Abstract
Previously, we showed that truncated soluble forms of herpes simplex virus (HSV) glycoprotein D (gDt) bound directly to a truncated soluble form of the herpesvirus entry mediator (HveAt, formerly HVEMt), a cellular receptor for HSV. The purpose of the present study was to determine the affinity of gDt for HveAt by surface plasmon resonance and to compare and contrast the kinetics of an expanded panel of gDt variants in binding to HveAt in an effort to better understand the mechanism of receptor binding and virus entry. Both HveAt and gDt are dimers in solution and interact with a 2:1 stoichiometry. With HveAt, gD1(306t) (from the KOS strain of HSV-1) had a dissociation constant (KD) of 3.2 × 10−6 M and gD2(306t) had a KD of 1.5 × 10−6 M. The interaction between gDt and HveAt fits a 1:1 Langmuir binding model, i.e., two dimers of HveAt may act as one binding unit to interact with one dimer of gDt as the second binding unit. A gD variant lacking all signals for N-linked oligosaccharides had an affinity for HveAt similar to that of gD1(306t). A variant lacking the bond from cysteine 1 to cysteine 5 had an affinity for HveAt that did not differ from that of the wild type. However, variants with double cysteine mutations that eliminated either of the other two disulfide bonds showed decreased affinity for HveAt. This result suggests that two of the three disulfide bonds of gD are important for receptor binding. Four nonfunctional gDt variants, each representing one functional domain of gD, were also studied. Mutations in functional regions I and II drastically decreased the affinity of gDt for HveAt. Surprisingly, a variant with an insertion in functional region III had a wild-type level of affinity for HveAt, suggesting that this domain may function in virus entry at a step other than receptor binding. A variant with a deletion in functional region IV [gD1(Δ290-299t)] exhibited a 100-fold enhancement in affinity for HveAt (KD = 3.3 × 10−8 M) due mainly to a 40-fold increase in its kinetic on rate. This agrees with the results of other studies showing the enhanced ability of gD1(Δ290-299t) to block infection. Interestingly, all the variants with decreased affinities for HveAt exhibited decreased kinetic on rates but only minor changes in their kinetic off rates. The results suggest that once the complex between gDt and HveAt forms, its stability is unaffected by a variety of changes in gD.
Herpes simplex virus (HSV) glycoprotein D (gD) is a viral envelope glycoprotein that has been studied extensively by immunological, biochemical, and genetic approaches (5, 9, 23, 31, 33, 50). It is one of the essential glycoproteins for virus entry, and much evidence indicates that it functions by interacting with a cellular receptor (3, 16, 17, 21, 25, 50).
Recently, expression cloning was used to isolate and identify a HeLa cell gene product that allows for entry of many HSV strains when it is expressed in normally nonpermissive Chinese hamster ovary (CHO) cells (25). This gene product, the herpesvirus entry mediator (HveA, formerly HVEM), is a type I integral membrane protein. Two other cellular receptors for HSV entry that are not related structurally to HVEM/HveA have been identified (12, 19, 49). Therefore, the nomenclature for naming HSV receptors has been unified (49). Truncated HveA (HveAt) contains a motif of four cysteine-rich pseudorepeat sequences and is a member of the tumor necrosis factor receptor superfamily (1, 24, 25, 37). HveA can be considered a cellular receptor for HSV, and gD can be considered the receptor binding protein because (i) HveA allows virus entry into normally nonpermissive cells (25), (ii) antibody against HveA blocks virus entry (25), (iii) soluble HveAt binds specifically to gD in purified virus and blocks entry (32, 50), and (iv) soluble truncated gD (gDt) interacts specifically with HveAt in vitro (50). Furthermore, the native conformation of gD is critical for this interaction whereas the N-linked oligosaccharides (N-CHO) are not (50), as was predicted by previous structure-function studies (reviewed in reference 7).
Our approach to defining the relationship between the structure and function of gD has been to generate and examine a panel of mutations (5, 22, 23, 27, 28, 33, 38–40). The genes for a number of these variant proteins have been cloned into a baculovirus expression system to produce truncated, soluble forms of the proteins that can be purified in the absence of detergent and easily studied (31, 33, 36, 50).
We have compared the abilities of the different gDt variant proteins to bind to soluble HveAt by enzyme-linked immunosorbent assay (ELISA) (25, 50). In particular, a gD-1 variant with a deletion in functional region IV, gD1(Δ290-299t), was able to bind to HveAt by ELISA and block virus entry 100-fold better than the wild-type protein, gD1(306t) (33, 50). While ELISA has yielded valuable information on the conditions and specificity of the gD-HveA interaction, we are unable to determine the kinetics of binding by this technique. Furthermore, ELISA detects only those molecules that remain bound to receptor after multiple washes.
To characterize the binding of gDt to HveAt in real time, optical biosensor technology was used. Biomolecular interaction analysis was used to quantitate binding affinities (dissociation constants [KDs]) and obtain values for kinetic on (kon) and off (koff) rates. The biosensor uses surface plasmon resonance (SPR) and changes in refractive index to measure interactions between biomolecules at the surface of a sensor chip (18, 29). Binding data are obtained in real time and in the absence of labeled protein. SPR has been used to study the binding of influenza virus hemagglutinin to sialic acid receptors (42), soluble intercellular cell adhesion molecule to human rhinovirus (4), and HSV gB to glycosaminoglycans (52).
The purpose of the present study was to determine the affinity of gDt for HveAt and to compare and contrast the kinetics of an expanded panel of gDt variants in binding to HveAt in an effort to better understand the mechanism of receptor binding and virus entry. Furthermore, we sought to explain why a series of insertion variants in functional regions I to IV of gD failed to complement the infectivity of a gD null virus in trans. Specifically, we looked at differences in affinities for HveAt among the gDt variants and determined if differences in KDs were due to changes in kon, the rate of association of the complex, koff, an indication of the stability of the complex, or a combination of both.
MATERIALS AND METHODS
Cells and viruses.
Sf9 (Spodoptera frugiperda) cells (GIBCO BRL), used for producing recombinant baculoviruses and recombinant glycoproteins, were propagated in Sf900II medium (GIBCO BRL) (53).
Construction of recombinant baculoviruses expressing the cysteine variants.
DNA fragments were generated by PCR with plasmids pWW201 (Cys 1,5), pDL143 (Cys 2,6), and pWW220 (Cys 3,4) (23) as templates and with the primers used to construct bac-gD1 (306t) (36). Each PCR product encoded a gD1 variant with one disulfide bond completely removed, with a truncation at amino acid 306 before the transmembrane region, and with a six-histidine C-terminal tail. The PCR products constructed with pWW201 (Cys 1,5), pDL143 (Cys 2,6), and pWW220 (Cys 3,4) were each ligated into the transfer vector pVTBac (33, 36, 53) to produce plasmids pCP262, pCP261, and pCP263, respectively, and recombined into baculovirus (33, 36, 53). The viruses were designated bac-gD1 (cys1,5), bac-gD1 (cys2,6), and bac-gD1 (cys3,4), and the proteins were designated gD1 (cys1,5), gD1 (cys2,6), and gD1 (cys3,4). gD1 (cys1,5) has Ser replacing Cys at positions 66 and 189, gD1 (cys2,6) has Ser replacing Cys at positions 106 and 202, and gD1 (cys3,4) has Ser replacing Cys at positions 118 and 127.
Production and purification of gDt.
Detailed protocols exist elsewhere for purification of gDt (36, 53). Briefly, Sf9 insect cells (GIBCO BRL) were grown in 3 liters of suspension cultures, infected with recombinant viruses at a multiplicity of infection of 4, and cultured at 27°C in a Celligen Bioreactor (New Brunswick Scientific) for 48 h. Cells were removed by centrifugation, and the clarified medium was concentrated to 1 liter with a Pellicon Cassette Filter (Millipore) with a 10,000-molecular-weight cutoff. The concentrated supernatant was then exchanged with 6 liters of phosphate-buffered saline (PBS) with the same tangential-flow system. The exchanged medium was passed over a DL6 immunosorbent column and eluted as previously described (33, 34, 36). Proteins were concentrated, dialyzed against PBS, and stored at −80°C.
Production and purification of HveAt.
Mature HveA is 245 amino acids long (25). A soluble form of HveA truncated at amino acid 200, just prior to the transmembrane region (HveAt), was produced from recombinant baculovirus-infected insect cells and purified by nickel-affinity chromatography as previously described (50).
Molecular weight analysis. (i) Gel filtration.
Dextran blue was used to determine the void volume of a Superdex 75 column and a Superdex 200 column (HR 10/30; Pharmacia), and the columns were calibrated with High and Low Molecular Weight Gel Filtration Calibration Kits (Pharmacia Biotech) with PBS as the running buffer. Two hundred microliters of a 1-mg/ml solution of gD1 (306t), gD2 (306t), and gD1 (QAAt) was eluted from the Superdex 75 column, and the retention time was measured. gD1 (Δ290-299t) was eluted from the Superdex 200 column. The molecular weights of the proteins were calculated from a standard plot according to the directions of the calibration kits.
(ii) Mass spectrometry.
To determine the mass of gD1 (306t), a 30-μg/ml sample was analyzed by matrix-assisted laser-desorption ionization and time of flight mass spectrometry (Fisons Instruments VG TofSpec) (15) as previously described (34, 50).
(iii) STEM.
A sample of gD1 (306t) in PBS was diluted to 5 μg/ml with 50 mM ammonium acetate and analyzed by scanning transmission electron microscopy (STEM). STEM was performed at the Brookhaven National Laboratory (46). The sample was processed and analyzed as previously described (45).
ELISA.
ELISA was used to monitor the binding of gDt to HveAt as previously described (50).
Binding of gDt to HveAt as detected with an optical biosensor.
SPR experiments were carried out on a BIACORE X or BIACORE 2000 optical biosensor (Biacore AB) at 25°C. The running buffer for the experiments was PBS (0.1 M sodium phosphate, 0.15 M NaCl; Pierce) containing 0.005% Tween 20 (PBS-T; pH 7.0). Approximately 2,000 response units (RU) of purified HveAt was coupled to flow cell 1 (Fc1) of a CM5 sensor chip via primary amines according to the manufacturer’s specifications. Fc2 was activated and blocked without the addition of protein. To characterize the binding of gDt variants to HveAt, the flow path was set to include both flow cells, the flow rate was 50 μl/min, and the data collection rate was set to high. Protein samples were serially diluted in PBS-T. Binding of each gDt sample was allowed to occur for 2 min, with the wash delay set for an additional 2 min to allow for a smooth dissociation curve. The chip surface was regenerated by injecting brief pulses of 0.2 M sodium carbonate, pH 9.5, until the response signal returned to baseline. SPR data were analyzed with BIAevaluation, version 3.0, software, which employs global fitting. Sensorgrams were corrected for nonspecific binding by subtracting the control sensorgram (Fc2) from the HveAt surface sensorgram (Fc1). Model curve fitting was done with a 1:1 Langmuir binding model with drifting baseline. This is the simplest model for the interaction between receptor and ligand; it follows the equation A + B ⇆ AB. The rate of association (kon) is measured from the forward reaction, while koff is measured from the reverse reaction (2).
RESULTS
Molecular weight and oligomeric state analysis.
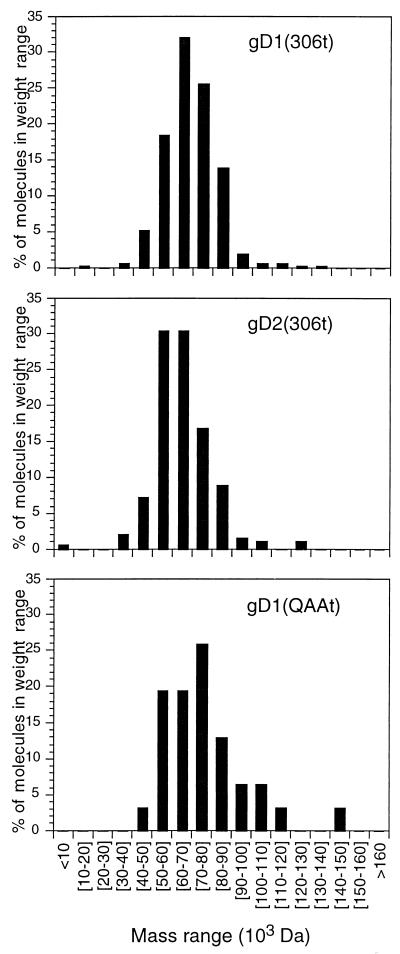
To calculate kinetic and affinity constants, the oligomeric state and accurate molecular weight of the protein flowing over the sensor chip surface need to be known. Two observations suggest that full-length gD is a dimer: (i) preparations of immunoaffinity-purified full-length gD1 contain a dimer that is resistant to sodium dodecyl sulfate (SDS) and reducing conditions (8) and (ii) gD can be detected as dimers and trimers in virions (14). On native SDS-polyacrylamide gel, gDt is present primarily as a monomer, although various amounts of dimer were routinely observed (33, 36). However, the native gel system may not have given an accurate representation of the amount of dimer present because the small amounts of SDS present in the system probably disrupted a portion of the noncovalently bound dimer. In this study we estimated the molecular size of gDt by mass spectrometry, gel filtration chromatography, and STEM.
Truncated forms of gD from HSV-1 and -2 [gD1 (306t) and gD2 (306t), respectively], a variant lacking the three N-CHO [gD1 (QAAt)], and a functional region IV variant [gD1 (Δ290-299t)] were used (Fig. 1; Table 1). The molecular mass of gD1 (QAAt) determined by mass spectrometry is close to the formula mass. The mass spectrometry values for gD1 (306t), gD2 (306t), and gD1 (Δ290-299t) are each 2,000 to 3,000 Da higher than that for gD1 (QAAt). This is consistent with the addition of two to three N-CHO, each with a mass of 1,000 Da (34). For all four proteins, the masses estimated by gel filtration were approximately twice those obtained by mass spectrometry. The gel filtration experiment was repeated multiple times for gD1 (306t) and gD1 (Δ290-299t). Because mass spectrometry disrupts protein oligomers that are not covalently bound, the gel filtration data indicated that each protein exists as a noncovalent dimer in solution. To confirm this, samples of gD1 (306t), gD2 (306t), and gD1 (QAAt) were submitted for STEM analysis. Individual molecules of protein were directly viewed by this technique, and masses were estimated by comparison to a standard. Because this technique does not disrupt noncovalent interactions, it is useful in determining the oligomeric state of individual molecules (45, 47). The range of masses for gD1 (306t), gD2 (306t), and gD1 (QAAt) was plotted on a histogram (Fig. 2), and the average masses are reported in Table 1. These values agree with the gel filtration values. Because mass spectrometry is most accurate for mass determination, protein concentrations were calculated with dimeric sizes for variants based on these values [e.g., 74,400 Da for gD1 (306t), which is equal to two times the mass spectrometry value].
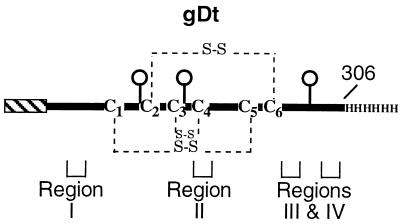
FIG. 1.
Schematic representation of truncated HSV gD produced by baculovirus-infected insect cells (33, 36). The protein has the honeybee melittin signal peptide (hatched box) in place of the wild-type gD signal for efficient translocation into the lumen of the endoplasmic reticulum, two extra amino acids (D and P) at the N terminus, and a histidine tag at the C terminus (33, 36, 44). The protein was truncated prior to the transmembrane region and has three N-CHO (balloons). The disulfide bonds are indicated by dashed lines. The functional regions of gD encompass the following amino acids: 27 to 43 for region I, 126 to 131 for region II, 225 to 246 for region III, and 277 to 300 for region IV (5).
TABLE 1.
Summary of mass analysis data
| Variant protein | Formula mass (Da)a | Mean mass (Da) ± SD by:
|
||
|---|---|---|---|---|
| Mass spectrometryb | Gel filtrationc | STEM (no. of molecules) | ||
| gD1(306t) | 34,928 | 37,200 ± 200 | 64,100 ± 8,550 | 70,900 ± 15,200 (309) |
| gD2 (306t) | 34,933 | 36,200 ± 200 | 62,000 | 65,900 ± 14,500 (262) |
| gD1 (QAAt) | 34,909 | 34,200 ± 200 | 65,400 | 79,100 ± 21,100 (31) |
| gD1 (Δ290-299t) | 34,430 | 36,400 ± 200 | 67,700 ± 10,500 | NDd |
Based on amino acid composition; mass of N-CHO not included.
Error estimated from broadness of peaks due to sugar heterogeneity.
Standard deviations derive from results of at least three experiments. The large errors in molecular mass determinations by gel filtration were due largely to the broadness of the peaks caused by the presence of N-CHO.
ND, not done.
FIG. 2.
STEM. Individual molecules of protein were directly viewed, and molecular masses were calculated by comparison to a tobacco mosaic virus standard. The range of molecular masses for each protein was plotted on a histogram as percentages of the total numbers of particles counted. The average molecular masses are reported in Table 1.
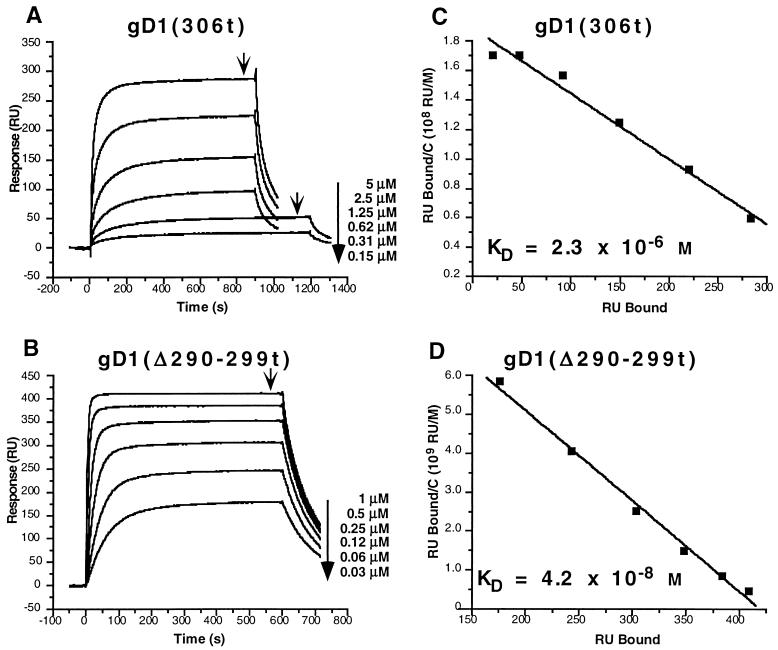
Binding of gD1 (306t) and gD1 (Δ290-299t) to immobilized HveAt.
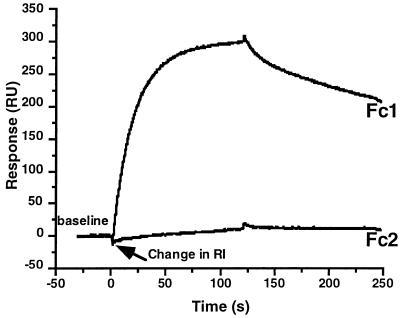
Previous studies showed that HveAt interacts with gDt (25, 32, 50). To examine the kinetics of binding of gDt to HveAt by SPR, HveAt was coupled to a sensor chip surface. Figure 3 shows an example of the raw sensorgram data. In this experiment, gD1 (Δ290-299t) at a concentration of 0.5 μM in PBS-T was allowed to flow over the chip surface. Fc1 contained the immobilized HveAt, while Fc2 was activated and blocked without the addition of protein. In the initial 50 s of each sensorgram, a buffer baseline was established. Sample was injected, and the association of gDt and HveAt followed for 2 min. At 120 s, the sample was replaced with buffer, and the dissociation of complex followed for 2 min. The response on the y axis is measured in response units. There was very little nonspecific binding of protein to the activated and blocked carboxymethyl dextran surface on Fc2. In the SPR experiments that followed, data from Fc2 were subtracted from the data from Fc1 to correct for changes in bulk refractive index (RI; represents a difference in solution composition between the buffer and the protein solution) and nonspecific binding to the sensor chip surface.
FIG. 3.
Example of raw sensorgram data. gD1 (Δ290-299t) at a concentration of 0.5 μM was injected over a surface containing 2,000 RU of HveAt (Fc1) in series with a blank surface (Fc2). At 120 s, the sample was replaced with buffer, and dissociation followed for 2 min. The change in refractive index (RI) represents a difference in solution composition between the buffer and the protein solution.
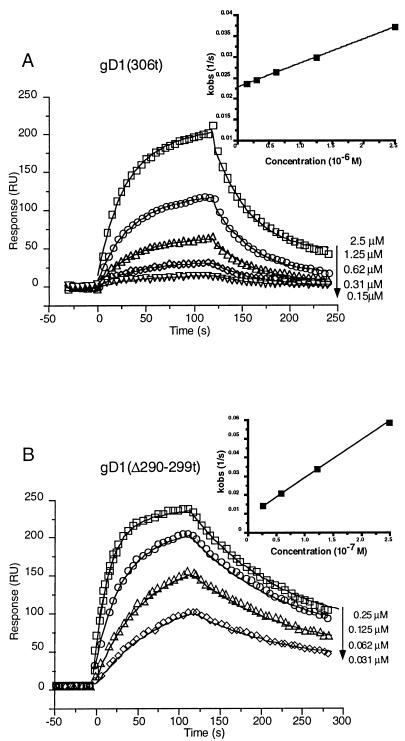
In the next experiment (Fig. 4), the binding kinetics of gD1 (306t) were compared with those of gD1 (Δ290-299t). The data were overlaid with the result of the global fitting analysis and are shown in Fig. 4. The global fitting analysis allows simultaneous fitting of both the association and dissociation phases of the sensorgram to all curves in the working set. This improves the robustness and stability of the fitting procedure (2). The residual plots for the fitted data (the difference between the experimental and fitted data [not shown]) were all within ±3 RU, and the chi-square values (a standard statistical measure of the closeness of the fit; values below 10 are acceptable) were all below 4.
FIG. 4.
Corrected sensorgram overlays for the gDt-HveAt interaction. (A) Repeat injections of gD1 (306t) at the indicated concentrations are represented by the small symbols. Data were collected at 5 Hz, but for clarity, one point per second is shown. The solid lines represent the best fits found by the global software analysis. The inset represents a plot of kobs versus concentration, the negative slope of which is equal to kon. The R2 for the linear fit of the data was 0.999. (B) Repeat injections of gD1 (Δ290-299t) at the indicated concentrations. Data collection and analysis were the same as described for panel A. The R2 for the linear fit of the data in the plot of kobs versus concentration (inset) was 0.999.
Based on ELISA, the affinity of gD1 (Δ290-299t) for HveAt was predicted to be 100-fold higher than the affinity of gD1 (306t) for HveAt (50). The values for KD calculated from the SPR data were 3.2 × 10−6 M for gD1 (306t) and 3.3 × 10−8 M for gD1 (Δ290-299t). The 100-fold higher affinity (lower KD) of gD1 (Δ290-299t) for HveAt than that of gD1 (306t) was due mainly to a 40-fold increase in kon, the rate of formation of complex (Table 2). There was also a twofold decrease in koff for gD1 (Δ290-299t) compared to that of gD1 (306t), which contributed to the increased affinity. Thus, the SPR data agree with and explain the predictions from ELISA data.
TABLE 2.
Kinetic and affinity values for gDt-HveAt complex formation
| Immobi- lized ligand | Analyte | kon (103 s−1 M−1) | koff (10−2 s−1) | KD (10−6 M)e |
|---|---|---|---|---|
| HveAt | gD1 (306t)a | 6.1 ± 0.6d | 2.0 ± 0.2 | 3.2 ± 0.6 |
| HveAt | gD1 (Δ290-299t)a | 240 ± 70 | 0.78 ± 0.1 | 0.033 ± 0.01 |
| HveAt | gD2 (306t)a | 8.3 | 1.2 | 1.5 |
| HveAt | gD1 (QAAt)a | 6.4 | 1.1 | 1.8 |
| HveAt | gD1 (∇34t)b | Does not bind | ||
| HveAt | gD1 (∇126t)b | 1.2 | 2.6 | 22 |
| HveAt | gD1 (∇243t)b | 11 | 1.5 | 1.3 |
| HveAt | gD1 (cys1,5)b | 2.9 | 1.6 | 5.4 |
| HveAt | gD1 (cys2,6)b | 2.2 | 2.4 | 11 |
| HveAt | gD1 (cys3,4)b | 1.1 | 1.7 | 16 |
| gD1 (306t) | HveAtc | 7.1 | 1.7 | 2.3 |
| gD1 (Δ290299t) | HveAt | Data do not fit |
Concentrations for data analysis were calculated by assuming that gDt is a dimer in solution and by using twice the mass spectrometry values for molecular mass (Table 1).
Concentrations were calculated as described in footnote a with the mass spectrometry values for gD1 (306t). Small differences in amino acid composition do not affect concentration calculations significantly.
Concentrations for data analysis were calculated by assuming that HveAt is a dimer in solution and by using twice the mass spectrometry values for molecular mass (50).
Such values are means ± the standard deviations of results from at least three separate experiments.
Chi-square values for the global fits ranged from 0.93 to 3.74.
Plots of the concentration dependent on rate (kobs) versus concentration were done to verify the values obtained for kon from the global analysis (Fig. 4, insets). The values for kon from these plots are the same as those calculated with global fitting software [6.1 × 103 for gD1 (306t) and 2.4 × 105 for gD1 (Δ290-299t)] (Table 2).
Equilibrium binding of gD1 (306t) and gD1 (Δ290-299t) as determined by Scatchard analysis.
The binding assays of gD1 (306t) and gD1 (Δ290-299t) to HveAt were repeated under equilibrium conditions to confirm the affinity constants calculated by global analysis. Binding of gD1 (306t) to HveAt was monitored for 10 to 15 min, until equilibrium was reached. Binding of gD1 (Δ290-299t) to HveAt was monitored for 10 min. Dissociation for each was monitored for 2 min. The flow rate for the experiment was 5 μl/min. We tested a range of concentrations of each protein from 5 to 0.15 μM for gD1 (306t) and from 1 to 0.03 μM for gD1 (Δ290-299t). Equilibrium was reached when the association portion of the curve flattened (Fig. 5A and B). Scatchard analysis of the data (Fig. 5C and D) yielded values for KD of 2.3 × 10−6 M and 4.2 × 10−8 M for gD1 (306t) and gD1 (Δ290-299t), respectively. These values are essentially the same as those obtained from the global analysis (Table 2).
FIG. 5.
Equilibrium binding sensorgrams and Scatchard analysis of the binding of gD1 (306t) and gD1 (Δ290-299t) to immobilized HveAt. (A) Binding of gD1 (306t) to HveAt was monitored for 15 min for the 5, 2.5, 1.25, and 0.62 μM injections and for 20 min for the 0.31 and 0.15 μM injections. The arrows indicate the time points used for the Scatchard analysis. (B) Binding of gD1 (Δ290-299t) to HveAt was monitored for 10 min for all of the injections. The arrows indicates the time points used for the Scatchard analysis. (C and D) Scatchard analysis. C is the concentration of gDt that flowed across the surface. The negative slope of each line is equal to the association constant (the reciprocal is KD). The R2 for the linear fit of the data in panel C was 0.98. The R2 for the linear fit of the data in panel D was 0.99.
Binding of HveAt to immobilized gD1 (306t) and gD1 (Δ290-299t).
In order to determine if the kinetics of binding between gDt and HveAt were dependent on the orientation of the complex, the reverse experiment was performed. Two chips were made analogously to the HveAt chip surface, one with gD1 (306t) covalently linked and the other with gD1 (Δ290-299t) covalently linked. HveAt was allowed to flow over each chip individually. The signal obtained from the gD1 (306t) chip surface was very noisy (data not shown). Analysis of the gD1 (306t) data, however, indicated that the binding kinetics for the formation of complex were the same as when the molecules were in the reverse orientation (Table 2). In contrast, HveAt bound to and dissociated from the immobilized gD1 (Δ290-299t), but the data did not fit any of the available models (data not shown). Thus, the quality of data obtained was higher when HveAt was immobilized on the chip surface than when either gD1 (306t) or gD1 (Δ290-299t) was immobilized. We previously found that chemical modification of gDt alters its biological activity (30, 51). Therefore, HveAt is more amenable to covalent linkage through primary amines than gDt.
Binding of gDt variants to immobilized HveAt.
Our approach to define the relationship between the structure of gD and its function has been to generate a panel of mutations within gD. With the discovery of HveA, a cellular receptor for HSV that binds gD, these variants become important once again to help determine the mechanism by which virus binds HveA and enters cells.
(i) gD1 versus gD2.
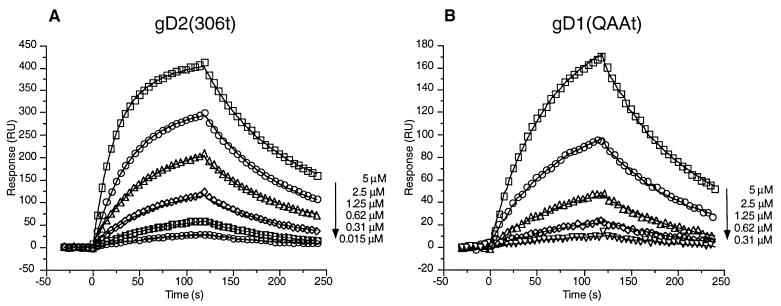
Whitbeck et al. (50) reported that gD2 (306t) bound to HveAt with an affinity similar to that of gD1 (306t) as measured by ELISA. The kinetics of binding of gD2 (306t) to HveAt were compared with those of gD1 (306t) to see if differences could be detected in kon and/or koff. The fitted data resulted in values for kon, koff, and KD for gD2 (306t) that are essentially the same as those for gD1 (306t) (Fig. 6A; Table 2).
FIG. 6.
Corrected sensorgram overlays for the interaction between gD2 (306t) (A) and gD1 (QAAt) (B) in binding to immobilized HveAt. Sensorgrams are presented as described in the legend to Fig. 4.
(ii) N-CHO.
Glycosylation of gD does not play a role in the binding of gDt to HveAt as measured by ELISA (50), and the N-CHO on gD are not important for the infectivity of the virus (39, 43). gD1 (QAAt) (36), has three mutations which ablate the signals for addition of all three N-CHO. As expected, gD1 (QAAt) bound to HveAt with the same affinity as gD1 (306t) (Fig. 6B; Table 2). In addition, the absence of N-CHO did not alter kon or koff.
(iii) Disulfide bonds.
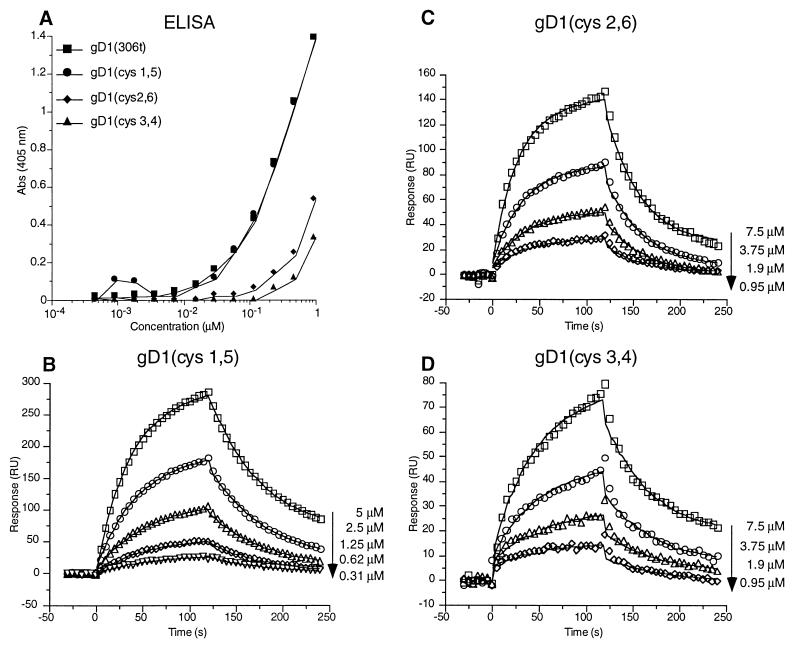
gD has three disulfide bonds that are necessary for maintaining the native structure and stability of the molecule; the pairings consist of Cys 1 to 5, Cys 2 to 6, and Cys 3 to 4 (Fig. 1) (23). We previously found that double cysteine mutations in gD that removed any one disulfide bond resulted in proteins that were able to bind conformation-dependent monoclonal antibodies (MAbs) and that were also able to complement a gD null virus, but only in a temperature-sensitive manner (23). Three cysteine variants were cloned into a baculovirus expression system. Each variant contained mutations of two cysteine residues that eliminated one disulfide bond in gD (Fig. 1). First, the variants were tested for their abilities to bind conformation-dependent MAbs by native Western and dot blot analyses (data not shown). The binding profiles were the same as those measured with the full-length proteins (23). Next, ELISA was used to examine the abilities of the variants to bind HveAt both by ELISA (Fig. 7A) and SPR (Fig. 7B to D). By ELISA, gD1 (cys1,5) bound to HveAt as well as gD1 (306t) did (Fig. 7A). The other two variants, gD1 (cys2,6) and gD1 (cys3,4), bound to HveAt less well than gD1 (306t) did by ELISA. These results parallel the ability of the full-length variants to complement a gD null virus in trans (23). SPR experiments were done to quantitatively measure the magnitudes of binding differences among the variants. gD1 (cys1,5) had the highest affinity for HveAt, with values for kon, koff, and KD similar to those of gD1 (306t) (Table 2). gD1 (cys2,6) had an affinity for HveAt that was one-third that of gD1 (306t), due entirely to a lower kon (Table 2). Finally, the affinity of gD1 (cys3,4) for HveAt was one-fifth that of gD1 (306t) due to a corresponding decrease in kon (Table 2).
FIG. 7.
Binding of the cysteine variants to immobilized HveAt. (A) Binding of the gDt cysteine variants to HveAt by ELISA. An ELISA plate was coated with 200 nM HveAt in PBS, blocked, and incubated with various concentrations of gDt. Bound gDt was detected with antiserum R7 and then with peroxidase-conjugated secondary antibody and substrate. The data are averages of results from duplicate wells, and each experiment was repeated twice with similar results. Abs, absorbance. (B to D) Binding of gD1 (cys1,5), gD1 (cys2,6), and gD1 (cys3,4), respectively, to immobilized HveAt by SPR. Data are plotted and analyzed as described for Fig. 4.
Binding of nonfunctional variants of gDt to HveAt.
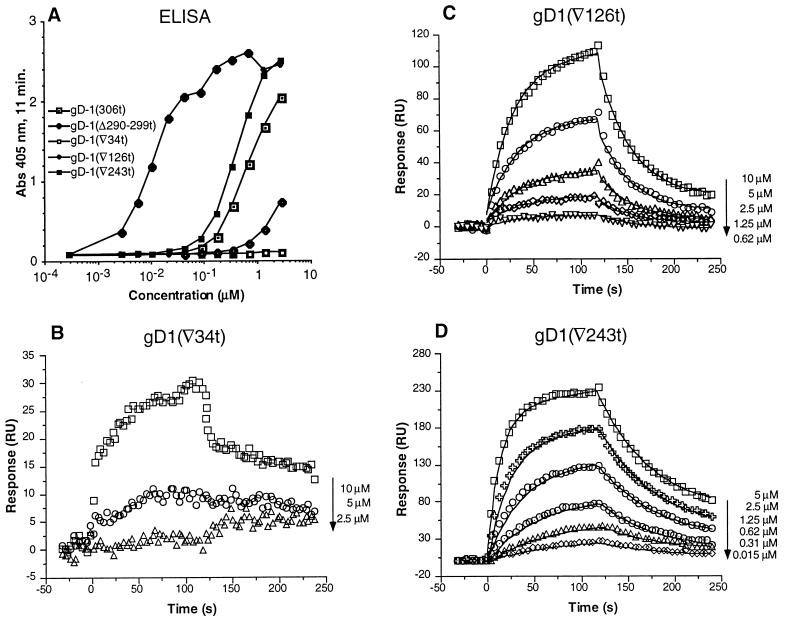
gD has four functional regions (I to IV) (Fig. 1) that are important for virus entry (5, 28). Soluble forms of a representative nonfunctional variant from each region have been expressed in a baculovirus expression system and previously characterized (31, 33). The functional region I variant, gD1 (∇34t), did not inhibit HSV plaque formation or cell-to-cell spread (33). gD1 (∇34t) did not bind to HveAt by ELISA (Fig. 8A). By SPR at the concentrations examined, the binding of this variant to HveAt was poor (Fig. 8B) and the data did not fit a 1:1 model. Therefore, values for kon, koff, and KD were not calculated for gD1 (∇34t). The functional region II variant, gD1 (∇126t), exhibited poor inhibition of plaque formation (33) and bound weakly to HveAt by ELISA (Fig. 8A). The overall affinity of gD1 (∇126t) for HveAt was seven times less than the affinity of gD1 (306t) for HveAt due to a lower kon (Table 2). However, the koff was similar to that of gD1 (306t), indicating that once the complex of gD1 (∇126t) and HveAt is formed, it is as stable as the complex of gD1 (306t) and HveAt.
FIG. 8.
Binding of the functional region variants to immobilized HveAt. (A) Binding of the gDt functional region variants to HveAt by ELISA. ELISA was done as described in the legend to Fig. 7. Abs, absorbance. (B to D) Binding of gD1 (∇34t), gD1 (∇126t), and gD1 (∇243t), respectively, to immobilized HveAt by SPR. Data are plotted and analyzed as described for Fig. 4.
The functional region III variant, gD1 (∇243t), bound to HveAt at a level similar to that of gD1 (306t) by ELISA (Fig. 8A). The fitted SPR data resulted in values for kon, koff, and KD similar to those of gD1 (306t) (Table 2). Although this mutant was nonfunctional by complementation analysis (5), the soluble protein was able to block HSV infection (33). Thus, the binding data agree with the blocking data but differ from the results of complementation analysis. In contrast, gD1 (Δ290-299t), the prototype variant from functional region IV, showed enhanced binding to HveAt, as was characterized above (50).
DISCUSSION
The mechanism of HSV entry is complicated, as it involves at least five envelope glycoproteins (41). A main function of gD is to interact with specific cellular receptors (3, 16, 17, 21, 25, 50). One of these, called herpesvirus entry mediator or HveA, is a member of the tumor necrosis factor receptor superfamily of membrane proteins and is found primarily on T cells and other cells of the immune system (25, 50). It is clear that (i) gDt and HveAt form a complex in solution (50), (ii) HveAt binds to gD on virions (32), and (iii) gDt interacts with immobilized HveAt (reference 50 and this report). The next step to complete the study of the interaction is a means to study the interaction of gD in the virus with HveA on the cell surface. The goal of this study was to document the binding capacity of gDt to HveAt and to begin to understand the characteristics of the interaction.
Kinetics of binding.
SPR was done to characterize the binding between HveA, a receptor for HSV entry, and gD, the HSV receptor-binding protein (25, 32, 50). We recently showed that two forms of gDt appear to bind to HveAt with different affinities (50). The first, gD1 (306t), is considered the wild-type form of the protein, while the second, gD1 (Δ290-299t), is a functional region IV variant that shows enhanced inhibition of virus entry on Vero cells (33) and enhanced binding to HveAt (25, 50). The SPR data showed that the 100-fold increase in the affinity of gD1 (Δ290-299t) for HveAt was due primarily to a 40-fold increase in the kon (Table 3). Therefore, the rate of association between gD1 (Δ290-299t) and HveAt is higher than the rate of association between gD1 (306t) and HveAt. However, the stability of the gD1 (Δ290-299t) HveAt complex once formed is similar to that of the gD1 (306t)-HveAt complex (Table 3). Furthermore, the difference in affinity between the two proteins was evident when affinities were measured under both kinetic (short association time and high flow rate) and equilibrium (long association time and low flow rate) conditions of binding.
TABLE 3.
Changes in kon and koff relative to values for gD1 (306t)
| Protein | Relative change in:
|
|
|---|---|---|
| kona | koffb | |
| gD1 (306t) | 1.0 | 1.0 |
| gD1 (Δ290-299t) | 40 | 0.4 |
| gD2 (306t) | 1.4 | 0.6 |
| gD1 (QAAt) | 1.0 | 0.6 |
| gD1 (∇126t) | 0.2 | 1.3 |
| gD1 (∇243t) | 1.8 | 0.8 |
| gD1 (cys1,5) | 0.5 | 0.8 |
| gD1 (cys2,6) | 0.4 | 1.2 |
| gD1 (cys3,4) | 0.2 | 0.9 |
The relative change in kon was calculated from the equation kon of gDt/kon of gD1 (306t).
The relative change in koff was calculated from the equation koff of gDt/koff of gD1 (306t).
Many mutations in gD have been made and tested for the ability to complement the infectivity of a gD null virus in trans (5, 22, 23, 27, 39). The most interesting variants are those that are structurally intact but fail to complement. Whitbeck et al. (50) compared the levels of binding of several variant forms of gDt to HveAt by ELISA. Most of the variants in the previous study were functional in the complementation assay. Our work extended that study to include other functional and nonfunctional variants and to examine the kinetics of binding by SPR.
(i) Affinity of gD2 (306t) and gD1 (QAAt) for HveAt.
Neither gD2 (306t) nor gD1 (QAAt) showed binding kinetics different from those of gD1 (306t) (Table 3), indicating that gDt from HSV-2 interacts with HveAt in the same fashion as gDt from HSV-1. Furthermore, N-CHO on gD1 do not play a role in this interaction.
(ii) Contribution of disulfide bonds to the binding of gDt to HveAt.
Long et al. (23) showed that three variants of gD, each lacking one disulfide bond, can fold into a structure that resembles that of native gD. Of the three double cysteine variants, gD1(cys1,5) was clearly the least damaged, and this protein had the greatest capacity to complement the infectivity of a gD null virus. Though the three double-Cys variants did show some proper antigenic conformation, the ability of each protein to fold and to function was temperature sensitive. This emphasized the importance of all three bonds for the proper structure and function of gD. Here we produced truncated forms of the three double-Cys variant proteins. We took advantage of the fact that the insect cells produced the proteins at a (presumably) permissive temperature of 27°C. Antigenic analysis of the truncated proteins showed the same profiles seen with the full-length viral forms of the proteins. Binding to HveAt followed the trends seen in each full-length protein’s ability to complement a gD null virus (23); gD1 (cys1,5) complements better than gD1 (cys2,6) and much better than gD1 (cys3,4). The affinity of gD1 (cys1,5) for HveAt was similar to that of gD1 (306t). However, the affinity of the gD1 (cys2,6) and gD1 (cys3,4) variants for HveAt were significantly lower. For both of these variants, the decrease in affinity was primarily due to a decrease in kon (Table 3). The koff values of all three variants were similar to that of wild-type gDt, indicating that the stability of the complex was not affected by the mutations removing any of the disulfide bonds. Thus, the disulfide bond between Cys 1 and 5 is not necessary for gDt binding to HveAt while the disulfide bonds between Cys 2 and 6 and Cys 3 and 4 are more important for the conformation of gDt that is necessary for binding to HveAt.
(iii) Affinity of nonfunctional variants for HveAt.
Four functional regions in gD (Fig. 1) were identified by studying insertion and deletion variants that retained antigenic conformation but lost the ability to complement a gD null virus (5, 33). In the binding experiments reported here, it appears that the mutations in functional regions I and IV have the biggest effect on binding of gDt to HveAt. The mutation within functional region I resulted in a loss of HveAt binding ability. Nicola et al. (32) recently reported that two groups of MAbs representing different epitopes on gD (sites Ib and VII) are important for HveAt binding to gD in the virus. Antigenic region VII (residues 11 to 19) (10, 20) is in close proximity to functional region I. Thus, the gD1 (∇34t) insertion may disrupt this region and diminish binding to HveAt. The functional region IV variant exhibited enhanced HveAt binding ability (reference 50 and this report). This variant, gD1 (Δ290-299t), may be nonfunctional because of its high kon, which leads to a high affinity for receptor (50). One possibility is that the conformation of the complex is altered such that later events in virus-cell fusion that occur after the interaction of gD1 (Δ290-299) with HveA are not triggered. The characterization of binding of other functional region IV variants as well as truncation variants of gDt to HveAt are presented in a paper by Rux et al. (35).
The functional region II variant, gD1 (∇126t), showed a marked decrease in binding to HveAt by both ELISA and SPR, which perhaps accounts for its nonfunctional phenotype in complementation assays. In contrast, the functional region III variant, gD1 (∇243t), showed a wild-type level of binding to HveAt. This result led us to ask why the full-length protein is nonfunctional in a complementation assay. Nicola et al. (33) presented data consistent with the idea that gD functions at more than one step in virus entry. Handler et al. (13) showed that oligomeric associations among the glycoproteins changed during HSV entry, and at least one study of HSV entry suggested that individual glycoproteins function at distinct and different points in entry in a sequential, cascade-like mechanism (11). Perhaps the insertion in functional region III at residue 243 alters the ability of gD to participate in a step in entry that occurs after receptor binding.
Johnson et al. (16) showed by Scatchard analysis that gD1 truncated at amino acid 275 from a mammalian expression system bound to the cell surface with an affinity of 0.26 × 10−6 M. This value is intermediate between that of gD1 (306t) and gD1 (Δ290-299t) (Table 2), possibly because (i) the gDt used for our study was produced in insect cells while that used by Johnson et al. (16) was from a mammalian system and/or (ii) Johnson et al. looked at binding of gDt to the cell surface, which may contain multiple receptors for gD (25, 50). In this study, we looked at direct binding between gDt and HveAt.
Significance of a higher kon with little to no change in koff.
Three recent reports look at the effects of point mutations on kon and koff in the binding of various proteins to receptors (6, 26, 48). Morton et al. (26) looked at mutations in human interleukin 5 (hIL-5) believed to be contact residues in the binding of hIL-5 to the α chain of its receptor. They reported up to 5-fold changes in kon and up to 10-fold changes in koff. Some of the mutations affected both kon and koff significantly. Cunningham and Wells (6) looked at the 31 side chains of residues buried at the interface between human growth hormone and the extracellular binding domain of its receptor. They found that mutations in one-quarter of the buried interface side chains led to a decrease in the koff, which stabilized the interaction between the two proteins. Up to 25-fold increases in koff were measured for some of the mutations, while the largest change in kon was 3-fold. Wang et al. (48) looked at mutations in proposed binding residues of hIL-4 with its cellular receptor. The mutations helped to define a koff epitope, i.e., residues that are involved in stabilizing the formed complex. Mutations that reduced the charge of this epitope increased koff by up to 360- fold, while the largest change in kon was a 5-fold decrease.
Unlike the results of the studies described above, the variants studied here had a limited effect on koff (up to 2.5-fold differences) but a more significant effect on kon (5- to 40-fold differences) (Table 3). This suggests that complex formation is sensitive to changes in gD but that once a gDt-HveAt complex is formed, slight differences in side chain composition and antigenic conformation have little effect on the stability of the complex. It is probable that examination of more variants will reveal portions of gD necessary for maintaining a stable complex with HveA (35).
Oligomerization of gDt and the gDt-HveAt complex.
A number of reports have indicated that full-length gD1 may oligomerize into dimers or trimers in the virion, on the surfaces of HSV-infected cells, and when purified (8, 14). There are no reports that gD from HSV-2 forms dimers. Here we show that both soluble gD1 and gD2 form noncovalent dimers in solution. Furthermore, N-CHO on gD1 do not play a role in dimer formation. We first used mass spectrometry to obtain accurate molecular masses for the monomeric forms of the proteins (34) and then gel filtration chromatography and STEM to show that gD1 (306t), gD2 (306t), gD1 (QAAt), and gD1 (Δ290-299t) (50) form dimers in solution.
Whitbeck et al. (50) showed by gel filtration that HveAt is a dimer in solution and proposed that the stoichiometry of the gD1 (Δ290-299t)–HveAt complex is 1:2. If gDt interacts in the complex as a dimer, then HveAt interacts in the complex as two dimers. The fact that the complex fits a 1:1 binding model suggests that the two dimers of HveAt act as one binding unit and that the gDt dimer acts as one binding unit. Thus, there is a 1:1 interaction between binding units. Alternatively, even though gDt is present as a dimer in solution, it may dissociate and interact as a monomer with one dimer of HveAt to yield a 1:1 binding interaction. When other binding models depicting more complicated binding schemes were tried, none fit the data as well as the 1:1 Langmuir binding model.
It is possible to determine the stoichiometry of the gDt-HveAt complex being formed on the surface of the chip, provided that all of the immobilized HveAt is active (2). However, in our system, not all of the immobilized HveAt was active. Therefore, various methods to uniformly orient the coupling of HveAt to the chip surface are being investigated. Analytical ultracentrifugation experiments are also in progress to determine the stoichiometries of the different gDt-HveAt complexes.
ACKNOWLEDGMENTS
This investigation was supported by Public Health Service grants AI-18289 from the National Institute of Allergy and Infectious Diseases and NS-36731 and NS-30606 from the National Institute of Neurological Diseases and Stroke. S.H.W. and A.H.R. received support from Public Health Service grant AI-07324, and A.V.N. received support from Public Health Service grant AI-07325. We thank the Schools of Dental and Veterinary Medicine at the University of Pennsylvania for supplying funds for the purchase of a BIACORE X.
We thank John D. Lambris and William T. Moore of the Protein Chemistry Laboratory of the School of Medicine, University of Pennsylvania, for the mass spectrometry data. We thank Beth Lin for STEM sample preparation and Martha Simon and Joseph Wall of the Biology Department at the Brookhaven National Laboratory, Upton, N.Y., for collecting and analyzing the STEM data. (The Brookhaven STEM is supported by NIH grant P41-RR01777 and by the U.S. Department of Energy.) We thank Gabriela Canziani, manager of the Biosensor/Interaction Analysis Core Facility, and Irwin Chaiken and Sheng-jiun Wu at the School of Medicine of the University of Pennsylvania for help with SPR training and data analysis. We also thank Claude Krummenacher for a critical reading of the manuscript.
REFERENCES
- 1.Baker S J, Reddy E P. Transducers of life and death: TNF receptor superfamily and associated proteins. Oncogene. 1996;12:1–9. [PubMed] [Google Scholar]
- 2.Biacore A B. BIAevaluation software handbook, version 3.0. Uppsala, Sweden: Biacore AB; 1997. [Google Scholar]
- 3.Campadelli-Fiume G, Arsenakis M, Farabegoli F, Roizman B. Entry of herpes simplex virus 1 in BJ cells that constitutively express viral glycoprotein D is by endocytosis and results in degradation of the virus. J Virol. 1988;62:159–167. doi: 10.1128/jvi.62.1.159-167.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Casasnovas J M, Springer T A. Kinetics and thermodynamics of virus binding to receptor. J Biol Chem. 1995;270:13216–13224. doi: 10.1074/jbc.270.22.13216. [DOI] [PubMed] [Google Scholar]
- 5.Chiang H-Y, Cohen G H, Eisenberg R J. Identification of functional regions of herpes simplex virus glycoprotein gD by using linker-insertion mutagenesis. J Virol. 1994;68:2529–2543. doi: 10.1128/jvi.68.4.2529-2543.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cunningham B C, Wells J A. Comparison of a structural and a functional epitope. J Mol Biol. 1993;234:554–563. doi: 10.1006/jmbi.1993.1611. [DOI] [PubMed] [Google Scholar]
- 7.Eisenberg R J, Long D, Sodora D L, Chiang H-Y, Wilcox W C, Abrams W R, Muggeridge M I, Cohen G H. Structure and function of glycoprotein D of herpes simplex virus. In: Becker Y, Darai G, editors. Frontiers in virology. Vol. 3. Heidelberg, Germany: Springer Verlag; 1994. pp. 43–65. [Google Scholar]
- 8.Eisenberg R J, Ponce de Leon M, Pereira L, Long D, Cohen G H. Purification of glycoprotein gD of herpes simplex virus types 1 and 2 by use of monoclonal antibody. J Virol. 1982;41:1099–1104. doi: 10.1128/jvi.41.3.1099-1104.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feenstra V, Hodaie M, Johnson D C. Deletions in herpes simplex virus glycoprotein D define nonessential and essential domains. J Virol. 1990;64:2096–2102. doi: 10.1128/jvi.64.5.2096-2102.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Friedman H M, Cohen G H, Eisenberg R J, Seidel C A, Cines D B. Glycoprotein C of herpes simplex virus 1 acts as a receptor for the C3b complement component on infected cells. Nature (London) 1984;309:633–635. doi: 10.1038/309633a0. [DOI] [PubMed] [Google Scholar]
- 11.Fuller A O, Lee W C. Herpes simplex virus type 1 entry through a cascade of virus-cell interactions requires different roles of gD and gH in penetration. J Virol. 1992;66:5002–5012. doi: 10.1128/jvi.66.8.5002-5012.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Geraghty, R. J., C. Krummenacher, R. J. Eisenberg, G. H. Cohen, and P. G. Spear. Entry of alphaherpesviruses mediated by poliovirus receptor related protein 1 and poliovirus receptor. Science, in press. [DOI] [PubMed]
- 13.Handler C G, Cohen G H, Eisenberg R J. Cross-linking of glycoprotein oligomers during herpes simplex virus type 1 entry. J Virol. 1996;70:6076–6082. doi: 10.1128/jvi.70.9.6076-6082.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Handler C G, Eisenberg R J, Cohen G H. Oligomeric structure of glycoproteins in herpes simplex virus type 1. J Virol. 1996;70:6067–6075. doi: 10.1128/jvi.70.9.6067-6070.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hillenkamp F, Karas M, Beavis R C, Chait B T. Matrix-assisted laser desorption/ionization mass spectrometry of biopolymers. Anal Chem. 1991;63:1193A–1203A. doi: 10.1021/ac00024a002. [DOI] [PubMed] [Google Scholar]
- 16.Johnson D C, Burke R L, Gregory T. Soluble forms of herpes simplex virus glycoprotein D bind to a limited number of cell surface receptors and inhibit virus entry into cells. J Virol. 1990;64:2569–2576. doi: 10.1128/jvi.64.6.2569-2576.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson D C, Ligas M W. Herpes simplex viruses lacking glycoprotein D are unable to inhibit virus penetration: quantitative evidence for virus-specific cell surface receptors. J Virol. 1988;62:4605–4612. doi: 10.1128/jvi.62.12.4605-4612.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karlsson R, Michaelsson A, Mattson A. Kinetic analysis of monoclonal antibody-antigen interactions with a new biosensor based analytical system. J Immunol Methods. 1991;145:229–240. doi: 10.1016/0022-1759(91)90331-9. [DOI] [PubMed] [Google Scholar]
- 19.Krummenacher, C., A. V. Nicola, J. C. Whitbeck, H. Lou, W. Hou, J. D. Lambris, R. J. Geraghty, P. G. Spear, G. H. Cohen, and R. J. Eisenberg. Herpes simplex virus glycoprotein D can bind to poliovirus receptor related protein 1 (PRR1/HveC) or herpes virus entry mediator (HVEM/HveA), two structurally unrelated mediators of virus entry. Submitted for publication. [DOI] [PMC free article] [PubMed]
- 20.Lasonder E, Schellenkens G A, Koedijk D G, Damhof R A, Welling-Wester S, Feijlbrief M, Scheffer A J, Welling G W. Kinetic analysis of synthetic analogues of linear-epitope peptides of glycoprotein D of herpes simplex virus type 1 by surface plasmon resonance. Eur J Biochem. 1996;240:209–214. doi: 10.1111/j.1432-1033.1996.0209h.x. [DOI] [PubMed] [Google Scholar]
- 21.Lee W C, Fuller A O. Herpes simplex virus type 1 and pseudorabies virus bind to a common saturable receptor on Vero cells that is not heparan sulfate. J Virol. 1993;67:5088–5097. doi: 10.1128/jvi.67.9.5088-5097.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Long D, Cohen G H, Muggeridge M I, Eisenberg R J. Cysteine mutants of herpes simplex virus type 1 glycoprotein D exhibit temperature-sensitive properties in structure and function. J Virol. 1990;64:5542–5552. doi: 10.1128/jvi.64.11.5542-5552.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Long D, Wilcox W C, Abrams W R, Cohen G H, Eisenberg R J. Disulfide bond structure of glycoprotein D of herpes simplex virus types 1 and 2. J Virol. 1992;66:6668–6685. doi: 10.1128/jvi.66.11.6668-6685.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mauri D N, Ebner R, Kochel K D, Montgomery R I, Cheung T C, Yu G-L, Murphy M, Eisenberg R J, Cohen G H, Spear P G, Ware C F. LIGHT, a new member of the TNF superfamily, and lymphotoxin (LT) α are ligands for herpesvirus entry mediator (HVEM) Immunity. 1998;8:21–30. doi: 10.1016/s1074-7613(00)80455-0. [DOI] [PubMed] [Google Scholar]
- 25.Montgomery R I, Warner M S, Lum B J, Spear P G. Herpes simplex virus-1 entry into cells mediated by a novel member of the TNF/NGF receptor family. Cell. 1996;87:427–436. doi: 10.1016/s0092-8674(00)81363-x. [DOI] [PubMed] [Google Scholar]
- 26.Morton T A, Li J, Cook R, Chaiken I M. Mutagenesis in the C-terminal region of human interleukin 5 reveals a central patch for receptor α chain recognition. Proc Natl Acad Sci USA. 1995;92:10879–10883. doi: 10.1073/pnas.92.24.10879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muggeridge M I, Chiang H-Y, Cohen G H, Eisenberg R J. Mapping of a functional site on herpes simplex virus glycoprotein D. J Cell Biochem Suppl. 1992;16C:132. [Google Scholar]
- 28.Muggeridge M I, Wilcox W C, Cohen G H, Eisenberg R J. Identification of a site on herpes simplex virus type 1 gD that is essential for infectivity. J Virol. 1990;64:3617–3626. doi: 10.1128/jvi.64.8.3617-3626.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Myszka D G. Kinetic analysis of macromolecular interactions using surface plasmon resonance biosensors. Curr Opin Biotechnol. 1997;8:50–57. doi: 10.1016/s0958-1669(97)80157-7. [DOI] [PubMed] [Google Scholar]
- 30.Nicola, A. V., R. J. Eisenberg, and G. H. Cohen. 1997. Unpublished data.
- 31.Nicola A V, Peng C, Lou H, Cohen G H, Eisenberg R J. Antigenic structure of soluble herpes simplex virus glycoprotein D correlates with inhibition of HSV infection. J Virol. 1997;71:2940–2946. doi: 10.1128/jvi.71.4.2940-2946.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nicola A V, Ponce de Leon M, Xu R, Hou W, Whitbeck J C, Krummenacher C, Montgomery R I, Spear P G, Eisenberg R J, Cohen G H. Monoclonal antibodies to distinct sites on the herpes simplex virus (HSV) glycoprotein D block HSV binding to HVEM. J Virol. 1998;72:3595–3601. doi: 10.1128/jvi.72.5.3595-3601.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nicola A V, Willis S H, Naidoo N N, Eisenberg R J, Cohen G H. Structure-function analysis of soluble forms of herpes simplex virus glycoprotein D. J Virol. 1996;70:3815–3822. doi: 10.1128/jvi.70.6.3815-3822.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rux A H, Moore W T, Lambris J D, Abrams W R, Peng C, Friedman H M, Cohen G H, Eisenberg R J. Disulfide bond structure determination and biochemical analysis of glycoprotein C from herpes simplex virus. J Virol. 1996;70:5455–5465. doi: 10.1128/jvi.70.8.5455-5465.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rux, A. H., S. H. Willis, A. V. Nicola, W. Hou, H. Lou, C. Peng, G. H. Cohen, and R. J. Eisenberg. Functional region IV of glycoprotein D from herpes simplex virus modulates glycoprotein binding to the herpes simplex virus entry mediator (HVEM/HveA). Submitted for publication. [DOI] [PMC free article] [PubMed]
- 36.Sisk W P, Bradley J D, Leipold R J, Stoltzfus A M, Ponce de Leon M, Hilf M, Peng C, Cohen G H, Eisenberg R J. High-level expression and purification of secreted forms of herpes simplex virus type 1 glycoprotein gD synthesized by baculovirus-infected insect cells. J Virol. 1994;68:766–775. doi: 10.1128/jvi.68.2.766-775.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith C A, Farrah T, Goodwin R G. The TNF receptor superfamily of cellular and viral proteins: activation, costimulation, and death. Cell. 1994;76:959–962. doi: 10.1016/0092-8674(94)90372-7. [DOI] [PubMed] [Google Scholar]
- 38.Sodora D L, Cohen G H, Eisenberg R J. Influence of asparagine-linked oligosaccharides on antigenicity, processing, and cell surface expression of herpes simplex virus type 1 glycoprotein D. J Virol. 1989;63:5184–5193. doi: 10.1128/jvi.63.12.5184-5193.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sodora D L, Cohen G H, Muggeridge M I, Eisenberg R J. Absence of asparagine-linked oligosaccharides from glycoprotein D of herpes simplex virus type 1 results in a structurally altered but biologically active protein. J Virol. 1991;65:4424–4431. doi: 10.1128/jvi.65.8.4424-4431.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sodora D L, Eisenberg R J, Cohen G H. Characterization of a recombinant herpes simplex virus which expresses a glycoprotein D lacking asparagine-linked oligosaccharides. J Virol. 1991;65:4432–4441. doi: 10.1128/jvi.65.8.4432-4441.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Spear P G. Membrane fusion induced by herpes simplex virus. In: Bentz J, editor. Viral fusion mechanisms. Boca Raton, Fla: CRC Press, Inc.; 1993. pp. 201–232. [Google Scholar]
- 42.Takemoto D K, Skehel J J, Wiley D C. A surface plasmon resonance assay for the binding of influenza virus hemagglutinin to its sialic acid receptor. Virology. 1996;217:452–458. doi: 10.1006/viro.1996.0139. [DOI] [PubMed] [Google Scholar]
- 43.Tal-Singer R, Eisenberg R J, Valyi-Nagy T, Fraser N W, Cohen G H. N-linked oligosaccharides on herpes simplex virus glycoprotein gD are not essential for establishment of viral latency or reactivation in the mouse eye model. Virology. 1994;202:1050–1053. doi: 10.1006/viro.1994.1437. [DOI] [PubMed] [Google Scholar]
- 44.Tessier D C, Thomas D Y, Khouri H E, Laliberte F, Vernet T. Enhanced secretion from insect cells of a foreign protein fused to the honeybee melittin signal peptide. Gene. 1991;98:177–183. doi: 10.1016/0378-1119(91)90171-7. [DOI] [PubMed] [Google Scholar]
- 45.Thomas D J, Wall J S, Hainfield J F, Kaczorek M, Booy R P, Trus B L, Eiserling F A, Stevens A C. gp160, the envelope glycoprotein of human immunodeficiency virus type 1, is a dimer of 125-kilodalton subunits stabilized through interactions between their gp41 domains. J Virol. 1991;65:3797–3803. doi: 10.1128/jvi.65.7.3797-3803.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wall J S. Biological scanning transmission electron microscopy. In: Hren J J, Goldstein J I, Joy D C, editors. Introduction to analytical electron microscopy. New York, N.Y: Plenum Publishing Corp.; 1979. pp. 333–342. [Google Scholar]
- 47.Wall J S, Hainfeld J F. Mass mapping with the scanning transmission electron microscope. Annu Rev Biophys Chem. 1986;15:355–376. doi: 10.1146/annurev.bb.15.060186.002035. [DOI] [PubMed] [Google Scholar]
- 48.Wang Y, Shen B-J, Sebalk W. A mixed-charge pair in human interleukin 4 dominates high-affinity interaction with the receptor α chain. Proc Natl Acad Sci USA. 1997;94:1657–1662. doi: 10.1073/pnas.94.5.1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Warner, M. S., W. Martinez, R. J. Geraghty, R. I. Montgomery, J. C. Whitbeck, R. Xu, R. J. Eisenberg, G. H. Cohen, and P. G. Spear. A cell surface protein with herpesvirus entry activity (HveB) confers susceptibility to infection by herpes simplex virus type 2, mutants of herpes simplex virus type 1 and pseudorabies virus. Virology, in press. [DOI] [PubMed]
- 50.Whitbeck J C, Peng C, Lou H, Xu R, Willis S H, Ponce de Leon M, Peng T, Nicola A V, Montgomery R I, Warner M S, Soulika A M, Spruce L A, Moore W T, Lambris J D, Spear P G, Cohen G H, Eisenberg R J. Glycoprotein D of herpes simplex virus (HSV) binds directly to HVEM, a member of the tumor necrosis factor receptor superfamily and a mediator of HSV entry. J Virol. 1997;71:6083–6093. doi: 10.1128/jvi.71.8.6083-6093.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Whitbeck, J. C., C. P. Peng, G. H. Cohen, and R. J. Eisenberg. 1997. Unpublished data.
- 52.Williams R K, Straus S E. Specificity and affinity of binding of herpes simplex virus type 2 glycoprotein B to glycosaminoglycans. J Virol. 1997;71:1375–1380. doi: 10.1128/jvi.71.2.1375-1380.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Willis S H, Peng C, Ponce de Leon M, Nicola A V, Rux A H, Cohen G H, Eisenberg R J. Expression and purification of secreted forms of HSV glycoproteins from baculovirus-infected insect cells. In: Brown S M, MacLean A R, editors. Methods in molecular medicine. 10, Herpes simplex virus protocols. Totowa, N.J: Humana Press Inc.; 1998. [DOI] [PubMed] [Google Scholar]