Abstract
A number of specific point mutations in the human cytomegalovirus (HCMV) DNA polymerase (UL54) gene have been tentatively associated with decreased susceptibility to antiviral agents and consequently with clinical failure. To precisely determine the roles of UL54 mutations in HCMV drug resistance, recombinant UL54 mutant viruses were generated by using cotransfection of nine overlapping HCMV DNA fragments into permissive fibroblasts, and their drug susceptibility profiles were determined. Amino acid substitutions located in UL54 conserved region IV (N408D, F412C, and F412V), region V (A987G), and δ-region C (L501I, K513E, P522S, and L545S) conferred various levels of resistance to cidofovir and ganciclovir. Mutations in region II (T700A and V715M) and region VI (V781I) were associated with resistance to foscarnet and adefovir. The region II mutations also conferred moderate resistance to lobucavir. In contrast to mutations in other UL54 conserved regions, those residing specifically in region III (L802M, K805Q, and T821I) were associated with various drug susceptibility profiles. Mutations located outside the known UL54 conserved regions (S676G and V759M) did not confer any significant changes in HCMV drug susceptibility. Predominantly an additive effect of multiple UL54 mutations with respect to the final drug resistance phenotype was demonstrated. Finally, the influence of selected UL54 mutations on the susceptibility of viral DNA replication to antiviral drugs was characterized by using a transient-transfection-plus-infection assay. Results of this work exemplify specific roles of the UL54 conserved regions in the development of HCMV drug resistance and may help guide optimization of HCMV therapy.
Despite the recent decline in the incidence of human cytomegalovirus (HCMV) infections in AIDS patients due to the implementation of highly active antiretroviral therapy, HCMV still remains a significant pathogen in human immunodeficiency virus-infected individuals (26, 30). In addition, immunocompromised patients undergoing bone marrow or solid organ transplantations belong to a group at high risk for HCMV infections (42). Ganciclovir (GCV), cidofovir (CDV), and foscarnet (PFA) are currently used for the management of HCMV disease (14, 19, 32). All three agents target viral DNA replication. GCV is a nucleoside analog that requires triphosphorylation to its active form. The first phosphorylation step is dependent on the viral phosphotransferase encoded by the UL97 gene (35, 49). CDV is a nucleoside monophosphate analog, and its activation to CDV-diphosphate does not require any product of viral infection (15). The active metabolites of both CDV and GCV act as alternative substrates and competitive inhibitors of the HCMV DNA polymerase (DNA Pol) encoded by the UL54 gene (28, 38). PFA, on the other hand, is an analog of inorganic pyrophosphate and as such functions directly as a noncompetitive inhibitor of the UL54 polymerase (39).
Several additional anti-HCMV drugs are currently under clinical evaluation. Among others, adefovir (ADV) [9-(2-phosphonomethoxyethyl)adenine] and lobucavir (LBV) (cyclobut-G) have been identified as highly effective inhibitors of HCMV replication in vitro. Similar to the case for CDV and GCV, their active forms, ADV-diphosphate and LBV-triphosphate, are potent competitive inhibitors of HCMV DNA Pol (51, 55).
HCMV strains with decreased drug susceptibility can be selected in cell culture as well as in patients undergoing anti-HCMV therapy. It has been suggested that the development of drug resistance in patients may result in clinical disease progression (22, 45, 53). Resistance to GCV is associated with specific sequence alterations in the UL97 gene alone (3, 11, 37) or in combination with UL54 mutations (6, 44, 50). GCV-resistant strains expressing only UL54 mutations are rare. Recently, it has been demonstrated that UL97 mutations are usually selected after shorter periods of GCV treatment and confer low-level GCV resistance, while the combination of UL97 and UL54 alterations arises predominantly during extended GCV therapy and such isolates exhibit high-level GCV resistance and cross-resistance to CDV in vitro (44). PFA-resistant viruses have not been detected when PFA is administered as first-line treatment (47). However, PFA-resistant strains can arise when PFA treatment follows extensive GCV therapy (13, 43, 44). Selection of CDV-resistant strains during CDV therapy has not yet been reported (9).
A large number of UL54 mutations have been detected in drug-resistant clinical strains (21, 44). The majority of these alterations are located within UL54 conserved regions. Eight conserved regions (designated I to VII and δ-region C) with significant amino acid sequence homology with other DNA-dependent DNA polymerases have been identified in UL54 (29, 54, 58). Specific amino acid residues located within regions I, II, and III have been shown to directly participate in the binding of deoxynucleoside triphosphates, chelating the Mg2+ ion, and interacting with primer and template (52, 57). Involvement in similar types of interactions is also expected for regions V, VI, and VII. On the other hand, it can be concluded from sequence homology with other α-like DNA polymerases that specific domains located within conserved region IV and δ-region C are probably involved in the 3′-5′ exonuclease function of HCMV DNA Pol (5, 7).
Based on the characterization of drug resistance-associated mutations in other herpesviruses, namely, herpes simplex virus type 1 (HSV-1), the UL54 conserved regions are expected to participate in interactions with antiviral inhibitors. However, obtaining absolute proof of the association of UL54 mutations with drug resistance has been difficult due to limitations of marker transfer as the only technique available for generation of HCMV recombinants expressing UL54 mutations (36, 50). Only a limited number of recombinants have been constructed by using this laborious, inefficient, and time-consuming approach, and these have shown that UL54 alterations L501I and A987G confer decreased susceptibility to GCV and CDV (36, 50). Similarly, the T700A and V715M mutations have been associated with PFA resistance (4). The same technique was recently used to prove the role of the F412C and L802M mutations in CDV-GCV and GCV-PFA resistance, respectively (13).
A more detailed knowledge of drug resistance phenotypes associated with specific UL54 alterations could be helpful in optimizing drug regimens to achieve more effective treatment of HCMV infections. Therefore, we developed a strategy for the efficient construction of UL54 mutant recombinants which is derived from the previously described cosmid cotransfection approach (31). Due to the large number and diverse character of the mutations studied, we were able to define the roles played by the UL54 conserved regions in HCMV drug resistance.
MATERIALS AND METHODS
Cells and viruses.
HFL-1 (ATCC CCL-153) and HEL-299 (ATCC CCL-137) human diploid lung fibroblasts and normal human dermal fibroblasts (NHDF) (Clonetics) were propagated in Eagle’s minimum essential medium supplemented with 10% fetal calf serum, nonessential amino acids, 100 U of penicillin per ml, and 100 μg of streptomycin per ml. The cells were not used beyond 10 passages. HCMV strain AD169 and strain Towne were grown in NHDF cells.
DNA constructs.
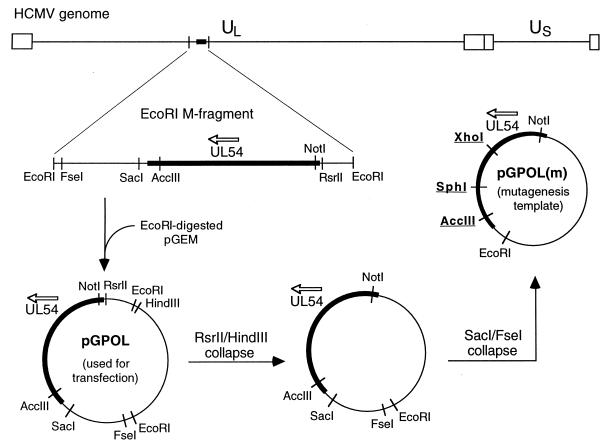
Construction of the cosmids containing Towne strain DNA fragments (Tn46, Tn45, Tn47, Tn44, Tn26, Tn15, and Tn20), as well as their sizes and locations in the HCMV genome, has been described previously (31). The viral fragment of Tn24 corresponded to the region between nucleotides (nt) 39479 and 77110 of the AD169 genome. Cloning of the EcoRI M fragment from the AD169 genome into pGEM-3Z (Promega) to generate pGPOL is described elsewhere (16), and the construction of pGPOL(m) from pGPOL is shown in Fig. 1. Plasmid pCOS47 was generated by cloning a 12-kb fragment (nucleotides 80526 to 92898) from a NotI-digested Tn47 cosmid into the NotI site of pGEM-3Z. Plasmid pSP50, used for the transient-transfection and -infection assay, was a generous gift of Greg Pari, and its construction is described elsewhere (1).
FIG. 1.
Construction of the pGPOL and pGPOL(m) plasmids. The key restriction sites used for UL54 mutagenesis are underlined.
Site-directed mutagenesis of the UL54 gene.
Mutagenesis was performed by a two-step PCR amplification procedure (27) with the Expand High Fidelity PCR System (Boehringer Mannheim) and pGPOL(m) as the template. Depending on the location of the mutation within UL54, either the XhoI-SphI or SphI-AccIII region of pGPOL(m) was amplified (Fig. 1). Each mutation was introduced by using a specific pair of 30-mer mutagenic primers. Oligonucleotide pairs 5′GA CGTGGACGTCTACGAGTTCCCT3′-5′CGCTCTAGCATGTCGCGACCG ATG3′ and 5′TACCAGGGCGCCACGGTGTTTGAGCCC3′-5′GCCGTGCTCGCGCACGTAGCTCGG3′ were used as outer primers for amplification of the XhoI-SphI and SphI-AccIII regions, respectively. After linearization of the template by HindIII digestion, the two products of the first PCR step were purified by agarose gel electrophoresis and joined by the second PCR amplification with the corresponding outer primers. The final PCR product was digested with either XhoI-SphI or SphI-AccIII restriction enzymes and subcloned back into pGPOL(m). The correct nucleotide sequence of the entire amplified region was verified by sequence analysis of both DNA strands. After digestion of pGPOL(m) with NotI-AccIII, the resulting 3.1-kb fragment carrying the desired UL54 mutation was subcloned into pGPOL, which was subsequently used for generation of the mutant recombinant viruses.
Generation of UL54 mutant viruses.
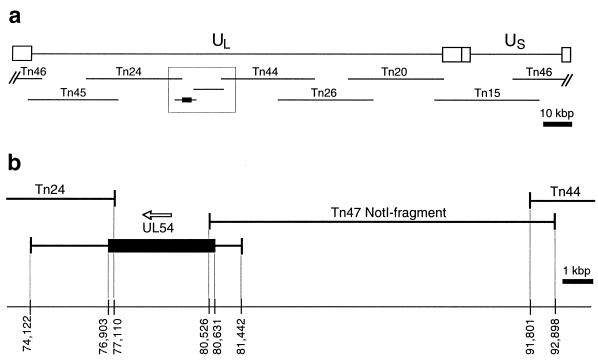
The viruses were generated by cotransfection of nine overlapping viral DNA fragments (Fig. 2) into HEL-299 or HFL-1 cells. The HCMV DNA fragments Tn46, Tn45, Tn24, Tn44, Tn26, Tn20, and Tn15 were released from the cosmid vector by digestion of 6 μg of each cosmid with PacI and mixed with 3 μg of EcoRI-digested pGPOL carrying the respective UL54 mutation and 3 μg of NotI-digested pCOS47. The mixture was ethanol precipitated and resuspended in 120 μl of TE buffer (10 mM Tris-HCl [pH 8.0] and 1 mM EDTA). The transfection of CaPO4 precipitation was performed for each virus in triplicate as described previously (31) with minor modifications. Briefly, 10 μg of the DNA mixture in 0.5 ml of 250 mM CaCl2 was mixed with the same volume of 2× HBS buffer (280 mM NaCl, 10 mM KCl, 1.4 mM Na2HPO4, 5.6 mM glucose, and 20 mM HEPES [pH 7.1]) and immediately added to 1.5 × 106 cells in a 25-cm2 flask seeded 3 to 4 h earlier. After 30 min at 37°C with occasional rocking, 2 ml of fresh medium was added, and the cells were incubated for additional 4 to 5 h. Then, the medium was removed, and cells were layered with 3 ml of 1× HBS–15% glycerol for 3 min, washed twice with 4 ml of medium, and fed with fresh medium supplemented with 15% fetal calf serum. Transfected cultures were refed every 3 to 4 days, and virus plaques were usually detected 10 to 16 days following transfection. Stocks of mutant viruses were prepared from infected cells and stored in growth medium with 10% dimethyl sulfoxide. To verify the size of the recombined UL54 gene and the presence of the desired mutation in each virus, DNA was purified from 105 infected cells by using the QIAamp Tissue Kit (Qiagen), the full-length UL54 gene was amplified by PCR with primers 5′AACTGGATATCTAGGTGCTGCATG3′ and 5′CTCAGTCTCAGCAGCATCATCACC3′, and sequence analysis of the PCR product was performed.
FIG. 2.
DNA fragments used for construction of UL54 mutant recombinant viruses. (a) Locations of all nine DNA fragments in the HCMV genome; (b) a detailed view of the region surrounding the UL54 gene. The numbers indicate positions of fragment termini according to the nucleotide numbering of the HCMV AD169 genome.
Structural analysis of viral genomes.
To purify DNA from recombinant viruses, extracellular virions were collected from 10 ml of culture medium by centrifugation at 100,000 × g for 90 min. The virus pellets were resuspended in 0.5 ml of TNE-sodium dodecyl sulfate (SDS) buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 10 mM EDTA, and 1% SDS) and incubated with 0.2 mg of proteinase K for 3 h at 50°C. DNA was extracted twice with phenol-chloroform-isoamylalcohol (25:24:1) and once with chloroform-isoamylalcohol (24:1), precipitated with ethanol, and resuspended in TE buffer. After digestion with EcoRI and separation on a 0.4% agarose gel, DNA was transferred onto a Hybond-N+ membrane (Amersham) by overnight capillary blotting in 20× SSC (300 mM Na3-citrate, 3 M NaCl). The membrane was then probed with various cosmids labeled by the ECL direct nucleic acid labeling system (Amersham). Labeling of probes, membrane hybridization, washing, and detection were carried out according to manufacturer’s instructions.
Drug susceptibility assays.
Sensitivities of recombinant viruses to CDV (Gilead Sciences), GCV (Hoffmann-La Roche), PFA (Sigma), ADV (Gilead Sciences), and LBV (Bristol-Myers Squibb) were determined by the plaque reduction assay (10). Freshly confluent NHDF cells in 24-well plates were infected with 30 to 60 PFU per well and incubated in either the absence or presence of each drug at various concentrations. Duplicate wells were maintained with each drug concentration. After 5 to 7 days, the plates were stained with 0.1% crystal violet in 20% methanol, and the number of plaques in each well was visually determined with a dissecting microscope and expressed as a percentage of the number of plaques detected in the absence of the drug. The 50% drug inhibitory concentration (IC50) was determined from the semilogarithmic plot of plaque percentage versus drug concentration. The IC50s for each mutant virus were determined at least three times with two independently generated recombinants. As a control, the UL54 wild-type recombinant virus was included in all experiments, and the susceptibility of mutant viruses to each drug was expressed as the fold change in IC50 relative to that for the UL54 wild-type virus.
Transient-transfection-plus-infection assay.
A modification of a previously described procedure (41) was used. NHDF cells were seeded into six-well plates at a density of 4 × 105 cells per well, and 24 h later the medium was replaced with 1.5 ml of fresh minimum essential medium. After further incubation for 2 to 3 h, transfection by CaPO4 precipitation was performed. Briefly, 2 μg of plasmid pSP50 in 125 μl of 250 mM CaCl2 was mixed with an equal volume of 2× BBS [50 mM N,N-bis(2-hydroxyethyl)-2-aminoethanesulfonic acid (BES), 280 mM NaCl, 1.5 mM Na2HPO4 (pH 6.95)] and incubated for 20 min at room temperature. This mixture was added dropwise to each well, and the cells were incubated at 37°C and 3% CO2 for 24 h. Each well was then washed twice with 5 ml of medium, and 3 ml of fresh medium was added. After an additional 24 h of incubation at 37°C and 5% CO2, cells were infected with either the wild-type or mutant recombinant virus at 10 PFU per cell for 3 h at 37°C. The wells were extensively washed with phosphate-buffered saline and incubated in either the absence or presence of various concentrations of CDV, GCV, or PFA. Total DNA was purified from each well 120 h after infection. Cells were washed twice with 5 ml of phosphate-buffered saline and directly solubilized in 0.5 ml of lysis buffer (10 mM Tris-HCl [pH 8.0], 10 mM EDTA, and 2% SDS). The lysates were incubated with 0.1 mg of proteinase K for 3 h at 50°C, and DNA was extracted as described above, ethanol precipitated, and resuspended in 150 μl of TE buffer. Digestion of 2 μg of purified DNA with 15 U of EcoRI and 15 U of DpnI in 20 μl was performed for 24 h, and half of the digest was separated on a 0.7% agarose gel, transferred onto a Hybond-N+ membrane (Amersham) by overnight capillary blotting in 20× SSC, and probed with pGEM-3Z which was labeled with [α-32P]dATP (Amersham) (3,000 Ci/mmol) by using the Prime-It II random priming kit (Stratagene). The membrane was prehybridized and hybridized with the probe in Gold hybridization buffer (Amersham) for 2 and 16 h, respectively, at 42°C and washed twice with 0.2× SSC–0.2% SDS at 55°C for 30 min and twice with 1× SSC at room temperature for 5 min. The membrane was first exposed to X-ray film at −70°C for 16 to 24 h and then scanned by using the Storm 860 PhosphorImager system (Molecular Dynamics). The bands corresponding to intracellularly replicated full-length pSP50 were quantified and expressed as a percentage of plasmid replication in the absence of drug. The IC50 was determined for each drug from the semilogarithmic plot of plasmid replication versus drug concentration.
RESULTS
Recombination of viral DNA fragments as an efficient strategy for construction of UL54 mutant viruses.
Recently, a system for construction of recombinant HCMVs via intracellular homologous recombination of eight overlapping viral DNA fragments was developed (31). The Towne strain DNA fragments were cloned into cosmid vectors and used successfully to create large-scale modifications of the HCMV genome. The sizes of these fragments, however, did not allow for efficient introduction of point mutations into the viral genome. To circumvent this obstacle, the original Tn47 fragment containing the UL54 gene was replaced by two smaller DNA fragments cloned into plasmid vectors (Fig. 2). Plasmid pGPOL contained the UL54 gene within the 7.4-kbp EcoRI M fragment cloned from the AD169 genome. The UL54 gene from the AD169 strain was chosen because its sequence represents a standard consensus most frequently used for comparative sequence analysis of drug-resistant HCMV strains (4, 44). A gap between the M fragment and an adjacent Tn44 fragment was spanned by a 12-kbp NotI fragment from the pCOS47 plasmid. Cotransfection into human lung fibroblasts of seven PacI-digested cosmids together with EcoRI-digested pGPOL and NotI-digested pCOS47 generated infectious recombinant virus, with plaques usually detectable within 10 to 16 days. This suggests that the recombination of nine viral DNA fragments occurred at an efficiency comparable to that of the recombination of the original eight cosmid-derived fragments despite significantly shorter overlapping regions between pGPOL, pCOS47, and the neighboring cosmids.
The construction of mutant viruses was carried out by using pGPOL plasmids which contained the UL54 mutation(s) of interest. Since all the mutations studied were located in the nonoverlapping region of the M fragment, the virus progeny generated by the cotransfection always represented a homogeneous population of the mutant strain, and no further selection and purification of the viruses were required.
The UL54 wild-type recombinant virus was tested for its susceptibility to five antiviral drugs. By using the plaque reduction assay, mean IC50s of 0.75, 3.5, 45, 42, and 3.8 μM were determined for CDV, GCV, PFA, ADV, and LBV, respectively, from at least three independent experiments. The UL54 wild-type recombinant virus exhibited drug susceptibilities identical to those of Towne and AD169 (data not shown).
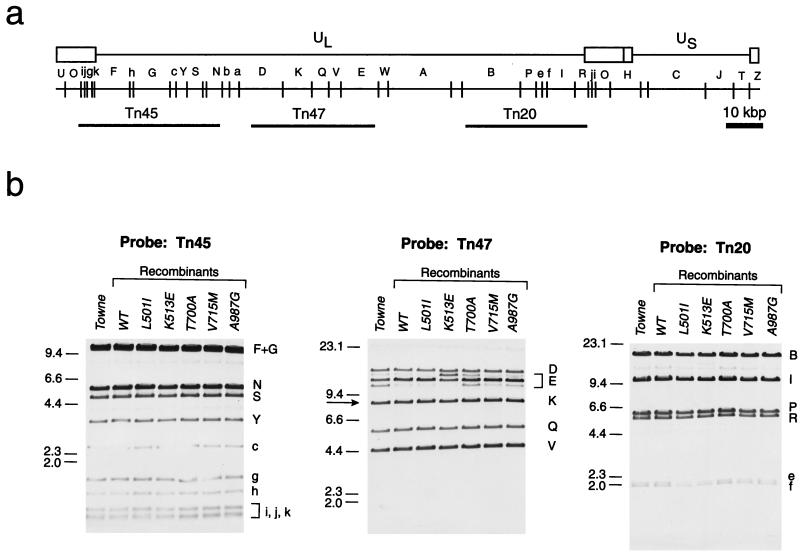
To verify the genomic structure of the recombinant viruses, especially in the region surrounding the UL54 gene, we purified DNAs from the UL54 wild-type and selected mutant recombinant viruses and performed a Southern blot analysis of EcoRI-digested DNA with independent cosmids as hybridization probes. Figure 3 shows representative results with three different probes. Overall, this analysis with the set of six cosmids revealed no structural differences compared to the genome of the parental Towne strain except in the heterogeneity of the EcoRI E fragment. However, several prevalent forms of the E fragment have been previously described for the Towne genome, and they reflect the heterogeneity of the region surrounding the origin of replication (31).
FIG. 3.
Structure of recombinant viral genomes. (a) EcoRI restriction map of the HCMV Towne genome and locations of the cosmid probes Tn45, Tn47, and Tn20 (adapted from reference 31). (b) Southern analyses with the corresponding cosmid probes. An arrow in the Tn47 panel indicates the EcoRI K fragment harboring the full-length UL54 gene, and the letters to the right of each panel correspond to the alphabetical designations of Towne EcoRI fragments. Numbers to the left indicate molecular sizes in kilobase pairs. WT, wild type. The analyses were performed as described in Materials and Methods.
Mutations associated with cross-resistance to CDV and GCV are located in the N- and C-terminal conserved regions of the UL54 gene.
In total, 17 recombinant viruses expressing various single point mutations in the UL54 gene were generated, and their drug susceptibilities were characterized. Fourteen of these alterations were previously observed in drug-resistant clinical isolates from patients treated with one or more anti-HCMV drugs, while the remaining three were identified in strains selected in vitro in the presence of GCV.
The in vitro susceptibility data for all 17 recombinant viruses with five anti-HCMV agents are summarized in Table 1. Nine mutant recombinants showed decreased susceptibility to GCV. Notably, eight of these viruses also exhibited a coincident decrease in the susceptibility to CDV. Amino acid substitutions N408D, F412C, and F412V, located in the most N-terminal conserved region IV, as well as substitutions L501I, K513E, P522S, and L545S, residing in the adjacent downstream δ-region C, conferred various levels of decreased susceptibility to both drugs. It is true for most of these CDV-GCV-resistant viruses that their susceptibility to CDV was decreased more substantially than that to GCV. The only exceptions were viruses with mutations N408D and P552S, which exhibited similar levels of resistance to both drugs. Interestingly, adjacent mutations located in the same conserved region conferred significantly different levels of resistance to CDV. For example, mutations F412C and K513E were associated with a threefold higher resistance to CDV than the N408D and P522S substitutions. On the other hand, both the Phe-to-Cys and Phe-to-Val substitutions at codon 412 resulted in almost identical levels of resistance to CDV and GCV.
TABLE 1.
Drug susceptibilities of recombinant HCMVs expressing various mutations in the UL54 gene
| Region | UL54 alteration | Reference(s)b | Selection drug(s)c | Fold change in IC50 relative to that for the wild-type recombinanta
|
||||
|---|---|---|---|---|---|---|---|---|
| GCV | CDV | PFA | ADV | LBV | ||||
| IV | N408D | 44 | GCV, PFA | 4.9 | 5.6 | 1.3 | −4.0 | 1.6 |
| F412C | 13, 44 | GCV, PFA | 4.2 | 18.0 | 1.2 | −4.2 | 1.2 | |
| F412Vd | 36 | GCV | 4.3 | 15.5 | 1.1 | −4.2 | 1.0 | |
| δ C | L501Id | 36 | GCV | 6.0 | 9.1 | 1.4 | −2.0 | 1.4 |
| K513E | 44 | GCV, PFA | 5.0 | 9.1 | 1.4 | −2.0 | 1.2 | |
| P522S | 8 | GCV, PFA, CDV | 3.1 | 3.6 | 1.1 | −2.5 | 1.1 | |
| L545S | 9 | GCV, PFA, CDV | 3.5 | 9.1 | 1.2 | −5.9 | 1.2 | |
| D588E | 44 | GCV, PFA | 1.3 | 1.1 | 2.3 | 2.0 | 1.1 | |
| II | T700A | 2, 4 | GCV, PFA | 1.2 | 1.3 | 5.8 | 5.8 | 3.1 |
| V715M | 2, 4 | GCV, PFA | 1.3 | 1.1 | 9.5 | 6.0 | 3.1 | |
| VI | V781I | 2 | GCV, PFA | 1.0 | 1.2 | 5.2 | 3.0 | 1.8 |
| III | L802M | 13, 44 | GCV, PFA | 1.1 | 0.9 | 3.2 | 2.8 | 1.3 |
| K805Q | 44 | GCV, PFA | 1.0 | 2.2 | −5.6 | −4.8 | −4.1 | |
| T821I | 44 | GCV, PFA | 4.5 | 1.9 | 21 | 6.4 | 3.6 | |
| V | A987Gd | 50 | GCV | 5.3 | 11.3 | 1.2 | 1.1 | 1.1 |
| Other | S676G | 44 | GCV, PFA | 1.1 | 1.2 | 0.9 | 1.4 | 1.0 |
| V759M | 8 | CDV | 1.5 | 1.1 | 1.1 | 1.2 | 1.3 | |
Values are averages from at least three determinations with two independently generated recombinant viruses. In all cases, the standard error was less than 35%. Boldface numbers indicate significant (i.e., at least twofold) changes in HCMV drug susceptibility. Positive and negative values represent fold increases and decreases, respectively, in IC50 relative to that of the UL54 wild-type recombinant virus.
Reference(s) describing the original identification of the UL54 alteration.
Therapy after which the mutation in clinical isolate was identified or drug used for selection in cell culture.
UL54 alteration selected in cell culture.
Substitutions located in the carboxy portion of the UL54 polypeptide can also be associated with decreased sensitivity to CDV and GCV. For example, a recombinant virus with the in vitro GCV-selected A987G alteration residing in region V, the most C-terminal UL54 conserved domain, exhibited 11- and 5-fold decreased susceptibilities to CDV and GCV, respectively, compared to the wild-type UL54 recombinant virus. These findings are in agreement with previous characterization of the A987G recombinant virus constructed by marker transfer (50). Interestingly, this mutation has not yet been identified in any of the CDV-GCV-resistant clinical isolates.
It is noteworthy that in addition to being associated with CDV-GCV resistance, all alterations in region IV and δ-region C also conferred hypersensitivity to ADV. The recombinant viruses expressing these UL54 mutations exhibited ADV IC50s two- to sixfold lower than that of the UL54 wild-type virus. This indicates that CDV and ADV, despite being members of the same class of compounds, exert their anti-HCMV activity by different molecular mechanisms.
PFA resistance and cross-resistance to ADV map to the central part of the UL54 polypeptide.
It was previously demonstrated by marker transfer that mutations T700A and V715M, residing in the UL54 conserved region II, resulted in approximately a fivefold decrease in sensitivity to PFA (4). In that study, the UL54 gene of the T700A mutant virus contained additional sequence modifications which were considered to be unrelated to the PFA-resistant phenotype. By using cotransfection of viral DNA fragments, recombinant viruses that independently expressed the T700A or V715M alterations without additional changes in the UL54 sequence were generated. These mutations were associated with six- and ninefold elevations in the PFA IC50, respectively, compared to the wild-type virus (Table 1). In addition, both mutations also conferred approximately a sixfold increase in the ADV IC50 and a threefold increase in the LBV IC50. Similarly, the V781I mutation, located in the neighboring region VI, was found to be associated with decreased susceptibility to PFA and ADV, at levels comparable to those determined for the T700A mutant virus.
The D588E substitution, located at the very C terminus of δ-region C, also conferred a two- to threefold decrease in the sensitivity to PFA and ADV. Although amino acid residue 588 is not conserved among other herpesviruses, it resides within a region of sequence homology conserved among herpesvirus polymerases previously designated domain A (25).
Mutations in UL54 conserved region III are associated with various changes in HCMV drug susceptibility.
Several novel mutations within UL54 region III and in its immediate proximity were recently identified in HCMV drug-resistant clinical isolates (44). To characterize these genotypic changes, recombinant viruses independently expressing L802M, K805Q, and T821I alterations were constructed. Drug susceptibility assays revealed that the T821I mutation was associated with measurable elevations in GCV, ADV, and LBV IC50s but with a very significant, over 20-fold, increase in the PFA IC50 (Table 1). The recombinant virus expressing the K805Q mutation exhibited moderate CDV resistance together with marked hypersensitivity to PFA, ADV, and LBV. The L802M substitution, located in immediate vicinity of the N terminus of region III, conferred a moderate decrease in susceptibility only to PFA and ADV. A recombinant virus expressing the L802M mutation was recently generated also by marker transfer. In contrast to our observations, this virus was shown to exhibit not only PFA resistance but also mild GCV resistance (13). However, in that study, two additional UL54 genetic changes were transferred along with the L802M mutation into the resulting recombinant virus, which may have influenced its final drug resistance phenotype.
Mutations located outside the UL54 conserved domains do not confer significant changes in drug susceptibility.
Mutations at codons 676 and 759, residing outside the known conserved domains of the UL54 gene, were also investigated by using recombinant viruses. The substitution S676G was identified in a clinical strain recovered from a retinitis patient treated with GCV and PFA and was tentatively associated with decreased susceptibility to GCV and CDV (44). The amino acid change V759M was found in the UL54 gene after its amplification from a blood sample of a patient receiving CDV therapy (8). However, neither S676G nor V759M conferred any significant change in drug susceptibility, suggesting that alterations located outside the defined UL54 conserved domains may not play a significant role in HCMV drug resistance.
Multiple UL54 mutations are predominantly additive with respect to the final level and profile of drug resistance.
Recent analysis of HCMV clinical isolates from AIDS patients treated sequentially with GCV and PFA revealed several strains with two mutations in the UL54 gene. These isolates showed increases in CDV, GCV, and PFA IC50s and therefore were considered to be multidrug resistant (13, 44). To elucidate the mutual effect of two UL54 mutations and their contributions to the final drug resistance phenotypes, each mutation was studied separately and three recombinant viruses expressing two UL54 mutations identified in resistant isolates were also generated and characterized. These three recombinants with double UL54 mutations exhibited multidrug resistance phenotypes, as did their corresponding clinical strains. As shown in Table 2, the susceptibility of the F412C-L802M mutant virus to CDV and GCV was similar to that of the virus expressing the F412C mutation alone. In addition, the threefold decrease in PFA susceptibility of the F412C-L802M mutant corresponded to that determined for the L802M virus. Similarly, the final levels of CDV-GCV and PFA resistance for the K513E-D588E mutant virus were comparable to those of independent K513E and D588E mutants, respectively. Interestingly, an additive effect was observed also with respect to final ADV susceptibility. The data show that the hypersensitivity to ADV due to the presence of the F412C or K513E mutation could be diminished or completely reversed by the D588E or L802M mutation conferring ADV resistance.
TABLE 2.
Drug susceptibilities of recombinant HCMVs expressing single and double mutations in the UL54 gene
| UL54 genotype | IC50 (μM)a
|
|||
|---|---|---|---|---|
| GCV | CDV | PFA | ADV | |
| Wild type | 3.5 | 0.75 | 45 | 42 |
| F412C | 15 (4.3) | 13 (17) | 54 (1.2) | 10 (−4.2) |
| L802M | 3.9 (1.1) | 0.7 (0.9) | 140 (3.1) | 120 (2.8) |
| F412C L802M | 22 (6.2) | 12 (16) | 140 (3.1) | 25 (−1.7) |
| K513E | 17.5 (5.0) | 6.9 (9.2) | 50 (1.1) | 20 (−1.9) |
| D588E | 4.2 (1.2) | 0.8 (1.1) | 110 (2.4) | 84 (2.0) |
| K513E D558E | 23 (6.6) | 8.3 (11.1) | 120 (2.7) | 33 (−1.3) |
| K805Q | 3.5 (1.0) | 1.7 (2.2) | 8.0 (−5.6) | 8.9 (−4.8) |
| T821I | 15.8 (4.5) | 1.4 (1.9) | 950 (21) | 260 (6.4) |
| K805Q T821I | 15.1 (4.3) | 1.6 (2.1) | 455 (10.1) | 170 (4.0) |
The IC50 values represent averages from at least three determinations with two independently generated recombinant viruses. In all cases, the standard error was less than 35%. The boldface numbers indicate a significant (i.e., at least twofold) change in HCMV drug susceptibility. Positive and negative numbers in parentheses represent fold increases and decreases, respectively, in the IC50 values of the mutant virus relative to that for the UL54 wild-type recombinant virus.
An additive contribution of the K805Q and T821I alterations to the final GCV resistance of the K805Q-T821I mutant virus was also determined. However, despite the finding that the T821I mutation alone conferred an approximately 20-fold decrease in susceptibility to PFA, the PFA IC50 for the K805Q- T821I mutant virus was more than 50-fold higher than that for the virus expressing K805Q alone. This suggests a nonadditive effect of these two mutations with respect to the final level of PFA susceptibility. A similar effect of these two mutations was observed also for the susceptibility to ADV.
Resistant viruses exhibit decreased sensitivity of viral DNA replication towards corresponding inhibitors.
The UL54 catalytic subunit of HCMV DNA Pol is the cornerstone of the viral DNA replication machinery and as such is the target for antiviral therapy. However, evidence that UL54 drug resistance-associated mutations directly diminish the sensitivity of viral DNA replication to antiviral drugs has not yet been presented. A transient-transfection-plus-infection assay was used to evaluate the correlation between the drug susceptibility of mutant viruses and the sensitivity of their DNA replication to corresponding inhibitors. In this type of assay, which was originally used for identification of the origin of HCMV lytic-phase DNA replication (oriLyt), intracellular replication of a plasmid containing the oriLyt sequence is driven by a complex of viral replication proteins provided in trans by viral infection (1). After its replication in infected cells, the plasmid is no longer methylated at specific sites and becomes resistant to DpnI digestion. Based on this fact, the intracellular replication of the plasmid can be quantitatively determined. Although the transient-transfection-plus-infection approach should not generate data significantly different from the direct measurement of viral DNA synthesis by a dot blot hybridization assay (48), it allows for more precise standardization based on the quantitation of the input of the transfected DNA. In addition, since the digested oriLyt plasmid is size fractioned on an agarose gel, the assay offers a possibility of detecting replication intermediates which may result from the inhibition of plasmid replication.
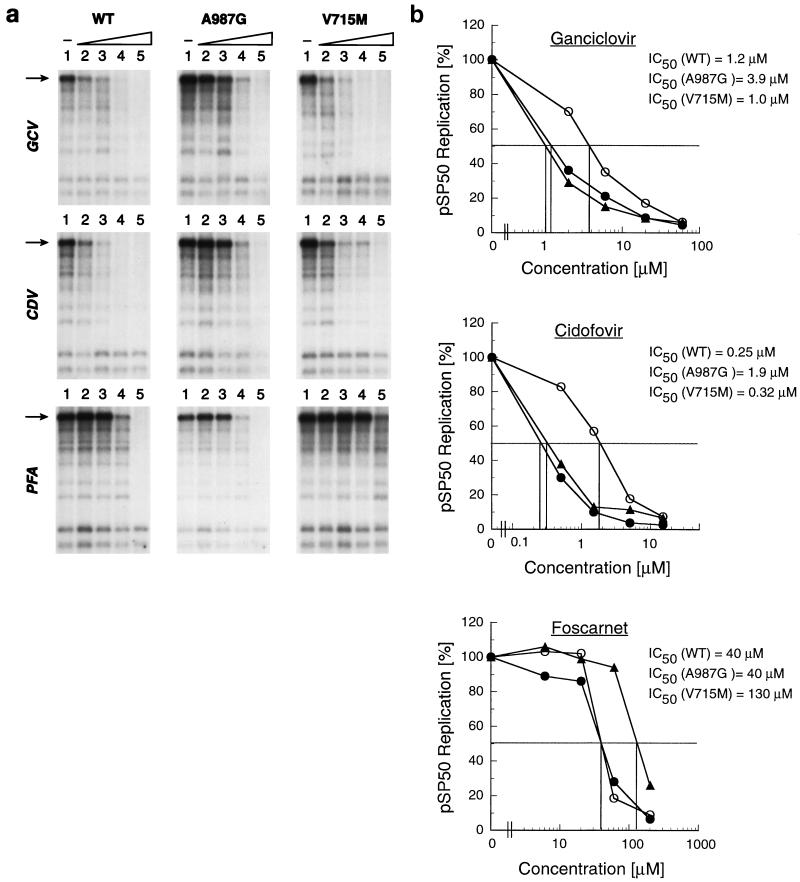
The sensitivity of oriLyt plasmid replication to high PFA concentrations was previously demonstrated (41). Our data indicate that the plasmid replication can also be blocked by CDV and GCV. To determine whether there was a difference in the response to antiviral agents of plasmid replication when driven either by wild-type polymerase or polymerase containing drug resistance-associated mutations, confluent human fibroblasts were transfected with plasmid pSP50 containing oriLyt and 24 h later were infected with either the wild-type or mutant recombinant virus. The A987G and V715M mutants, representing the CDV-GCV or PFA resistance phenotype, respectively, were selected for the experiment. Following infection, inhibitors were added at different concentrations, and the replication of pSP50 was determined after a 120-h incubation as described in Materials and Methods. Figure 4a shows the levels of pSP50 replication in the presence of CDV, GCV, and PFA after infection with the UL54 wild-type or mutant viruses. The IC50 for plasmid replication was determined for each antiviral drug from the dose-response curves (Fig. 4b). pSP50 replication after infection with the A987G mutant was six- and threefold less susceptible to CDV and GCV, respectively, than plasmid replication after infection with either the UL54 wild-type or V715M mutant virus. Comparatively, the susceptibility of pSP50 replication to PFA when driven by the V715M mutant virus was more than threefold lower than that after the infection with either the UL54 wild-type or A987G mutant virus. These data indicate a correlation between the drug susceptibility of the virus and the response of its DNA replication to antiviral drugs.
FIG. 4.
Drug sensitivity of HCMV oriLyt (pSP50) plasmid replication driven by the wild-type (WT) and drug-resistant UL54 mutant viruses. (a) The levels of pSP50 plasmid replication in the presence of various concentrations of GCV (lane 1, no drug; lanes 2 to 5, 2, 6, 20, and 60 μM, respectively), CDV (lane 1, no drug; lanes 2 to 5, 0.5, 1.5, 5, and 15 μM, respectively), and PFA (lane 1, no drug; lanes 2 to 5, 6, 20, 60, and 200 μM, respectively) were determined by the transient-transfection- plus-infection assay as described in Materials and Methods. The arrows indicate the intracellularly replicated to pSP50 plasmid resistant to DpnI digestion. (b) Drug dose-response curves and IC50s for inhibition of pSP50 replication after infection with the UL54 wild-type (solid circles), A987G (open circles), and V715M (solid triangles) recombinant viruses.
DISCUSSION
Treatment of HCMV infections in immunocompromised patients with antiviral drugs that target viral DNA Pol can lead to the development of specific sequence alterations in the UL54 gene which decrease the drug susceptibility of the virus. To study these alterations, we developed a novel approach for the construction of recombinant HCMVs expressing UL54 mutations. This method offers significant advantages over the marker transfer technique. Since homogeneous progeny of mutant virus is generated from the overlapping fragments, there is no need for further selection in the presence of drug or for subsequent extensive plaque purification. Consequently, the possible development of additional, independent mutations during the selection step after marker transfer is eliminated by using the cotransfection strategy.
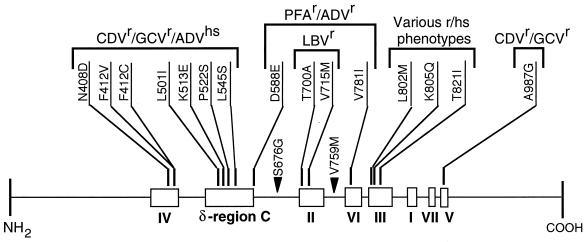
The study of 17 single-amino-acid substitutions in HCMV DNA Pol revealed two major distinct cross-resistance profiles. The CDV-GCV and PFA-ADV cross-resistance phenotypes are consistent with previous characterization of HCMV drug-resistant strains selected in vitro in the presence of various antiviral drugs (46). As shown in Fig. 5, most of the mutations conferring the same drug resistance phenotype cluster together in specific conserved regions. The overwhelming majority of the CDV-GCV resistance-associated alterations reside in region IV and in the N-terminal portion of δ-region C. The only exception is the mutation A987G located in region V. Since the functions predicted for the N- and C-terminal conserved regions are distinct from one another (7, 54), the molecular mechanisms by which drug susceptibility is altered may not be identical for the amino acid alterations residing in different conserved regions. The molecular mechanisms of action of CDV and GCV may consist of several independent steps. Active metabolites of both compounds compete with natural substrates for the deoxynucleoside triphosphate-binding site and also can be incorporated into the nascent chain during viral DNA replication. It has been shown in primer extension experiments with purified HCMV polymerase that the incorporation of CDV slows further elongation of the primer and that the enzyme is very inefficient in utilizing a template containing internally incorporated CDV molecules (56). Thus, based on the function assigned to the conserved domains, it could be expected that the alterations in regions III and V may decrease the enzyme affinity to the inhibitor. Those located in N-terminal conserved regions within or near the 3′-5′ exonuclease domains may modify the efficiency of incorporation of the inhibitor into and its removal from the 3′ end of the primer and/or the ability of the enzyme to utilize either primer or template with the inhibitor structure incorporated. A difference in the molecular mechanisms of CDV-GCV resistance conferred by the alterations located in the N-terminal and C-terminal conserved regions is supported also by the fact that region IV and δ-region C mutations, but not the region V A987G mutation, result in increased susceptibility of HCMV to ADV.
FIG. 5.
Conserved regions in HCMV DNA Pol and general locations of the drug resistance (r)- and drug hypersensitivity (hs)-associated mutations. The drugs for which the susceptibility has not changed are not listed. Solid triangles indicate mutations not associated with significant modifications of HCMV drug susceptibility. The boundaries of the conserved regions have been defined by Biron and Baldanti (6) and by Zhang et al. (58) as follows (amino acid numbering): IV, 379 to 421; δ-region C, 492 to 588; II, 696 to 742; VI, 771 to 790; III, 805 to 845; I, 905 to 919; VII, 962 to 970; and V, 978 to 988.
Conserved regions II and VI, located in the central part of the UL54 polypeptide, harbor amino acid substitutions conferring PFA-ADV resistance. The same phenotype was demonstrated also for the D588E mutation residing in δ-region C. In addition, mutations in region III significantly modify HCMV susceptibility to PFA. Such a wide distribution of PFA resistance-associated mutations across the central part of the UL54 polypeptide suggests that the binding site of the pyrophosphate moiety may be formed by a complex folding of several UL54 conserved regions. Based on the distribution of drug resistance-associated mutations, the involvement of remote amino acid residues in substrate binding has been previously proposed for HSV DNA Pol (18).
Despite being located in the same region, region III, mutations K805Q and T821I confer completely different drug susceptibility phenotypes. The K805Q mutation is associated with marked hypersensitivity to PFA, while the T821I substitution results in the highest PFA resistance ever reported among HCMV strains. Similarly, specific mutations in region III of HSV-1 DNA Pol could also confer either resistance (25) or hypersensitivity (33) to pyrophosphate analogs. Recently, an additional region III substitution, A809V, has been shown to confer 2.5- and 6-fold decreases in HCMV susceptibility to GCV and PFA, respectively (12). This phenotype differs from those associated with any of the above-mentioned two mutations. Thus, contrary to the case for mutations found in other UL54 conserved domains, those residing in region III are not strictly associated with either CDV-GCV or PFA-ADV resistance, and they confer various HCMV drug susceptibility profiles.
Surprisingly, alterations T700A, V715M, and T821I were the only ones observed which conferred some level of resistance to LBV. To date, the role of UL54 mutations in LBV resistance has not been established, and these findings represent the first identification of UL54 mutations associated with minor LBV resistance. It has been recently suggested that UL97 phosphotransferase does not participate in LBV phosphorylation. Thus, HCMV strains resistant to GCV due to the UL97 mutations still remain fully sensitive to LBV (51). Together, these data indicate that LBV is an anti-HCMV drug with a unique resistance profile.
The important roles of the UL54 conserved regions in the development of HCMV drug resistance are also illustrated by the findings that the substitutions S676G and V759M, located outside the known conserved regions, do not modify HCMV drug susceptibility. Interestingly, no mutation conferring an HCMV drug resistance phenotype has been identified to date in UL54 conserved region I or VII. Region I is the most conserved domain among the α-like DNA polymerases. Although region I contains a stretch of highly invariable amino acids (54), which might be less tolerant to substitutions, HSV-1 and varicella-zoster virus strains resistant to acyclovir and pyrophosphate analogs due to mutations in region I have been identified (17, 23). A mutation in HSV-1 region VII conferring PFA and acyclovir resistance has also been described (29). This suggests that analogous mutations might also exist in HCMV drug-resistant strains.
Recent genotypic analysis of numerous clinical strains exhibiting decreased susceptibility to GCV, CDV, and PFA revealed various combinations of two mutations in their UL54 genes (44). Similar to what has been observed with alterations in human immunodeficiency virus reverse transcriptase (34), the final drug resistance phenotype of the virus with multiple UL54 alterations may result from mutual additive, synergistic, or antagonistic interactions between the single alterations. Furthermore, it is possible that certain combinations of alterations may generate a novel, unexpected drug resistance profile. Therefore, three recombinant viruses expressing specific combinations of two UL54 alterations observed in clinical isolates were constructed. Their characterization revealed predominantly an additive effect of these mutations with respect to the final profile and level of HCMV drug resistance.
It has been reported that HSV-1 DNA polymerase purified from cells infected with drug-resistant strains was less sensitive to corresponding inhibitors (20, 24). However, this correlation has not yet been extended to HCMV. Since at least six virus-encoded proteins physically participate in the replication of HCMV DNA (40), the DNA Pol alone may not precisely reflect the functionality of the entire virus replication complex and its susceptibility to inhibitors. To circumvent this possible limitation, a transient-transfection and -infection assay was used to show that replication of a plasmid containing HCMV oriLyt was inhibited less efficiently in cells infected with drug-resistant strains than in cells infected with the wild-type virus. The data shown here clearly demonstrate that drug resistance-associated amino acid substitutions in HCMV DNA Pol diminish the sensitivity of viral DNA synthesis to inhibition by the corresponding antiviral agents.
This study significantly expands the limited information previously available concerning the role of HCMV DNA Pol mutations in drug resistance and contributes to the general understanding of the molecular mechanisms of HCMV drug resistance. Development of drug resistance during the treatment of HCMV infections has been associated with clinical progression of disease; therefore, such information may be helpful in further optimizing therapy for HCMV infections.
ACKNOWLEDGMENTS
We are exceptionally grateful to George Kemble from Aviron for the set of HCMV cosmids and for many helpful comments and discussions. We also thank Greg Pari from Hybridon for the kind gift of the pSP50 plasmid and Jay Toole and Mick Hitchcock from Gilead Sciences for critical reading of the manuscript.
REFERENCES
- 1.Anders D G, Kacica M A, Pari G, Punturieri S M. Boundaries and structure of human cytomegalovirus orilyt, a complex origin for lytic-phase DNA replication. J Virol. 1992;66:3373–3384. doi: 10.1128/jvi.66.6.3373-3384.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baldanti F, Sarasini A, Silini E, Barbi M, Lazzarin A, Biron K K, Gerna G. Four dually resistant human cytomegalovirus strains from AIDS patients: single mutations in UL97 and UL54 open reading frames are responsible for ganciclovir- and foscarnet-specific resistance, respectively. Scand J Infect Dis Supl. 1995;99:103–104. [PubMed] [Google Scholar]
- 3.Baldanti F, Silini E, Sarasini A, Talarico C L, Stanat S C, Biron K K, Furione M, Bono F, Palu G, Gerna G. A three-nucleotide deletion in the UL97 open reading frame is responsible for the ganciclovir resistance of a human cytomegalovirus clinical isolate. J Virol. 1995;69:796–800. doi: 10.1128/jvi.69.2.796-800.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baldanti F, Underwood M R, Stanat S C, Biron K K, Chou S, Sarasini A, Silini E, Gerna G. Single amino acid changes in the DNA polymerase confer foscarnet resistance and slow-growth phenotype, while mutations in the UL97-encoded phosphotransferase confer ganciclovir resistance in three double-resistant human cytomegalovirus strains recovered from patients with AIDS. J Virol. 1996;70:1390–1395. doi: 10.1128/jvi.70.3.1390-1395.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bernad A, Blanco L, Lazaro J M, Martin G, Salas M. A conserved 3′-5′ exonuclease active site in prokaryotic and eukaryotic DNA polymerases. Cell. 1989;59:219–228. doi: 10.1016/0092-8674(89)90883-0. [DOI] [PubMed] [Google Scholar]
- 6.Biron K K, Baldanti F. Nucleosides and foscarnet—mechanisms. In: Richman D D, editor. Antiviral drug resistance. New York, N.Y: John Wiley & Sons Ltd.; 1996. pp. 125–152. [Google Scholar]
- 7.Blanco L, Bernad A, Blasco M A, Salas M. A general structure for DNA-dependent DNA polymerases. Gene. 1991;100:27–38. doi: 10.1016/0378-1119(91)90346-d. [DOI] [PubMed] [Google Scholar]
- 8.Cherrington, J. M., et al. 1997. Unpublished data.
- 9.Cherrington, J. M., M. D. Fuller, P. D. Lamy, R. Miner, J. P. Lalezari, S. Nuessle, and W. L. Drew. In vitro antiviral susceptibilities of isolates from CMV retinitis patients receiving first or second line cidofovir therapy: relationship to clinical outcome. Submitted for publication. [DOI] [PubMed]
- 10.Cherrington J M, Miner R, Hitchcock M J M, Lalezari J P, Drew W L. Susceptibility of human cytomegalovirus (HCMV) to cidofovir is unchanged after limited in vivo exposure to various clinical regimens of drug. J Infect Dis. 1996;173:987–992. doi: 10.1093/infdis/173.4.987. [DOI] [PubMed] [Google Scholar]
- 11.Chou S, Erice A, Jordan M C, Vercellotti G M, Michels K R, Talarico C L, Stanat S C, Biron K K. Analysis of the UL97 phosphotransferase coding sequence in clinical cytomegalovirus isolates and identification of mutations conferring ganciclovir resistance. J Infect Dis. 1995;171:576–583. doi: 10.1093/infdis/171.3.576. [DOI] [PubMed] [Google Scholar]
- 12.Chou S, Marousek G, Parenti D, Lalezari J, Miner R, Drew L. Abstracts from the 35th Annual Meeting of Infectious Diseases Society of America. 1997. Mutation in region III of the DNA polymerase gene associated with dual ganciclovir-foscarnet resistance in cytomegalovirus (CMV) isolates from three subjects receiving prolonged antiviral therapy, abstr. 510; p. 166. [Google Scholar]
- 13.Chou S, Marousek G, Guentzel S, Follansbee S E, Poscher M E, Lalezari J P, Miner R C, Drew W L. Evolution of mutations conferring multidrug resistance during prophylaxis and therapy for cytomegalovirus disease. J Infect Dis. 1997;176:786–789. doi: 10.1086/517302. [DOI] [PubMed] [Google Scholar]
- 14.Chrisp P, Clissold S P. Foscarnet: a review of its antiviral activity, pharmacokinetic properties and therapeutic use in immunocompromised patients with cytomegalovirus retinitis. Drugs. 1991;41:104–129. doi: 10.2165/00003495-199141010-00009. [DOI] [PubMed] [Google Scholar]
- 15.Cihlar T, Chen M S. Identification of enzymes catalyzing two-step phosphorylation of cidofovir and the effect of cytomegalovirus infection on their activities in host cells. Mol Pharmacol. 1996;50:1502–1510. [PubMed] [Google Scholar]
- 16.Cihlar T, Fuller M D, Cherrington J M. Expression of the catalytic subunit (UL54) and the accessory protein (UL44) of human cytomegalovirus DNA polymerase in a coupled in vitro transcription/transplantation system. Protein Exp Purif. 1997;11:209–218. doi: 10.1006/prep.1997.0781. [DOI] [PubMed] [Google Scholar]
- 17.Coen D M. Viral DNA polymerases. In: DePamphilis M L, editor. DNA replication in eucaryotic cells. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 495–523. [Google Scholar]
- 18.Coen D M. Nucleosides and foscarnet—mechanisms. In: Richman D D, editor. Antiviral drug resistance. New York, N.Y: John Wiley & Sons Ltd.; 1996. pp. 81–102. [Google Scholar]
- 19.Crumpacker C S. Ganciclovir. Drug Therapy. 1996;335:721–729. doi: 10.1056/NEJM199609053351007. [DOI] [PubMed] [Google Scholar]
- 20.Derse D, Bastow K F, Cheng Y-C. Characterization of the DNA polymerases induced by a group of herpes simplex virus type 1 variants selected for growth in the presence of phosphonoformic acid. J Biol Chem. 1982;257:10251–10260. [PubMed] [Google Scholar]
- 21.Erice A, Gil-Roda C, Pérez J, Balfour Jr H H, Sannerud K J, Hanson M N, Boivin G, Chou S. Antiviral susceptibilities and analysis of UL97 and DNA polymerase sequences of clinical cytomegalovirus isolates from immunocompromised patients. J Infect Dis. 1997;175:1087–1092. doi: 10.1086/516446. [DOI] [PubMed] [Google Scholar]
- 22.Erice A, Chou S, Biron K K, Stanat S C, Balfour H H, Jordan M C. Progressive disease due to ganciclovir-resistant cytomegalovirus in immunocompromised patients. N Engl J Med. 1989;320:289–293. doi: 10.1056/NEJM198902023200505. [DOI] [PubMed] [Google Scholar]
- 23.Field A K, Biron K K. “The end of innocence” revisited: resistance of herpesviruses to antiviral drugs. Clin Microbiol Rev. 1994;7:1–13. doi: 10.1128/cmr.7.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Foster S A, Cerny J, Cheng Y-C. Herpes simplex virus-specified DNA polymerase is the target for the antiviral action of 9-(2-phosphonylmethoxyethyl)adenine. J Biol Chem. 1991;266:238–244. [PubMed] [Google Scholar]
- 25.Gibbs J S, Chiou H C, Bastow K F, Cheng Y-C, Coen D M. Identification of amino acids in herpes simplex virus DNA polymerase involved in substrate and drug recognition. Proc Natl Acad Sci USA. 1988;85:6672–6676. doi: 10.1073/pnas.85.18.6672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gilguin J, Piketty C, Thomas V, Gonzales-Canali G, Belec L, Kazatchkine M D. Acute cytomegalovirus infection in AIDS patients with CD4 counts above 100 cells/ml following combination antiretroviral therapy including protease inhibitors. AIDS. 1997;11:1659–1660. [PubMed] [Google Scholar]
- 27.Higuchi R, Krummel B, Saiki R K. A general method of in vitro preparation and specific mutagenesis of DNA fragments: study of protein and DNA interactions. Nucleic Acids Res. 1988;16:7351–7367. doi: 10.1093/nar/16.15.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hitchcock M J M, Jaffe H S, Martin J C, Stagg R J. Cidofovir, a new agent with potent anti-herpesvirus activity. Antiviral Chem Chemother. 1996;7:115–127. [Google Scholar]
- 29.Hwang C B C, Ruffner K L, Coen D M. A point mutation within the distinct conserved region of the herpes simplex virus DNA polymerase gene confers drug resistance. J Virol. 1992;66:1774–1776. doi: 10.1128/jvi.66.3.1774-1776.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jacobson M A, Zegans N, Pavan P R, O’Donnell J J, Sattler F, Owens S, Pollard R. Cytomegalovirus retinitis after initiation of highly active antiretroviral therapy. Lancet. 1997;349:1443–1445. doi: 10.1016/S0140-6736(96)11431-8. [DOI] [PubMed] [Google Scholar]
- 31.Kemble G, Duke G, Winter R, Spaete R. Defined large-scale alternation of the human cytomegalovirus genome constructed by cotransfection of overlapping cosmids. J Virol. 1996;70:2044–2048. doi: 10.1128/jvi.70.3.2044-2048.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lalezari J P. Cidofovir: a new therapy for cytomegalovirus retinitis. J Acquired Immune Defic Syndr Hum Retrovirol. 1997;14:S22–S26. doi: 10.1097/00042560-199700001-00005. [DOI] [PubMed] [Google Scholar]
- 33.Larder B A, Kemp S D, Darby G. Related functional domains in virus DNA polymerases. EMBO J. 1987;6:169–175. doi: 10.1002/j.1460-2075.1987.tb04735.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Larder B A, Kemp S D, Harrigan P R. Potential mechanism for sustained antiretroviral efficacy of AZT-3TC combination therapy. Science. 1995;269:696–699. doi: 10.1126/science.7542804. [DOI] [PubMed] [Google Scholar]
- 35.Littler E, Stuart A D, Chee M S. Human cytomegalovirus UL97 open reading frame encodes a protein that phosphorylates the antiviral nucleoside analogue ganciclovir. Nature. 1992;358:160–162. doi: 10.1038/358160a0. [DOI] [PubMed] [Google Scholar]
- 36.Lurain N S, Thompson K D, Holmes E W, Read G S. Point mutations in the DNA polymerase gene of human cytomegalovirus that result in resistance to antiviral agents. J Virol. 1992;66:7146–7152. doi: 10.1128/jvi.66.12.7146-7152.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lurain N S, Spafford L E, Thompson K D. Mutation in the UL97 open reading frame of human cytomegalovirus strains resistant to ganciclovir. J Virol. 1994;68:4427–4431. doi: 10.1128/jvi.68.7.4427-4431.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mar E-C, Chiou J-F, Cheng Y-C, Huang E-S. Inhibition of cellular DNA polymerase a and human cytomegalovirus-induced DNA polymerase by the triphosphates of 9-(2-hydroxyethoxymethyl)guanine and 9-(1,3-dihydroxy-2-propoxymethyl)guanine. J Virol. 1985;53:776–780. doi: 10.1128/jvi.53.3.776-780.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Öberg B. Antiviral effects of phosphonoformate (PFA, foscarnet sodium) Pharmacol Ther. 1989;40:213–285. doi: 10.1016/0163-7258(89)90097-1. [DOI] [PubMed] [Google Scholar]
- 40.Pari G S, Anders D G. Eleven loci encoding trans-acting factors are required for transient complementation of human cytomegalovirus oriLyt-dependent DNA replication. J Virol. 1993;67:6979–6988. doi: 10.1128/jvi.67.12.6979-6988.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pari G S, Kacica M A, Anders D G. Open reading frames UL44, IRS1/TRS1, and UL36-38 are required for transient complementation of human cytomegalovirus oriLyt-dependent DNA synthesis. J Virol. 1993;67:2575–2582. doi: 10.1128/jvi.67.5.2575-2582.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rubin R H. Impact of cytomegalovirus infection on organ transplant recipients. Rev Infect Dis. 1990;12:S754–S766. doi: 10.1093/clinids/12.supplement_7.s754. [DOI] [PubMed] [Google Scholar]
- 43.Sarasini A, Baldanti F, Furione M, Percivalle E, Brerra R, Barbi M, Gerna G. Double resistance to ganciclovir and foscarnet of four human cytomegalovirus strains recovered from AIDS patients. J Med Virol. 1995;47:237–244. doi: 10.1002/jmv.1890470309. [DOI] [PubMed] [Google Scholar]
- 44.Smith I L, Cherrington J M, Jiles R E, Fuller M D, Freeman W R, Spector S A. High-level resistance of cytomegalovirus to ganciclovir is associated with alterations in both the UL97 and DNA polymerase genes. J Infect Dis. 1997;176:69–77. doi: 10.1086/514041. [DOI] [PubMed] [Google Scholar]
- 45.Smith I L, Flores-Aguilar M, Taskituna I, Jiles R E, Freeman W R, Spector S A. Abstracts of the 36th Interscience Conference on Antimicrobial Agents and Chemotherapy. 1996. Cytomegalovirus resistance is associated with clinical failure in AIDS patients receiving ganciclovir treatment for retinitis, abstr. H30; p. 168. [Google Scholar]
- 46.Snoeck R, Andrei G, De Clercq E. Patterns of resistance and sensitivity to antiviral compounds of drug resistant strains of human cytomegalovirus selected in vitro. Eur J Clin Microbiol Infect Dis. 1996;15:574–579. doi: 10.1007/BF01709366. [DOI] [PubMed] [Google Scholar]
- 47.Studies of Ocular Complications of AIDS (SOCA) in collaboration with the AIDS Clinical Trial Group. Cytomegalovirus (CMV) culture results, drug resistance, and clinical outcome in patients with AIDS and CMV retinitis treated with foscarnet and ganciclovir. J Infect Dis. 1997;176:50–58. doi: 10.1086/514039. [DOI] [PubMed] [Google Scholar]
- 48.Sullivan V, Coen D M. Isolation of foscarnet-resistant human cytomegalovirus patterns of resistance and sensitivity to other antiviral drugs. J Infect Dis. 1991;164:781–784. doi: 10.1093/infdis/164.4.781. [DOI] [PubMed] [Google Scholar]
- 49.Sullivan V, Talarico C L, Stanat S C, Davis M, Coen D M, Biron K K. A protein kinase homologue controls phosphorylation of ganciclovir in cytomegalovirus-infected cells. Nature. 1992;358:162–164. doi: 10.1038/358162a0. [DOI] [PubMed] [Google Scholar]
- 50.Sullivan V, Biron K K, Talarico C L, Stanat S C, Davis M, Pozzi L M, Coen D M. A point mutation in the human cytomegalovirus DNA polymerase gene confers resistance to ganciclovir and phosphonylmethoxyalkyl derivatives. Antimicrob Agents Chemother. 1993;37:19–25. doi: 10.1128/aac.37.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tenney D J, Yamanaka G, Voss S M, Cianci C W, Tuomari A V, Sheaffer A K, Alam M, Colonno R J. Lobucavir is phosphorylated in human cytomegalovirus-infected and -uninfected cells and inhibits the viral DNA polymerase. Antimicrob Agents Chemother. 1997;41:2680–2685. doi: 10.1128/aac.41.12.2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang T S-F. Enzymatic properties and characteristics that distinguish each DNA polymerase. In: DePamphilis M L, editor. DNA replication in eucaryotic cells. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 461–493. [Google Scholar]
- 53.Wolf D G, Smith I L, Lee D J, Freeman W R, Flores-Aguilar M, Spector S A. Mutations in human cytomegalovirus UL97 gene confer clinical resistance to ganciclovir and can be detected directly in patient plasma. J Clin Invest. 1995;95:257–263. doi: 10.1172/JCI117648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wong S W, Wahl A F, Yuan P-M, Arai N, Pearson B E, Arai K-I, Korn D, Hunkapiller M W, Wang T S-F. Human DNA polymerase α gene expression is cell proliferation dependent and its primary structure is similar to both prokaryotic and eukaryotic replicative DNA polymerases. EMBO J. 1988;7:37–47. doi: 10.1002/j.1460-2075.1988.tb02781.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xiong X, Flores C, Fuller M D, Mendel D B, Mulato A S, Moon K, Chen M S, Cherrington J M. In vitro characterization of the anti-human cytomegalovirus activity of PMEA (Adefovir) Antiviral Res. 1997;36:131–137. doi: 10.1016/s0166-3542(97)00050-8. [DOI] [PubMed] [Google Scholar]
- 56.Xiong X, Smith J L, Chen M S. Effect of incorporation of cidofovir into DNA by human cytomegalovirus DNA polymerase on DNA elongation. Antimicrob Agents Chemother. 1997;41:594–599. doi: 10.1128/aac.41.3.594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ye L-B, Huang E-S. In vitro expression of the human cytomegalovirus DNA polymerase gene: effects of sequence alterations on enzyme activity. J Virol. 1993;67:6339–6347. doi: 10.1128/jvi.67.11.6339-6347.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang J, Chung D W, Tan C-K, Downey K M, Davie E W, So A G. Primary structure of the catalytic subunit of calf thymus DNA polymerase δ: sequence similarities with other DNA polymerases. Biochemistry. 1991;30:11742–11750. doi: 10.1021/bi00115a002. [DOI] [PubMed] [Google Scholar]