Highlights
-
•
PDL1, CD155, PD-1 and TIGIT are upregulated in liver tissues and PBMCs of HCC patients, respectively.
-
•
CCAT-1, H19, and MALAT-1 are significantly upregulated in the sera, PBMCs, and tissues of HCC patients compared to HCV patients and healthy controls.
-
•
miR-944–5p, miR-105–5p, miR-486–5p, miR-506–5p, and miR-30a-5p are downregulated in the sera and liver tissues of HCC patients.
-
•
Knocking down of CCAT-1, MALAT-1, or H19 markedly repressed the co-expression of PD-L1 and CD155 and accordingly induced the cytotoxic profile of co-cultured immune cells.
-
•
Ectopic expression of miR-486–5p, miR-506–5p, and miR-30a-5p in PBMCs significantly repressed the PD-1.
-
•
Ectopic expression of miR-944–5p and miR-105–5p in PBMCs dramatically reduced the co-expression of PD-1 and TIGIT.
Keywords: Immune checkpoint inhibitors (ICI), Programmed death-ligand 1 (PD-L1), T cell immunoreceptor with Ig and ITIM domains (TIGIT), MALAT-1, H19, CCAT-1, HCC
Abstract
Tumor microenvironment is an intricate web of stromal and immune cells creating an immune suppressive cordon around the tumor. In hepatocellular carcinoma (HCC), Tumor microenvironment is a formidable barrier towards novel immune therapeutic approaches recently evading the oncology field. In this study, the main aim was to identify the intricate immune evasion tactics mediated by HCC cells and to study the epigenetic modulation of the immune checkpoints; Programmed death-1 (PD-1)/ Programmed death-Ligand 1 (PD-L1) and T cell immunoreceptor with Ig and ITIM domains (TIGIT)/Cluster of Differentiation 155 (CD155) at the tumor-immune synapse. Thus, liver tissues, PBMCs and sera were collected from Hepatitis C Virus (HCV), HCC as well as healthy individuals. Screening was performed to PD-L1/PD-1 and CD155/TIGIT axes in HCC patients. PDL1, CD155, PD-1 and TIGIT were found to be significantly upregulated in liver tissues and peripheral blood mononuclear cells (PBMCs) of HCC patients. An array of long non-coding RNAs (lncRNAs) and microRNAs validated to regulate such immune checkpoints were screened. The lncRNAs; CCAT-1, H19, and MALAT-1 were all significantly upregulated in the sera, PBMCs, and tissues of HCC patients as compared to HCV patients and healthy controls. However, miR-944–5p, miR-105–5p, miR-486–5p, miR-506–5p, and miR-30a-5p were downregulated in the sera and liver tissues of HCC patients. On the tumor cell side, knocking down of lncRNAs—CCAT-1, MALAT-1, or H19—markedly repressed the co-expression of PD-L1 and CD155 and accordingly induced the cytotoxicity of co-cultured primary immune cells. On the immune side, ectopic expression of the under-expressed microRNAs; miR-486–5p, miR-506–5p, and miR-30a-5p significantly decreased the transcript levels of PD-1 in PBMCs with no effect on TIGIT. On the other hand, ectopic expression of miR-944–5p and miR-105–5p in PBMCs dramatically reduced the co-expression of PD-1 and TIGIT. Finally, all studied miRNAs enhanced the cytotoxic effects of PBMCs against Huh7 cells. However, miR-105–5p showed the highest augmentation for PBMCs cytotoxicity against HCC cells. In conclusion, this study highlights a novel co-targeting strategy using miR-105–5p mimics, MALAT-1, CCAT-1 and H19 siRNAs to efficiently hampers the immune checkpoints; PD-L1/PD-1 and CD155/TIGIT immune evasion properties in HCC.
Graphical abstract
Introduction
Hepatocellular carcinoma (HCC) represents a global health threat with escalating incidence rates [1]. HCC ranks as the 2nd most common cause of cancer-related mortalities with 5-year survival rate as only 18% [2]. Moreover, the estimated number of HCC cases in both genders and among all age groups is expected to increase from 841,080 in 2018 to 1361,836 in 2040 [3]. HCC patients are commonly diagnosed at late stages of the disease, a status that hampers most of the conventional therapeutic approaches success rates [4,5].
Geographically North Africa and Middle East countries including Egypt, Hepatitis C virus (HCV) contributes for 44% of HCC-related cases [6]. Nonetheless, a recent study reported that increasing trends in HCC incidence across all etiology groups have been witnessed where HBV-related HCC incidence rates increased by 42% while HCV-related HCC incidence increased by 114% [7].
Harnessing the power of the immune system to treat malignancies have gained significant momentum in the last decade [8], [9], [10], [11]. However, in context of HCC tumor microenvironment (TME) a lot of immune-related puzzling questions are unanswered [12]. Immune cells at the TME have a fundamental role in the development and progression of HCC [13]. Immune escape phenomena of HCC have recently been associated with a lot of obstacles and failures when it comes to the incorporation of the novel immune therapeutic approach in the treatment protocols of HCC patients [14]. Tumor cells dodge the normal immune screening and killing capacities through intricate genetic and epigenetic manipulations [15,16]. Moreover, tumor cells follow other tactics through the circulating tumor cells to directly repress the immune system activity inducing the initiation of immune suppression phenomena within the whole body [17].
Among the most imperative tactics tumor cells use to evade the immune system is the up-regulation of the immune checkpoints such as programmed cell death ligand 1 (PD-L1) and the up-regulation of poliovirus receptor (PVR, CD155), which has been recently reported in several malignancies [8,18,19]. Yet, its expression in HCC tumor cells is not well-investigated [20]. At the tumor-immune cell synapse in the TME, PD-L1 and CD155 act as ligands for programmed cell death receptor 1 (PD-1) and T cell immune-receptor with Ig and ITIM domains (TIGIT), respectively. Upon the interaction between CD155/TIGIT and PD-L1/PD-1 abolishing of the cytotoxic potential of T cells and natural killer (NK) cells occurs [21,22].
Recently, we and others have unraveled a novel potential role of non-coding RNAs (ncRNAs) in re-sensitizing the immune system to eradicate tumor cells [23], [24], [25], [26]. This is mainly performed by re-tweaking the immune suppressive tactics and altered expression of immune checkpoints employed by tumor cells through immunomodulatory ncRNAs [11,27]. Several ncRNAs including microRNAs (miRNAs) and long non-coding RNAs (lncRNAs) have recently been implicated in the epigenetic-mediated regulation of the immune checkpoints PD-1/PD-L1 and TIGIT/CD155 in several malignancies, including hepatoma and pancreatic cancer [28,29].
For instance, MALAT-1 has recently been reported to act as an immunomodulatory lncRNA in several malignancies that have the ability to modulate the expression of PD-L1 and other tumor-secreted immune suppressive cytokines such as IL-10 and TNF-α [30]. Nonetheless, MALAT-1 has been directly linked to tumor growth and suggested to act as a new predictor of tumor recurrence following liver transplantation and a promising therapeutic target [31]. However, the immunomodulatory role of MALAT-1 in HCC has not been extensively investigated.
LncRNA H19 is another lncRNA that has been recently added to the list of emerging immunomodulatory lncRNAs through manipulating the expression profile of NK activating ligands on tumor cells and CD155 expression in several malignancies [11,30,[32], [33], [34], [35]]. Furthermore, H19 has also been linked to tumor cell development, hypoxic stress, macrophage activation, chemotaxis, and differentiation [36,37]. It also serves as a diagnostic and prognostic biomarker for HCC [36]. Yet, its immune-oncological role in HCC remains debatable [38,39].
CCAT-1 is a recent under-investigated lncRNA [40]. However, our research group has recently reported its fundamental role in modulating PD-L1 expression in triple negative breast cancer patients [41]. CCAT-1 has been previously linked to HCC tumor growth, proliferation, and invasion and characterized as an oncogenic mediator in HCC, through modulating an array of oncogenic signaling pathways in HCC cell lines [42,43]. Yet, the CCAT-1 immuno-oncological role in HCC patients and cell lines has been scarcely discussed.
Similar to lncRNAs, miRNAs have been reported to act as functional players in the immunoregulatory portrait of tumor cells [16,26,[44], [45], [46]]. Specifically, miR-506–5p [47], miR-486–5p [48], miR-30a-5p [49], miR-944–5p [50] and miR-105–5p [51] are considered immune regulatory miRNAs with a fundamental role in tumor progression and metastasis in several solid malignancies. However, their immune modulatory role in HCC patients and cell lines remains to be elucidated. Therefore, this study aimed to investigate an array of ncRNAs that could function as upstream modulators of the immune checkpoints axes CD155/TIGIT and PD-1/PD-L1 at the tumor-immune synapse in HCC patients and cell lines.
Participants and methods
Study participants
The current study involved 45 HCC patients with a male-to-female ratio of 4:1, 35 hepatitis C virus (HCV) patients with a male-to-female ratio of 3:2, and 30 healthy controls with a male-to-female ratio of 3:2. Whole blood, sera, and liver tissues were collected from the involved participants. However, due to the scarcity of normal liver tissues, only 13 normal liver biopsies were collected from healthy donors during the liver transplantation procedures. All patients with HCC and HCV, as well as healthy controls, were age matched. All patients with HCC were virologically and serologically positive for HCV. All subjects were negative for HIV and HBV. Healthy controls were non-diabetic, non-hypertensive, and not suffering from any chronic or autoimmune diseases. All subjects gave their written informed consent before the collection of the samples. All the experiments were carried out in compliance with the guidelines set forth by the declaration of Helsinki. The study has been approved by the Institutional Review Board of Ain-Shams University, National Liver Instiute, the German University in Cairo (Ethical Approval Number: HCC-RA-2016-04) and the German International University (Ethical Approval Number: BIOT-HCC-01–2023).
PBMCs isolation and serum collection
Approximately, 8–10 ml of peripheral venous blood were withdrawn from each participant for PBMCs isolation, while another 4 ml peripheral venous blood samples were collected from the participants for serum separation. Within 4–6 hrs of sample collection, PBMCs were isolated using the Ficoll-density gradient centrifugation method. Briefly, peripheral venous blood was diluted in an equal volume of wash mix (Roswell Park Memorial Institute (RPMI) 94%, fetal bovine serum (FBS) 5%, penicillin/ streptomycin 1%). Diluted blood, approximately 6 ml, was carefully layered on a 3 ml Ficoll reagent, followed by 30 min of centrifugation at 1000 rpm. The buffy layer containing the PBMCs was collected and washed twice with the wash mix. The PBMCs of each sample were cryopreserved and stored in a −80 °C freezer for later use as previously described [15,17]. Serum separation was done by centrifuging at 1500 rcf for 10 min, followed by the collection of the upper serum phase that was stored at −80 °C until further use.
Culturing of PBMCs and Huh7 cells
Primary PBMCs were cultured in RPMI media whereas Huh7 cells were cultured in Dulbecco's Modified Eagle's Medium (DMEM) media, all supplemented with l-glutamine, 10% FBS, 100 mg/ml of streptomycin, and 100 IU/ml of penicillin (Lonza, Basel, Switzerland). The cells were incubated at 37 °C in 5% CO2 as previously described [52,53].
Delivery of oligonucleotides into primary PBMCs and Huh7 cells
Primary PBMCs isolated from HCC patients or healthy controls and Huh7 cells were transfected with miR-486–5p, miR-506–3p, miR-30a-5p, miR-944–5p and miR-105–5p mimics (Qiagen, Germany). CCAT-1, H19, and MALAT-1 siRNAs were also used for the transfection experiments (Qiagen, Germany). A day before transfection, 6 × 104 PBMCs or 3 × 105 Huh7 cells were seeded in a 96-well plate or a 24-well plate, respectively. All transfection experiments were carried out in triplicates using HiPerfect Transfection Reagent (Qiagen, Germany) according to the manufacturer's protocol, and experiments were repeated at least three times as previously described [10,11,48,54]. Mock cells were only exposed to transfection reagent while cells transfected with scrambled miRNAs or scrambled siRNAs were designated as Scr-miRNAs or Scr-siRNAs, respectively. Co-culturing experiments or RNA extraction were performed 48 hr post-transfection.
Total RNA and miRNA extraction
mRNAs and miRNAs were extracted from sera and PBMCs of HCC patients and healthy controls using Qiazol RNA extraction according to the manufacturer's protocol. RNA isolation was performed from liver tissues and Huh7 cells using Biozol (BIOR, China) as previously described [54], [55], [56], [57].
mRNA and miRNA quantification
The extracted miRNAs were reverse transcribed into single-stranded complementary DNA (cDNA) using TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA) and specific primers for hsa-miR-486–5p, hsa-miR-506–5p, hsa-miR-30a-5p, hsa-miR-944–5p, hsa-miR-105–5p, and RNU6B. PD-1, PD-L1, TIGIT, CD155, CCAT-1, H19, and MALAT-1 and β-actin were reverse transcribed into cDNA using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA) according to the manufacturer's instructions. Relative expression of miRNAs of choice and RNU6B, as well as PD-1, PD-L1, TIGIT, CD155, CCAT-1, H19, MALAT-1, and β-actin were quantified using TaqMan real-time qPCR using Step One System (Applied Biosystems, Foster City, CA, USA). Normalization of miRNAs expression was done using RNU6B, while normalization of mRNA and lncRNA expression was done using β-actin. Relative expression was calculated using the 2−ΔΔCt method as previously described in [55,58]. All qPCR reactions including controls were run in triplicate reactions.
Co-culturing of Huh7 cells and PBMCs isolated from HCC patients
Huh7 cells were seeded 2 hrs prior to co-culturing in a flat-bottomed 96-well plate. Transfected primary PBMCs isolated from HCC patients or healthy controls were incubated with target Huh7 cells at different effector-to-target (E:T) ratios for 6–8 hrs at 37 °C and 5% CO2. Upon optimization, the E:T ratio 5:1 was chosen, where 15 × 103 Huh7 cells/well were cocultured with 75 × 103 primary PBMCs/well as previously described in [59].
Lactate dehydrogenase (LDH) cytotoxicity assay
In vitro cytolytic activity of PBMCs of HCC patients and healthy controls against their target Huh7 cells was assessed using the Lactate Dehydrogenase (LDH) Activity Assay kit (Lactate dehydrogenase (LDH)-Liquizyme (4+1) E.C.1.1.1.27., Spectrum, Egypt) according to the manufacturer's instructions. Briefly, centrifugation at 2500 rpm for 15 min at room temperature was performed, and the supernatant was collected for LDH measurement. Maximum release was determined by complete lysis of non-co-cultured target cells. The LDH activity was measured and then the percentage of lysis was calculated as follows: percentage (%) cytotoxicity=(target maximum release- maximum release experimental release)/(target maximum release) x 100 as previously described in [60]. All experiments were done in triplicates and repeated at least three times.
Preparation of siRNAs oligonucleotide liposomal formulations
DSPC, DSPE, MPEG-2000-DSPE, and cholesterol were obtained from Corden Pharma (Plankstdt, Germany). HPLC-grade acetonitrile and dichloromethane (DCM) were purchased from Sigma-Aldrich (Hamburg, Germany). Liposomes composed of DSPC/DSPE/MPEG-2000-DSPE/cholesterol in a molar ratio of 1:0.1:0.1:0.8 were fabricated using the thin-film hydration approach as previously detailed with some modifications [61]. Briefly, 0.0412 mmol lipids and cholesterol were dissolved in dichloromethane in a round bottom flask. Then the organic solvent was evaporated under vacuum in a rotary evaporator operating at 130 rpm at 55 °C (the transition temperature of the lipids), forming a thin lipid film. This film was rehydrated using 3 mL of PBS of pH 7.4 at the same temperature for 1 hour. The obtained suspension was extruded for 10 cycles (at 55 °C) utilizing 0.1 µm polycarbonate membranes fixed in an Avanti Mini extruder (Avanti polar lipids Inc., Alabama, AL, USA) to generate uni-lamellar liposomes with a narrow polydispersity index. The liposomal formulations were stored at 4 °C until use. The oligonucleotide (3 siRNAs) liposomal formulation was designed by the dropwise addition of siRNAs to liposomal suspension at 4 °C while stirring. Then, the mixture was kept under stirring for 4 hr at 4 °C.
Colony Forming Assay
Forty-Eight hourse post-transfection with CCAT-1, H19, and MALAT-1 siRNAs and miR-105-5p mimics, cells were counted and seeded in 6-well plate (1000 cells/well). Cells were incubated for 1-3 weeks under normal growth conditions (5% CO2, 37 οC). Then, fixation and staining of the colonies were performed using 6% Glutaraldehyde and 0.5% crystal violet, respectively. Then, colonies were manually counted.
Transwell migration and invasion
Transwell migration assay was performed in 24-well plate with 8 μm pore-size (cellQART®, Germany). Transwell invasion assay was performed using 8 μm pore-size coated with collagen. Transfected cells were seeded in upper chamber and suspended in 1% FBS media. The cells were allowed to migrate toward the lower chamber, where 10% FBS media was placed 24 hr. The migrated/invaded cells were then fixed using 6% glutaraldehyde and stained with 0.4% crystal violet solution, followed by capturing and counting migrated or invaded cells using an optical microscope. The cells were counted in five photographed fields [60].
Statistical analysis
All data are presented as mean ± standard error of the mean (SEM). Statistical analysis was based on non-parametric unpaired Student's t-test or one-way analysis of variance with post hoc analysis for multiple comparisons. The correlation analysis was performed by Spearman analysis. A p-value less than 0.05 was considered statistically significant. ***p < 0.001, **p < 0.01, *p < 0.05 and ns (statistically not significant). All the data were statistically analyzed using GraphPad Prism version 5.0 software (La Jolla, CA, USA).
Results
Differential expression patterns of PD-1/PD-L1 and TIGIT/CD155 in PBMCs and liver tissues from HCV and HCC patients
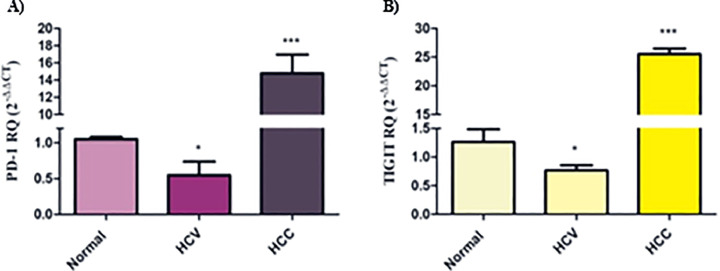
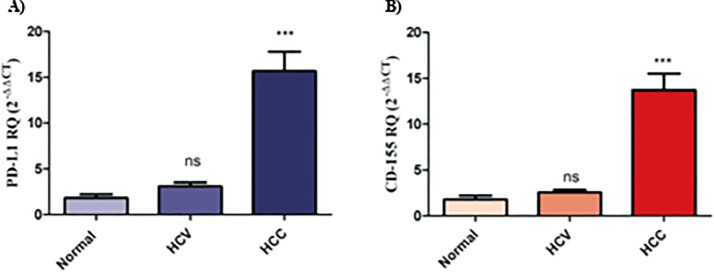
First, it was crucial to investigate the expression of immune checkpoint axes PD-1/PD-L1 and TIGIT/CD155 in PBMCs and liver tissues of HCV and HCC patients to identify the alterations upon tumor progression. The PD-1 transcript level in PBMCs isolated from HCV patients was found to be down-regulated compared to normal individuals (p < 0.05). On the contrary, PBMCs isolated from HCC patients presented a significant up-regulation in PD-1 expression (p < 0.001) (Fig. 1A). Similarly, TIGIT expression was found to be repressed in PBMCs isolated from HCV patients (p < 0.05), while its expression was significantly elevated in PBMCs isolated from HCC patients (p < 0.001) (Fig. 1B). Conversely, their respective ligands were probed in liver tissues of HCV and HCC patients. It was shown that there was no significant difference in the expression of PD-L1 and CD155 in the liver tissues of HCV patients. However, there was a highly significant up-regulation in PD-L1 expression level in the liver tissues of HCC patients (p < 0.001, Fig. 2A). Likewise, CD155 was found to be significantly up-regulated in the liver tissues of HCC patients (p < 0.001, Fig. 2B).
Fig. 1.
PD-1 and TIGIT expression profile in peripheral blood mononuclear cells (PBMCs) of HCV and HCC patients. (A) PD-1 and (B) TIGIT expression was down-regulated in PBMCs of HCV patients, while their expression was up-regulated in HCC patients. *p < 0.05,***p < 0.001.
Fig. 2.
PD-L1 and CD155 expression profile in liver tissues of HCV and HCC patients. (A) PD-L1 and (B) CD155 expression were unaltered in liver tissues of HCV patients, while their expression was significantly up-regulated in HCC patients. ns=not significant, ***p < 0.001.
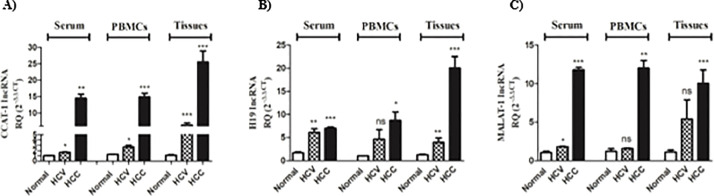
Differential expression patterns of CCAT-1, H19, and MALAT-1 lncRNAs in HCV and HCC sera, PBMCs, and tissues
Next, it was interesting to explore the expression of the lncRNAs validated to regulate immune checkpoints axes of interest in serum, PBMCs, and liver tissues of HCV and HCC patients. As shown in Fig. 3A, CCAT-1 expression was higher in the serum, PBMCs, and liver tissues of HCV patients (p < 0.05, p < 0.05, and p < 0.001, respectively) with a further increase in the serum, PBMCs, and liver tissue of HCC patients (p < 0.01, p < 0.001 and p < 0.001, respectively). Serum and liver tissues of HCV patients revealed a high expression of H19 (p < 0.01); however, PBMCs of HCV patients only showed a slight increase in H19 expression that was not significant. In HCC patients’ serum, PBMCs, and liver tissues showed elevated levels of H19 expression (p < 0.001, p < 0.05, p < 0.001, respectively) with the highest expression profile being in HCC liver tissues (Fig. 3B). Looking at MALAT-1 expression, only the serum of HCV patients showed a significant increase in its expression compared to healthy controls (p < 0.05). While HCC patients showed a robust significant elevation in MALAT-1 expression in their sera (p < 0.001), PBMCs (p < 0.01), and liver tissues (p < 0.001) (Fig. 3C).
Fig. 3.
Expression Profile of CCAT-1, H19 and MALAT-1 in serum, PBMCs, and liver tissues of HCV and HCC patients. (A) CCAT-1, (B) H19, (C) MALAT-1 expression was assessed using real-time qPCR in the serum, PBMCs, and liver tissues of HCV and HCC patients, and compared to healthy controls. ns=not significant, *p < 0.05, **p < 0.01, ***p < 0.001.
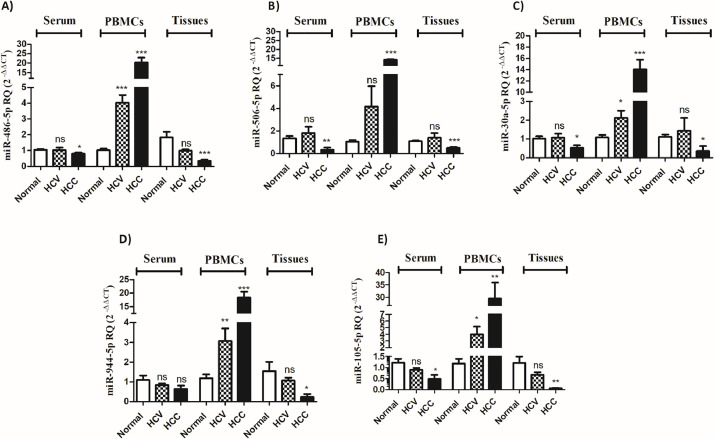
Distinct expression signatures of selected miRNAs in HCV and HCC sera, PBMCs, and tissues
Based on bioinformatics analysis and extensive literature review, the selected upstream miRNAs were screened. The selected miR-486–5p, miR-506–5p, miR-30a-5p, miR-944, and miR-105–5p demonstrated diverse patterns of expression in the different tissues investigated. For instance, miR-486–5p showed a significant reduction in its expression in the serum and liver tissues of HCC patients (p < 0.05, p < 0.001, respectively) but not in HCV patients, when compared to the control. Of note, the latter did not exhibit any statistical significance. On the contrary, there was a significant increase in the expression of miR-486–5p in the PBMCs of HCV and HCC patients (p < 0.001, Fig. 4A). miR-506–5p expression was found to be reduced in the serum and liver tissues of HCC patients (p < 0.01, p < 0.001, respectively), whereas it was significantly increased in the PBMCs of HCC patients (p < 0.001, Fig. 4B). Similar to miR-486–5p, miR-30a-5p expression showed a significant reduction in the serum and liver tissues of HCC but not HCV patients (Fig. 4C). Only PBMCs of HCV patients showed a significant increase in miR-30a-5p expression (p < 0.05) with a further increase in the PBMCs of HCC patients (p < 0.001).On the contrary, miR-944 did not show any difference in its expression in the serum of HCV and HCC patients. There was a significant reduction in the expression of miR-944 in HCC liver tissues compared to healthy controls (p < 0.05). Interestingly, miR-944 was found to be increased in the PBMCs of HCV patients (p < 0.01) with a further increase in PBMCs of HCC patients (p < 0.001) (Fig. 4D). Like miR-486–5p and miR-30a-5p, miR-105–5p showed a similar pattern of expression with a significant reduction noted in the serum and liver tissues of HCC patients only and not in HCV patients (p < 0.05, p < 0.01, respectively). The PBMCs of HCV and HCC patients showed a significantly high expression of miR-105–5p compared to healthy controls (p < 0.05, p < 0.01, respectively) (Fig. 4E).
Fig. 4.
Selected miRNA expression in the serum, PBMCs, and liver tissues of HCV and HCC patients. The expression of (A) miR-486–5p, (B) miR-506–5p, (C) miR-30a-5p, (D) miR-944, (E) miR-105–5p was explored in the serum, PBMCs and liver tissues of HCV and HCC patients compared to healthy controls. ns=not significant, *p < 0.05, **p < 0.01, ***p < 0.001.
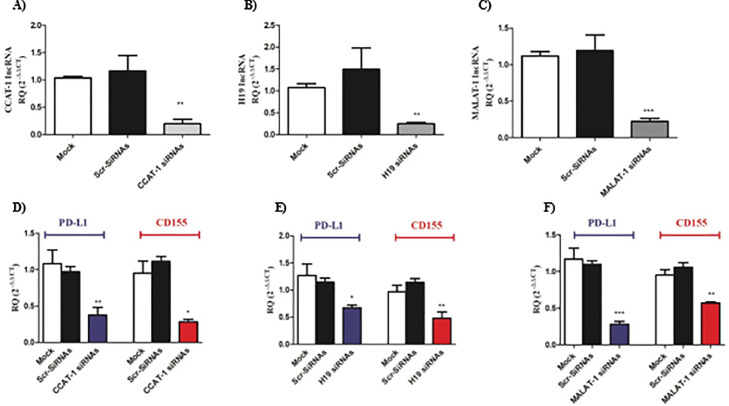
CCAT-1, H19, and MALAT-1 siRNAs repressed the co-expression of PD-L1 and CD155 in Huh-7 cells
After the screening of PD-L1 and CD155 in liver tissues of HCV and HCC patients and their upstream lncRNAs, it was critical to investigate the effect of silencing of the candidate lncRNAs on the expression of PD-L1 and CD155 in Huh7 cell lines. As illustrated in Fig. 5A-C, upon the transfection of different siRNAs, expression of lncRNAs was found to show a significant reduction by more than 80% compared to mock and scrambled siRNAs transfected Huh-7 cells. Moreover, silencing of CCAT-1 led to a significant reduction in the expression of PD-L1 and CD155 (Fig. 5D) (p < 0.01 and p < 0.05, respectively). Likewise, PD-L1 and CD155 expression level was significantly repressed upon silencing of H19 in Huh7 cell lines (Fig. 5E) (p < 0.05 and p < 0.01, respectively). Similarly, PD-L1 and CD155 expression were dramatically reduced upon silencing of MALAT-1 in Huh7 cell lines (p < 0.001 and p < 0.01, respectively) (Fig. 5F).
Fig. 5.
Impact of silencing of lncRNAs on PD-L1 and CD155 expression in Huh7 cell lines. Efficient knockdown of lncRNAs was evaluated, where (A) CCAT-1, (B) H19, and (C) MALAT-1 were markedly repressed in Huh7 cells transfected with respective lncRNA siRNA to ensure transfection efficiency. PD-L1 and CD155 was assessed upon knocking down of (D) CCAT-1, (E) H19, and (F) MALAT-1 in Huh7 cells compared to mock and scrambled mock cells. *p < 0.05, **p < 0.01, ***p < 0.001.
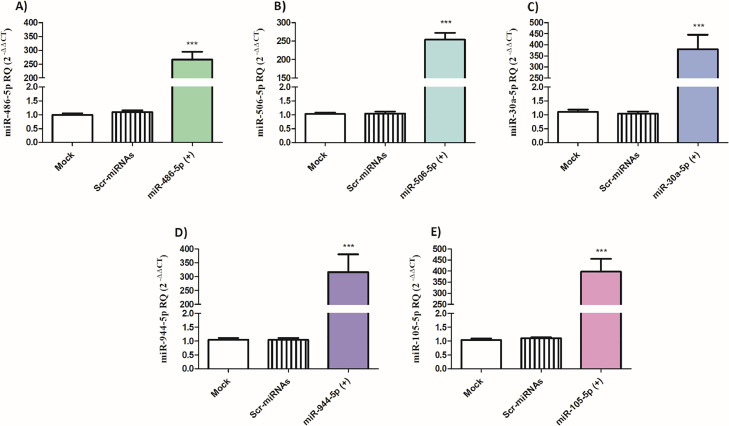
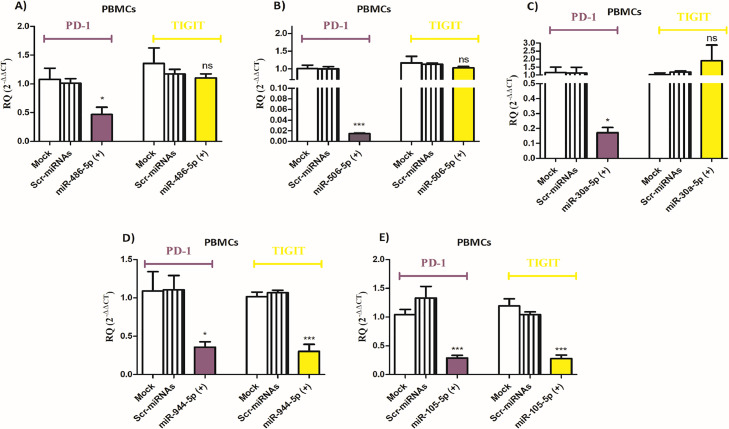
PD-1 and TIGIT expression levels are differentially altered by miR-486–5p, miR-506–5p, miR-30a-5p, miR-944, and miR-105–5p in PBMCs isolated from HCC patients
Next, it was important to explore the impact of the selected miRNAs on the expression of PD-1 and TIGIT in the PBMCs of HCC patients. First, the expression of miR-486–5p, miR-506–5p, miR-30a-5p, miR-944, and miR-105–5p was assessed after transfection of respective miRNA mimics, where the expression increased by more than 200-fold compared to mock cells and Scr-miRNAs transfected cells ensuring efficient transfection procedure (Fig. 6A-E). Ectopic expression of either miR-486–5p or miR-506–5p or miR-30a-5p led to a significant reduction in PD-1 expression in primary PBMCs of HCC patients (p < 0.05, p < 0.001, and p < 0.05, respectively). However, induced-expression of these miRNAs did not affect TIGIT expression in PBMCs of HCC patients (Fig. 7A-C). On the other hand, transfection of miR-944 and miR-105–5p led to a significant decrease in PD-1 expression (p < 0.05 and p < 0.001, respectively) as well as TIGIT expression (p < 0.001) in the primary PBMCs of HCC patients (Fig. 7D-E).
Fig. 6.
Transfection efficiency of miR-486–5p, miR-506–5p, miR-30a-5p, miR-944–5p and miR-105–5p mimics in PBMCs of HCC patients. (A) miR-486–5p, (B) miR-506–5p, (C) miR-30a-5p, (D) miR-944–5p, and (E) miR-105–5p expression was measured in primary PBMCs of HCC patients after transfection of the respective miRNA mimics, compared to mock cells and cells transfected with scrambled miRNAs. ***p < 0.001.
Fig. 7.
Impact of overexpression of miRNAs on PD-1 and TIGIT expression in the PBMCs of HCC patients. PD-1 and TIGIT expression was assessed in the PBMCs of HCC patients after the transfection of mimics of the selected miRNAs: (A) miR-486–5p, (B) miR-506–5p, (C) miR-30a-5p, (D) miR-944, and (E) miR-105–5p, in comparison to mock cells and cells transfected with scrambled miRNAs. ns=not significant, *p < 0.05, ***p < 0.001.
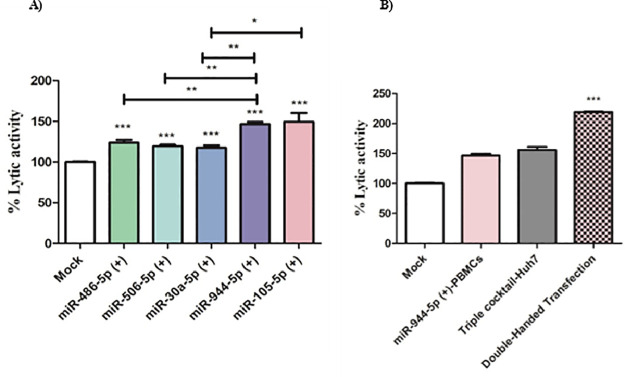
CCAT-1, H19, and MALAT-1 lncRNAs modulate the immunogenic profile of Huh7 cells and induces the cytotoxic potential of primary PBMCs against transfected Huh7 cells
As a consequence of the observed reduction in PD-L1 and CD155 expression in Huh7 cells, it was essential to explore the net impact of silencing CCAT-1, H19, and MALAT-1 on the cytotoxic potential of primary PBMCs isolated from healthy controls. As shown in Fig. 8, silencing of CCAT-1, H19, and MALAT-1 in Huh7 cells augmented the cytotoxic potential of primary healthy PBMCs. Furthermore, upon nano-encapsulation of the 3 siRNAs forming a triple cocktail of siRNAs in Huh7 target cells, there was a marked increase in the lytic activity of PBMCs compared to mock cells (p < 0.001). Also, it is worth mentioning, the transfection of the triple liposomal cocktail of siRNAs led to an increase in the lytic activity of PBMCs compared to the transfection of each of the siRNAs separately (p < 0.05).
Fig. 8.
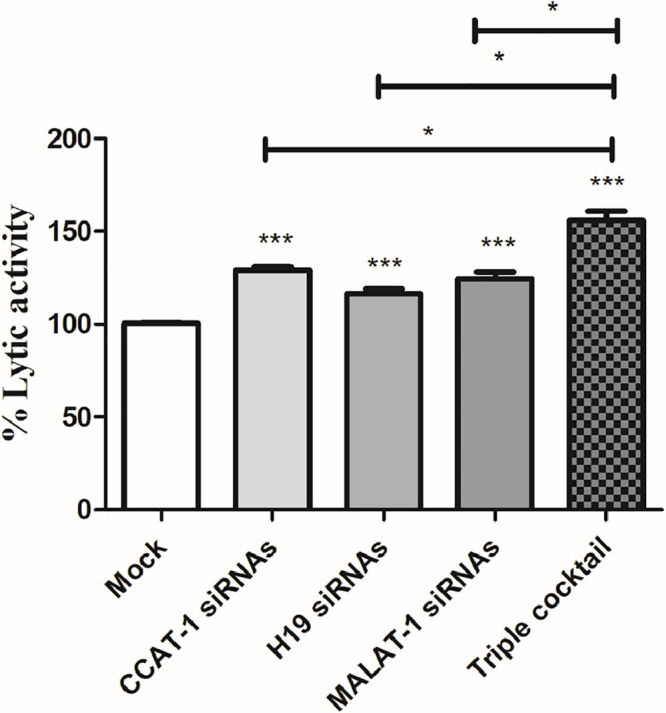
Cytotoxicity of primary healthy PBMCs against CCAT-1, H19, and MALAT-1 silenced Huh7 cell lines. The cytolytic activity of PBMCs was investigated against Huh7 cells silenced with either CCAT-1 or H19 or MALAT-1 separately compared to mock cells. Also, Huh7 cells transfected by triple cocktail were more prone to the lytic potential of primary PBMCs. *p < 0.05, ***p < 0.001.
miR-944–5p and miR-105–5p showed the highest increase in the cytolytic potential of primary PBMCs isolated from HCC patients against Huh7 cells
On another note, it was of great interest to investigate the cytotoxic potential of PBMCs ectopically expressing one of the candidate miRNAs selected in this study. As shown in Fig. 9A, PBMCs transfected with miR-486–5p, miR-506–5p, miR-30a-5p, miR-944, and miR-105–5p showed a significant increase in the cytolytic activity of primary PBMCs isolated from HCC patients against Huh7 cells compared to mock PBMCs (p < 0.001). However, miR-944–5p and miR-105–5p was shown to have the highest cytolytic activity compared to miR-486–5p, miR-506–5p, and miR-30a-5p (p < 0.01). Furthermore, double-handed transfection of primary PBMCs isolated from HCC patients with miR-105–5p and Huh7 cells with the triple liposomal cocktail of siRNAs for CCAT-1, H19, and MALAT-1 led to a more than 100-fold increase in the lytic activity of PBMCs (p < 0.001) compared to mock PBMCs and Huh7 cells.
Fig. 9.
Cytotoxicity of PBMCs transfected with the selected miRNAs against Huh7 cells. (A) Primary PBMCs isolated from HCC patients were transfected with miRNA mimics for each of the selected miRNAs: miR-486–5p, miR-506–5p, miR-30a-5p, miR-944, and miR-105–5p and used to assess for the cytotoxicity against Huh7 cell line.(B) Cytolytic activity was investigated in double-handed transfected primary PBMCs isolated from HCC patients using miR-944 mimics against Huh7 cells transfected using the triple cocktail (CCAT-1, H19, and MALAT-1 siRNAs) and showed a more than 100-fold increase compared to mock cells. *p < 0.05, **p < 0.01, ***p < 0.001.
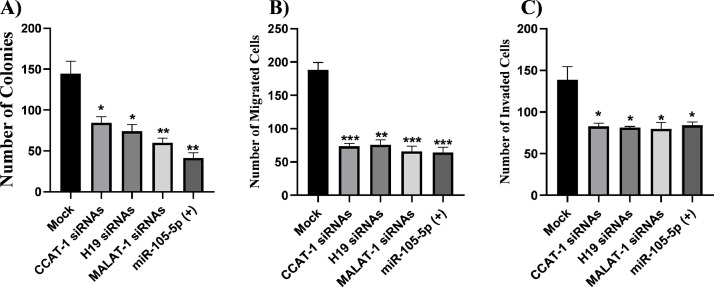
CCAT-1, H19, and MALAT-1 siRNAs and miR-105–5p mimics abrogated the Huh7 colony forming ability, migration and invasion capacities
To validate the therapeutic potential and the long term effects of of CCAT-1, H19, MALAT-1 siRNAs and miR-105–5p mimics on tumor:immune synapse at HCC TME, it was elementary to investigate their impact on Huh7 colony forming ability, cellular migration, and cellular invasion capacities. Knock down of CCAT-1 (p = 0.024), H19 (p = 0.0152), and MALAT-1 (p = 0.0065) significantly decreased the number of colonies in Huh7 cells. Similar results were observed when miR-105–5p was ectopically expressed in Huh7 cells (p = 0.0034; Fig. 10A). Moreover, the cellular migration and invasion of Huh7 cells were significantly halted by siRNAs against CCAT-1 (p = 0.0006 and p = 0.0260, respectively), H19 (p = 0.0011 and p = 0.0223, respectively), and MALAT-1 (p = 0.0008 and p = 0.0282, respectively), and miR-105–5p mimics (p = 0.0008 and p = 0.0283, respectively; Fig. 10B–C).
Fig. 10.
CCAT-1, H19, and MALAT-1 siRNAs and miR-105–5p mimics attenuated the colony forming ability, cellular migration, and cellular invasion capacities in Huh7 cells. (A) Huh7 cells were transfected with CCAT-1, H19, MALAT-1 siRNAs and miR-105–5p to validating their impact on colony-forming ability. Knock down of CCAT-1, H19, and MALAT-1 significantly decreased the number of colonies in Huh7 cells. Similar results were observed when miR-105–5p was ectopically expressed in Huh7 cells. (B) Huh7 cells were transfected with CCAT-1, H19, MALAT-1 siRNAs and miR-105–5p to validating their impact on cellular migration. The cellular migration of Huh7 cells was significantly halted by siRNAs against CCAT-1, H19, and MALAT-1, and miR-105–5p mimics. (C) Huh7 cells were transfected with CCAT-1, H19, MALAT-1 siRNAs and miR-105–5p to validating their impact on cellular invasion. The cellular invasion of Huh7 cells was significantly halted by siRNAs against CCAT-1, H19, and MALAT-1, and miR-105–5p mimics. *p < 0.05, **p < 0.01, ***p < 0.001.
Discussion
HCC tumors have a unique nature of their TME, several convoluted tactics of immune escape phenomena orchestrated by the HCC tumor cells have been validated. In this study, the authors shed the light on the tumor-immune synapse at the TME of HCC patients to unravel the immune evasion tactics orchestrated by HCC tumor cells. In addition, we proposed an epigenetic-immune modulatory strategy to re-sensitize the immune cells to eradicate HCC tumor cells.
Immune checkpoints such as PD-1 and TIGIT are co-inhibitory molecules expressed by effector immune cells to prevent their overactivation. HCC cancer cells exploit this physiological phenomenon to evade immune surveillance phenomena by expressing the corresponding ligands such as PDL1 and CD155 on tumor cells at the TME. Such immune checkpoints axes inhibit immune effector functions and lead to exhaustion of immune cells’ screening and killing capacities [62]. The results showed that PD-L1 and CD155 are highly upregulated in HCC tissues compared to normal tissues with more than 15 folds and 12 folds increase HCC tissues, respectively. This goes in line with a previous study highlighting the immune evasion properties of HCC patients through the induced expression of immune checkpoints ligands [63]. Another recent study reported that PD-L1 is highly upregulated in Chinese HBV-induced HCC patients [64] which underscores the importance of PD-L1 elevation in hampering the immune evasion status of HCC regardless the etiology of the disease. It is also worth mentioning that several studies predict PD-L1 over-expression as a poor prognostic factor for HCC patients [65,66].
On the other hand, CD155 over-expression has also been recently reported in several hematological and solid malignancies such as malignant glioma, colorectal carcinoma, myeloid leukemias, and neuroblastoma, whereas its expression is low or absent in most healthy tissues [67], [68], [69], [70], [71]. Nonetheless, a study has recently unraveled the elevated expression of CD155 in the cancerous tissues of Chinese HBV-induced HCC patients compared to the adjacent healthy tissues [72]. However, to the best of our knowledge, this study is the first to unravel the expression of CD155 in liver tissues resected from Egyptian HCV-induced HCC patients. Previously, CD155 overexpression has been directly linked to induced tumor cell migration, invasion, and proliferation capacities and was associated with a poor prognosis and enhanced tumor progression [73,74].
From the immune side, PD-1 and TIGIT were found to be significantly upregulated in HCC patients while being downregulated in HCV patients compared to healthy controls. This goes in line with a recent study reporting that the co-elevated expression of TIGIT and PD-1 on circulating immune cells is negatively correlated with immune cells exhaustion and reduced overall survival rate and progression-free survival rates in patients with HBV-induced HCC patients [75]. A recent report suggested that a single CD-155/TIGIT or PD-1/PD-L1 blockade has limited anti-tumor efficacy [76]. Yet, pre-clinical studies indicate that co-blockade of CD-155/TIGIT and PD-1/PD-L1 pathway leads to efficient tumor immune eradication even in anti-PD-1 resistant tumor models [77,78]. This is mainly attributed to the unique consequences of TIGIT blockade as it does not only target anti-tumor effector T-cell responses, but also an induction in NK-cell killing capacities and reduction in the suppressive capacity of regulatory T cells. For that reason, we ought to reveal the mechanism underpinning ncRNA-mediated immuno-modulation of the PD-1/PD-L1 and TIGIT/CD-155 axes. In order to address both sides of the coin, we performed a detailed literature survey and bioinformatic analysis to choose an array of ncRNAs that can concurrently target PD-L1 and CD-155 on tumor cells and PD-1 and TIGIT on effect or immune cells. The lncRNAs expected to target PD-L1 and CD-155 transcripts were CCAT-1, H19, and MALAT-1. Screening of CCAT-1, H19, and MALAT-1 were found to be significantly upregulated in the sera, PBMCs, and tissues of HCC patients compared to HCV patients and healthy controls. Our screening results are consistent with other studies in which primary HCC tissues had significantly higher expression of CCAT1 than controls [43,79]. Same for MALAT-1 and H19 expression levels, our results were in agreement with prior studies confirming the same screening profile in HCC patients [31,80].
In this study, the aim was extended to unravel the immunological role of MALAT-1, H19 and CCAT-1 lncRNAs in HCC cell lines. The results showed that knocking down of lncRNAs—CCAT-1, MALAT-1, or H19—markedly decreased the co-expression of PD-L1 and CD155. Moreover, this led in a significant increase in primary immune cells cytotoxicity upon co-culturing with transfected Huh7 cells. Our results were consistent with recent reports highlighting the immunomodulatory role of CCAT-1 [81], H19 [32] and MALAT-1 [49] in several solid malignancies.
To modulate the immune side on the tumor-immune synapse, microRNAs; miR-944–5p, miR-105–5p, miR-486–5p, miR-506–5p, and miR-30a-5p were screened and probed for their regulatory role in modulating PD-1 and TIGIT on primary immune effector cells. It is interesting to note that in the sera and tissues of HCC patients, all five miRNAs showed a considerable down regulation. However, the candidate miRNAs were found to be significantly upregulated in HCC patients similar to the pattern seen with PD-1 and TIGIT transcripts in the PBMCs in the same pool of patients. This goes in line with a previous study that reported elevated expression of miR-506–5p, PD-1, and TIGIT in the PBMCs of HCC patients [82]. However, to the best of our knowledge this could be the first study to screen those specific five miRNAs in sera, PBMCs and tissues correlated with PD-1 and TIGIT expression in the same pool of patients.
Mechanistically, ectopic expression of miR-486–5p, miR-506–5p, and miR-30a-5p significantly decreased the transcript levels of PD-1 in PBMCs which is consistent with the aforementioned study reporting that miR-506–5p mimics led to more than 90% decrease in PD-1 expression in the PBMCs of HCC patients [82]. According to another study, the lncRNA KCNQ1OT1, which is produced from tumor exosomes, controls the ubiquitination of PD-L1 through miR-30a-5p/USP22 and inhibits the CD8+ T cell response, hence encouraging immunological escape and progression of colorectal cancer [83]. This draw attention to the possibility that miR-30a-5p could be used as a potential target to alleviate the immune suppression shown in HCC. On the other hand, our findings demonstrated that ectopic expression of miR-944–5p and miR-105–5p in PBMCs dramatically reduced the co-expression of PD-1 and TIGIT. According to Miliotis and Slack, miR-105–5p binds to the 3′UTR of PD-L1 and negatively correlates with its expression. This miRNA is dysregulated in several cancer types. It has also been demonstrated that miR-105–5p controls tumor immunogenicity in gastric cancer [84]. To our knowledge, this is the first investigation into how miR-944–5p and miR-486–5p affect PD-1 regulation. Functional analysis showed that all miRNAs under investigation enhanced the cytotoxic effects of PBMCs against Huh7 cells, possibly by targeting PD-1 and/or TIGIT and restricting the tumor cells from evading the immune system. Interestingly, transfection of miR-105–5p led to a more than 50% increase in the cytotoxic effects of PBMCs. This could be attributed to their combined/synergistic effects on reducing PD-1 and TIGIT levels, which in turn causes to the release of their inhibitory effects on the immunological checkpoints PD-1/PD-L1 and TIGIT/CD155. Our results go in line with a very recent study that concluded that co-blockade of TGIT/PD1 improves functionality of CD8+ cells in HCC patients [85]. Furthermore, it was crucial to validate the anti-cancer properties of CCAT-1, MALAT-1, and H19 siRNAs and miR-105–5p mimics on Huh7 cells. Significant repression of cellular migration, cellular invasion, and colony forming abilities were observed in Huh7 cells. These findings validate the potential deployment of the lncRNAs CCAT-1, MALAT-1, and H19 and miR-105–5p be used as promising theranostic agents for HCC patients.
Conclusion
In this study, the authors shed the light on the tumor-immune synapse in the TME of HCC patients. Through examining an array of ncRNAs targeting PD-1 and TIGIT, PDL1 and CD155, an epigenetic-immune modulation strategy was proposed to re-sensitize the immune cells to kill HCC cells. LncRNAs; CCAT-1, H19, and MALAT-1 and microRNAs; miR-944–5p, miR-105–5p were found to dually target CD-155/TIGIT and PD-1/PD-L1 axes, unleashing the immune brakes employed by HCC cells to escape immune-surveillance.
CRediT authorship contribution statement
Reem A. Assal: Writing – original draft, Methodology, Formal analysis, Data curation. Noha M. Elemam: Writing – review & editing, Validation, Formal analysis. Radwa Y. Mekky: Writing – review & editing, Data curation. Abdelrahman A. Attia: Validation, Methodology, Investigation. Aya Hesham Soliman: Writing – review & editing, Methodology. Asmaa Ibrahim Gomaa: Resources, Methodology. Eleni K. Efthimiadou: Writing – review & editing, Validation. Maria Braoudaki: Writing – review & editing, Validation. Sherif Ashraf Fahmy: Methodology, Resources, Writing – original draft. Rana A. Youness: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Validation, Writing – original draft, Writing – review & editing.
Declaration of competing interest
The authors declare no conflict of interest
References
- 1.Rumgay H., et al. Global burden of primary liver cancer in 2020 and predictions to 2040. J. Hepatol. 2022;77(6):1598–1606. doi: 10.1016/j.jhep.2022.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Villanueva A. Hepatocellular carcinoma. N. Engl. J. Med. 2019;380(15):1450–1462. doi: 10.1056/NEJMra1713263. [DOI] [PubMed] [Google Scholar]
- 3.Rawla P., et al. Update in global trends and aetiology of hepatocellular carcinoma. Contemp. Oncol. (Pozn) 2018;22(3):141–150. doi: 10.5114/wo.2018.78941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hao X., et al. Targeting Immune Cells in the Tumor Microenvironment of HCC: new Opportunities and Challenges. Front. Cell Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.775462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dawoud A., et al. Involvement of CircRNAs in regulating The "New Generation of Cancer Hallmarks": a Special Depiction on Hepatocellular Carcinoma. Crit. Rev. Oncol. Hematol. 2024;196 doi: 10.1016/j.critrevonc.2024.104312. [DOI] [PubMed] [Google Scholar]
- 6.Zhuo Y., Chen Q., Chhatwal J. Changing epidemiology of hepatocellular carcinoma and role of surveillance. Hepatocellular Carcinoma: Translational Precision Medicine Approaches. 2019:53–67. [Google Scholar]
- 7.Akinyemiju T., et al. The Burden of Primary Liver Cancer and Underlying Etiologies From 1990 to 2015 at the Global, Regional, and National Level: results From the Global Burden of Disease Study 2015. JAMa Oncol. 2017;3(12):1683–1691. doi: 10.1001/jamaoncol.2017.3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abdel-Latif M., Youness R.A. Why natural killer cells in triple negative breast cancer? World J. Clin. Oncol. 2020;11(7):464–476. doi: 10.5306/wjco.v11.i7.464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Youness R.A., et al. Heat Shock Proteins: central Players in Oncological and Immuno-Oncological Tracks. Adv. Exp. Med. Biol. 2022 doi: 10.1007/5584_2022_736. [DOI] [PubMed] [Google Scholar]
- 10.Abdel-Latif M., et al. MALAT-1/p53/miR-155/miR-146a ceRNA circuit tuned by methoxylated quercitin glycoside alters immunogenic and oncogenic profiles of breast cancer. Mol. Cell Biochem. 2022;477(4):1281–1293. doi: 10.1007/s11010-022-04378-4. [DOI] [PubMed] [Google Scholar]
- 11.Abdallah R.M., et al. Hindering the Synchronization Between miR-486-5p and H19 lncRNA by Hesperetin Halts Breast Cancer Aggressiveness Through Tuning ICAM-1. AntiCancer Agents Med. Chem. 2022;22(3):586–595. doi: 10.2174/1871520621666210419093652. [DOI] [PubMed] [Google Scholar]
- 12.Elemam N.M., et al. Pharmacogenomic and epigenomic approaches to untangle the enigma of IL-10 blockade in oncology. Expert. Rev. Mol. Med. 2024;26:e1. doi: 10.1017/erm.2023.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abaza T., et al. Emerging Role of Circular RNAs in Hepatocellular Carcinoma Immunotherapy. Int. J. Mol. Sci. 2023;24(22) doi: 10.3390/ijms242216484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.El-Aziz M.K.A., et al. Decoding hepatocarcinogenesis from a noncoding RNAs perspective. J. Cell Physiol. 2023;238(9):1982–2009. doi: 10.1002/jcp.31076. [DOI] [PubMed] [Google Scholar]
- 15.Rahmoon M.A., et al. MiR-615-5p depresses natural killer cells cytotoxicity through repressing IGF-1R in hepatocellular carcinoma patients. Growth Factors. 2017;35(2–3):76–87. doi: 10.1080/08977194.2017.1354859. [DOI] [PubMed] [Google Scholar]
- 16.Fahmy S.A., et al. Molecular Engines, Therapeutic Targets, and Challenges in Pediatric Brain Tumors: a Special Emphasis on Hydrogen Sulfide and RNA-Based Nano-Delivery. Cancers. (Basel) 2022;14(21) doi: 10.3390/cancers14215244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Youness R.A., et al. Contradicting interplay between insulin-like growth factor-1 and miR-486-5p in primary NK cells and hepatoma cell lines with a contemporary inhibitory impact on HCC tumor progression. Growth Factors. 2016;34(3–4):128–140. doi: 10.1080/08977194.2016.1200571. [DOI] [PubMed] [Google Scholar]
- 18.Zhang H., et al. Recombinant adenovirus expressing the fusion protein PD1PVR improves CD8(+) T cell-mediated antitumor efficacy with long-term tumor-specific immune surveillance in hepatocellular carcinoma. Cell Oncol. (Dordr) 2021;44(6):1243–1255. doi: 10.1007/s13402-021-00633-w. [DOI] [PubMed] [Google Scholar]
- 19.Mekky R.Y., et al. MALAT-1: immunomodulatory lncRNA hampering the innate and the adaptive immune arms in triple negative breast cancer. Transl. Oncol. 2023;31 doi: 10.1016/j.tranon.2023.101653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang C., et al. TIGIT Can Exert Immunosuppressive Effects on CD8+ T Cells by the CD155/TIGIT Signaling Pathway for Hepatocellular Carcinoma In Vitro. J. ImmunOther. 2020;43(8):236–243. doi: 10.1097/CJI.0000000000000330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harjunpaa H., Guillerey C. TIGIT as an emerging immune checkpoint. Clin. Exp. Immunol. 2020;200(2):108–119. doi: 10.1111/cei.13407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun H., et al. Human CD96 Correlates to Natural Killer Cell Exhaustion and Predicts the Prognosis of Human Hepatocellular Carcinoma. Hepatology. 2019;70(1):168–183. doi: 10.1002/hep.30347. [DOI] [PubMed] [Google Scholar]
- 23.Rashwan H.H., et al. Harnessing the supremacy of MEG3 LncRNA to defeat gastrointestinal malignancies. Pathol. Res. Pract. 2024;256 doi: 10.1016/j.prp.2024.155223. [DOI] [PubMed] [Google Scholar]
- 24.Youness R.A., et al. Role of Hydrogen Sulfide in Oncological and Non-Oncological Disorders and Its Regulation by Non-Coding RNAs: a Comprehensive Review. Noncoding. RNa. 2024;10(1) doi: 10.3390/ncrna10010007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Elmasri R.A., et al. Puzzling out the role of MIAT LncRNA in hepatocellular carcinoma. Noncoding. RNa Res. 2024;9(2):547–559. doi: 10.1016/j.ncrna.2024.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.ZeinElAbdeen Y.A., AbdAlSeed A., Youness R.A. Decoding Insulin-Like Growth Factor Signaling Pathway From a Non-coding RNAs Perspective: a Step Towards Precision Oncology in Breast Cancer. J. Mammary. Gland. Biol. Neoplasia. 2022;27(1):79–99. doi: 10.1007/s10911-022-09511-z. [DOI] [PubMed] [Google Scholar]
- 27.Dawoud A., et al. Circular RNAs: new layer of complexity evading breast cancer heterogeneity. Noncoding. RNa Res. 2023;8(1):60–74. doi: 10.1016/j.ncrna.2022.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang T., et al. TIGIT/PVR and LncRNA ANRIL dual-targetable PAMAM polymeric nanoparticles efficiently inhibited the hepatoma carcinoma by combination of immunotherapy and gene therapy. J. Drug Target. 2021;29(7):783–791. doi: 10.1080/1061186X.2021.1879088. [DOI] [PubMed] [Google Scholar]
- 29.Yao K., et al. A competing endogenous RNA network identifies novel mRNA, miRNA and lncRNA markers for the prognosis of diabetic pancreatic cancer. Tumour. Biol. 2017;39(6) doi: 10.1177/1010428317707882. [DOI] [PubMed] [Google Scholar]
- 30.SOLIMAN R.-A., et al. Uncoupling tumor necrosis factor-α and interleukin-10 at tumor immune microenvironment of breast cancer through miR-17-5p/MALAT-1/H19 circuit. Biocell. 2022;46(3):769–783. [Google Scholar]
- 31.Lai M.C., et al. Long non-coding RNA MALAT-1 overexpression predicts tumor recurrence of hepatocellular carcinoma after liver transplantation. Med. Oncol. 2012;29(3):1810–1816. doi: 10.1007/s12032-011-0004-z. [DOI] [PubMed] [Google Scholar]
- 32.Elkhouly A., Soliman R.A., Youness R.A. 11P Convoluted role of H19 long non-coding RNA in regulating ICAM-1 and PVR in breast cancer patients. Annals of Oncology. 2021;32:S6. [Google Scholar]
- 33.Soliman R.A., et al. Interplay between miR-17-5p and MALAT-1 shapes the cytokine storm in triple negative breast cancer (TNBC) tumor microenvironment. Annals of Oncology. 2019;30:v769. [Google Scholar]
- 34.Zeinelabdeen Y., Soliman R.A., Youness R.A. 24P miR-17-5p: a potential activator of natural killer cells through tuning STAT3/H19/ULBP2 axis in breast cancer. Annals of Oncology. 2021;32:S10–S11. [Google Scholar]
- 35.Soliman A.H., et al. Phytochemical-derived tumor-associated macrophage remodeling strategy using Phoenix dactylifera L. boosted photodynamic therapy in melanoma via H19/iNOS/PD-L1 axis. Photodiagnosis. Photodyn. Ther. 2023;44 doi: 10.1016/j.pdpdt.2023.103792. [DOI] [PubMed] [Google Scholar]
- 36.Rojas A., et al. Long non-coding RNA H19 as a biomarker for hepatocellular carcinoma. Liver. Int. 2022;42(6):1410–1422. doi: 10.1111/liv.15230. [DOI] [PubMed] [Google Scholar]
- 37.Tietze L., Kessler S.M. The Good, the Bad, the Question-H19 in Hepatocellular Carcinoma. Cancers. (Basel) 2020;12(5) doi: 10.3390/cancers12051261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li X., et al. Cholangiocyte-derived exosomal long noncoding RNA H19 promotes cholestatic liver injury in mouse and humans. Hepatology. 2018;68(2):599–615. doi: 10.1002/hep.29838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raveh E., et al. The H19 Long non-coding RNA in cancer initiation, progression and metastasis - a proposed unifying theory. Mol. Cancer. 2015;14:184. doi: 10.1186/s12943-015-0458-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Selem N.A., Youness R.A., Gad M.Z. What is beyond LncRNAs in breast cancer: a special focus on colon cancer-associated Transcript-1 (CCAT-1) Noncoding. RNa Res. 2021;6(4):174–186. doi: 10.1016/j.ncrna.2021.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Selem N.A., et al. Let-7a/cMyc/CCAT1/miR-17-5p Circuit Re-sensitizes Atezolizumab Resistance in Triple Negative Breast Cancer through Modulating PD-L1. Pathol. Res. Pract. 2023;248 doi: 10.1016/j.prp.2023.154579. [DOI] [PubMed] [Google Scholar]
- 42.Deng L., et al. Long noncoding RNA CCAT1 promotes hepatocellular carcinoma progression by functioning as let-7 sponge. J. Exp. Clin. Cancer Res. 2015;34(1):18. doi: 10.1186/s13046-015-0136-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhu H., et al. CCAT1 promotes hepatocellular carcinoma cell proliferation and invasion. Int. J. Clin. Exp. Pathol. 2015;8(5):5427–5434. [PMC free article] [PubMed] [Google Scholar]
- 44.Youness R.A., Gad M.Z. Long non-coding RNAs: functional regulatory players in breast cancer. Noncoding. RNa Res. 2019;4(1):36–44. doi: 10.1016/j.ncrna.2019.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.ElKhouly A.M., Youness R.A., Gad M.Z. MicroRNA-486-5p and microRNA-486-3p: multifaceted pleiotropic mediators in oncological and non-oncological conditions. Noncoding. RNa Res. 2020;5(1):11–21. doi: 10.1016/j.ncrna.2020.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Youssef S.S., et al. miR-516a-3P, a potential circulating biomarker in hepatocellular carcinoma, correlated with rs738409 polymorphism in PNPLA3. Per. Med. 2022;19(6):483–493. doi: 10.2217/pme-2022-0005. [DOI] [PubMed] [Google Scholar]
- 47.El Din G.S., et al. miRNA-506-3p Directly Regulates rs10754339 (A/G) in the Immune Checkpoint Protein B7-H4 in Breast Cancer. Microrna. 2020;9(5):346–353. doi: 10.2174/2211536609666201209152949. [DOI] [PubMed] [Google Scholar]
- 48.Youness R.A., et al. MicroRNA-486-5p enhances hepatocellular carcinoma tumor suppression through repression of IGF-1R and its downstream mTOR, STAT3 and c-Myc. Oncol. Lett. 2016;12(4):2567–2573. doi: 10.3892/ol.2016.4914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ramzy A., et al. 40P MALAT-1/miR-30a-5p competing endogenous (ceRNA) network releases the brakes of immune surveillance in breast cancer through its quadruple targets: PD-L1, MIF, IL-10 and TNF-α. Annals of Oncology. 2021;32:S1357–S1358. [Google Scholar]
- 50.Lin J., et al. Circ_0031242 regulates the functional properties of hepatocellular carcinoma cells through the miR-944/MAD2L1 axis. Histol. Histopathol. 2022:18519. doi: 10.14670/HH-18-519. [DOI] [PubMed] [Google Scholar]
- 51.Fang S.S., et al. A P53-related microRNA model for predicting the prognosis of hepatocellular carcinoma patients. J. Cell Physiol. 2020;235(4):3569–3578. doi: 10.1002/jcp.29245. [DOI] [PubMed] [Google Scholar]
- 52.El Kilany F.H., et al. miR-744/eNOS/NO axis: a novel target to halt triple negative breast cancer progression. Breast. Dis. 2021;40(3):161–169. doi: 10.3233/BD-200454. [DOI] [PubMed] [Google Scholar]
- 53.Nafea H., et al. LncRNA HEIH/miR-939-5p interplay modulates triple-negative breast cancer progression through NOS2-induced nitric oxide production. J. Cell Physiol. 2021;236(7):5362–5372. doi: 10.1002/jcp.30234. [DOI] [PubMed] [Google Scholar]
- 54.Youness R.A., et al. The long noncoding RNA sONE represses triple-negative breast cancer aggressiveness through inducing the expression of miR-34a, miR-15a, miR-16, and let-7a. J. Cell Physiol. 2019;234(11):20286–20297. doi: 10.1002/jcp.28629. [DOI] [PubMed] [Google Scholar]
- 55.Mekky R.Y., et al. Epigallocatechin gallate (EGCG) and miR-548m reduce HCV entry through repression of CD81 receptor in HCV cell models. Arch. Virol. 2019;164(6):1587–1595. doi: 10.1007/s00705-019-04232-x. [DOI] [PubMed] [Google Scholar]
- 56.Youness R.A., et al. A novel role of sONE/NOS3/NO signaling cascade in mediating hydrogen sulphide bilateral effects on triple negative breast cancer progression. Nitric. Oxide. 2018;80:12–23. doi: 10.1016/j.niox.2018.07.004. [DOI] [PubMed] [Google Scholar]
- 57.Youssef S.S., et al. PNPLA3 and IL 28B signature for predicting susceptibility to chronic hepatitis C infection and fibrosis progression. Arch. Physiol. Biochem. 2022;128(2):483–489. doi: 10.1080/13813455.2019.1694039. [DOI] [PubMed] [Google Scholar]
- 58.Ahmed Youness R., et al. A methoxylated quercetin glycoside harnesses HCC tumor progression in a TP53/miR-15/miR-16 dependent manner. Nat. Prod. Res. 2020;34(10):1475–1480. doi: 10.1080/14786419.2018.1509326. [DOI] [PubMed] [Google Scholar]
- 59.Aboelenein H.R., et al. Reduction of CD19 autoimmunity marker on B cells of paediatric SLE patients through repressing PU.1/TNF-α/BAFF axis pathway by miR-155. Growth Factors. 2017;35(2–3):49–60. doi: 10.1080/08977194.2017.1345900. [DOI] [PubMed] [Google Scholar]
- 60.Youness R.A., et al. Targeting hydrogen sulphide signaling in breast cancer. J. Adv. Res. 2021;27:177–190. doi: 10.1016/j.jare.2020.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.El-Shafie S., et al. Encapsulation of Nedaplatin in Novel PEGylated Liposomes Increases Its Cytotoxicity and Genotoxicity against A549 and U2OS Human Cancer Cells. Pharmaceutics. 2020;12(9) doi: 10.3390/pharmaceutics12090863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sangro B., et al. Advances in immunotherapy for hepatocellular carcinoma. Nat. Rev. Gastroenterol. Hepatol. 2021;18(8):525–543. doi: 10.1038/s41575-021-00438-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tan Y., et al. Overexpression of PD-L1 is an Independent Predictor for Recurrence in HCC Patients Who Receive Sorafenib Treatment After Surgical Resection. Front. Oncol. 2022;11 doi: 10.3389/fonc.2021.783335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xu H., et al. The landscape of PD-L1 expression and somatic mutations in hepatocellular carcinoma. J. Gastrointest. Oncol. 2021;12(3):1132–1140. doi: 10.21037/jgo-21-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Winograd P., et al. Hepatocellular Carcinoma-Circulating Tumor Cells Expressing PD-L1 Are Prognostic and Potentially Associated With Response to Checkpoint Inhibitors. Hepatol. Commun. 2020;4(10):1527–1540. doi: 10.1002/hep4.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhu A.X., et al. Pembrolizumab in patients with advanced hepatocellular carcinoma previously treated with sorafenib (KEYNOTE-224): a non-randomised, open-label phase 2 trial. Lancet Oncol. 2018;19(7):940–952. doi: 10.1016/S1470-2045(18)30351-6. [DOI] [PubMed] [Google Scholar]
- 67.Carlsten M., et al. Primary human tumor cells expressing CD155 impair tumor targeting by down-regulating DNAM-1 on NK cells. J. Immunol. 2009;183(8):4921–4930. doi: 10.4049/jimmunol.0901226. [DOI] [PubMed] [Google Scholar]
- 68.Castriconi R., et al. Natural killer cell-mediated killing of freshly isolated neuroblastoma cells: critical role of DNAX accessory molecule-1-poliovirus receptor interaction. Cancer Res. 2004;64(24):9180–9184. doi: 10.1158/0008-5472.CAN-04-2682. [DOI] [PubMed] [Google Scholar]
- 69.Gromeier M., et al. Intergeneric poliovirus recombinants for the treatment of malignant glioma. Proc. Natl. Acad. Sci. u S. a. 2000;97(12):6803–6808. doi: 10.1073/pnas.97.12.6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Masson D., et al. Overexpression of the CD155 gene in human colorectal carcinoma. Gut. 2001;49(2):236–240. doi: 10.1136/gut.49.2.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pende D., et al. Analysis of the receptor-ligand interactions in the natural killer-mediated lysis of freshly isolated myeloid or lymphoblastic leukemias: evidence for the involvement of the Poliovirus receptor (CD155) and Nectin-2 (CD112) Blood. 2005;105(5):2066–2073. doi: 10.1182/blood-2004-09-3548. [DOI] [PubMed] [Google Scholar]
- 72.Duan X., et al. Expression of TIGIT/CD155 and correlations with clinical pathological features in human hepatocellular carcinoma. Mol. Med. Rep. 2019;20(4):3773–3781. doi: 10.3892/mmr.2019.10641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nishiwada S., et al. Clinical significance of CD155 expression in human pancreatic cancer. Anticancer Res. 2015;35(4):2287–2297. [PubMed] [Google Scholar]
- 74.Sloan K.E., et al. CD155/PVR enhances glioma cell dispersal by regulating adhesion signaling and focal adhesion dynamics. Cancer Res. 2005;65(23):10930–10937. doi: 10.1158/0008-5472.CAN-05-1890. [DOI] [PubMed] [Google Scholar]
- 75.Yu L., et al. TIGIT(+) TIM-3(+) NK cells are correlated with NK cell exhaustion and disease progression in patients with hepatitis B virusrelated hepatocellular carcinoma. Oncoimmunology. 2021;10(1) doi: 10.1080/2162402X.2021.1942673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Selem N.A., et al. Let-7a/cMyc/CCAT1/miR-17-5p Circuit Re-sensitizes Atezolizumab Resistance in Triple Negative Breast Cancer through Modulating PD-L1. Pathology - Research and Practice. 2023 doi: 10.1016/j.prp.2023.154579. [DOI] [PubMed] [Google Scholar]
- 77.Ge Z., et al. TIGIT, the Next Step Towards Successful Combination Immune Checkpoint Therapy in Cancer. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.699895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chiang E.Y., Mellman I. TIGIT-CD226-PVR axis: advancing immune checkpoint blockade for cancer immunotherapy. J. ImmunOther Cancer. 2022;10(4) doi: 10.1136/jitc-2022-004711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhu H.Q., et al. Aberrant Expression of CCAT1 Regulated by c-Myc Predicts the Prognosis of Hepatocellular Carcinoma. Asian Pac. J. Cancer Prev. 2015;16(13):5181–5185. doi: 10.7314/apjcp.2015.16.13.5181. [DOI] [PubMed] [Google Scholar]
- 80.Rojas Á., et al. Long non-coding RNA H19 as a biomarker for hepatocellular carcinoma. Liver. Int. 2022;42(6):1410–1422. doi: 10.1111/liv.15230. [DOI] [PubMed] [Google Scholar]
- 81.Selem N., et al. 32P Immunoregulatory loop between let-7a and CCAT1 lncRNA coordinated by c-Myc underlies the PD-1/PD-L1 immunoresistance in triple negative breast cancer patients. Ann. Oncol. 2021;32:S1355. [Google Scholar]
- 82.Zhong S., et al. miRNAs in lung cancer. A systematic review identifies predictive and prognostic miRNA candidates for precision medicine in lung cancer. Transl. Res. 2021;230:164–196. doi: 10.1016/j.trsl.2020.11.012. [DOI] [PubMed] [Google Scholar]
- 83.Xian D., et al. LncRNA KCNQ1OT1 Secreted by Tumor Cell-Derived Exosomes Mediates Immune Escape in Colorectal Cancer by Regulating PD-L1 Ubiquitination via MiR-30a-5p/USP22. Front. Cell Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.653808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Miliotis C., Slack F.J. miR-105-5p regulates PD-L1 expression and tumor immunogenicity in gastric cancer. Cancer Lett. 2021;518:115–126. doi: 10.1016/j.canlet.2021.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ge Z., et al. TIGIT and PD1 Co-blockade Restores ex vivo Functions of Human Tumor-Infiltrating CD8+ T Cells in Hepatocellular Carcinoma. Cell Mol. Gastroenterol. Hepatol. 2021;12(2):443–464. doi: 10.1016/j.jcmgh.2021.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]