Abstract
CCCTC-binding factor (CTCF) binding sites are hotspots of genome instability. Although many factors have been associated with CTCF binding site fragility, no study has integrated all fragility-related factors to understand the mechanism(s) of how they work together. Using an unbiased, genome-wide approach, we found that DNA double-strand breaks (DSBs) are enriched at strong, but not weak, CTCF binding sites in five human cell types. Energetically favorable alternative DNA secondary structures underlie strong CTCF binding sites. These structures coincided with the location of topoisomerase II (TOP2) cleavage complex, suggesting that DNA secondary structure acts as a recognition sequence for TOP2 binding and cleavage at CTCF binding sites. Furthermore, CTCF knockdown significantly increased DSBs at strong CTCF binding sites and at CTCF sites that are located at topologically associated domain (TAD) boundaries. TAD boundary-associated CTCF sites that lost CTCF upon knockdown displayed increased DSBs when compared to the gained sites, and those lost sites are overrepresented with G-quadruplexes, suggesting that the structures act as boundary insulators in the absence of CTCF, and contribute to increased DSBs. These results model how alternative DNA secondary structures facilitate recruitment of TOP2 to CTCF binding sites, providing mechanistic insight into DNA fragility at CTCF binding sites.
Graphical Abstract
Graphical Abstract.
Introduction
Genomic DNA fragility, particularly at regions of the genome called common fragile sites, is associated with cancer risk (1,2). Importantly, DNA fragility contributes to chromosomal abnormality formation in cells, and we have previously shown that over half of the translocation breakpoints in at least one gene of gene pairs resided within a fragile site (3). Fragile sites have also been associated with focal deletions and amplifications of oncogenes (4–6), further demonstrating that breakage within these regions has a role in promoting the formation of cancer-associated chromosomal rearrangements and abnormalities. Recent bioinformatic approaches have begun to identify and characterize the features associated with the fragile site location across the genome. Histone modifications, CCCTC-binding factor (CTCF) binding sites and DNA flexibility were positively associated with common fragile sites (7,8). Exploring these features in depth could provide further insight into the mechanisms contributing to genomic instability, thus enabling better prediction of DNA fragility and identification of individuals most susceptible to cancer-promoting mutations. Alternative DNA secondary structures (9) and topoisomerase II (TOP2) (9–12) were also characterized as mediators of DNA fragility. These results indicate that particular genomic features and proteins are associated with DNA double-strand break (DSB) incidence. These findings are consistent with our previous work in which fragile sites have the potential to form more energetically favorable alternative DNA secondary structures (13,14). Additionally, we and others have shown that these alternative DNA secondary structures can act as a recognition signal for TOP2, facilitating TOP2 binding and eventual cleavage (9,15–20).
CTCF acts as the anchoring protein that demarcates boundaries between topologically associated domains (TADs) (21,22), which are generated through cohesin-mediated loop extrusion (23–26). TADs are regions wherein DNA within a TAD is more likely and frequently in contact with DNA in the same TAD (27) and regulate gene expression, replication timing and DNA repair domains (28–31). CTCF binding sites are enriched for DSBs (9–12,32). However, there are over 600 000 CTCF binding sites in the human genome, which are differentially used and bound with varying strength and frequency depending on cell type (33); yet, the mechanisms driving cell-type-specific CTCF usage are still not completely understood. Two studies have indicated that TOP2B and CTCF/cohesin binding sites overlap with each other (34,35), suggesting that these proteins may function together to contribute to DNA fragility at these sites. The overlap of TOP2B and CTCF/cohesin binding sites also indicates that TAD boundaries are involved with this fragility as they are sites of high topological stress. The topological stress of cohesin-mediated loop extrusion at TAD boundaries is in part regulated by transcription-coupled supercoiling (36). Interestingly, a core feature of the common fragile sites in the genome is that they are in large, expressed, late-replicating genes that span TAD boundaries (37). No studies have examined how CTCF binding, alternative DNA secondary structure, TOP2 and TAD boundaries function in tandem to mediate DSBs.
Here, we examined how these key features participate in CTCF-mediated DNA fragility, to better understand the role of CTCF in genomic instability. We found that strong and weak CTCF binding sites exhibit different DNA fragility, with more DSBs at strong sites. Analysis of strong CTCF binding sites revealed a potential to form more stable alternative DNA secondary structures, enrichment of TOP2 cleavage complexes (TOP2ccs) and reduction of TOP2ccs upon TOP2B knockout, when compared to weak CTCF binding sites. Knockdown of CTCF protein in nonmalignant MCF10A cells revealed that strong CTCF binding sites are preferentially sensitive to DSBs after CTCF knockdown. Since strong CTCF binding sites are enriched for TAD boundary-associated CTCF binding sites, we evaluated DSBs at altered CTCF binding sites that were either TAD boundary or loop associated. This uncovered that all TAD boundary-associated CTCF binding sites increased DSBs in response to CTCF knockdown regardless of how binding may have changed (lost, gained or unchanged), while loop-associated CTCF binding sites have no significant changes in DSBs. We then assessed secondary structure formation at altered CTCF binding sites and found TAD boundary-associated CTCF binding sites that recused binding had significantly more stable secondary structures than either gained or unchanged CTCF binding sites. Furthermore, these TAD boundary-associated CTCF lost sites were enriched for G-quadruplex (G4) structures, and had more DSBs upon CTCF knockdown, supporting a model of these structures acting as insulating factors when CTCF is limited, and contributing to the increased fragility at TAD boundary-associated lost sites. Overall, TAD boundary association drives the fragility patterns at CTCF binding sites and co-occurs with stronger CTCF binding sites, more stable alternative DNA secondary structures and greater TOP2B binding. These factors along with the topological stress at TAD boundaries dictate DNA fragility of these regions.
Materials and methods
Cell culture
Jurkat cells (GenScript) were grown in RPMI 1640 medium (Gibco), supplemented with 10% fetal bovine serum (FBS, Gibco 16000044). MCF10A cells (ATCC) were grown in MEGM media (Lonza, CC-3150) without FBS and treated with etoposide (Sigma, E1383) for 24 h. Jurkat and MCF10A cells were verified using the short tandem repeat profiling and showed 100% and 98% match, respectively, to the ATCC database profile. Both cell lines were tested negative for mycoplasma by employing the LookOut Mycoplasma qPCR Detection Kit (Sigma).
shRNA construction, transduction and expression
Short hairpin RNA (shRNA) targeting sequences from the RNAi Consortium (38) were cloned into tet-pLKO.1 puro (39) (Addgene #21915): shCTCF (TRCN0000218498), shLuc (40) (Addgene #136587). Briefly, targeting sequences from the RNAi Consortium were modified to change the XhoI restriction site in the shRNA loop to a PstI site. Oligonucleotides were annealed at 95°C in annealing buffer [10 mM Tris–HCl, 100 mM NaCl and 1 mM ethylenediaminetetraacetic acid (EDTA)] for 5 min on a thermocycler and cooled slowly to room temperature. Annealed primers were phosphorylated in vitro with T4 polynucleotide kinase (NEB) and then cloned into tet-pLKO.1 puro that had been digested with EcoRI and AgeI. All constructs were confirmed by Sanger sequencing. shCTCF designed sequence: CCGGTATGATTTCCCATCGACATTTCTGCAGAAATGTCGATGGGAAA TCATATTTTTG.
Production of viral supernatant and transduction of MCF10A cells were conducted using previously published protocols (41). To induce shRNA expression, shLuc and shCTCF MCF10A cells were treated with 1 μg/ml of doxycycline (Sigma, cat. D9891) for 48 h, and processed for DNA break mapping, western blotting and RNA extraction.
Western blot
Cells were lysed in an ice-cold RIPA buffer [50 mM Tris–HCl, pH 8.0, 150 mM NaCl,1% NP-40, 0.1% sodium dodecyl sulfate (SDS), 1× protease inhibitors (cOmplete mini EDTA-free, Roche) and 1× phosphatase inhibitors (PhosStop, Roche)] and protein concentrations of the lysates were determined by Pierce Micro BCA assay (cat. 22225). Equal total protein amount was loaded into stacking and resolving SDS–polyacrylamide gel electrophoresis (PAGE) gel. Using a wet transfer system, proteins were transferred to polyvinylidene fluoride membranes. Membranes were blocked with 5% non-fat milk in 1× TBST buffer (20 mM Tris, pH 7.5, 150 mM NaCl, 0.05% Tween 20). Primary antibodies were incubated at 4°C overnight [CTCF (BD, 1:1000, cat. 612149), GAPDH (Cell Signaling, 1:10 000, cat. 5174), p53 (Santa Cruz, 1:500, cat. sc-126) and γH2AX (Ser139) (Abcam, 1:5000, cat. ab11174)]. Membranes were washed and then incubated with horseradish peroxidase (HRP)-conjugated secondary antibodies, anti-mouse (Bio-Rad, 1:500, cat. 170-5047) and anti-rabbit (Bio-Rad, 1:500, cat. 170-5046), respectively. Pierce™ ECL (cat. 32109) kit was used for HRP substrate detection. Images were captured by Bio-Rad ChemiDoc Imaging System and signal intensity was quantified by Bio-Rad Image Lab Software.
MTS proliferation assay
MCF10A cells were analyzed using CellTiter 96® AQueous One Solution Cell Proliferation Assay (Promega, cat. G3580) following the manufacturer’s protocol.
Reverse transcription and real-time RT-PCR
Total RNA from MCF10A cells was obtained by organic extraction using TRIzol reagent according to the manufacturer’s protocol (Life Technologies, cat. 15596018). Reverse transcription reaction was performed following the manufacturer’s protocol (Invitrogen, cat. 18091050). Bio-Rad CFX384 Touch Real-Time PCR Detection System was used for real-time polymerase chain reaction (PCR). To avoid genomic DNA contamination, all primers were designed to span across exon junctions. CTCF forward sequence: ACCAGTGGAGAATTGGTTCG; CTCF reverse sequence: GTGTCCCTGCTGGCATAACT. GAPDH forward sequence: ACATCGCTCAGACACCATG; GAPDH reverse sequence: TGTAGTTGAGGTCAATGAAGGG. The messenger RNA (mRNA) expression level of CTCF was normalized to the GAPDH.
RNA sequencing
Total RNA from Jurkat cells was extracted as described above, enriched for poly-A fragments, followed by fragmentation, reverse transcription and second strand complementary DNA (cDNA) synthesis. Double-stranded cDNA was purified with AMPure XP beads and ends were repaired and then ligated with Illumina sequencing adapters followed by PCR amplification (Novogene, Inc.). Libraries were then subjected to 150-bp paired-end sequencing with the Illumina NovaSeq 6000.
Genome-wide break mapping and sequencing
Detection of DSBs using either purified genomic DNA or fixed nuclei was performed as described (9,42,43). Briefly, fixed nuclei were subjected to blunting/A-tailing reactions and Illumina P5 adapter ligation to capture DSB ends. Genomic DNA was then purified and fragmented by sonication and subsequently ligated to the Illumina P7 adapter, and the libraries were PCR amplified. To map DSBs from purified genomic DNA, the genomic DNA was subjected to blunting/A-tailing reactions and Illumina P5 adapter ligation to capture DSB ends. The excess adapter was removed and then DNA was fragmented by sonication, and subsequently ligated to Illumina P7 adapter, and the libraries were PCR amplified. Prepared libraries were then subjected to whole-genome, 75- and 150-bp paired-end sequencing with the Illumina NextSeq 500 and HiSeq X Ten platforms, respectively.
Processing of DSB reads
Sequencing reads were aligned to the human genome (GRCh38/hg38) with bowtie2 (v. 2.3.4.1) aligner running in high-sensitivity mode (--very-sensitive). Restriction on the fragment length from 100 to 2000 nt (-X 2000 -I 100 options) was imposed. Unmapped, nonprimary, supplementary and low-quality reads were filtered out with SAMtools (v. 1.7) (-F 2820). Further, PCR duplicates were marked with picard-tools (v. 1.95) MarkDuplicates, and finally, the first mate of nonduplicated pairs (-f 67 -F 1024) was filtered with SAMtools for downstream analysis. For each detected break, the most 5′ nucleotide of the first mate defined the DNA break position. Sequencing and alignment statistics for the DSB mapping/sequencing libraries prepared from MCF10A and Jurkat cells are listed in Supplementary Tables S1 and S2, respectively. Biological repeats of each sample that showed very high reproducibility of genomic coverage (Pearson’s correlation r = 0.803–0.942 for MCF10A, r = 0.926–0.989 for Jurkat, P ∼ 0, Supplementary Figure S1) were combined for downstream data analysis. This strong correlation confirms that the break mapping procedure does not introduce significant amounts of random DNA breaks that could convert single-stranded nicks into DSBs. The DSB data from GM13069 cells (42), neural progenitor cells (NPCs) (9) and HeLa cells (44) were previously published.
Processing RNA-seq and ChIP-seq data
High-throughput sequencing data used in this study were downloaded from Gene Expression Omnibus through Sequence Read Archive or from the ENCODE project (45); all datasets used in this study are listed in Supplementary Table S3. The publicly available RNA sequencing (RNA-seq) data from HeLa (46), GM12878 (47), MCF10A (48) and NPCs (49) were aligned to the GRCh38/hg38 genome using HISAT2 aligner (50), and the gene expression [fragments per kilobase million (FPKM) values] was quantified using StringTie (51). The RNA-seq data from Jurkat cells generated from this study were aligned to the same human genome assembly using STAR aligner (v. 2.7.9) with RefGene annotation, downloaded from the University of California, Santa Cruz (UCSC) browser, and transcript quantification was performed with RSEM (v. 1.3.0) (52,53).
The publicly available data for CTCF chromatin immunoprecipitation sequencing (ChIP-seq) from HeLa (45), GM12878 (45), MCF10A (54,55), Jurkat (56), NPCs (45) and RPE-1 (45), and each associated input control data, were downloaded and aligned to the GRCh38/hg38 genome using bowtie2 (v. 2.3.4.1). Binding peaks were called by the macs2 tool (v. 2.2.9.1) using the default setting with each dataset controlled for the matching input data (-c). Using BEDtools (v. 2.27.1) intersect between called CTCF ChIP-seq peaks and a list of determined genome-wide CTCF motifs (n = 887 981) (33), CTCF binding peaks with CTCF motifs were refined by excluding the binding sites that lack CTCF motifs (Supplementary Table S4). Peak strength as defined by macs2 score that reflects the significance of each called peak was used to group into bins for further analyses. Peak summits were used to center regions of interest for all analyses. The publicly available data for BG4 ChIP-seq from NHEK (57), HaCaT (58) and K562 (59), BG4 CUT&Tag from U2OS (60) and TOP2B ChIP-seq from MCF10A were processed similarly.
Single-nucleotide cumulative plots at CTCF binding sites
The strongest (top 10%) and weakest (bottom 10%) CTCF binding sites were determined based on macs2 score of CTCF ChIP-seq data: for all binding peaks, n = 6011, 4019, 6911, 9841 or 6820 each for MCF10A, GM12878, HeLa, NPCs and Jurkat, respectively; for binding peaks with CTCF motifs, n = 4878, 3529, 5386, 6912 or 5593 each for MCF10A, GM12878, HeLa, NPCs and Jurkat, respectively. DSB coverage at these regions was determined using BEDtools (v. 2.27.1) coverage reporting the depth at each position in the reference regions (-d), and then the merge function was used to compile each region’s coverage into a single line readable to python3. Using python3 (v. 3.6.5) with matplotlib (v. 2.2.2), numpy (v. 1.15.0) and pandas (v. 0.23.3), the cumulative single-nucleotide break profiles were plotted over the relative nucleotide position to the CTCF ChIP-seq peak summit and in the ±500 bp flanking regions with read normalization (reads per million, RPM), and boxplots were plotted over the relative nucleotide position to the CTCF ChIP-seq peak summit and in the ±150 bp flanking regions with read normalization (RPM). Statistical tests were performed using python3 (v. 3.6.5) with scipy stats (v. 0.19.1).
Differential binding analysis of CTCF ChIP-seq data
CTCF ChIP-seq data from MCF10A CTCF wild-type (WT) and knockdown (KD) cells (55) were downloaded and processed as described earlier. Binding peaks with CTCF motifs were used for further analysis to identify differentially binding regions using DiffBind 3.0 with the same parameters as described in Lebeau et al. (55). Briefly, bam and narrowPeak files for each sample and bam files of the associated input were used. Normalization and analysis of CTCF binding peaks was carried out using the parameters as follows: normalize = DBA_NORM_DEFAULT, library = DBA_LIBSIZE_PEAKREADS, background = False, bREtrieve = False. The threshold for significance was set at false discovery rate (FDR) ≤ 0.01 and abs(log2FC) ≥ 1 in all conditions. Differential peak sets conserved between replicates and conditions were used for downstream analysis. Heatmaps of the differential peaks identified by DiffBind were generated using deepTools (61).
ChIP-qPCR assay
ChIP-qPCR procedures were followed as described in Khoury et al. (62) with minor modifications as follows. MCF10A shLuc and shCTCF cells were harvested at 80–90% confluence. Chromatin was measured by the BCA assay, and 600 μg chromatin was used for each ChIP reaction, and 2% of the chromatin was aliquoted as an input. Chromatin pre-cleared with 30 μl of Salmon Sperm DNA/Protein G Agarose–50% Slurry (#16-201, Millipore) was incubated with CTCF antibody (Active Motif, cat. 91286) or no antibody at 4°C overnight. ChIP-DNA was purified and suspended in 50 μl solution of 10 mM Tris–HCl (pH 8.0) and 0.1 mM EDTA. The Bio-Rad CFX384 Touch Real-Time PCR Detection System was used for qPCR reactions. Primers used in qPCR reactions are listed in Supplementary Figure S8A. A two-step ΔΔCT equation was used for quantification. The value was then normalized to a 2% chromatin input for each sample and further normalized against the no antibody control to calculate the fold enrichment.
Identifying consensus G4-forming regions
To generate G4 consensus sites, publicly available BG4 ChIP-seq data from NHEK, K562 and HaCaT cells and CUT&Tag data from U2O2 cells were downloaded through Sequence Read Archive and aligned to the human genome assembly GRCh38/hg38 using bowite2. Biological replicates were merged. Low-quality, secondary and supplementary alignments, and unmapped reads were filtered out. Peaks were called by macs2 tool (v. 2.2.9.1) individually for each cell line controlled by input or IgG control (-c). Using BEDtools (v. 2.27.1), merged union list of BG4 binding peaks was created (n = 57 673) and used for intersection and reporting number of peaks that were present at the same genomic location in at least three out of four cell lines; these sites were extracted and defined as G4 consensus sites (n = 8250). To generate G4 random shuffle control, BEDtools (v. 2.27.1) was used to shuffle with -chrom option to randomly choose genomic location and keep features of G4 consensus sites on the same chromosome.
Analysis of CC-seq data
The CC-seq data from Gittens et al.(63) were downloaded and aligned to the human genome (GRCh38/hg38) following the same processing as break data (as detailed in the ‘Processing of DSB reads’ section). The matched sets of etoposide-treated WT and TOP2B−/− RPE-1 cells in both asynchronous and G1 arrested cells had replicates merged, respectively, and the coverage from each was calculated in the preferential DNA break sites defined above.
Genomic region definitions
The GRCh38/hg38 build RefSeq genes were downloaded from the UCSC browser. The definitions used for each genomic feature are as follows: promoter region ranging from transcription start site −1000 to −250 nt, transcription start site region ranging from transcription start site −250 to +250 nt, gene body region ranging from transcription start site +250 nt to transcription termination site −250 nt and transcription termination site regions ranging from transcription termination site −250 to +250 nt.
DNA secondary structure analysis
We previously described how to use DNA secondary structure calculation programs to examine the energetic potential for secondary structure formation across the human genome (9,64). Here, we applied the same analysis [ViennaRNA programs with parameters for analyzing DNA (65)] to sequences of regions of interest in the human genome assembly GRCh38/hg38. Energetic potential calculations were performed with a window of 30 nt and a step of 1 nt.
Mutation depletion score analysis
To determine the level of mutation constraint on the consensus G4-forming regions, we used the recently published UK Biobank depletion rank score (66). To calculate the score, Halldorsson et al. (66) tabulated the number of UK Biobank variants in each 500-bp window of the genome. Then, this number was compared to an expected number, given the heptamer nucleotide composition of the window, and the fraction of heptamers with a sequence variant across the genome and their mutational classes. The depletion score was assigned from 0 (most depletion) to 1 (least depletion) to each 500-bp window with a 50-bp step size and showed that the windows with low depletion scores were enriched for cis-regulatory regions and variants identified in genome-wide association studies. We identified all 500-bp windows scored by Halldorsson et al. that had an overlap of over 250 bp with the consensus G4 regions.
Statistical analysis
Statistical analysis was carried out using scipy stats (v. 0.19.1) and R (v. 4.2.0). Tests are specified in figure legends, and statistical significance is denoted by asterisks: *P < 0.05, **P < 0.01, ***P < 0.001 and ****P ∼ 0, unless stated otherwise.
Results
DNA DSBs and stable alternative DNA secondary structures are signatures of strong CTCF binding sites
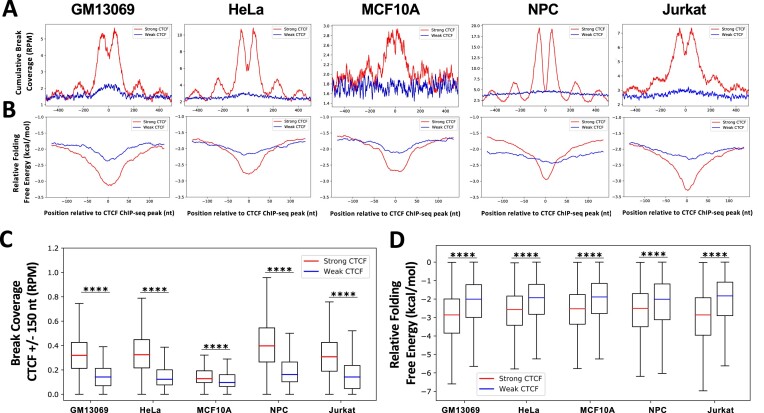
CTCF binding strength has been correlated with sequence conservation across mammalian species (67). To examine the effect of CTCF binding strength on the degree of DNA breakage, genome-wide DSBs were sequenced/mapped with single-nucleotide resolution in five untreated cell lines: GM13069 (nonmalignant lymphoblastoid), HeLa (malignant cervical epithelial), MCF10A (nonmalignant breast epithelial), NPCs (from apparently healthy individual) and Jurkat (malignant, T lymphocytes) (9,44). DNA breaks were assessed for each cell line at all CTCF binding sites, grouped into 10 equal size bins based on CTCF protein binding strength derived from ChIP-seq signal (45,54,56). Mapped DSBs in five cell lines are enriched at stronger CTCF binding sites and DSB coverage medians decrease with decreasing CTCF binding strength (Supplementary Figure S2A). Significantly more DSBs were observed at the strongest (top 10%) CTCF binding sites in each cell line than at the weakest (bottom 10%) CTCF binding sites (P ∼ 0, two-sample, Kolmogorov–Smirnov test) (Supplementary Figure S2B and D). Also, a periodic DSB signal flanking the strong CTCF binding sites was observed, consistent with our previous reports (9,43), which results from the strong nucleosome positioning capability of CTCF (68). Next, we refined the CTCF binding sites by only including the binding peaks that contain CTCF binding motifs (Supplementary Table S4), and again used 10 equally sized bins to define the top 10% strongest and the 10% weakest CTCF binding sites. Consistent with our earlier observation, the strong CTCF binding sites are significantly enriched with DSBs when compared to the weakest CTCF binding sites (P ∼ 0, two-sample, Kolmogorov–Smirnov test) (Figure 1A and C).
Figure 1.
DSBs and highly stable alternative DNA secondary structures are significantly enriched at strong CTCF binding sites in multiple cell types. (A) DSBs were mapped in three untreated nonmalignant (MCF10A, GM13039 and NPC) and two untreated cancer cell lines (HeLa and Jurkat). CTCF binding strength was defined based on ChIP-seq signal for each cell line, and binding sites with CTCF motifs were identified and used here. DSBs are enriched at the top 10% strongest CTCF binding sites (strong, red), but not at the 10% weakest CTCF binding sites (weak, blue) in untreated GM13069 (n = 3529), HeLa (n = 5386), MCF10A (n = 4878), NPC (n = 6912) and Jurkat (n = 5593) cells, as demonstrated by cumulative DSB coverage (RPM) (±500 nt) at these sites. (B) DNA sequences around strong CTCF binding sites (red) (±150 nt) form more energetically favorable DNA secondary structures (ΔG, kcal/mol) than sequences around weak sites (blue), as determined by folding predictions of single-stranded DNA using ViennaRNA with DNA thermodynamic parameters and a 30-nt sliding window with a 1-nt step; a low ΔG (kcal/mol) indicates that sequences are more favorable to form the structure. (C) Read-normalized DSB coverage (RPM) was significantly greater at the strong (red) versus the weak (blue) CTCF binding peak sites (±150 nt) for each cell line. (D) Relative folding free energy was significantly more stable at the strong (red) versus the weak (blue) CTCF binding peaks (±150 nt) for each cell line. Boxes denote 25th and 75th percentiles, middle lines show medians and whiskers span from 5% to 95%; ****P ∼ 0, two-sample, Kolmogorov–Smirnov test.
To analyze all CTCF binding sites across five cell lines, we compared DSBs across the union set of CTCF sites and found that median normalized DSB coverage decreased with diminishing CTCF binding strength across all lines (Supplementary Figure S3A). Furthermore, we evaluated how common the strongest CTCF binding sites were between lines and found that of the 6884 strongest CTCF binding sites in each cell line, 2100 were common to all five. When the common strong CTCF binding sites were compared to the strong sites not shared in each line, we found that the shared sites were significantly more enriched in DSBs in each cell line (P < 0.001, Kruskal–Wallis, post-hoc Dunn test) (Supplementary Figure S3B). Together, this demonstrates that DSB enrichment is a common signature at strong CTCF binding sites and the fragility of strong CTCF binding sites is driven by features intrinsic to these sites.
We previously demonstrated that sequences that can form highly stable DNA secondary structures are enriched at CTCF binding sites, and secondary structure-containing CTCF binding sites accumulate significantly more DSBs than CTCF binding sites that do not contain alternative DNA secondary structures (9). To examine whether there is a difference in the potential to form DNA secondary structures at strong and weak CTCF binding sites, the relative folding free energy (ΔG, kcal/mol) at these sites was calculated using ViennaRNA with DNA thermodynamic parameters to determine the extent of DNA secondary structure formation potential from single-stranded DNA in five cell lines. We found that DNA at and around strong CTCF binding sites (peak summit ± 75 nt) had the potential to form more energetically favorable DNA secondary structures than DNA at weak CTCF binding sites in five cell types (P ∼ 0, two-sample, Kolmogorov–Smirnov test) (for CTCF binding sites with CTCF motifs only, see Figure 1B and D; for all CTCF binding sites, see Supplementary Figure S2C and E). This suggests that alternative DNA secondary structure stability is one intrinsic feature of strong CTCF binding that mediates fragility.
Because CTCF is also classified as a transcription factor (69,70), and transcription start sites are known to accumulate DSBs dependent on the expression level (9,10,12,44), we assessed whether the presence of transcription start sites correlates with CTCF binding strength, which could explain the difference in DSB accumulation observed. Interestingly, there was no preferential distribution of strong CTCF binding sites at transcription start sites, or genic regions in general, compared to weak CTCF binding sites for the five cell lines (Supplementary Figure S4), but we observed a trend of weak CTCF binding sites having increased abundance in genic regions. To determine whether gene expression at genic CTCF binding sites underlies the difference in fragility, we evaluated the gene expression of all genic CTCF binding sites by grouping them based on their CTCF binding strength and found that the strong CTCF binding sites did not have higher expression levels than the weak CTCF binding sites (Supplementary Figure S5A). Furthermore, when breaks were assessed across each decile of CTCF binding site strength in all cell lines, there was no significant difference between DSBs of the top strongest bins based on genic or intergenic status (Supplementary Figure S5B). Interestingly, among the weaker CTCF binding sites there were increased levels of DNA breaks in the genic CTCF sites compared to the intergenic CTCF sites. However, these levels of DNA breaks were still lower than DSBs at the strongest CTCF binding sites. Altogether, this suggests that while transcription plays a role in the breaks at weaker CTCF binding sites, the level of expression does not explain the differences in fragility between the strong and weak CTCF binding sites; instead, alternative DNA secondary structure stability is likely to underlie this difference.
DNA breaks at strong CTCF binding sites are caused by TOP2 cleavage
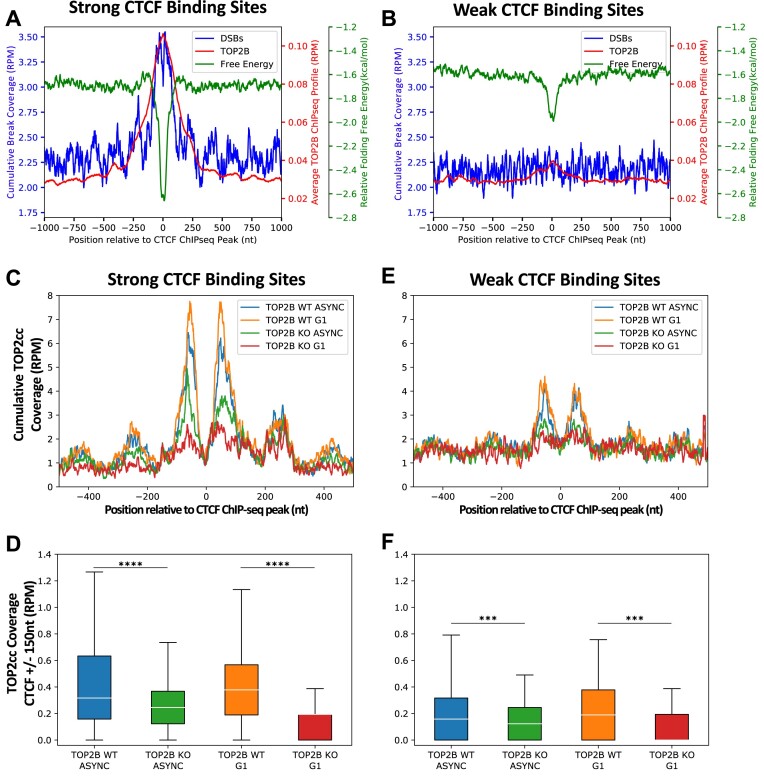
Alternative DNA secondary structures can arise from single-stranded DNA when duplex DNA is unwound such as in the presence of negative supercoiling, and these alternative DNA secondary structures can be recognized by TOP2, enhancing TOP2 binding and eventual cleavage (9,15–20). Furthermore, TOP2B has been shown to be located at CTCF binding sites (34,35) and CTCF binding sites display increased DNA breakage following exposure to the TOP2 poison etoposide (9–12). To further characterize the difference between strong and weak CTCF binding sites, we evaluated TOP2B binding at the strongest and weakest CTCF binding sites in MCF10A cells and found that TOP2B bound significantly more at the strong CTCF binding site (±150 nt, P ∼ 0, two-sample, Kolmogorov–Smirnov test), which matches with significantly more stable alternative DNA secondary structure formation and higher DSB frequency, compared to the weak sites (Figure 2A and B, P ∼ 0, two-sample, Kolmogorov–Smirnov test). This suggested that differential targeting of CTCF binding sites by TOP2B could underlie the pattern of differential DSB accumulation. Furthermore, we have shown that DSBs significantly increase in an etoposide dose-dependent manner at the strongest CTCF binding sites in GM13069 cells, but not at weak CTCF binding sites (9). Here, a significant dose-dependent increase of DSBs after exposure to etoposide was also observed in MCF10A and HeLa cells at strong CTCF binding sites (Supplementary Figure S6) (P< 0.001, Kolmogorov–Smirnov test). Etoposide traps TOP2ccs, preventing religation of the DNA strands (71); therefore, these results further suggest that TOP2 is significantly more present at strong CTCF binding sites and could contribute to increased DNA breakage at these sites.
Figure 2.
TOP2 preferentially binds and cleaves DNA at strong CTCF binding sites. (A) Cumulative DSBs from MCF10A (RPM), average TOP2B binding from MCF10A (RPM) and relative free energy of alternative DNA secondary structure formation (kcal/mol) were assessed at the strong CTCF binding sites in MCF10A (n = 6011) showing an overlap of TOP2B binding, DSB accumulation and energetically favorable structures at these CTCF binding sites. (B) Cumulative DSBs from MCF10A (RPM), average TOP2B binding from MCF10A (RPM) and relative free energy of alternative DNA secondary structure formation (kcal/mol) were assessed at the weak CTCF binding sites in MCF10A (n = 6011) showing a low degree of overlap between TOP2B binding secondary structure formation at these CTCF binding sites and low DSB accumulation. (C) Cumulative TOP2cc coverage (RPM) was calculated at single-nucleotide positions of the top 10% strong CTCF binding sites (n = 1973; ±500 nt) for asynchronous WT (blue), asynchronous TOP2B knockout (green), G1 WT (orange) and G1 TOP2B knockout (red) RPE-1 cells. (D) Quantification of TOP2cc coverage at strong CTCF binding sites (n = 1973; ±150 nt) demonstrates that TOP2B knockout, in asynchronous and G1 cells, significantly reduced TOP2cc accumulation. (E) Cumulative TOP2cc coverage (RPM) was calculated at single-nucleotide positions of the weak (bottom 10%) CTCF binding sites (n = 1973; ±500 nt) for asynchronous WT (blue), asynchronous TOP2B knockout (green), G1 WT (orange) and G1 TOP2B knockout (red) RPE-1 cells. (F) Quantification of TOP2cc coverage at weak CTCF binding sites (n = 1973; ±150 nt). Boxes denote 25th and 75th percentiles, middle lines show medians and whiskers span from 5% to 95%; ***P< 0.001 and ****P ∼ 0, Wilcoxon rank sum test.
While etoposide treatment demonstrates TOP2-mediated fragility, we next examined the direct involvement of TOP2 activity at CTCF binding sites. Using the CC-seq from Gittens et al. (63), a direct measurement of TOP2 activity by mapping TOP2ccs, we evaluated TOP2 at CTCF binding sites from RPE-1 cells (45). We found that the strongest CTCF binding sites were sensitive to TOP2 activity, as indicated by the significant reduction of TOP2cc coverage observed in the TOP2B knockout condition when compared to WT cells in both G1 and asynchronous states (P ∼ 0, Wilcoxon rank sum test) (Figure 2C and D). TOP2B is the predominant TOP2 isoform present during G1, whereas asynchronous cells can utilize both TOP2A and TOP2B (72). Therefore, the significant reduction of TOP2ccs observed in TOP2B knockout G1 cells at strong CTCF binding sites indicates that TOP2B directly contributes to DNA breaks observed at these sites (Figure 2C and D). When TOP2cc coverage was similarly analyzed at weak CTCF binding sites (Figure 2E and F), we found that while there was a significant reduction of TOP2ccs after TOP2B knockout for both asynchronous and G1 phase cells (P< 0.001, Wilcoxon rank sum test), TOP2ccs were not enriched to the same extent at these sites as compared to the strong CTCF binding sites. Altogether, this demonstrates that TOP2B preferentially binds and cuts at the strong CTCF binding sites compared to the weak CTCF binding sites, leading to the differential breakage at strong CTCF binding sites.
Loss of CTCF increases DSBs at strong CTCF binding sites
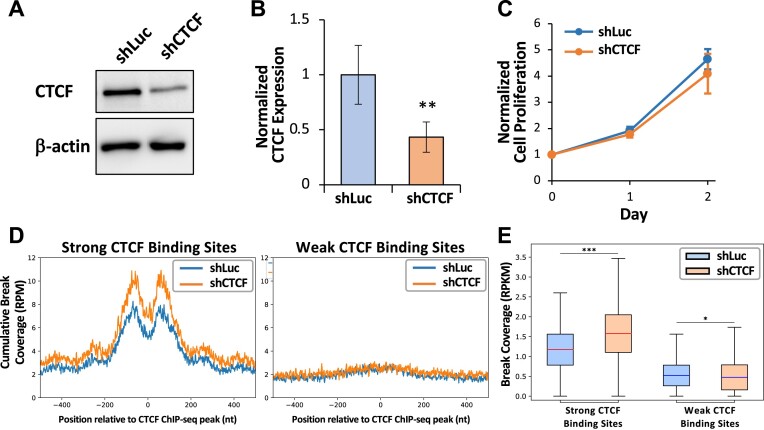
We have identified TOP2 cleavage as a source of increased fragility at strong CTCF binding sites and that etoposide treatment further increases DSBs at strong CTCF binding sites. Next, we assessed the CTCF protein level in MCF10A cells treated with etoposide and found that there is a dose-dependent decrease in CTCF protein (P < 0.05, UT to 0.15 μM and 0.15 to 15 μM, Student’s t-test), while classical markers of DNA damage, γH2AX and p53, showed expected increases (Supplementary Figure S7A and B). We confirmed this decreased CTCF expression by RT-qPCR that also exhibited a dose-dependent decrease in CTCF mRNA level (P < 0.01, UT to 0.15 μM, P < 0.05, 0.15 to 1.5 μM, P < 0.01, 0.15 to 15 μM, Student’s t-test) (Supplementary Figure S7C). This demonstrates that increased DSBs at strong CTCF binding sites are co-incident with the decreased overall expression of CTCF. To determine whether the loss of CTCF directly influences the formation of DSBs at strong CTCF binding sites, we knocked down CTCF protein in MCF10A cells using lentivirus to introduce either a control (shLuc) or inducible shRNA construct against CTCF (shCTCF). Following selection for stable cell lines from bulk populations, we confirmed CTCF was significantly reduced by 60% in the shCTCF population following induction with doxycycline for 48 h (P< 0.001, Student’s t-test) (Figure 3A and B), and this inducible knockdown did not change cell proliferation (Figure 3C). Following DSB mapping, we found that knockdown of CTCF led to a significant increase in DNA breaks at strong, but not weak, CTCF binding sites (P< 0.001, Wilcoxon rank sum test) (Figure 3D and E). This is consistent with the etoposide-induced DNA breaks with a dose-dependent increase at strong CTCF binding sites (Supplementary Figure S6) and implicates the loss of CTCF promoting increased fragility at strong CTCF binding sites. However, this seems unexpected based on the observations in Figure 1 and Supplementary Figures S2 and S3, where DSB enrichment was present at strong CTCF binding sites.
Figure 3.
CTCF knockdown significantly increases DSB enrichment at the strong CTCF binding sites in MCF10A cells. (A) CTCF is knocked down in shCTCF MCF10A cells compared to shLuc (top, CTCF; bottom, β-actin loading control). (B) Knockdown of CTCF in MCF10A cells is significantly reduced by 60%, normalized to β-actin (**P< 0.01, Student’s t-test). (C) Knockdown of CTCF in MCF10A cells does not alter cell proliferation as measured by the MTS assay. (D) CTCF knockdown increased DSBs at strong CTCF binding sites (left, n = 6011) but not at weak CTCF binding sites (right, n = 6011), as demonstrated by cumulative read-normalized coverage (RPM) of DSBs mapped in MCF10A shLuc (blue) and shCTCF (orange) cells. (E) Quantification of DSB coverage at the 3862 CTCF binding sites (±150 nt) after eliminating sites at which both samples had zero DSB coverage among the 6011 strong CTCF binding sites. Bars indicate means and error bars show ±standard deviation. Boxes denote 25th and 75th percentiles, middle lines show medians and whiskers span from 5% to 95%; *P< 0.05 and ***P< 0.001, Wilcoxon rank sum test.
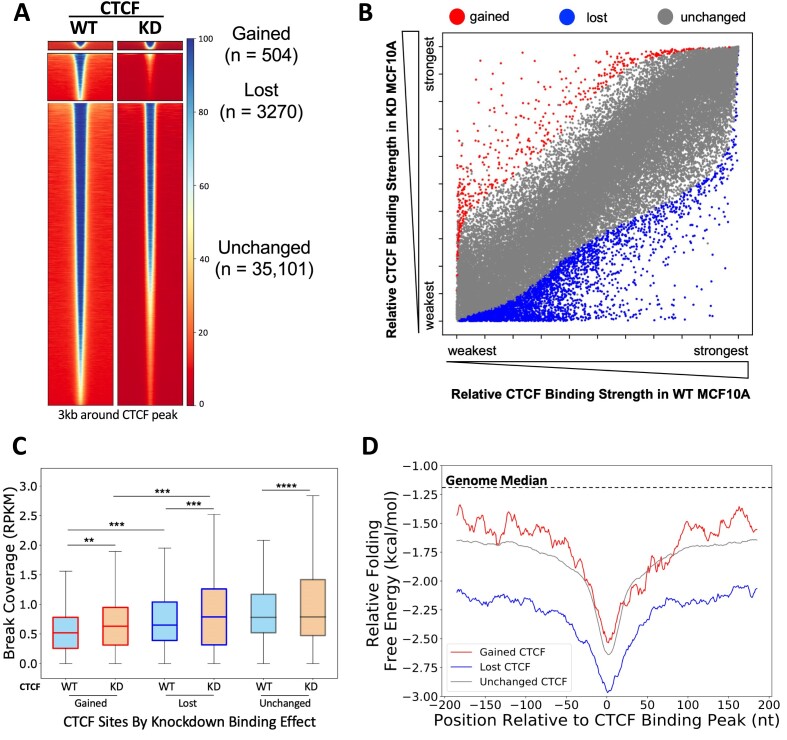
To understand the interplay between CTCF expression and binding, we analyzed the publicly available CTCF ChIP-seq data of CTCF knockdown in MCF10A from Lebeau et al. (55), in which they showed a 50–60% reduction in CTCF protein, and the overall CTCF binding of the cells remained largely unchanged, with a small number of lost CTCF binding sites, and even smaller number of gained CTCF binding sites (55). Reanalyzing their CTCF ChIP-seq data (alignment to the human GRCh38/hg38 build genome) and filtering out non-CTCF motif-containing binding sites, we found very similar results with a total of 38 875 CTCF peaks, of which 3270 were significantly lost (FDR < 0.01, log2FC < −1) and 504 CTCF peaks significantly gained (FDR < 0.01, log2FC > 1) in the CTCF knockdown cells, while the rest of CTCF binding peaks (n = 35 101) remained statistically unchanged (55) (Figure 4A). Six randomly selected sites that are defined as lost, gained and unchanged (two each) were validated by CTCF ChIP-qPCR in the shCTCF MCF10A cells and the control shLuc cells (Supplementary Figure S8). Additionally, we assessed how the relative binding strengths changed between WT and knockdown, and found that a larger number of CTCF binding sites altered their relative rankings but did not meet the stringent thresholds of significance (Figure 4B). This suggests that changes in the overall CTCF binding profile are more prominent than depicted by binding strength significance alone and could also contribute to the observed phenotypes.
Figure 4.
CTCF knockdown altered binding at CTCF binding sites and differential DSBs and DNA secondary structure folding potentials at gained, lost and unchanged CTCF binding sites in MCF10A cells. (A) Heatmaps show CTCF binding at significantly gained (n = 504), lost (n = 3270) and unchanged (n = 35 101) CTCF binding sites between WT and CTCF knockdown MCF10A cells. Binding peaks with CTCF motifs only were used for the analysis, and differentially binding sites were calculated and identified using DiffBind 3.0 (55). Heatmaps were generated using deepTools (61) at the summit ± 1.5 kb regions of the differential peaks identified by DiffBind. (B) Scatter plot of each CTCF binding site based on relative binding strength rank in WT and CTCF knockdown conditions (gained, red; lost, blue; unchanged, gray). (C) DSBs were mapped in shLuc and shCTCF MCF10A lines. DSBs are significantly increased at gained and lost CTCF binding sites following CTCF knockdown (reads per kilobase million, RPKM). (D) DNA sequences around lost (blue) CTCF binding sites form more energetically favorable alternative DNA secondary structures than unchanged (gray) or gained (red) CTCF binding sites. Boxes denote 25th and 75th percentiles, middle lines show medians and whiskers span from 5% to 95%; **P < 0.01, ***P< 0.001 and ****P ∼ 0; pairwise comparisons, Wilcoxon rank sum test; cross-group comparisons, Kolmogorov–Smirnov test.
We then examined DSBs at the gained, lost and unchanged CTCF binding sites in the shLuc and shCTCF MCF10A cells, and found that the level of DNA breaks was significantly increased upon CTCF knockdown in all groups (gained: P < 0.01, lost: P< 0.001, unchanged: P ∼ 0, Wilcoxon rank sum test) (Figure 4C). Interestingly, we observed that lost sites were more enriched in DSBs in WT cells compared to gained sites, and this remained true after CTCF knockdown (WT, P < 0.001; knockdown, P < 0.001, Kolmogorov–Smirnov test). This is intriguing, as we showed that the binding strength of CTCF correlates with DSBs (Figure 1 and Supplementary Figures S2 and S3), and we would have predicted that lost CTCF binding sites would show a reduction in DSBs since binding was decreasing. This suggests that there are other contributors to the fragility at the lost CTCF binding sites. Therefore, we investigated the alternative DNA secondary structure forming potential at these three CTCF binding site groups, as we have shown that alternative DNA structure forming potential contributes to increased fragility. We found that the lost CTCF binding sites were defined by more favorable DNA secondary structure formation than either the unchanged or gained sites (Figure 4D). More interestingly, the more favorable DNA secondary structure formation is both in the immediate CTCF binding summit and in the flanking sequence for the lost sites. This demonstrates that lost CTCF binding sites are defined by regions of highly stable DNA secondary structure. Furthermore, this suggests that the ability of these sites to form highly stable secondary structures could predispose these sites for decreased binding under CTCF-limiting conditions, while still maintaining TAD boundaries.
Alternative DNA secondary structures, G4s act as backup boundary elements upon CTCF loss and contribute to increased DSBs
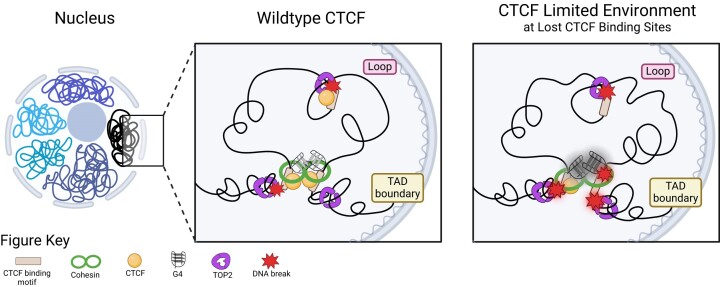
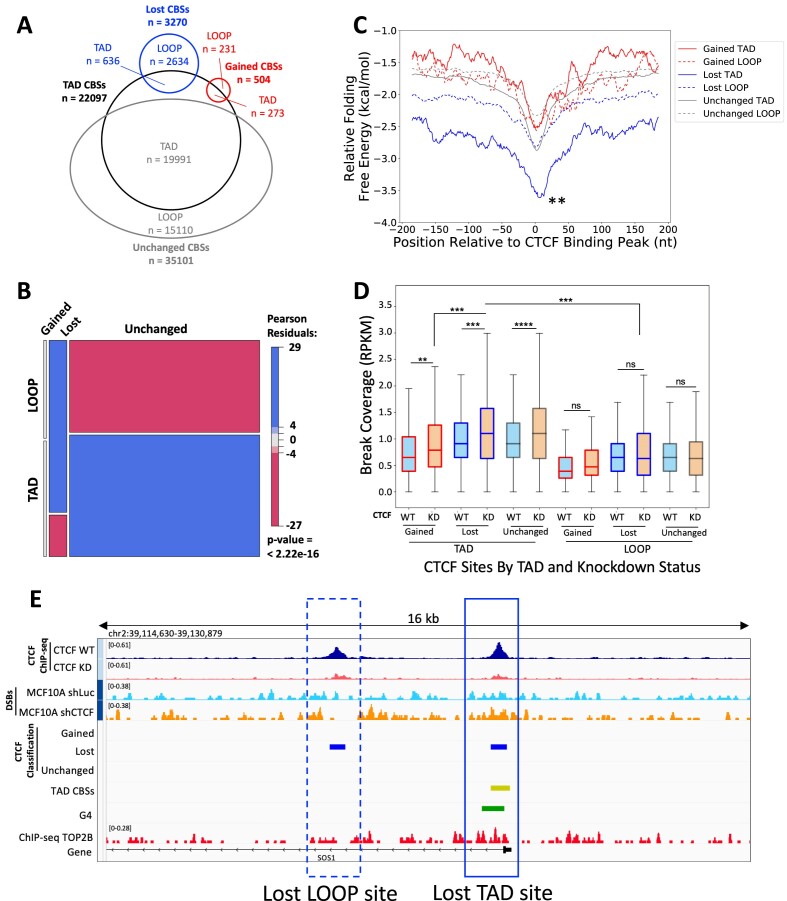
In support of our hypothesis that highly stable DNA secondary structure formation could predispose CTCF binding sites for CTCF loss but maintain TAD boundaries, Hou et al. showed G4s were enriched at TAD boundaries and CTCF-distal G4s were able to independently insulate chromatin (73). Indeed, Lebeau et al. found that the lost CTCF binding sites that occurred at TAD boundaries did not significantly alter insulation when compared to WT and CTCF knockdown cells (55). To determine whether the lost CTCF binding sites at TAD boundaries had a higher potential to form DNA secondary structure than those within TADs (loop-associated), we evaluated the lost, gained and unchanged CTCF binding sites, by intersecting these sites with constitutive CTCF binding sites (33), as these sites are associated with TAD boundaries across different cell types (74) (Figure 5A). Lost sites were significantly underrepresented for TAD boundaries, and unchanged sites were overrepresented among TAD boundaries (P < 2.22 × 10−16, chi-square test) (Figure 5B), suggesting overall protection or preservation of the TAD boundaries in the CTCF-limiting environment. We then examined the ability to form alternative DNA secondary structures at the TAD boundary versus loop-associated CTCF binding sites and found that the lost CTCF binding sites at TAD boundaries are predicted to form significantly more stable DNA secondary structures than either loop-associated lost CTCF sites or gained or unchanged TAD boundary sites (Figure 5C, P < 0.01, Kruskal–Wallis, post-hoc Dunn test). Overall, this supports the model in which alternative DNA secondary structures, such as G4s, maintain boundaries in the absence of CTCF.
Figure 5.
TAD boundary-associated CTCF binding sites are enriched for DSBs and increase fragility upon CTCF knockdown. (A) Venn diagram shows the division of gained, lost and unchanged CTCF binding sites (CBSs) among constitutive CTCF binding sites (n = 22 097). (B) Mosaic plot shows the underrepresentation of lost CTCF binding sites for TAD boundaries and being overrepresented in loop-associated sites. Meanwhile, unchanged CTCF binding sites are overrepresented among TAD boundaries and underrepresented among loop-associated sites (P = 2.22 × 10−16, chi-square test). (C) DNA sequences around lost TAD boundary-associated CTCF binding sites (solid blue) form the most energetically stable alternative DNA secondary structures among altered CTCF binding sites (±150 bp). **P < 0.01, Kruskal–Wallis, post-hoc Dunn test. (D) DSBs are enriched at all TAD boundary-associated CTCF binding sites and significantly increased upon CTCF knockdown. Loop-associated CTCF binding sites show changes in DSBs in the same direction as binding changes (RPKM). (E) A representative view of a lost TAD boundary-associated CTCF binding site and a lost loop-associated CTCF binding site near the gene SOS1 demonstrates the selective increase in DSBs at lost sites associated with TADs. CTCF ChIP-seq from CTCF WT and knockdown cells (dark blue and maroon, respectively), DSBs from CTCF shRNA control WT and CTCF knockdown (blue and orange, respectively), along with the CTCF classifications (gained; lost; unchanged), TAD CTCF binding sites (yellow), consensus G4 sites (green) and TOP2B binding (red). Boxes denote 25th and 75th percentiles, middle lines show medians and whiskers span from 5% to 95%; **P < 0.01, ***P< 0.001, ****P ∼ 0, and ns, not significant; pairwise comparisons, Wilcoxon rank sum test; cross-group comparisons, Kruskal–Wallis with post-hoc Dunn test.
Next, we investigated whether the classification as a TAD boundary- or loop-associated CTCF binding site influenced the DSB accumulation at these sites. We found that among all lost CTCF binding sites, those associated with TAD boundaries are more enriched for DSBs in WT, and upon CTCF knockdown when compared to lost sites located at loops (P < 0.001, Kruskal–Wallis, post-hoc Dunn test) (Figure 5D). More interestingly, all TAD boundary-associated sites significantly increased breaks upon CTCF knockdown (gained: P < 0.01, lost: P < 0.001, unchanged: P ∼ 0, Wilcoxon rank sum test); meanwhile, all loop-associated sites had no significant change in DSBs following CTCF knockdown (Figure 5D). The increased DSBs at the TAD boundary-associated sites were positively in agreement with the TOP2B and G4 abundance at these sites; both G4 and TOP2B are significantly enriched at TAD boundary-associated sites of all three types (lost, gained and unchanged) compared to the loop-associated sites (Supplementary Figure S9; P < 0.001, Kruskal–Wallis, post-hoc Dunn test). In Figure 5E, an example of neighboring lost TAD boundary- and loop-associated sites illustrates the differential DSBs at these two classes of lost CTCF sites, and G4 and TOP2B enrichments can be seen in the TAD boundary-associated lost sites.
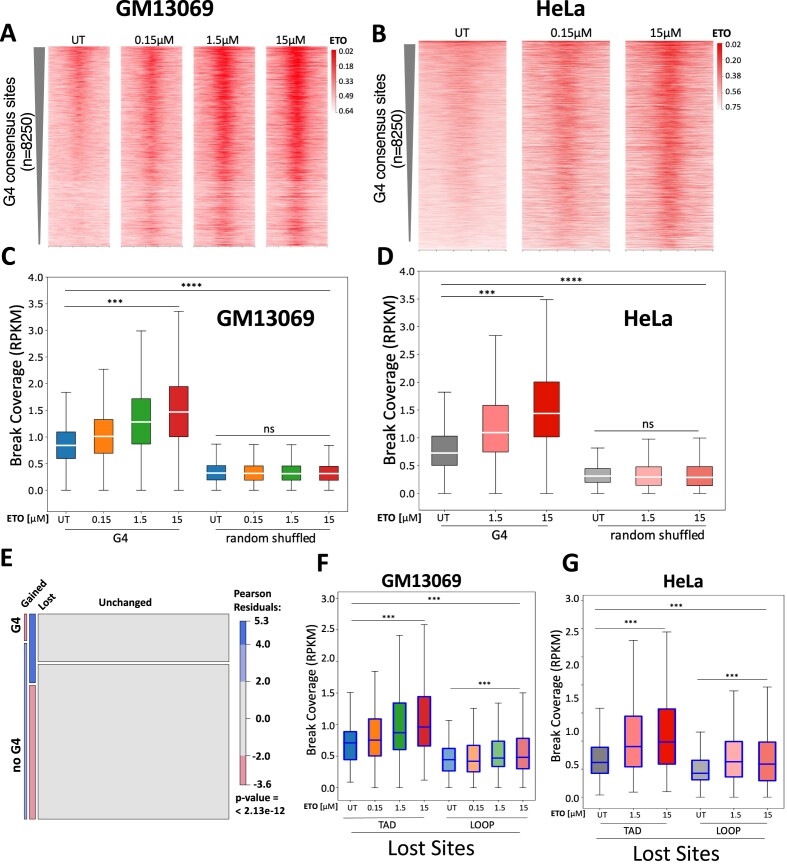
We then asked whether G4s can be recognized and cleaved by TOP2. We curated a consensus set of G4-forming regions (n = 8250) (see the ‘Materials and methods’ section), based on publicly available ChIP-seq/CUT&Tag data using BG4 (57–60), an antibody specific for G4 structure (75), and evaluated the presence of etoposide-induced DNA breaks. Almost all G4 consensus sites (94%) overlap with CTCF binding sites, and etoposide treatment revealed that there are significantly increased DSBs at G4 sites as compared to the untreated, and the intensity of DSBs significantly increases with increasing etoposide concentrations (Figure 6A and C for GM13069 and Figure 6B and D for HeLa, P < 0.001, Wilcoxon rank sum test). The same dose-dependent changes in the DSB level upon etoposide treatment were not observed at the randomly shuffled G4 sites (Figure 6C and D). More importantly, endogenous DSBs at the G4 consensus sites in untreated cells displayed an enrichment compared to shuffled sites (Figure 6C and D, P < 0.001, ns: not significant, Kruskal–Wallis, post-hoc Dunn test). In addition, heatmaps of DSB coverage over the G4 consensus sites (Figure 6A and B) also revealed that a snapshot of endogenous breaks was captured in untreated cells, and etoposide traps these breaks as the concentration increases. This notion is supported by the study of Hoa et al. (76) in which TOP2 was shown to frequently fail to religate endogenous, transiently cleaved products even without the presence of inhibitors, which can then be processed into persistent DSBs. Thus, our result indicates that TOP2-mediated DSBs are preferentially present at G4 structure regions and contribute to endogenous DSBs at these regions.
Figure 6.
TOP2-mediated DSBs preferentially present at G4 structure regions and at the lost TAD boundary-associated CTCF binding sites. Etoposide (ETO) treatment significantly increases DSB enrichment at G4 consensus regions in GM13069 (A) and HeLa cells (B). Boxplot illustration of DSB coverage at the consensus G4 regions in GM13069 (C) and HeLa cells (D). (E) Mosaic plot shows lost CTCF binding sites at TAD boundaries are overrepresented among shared G4 CTCF binding sites, while gained CTCF binding sites are underrepresented among G4 CTCF binding sites (P = 2.13 × 10−12, chi-square test). The lost CTCF binding sites at TAD boundaries, when compared to those at loops, are significantly more enriched with etoposide-induced DSBs in GM13069 (F) and HeLa cells (G), and maintain the strong DSB increase corresponding to the increased concentrations of etoposide. Boxes denote 25th and 75th percentiles, middle lines show medians and whiskers span from 5% to 95%; ***P < 0.001, ****P ∼ 0, and ns, not significant, Kruskal–Wallis followed by post-hoc Dunn test.
As shown in Figure 5C, the lost CTCF binding sites at TAD boundaries possess significantly more stable predicted secondary structures than lost loop sites. Also, among the TAD-associated CTCF binding sites, we found that the lost sites were overrepresented for overlap with G4s (P < 2.13 × 10−12, chi-square test) (Figure 6E). We then examined lost TAD boundary- and lost loop-associated CTCF binding sites for DNA breaks in response to etoposide treatment in both GM13069 and HeLa cells, different from MCF10A to assess the role of DNA secondary structure. Figure 6F and G reveals that lost sites at TAD boundaries, when compared to those at loops, were significantly more enriched with etoposide-induced DSBs (P < 0.001, Kruskal–Wallis, post-hoc Dunn test) and maintained the strong increase in breaks corresponding to the increased concentrations of etoposide. This result demonstrates that the lost TAD boundary-associated CTCF binding sites can be recognized by TOP2, generating endogenous and etoposide-induced DSBs.
G4 structures overlapping TAD boundary-associated CTCF binding sites are under high mutational constraint
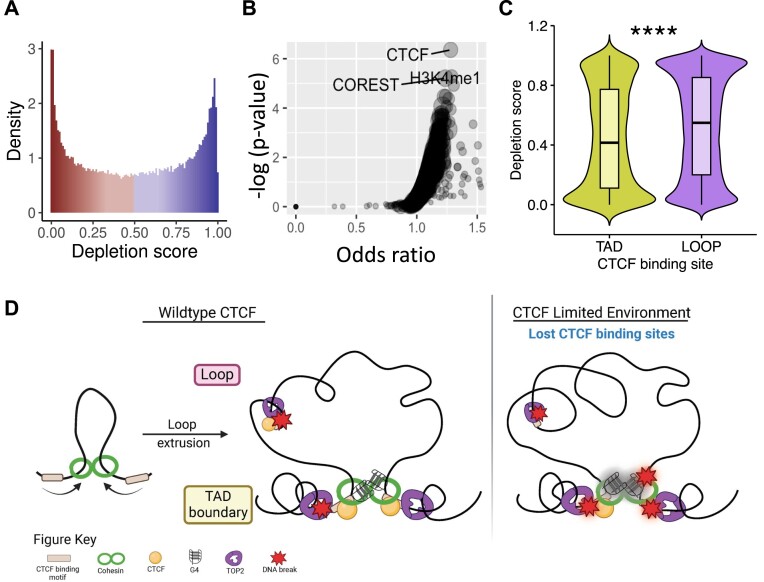
A recent study by Wulfridge et al. demonstrated that G4s enhance CTCF binding and stabilize TAD formation (77). To further determine whether there was a critical role for the G4 structure at TAD boundary-associated CTCF binding sites, we evaluated whether there was a selective pressure to preserve G4 sequences at CTCF binding sites. Across the genome, the rate of polymorphism is higher at non-B DNA loci, such as G4s, compared to other loci (78). However, if a subset of these structures is integral to regulating CTCF binding site dynamics with downstream consequences, we would expect those loci to be under purifying selection to combat this high mutability. We examined the consensus set of G4-forming regions (n = 8250) using the recently published UK Biobank depletion rank score by Halldorsson et al. (66), to determine the level of constraint on these G4 structures. We identified all 500-bp windows scored by Halldorsson et al. that had an overlap of over 250 bp with the consensus G4 regions. The distribution of the depletion scores (0, most depletion, to 1, least depletion) for the windows overlapping the G4 structures in our dataset demonstrates that not all G4 sequences are under the same mutational constraint (Figure 7A). Limiting the analysis to the G4 structures with the lowest depletion scores (depletion score <0.05), we compared the overlapping regions to ChIP-seq peaks for various transcription factors and histone marks sequenced by ENCODE. Using locus overlap analysis (79), we observed that these highly constrained regions overlapping G4 structures are significantly enriched for CTCF binding sites (P < 10−6, odds ratio = 1.28) (Figure 7B). We also found enrichment of H3K4me1 (P < 10−5, odds ratio = 1.29), which is often associated with enhancers, and CoREST (P < 10−5, odds ratio = 1.24), which has been identified as a subunit for several protein complexes in these regions. Importantly, depletion scores of G4 structures at TAD boundary-associated CTCF binding sites are significantly lower than the depletion scores of G4 structures at loop-associated CTCF binding sites (Figure 7C, P < 2.2 × 10−16, Wilcoxon rank sum test). These analyses indicate that G4 structures overlapping TAD boundary-associated CTCF binding sites are under high mutational constraint and might be functional, as previously suggested (78,80). TAD boundaries that are stable across cell types also show enriched heritability and evolutionary constraint (81), further supporting a role for a sequence/structure-dependent function at TAD boundaries.
Figure 7.
G4s have functional and conserved roles at TAD boundary-associated CTCF binding sites. (A) The mutational constraint on the observed G4 structures varies throughout the genome, as shown by the distribution of the depletion scores (0, most depleted, to 1, least depleted). (B) The most constrained G4s are enriched for CTCF binding along with H3K4me1 and CoREST. (C) The G4 structures with at least one overlapping TAD boundary-associated CTCF binding site are more constrained compared to structures that overlap with loop-associated CTCF binding sites (****P < 2.2 × 10−16, Wilcoxon rank sum test). (D) Model of DNA fragility at CTCF binding sites driven by TAD and loop associations, TOP2B activity, G4s and CTCF binding.
Overall, this suggests that G4s are conserved at CTCF binding sites, presumably to act as backup insulating factors, particularly at TAD boundary-associated CTCF binding sites. The strong structure formation at certain sites then allows for the sites to reduce CTCF occupancy in a CTCF-limiting environment. Furthermore, the topological stress at TAD boundaries creates an active region for local DNA melting to form alternative secondary structures driving a higher DSB environment from TOP2B activity. Then in a CTCF-limiting environment, the stress of global topological changes across all TAD boundary-associated sites leads to increased DSBs in CTCF knockdown at these sites. At all sites, the differences in alternative secondary structure strength drive the baseline DNA break levels at CTCF binding sites (Supplementary Figure S9). The new model described here is depicted in Figure 7D.
Further validating this model, we uncovered that in five cell lines, the stronger CTCF binding sites were enriched for TAD boundary-associated CTCF binding sites, while weak sites were depleted (Supplementary Figure S10A). We then examined DSBs in all cell lines across the stratified CTCF binding site bins, and found that DSBs were enriched in TAD boundary-associated CTCF binding sites compared to loop-associated sites across all decile strengths in five cell lines. This indicates that TAD boundary-associated CTCF binding sites were the primary drivers of the binding strength-mediated breaks, not CTCF binding per se (Supplementary Figure S10B). This supports the results from the CTCF knockdown, where DNA breaks at CTCF binding sites are shaped by the formation of strong alternative DNA secondary structure derived from topological stress at TAD boundaries, and are mediated by TOP2 activity.
Discussion
Here, we investigated factors underlying DNA fragility at CTCF binding sites and how this fragility acts within the broader context of CTCF’s role in 3D chromatin organization. We found that both alternative DNA secondary structures such as G4s and TOP2 cleavage contribute to the differential DNA fragility at CTCF binding sites. Using an unbiased, genome-wide DNA break mapping approach, we first determined that DSBs are preferentially enriched at the strong CTCF binding sites in multiple cell types, as compared to the weak binding sites. This led us to reveal strong CTCF binding sites dominated by TAD boundary-associated sites, and TAD boundary-associated sites contain significantly more G4s and TOP2 binding than loop-associated sites. Upon CTCF knockdown, TAD boundary-associated CTCF binding sites significantly increased DNA breaks, while loop-associated sites had no significant DSB change. The TAD boundary-associated lost sites that displayed significantly reduced CTCF binding upon CTCF knockdown are overrepresented with G4s and sufficiently maintained TAD boundaries even in the absence of CTCF.
CTCF has been classified as a tumor suppressor gene, as demonstrated by increased susceptibility to both hematological and solid cancers in Ctcf hemizygous mice. These Ctcf+/– tumors showed a more invasive and metastatic phenotype, leading to an overall worse prognosis/outcome in these mice (82). Analysis of human breast tumors in the Cancer Genome Atlas database showed that 60% of breast cancers possess CTCF copy number loss (83). In addition to deletions, mutations within CTCF either subsequently inactivating CTCF protein or decreasing CTCF gene expression have been demonstrated to be significantly associated with cancer progression and prognosis (84–86). Studies have begun to provide explanations for how reduced functional CTCF protein levels promote oncogenesis. Reduction of CTCF protein levels has direct effects on CpG and CTCF binding site methylation (82,83), resulting in gene expression changes of cancer progression-associated pathways such as stress response (i.e. hypoxia and hormones) and cell motility (83). Lebeau et al. showed that altered sub-TAD architectures in single allele knockout CTCF cells resulted in novel promoter–enhancer interaction leading to oncogenic activation (55). Similarly, mutations within CTCF binding sites are observed in many cancer types and can diminish CTCF binding to specific DNA sites (87–89). Cancers with these CTCF binding site mutations are associated with chromatin architecture and gene expression changes (33), and have also been shown to display chromosomal instability (89). Recent work by Lambuta et al. showed that whole genome doubling events caused a reduction of CTCF protein, and resulted in loss of proper TAD establishment and increased accumulation of copy number variants (90). CTCF levels in these cells are at 50% of WT cells, meaning it is only 25% of that normally required for the cellular DNA content. This would create an even more aberrant binding landscape. They also found CTCF binding sites with both lost and gained CTCF binding (90), which likely follows a similar reprioritization as we propose here. Though they suggest the loss of CTCF binding is stochastic across CTCF binding sites, our evidence suggests that it is driven by the DNA secondary structure forming ability of the CTCF binding sites. These highly stable DNA secondary structures being more persistent would in turn drive increased fragility at these sites and could explain their observation of recurrent copy number variants in the whole genome doubling model compared to the diploid control cells. Furthermore, DNA breaks at a putative TAD boundary-associated CTCF binding site within the KMT2A gene can lead to the generation of KMT2A rearrangements often found in acute myeloid leukemias and acute lymphoblastic leukemias (10,12). Gothe et al. identified that TOP2-mediated DNA breakage at CTCF binding sites is associated with rearrangement partners, particularly known in therapy-related acute myeloid leukemias, and that breakage and KMT2A rearrangement events are increased with etoposide treatment (12).
A strong presence of G4s at regulatory regions of the human genome has been well established (57,91–93). G4s can lead to the accumulation of DNA damage during DNA replication and transcription by acting as a barrier to impede these processes (94–96). We and others have shown that TOP2 and alternative DNA secondary structures are associated with DNA fragility (9,10,12,13,15), and that TOP2B-mediated DNA breaks are preferentially located at CTCF/cohesin binding sites. Therefore, TOP2, when failing to religate its cleaved products (76), can promote the generation of endogenous DSBs at CTCF/cohesin binding sites. TOP2B interacts with CTCF and cohesin, and is colocalized with CTCF and cohesin at TAD boundaries (34,35). TOP2B displayed an orientation-specific binding relative to the position of CTCF and cohesin, and the binding order positions TOP2B at the base/outside of the TAD loop and cohesin is located inside the loop (35). Moreover, cohesin, a ring structured complex, uses an ATP-dependent loop extrusion mechanism to guide TAD formation (23–26), and its narrow opening limits the rotation of chromatin (97,98). The strategic positioning of TOP2B at TAD boundaries is suggested to resolve topological stress and DNA entanglements caused by active DNA extrusion when chromatin is passed through the cohesin rings (97,98). Analysis of biotinylated 4,5,8-trimethylpsoralen (TMP) incorporation revealed TOP2-mediated negative supercoiling at CTCF/cohesin binding sites (35,99). In addition to DNA supercoiling in recruiting TOP2B to the TAD boundaries, we showed that TAD boundary sites are enriched with G4s, and G4s can be recognized and cleaved by TOP2. Recent studies have demonstrated that G4s have a direct role in the recruitment of CTCF to DNA (77) and that the G4 stabilizing ligands (pyridostatin and CX5461) generate DSBs through a TOP2 poisoning mechanism, like etoposide (100,101). Here, we found that the TAD boundary-associated CTCF binding sites had the potential to form highly stable DNA secondary structures such as G4s, leading to TOP2 cleavage, and contributing to the differential DNA fragility at these sites under an abundance of CTCF.
Upon reduction of CTCF protein, DSBs further increase significantly at the TAD boundary-associated CTCF binding sites, regardless of gained, lost or unchanged sites, although the increase is less in the gained sites, which correlated with significantly less G4s and TOP2 binding compared to the lost and the unchanged sites. In contrast, this significant increase was not observed in all three types of the loop-associated sites. Recent evidence points to CTCF’s direct role in DSB repair, specifically in promoting homologous recombination by directly interacting with CtIP, BRCA2 and Rad51, and recruiting them to the broken ends (102–104). Particularly, CtIP is required for the removal of TOP2cc by regulating the nuclease activity of MRE11 (105), and CTCF-depleted cells showed an increase in γH2AX foci (103) and displayed high sensitivity to etoposide treatment (102,103). Importantly, using γH2AX ChIP-qPCR, Lang et al. revealed that six genomic regions (co-mapped to the unchanged TAD boundary-associated sites in our study) have increased DNA breaks in CTCF knockdown cells (103). This increase in DNA breaks at unchanged TAD sites agrees with our observation. Interestingly, the CTCF-depleted cells were more sensitive to etoposide treatment than the CtIP-depleted cells, even though both have impaired homologous recombination and other DNA repair pathways in a comparable manner (102). These studies indicate that the limited pool of CTCF causes more TOP2cc damage, likely due to diminishing the general repair of TOP2cc and/or promoting the formation of TOP2cc at TAD boundary-associated CTCF binding sites. Furthermore, CTCF’s binding is highly dynamic with a residence time of 1–2 min in human cells, and the reduced level of CTCF can slow down the search time for the next binding sites (106), which would prolong cohesin extrusion and promote G4 formation and TOP2 binding.
G4s are enriched at TAD boundaries (73) and are shown to recruit CTCF to TAD-associated binding sites and reinforce TAD formation (107). In addition to our finding that the lost CTCF sites at TAD boundaries have increased DSBs and were overrepresented with G4s, Lebeau et al. (55) demonstrated that even with a 50–60% reduction in CTCF protein, these sites can maintain TAD domain insulation ability, suggesting that G4s can act as boundary stabilizers in the absence or limiting of CTCF. The polarity of CTCF binding is critical for the regulation of TAD organization and a convergent orientation of CTCF binding sites has been shown to serve as anchors for chromatin loops (22,27,108–110). Interestingly, Hou et al. showed the strand orientation of the G4s at CTCF binding sites strongly correlated with the orientation of the CTCF motifs, suggesting the involvement of G4s in establishing the orientation of CTCF binding and possibly enhancing CTCF function (73). Moreover, TAD boundaries with CTCF and G4s are stronger and more insulating than those without G4s, and modeling studies indicate that G4s, independent of CTCF binding, behave as strong insulators (73). These studies lend support to the observation that the lost sites at TAD boundaries, despite reduction in CTCF binding, preserve TAD insulation, and could be due to the occurrence of G4s to safeguard the TAD boundaries. However, it comes at the expense of increased DSB at these sites, possibly leading to an increased formation of chromosomal abnormalities.
We present a mechanistic model in which alternative DNA secondary structures, especially at TAD-associated CTCF binding sites, facilitate the recruitment of TOP2 and lead to increased DNA fragility (Figure 7D). Furthermore, our results support the model in which G4s act as backup boundary elements at CTCF boundary sites. Altogether, this offers a unique insight into the mechanisms for how reduced CTCF protein levels and/or alterations to CTCF binding sites contribute to the genomic instability and processes that facilitate the formation of oncogenic chromosomal abnormalities, thus providing a better understanding of cancer susceptibility.
Supplementary Material
Acknowledgements
We acknowledge the UVA Genome Analysis and Technology Core, RRID:SCR_018883 (supported by the NCI Cancer Center Support Grant 5P30CA044579), for sequencing on the Illumina NextSeq 500. We would also like to thank Sandeep Singh for the preliminary data analysis, Kyubin Lee for revision analysis and Kevin Janes for supplying the shLuc and shCTCF plasmid constructs.
Author contributions: H.M.R.Y., P.-C.H., N.D.A. and Y.-H.W. designed the study. P.-C.H., N.D.A., A.R.B. and L.W. performed experiments. H.M.R.Y., A.R.B., Z.W., C.Z. and A.R. performed data analysis. H.M.R.Y., A.R.B. and Y.-H.W. drafted the paper. All authors provided critical review and feedback on the final submission.
Contributor Information
Heather M Raimer Young, Department of Biochemistry and Molecular Genetics, University of Virginia School of Medicine, Charlottesville, VA 22908-0733, USA.
Pei-Chi Hou, Department of Biochemistry and Molecular Genetics, University of Virginia School of Medicine, Charlottesville, VA 22908-0733, USA.
Anna R Bartosik, Department of Biochemistry and Molecular Genetics, University of Virginia School of Medicine, Charlottesville, VA 22908-0733, USA.
Naomi D Atkin, Department of Biochemistry and Molecular Genetics, University of Virginia School of Medicine, Charlottesville, VA 22908-0733, USA.
Lixin Wang, Department of Biomedical Engineering, University of Virginia, Charlottesville, VA 22908, USA.
Zhenjia Wang, Center for Public Health Genomics, University of Virginia School of Medicine, Charlottesville, VA 22908-0717, USA.
Aakrosh Ratan, Department of Biochemistry and Molecular Genetics, University of Virginia School of Medicine, Charlottesville, VA 22908-0733, USA; Center for Public Health Genomics, University of Virginia School of Medicine, Charlottesville, VA 22908-0717, USA; Department of Public Health Sciences, University of Virginia School of Medicine, Charlottesville, VA 22908, USA; University of Virginia Comprehensive Cancer Center, Charlottesville, VA 22908, USA.
Chongzhi Zang, Department of Biochemistry and Molecular Genetics, University of Virginia School of Medicine, Charlottesville, VA 22908-0733, USA; Center for Public Health Genomics, University of Virginia School of Medicine, Charlottesville, VA 22908-0717, USA; Department of Public Health Sciences, University of Virginia School of Medicine, Charlottesville, VA 22908, USA; University of Virginia Comprehensive Cancer Center, Charlottesville, VA 22908, USA.
Yuh-Hwa Wang, Department of Biochemistry and Molecular Genetics, University of Virginia School of Medicine, Charlottesville, VA 22908-0733, USA; University of Virginia Comprehensive Cancer Center, Charlottesville, VA 22908, USA.
Data availability
DSB mapping/sequencing datasets and RNA-seq of Jurkat cells generated in this study can be accessed at Sequence Read Archive: PRJNA795482. The publicly available datasets used in this study are listed in Supplementary Table S3.
Supplementary data
Supplementary Data are available at NAR Online.
Funding
National Cancer Institute [T32CA009109 to N.D.A., R50CA265089 to L.W.]; National Institute of General Medical Sciences [T32GM008136 to H.M.R.Y., R35GM133712 to C.Z., RO1GM101192 to Y.-H.W.]; University of Virginia Farrow Fellowship to A.R.B.; Virginia Commonwealth Health Research Board [CHRB 207-12-20 to C.Z.]. Funding for open access charge: National Institute of General Medical Sciences [RO1GM101192].
Conflict of interest statement. None declared.
References
- 1. Dillon L.W., Burrow A.A., Wang Y.H. DNA instability at chromosomal fragile sites in cancer. Curr. Genomics. 2010; 11:326–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. O’Keefe L.V., Richards R.I. Common chromosomal fragile sites and cancer: focus on FRA16D. Cancer Lett. 2006; 232:37–47. [DOI] [PubMed] [Google Scholar]
- 3. Burrow A.A., Williams L.E., Pierce L.C., Wang Y.H. Over half of breakpoints in gene pairs involved in cancer-specific recurrent translocations are mapped to human chromosomal fragile sites. BMC Genomics. 2009; 10:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Glover T.W., Wilson T.E., Arlt M.F. Fragile sites in cancer: more than meets the eye. Nat. Rev. Cancer. 2017; 17:489–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ciullo M., Debily M.A., Rozier L., Autiero M., Billault A., Mayau V., El Marhomy S., Guardiola J., Bernheim A., Coullin P. et al. Initiation of the breakage–fusion–bridge mechanism through common fragile site activation in human breast cancer cells: the model of PIP gene duplication from a break at FRA7I. Hum. Mol. Genet. 2002; 11:2887–2894. [DOI] [PubMed] [Google Scholar]
- 6. Hellman A., Zlotorynski E., Scherer S.W., Cheung J., Vincent J.B., Smith D.I., Trakhtenbrot L., Kerem B. A role for common fragile site induction in amplification of human oncogenes. Cancer Cell. 2002; 1:89–97. [DOI] [PubMed] [Google Scholar]
- 7. Fungtammasan A., Walsh E., Chiaromonte F., Eckert K.A., Makova K.D. A genome-wide analysis of common fragile sites: what features determine chromosomal instability in the human genome?. Genome Res. 2012; 22:993–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Georgakilas A.G., Tsantoulis P., Kotsinas A., Michalopoulos I., Townsend P., Gorgoulis V.G. Are common fragile sites merely structural domains or highly organized “functional” units susceptible to oncogenic stress?. Cell. Mol. Life Sci. 2014; 71:4519–4544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Szlachta K., Manukyan A., Raimer H.M., Singh S., Salamon A., Guo W., Lobachev K.S., Wang Y.H. Topoisomerase II contributes to DNA secondary structure-mediated double-stranded breaks. Nucleic Acids Res. 2020; 48:6654–6671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Canela A., Maman Y., Huang S.N., Wutz G., Tang W., Zagnoli-Vieira G., Callen E., Wong N., Day A., Peters J.M. et al. Topoisomerase II-induced chromosome breakage and translocation is determined by chromosome architecture and transcriptional activity. Mol. Cell. 2019; 75:252–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Canela A., Maman Y., Jung S., Wong N., Callen E., Day A., Kieffer-Kwon K.R., Pekowska A., Zhang H., Rao S.S.P. et al. Genome organization drives chromosome fragility. Cell. 2017; 170:507–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gothe H.J., Bouwman B.A.M., Gusmao E.G., Piccinno R., Petrosino G., Sayols S., Drechsel O., Minneker V., Josipovic N., Mizi A. et al. Spatial chromosome folding and active transcription drive DNA fragility and formation of oncogenic MLL translocations. Mol. Cell. 2019; 75:267–283. [DOI] [PubMed] [Google Scholar]
- 13. Dillon L.W., Pierce L.C., Ng M.C., Wang Y.H. Role of DNA secondary structures in fragile site breakage along human chromosome 10. Hum. Mol. Genet. 2013; 22:1443–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Thys R.G., Lehman C.E., Pierce L.C., Wang Y.H. DNA secondary structure at chromosomal fragile sites in human disease. Curr. Genomics. 2015; 16:60–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dillon L.W., Pierce L.C., Lehman C.E., Nikiforov Y.E., Wang Y.H. DNA topoisomerases participate in fragility of the oncogene RET. PLoS One. 2013; 8:e75741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Froelich-Ammon S.J., Gale K.C., Osheroff N. Site-specific cleavage of a DNA hairpin by topoisomerase II. DNA secondary structure as a determinant of enzyme recognition/cleavage. J. Biol. Chem. 1994; 269:7719–7725. [PubMed] [Google Scholar]
- 17. Jonstrup A.T., Thomsen T., Wang Y., Knudsen B.R., Koch J., Andersen A.H. Hairpin structures formed by alpha satellite DNA of human centromeres are cleaved by human topoisomerase IIalpha. Nucleic Acids Res. 2008; 36:6165–6174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mills W.E., Spence J.M., Fukagawa T., Farr C.J. Site-specific cleavage by topoisomerase 2: a mark of the core centromere. Int. J. Mol. Sci. 2018; 19:534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. West K.L., Austin C.A. Human DNA topoisomerase IIbeta binds and cleaves four-way junction DNA in vitro. Nucleic Acids Res. 1999; 27:984–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Le H., Singh S., Shih S.J., Du N., Schnyder S., Loredo G.A., Bien C., Michaelis L., Toor A., Diaz M.O. et al. Rearrangements of the MLL gene are influenced by DNA secondary structure, potentially mediated by topoisomerase II binding. Genes Chromosomes Cancer. 2009; 48:806–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hansen A.S. CTCF as a boundary factor for cohesin-mediated loop extrusion: evidence for a multi-step mechanism. Nucleus. 2020; 11:132–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rao S.S., Huntley M.H., Durand N.C., Stamenova E.K., Bochkov I.D., Robinson J.T., Sanborn A.L., Machol I., Omer A.D., Lander E.S. et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014; 159:1665–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Orlandini E., Marenduzzo D., Michieletto D Synergy of topoisomerase and structural-maintenance-of-chromosomes proteins creates a universal pathway to simplify genome topology. Proc. Natl Acad. Sci. U.S.A. 2019; 116:8149–8154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bauer B.W., Davidson I.F., Canena D., Wutz G., Tang W., Litos G., Horn S., Hinterdorfer P., Peters J.M. Cohesin mediates DNA loop extrusion by a “swing and clamp” mechanism. Cell. 2021; 184:5448–5464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ruskova R., Racko D Entropic competition between supercoiled and torsionally relaxed chromatin fibers drives loop extrusion through pseudo-topologically bound cohesin. Biology (Basel). 2021; 10:130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Davidson I.F., Peters J.M. Genome folding through loop extrusion by SMC complexes. Nat. Rev. Mol. Cell Biol. 2021; 22:445–464. [DOI] [PubMed] [Google Scholar]
- 27. Dixon J.R., Gorkin D.U., Ren B. Chromatin domains: the unit of chromosome organization. Mol. Cell. 2016; 62:668–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lupianez D.G., Kraft K., Heinrich V., Krawitz P., Brancati F., Klopocki E., Horn D., Kayserili H., Opitz J.M., Laxova R. et al. Disruptions of topological chromatin domains cause pathogenic rewiring of gene–enhancer interactions. Cell. 2015; 161:1012–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pope B.D., Ryba T., Dileep V., Yue F., Wu W., Denas O., Vera D.L., Wang Y., Hansen R.S., Canfield T.K. et al. Topologically associating domains are stable units of replication-timing regulation. Nature. 2014; 515:402–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Arnould C., Rocher V., Finoux A.L., Clouaire T., Li K., Zhou F., Caron P., Mangeot P.E., Ricci E.P., Mourad R. et al. Loop extrusion as a mechanism for formation of DNA damage repair foci. Nature. 2021; 590:660–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Emerson D.J., Zhao P.A., Cook A.L., Barnett R.J., Klein K.N., Saulebekova D., Ge C., Zhou L., Simandi Z., Minsk M.K. et al. Cohesin-mediated loop anchors confine the locations of human replication origins. Nature. 2022; 606:812–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lensing S.V., Marsico G., Hansel-Hertsch R., Lam E.Y., Tannahill D., Balasubramanian S. DSBCapture: in situ capture and sequencing of DNA breaks. Nat. Methods. 2016; 13:855–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fang C., Wang Z., Han C., Safgren S.L., Helmin K.A., Adelman E.R., Serafin V., Basso G., Eagen K.P., Gaspar-Maia A. et al. Cancer-specific CTCF binding facilitates oncogenic transcriptional dysregulation. Genome Biol. 2020; 21:247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Martinez-Garcia P.M., Garcia-Torres M., Divina F., Terron-Bautista J., Delgado-Sainz I., Gomez-Vela F., Cortes-Ledesma F. Genome-wide prediction of topoisomerase IIbeta binding by architectural factors and chromatin accessibility. PLoS Comput. Biol. 2021; 17:e1007814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Uuskula-Reimand L., Hou H., Samavarchi-Tehrani P., Rudan M.V., Liang M., Medina-Rivera A., Mohammed H., Schmidt D., Schwalie P., Young E.J. et al. Topoisomerase II beta interacts with cohesin and CTCF at topological domain borders. Genome Biol. 2016; 17:182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Neguembor M.V., Martin L., Castells-Garcia A., Gomez-Garcia P.A., Vicario C., Carnevali D., AlHaj Abed J., Granados A., Sebastian-Perez R., Sottile F. et al. Transcription-mediated supercoiling regulates genome folding and loop formation. Mol. Cell. 2021; 81:3065–3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sarni D., Sasaki T., Irony Tur-Sinai M., Miron K., Rivera-Mulia J.C., Magnuson B., Ljungman M., Gilbert D.M., Kerem B. 3D genome organization contributes to genome instability at fragile sites. Nat. Commun. 2020; 11:3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Moffat J., Grueneberg D.A., Yang X., Kim S.Y., Kloepfer A.M., Hinkle G., Piqani B., Eisenhaure T.M., Luo B., Grenier J.K. et al. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell. 2006; 124:1283–1298. [DOI] [PubMed] [Google Scholar]
- 39. Wiederschain D., Wee S., Chen L., Loo A., Yang G., Huang A., Chen Y., Caponigro G., Yao Y.M., Lengauer C. et al. Single-vector inducible lentiviral RNAi system for oncology target validation. Cell Cycle. 2009; 8:498–504. [DOI] [PubMed] [Google Scholar]
- 40. Pereira E.J., Burns J.S., Lee C.Y., Marohl T., Calderon D., Wang L., Atkins K.A., Wang C.-C., Janes K.A. Sporadic activation of an oxidative stress–dependent NRF2-p53 signaling network in breast epithelial spheroids and premalignancies. Sci. Signal. 2020; 13:eaba4200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wang L., Brugge J.S., Janes K.A. Intersection of FOXO- and RUNX1-mediated gene expression programs in single breast epithelial cells during morphogenesis and tumor progression. Proc. Natl Acad. Sci. U.S.A. 2011; 108:E803–E812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Szlachta K., Raimer H.M., Comeau L.D., Wang Y.H. CNCC: an analysis tool to determine genome-wide DNA break end structure at single-nucleotide resolution. BMC Genomics. 2020; 21:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Atkin N.D., Raimer H.M., Wang Z., Zang C., Wang Y.H. Assessing acute myeloid leukemia susceptibility in rearrangement-driven patients by DNA breakage at topoisomerase II and CCCTC-binding factor/cohesin binding sites. Genes Chromosomes Cancer. 2021; 60:808–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Singh S., Szlachta K., Manukyan A., Raimer H.M., Dinda M., Bekiranov S., Wang Y.H. Pausing sites of RNA polymerase II on actively transcribed genes are enriched in DNA double-stranded breaks. J. Biol. Chem. 2020; 295:3990–4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Encode ProjectConsortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012; 489:57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tchasovnikarova I.A., Timms R.T., Douse C.H., Roberts R.C., Dougan G., Kingston R.E., Modis Y., Lehner P.J. Hyperactivation of HUSH complex function by Charcot–Marie–Tooth disease mutation in MORC2. Nat. Genet. 2017; 49:1035–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tilgner H., Grubert F., Sharon D., Snyder M.P. Defining a personal, allele-specific, and single-molecule long-read transcriptome. Proc. Natl Acad. Sci. U.S.A. 2014; 111:9869–9874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kang B.H., Jensen K.J., Hatch J.A., Janes K.A. Simultaneous profiling of 194 distinct receptor transcripts in human cells. Sci. Signal. 2013; 6:rs13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Michel N., Raimer Young H.M., Atkin N.D., Arshad U., Al-Humadi R., Singh S., Manukyan A., Gore L., Burbulis I.E., Wang Y.H. et al. Transcription-associated DNA DSBs activate p53 during hiPSC-based neurogenesis. Sci. Rep. 2022; 12:12156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kim D., Langmead B., Salzberg S.L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods. 2015; 12:357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Pertea M., Pertea G.M., Antonescu C.M., Chang T.C., Mendell J.T., Salzberg S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015; 33:290–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Li B., Dewey C.N. RSEM: accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinformatics. 2011; 12:323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013; 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Fritz A.J., Ghule P.N., Boyd J.R., Tye C.E., Page N.A., Hong D., Shirley D.J., Weinheimer A.S., Barutcu A.R., Gerrard D.L. et al. Intranuclear and higher-order chromatin organization of the major histone gene cluster in breast cancer. J. Cell. Physiol. 2018; 233:1278–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lebeau B., Jangal M., Zhao T., Wong C.K., Wong N., Canedo E.C., Hebert S., Aguilar-Mahecha A., Chabot C., Buchanan M. et al. 3D chromatin remodeling potentiates transcriptional programs driving cell invasion. Proc. Natl Acad. Sci. U.S.A. 2022; 119:e2203452119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Hnisz D., Weintraub A.S., Day D.S., Valton A.L., Bak R.O., Li C.H., Goldmann J., Lajoie B.R., Fan Z.P., Sigova A.A. et al. Activation of proto-oncogenes by disruption of chromosome neighborhoods. Science. 2016; 351:1454–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hansel-Hertsch R., Beraldi D., Lensing S.V., Marsico G., Zyner K., Parry A., Di Antonio M., Pike J., Kimura H., Narita M. et al. G-quadruplex structures mark human regulatory chromatin. Nat. Genet. 2016; 48:1267–1272. [DOI] [PubMed] [Google Scholar]
- 58. Hansel-Hertsch R., Spiegel J., Marsico G., Tannahill D., Balasubramanian S. Genome-wide mapping of endogenous G-quadruplex DNA structures by chromatin immunoprecipitation and high-throughput sequencing. Nat. Protoc. 2018; 13:551–564. [DOI] [PubMed] [Google Scholar]
- 59. Mao S.Q., Ghanbarian A.T., Spiegel J., Martinez Cuesta S., Beraldi D., Di Antonio M., Marsico G., Hansel-Hertsch R., Tannahill D., Balasubramanian S. DNA G-quadruplex structures mold the DNA methylome. Nat. Struct. Mol. Biol. 2018; 25:951–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Hui W.W.I., Simeone A., Zyner K.G., Tannahill D., Balasubramanian S. Single-cell mapping of DNA G-quadruplex structures in human cancer cells. Sci. Rep. 2021; 11:23641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ramirez F., Dundar F., Diehl S., Gruning B.A., Manke T. deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 2014; 42:W187–W191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Khoury A., Achinger-Kawecka J., Bert S.A., Smith G.C., French H.J., Luu P.L., Peters T.J., Du Q., Parry A.J., Valdes-Mora F. et al. Constitutively bound CTCF sites maintain 3D chromatin architecture and long-range epigenetically regulated domains. Nat. Commun. 2020; 11:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Gittens W.H., Johnson D.J., Allison R.M., Cooper T.J., Thomas H., Neale M.J. A nucleotide resolution map of Top2-linked DNA breaks in the yeast and human genome. Nat. Commun. 2019; 10:4846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Szlachta K., Thys R.G., Atkin N.D., Pierce L.C.T., Bekiranov S., Wang Y.H. Alternative DNA secondary structure formation affects RNA polymerase II promoter-proximal pausing in human. Genome Biol. 2018; 19:89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Lorenz R., Bernhart S.H., Honer Zu Siederdissen C., Tafer H., Flamm C., Stadler P.F., Hofacker I.L. ViennaRNA package 2.0. Algorithms Mol. Biol. 2011; 6:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Halldorsson B.V., Eggertsson H.P., Moore K.H.S., Hauswedell H., Eiriksson O., Ulfarsson M.O., Palsson G., Hardarson M.T., Oddsson A., Jensson B.O. et al. The sequences of 150,119 genomes in the UK Biobank. Nature. 2022; 607:732–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Schmidt D., Schwalie P.C., Wilson M.D., Ballester B., Goncalves A., Kutter C., Brown G.D., Marshall A., Flicek P., Odom D.T. Waves of retrotransposon expansion remodel genome organization and CTCF binding in multiple mammalian lineages. Cell. 2012; 148:335–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Fu Y., Sinha M., Peterson C.L., Weng Z. The insulator binding protein CTCF positions 20 nucleosomes around its binding sites across the human genome. PLoS Genet. 2008; 4:e1000138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Kim S., Yu N.K., Kaang B.K. CTCF as a multifunctional protein in genome regulation and gene expression. Exp. Mol. Med. 2015; 47:e166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Ong C.T., Corces V.G. CTCF: an architectural protein bridging genome topology and function. Nat. Rev. Genet. 2014; 15:234–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Wu C.C., Li T.K., Farh L., Lin L.Y., Lin T.S., Yu Y.J., Yen T.J., Chiang C.W., Chan N.L. Structural basis of type II topoisomerase inhibition by the anticancer drug etoposide. Science. 2011; 333:459–462. [DOI] [PubMed] [Google Scholar]
- 72. Nitiss J.L. DNA topoisomerase II and its growing repertoire of biological functions. Nat. Rev. Cancer. 2009; 9:327–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Hou Y., Li F., Zhang R., Li S., Liu H., Qin Z.S., Sun X. Integrative characterization of G-quadruplexes in the three-dimensional chromatin structure. Epigenetics. 2019; 14:894–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Wang H., Maurano M.T., Qu H., Varley K.E., Gertz J., Pauli F., Lee K., Canfield T., Weaver M., Sandstrom R. Widespread plasticity in CTCF occupancy linked to DNA methylation. Genome Res. 2012; 22:1680–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Biffi G., Tannahill D., McCafferty J., Balasubramanian S. Quantitative visualization of DNA G-quadruplex structures in human cells. Nat. Chem. 2013; 5:182–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Hoa N.N., Shimizu T., Zhou Z.W., Wang Z.Q., Deshpande R.A., Paull T.T., Akter S., Tsuda M., Furuta R., Tsutsui K. et al. Mre11 Is essential for the removal of lethal topoisomerase 2 covalent cleavage complexes. Mol. Cell. 2016; 64:580–592. [DOI] [PubMed] [Google Scholar]
- 77. Wulfridge P., Yan Q., Rell N., Doherty J., Jacobson S., Offley S., Deliard S., Feng K., Phillips-Cremins J.E., Gardini A. et al. G-quadruplexes associated with R-loops promote CTCF binding. Mol. Cell. 2023; 83:3064–3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Guiblet W.M., Cremona M.A., Harris R.S., Chen D., Eckert K.A., Chiaromonte F., Huang Y.-F., Makova K.D. Non-B DNA: a major contributor to small- and large-scale variation in nucleotide substitution frequencies across the genome. Nucleic Acids Res. 2021; 49:1497–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Sheffield N.C., Bock C. LOLA: enrichment analysis for genomic region sets and regulatory elements in R and Bioconductor. Bioinformatics. 2016; 32:587–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Gong J.-Y., Wen C.-J., Tang M.-L., Duan R.-F., Chen J.-N., Zhang J.-Y., Zheng K.-W., He Y.-D., Hao Y.-H., Yu Q. et al. G-quadruplex structural variations in human genome associated with single-nucleotide variations and their impact on gene activity. Proc. Natl Acad. Sci. U.S.A. 2021; 118:e2013230118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. McArthur E., Capra J.A. Topologically associating domain boundaries that are stable across diverse cell types are evolutionarily constrained and enriched for heritability. Am. J. Hum. Genet. 2021; 108:269–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Kemp C.J., Moore J.M., Moser R., Bernard B., Teater M., Smith L.E., Rabaia N.A., Gurley K.E., Guinney J., Busch S.E. et al. CTCF haploinsufficiency destabilizes DNA methylation and predisposes to cancer. Cell Rep. 2014; 7:1020–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Damaschke N.A., Gawdzik J., Avilla M., Yang B., Svaren J., Roopra A., Luo J.H., Yu Y.P., Keles S., Jarrard D.F. CTCF loss mediates unique DNA hypermethylation landscapes in human cancers. Clin. Epigenetics. 2020; 12:80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Akhtar M.S., Akhter N., Najm M.Z., Deo S.V.S., Shukla N.K., Almalki S.S.R., Alharbi R.A., Sindi A.A.A., Alruwetei A., Ahmad A. et al. Association of mutation and low expression of the CTCF gene with breast cancer progression. Saudi Pharm. J. 2020; 28:607–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Oh S., Oh C., Yoo K.H. Functional roles of CTCF in breast cancer. BMB Rep. 2017; 50:445–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Docquier F., Kita G.X., Farrar D., Jat P., O’Hare M., Chernukhin I., Gretton S., Mandal A., Alldridge L., Klenova E. Decreased poly(ADP-ribosyl)ation of CTCF, a transcription factor, is associated with breast cancer phenotype and cell proliferation. Clin. Cancer Res. 2009; 15:5762–5771. [DOI] [PubMed] [Google Scholar]
- 87. Katainen R., Dave K., Pitkanen E., Palin K., Kivioja T., Valimaki N., Gylfe A.E., Ristolainen H., Hanninen U.A., Cajuso T. et al. CTCF/cohesin-binding sites are frequently mutated in cancer. Nat. Genet. 2015; 47:818–821. [DOI] [PubMed] [Google Scholar]
- 88. Song S.H., Kim T.Y. CTCF, cohesin, and chromatin in human cancer. Genomics Inform. 2017; 15:114–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Guo Y.A., Chang M.M., Huang W., Ooi W.F., Xing M., Tan P., Skanderup A.J. Mutation hotspots at CTCF binding sites coupled to chromosomal instability in gastrointestinal cancers. Nat. Commun. 2018; 9:1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Lambuta R.A., Nanni L., Liu Y., Diaz-Miyar J., Iyer A., Tavernari D., Katanayeva N., Ciriello G., Oricchio E. Whole-genome doubling drives oncogenic loss of chromatin segregation. Nature. 2023; 615:925–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Chambers V.S., Marsico G., Boutell J.M., Di Antonio M., Smith G.P., Balasubramanian S. High-throughput sequencing of DNA G-quadruplex structures in the human genome. Nat. Biotechnol. 2015; 33:877–881. [DOI] [PubMed] [Google Scholar]
- 92. Huppert J.L., Balasubramanian S. G-quadruplexes in promoters throughout the human genome. Nucleic Acids Res. 2007; 35:406–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Marsico G., Chambers V.S., Sahakyan A.B., McCauley P., Boutell J.M., Antonio M.D., Balasubramanian S. Whole genome experimental maps of DNA G-quadruplexes in multiple species. Nucleic Acids Res. 2019; 47:3862–3874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Rhodes D., Lipps H.J. G-quadruplexes and their regulatory roles in biology. Nucleic Acids Res. 2015; 43:8627–8637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Varshney D., Spiegel J., Zyner K., Tannahill D., Balasubramanian S. The regulation and functions of DNA and RNA G-quadruplexes. Nat. Rev. Mol. Cell Biol. 2020; 21:459–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Pavlova A.V., Kubareva E.A., Monakhova M.V., Zvereva M.I., Dolinnaya N.G. Impact of G-quadruplexes on the regulation of genome integrity, DNA damage and repair. Biomolecules. 2021; 11:1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Stigler J., Camdere G.O., Koshland D.E., Greene E.C. Single-molecule imaging reveals a collapsed conformational state for DNA-bound cohesin. Cell Rep. 2016; 15:988–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Racko D., Benedetti F., Dorier J., Stasiak A. Transcription-induced supercoiling as the driving force of chromatin loop extrusion during formation of TADs in interphase chromosomes. Nucleic Acids Res. 2018; 46:1648–1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Naughton C., Avlonitis N., Corless S., Prendergast J.G., Mati I.K., Eijk P.P., Cockroft S.L., Bradley M., Ylstra B., Gilbert N. Transcription forms and remodels supercoiling domains unfolding large-scale chromatin structures. Nat. Struct. Mol. Biol. 2013; 20:387–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Olivieri M., Cho T., Alvarez-Quilon A., Li K., Schellenberg M.J., Zimmermann M., Hustedt N., Rossi S.E., Adam S., Melo H. et al. A genetic map of the response to DNA damage in human cells. Cell. 2020; 182:481–496.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Bossaert M., Pipier A., Riou J.F., Noirot C., Nguyen L.T., Serre R.F., Bouchez O., Defrancq E., Calsou P., Britton S. et al. Transcription-associated topoisomerase 2alpha (TOP2A) activity is a major effector of cytotoxicity induced by G-quadruplex ligands. eLife. 2021; 10:e65184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Hwang S.Y., Kang M.A., Baik C.J., Lee Y., Hang N.T., Kim B.G., Han J.S., Jeong J.H., Park D., Myung K. et al. CTCF cooperates with CtIP to drive homologous recombination repair of double-strand breaks. Nucleic Acids Res. 2019; 47:9160–9179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Lang F., Li X., Zheng W., Li Z., Lu D., Chen G., Gong D., Yang L., Fu J., Shi P. et al. CTCF prevents genomic instability by promoting homologous recombination-directed DNA double-strand break repair. Proc. Natl Acad. Sci. U.S.A. 2017; 114:10912–10917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Hilmi K., Jangal M., Marques M., Zhao T., Saad A., Zhang C., Luo V.M., Syme A., Rejon C., Yu Z. et al. CTCF facilitates DNA double-strand break repair by enhancing homologous recombination repair. Sci. Adv. 2017; 3:e1601898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Aparicio T., Baer R., Gottesman M., Gautier J. MRN, CtIP, and BRCA1 mediate repair of topoisomerase II–DNA adducts. J. Cell Biol. 2016; 212:399–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Hansen A.S., Pustova I., Cattoglio C., Tjian R., Darzacq X. CTCF and cohesin regulate chromatin loop stability with distinct dynamics. eLife. 2017; 6:e25776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Tikhonova P., Pavlova I., Isaakova E., Tsvetkov V., Bogomazova A., Vedekhina T., Luzhin A.V., Sultanov R., Severov V., Klimina K. et al. DNA G-quadruplexes contribute to CTCF recruitment. Int. J. Mol. Sci. 2021; 22:7090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. de Wit E., Vos E.S., Holwerda S.J., Valdes-Quezada C., Verstegen M.J., Teunissen H., Splinter E., Wijchers P.J., Krijger P.H., de Laat W. CTCF binding polarity determines chromatin looping. Mol. Cell. 2015; 60:676–684. [DOI] [PubMed] [Google Scholar]
- 109. Nora E.P., Caccianini L., Fudenberg G., So K., Kameswaran V., Nagle A., Uebersohn A., Hajj B., Saux A.L., Coulon A. et al. Molecular basis of CTCF binding polarity in genome folding. Nat. Commun. 2020; 11:5612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Li Y., Haarhuis J.H.I., Sedeno Cacciatore A., Oldenkamp R., van Ruiten M.S., Willems L., Teunissen H., Muir K.W., de Wit E., Rowland B.D. et al. The structural basis for cohesin–CTCF-anchored loops. Nature. 2020; 578:472–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
DSB mapping/sequencing datasets and RNA-seq of Jurkat cells generated in this study can be accessed at Sequence Read Archive: PRJNA795482. The publicly available datasets used in this study are listed in Supplementary Table S3.