viral encapsidation in equivalent (RT-equalized) quantities of viral particles. Viral replication assays are shown in Fig. 7. Replication was monitored for up to 30 days. WT virus replication showed a peak value at about day 16. RPAs of cellular and viral RNA are shown in Fig. 8 and 9. These assays distinguish spliced and unspliced species. The relative efficiencies of viral RNA packaging are shown in Fig. 10.
FIG. 7.
RT activity in the supernatants of Jurkat-tat cells infected with the WT and mutant proviruses (counts per second of [35S]TTP incorporation per 10 μl) plotted against days postinfection. (A) Mutations to SL1; (B) mutations to SL2; (C) mutations to SL3; (D) mutations to purine motifs.
FIG. 8.
RPAs of RNAs extracted from virions in the supernatants of chronically infected Jurkat-tat cells. Abbreviations: M, RNA size markers; P, undigested riboprobe; D, digested riboprobe; U, uninfected cells; LTR, long terminal repeat. Lanes containing relevant mutants are indicated.
FIG. 9.
RPAs of RNAs extracted from the cytoplasms of chronically infected Jurkat-tat cells. Abbreviations: M, RNA size markers; P, undigested riboprobe; D, digested riboprobe; U, uninfected cells; LTR, long terminal repeat. Lanes containing relevant mutants are indicated.
FIG. 10.
Intensity of protected RNA bands, after RPAs with virion RNAs, shown as a percentage of the intensity of the band from WT RNA.
Replication and encapsidation closely mirrored each other. A2 (SL1) and A11 (SL1) severely impaired replication and encapsidation, the less severe truncation of SL1 (A3) having a less marked effect, and A13, the compensatory mutation for A11, restored WT levels of replication and encapsidation. This is likely due to the reformation of the base pairing in SL1, albeit with an altered sequence. A8 (SL3) and ΔP2 (SL3) both profoundly reduced encapsidation and packaging, and A10, the compensatory mutant of A8, restored WT levels of both. None of the purine motif mutants D1 to D4 significantly affected viral replication or RNA encapsidation. Disruption of SL2 (A5 and A6) had little effect, but combined mutations of both sides of the helix, which maintained structure but altered sequence (A7), impaired RNA encapsidation and viral replication.
DISCUSSION
This study has documented the effects of a large number of mutations in the packaging signal region of HIV-1 on encapsidation and revealed the functional importance of SL1 and SL3. Some of these mutations also had effects on viral protein and particle production. This might be secondary to the effects on RNA stability, or, in some cases, there may have been minor effects on splicing efficiency. Such effects may contribute to the rate of viral spread in a culture. However, all of the encapsidation data presented have been obtained from equivalent quantities of viral particles; thus, clear conclusions on the effects of the mutations on encapsidation alone and on the role of the particular structures within this region on retroviral RNA packaging can be drawn.
SL1.
The functionally important part of SL1 in HXB2 probably lies between bases 243A and 277G (Fig. 2) even though the stem-loop has the potential to form three more GC base pairs. This is because the equivalent structure in chimpanzee immunodeficiency virus (CIV) (23) and the clade O isolates ANT70C and MVP5180 (15, 17) (Fig. 11) starts at base 243A in HXB2 and ends at base 277G (Fig. 2). Kim et al. (28) deleted bases 216 to 278 in the isolate HIV-1LAI and found that replication was delayed by 4 days in MT4 cells. Paillart et al. (50) deleted bases 243 to 277 and also bases 248 to 270 and found low levels of infectivity (the pattern of replication was not reported).
FIG. 11.
RNA secondary-structure predictions done by use of the GCG program MFold. Bases which differ from those of HIV-1 HXB2 are in lowercase letters. (a) The divergent HIV-1 isolate ANT70C. Bases in MVP5180 which differ from those of ANT70C are shown in boxes. (b) CIV.
The kissing hairpin is thought to be the signal for initiation of dimerization of genomic RNAs (7, 14, 16, 31, 32, 34, 40, 49–51, 60). Our deletion mutants A2 and A3 both maintained palindromes at the point where bases were removed; therefore, it could be argued that these novel palindromes substituted for the absence of the WT palindrome. However, the deletion mutants Δ248–270 and Δ243–277 of Paillart et al. (50) did not contain novel palindromes yet retained some ability to infect permissive cells, albeit at a level much lower than that of WT. This suggests that in the absence of the palindrome, an alternative method of initiation of dimerization occurs or that monomer RNA can be encapsidated. The fact that alterations to palindromic sequences in the loop of the kissing hairpin decrease but do not abolish replication (7, 10, 16, 50) is consistent with the suggestion that the kissing hairpin is not an absolute requirement for either dimerization or encapsidation (33). It has been shown that sequences upstream of SL1 contribute to dimerization (7) and encapsidation (28, 42). Previous studies have also shown that retroviral dimers are linked at many locations within the 5′ end of the genomic RNA (39). The processes of encapsidation and dimerization may be tightly linked (14, 34). Such a tight linkage would explain why sequences upstream of the splice donor (SD) sequence are required for encapsidation of genomic RNAs since there are dimerization signals upstream of the SD. The WT phenotype of the compensatory mutant A13 shows that the structure of the stem of the kissing hairpin rather than its sequence is critically important. McBride and Panganiban (41) made a disruption mutation in SL1 that was slightly different from that in our A12 mutant, changing 248C to 248A, 249U to 249A, and 251G to 251U. This disruption decreased RNA packaging in HeLa cells by about one-fifth compared to that of WT. The difference between our disruption mutants A11 and A12 may be due to the effect of these disruptions on RNA secondary structure. Computer predictions (24, 66) for the secondary structure of the disruption mutants A11 and A12 did not provide any explanation of these differences in terms of the maintenance of palindrome regions (data not shown). We note that mutations to the purine-rich bulge of SL1 (245G to 245U, 268A to 268U, 269G to 269U, and 270G to 270U) were shown to affect RNA packaging in nonpermissive cells (11).
SL2.
This stem-loop, which contains the major splice donor signal, has two possible structures: one is shown in Fig. 2, and the other, first documented by Sakaguchi et al. (57), is shown in Fig. 12 (see below). The alternative structure for this stem-loop places the SD at the end of a stem-loop. In this prediction, there is a stable stem with a loop containing the motif GGUGA. We were interested in addressing the question of whether either of these two possible structures has functional importance. Our disruption mutants (A5 and A6) and the compensatory mutations in this stem-loop were based on the structure shown in Fig. 2. Sequence comparisons between the HXB2 sequence and divergent HIV sequences show that the only part of this region which is conserved between HIV HXB2 (Fig. 2) and CIV (23) and the clade O isolates (ANT70C and MVP5180) (15, 17) is the sequence GGUGAG containing the SD signal (Fig. 11); even so, our mutant A5 altered the sequence motif from GGUGAG to AGUAAG with little effect on replication. The compensatory mutant in this stem (A7) (Fig. 2) did not have a WT phenotype; therefore, either our original suggested structure of this region is not functionally important or the primary sequence is more important than its secondary structure. Kim et al. (28) deleted bases 293A to 302A and found that replication of this virus was delayed slightly. RT activity from the mutant provirus in MT4 cells peaked at 9 days as opposed to 6 days for the WT. The predicted effects of mutants A5, A6, A7, and D1 on the Sakaguchi structure are shown in Fig. 12. Mutants A6 and A7 disrupt the Sakaguchi model for this region, whereas the A5 and D1 mutants still present the GU splice donor motif on the end of a stem-loop. This strongly suggests that the Sakaguchi prediction is the structure which has functional importance. We note that our mutant A1 also disrupts the Sakaguchi prediction for SL2. The Sakaguchi model of this region is conserved in CIV (Fig. 12), whereas our original prediction (18) is not. Neither structure is conserved in the equivalent region of the clade O isolates, in which the SD is predicted to be 3′ to a purine loop. The effect of sequence variations in the naturally occurring HIV-1 isolates RF and Z2Z6 is shown in Fig. 12. The lack of conservation of this sequence and structure in this region suggests that SL2 has less functional importance than SL1 and SL3.
FIG. 12.
Predictions of RNA secondary structures within SL2 of RNAs from mutant proviruses, based on the structure for this region first documented by Sakaguchi et al. (57), by using the output program MFold. Predictions of similar structures in the equivalent regions of the HIV-1 isolates RF, Z2Z6, ANT70C, and HIV-2 ROD are shown.
SL3.
Common to all of the secondary-structure predictions to date (3, 18, 57) is a stem-loop structure including bases 312C to 325G. The A at position 319 changes to U in CIV and the clade O isolates; there is also a compensatory mutation in the clade O viruses where bases 314A and 323U are changed to G and C, respectively. The loop of SL3 contains the conserved motif GGNGR. In CIV and the clade O HIV isolates, there is a third such motif, GGCGR, in the extra stem-loop that is found in these viruses (Fig. 11). This suggests that the GGNGR motif is important in primate lentiviruses either in the encapsidation process or in another essential part of the replication cycle. It is possible that the presence of just one GGNGR motif is sufficient for the normal life cycle to occur. The ΔP1 deletion (bases 298A to 319A inclusive) first demonstrated that this region is required for efficient packaging (35). A mutant with a larger (298G to 332A) deletion (ΔP2) replicated detectably after only 20 days in Jurkat-tat cells, similar to the A2 deletion mutant. Deletions in the same area assayed by other groups are not directly comparable, but they do indicate that this region is important for HIV-1 RNA encapsidation. Clavel and Orenstein (9) deleted bases 303A to 332G from the sequence of isolate HIVNL4-3 and found that while WT levels of particles were produced in SL480 cells by this mutant, these particles could not infect A3.01 CD4-positive lymphoid cells. Kim et al. (28) deleted bases 308U to 328G from the sequence of HIVLAI and found that replication was delayed by 5 days compared to that of WT in MT4 cells due to a packaging defect, as detected by slot blot analysis. A very similar deletion mutant, mutant pA4HXB, of Aldovini and Young (1) (with bases 295U to 315G deleted from isolate HXB2 sequence) did not replicate in H9 cells, whereas the present results show that replication of the deletion mutant ΔP1 (with bases 298A to 319A inclusive deleted) was delayed only in Jurkat-tat cells. This suggests that there are significant differences in the phenotypes of mutants when assayed in these two permissive lines, H9 and Jurkat-tat. Similarly, pA3HXB, the 39-bp deletion mutant of Aldovini and Young (from 295U to 333G deleted) did not replicate, whereas our ΔP2 deletion mutant (from 298G to 332A deleted) did so after a long delay. However, we note that the mutants were not identical.
Free-energy minimization calculations for the disruption mutant A8 predicted no stable secondary structure for the region between nt 310 and nt 320 in mutant A8 (data not shown), whereas the disruption mutant A9 could form a weak structure that presents the GGAGC motif at the end (data not shown). Perhaps the difference between the packaging efficiency and replication rates of disruption mutants A8 and A9 are due to the fact that A8 cannot present the GGAGG motif on the end of a stem whereas A9 can form a weak secondary structure which presents the motif GGAGC in a loop (data not shown). The compensatory mutant A10 showed that the structure of this stem is sufficient to restore the WT phenotype independent of its sequence. Sequences downstream of the gag initiation codon have been reported to be required for efficient RNA encapsidation (38), but apart from our D4 mutant, we have not investigated this region.
Purine-rich sequences.
Awang and Sen (2), Sundquist and Heaphy (61), and Marquet et al. (40) proposed that dimerization occurs as a result of highly stable purine-purine interactions. This work was carried out on synthetic RNAs in vitro in the absence of proteins and may not be directly relevant in vivo. Our proviral constructs D1 to D4 abolished four purine-rich sequences identified by Sundquist and Heaphy (61) but did not produce effects on replication that were consistent with interference with dimerization. A guanine-to-uridine change at base 365 introduced by Haddrick et al. (16), which interrupted a run of five G residues, did not have a deleterious effect on replication of isolate HIVNL4-3. Our alteration of the same run of five G’s to CUUGG (D4) had the greatest effect of all our purine motif mutants; however, a significant contributor to this effect may be the coding change of a glycine to a leucine in gag. Removal of a purine bulge in SL1 of HIV-1 was found to affect RNA encapsidation in a nonpermissive cell line (11). We conclude that the removal of any single G-rich purine motif in this region has little effect on encapsidation or replication. Similarly, dimer linkage in bovine leukemia virus and HIV-2 appears to be independent of purine motifs (6, 25). However, predicted loops and bulges within RNA structures of the primate immunodeficiency lentiviruses are very rich in purines (18a) and purines have previously been shown to be important for highly specific binding of protein to RNA in other systems. These include the HIV-1 Rev binding to a purine-rich bubble of the rev-responsive element (20, 52), yeast ribosomal binding protein L32 recognition of the 5′ end of its mRNA (36, 63), and recognition of an RNA hairpin by the MS2/R17 coat protein (55, 62). It seems likely that Gag may interact with unpaired purines such as GGNGR motifs during encapsidation.
We conclude that the stem-loop structures at the ends of SL1 and SL3 are both important for efficient encapsidation in permissive cells independent of their stem sequences. Our data support and augment those of McBride and Panganiban (41) for HeLa cells and of Clever and Parslow for human osteosarcoma cells and 293T cells (11); moreover, we have formally excluded the possibility of promiscuous RNA encapsidation, by analyzing encapsidation of actin mRNA in this permissive cell line. In contrast to McBride and Panganiban, we found that compensatory mutations to these stem-loops completely restored the WT phenotype. Like them, we found that in the absence of either SL1 or SL3, some RNA encapsidation still occurs and is highly specific, implying that the RNA encapsidation signal is multipartite and diffuse (8, 11, 41). We did not investigate the ability of our mutants to form dimers. Our data show that deletions to the kissing hairpin (SL1) affect RNA encapsidation. Other workers have shown that alterations to SL1 have an effect on dimerization (7, 14, 16, 31–34, 40, 46, 49–51, 60). It appears that mutations to SL1 affect both RNA dimerization and RNA encapsidation in the same way, which suggests that the processes of dimerization and RNA encapsidation are linked. A GGNGR motif is found in the loops of SL2 and SL3 and in an additional stem-loop found between the SD and the gag initiation codon in CIV and the known clade O isolates. Disruption of one copy of this motif has little effect on virus replication and RNA encapsidation, suggesting that multiple copies of the motif may confer a degree of functional redundancy or that they have a role other than encapsidation in the virus life cycle.
These data have led us to reevaluate structural predictions made by ourselves and others. The structure which we previously referred to as stem-loop 1 (18), in which bases 227C to 231C were predicted to be bound to bases 335G to 331G, is poorly conserved in CIV and is not conserved at all in the known clade O isolates (Fig. 11); therefore, it is probably not functionally important in HXB2. The structure labeled SL4 (stem 4) in Fig. 2 (38) is not conserved in CIV or the clade O isolates and therefore is probably not functionally important.
FIG. 5.
Western blots of proteins extracted from the cytoplasms (a, c, and e) and supernatants (b, d, and f) of Jurkat-tat cells, transfected with the WT and mutant HIV-1 proviruses A1 to A3 and A5 to A13, probed with a monoclonal antibody to gp120/160. Mock, nontransfected cells.
ACKNOWLEDGMENTS
We thank Teresa Barnes for secretarial work and J. Greatorex, S. Hassard, M. Sakalian, S. Heaphy, and W. Sundquist for helpful discussions.
This work was supported by the Medical Research Council (United Kingdom), The Sykes Trust, and EC Biomed grant BMH4-CT96-0675.
REFERENCES
- 1.Aldovini A, Young R A. Mutations of RNA and protein sequences involved in human immunodeficiency virus type 1 packaging result in production of noninfectious virus. J Virol. 1990;64:1920–1926. doi: 10.1128/jvi.64.5.1920-1926.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Awang G, Sen D. Mode of dimerization of HIV-1 genomic RNA. Biochemistry. 1993;32:11453–11457. doi: 10.1021/bi00093a024. [DOI] [PubMed] [Google Scholar]
- 3.Baudin F, Marquet R, Isel C, Darlix J-L, Ehresmann B, Ehresmann C. Functional sites in the 5′ region of HIV-1 RNA form defined structural domains. J Mol Biol. 1993;229:382–397. doi: 10.1006/jmbi.1993.1041. [DOI] [PubMed] [Google Scholar]
- 4.Bender W, Davidson N. Mapping of poly(A) sequences in the electron microscope reveals unusual structure of type C type oncornavirus RNA molecules. Cell. 1976;7:595–607. doi: 10.1016/0092-8674(76)90210-5. [DOI] [PubMed] [Google Scholar]
- 5.Berkhout B. Structure and function of the HIV leader RNA. Prog Nucleic Acid Res. 1997;54:1–34. doi: 10.1016/s0079-6603(08)60359-1. [DOI] [PubMed] [Google Scholar]
- 6.Berkhout B, Essink B B, Schoneveld I. In vitro dimerisation of HIV-2 leader RNA in the absence of PuGGAPuA motifs. FASEB J. 1993;7:181–187. doi: 10.1096/fasebj.7.1.8422965. [DOI] [PubMed] [Google Scholar]
- 7.Berkhout B, van Wamel J L B. Role of the DIS hairpin in replication of human immunodeficiency virus type 1. J Virol. 1996;70:6723–6732. doi: 10.1128/jvi.70.10.6723-6732.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berkowitz R D, Öhagen Å, Höglund S, Goff S P. Retroviral nucleocapsid domains mediate the specific recognition of genomic viral RNAs by chimeric Gag polyproteins during RNA packaging in vivo. J Virol. 1995;69:6445–6456. doi: 10.1128/jvi.69.10.6445-6456.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clavel F, Orenstein J M. A mutant of human immunodeficiency virus with reduced RNA packaging and abnormal particle morphology. J Virol. 1990;64:5230–5234. doi: 10.1128/jvi.64.10.5230-5234.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clever J L, Wong M L, Parslow T G. Requirements for kissing-loop-mediated dimerization of human immunodeficiency virus type 1 RNA. J Virol. 1996;70:5902–5908. doi: 10.1128/jvi.70.9.5902-5908.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clever J L, Parslow T G. Mutant human immunodeficiency virus type 1 genomes with defects in RNA dimerization or encapsidation. J Virol. 1997;71:3407–3414. doi: 10.1128/jvi.71.5.3407-3414.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Darlix J-L, Gabus C, Nugeyre M T, Clavel F, Barré-Sinoussi F. cis-elements and trans-acting factors involved in the RNA dimerisation of HIV-1. J Mol Biol. 1990;216:689–699. doi: 10.1016/0022-2836(90)90392-Y. [DOI] [PubMed] [Google Scholar]
- 13.Fisher A G, Collati E, Ratner L, Gallo R C, Wong-Staal F. A molecular clone of HTLV III with biological activity. Nature (London) 1985;316:262–265. doi: 10.1038/316262a0. [DOI] [PubMed] [Google Scholar]
- 14.Fu W, Gorelick R J, Rein A. Characterization of human immunodeficiency virus type 1 dimeric RNA from wild-type and protease-defective virions. J Virol. 1994;68:5013–5018. doi: 10.1128/jvi.68.8.5013-5018.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gürtler L G, Hauser P H, Eberle J, von Brunn A, Knapp S, Zekeng L, Tsague J M, Kaptue L. A new subtype of human immunodeficiency virus type 1 (MVP-5180) from Cameroon. J Virol. 1994;68:1581–1585. doi: 10.1128/jvi.68.3.1581-1585.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haddrick M, Lear A L, Cann A J, Heaphy S. Evidence that a kissing loop structure facilitates genomic RNA dimerisation in HIV-1. J Mol Biol. 1996;259:58–68. doi: 10.1006/jmbi.1996.0301. [DOI] [PubMed] [Google Scholar]
- 17.Haesevelde M V, Decourt J L, DeLeys R J, Vanderborght B, der Groen G, van Neuverswiyn H, Saman E. Genomic cloning and complete sequence analysis of a highly divergent African human immunodeficiency virus isolate. J Virol. 1994;68:1586–1596. doi: 10.1128/jvi.68.3.1586-1596.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harrison G P, Lever A M L. The 5′ packaging signal and major splice donor region of human immunodeficiency virus type 1 have a conserved stable secondary structure. J Virol. 1992;66:4144–4153. doi: 10.1128/jvi.66.7.4144-4153.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18a.Harrison, G. P., and A. M. L. Lever. Unpublished observations.
- 19.Harrison G P, Hunter E, Lever A M L. Secondary structure of the Mason-Pfizer monkey virus 5′ leader sequence: identification of a structural motif common to a variety of retroviruses. J Virol. 1995;69:2175–2186. doi: 10.1128/jvi.69.4.2175-2186.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heaphy S, Finch J T, Gait M, Karn J, Singh M. HIV-1 regulator of virion expression, rev, forms nucleoprotein filaments after binding to a purine-rich “bubble” located within the rev-responsive region of viral mRNAs. Proc Natl Acad Sci USA. 1991;88:7366–7370. doi: 10.1073/pnas.88.16.7366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hayashi T, Ueno Y, Okamoto T. RNA packaging signal of HIV-1. FEBS Lett. 1993;327:213–218. doi: 10.1016/0014-5793(93)80172-q. [DOI] [PubMed] [Google Scholar]
- 22.Höglund S, Öhagen Å, Goncalves J, Panganiban A T, Gabuzda D. Ultrastructure of HIV-1 genomic RNA. Virology. 1997;233:271–279. doi: 10.1006/viro.1997.8585. [DOI] [PubMed] [Google Scholar]
- 23.Huet T, Cheynier R, Meyerhans A, Roelants G, Wain-Hobson S. Genetic organization of a chimpanzee lentivirus related to HIV-1. Nature (London) 1990;345:356–359. doi: 10.1038/345356a0. [DOI] [PubMed] [Google Scholar]
- 24.Jaeger J A, Turner D H, Zucker M. Improved predictions of secondary structures for RNA. Proc Natl Acad Sci USA. 1989;86:7706–7710. doi: 10.1073/pnas.86.20.7706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Katoh I, Yasunaga T, Yoshinaka Y. Bovine leukemia virus RNA sequences involved in dimerization and specific gag protein binding: close relation to the packaging sites of avian, murine, and human retroviruses. J Virol. 1993;67:1830–1839. doi: 10.1128/jvi.67.4.1830-1839.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaye J F, Richardson J H, Lever A M L. cis-acting sequences involved in human immunodeficiency virus type 1 RNA packaging. J Virol. 1995;69:6588–6592. doi: 10.1128/jvi.69.10.6588-6592.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaye J F, Lever A M L. trans-acting proteins involved in RNA encapsidation and viral assembly in human immunodeficiency virus type 1. J Virol. 1996;70:880–886. doi: 10.1128/jvi.70.2.880-886.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim H-J, Lee K, O’Rear J J. A short sequence upstream of the 5′ major splice site is important for encapsidation of HIV-1 genomic RNA. Virology. 1994;198:336–340. doi: 10.1006/viro.1994.1037. [DOI] [PubMed] [Google Scholar]
- 29.Konings D A, Nash M A, Maizel J V, Arlinghaus R B. Novel GACG-hairpin motif in the 5′ untranslated region of type C retroviruses related to murine leukemia virus. J Virol. 1992;66:632–640. doi: 10.1128/jvi.66.2.632-640.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kunkel T A, Roberts J D, Zakour R A. Rapid and efficient site specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 31.Laughrea M, Jetté L. A 19 nucleotide sequence upstream of the major splice donor is part of the dimerization domain of HIV-1 genomic RNA. Biochemistry. 1994;33:13464–13474. doi: 10.1021/bi00249a035. [DOI] [PubMed] [Google Scholar]
- 32.Laughrea M, Jetté L. HIV-1 genomic dimerisation: formation kinetics and thermal stability of dimeric HIV-1 LAI RNAs are not improved by the 1–232 and 292–790 regions flanking the kissing loop domain. Biochemistry. 1996;35:9366–9374. doi: 10.1021/bi960395s. [DOI] [PubMed] [Google Scholar]
- 33.Laughrea M, Jetté L. Kissing-loop model of HIV-1 genome dimerization: HIV-1 RNAs can assume alternative dimeric forms, and all sequences upstream or downstream of hairpin 248–271 are dispensible for dimer formation. Biochemistry. 1996;35:1589–1598. doi: 10.1021/bi951838f. [DOI] [PubMed] [Google Scholar]
- 34.Laughrea M, Jetté L, Mak J, Kleiman L, Liang C, Wainberg M A. Mutations in the kissing loop hairpin of human immunodeficiency virus type 1 reduce viral infectivity as well as genomic RNA dimerization. J Virol. 1997;71:3397–3406. doi: 10.1128/jvi.71.5.3397-3406.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lever A M L, Gottlinger H, Haseltine W, Sodroski J. Identification of a sequence required for efficient packaging of human immunodeficiency virus type 1 RNA into virions. J Virol. 1989;63:4085–4087. doi: 10.1128/jvi.63.9.4085-4087.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li H, White S A. RNA aptamers for yeast ribosomal protein L32 have a conserved purine-rich internal loop. RNA. 1997;3:245–254. [PMC free article] [PubMed] [Google Scholar]
- 37.Linial M L, Miller A D. Retroviral RNA packaging: sequence requirements and implications. In: Swanstrom R, Vogt P K, editors. Retroviruses: strategies of replication. Berlin, Germany: Springer-Verlag; 1990. pp. 125–152. [DOI] [PubMed] [Google Scholar]
- 38.Luban J, Goff S P. Mutational analysis of cis acting packaging signals in HIV-1 RNA. J Virol. 1994;68:3784–3793. doi: 10.1128/jvi.68.6.3784-3793.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mangel W F, Delius H, Duesburg P H. Structure and molecular weight of the 60-70S RNA and the 30-40S RNA of Rous sarcoma virus. Proc Natl Acad Sci USA. 1994;71:4541–4545. doi: 10.1073/pnas.71.11.4541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marquet R, Paillart J-C, Skripkin E, Ehresmann C, Ehresmann B. Dimerization of HIV-1 RNA involves sequences located upstream of the splice donor site. Nucleic Acids Res. 1994;22:145–151. doi: 10.1093/nar/22.2.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McBride M S, Panganiban A T. The human immunodeficiency virus type 1 encapsidation site is a multipartite RNA element composed of functional hairpin structures. J Virol. 1996;70:2963–2973. doi: 10.1128/jvi.70.5.2963-2973.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McBride M S, Schwartz M D, Panganiban A T. Efficient encapsidation of human immunodeficiency virus type 1 vectors and further characterization of cis elements required for encapsidation. J Virol. 1997;71:4544–4554. doi: 10.1128/jvi.71.6.4544-4554.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McCann E, Lever A M L. Location of cis-acting signals important for RNA encapsidation in the leader sequence of human immunodeficiency virus type 2. J Virol. 1997;71:4133–4137. doi: 10.1128/jvi.71.5.4133-4137.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miele G, Mouland A, Harrison G P, Cohen E, Lever A M L. The human immunodeficiency virus type 1 5′ packaging signal structure affects translation but does not function as an internal ribosome entry site structure. J Virol. 1996;70:944–951. doi: 10.1128/jvi.70.2.944-951.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mougel M, Barklis E. A role for two hairpin structures as a core RNA encapsidation signal in murine leukemia virus virions. J Virol. 1997;71:8061–8065. doi: 10.1128/jvi.71.10.8061-8065.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Muriaux D, Girard P-M, Bonnet-Mathonière B, Paoletti J. Dimerization of HIV-1LAI at low ionic strength. An autocomplementary sequence in the 5′ leader region is evidenced by an antisense oligonucleotide. J Biol Chem. 1995;270:8209–8216. doi: 10.1074/jbc.270.14.8209. [DOI] [PubMed] [Google Scholar]
- 47.Murti K G, Bondurant M, Tereba A. Secondary structural features in the 70S RNAs of Moloney murine leukemia and Rous sarcoma viruses as observed by electron microscopy. J Virol. 1981;37:411–419. doi: 10.1128/jvi.37.1.411-419.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Osterburg G, Sommer R. Computer support of DNA sequence analysis. Comput Programs Biomed. 1981;13:101–109. doi: 10.1016/0010-468x(81)90088-x. [DOI] [PubMed] [Google Scholar]
- 49.Paillart J-C, Skripkin E, Ehresmann B, Ehresmann C, Marquet R. A loop-loop “kissing” complex is the essential part of the dimer linkage of genomic HIV-1 RNA. Proc Natl Acad Sci USA. 1996;93:5572–5577. doi: 10.1073/pnas.93.11.5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Paillart J-C, Berthoux L, Ottmann M, Darlix J-L, Marquet R, Ehresmann B, Ehresmann C. A dual role of the putative RNA dimerization initiation site of HIV-1 in genomic RNA packaging and proviral synthesis. J Virol. 1996;70:8348–8354. doi: 10.1128/jvi.70.12.8348-8354.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Paillart J-C, Westhof E, Ehresmann C, Ehresmann B, Marquet R. Noncanonical interactions in a kissing loop complex: the dimerisation initiation site of HIV-1 genomic RNA. J Mol Biol. 1997;270:36–49. doi: 10.1006/jmbi.1997.1096. [DOI] [PubMed] [Google Scholar]
- 52.Peterson R D, Feigon J. Structural change in rev responsive element RNA of HIV-1 on binding rev peptide. J Mol Biol. 1996;264:863–877. doi: 10.1006/jmbi.1996.0683. [DOI] [PubMed] [Google Scholar]
- 53.Potts B J. Mini RT assay. In: Aldovini A, Walker B D, editors. Techniques in HIV research. New York, N.Y: Stockton Press; 1990. pp. 103–106. [Google Scholar]
- 54.Rizvi T A, Panganiban A T. Simian immunodeficiency virus RNA is efficiently encapsidated by human immunodeficiency virus type 1 particles. J Virol. 1993;67:2681–2688. doi: 10.1128/jvi.67.5.2681-2688.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Romaniuk P J, Lowary P, Wu H N, Stormo G, Uhlenbeck O C. RNA binding site of R17 coat protein. Biochemistry. 1987;26:1563–1568. doi: 10.1021/bi00380a011. [DOI] [PubMed] [Google Scholar]
- 56.Rosen C A, Terwilliger A, Dayton J, Sodroski J G, Haseltine W A. Intragenic cis-acting ART responsive sequences of HIV-1. Proc Natl Acad Sci USA. 1988;85:2071–2085. doi: 10.1073/pnas.85.7.2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sakaguchi K, Zambrano N, Baldwin E T, Shapiro B A, Erickson J W, Omichinski J G, Clore G M, Gronenborn A M, Appella E. Identification of a binding site for the HIV-1 nucleocapsid protein. Proc Natl Acad Sci USA. 1993;90:5219–5223. doi: 10.1073/pnas.90.11.5219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schlesinger S, Makino S, Linial M L. cis-acting genomic elements and trans-acting proteins involved in the assembly of RNA viruses. Semin Virol. 1994;5:39–49. doi: 10.1006/smvy.1994.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Selden R F. Transfection using DEAE-dextran. In: Asubel R, Brent R E, Kingston D D, Moore J G, Seidman J A, Smith J A, Struhl K, editors. Current protocols in molecular biology. Vol. 1. New York, N.Y: Greene Publishing Associates and Wiley Interscience; 1987. pp. 9.2.1.–9.2.6. [Google Scholar]
- 60.Skripkin E, Paillart J-C, Marquet R, Ehresmann B, Ehresmann C. Identification of the primary site of HIV-1 RNA dimerization in vitro. Proc Natl Acad Sci USA. 1994;91:4945–4949. doi: 10.1073/pnas.91.11.4945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sundquist W I, Heaphy S. Evidence for interstrand quadruplex formation in the dimerization of HIV-1 genomic RNA. Proc Natl Acad Sci USA. 1993;90:3393–3397. doi: 10.1073/pnas.90.8.3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Valegard K, Murray J B, Stockley P G, Stonehouse N J, Lijas L. Crystal structure of an RNA bacteriophage coat protein-operator complex. Nature (London) 1994;371:623–626. doi: 10.1038/371623a0. [DOI] [PubMed] [Google Scholar]
- 63.White S A, Li H, Rauch M E. A thermodynamic and mutational analysis of an RNA purine loop as a protein binding site. J Biomol Struct Dyn. 1995;13:285–299. doi: 10.1080/07391102.1995.10508840. [DOI] [PubMed] [Google Scholar]
- 64.Williamson J R, Raghuraman M K, Cech T R. Monovalent cation-induced structure of telomeric DNA: the G-quartet model. Cell. 1989;59:871–880. doi: 10.1016/0092-8674(89)90610-7. [DOI] [PubMed] [Google Scholar]
- 65.Yang S, Temin H M. A double hairpin structure is necessary for the encapsidation of spleen necrosis virus retroviral RNA. EMBO J. 1994;13:713–726. doi: 10.1002/j.1460-2075.1994.tb06311.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zuker M. On finding all suboptimal foldings of an RNA molecule. Science. 1989;244:48–52. doi: 10.1126/science.2468181. [DOI] [PubMed] [Google Scholar]