Abstract
We used the PCR to screen for the presence of endogenous retroviruses within the genomes of 18 vertebrate orders across eight classes, concentrating on reptilian, amphibian, and piscine hosts. Thirty novel retroviral sequences were isolated and characterized by sequencing approximately 1 kb of their encoded protease and reverse transcriptase genes. Isolation of novel viruses from so many disparate hosts suggests that retroviruses are likely to be ubiquitous within all but the most basal vertebrate classes and, furthermore, gives a good indication of the overall retroviral diversity within vertebrates. Phylogenetic analysis demonstrated that viruses clustering with (but not necessarily closely related to) the spumaviruses and murine leukemia viruses are widespread and abundant in vertebrate genomes. In contrast, we were unable to identify any viruses from hosts outside of mammals and birds which grouped with the other five currently recognized retroviral genera: the lentiviruses, human T-cell leukemia-related viruses, avian leukemia virus-related retroviruses, type D retroviruses, and mammalian type B retroviruses. There was also some indication that viruses isolated from individual vertebrate classes tended to cluster together in phylogenetic reconstructions. This implies that the horizontal transmission of at least some retroviruses, between some vertebrate classes, occurs relatively infrequently. It is likely that many of the retroviral sequences described here are distinct enough from those of previously characterized viruses to represent novel retroviral genera.
Vertebrate genomes contain numerous parasitic genetic elements, many of which undergo vertical germ line transmission and are capable of remaining in the same locus for millions of years (40, 45). Some of the best studied of these elements are members of the Retroviridae which, as exogenous infectious viruses, cause neurological and immunological diseases, malignancies, and immunodeficiencies (4). Prior to integration within their host’s genome, retroviruses use virally encoded reverse transcriptase to copy their RNA genome into DNA. The low fidelity of this enzyme results in a high rate of mutation, with the subsequent result that retroviruses have highly divergent nucleotide sequences (36). However, there are regions of the retroviral genome, especially within the polymerase gene, which are reasonably well conserved when different isolates are compared, and this has enabled phylogenetic trees of the retroviruses to be constructed (6, 8, 45). The latest classification, based at least partly on this type of analysis, placed retroviruses into seven genera: the spumaviruses, murine leukemia-related viruses (MLVs), lentiviruses, human T-cell leukemia-related viruses (HTLVs), avian leukosis viruses (ALVs), type D viruses, and mammalian type B viruses (4, 9). With few exceptions, all of the retroviruses from these genera were isolated from mammalian (and a small number of avian) hosts, leaving questions pertaining to their origin, evolution, and distribution within other vertebrates that remain largely unanswered. Other questions of interest concern the host range boundary of the Retroviridae and whether additional retroviral genera remain to be discovered. With regard to the latter point, several recently reported novel retroviruses in reptiles, amphibians, and fish have suggested that this may indeed be the case (13, 15, 39).
One approach to answering these questions involves the amplification of endogenous retroviral sequences by PCR (34, 43), as there are several highly conserved motifs within retroviral proteins against which degenerate oligonucleotide primers can be designed (37, 41, 45). It should therefore be possible to obtain a good idea of the retroviral diversity within a range of organisms by using this methodology, in conjunction with multiple phylogenetic analyses of the resultant sequences.
Here we use this procedure to isolate and characterize novel endogenous retroviral sequences from a wide range of vertebrate hosts and to examine their relationships with previously described retroviruses.
MATERIALS AND METHODS
Nomenclature.
The term retrovirus is currently used to describe two different (but overlapping) sets of retroelements: (i) a member of the Retroviridae, in the sense that this family is monophyletic with respect to other retroelements, and (ii) an infectious retroelement, in the sense that (in addition to many members of the Retroviridae) the gypsy long terminal repeat (LTR) retrotransposon in Drosophila melanogaster can also be transmitted horizontally (16). For purposes of clarity we use the former definition in this report.
Primer design and amplification.
Four degenerate oligonucleotide primers were used in this study. One was designed against the active-site motif present within the protease protein and three were designed against the active site of the reverse transcriptase protein as follows: protease (PRO) 5′GTT/GTTIG/TTI GAT/CACIGGIG/TC3′ and reverse transcriptase, designated CT, 5′AGIAG GTCA/GTCIACA/GTAC/GTG3′, JO 5′ATIAGIAG/TA/GTCA/GTCIACA/G TA3′, and EM 5′ATIAGIAG/TA/GTCA/GTCCATA/GTA3′, where I = inosine. PCR conditions, which have been previously described in detail (37), consisted of 2 min at 80°C followed by 35 cycles of a 45 to 50°C annealing step for 30 s, polymerization at 74°C for 60 to 70 s, and denaturation at 94°C for 30 s and finally one cycle at 45 to 50°C for 3 min and 74°C for 10 min. Reaction conditions were as follows: 50 mM KCl, 10 mM Tris-HCl (pH 8.3), 1.5 mM MgCl2, 200 μM (each) deoxynucleoside triphosphate, 150 pmol of each primer, 100 to 500 ng of template DNA, and 2 U of Taq polymerase.
Sequencing and sequence analysis.
Amplification products were electrophoresed through 1.3% agarose gels, and products of between 600 and 1,200 bp were excised, purified, and cloned into the vector pCRII (Invitrogen). Cloned inserts were sequenced in both directions, either manually using a Sequenase kit (U.S. Biochemicals) or with an automated DNA sequencer (ABI 373 Stretch) and a dye-terminator kit (Perkin-Elmer Cetus). Retroviral sequences were initially identified by BLAST searches (1) or by screening CD-ROM sequence libraries.
Phylogenetic analysis.
The data matrix consisted of 190 amino acid residues, 160 derived from the reverse transcriptase protein (aligned as described in reference 45) and 30 from the protease [from 15 residues 5′ to 12 residues 3′ of the well-conserved GR(D/N) motif]. Phylogenetic trees were constructed by using the program PAUP4d-56–4d61 (written by D. L. Swofford) and utilized both the neighbor-joining and maximum parsimony approaches. All trees, except where stated, were generated by using amino acids and an unordered matrix. Six equally parsimonious trees were identified after the data set was subjected to a heuristic search comprising 100 random-addition replicates. The robustness of each node was assessed by bootstrap resampling with 1,000 replicates using neighbor joining or with 100 replicates using maximum parsimony (each of 25 random-addition sequences with all characters unordered). The group III sequences were investigated further by using a longer sequence alignment of 257 amino acid residues. The phylogeny of these sequences was reconstructed using the PROTPARS matrix and was subjected to 1,000 bootstrap replicates with neighbor-joining analyses or 100 bootstrap replicates (each with five random additions) in the case of maximum parsimony. The level of clustering of the group III sequences into clades derived from a single vertebrate class was examined by using the program MacClade (20). A multistate character representing the host class was scored for each taxon, and the minimum number of character state changes (i.e., shifts between host classes) required by the phylogeny was then calculated. This number was then compared to those obtained for each of 100 test replicates in which the host class characters were shuffled randomly between the viral taxa while maintaining the same tree. A smaller number of steps required to generate the fit of the real associations, when compared to the fit of the random associations, indicates that the virus-host associations are phylogenetically correlated.
Southern hybridization.
Southern hybridization analysis was performed by the method of Sambrook et al. (32). Hybridization of each of the fragments to 10 μg of host genomic DNA was carried out at 65°C, and the filters were washed down to 0.5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.5% sodium dodecyl sulfate at 65°C.
Sequence sources.
Sequences were obtained from sequence databases with the original sources as described in a previous report (38), in the text, or as follows: micropia (18), feline syncytial foamy virus (FeSFV) (14), human endogenous retrovirus HERV.L (5), murine endogenous retrovirus MuERV.L (2), endogenous retrovirus type 9 (ERV-9) (17), HERV.I (21), human cosmid U85A3 [H- cosmid(U85A3)] (25), jaagsiekte (46), HERV.K10 (26), and RSV (33). The sequences described here have been submitted to the EMBL/GenBank/DDLB databases with the following accession numbers: RV-common possum, AJ225211; RV-stripe faced dunnartI, AJ225230; RV-stripe faced dunnartII, AJ225231; RV-bower bird, AJ225208; RV-tinamou, AJ225235; RV-pit viper, AJ225222; RV-tuatara, AJ225236; RV-gharial, AJ225215; RV-slider turtleI, AJ225227; RV-slider turtleII, AJ225228; RV-palmate newtI, AJ225220; RV-palmate newtII, AJ225221; RV-tiger salamanderI, AJ225232; RV-tiger salamanderII, AJ225233; RV-tiger salamanderIII, AJ225234; RV-rhinatremid caecilianI, AJ225224; RV-rhinatremid caecilianII, AJ225225; RV-rocket frog, AJ225226; RV-edible frog, AJ225212; RV-leopard frog, AJ225218; RV-Iberian frog, AJ225217; RV-European frog, AJ225213; RV-African clawed toad, AJ225207; RV-painted frog, AJ225219; RV-stickleback, AJ225229; RV-brook trout, AJ225209; RV-brown trout, AJ225210; RV-freshwater houting, AJ225214; RV-puffer fish, AJ225223.
RESULTS
Retroviruses have highly variable nucleotide sequences (6, 45), and therefore, to ensure PCR amplification of as wide a range of retroviral sequences as possible, several degenerate oligonucleotide primers were used in this study. Table 1 shows the amino acid motifs on which the primers were based and those retroviral genera which they were predicted to amplify. The PRO primer, based on the protease protein, was used in combination with either CT, JO, or EM, which are complementary to the conserved motif within domain 5 of the reverse transcriptase protein. Virtually all described retroviruses encode either YVDD or YMDD in this position (Table 1), and thus, together, the reverse transcriptase primers are suitable for amplifying retroviruses from all seven genera. This is also the case for the protease primer, as all known retroviral proteases encode the motifs DTGA or DSGA (41), against which the PRO primer was designed. Furthermore, because many elements from a separate retroelement family, the gypsy LTR retrotransposons, also contain identical or very similar sequences, it seemed probable that even highly divergent retroviruses would be amplified by a PCR-based approach utilizing these primers. Although CT, JO, and EM were designed to amplify specific retroviral subgroups or genera (Table 1), some cross-amplification of nontarget genera was observed. Table 2 shows the reverse transcriptase primer used in each successful amplification, the host species, and the name of the isolate.
TABLE 1.
Oligonucleotide primer motifs and target genera
| Primer | Target site | Target genera |
|---|---|---|
| PROa | 5′V(F/L)(F/L/V)DTG(S/A)3′ (protease) | All |
| JOa | 5′YVDD(L/I/M)LI3′(RT)b | Spumaviruses |
| MLV-related viruses | ||
| CT | 5′(H/Q)YVDDLL3′(RT) | MLV-related viruses |
| EM | 5′YMDD(L/I/M)L(I/M)3′(RT) | HTLV-BLV group |
| ALV-related viruses | ||
| Type D viruses | ||
| Mammalian type B viruses | ||
| Lentiviruses |
PRO and JO are also capable of amplifying gypsy and copia LTR retrotransposons.
RT, reverse transcriptase.
TABLE 2.
Retroviral isolates from which sequence data is available, excluding those derived from placental mammals
| Host organism | Primer | Namea |
|---|---|---|
| Marsupials | ||
| Common possum (Trichosurus vulpecula) | JO | RV-common possum |
| Stripe-faced dunnart (Sminthopsis macroura) | EM | RV-stripe faced dunnartI |
| CT | RV-stripe faced dunnartII | |
| Rock wallaby (Petrogale godmani) | CT | RV-rock wallaby (21) |
| Birds | ||
| Order Galliformes | ||
| Chicken (Gallus domesticus) | RSV (32) | |
| Turkey (Meleagris gallopavo) | LDV (11), REV (30) | |
| Order Anseriformes | ||
| Duck | SNV (41) | |
| Order Passeriforms | ||
| House sparrow (Passer domesticus) | JO | RV-sparrow (21) |
| Regent bower bird (Sericulus bakeri) | EM | RV-bower bird |
| Order Tinamiformes | ||
| Elegant crested tinamou (Eudromia elegans) | JO | RV-tinamou |
| Reptiles | ||
| Order Squamata (snakes and lizards) | ||
| South American pit viper (Bothrops jararaca) | JO | RV-pit viper |
| Komodo dragon (Varanus komodoensis) | CT | RV-Komodo dragon (21) |
| Order Rhynchocephalia (tuatara) | ||
| Tuatara (Sphenodon sp.) | JO | SpeV (38) |
| CT | RV-tuatara | |
| Order Crocodylia | ||
| Gharial (Gavialis gangeticus) | CT | RV-gharialII |
| Order Chelonia (turtles) | ||
| Red-eared slider turtle (Chrysemys scripta) | JO | RV-slider turtleI |
| JO | RV-slider turtleII | |
| Amphibians | ||
| Order Urodela (salamanders and newts) | ||
| Palmate newt (Triturus helveticus) | JO | RV-palmate newtI |
| EM | RV-palmate newtII | |
| Tiger salamander (Ambystoma tigrinum) | JO | RV-tiger salamanderI |
| CT | RV-tiger salamanderII | |
| CT | RV-tiger salamanderIII | |
| Order Gymnophiona (caecilians) | ||
| Rhinatremid caecilian (Epicrionops marmoratus) | CT | RV-rhinatremid caecilianI |
| CT | RV-rhinatremid caecilianII | |
| Order Anura (frogs and toads) | ||
| Amazonian poison dart frog (Dendrobates ventrimaculatus) | JO | DevI (37) |
| JO | DevII (37) | |
| JO | DevIII (37) | |
| Rocket frog (Colostethus talamancae) | CT | RV-rocket frog |
| Edible frog (Rana esculenta) | JO | RV-edible frog |
| Leopard frog (Rana pipiens) | JO | RV-leopard frog |
| Iberian frog (Rana iberica) | CT | RV-Iberian frog |
| European common frog (Rana temporaria) | JO | RV-European frog |
| African clawed toad (Xenopus laevis) | JO | RV-African clawed toad |
| Painted frog (Discoglossus galganoi) | CT | RV-painted frog |
| Bony fish | ||
| Order Gasterosteiformes | ||
| Stickleback (Gasterosteus aculeatus) | CT | RV-stickleback |
| Order Salmoniformes | ||
| American brook trout (Salvelinus fontinalis) | CT | RV-brook trout |
| Brown trout (Salmo trutta) | JO | RV-brown trout |
| Freshwater houting (Corogonus lavaretus) | JO | RV-freshwater houting |
| Order Tetraodontiformes | ||
| Puffer fish (Fugu rubripes) | JO | RV-puffer fish |
| Order Perciformes | ||
| Walleye (Stizostedion vitreum) | WDSV (15) | |
| Snakehead (Ophicephalus striatus) | SnRV (13) | |
| Cartilaginous fish | ||
| Order Lamniformes | ||
| Lemon shark (Negaprion brevirostis) | CT | RV-lemon shark (21) |
References are provided for previously characterized viruses.
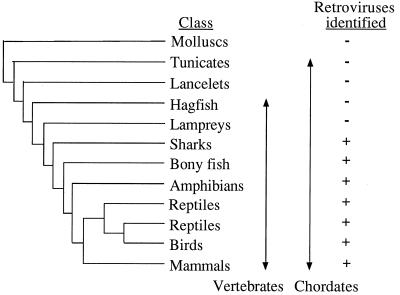
PCRs were performed on genomic DNA samples from more than 50 taxa (which were either obtained from other researchers or prepared from tissue samples). The taxa included members of eight vertebrate and three nonvertebrate classes (Fig. 1), but were mainly derived from reptiles, amphibians, and bony fish, as there have been numerous reports of retroviral particles in these organisms (24, 30). Retroviral sequences were identified in each of the vertebrate classes surveyed, with the exception of the lampreys (Cephalaspidomorphi, such as the river lamprey Lampetra fluviatilis) and the hagfish (Myxini, such as the Taiwanease hagfish Myxine yangi). Furthermore, no retroviral sequences were amplified from nonvertebrate hosts: we examined the lancelet Amphioxus floridae (Cephalochordata), the sea squirt Ciona intestinalis (Urochordata: Ascidiacea), and nine species of mollusc (including the soft-shelled clam Mya arenaria). Almost all the amplifications were performed in duplicate, using both the CT and JO primers, whereas the EM primer was used (generally in parallel with the other primers) with a somewhat smaller number (approximately 40) of taxa.
FIG. 1.
Distribution of retroviral sequences within vertebrates and other Metazoa. The phylogeny is based on that described by Young (47). Endogenous retroviral sequences identified by PCR screening are indicated by a +, whereas a − represents screened taxa from which retroviral sequences have yet to be identified.
Amplification often resulted in the isolation of more than one clone with homology to reverse transcriptase. However, these clones were usually very similar or identical to each other, and only in those cases where homology was less than 90% at the nucleotide level were both clones investigated further. Thirty novel endogenous retroviral fragments were characterized by sequencing the entire length of the amplified and cloned fragment. Of these, 3 were derived from nonplacental mammals (marsupials and monotremes), 2 were from birds, 5 were from reptiles, 14 were from amphibians, and 5 were from bony fish (Table 2). The retroviral isolates included representatives from each of the four orders of reptiles and the three orders of amphibians. Southern hybridization (32) of each of the fragments was performed to confirm its species of origin (data not shown). The lack of sufficient genomic DNA meant that it was not possible to accurately determine the copy number of many of the isolates.
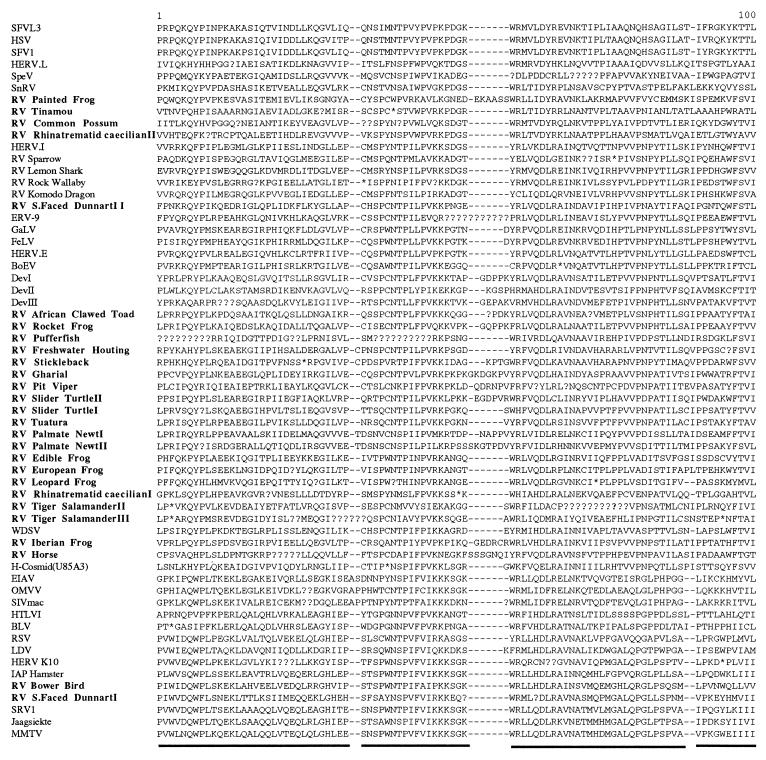
Conceptual translations of the DNA sequence derived from each isolate were performed, and a 160-amino-acid-residue region of the reverse transcriptase protein was then aligned to that of other previously described retroviruses, as described by Xiong and Eickbush (45). Many of the sequences contained stop codons and frameshifts, which required the insertion of one to several nucleotides (coded as unknown) in order to maintain the reading frame (Fig. 2). This alignment was used as the basis for the phylogenetic analyses. The majority of sequence information from outside of this region was not suitable for phylogenetic reconstruction due to the difficulty of aligning homologous amino acid positions, although it was possible to include a 30-amino-acid-residue region from the protease protein. Both neighbor-joining and maximum parsimony trees were generated from the alignment, with support for individual branches investigated by bootstrap analysis.
FIG. 2.
Amino acid alignment derived from retroviral reverse transcriptase proteins, based on that described by Xiong and Eickbush (45). Sequences identified in this study are indicated in bold type. The underlined regions of the alignment were used in subsequent phylogenetic analysis. Asterisks represent in-frame stop codons, and missing data (due to frameshifts or deletions) are indicated by question marks. Three isolates, RV-brook trout, RV-brown trout, and RV-tiger salamanderI, contained large deletions and are not shown.
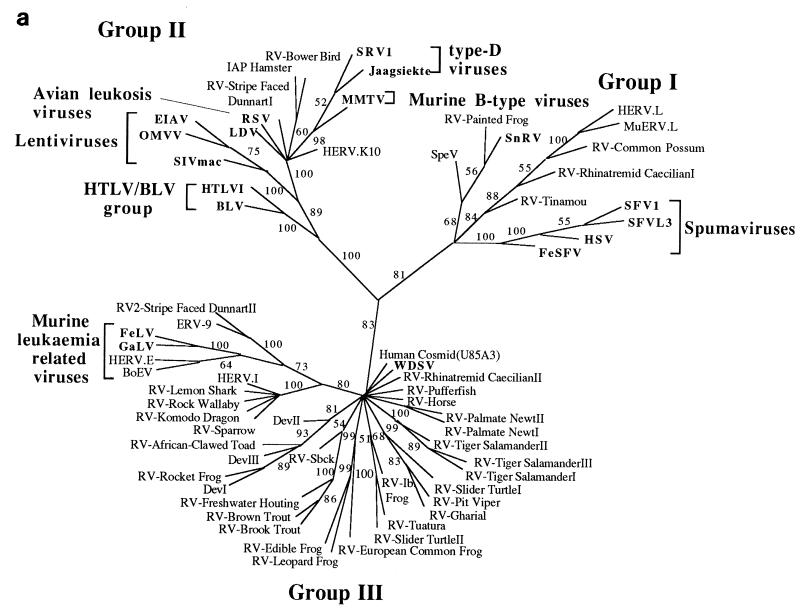
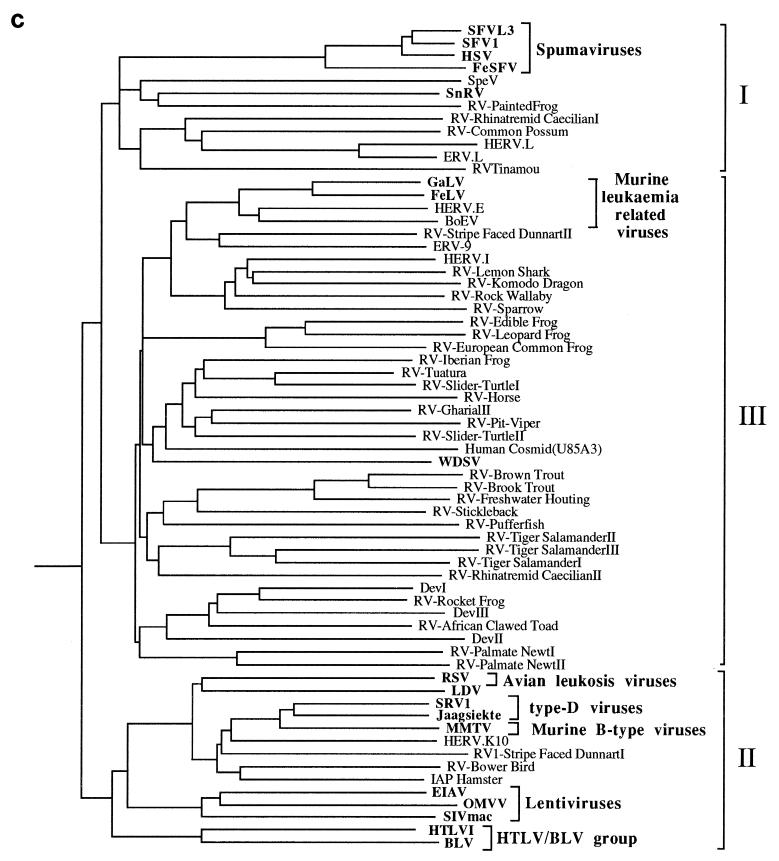
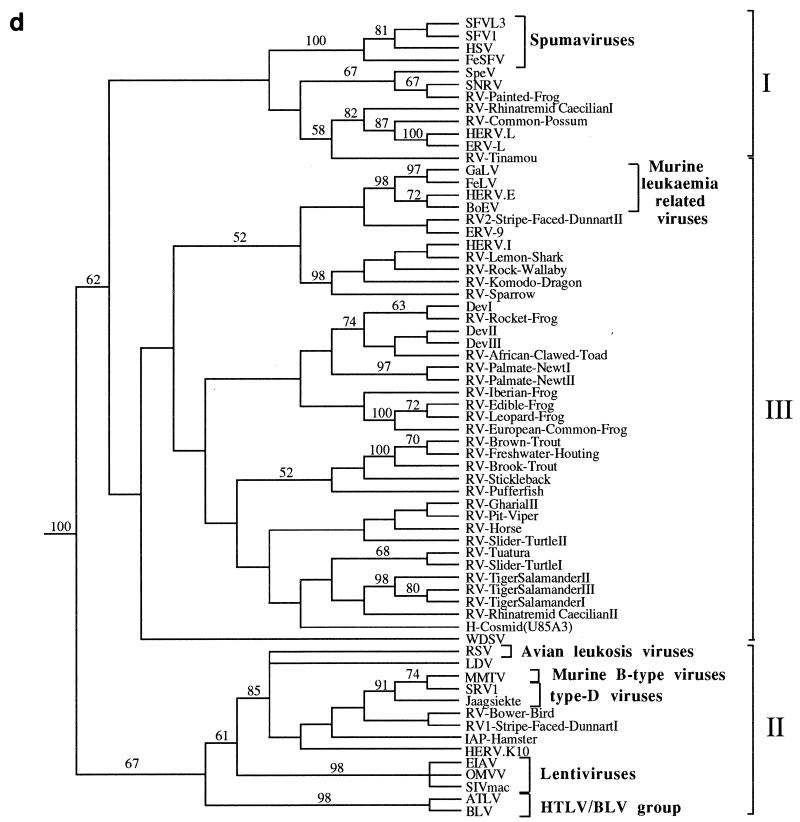
Figure 3a shows an unrooted, bootstrapped neighbor-joining tree of the isolates shown in Fig. 2 (groups not recovered in at least 50% of the bootstrap replicates were collapsed and are represented by polytomies). It was apparent from this analysis that the retroviral sequences clustered into three main groups, each with bootstrap support greater than 80%.
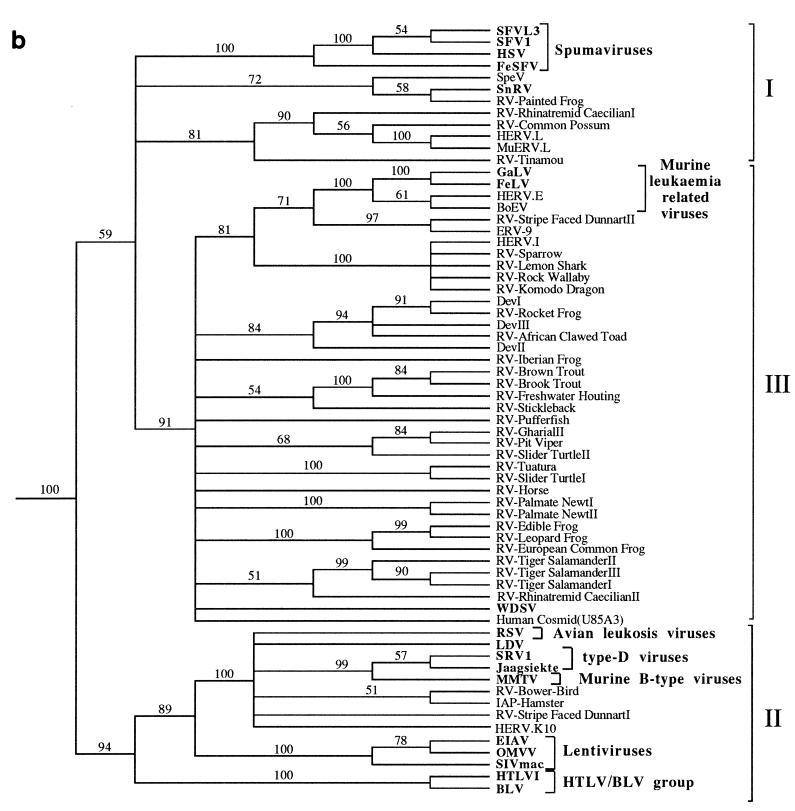
FIG. 3.
Phylogenetic trees of the retroviruses based on the alignment shown in Fig. 2, with the addition of 30 residues derived from the protease protein. Exogenous isolates are indicated by bold type. The genera to which particular retroviruses have been assigned are also shown. (a) Unrooted neighbor-joining bootstrap tree. The figures on each branch represent bootstrap support (from 1,000 replicates), with percentage values on only those branches with support greater than 50 being shown. Unresolved branches are shown as polytomies. (b) Neighbor-joining tree rooted on several gypsy LTR retrotransposon sequences (gypsy, Del, Ty3, and micropia), with figures on each branch again showing percentage bootstrap support (1,000 replicates). (c) Rooted neighbor-joining tree with branch lengths proportional to the genetic distance between the sequences. The tree is rooted as described in the legend to panel a. (d) Strict consensus of six equal-length trees identified by maximum parsimony. Bootstrap values were generated from 100 replicates (each of 25 random-sequence additions) by using the unordered matrix. The tree is rooted as described in the legend to panel a.
One group (group I) contained the spumaviruses (human spumavirus [HSV], simian foamy virus type 1 [SFV1], SFVL3, and FeSFV), the previously described Sphenodon endogenous retrovirus (SpeV, isolated from a reptile), snakehead fish retrovirus (SnRV), HERV.L (a recently identified endogenous human retrovirus), and MuERV.L (an endogenous murine retrovirus closely related to HERV.L), as well as several other novel sequences derived from marsupials, birds, and amphibians. In contrast, group II contained viruses derived exclusively from mammalian and avian hosts and included all members of five of the seven currently recognized retroviral genera (the lentiviruses, the HTLV-bovine leukemia virus [BLV] group, the ALVs, and the type B and D retroviruses), as well as several novel endogenous viruses. Investigation of 30 other amphibian, reptilian, and piscine hosts with the EM primer failed to reveal retroviral sequences related to this group. Group III contained the majority of the endogenous retroviral sequences isolated by our PCR screening, as well as all members of the murine leukemia virus (MLV) genus, exemplified by the feline leukemia virus (FeLV), gibbon ape leukemia virus (GaLV), human endogenous retrovirus type E (HERV.E), and bovine endogenous virus (BoEV). Members of the MLV genus have previously been identified in several species of birds and reptiles (4, 19, 29, 35, 42, 48) and, consistent with this, we also identified elements clustering with the FeLV/GaLV/BoEV/HERV.E isolates in both of these vertebrate classes (data not shown). Several other previously described viruses were also present within this lineage. These included the human endogenous retroviruses ERV-9 and HERV.I (44), the recently described walleye dermal sarcoma virus (WDSV) from fish, and the Dendrobates elements (DevI, DevII, and DevIII) from a poison dart frog. A BLAST search revealed that one further retroviral sequence should also be included. This element was present in a cosmid (accession no. U85A3) sequenced as part of the human genome mapping project (25). One further point to note is that the entire nucleotide sequence of the WDSV isolate has already been determined and contains at least two novel accessory genes (15). This, and other evidence, has led to the suggestion that it probably represents a novel retroviral subfamily or genus (15, 30).
Several gypsy LTR retrotransposon sequences (gypsy, del, Ty3, and micropia) were added to the alignment and used as outgroups to root the retrovirus tree. In rooted phylogenetic analyses, the group II sequences were recovered in a well-supported clade that is the sister group to all other retroviruses (Fig. 3b). Group III sequences also comprised a well-supported clade. However, the addition of the outgroup sequences abolished the bootstrap support for the group I spumaviruslike sequences shown in the unrooted analyses, breaking them up into three well-supported clades that form a polytomy with the group III sequences. Support for this basal polytomy (59%) is also uncompelling. Varying the composition of the outgroup sometimes resulted in weak bootstrap support (about 60%) for the monophyly of group I or an association of the spumaviruses with the SpeV, SnRV, RV-painted frog subgroup. Individual rooted neighbor-joining and parsimony trees did usually include the group I sequences as a clade (Fig. 3c and d), but this monophyly was also dependent on the composition of the outgroup. The uncertainty of the relationships of the group I spumaviruslike sequences close to the base of the retrovirus tree and the weak support for the clade comprising group I and group III sequences must also limit confidence in the placement of the root of the Retroviridae.
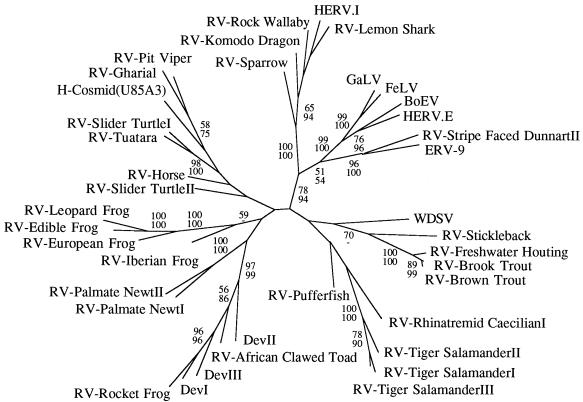
The complex and diverse nature of retroviruses clustering with the MLV genus is emphasized in Fig. 3b. Most of the relationships within this lineage were not well supported by bootstrap analysis, and the reason for this lack of resolution is apparent from Fig. 3c and d: the distance from the base of the group to the separation points of many of the taxa is small when compared to their overall diversity. Despite this, it did appear that there were several well-supported subgroups which contained members derived from only a single vertebrate class or order. For example, many of the fish isolates clustered together and groups of amphibian viruses were also observed. To determine whether this clustering was statistically significant and to try and increase the resolution of the various taxa, the group III viruses were then analyzed separately from the other retroviral sequences. This enabled trees to be generated from an extended sequence alignment (257 as opposed to 190 amino acids), as shown in Fig. 4. The degree to which viruses clustered on the basis of their host class of origin was assessed using the program MacClade (20). The number of steps required to generate the actual host class-virus associations shown in Fig. 4 (nine) was always significantly lower than that observed for each of 100 test replicates in which the host class characters were shuffled randomly between the viral taxa (range, 18 to 24 steps; P < 0.01).
FIG. 4.
Unrooted maximum parsimony tree of the group III viruses shown in Fig. 3. Branch lengths are proportional to the number of changes required to generate the observed variation. Numbers on each branch reflect percentage bootstrap support using maximum parsimony (upper value) or neighbor-joining (lower value).
DISCUSSION
Retroviral phylogeny and taxonomy has for a long time been based almost exclusively on viruses within mammalian and avian hosts, and with the exception of several MLV-related retroviruses within reptiles (for which no sequence data have been reported), all members of the seven currently recognized retroviral genera are present within one or both of these vertebrate classes (4, 9, 19, 48). We, and others, have previously obtained molecular data from a small number of reptilian, amphibian, and piscine retroviruses (13, 15, 22, 38, 39); those sequences, together with the ones in this report, create an emerging picture of the distribution and diversity of retroviruses within lower vertebrate taxa. Endogenous retroviral sequences have now been identified in more than 25 vertebrate orders across six classes, and this suggests that retroviruses may well be ubiquitous in many vertebrate taxa, although their exact host range remains to be determined. The most basal vertebrate from which a retroviral sequence has so far been identified is the lemon shark Negaprion brevirostris (22). We think it unlikely that this element represents a particularly primitive or basal virus for two reasons. First, it is closely related to viruses within several different vertebrate classes, including HERV.I within humans. Second, rooted topological constraint trees which placed RV-lemon shark (or all of the HERV.I-related retroviruses) basal to the other retroviruses required at least an additional 17 steps over the shortest tree in maximum parsimony analysis (data not shown). These points imply that the presence of HERV.I-related viruses in sharks may be the result of horizontal transfer from another vertebrate class. However, numerous additional members of this group will have to be identified and characterized before their mode of dispersal within vertebrates can be definitively answered.
We screened four other (more primitive) vertebrate and chordate classes for the presence of endogenous retroviruses without success (data not shown). However, the number of taxa investigated has so far been small; DNA samples were available from only two species of lamprey (Cephalaspidomorphi, Petromyzoniformes), one hagfish (Myxini, Myxiniformes), one lancelet (Cephalochordata, Branchiostomidae), and one sea squirt (Urochordata, Ascidiacea), and investigation of a much larger number of primitive chordates is required before the absence of retroviruses in these groups can be considered probable. Furthermore, there is some evidence to suggest that retroviruses may be present in nonvertebrate/chordate taxa: seasonal neoplasm in the soft-shelled clam (Mya arenaria) has been linked to exogenous type B retroviruslike particles identified in this species (27, 28). However, PCR screening of nine species of mollusc (including the soft-shelled clam) failed to isolate any endogenous retroviral sequences (data not shown).
Retroviral phylogenies have been reported previously by a number of workers (6–8, 45). Many were constructed using amino acid data sets derived from the 5′ end of the reverse transcriptase protein, and the PCR-amplified retroviral fragments described here contain most of this region. Phylogenetic reconstruction has generally resulted (when the trees are rooted) in the placement of the spumaviruses (here shown within group I) and MLVs (within group III) as the sister taxon to a (group II) lineage containing the other five retroviral genera (lentiviruses, HTLV-BLV group, ALVs, and type B and D retroviruses). Our phylogenetic analyses are consistent with this view of retroviral phylogeny but highlight the uncertainty of the interrelationships of the group I spumaviruslike sequences and the root of the retrovirus tree.
Despite screening a large number of lower vertebrate taxa, no endogenous retroviral sequences were identified which clustered with the lentiviruses (equine infectious anemia virus [EIAV], ovine maedi-visna virus [OMVV], and simian immunodeficiency virus SIVmac), the HTLV-BLV group, or the ALVs (Rous sarcoma virus [RSV]). Indeed, the only sequences which were placed with other group II viruses were basal to the murine type B (mouse mammary tumor virus [MMTV]) and mammalian type D (jaagsiekte and simian retrovirus type 1 [SRV1]) retroviruses, being placed in a polytomy with HERV. K10, RSV, lymphoproliferative disease virus (LDV), and intracisternal type-A retroviral particle (IAP-hamster). The absence of isolates derived from nonmammalian or avian hosts suggests that endogenous retroviruses related to these five genera are likely to be rare in lower vertebrate genomes. We think it unlikely that this distribution is simply the result of target sequences being missed during amplification for several reasons. (i) The EM primer was designed against a sequence motif conserved in each of the five genera, (ii) negative results using EM were obtained with more than 30 species of reptiles, amphibians, and fish, (iii) the EM primer gave positive results with mammalian and avian taxa, and (iv) the JO and CT primers have previously cross-amplified group II-related viruses from mammalian genomes (37) but failed to do so when used against lower vertebrate taxa. The distribution of the group II sequences in mammals and birds is interesting, particularly given their apparent absence from reptilian relatives of birds. This suggests that their distribution may involve either dispersal across significant taxonomic distances or possibly the extinction of some taxa.
The spumaviruses (human spumavirus [HSV], two simian foamy viruses [SFV and SFVL3], and FeSFV) are exogenous viruses of primates and other mammals (4). There have been previous reports of two endogenous retroviruses which cluster with this genus, SpeV from the reptile tuatara (39) and HERV.L, a recently identified human isolate (5). It has been suggested that HERV.L represents a possible ancestor of the exogenous mammalian spumaviruses (5). Our phylogenetic analyses suggest that viruses distantly related to the spumaviruses may be widespread in vertebrates: unrooted trees (Fig. 3a) showed strong support for a group containing the spumaviruses and novel isolates derived from birds, reptiles, amphibians, and fish. However, the spumaviruses are unlikely to have emerged from HERV.L or a closely related endogenous virus, because this element appears more closely related to several viruses derived from nonmammalian hosts. The piscine member of the spumaviruslike lineage, SnRV, is an exogenous retrovirus which was originally isolated from striped snakehead fish (10). It has previously been reported to group (albeit distantly) with the MLVs (13), but the inclusion of additional retroviral isolates (which were not available to previous workers) suggests this may not be correct. It is our opinion, however, that none of the viruses described here should be assigned to the spumavirus genus. Although a spumaviruslike group was well supported by bootstrap analysis in unrooted phylogenies, this was not the case when several gypsy LTR retrotransposon sequences were included for rooting purposes (Fig. 3b). Furthermore, the monophyly of this group within individual rooted neighbor-joining and maximum parsimony trees (Fig. 3c and d) was outgroup dependent; varying the composition of the outgroup taxa sometimes resulted in the placement of the spumaviruslike viruses into two or three separate groups. These factors suggest that the spumavirus genus is only distantly related to the other group I viruses described here. Consistent with this, SnRV, HERV.L, and the spumaviruses are known to have significant differences in genomic organization when compared to each other and to other retroviral genera (5, 13). We were also unable to detect obvious sequence homology between the accessory proteins of SnRV and those encoded by the spumaviruses (data not shown).
The vast majority of endogenous lower vertebrate viruses characterized during this study (group III) clustered with the MLV-related genus. MLV-like viruses are present in numerous mammalian species and have also been described in birds (such as the spleen necrosis virus [SNV] and reticuloendotheliosis virus [REV] of domestic fowl) and several reptiles, including the corn snake and Russell’s viper (3, 19, 29, 31, 48). Several other groups of endogenous viruses (such as the HERV.I-related viruses and ERV-9 [also present within humans]) cluster with these viruses (17, 21, 44). Thus, members of this retroviral genus are already known to be present within several vertebrate classes. However, the phylogenetic trees shown in Fig. 3 demonstrate that the lineage containing the MLV genus is extremely complex. A polytomy, consisting of 13 lineages (one of which contained MLV/ERV-9/HERV.I-related viruses), was left unresolved by bootstrap analyses. Several of these lineages contained a single retroviral sequence, such as the previously described WDSV of fish (15), whereas others contained groups with several members. We believe that many of these lineages are likely to represent novel retroviral genera. Although other factors, in addition to sequence divergence (such as the presence of novel genes and alternative primer binding site homology), are required for genus-level classification (4), these may well become apparent when the complete nucleotide sequences of these viruses are determined. For example, it has already been suggested that WDSV represents a novel retroviral genus because it encodes two accessory proteins which are absent from members of the MLV genus and (unusually) utilizes a primer binding site homologous to tRNAHis (15, 23).
Although most of the retroviruses clustering in this region of the tree (excluding those present within the MLV genus) were derived from reptiles, amphibians, and fish, it was apparent that two novel mammalian viruses were also present. We originally isolated the first of these (RV-horse) when screening mammalian genomes but were unable at the time to determine its phylogenetic relationship to other retroviruses. The second element, H-cosmid(U85A3), was identified from a BLAST search of the EMBL/GenBank/DDBJ data banks and was originally sequenced as part of the human genome project (25). Our sequence analysis of this element suggests it contains a full-length polymerase gene, part of the envelope region, and that the gag gene has probably been replaced by a later insertion (in the reverse orientation) of an HERV-H-like element (unpublished results). We also identified a second element (by BLAST searches), closely related to H-cosmid(U85A3) on the X chromosome within cosmid HS49L23 (12). These results imply that other highly divergent endogenous retroviruses remain to be discovered in both humans and other mammals.
Finally, the distribution and phylogeny of the sequences shown in Fig. 3 (especially those within group III) is intriguing as it suggests that some of the retroviral groups may be restricted to particular vertebrate classes, indicating that interclass horizontal transmission may be a rare event for certain types of retrovirus. Although interpretation of these trees should be cautious (due to the lack of robust support for many of the relationships), the overall tree topologies are consistent with this possibility. For example, the majority of endogenous piscine retroviruses appear to be monophyletic, even though their hosts are classified into several different taxonomic orders. This also appears to be the case for many of the reptilian and amphibian retroviruses. Tests using the program MacClade also demonstrated that the host class-virus associations shown in Fig. 4 are phylogenetically correlated. We are currently trying to obtain a better idea of interclass transmission rates by looking in detail at the phylogeny and distribution of the MLV genus.
ACKNOWLEDGMENTS
We are indebted to D. Swofford for permission to publish results using PAUP4. We thank M. Clark, A. Fergusson, A. Flavell, J. Gatesy, P. Holland, R. Kusmierski, D. R. Martin, N. Okada, G. Olbricht, J. Pino-Perez, and R. Waugh-O’Neill for providing some of the genomic DNA samples used in this study.
E.H. thanks Université Paris XI for a DEA scholarship. M.T. and J.C. are supported by the NERC Taxonomy Initiative and the Royal Society. This work was supported in part by an NERC grant (GST/02/852) to M.W.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D L. Basic alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Bénit L, de Parseval N, Casella J F, Callebaut I, Cordonnier A, Heidmann T. Cloning of a new murine endogenous retrovirus, MuERV- L, with strong similarity to the human HERV-L element and with a gag coding sequence closely related to the Fv1 restriction gene. J Virol. 1997;71:5652–5657. doi: 10.1128/jvi.71.7.5652-5657.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen Y C, Cui Z, Lee L F, Witter R L. Serological differences among non-defective reticuloendotheliosis viruses. Arch Virol. 1987;93:233–245. doi: 10.1007/BF01310977. [DOI] [PubMed] [Google Scholar]
- 4.Coffin J M. Structure and classification of retroviruses. In: Levy J A, editor. The Retroviridae. Vol. 1. New York, N.Y: Plenum Press; 1992. pp. 19–49. [Google Scholar]
- 5.Cordonnier A, Casella J-P, Heidmann T. Isolation of novel human endogenous retrovirus-like elements with foamy virus-related pol sequence. J Virol. 1995;69:5890–5897. doi: 10.1128/jvi.69.9.5890-5897.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doolittle R F, Feng D-F, Johnson M S, McClure M A. Origins and evolutionary relationships of retroviruses. Quart Rev Biol. 1989;64:1–30. doi: 10.1086/416128. [DOI] [PubMed] [Google Scholar]
- 7.Doolittle R F, Feng D-F, McClure M A, Johnson M S. Retrovirus phylogeny and evolution. Curr Top Microbiol Immunol. 1990;157:1–18. doi: 10.1007/978-3-642-75218-6_1. [DOI] [PubMed] [Google Scholar]
- 8.Eickbush T H. Origin and evolutionary relationships of retroelements. In: Morse S S, editor. The evolutionary biology of viruses. New York, N.Y: Raven Press; 1994. pp. 121–157. [Google Scholar]
- 9.Francki, R. I. B., C. M. Fauquet, D. L. Knudson, and F. Brown (ed.). 1991. Classification and nomenclature of viruses. Fifth report of the International Committee on Taxonomy of Viruses. Arch. Virol. 1991(Suppl. 2): 290–298.
- 10.Frerichs G N, Morgan D, Hart D, Skerrow C, Roberts R J, Onions D E. Spontaneously productive C-type retrovirus infection of fish cell lines. J Gen Virol. 1991;72:2537–2539. doi: 10.1099/0022-1317-72-10-2537. [DOI] [PubMed] [Google Scholar]
- 11.Gak E, Yanev A, Sherman L, Ianconescu M, Tronick S R, Gazit A. Lymphoproliferative disease virus of turkeys: sequence analysis and transcriptional activity of the long terminal repeat. Gene. 1991;99:157–162. doi: 10.1016/0378-1119(91)90122-r. [DOI] [PubMed] [Google Scholar]
- 12.Grafham, D. Unpublished data.
- 13.Hart D, Frerichs N, Rambaut A, Onions D E. Complete nucleotide sequence and transcriptional analysis of the snakehead fish retrovirus. J Virol. 1996;70:3606–3616. doi: 10.1128/jvi.70.6.3606-3616.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Helps, C. R., and D. A. Harbour. Unpublished data.
- 15.Holzschu D L, Martineau D, Fodor S K, Vogt V M, Bowser P R, Casey J W. Nucleotide sequence and protein analysis of a complex piscine retrovirus, walleye dermal sarcoma virus. J Virol. 1995;69:5320–5331. doi: 10.1128/jvi.69.9.5320-5331.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim A, Terzian C, Santamaria P, Pelisson A, Prud’homme N, Bucheton A. Retroviruses in invertebrates: the gypsy retrotransposon is apparently an infectious retrovirus of Drosophila melanogaster. Proc Natl Acad Sci USA. 1994;91:1265–1269. doi: 10.1073/pnas.91.4.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.La Mantia G, Maglione D, Pengue G, Di Cristofano A, Simeone A, Lanfrancone L, Lania L. Identification and characterization of novel human endogenous retroviral sequences preferentially expressed in undifferentiated embryonal carcinoma cells. Nucleic Acids Res. 1991;19:1513–1520. doi: 10.1093/nar/19.7.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lankenau D H, Huijser P, Jansen E, Miedema K, Hennig W. DNA sequence comparison of micropia transposable elements from Drosophila hydei and Drosophila melanogaster. Chromosoma. 1990;99:111–117. doi: 10.1007/BF01735326. [DOI] [PubMed] [Google Scholar]
- 19.Lunger P D, Hardy W D, Clark H F. C type virus particles in a reptilian tumor. J Natl Cancer Inst. 1974;52:1231–1235. doi: 10.1093/jnci/52.4.1231. [DOI] [PubMed] [Google Scholar]
- 20.Maddison W P, Maddison D R. MacClade: analysis of phylogeny and character evolution, version 3.0. Sunderland, Mass: Sinauer Associates; 1992. [Google Scholar]
- 21.Maeda N. Nucleotide sequence of the haptoglobin and haptoglobin-related gene pair. J Biol Chem. 1985;260:6698–6709. [PubMed] [Google Scholar]
- 22.Martin J, Herniou E, Cook J, Waugh O’Neill R, Tristem M. Human endogenous retrovirus type I-related viruses have an apparently widespread distribution within vertebrates. J Virol. 1997;71:437–443. doi: 10.1128/jvi.71.1.437-443.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martineau D, Bowser P R, Renshaw R R, Casey J W. Molecular characterization of a unique retrovirus associated with a fish tumor. J Virol. 1992;66:596–599. doi: 10.1128/jvi.66.1.596-599.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Masahito P, Nishioka M, Ueda H, Kato Y, Yamazaki I, Nomura K, Sugano H, Kitagawa T. Frequent development of pancreatic carcinomas in the Rana nigromaculata group. Cancer Res. 1995;55:3781–3784. [PubMed] [Google Scholar]
- 25.Odell, C. Unpublished data.
- 26.Ono M, Yasunaga T, Miyata T, Ushikubo H. Nucleotide sequence of human endogenous retrovirus genome related to the mouse mammary tumor virus genome. J Virol. 1986;60:589–598. doi: 10.1128/jvi.60.2.589-598.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oprandy J J, Chang P W. 5-Bromodeoxyuridine induction of hematopoietic neoplasia and retrovirus activation in the soft-shelled clam, Mya arenaria. J Invertebr Pathol. 1983;42:196–206. doi: 10.1016/0022-2011(83)90062-9. [DOI] [PubMed] [Google Scholar]
- 28.Oprandy J J, Chang P W, Pronovost A D, Cooper K R, Brown R S, Yates V J. Isolation of a viral agent causing hematopoietic neoplasia in the soft-shelled clam, Mya arenaria. J Invertebr Pathol. 1981;38:45–51. [Google Scholar]
- 29.Payne L N. Biology of avian retroviruses. In: Levy J A, editor. The Retroviridae. Vol. 1. New York, N.Y: Plenum Press; 1992. pp. 299–404. [Google Scholar]
- 30.Poulet F M, Bowser P R, Casey J W. Retroviruses of fish, reptiles and molluscs. In: Levy J A, editor. The Retroviridae. Vol. 3. New York, N.Y: Plenum Press; 1994. pp. 1–38. [Google Scholar]
- 31.Purchase H G, Ludford C, Nazerian K, Cox H W. A new group of oncogenic viruses: reticuloendotheliosis viruses, chick syncytial, duck infectious anemia, and spleen necrosis viruses. J Natl Cancer Inst. 1973;51:489–499. [PubMed] [Google Scholar]
- 32.Sambrook J, Fritsch E, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. New York, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 33.Schwartz D E, Tizard R, Gilbert W. Nucleotide sequence of Rous sarcoma virus. Cell. 1983;32:853–869. doi: 10.1016/0092-8674(83)90071-5. [DOI] [PubMed] [Google Scholar]
- 34.Shih A, Misra R, Rush M G. Detection of multiple, novel reverse transcriptase coding sequences in human nucleic acids: relation to primate retroviruses. J Virol. 1989;63:64–75. doi: 10.1128/jvi.63.1.64-75.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stephens R M, Rice N R, Hiebsch R R, Bose H R, Gilden R V. Nucleotide sequence of v-rel—the oncogene of reticuloendotheliosis virus. Proc Natl Acad Sci USA. 1983;80:6229–6233. doi: 10.1073/pnas.80.20.6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Temin H M. Retrovirus variation and evolution. Genome. 1989;31:17–22. doi: 10.1139/g89-007. [DOI] [PubMed] [Google Scholar]
- 37.Tristem M. Amplification of divergent retroelements by PCR. Biotechniques. 1996;20:608–612. doi: 10.2144/19962004608. [DOI] [PubMed] [Google Scholar]
- 38.Tristem M, Herniou E, Summers K, Cook J. Three retroviral sequences in amphibians are distinct from those in mammals and birds. J Virol. 1996;70:4864–4870. doi: 10.1128/jvi.70.7.4864-4870.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tristem M, Myles T, Hill F. A highly divergent retroviral sequence in the tuatara (Sphenodon) Virology. 1995;210:206–211. doi: 10.1006/viro.1995.1333. [DOI] [PubMed] [Google Scholar]
- 40.Varmus H, Brown P. Retroviruses. In: Berg D E, Howe M M, editors. Mobile DNA. Washington, D.C: American Society for Microbiology; 1989. pp. 53–108. [Google Scholar]
- 41.Wain-Hobson S. Is antigenic variation of HIV important for AIDS, and what might be expected in the future. In: Morse S S, editor. The evolutionary biology of viruses. New York, N.Y: Raven Press; 1994. pp. 185–209. [Google Scholar]
- 42.Weaver T A, Talbot K J, Panganiban A T. Spleen necrosis virus Gag polyprotein is necessary for particle assembly and release but not for proteolytic processing. J Virol. 1990;64:2642–2652. doi: 10.1128/jvi.64.6.2642-2652.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wichman H A, Van den Bussche R A. In search of retrotransposons—exploring the potential of the PCR. Biotechniques. 1992;13:258–264. [PubMed] [Google Scholar]
- 44.Wilkinson D A, Mager D L, Leong J-A C. Endogenous human retroviruses. In: Levy J A, editor. The Retroviridae. Vol. 3. New York, N.Y: Plenum Press; 1994. pp. 465–535. [Google Scholar]
- 45.Xiong Y, Eickbush T H. Origin and evolution of retroelements based upon their reverse transcriptase sequences. EMBO J. 1990;9:3353–3362. doi: 10.1002/j.1460-2075.1990.tb07536.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.York D F, Vigne R, Verwoerd D W, Querat G. Nucleotide sequence of the jaagsiekte retrovirus, an exogenous and endogenous type D and B retrovirus of sheep and goats. J Virol. 1992;66:4930–4939. doi: 10.1128/jvi.66.8.4930-4939.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Young A Z. The life of vertebrates. 3rd ed. Oxford, United Kingdom: Clarendon Press; 1981. [Google Scholar]
- 48.Ziegel R F, Clark H F. Electron microscopic observations on a “C”-type virus in cell cultures derived from a tumor bearing viper. J Natl Cancer Inst. 1969;43:1097–1101. [PubMed] [Google Scholar]