Abstract
We have previously shown that the nonconserved carboxy-terminal exon of the adenovirus type 2 E1A-289R protein contains two interchangeable sequence elements, auxiliary region (AR) 1 and AR2, that are required for efficient CR3-mediated transcriptional activation of the viral E4 promoter (M. Bondesson, C. Svensson, S. Linder, and G. Akusjärvi, EMBO J. 11:3347–3354, 1992). Here we show that CR3-mediated transactivation of all adenovirus early promoters and the HSP70 promoter requires the AR1 element. We further show that AR2 can substitute for AR1 only when artificially juxtaposed to CR3. AR1 consists of six tandem glutamic acid-proline (EP) repeats and is positioned immediately downstream of CR3. Genetic dissection of AR1 showed that the number of EP repeats in AR1 is critical for CR3 function. Thus, reducing or increasing the number of EP repeats reduces the CR3 transactivation capacity. Furthermore, the introduction of amino acid substitutions into AR1 suggested that the net negative charge in AR1 is of critical importance for its function as an enhancer of CR3-mediated transcriptional activation. Using an in vitro binding approach, we showed that the AR1 element is not part of the CR3 promoter localization signal mediating contact with the Sp1, ATF-2, or c-Jun upstream-binding transcription factors. Previous studies have suggested that the 49-amino-acid sequence constituting CR3 represents the minimal domain required for E1A-induced activation of viral early promoters. Since AR1 was required for efficient CR3-mediated transcriptional activation of all tested promoters, we suggest that the carboxy-terminal boundary for the CR3 transactivation domain should be extended to include the AR1 element.
The adenovirus E1A gene encodes a family of structurally related proteins that are required to activate all viral early promoters during lytic virus growth (for reviews, see references 11 and 29). The two major E1A proteins, of 289 and 243 amino acids (E1A-289R and E1A-243R, respectively), are translated from the two most abundant E1A mRNAs, the 13S and 12S mRNAs, respectively. The E1A-289R protein contains three conserved amino acid regions, designated CR1, CR2, and CR3 (18). Genetic studies have shown that these regions are responsible for most of the activities ascribed to E1A in transcriptional control and transformation (3, 29).
E1A activates transcription through several different mechanisms which involve all three conserved domains (for a review, see reference 2). However, the classical E1A transcription activation domain is contained within CR3 and is therefore unique to the E1A-289R protein. This domain has been shown to participate in transcription activation by physically interacting with both basal (i.e., TATA-binding protein [TBP]) and upstream-binding transcription factors such as the ATF family of transcription factors, c-Jun, Sp1, upstream stimulatory factor (USF), CCAAT box binding factor, and Oct4/Oct3 (1, 7, 13, 22, 23, 28). In addition, it has been demonstrated that several TBP-associated factors (Drosophila TAF110 and human TAF135, TAF250, and TAF55) complex with the CR3 region of E1A (8, 12, 24, 25). It has been suggested that once E1A is anchored to the promoter region via the promoter-targeting signal located at the carboxy terminus of CR3, the amino-terminal activation domain of CR3 triggers preinitiation complex formation through an interaction with TBP (20, 22, 23, 35). E1A would then make simultaneous contact with sequence-specific DNA-binding transcription factors and the basal transcription machinery. E1A-289R also activates transcription by inducing phosphorylation of certain cellular upstream-binding transcription factors, such as E4F (10, 26) and TFIIIC (17).
We previously showed that efficient E1A-289R transactivation of the viral E4 promoter requires one of the two interchangeable auxiliary regions encoded by the nonconserved C-terminal exon of E1A (designated AR1 and AR2 in reference 5). Here we show that AR1 is of general importance, being required for CR3-induced activation of all tested promoters. Thus, we propose that the carboxy-terminal boundary for the minimal CR3 transactivation domain should be extended to include AR1. A genetic dissection of the adenovirus type 2 (Ad2) AR1 element (six glutamic acid-proline [EP] repeats) demonstrated that the primary sequence of AR1 is not important for its function. Thus, changing EP repeats to DP repeats resulted in a functional E1A transactivator protein, whereas replacement of EP repeats with AP or KP repeats inactivated AR1. Furthermore, the prolines in the EP repeat can be replaced with glycine residues without having much of an effect on AR1 function. Most likely, the significant feature of AR1 is its presentation of a net negative charge. Although AR1 is juxtaposed to the CR3 promoter localization signal in the wild-type protein, it was found not to be required for E1A interaction with cellular upstream-binding transcription factors in vitro in glutathione S-transferase (GST) pull-down experiments.
MATERIALS AND METHODS
Plasmid DNA.
Plasmids pKGO-007SVRI (33) (referred to as wtE1A in Fig. 2 and 3) and E1A deletion mutants D̂X and D̂Dr (21), E/P−, and 6E/P (previously named E/P+ in reference 5) have been described previously. Mutants 4E/P, 8E/P, and 10E/P, which contain a reduced or increased number of EP repeats compared to the wild-type E1A protein (which has six EP repeats), were generated by PCR amplification with appropriately designed primers. In 4E/P, wtE1A residues 197 to 245 were replaced with the sequence ARG; in 8E/P and 10E/P, residues 201 to 245 were replaced with EPEPARG and EPEPEPEPARG, respectively. The PCR mutants 4D/P, 4E/G, 4A/P, and 4K/P contain amino acid substitutions in AR1. In 4D/P, wtE1A residues 193 to 245 were replaced with DPDPDPDPARG; in 4E/G, residues 193 to 245 were replaced with EGEGEGE GARG; and in 4A/P and 4K/P, residues 193 to 245 were replaced with APA PAPAPARG and KPKPKPKPARG, respectively. In AR1mut, residues 193 to 203 were replaced with APAPAPAPARG, generating a protein containing a mutated AR1 region in the context of the full-length E1A-289R protein.
FIG. 2.
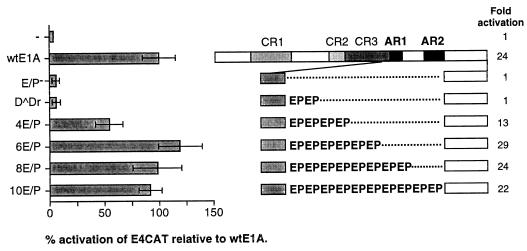
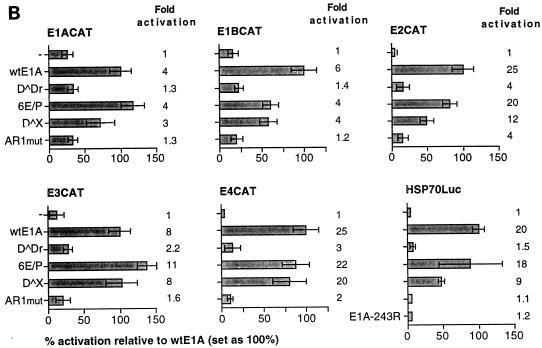
The number of EP repeats in AR1 is critical for function. HeLa cells were cotransfected with an Ad5 E4CAT reporter plasmid and either wtE1A or plasmids encoding E1A mutant proteins (schematically illustrated to the right). Relative CAT activities, based on the mean values of data from at least three independent experiments, are shown as percentages of wtE1A activation (set at 100%). Error bars show the standard derivations. The fold activation relative to basal reporter activity is marked to the right of each E1A protein.
FIG. 3.
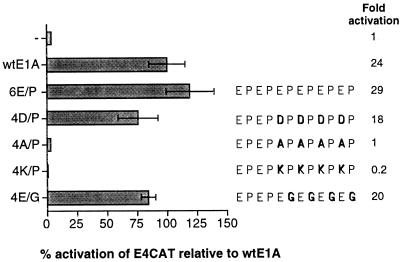
The negative charge of AR1 is important for its function. Levels of CAT activity in HeLa cells cotransfected with the E4CAT reporter and E1A activator plasmids were determined. Relative CAT activities, based on the mean values of data from at least three independent experiments, are shown as percentages of wtE1A activation (set at 100%). Error bars show the standard derivations. The fold activation relative to the basal reporter activity is marked to the right of each E1A mutant protein. The various conservative or nonconservative amino acid substitutions in the six EP repeats are schematically shown in boldface.
Mutants E/P−, 6E/P, and D̂X were also introduced into vector pML005 (referred to as wtE1A in Fig. 1). This plasmid is identical to pKGO-007SVRI (33) except that it lacks the E1B coding sequences and the simian virus 40 enhancer (4). The cDNA clone encoding the E1A-243R protein (previously named pML00512S) has been described elsewhere (30). The reporter plasmids E1ACAT (15), E1BCAT (9), E2CAT(p2CAT) (36), E3CAT (pKCAT23) (36), and E4CAT (20) have previously been described. The reporter plasmid HSP70Luc (kindly provided by C. Svensson) consists of the HSP70 promoter fused to a luciferase reporter gene.
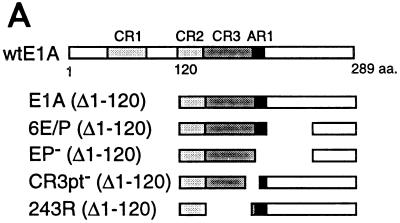
FIG. 1.
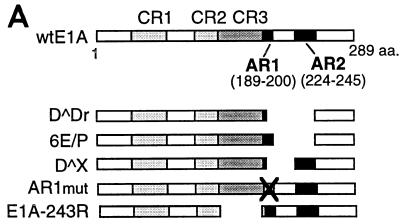
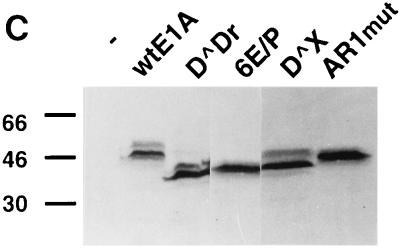
AR1 is essential for E1A transactivation of early viral promoters. (A) A schematic representation of the E1A deletions mutants. (B) Relative CAT and luciferase (HSP70) activities, based on the mean values of data from at least three independent experiments, are shown as percentages of wtE1A activation (set at 100%) with the respective reporter plasmids. Error bars show the standard derivations. The fold activation values relative to the basal reporter activity are marked to the right of each panel. (C) Western blot showing E1A protein expression in transfected HeLa cells. The positions of protein molecular size standards are marked on the left side (in kilodaltons).
GST/Sp1(DBD) (amino acids 612 to 778) was generated by deleting the BamHI fragment encoding Sp1 amino acids 1 to 611 from GST/Sp1 (31). Full-length cDNAs encoding c-Jun and ATF-2 were inserted into GST gene fusion vectors (Pharmacia), generating GST/c-Jun and GST/ATF-2.
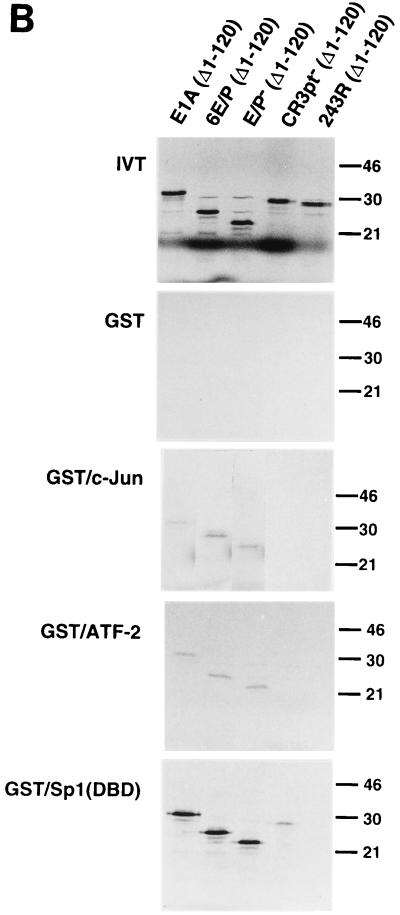
T7-E1A (Δ1–120), T7-6E/P (Δ1–120), T7-E/P− (Δ1–120), T7-243R (Δ1–120), and T7-CR3pt− (Δ1–120), encoding amino-terminally truncated E1A proteins, were generated by cDNA cloning, using an RNA PCR kit from Perkin-Elmer, or by standard PCR amplification with appropriately designed primers. The cDNAs were inserted into the pRSET-B vector (Invitrogen). The T7-CR3pt− (Δ1–120) mutant, which bears a deletion of 15 amino acids (CGMFVYSPVSEPEPE) covering the CR3 promoter-targeting region, was generated from E1a(Δ179–193) (22).
Transfections and CAT assay.
HeLa cells were maintained in Dulbecco modified Eagle medium supplemented with 10% newborn calf serum. One hour before the cells were transfected, the medium was changed to Dulbecco modified Eagle medium containing 10% fetal calf serum. Cells were cotransfected with 1 μg of reporter plasmid and 0.1 to 1 μg of activator plasmid by the calcium phosphate coprecipitation technique (37). The amount of E1A activator plasmid was titrated so that equal E1A protein expression was detected by Western blot analysis. Therefore, wild-type E1A (wtE1A), 8E/P, and 10E/P were transfected with 0.1 μg of plasmid DNA, whereas all other E1A mutants were transfected with 1 μg. pUC19 DNA was added to a total of 15 μg of DNA per 60-mm-diameter dish. Eight to 12 h after transfection, cells were treated with 15% glycerol in HBS buffer (160 mM NaCl, 25 mM HEPES [pH 7.1], and 0.75 mM NH2PO4). Cells were harvested 48 h posttransfection and lysed by three consecutive freeze-thawing cycles, and the extract was used to measure chloramphenicol acetyltransferase (CAT) activity (32). Quantitative results were obtained by PhosphorImager scanning.
Transfections and luciferase assay.
The effect of E1A mutants on HSP70 gene transcription was assayed on an HSP70-luciferase reporter plasmid (HSP70Luc). HeLa cells were grown in 35-mm-diameter petri dishes and cotransfected with 0.05 μg of reporter plasmid and 0.05 or 0.1 μg of activator plasmid, using the FuGENE6 transfection reagent (Boehringer-Mannheim). Forty-eight hours posttransfection, the cells were lysed and luciferase activity was assayed by using a luciferase assay system (Promega) as described by the manufacturer. Luciferase emissions were quantified with a luminometer (Labsystems Luminoscan).
GST protein binding analyses.
35S-labelled E1A proteins were synthesized by using the T7-E1A variant plasmids described above in a coupled in vitro transcription-translation reticulocyte lysate system (Promega). Synthesis conditions were those recommended by the manufacturer, except that 100 μM ZnCl2 was included. GST fusion proteins were expressed in Escherichia coli and bound to glutathione-Sepharose beads. Protein concentrations were estimated from Coomassie-stained sodium dodecyl sulfate (SDS)-polyacrylamide gels. Approximately 10 μg of each GST fusion protein was incubated with 10 μl of precleared 35S-labelled E1A protein. The binding reactions were performed in a total volume of 50 μl in binding buffer (10 mM Tris, 0.15 M NaCl, 1.5 mM MgCl2, 0.1% Nonidet P-40, 0.1 mM ZnCl2, and 20 mM β-mercaptoethanol) for 1 h on ice. The beads were washed three times with 1 ml of binding buffer, and interacting proteins were separated on an SDS–12% polyacrylamide gel and visualized by autoradiography.
Analysis of E1A protein expression by Western blotting.
Approximately one-third of each supernatant extract prepared from transfected cells (see above) were separated on an SDS–12% polyacrylamide gel and transferred to a Protran BA85 membrane (Schleicher & Schuell). Blots were probed with M73 monoclonal antibody (diluted 1:1,000; Oncogene Research Products), directed against the E1A carboxy terminus. E1A proteins were detected by the enhanced chemiluminescence technique as described by the manufacturer (New England BioLabs).
RESULTS
AR1 is essential for efficient E1A transactivation of early viral promoters.
It has previously been demonstrated that two interchangeable elements, AR1 and AR2, are essential for efficient CR3-mediated transcriptional activation of the viral E4 promoter (5). Here, we analyzed whether the AR elements are of general importance for E1A-289R transactivation of other adenovirus promoters. Therefore, the transactivation capacity of E1A mutant proteins (Fig. 1A), lacking one or both AR elements, was measured in a transient transfection assay in HeLa cells. As a reporter we used the CAT gene cloned under the transcriptional control of one of the adenovirus early promoters, E1A, E1B, E2, E3, or E4. To obtain accurate results reflecting the transactivation capacity of E1A wild-type and mutant proteins, much care was taken to normalize E1A protein expression by titrating the amount of plasmid required for equal E1A protein expression (Fig. 1C) (see Materials and Methods). In this and all subsequent experiments, the levels of E1A protein expression in transfected cells were similar.
As shown in Fig. 1B, the effect of wtE1A (pML005) cotransfection varied among the promoters tested. Thus, E1ACAT was the least responsive of the promoters (4-fold activation), and E2CAT and E4CAT were the most responsive (25-fold activation). Deletion of both AR elements (D̂Dr) resulted in a severe impairment of the capacity of E1A to activate transcription on all reporter constructs. The residual E1A transactivation capacity on the E1ACAT and E1BCAT reporter constructs was negligible. In contrast, cotransfection of plasmids encoding E1A mutant proteins that contain either AR1 (6E/P) or AR2 (D̂X) restored the activation capacity of E1A on all promoters tested. AR1 was consistently shown to rescue transactivation more efficiently than AR2, and in most cases AR1 was sufficient to restore wild-type E1A activity.
A question not answered in our previous work (5) was whether AR2 also functionally substitutes for AR1 at its natural position. Results of our present study showed that AR2 efficiently substitutes for AR1 when artificially juxtaposed to the CR3 domain (D̂X, Fig. 1B). AR2 has been mapped to a 23-amino-acid serine-threonine-rich region (5) and contains a major E1A phosphorylation site, serine 231 (34). To determine whether AR2 is also a functional homolog of AR1 in the context of a full-length protein, we destroyed AR1 in the wild-type protein by replacing four of the six EP repeats with AP repeats (see below). As shown in Fig. 1B, the E1A mutant protein AR1mut did not efficiently rescue E1A transactivation at the early viral promoters. In fact, AR1mut showed a phenotype similar to that of D̂Dr, which lacks both AR elements (Fig. 1B). Thus, we conclude that AR1 is the critical element in the nonconserved C-terminal exon enhancing CR3-mediated transactivation. Since AR1 is a necessary element required for efficient CR3 transactivation of all early viral promoters, we propose that it should be regarded as an integral part of the CR3 transactivation domain. Thus, the CR3 transactivation domain should be expanded to include AR1.
AR1 is essential for E1A-CR3 transactivation of the cellular HSP70 promoter.
To determine whether AR1 is of general significance for CR3-mediated transactivation, the capacity of E1A mutant proteins (Fig. 1A) to activate transcription of the HSP70Luc reporter plasmid was tested. This promoter has previously been shown to be activated by both the E1A-289R and E1A-243R proteins (1, 19, 38). However, under our experimental conditions, the E1A-243R protein did not activate HSP70 transcription (Fig. 1B). In contrast, wtE1A (pML005) cotransfection resulted in an approximately 20-fold stimulation of HSP70 transcription. Deletion of both AR elements (D̂Dr) completely abolished E1A activation. Cotransfection of plasmids encoding AR1 (6E/P) or AR2 (D̂X) restored the activation capacity of E1A, with AR1 rescuing transactivation with a much higher efficiency. Similar to our findings with the viral promoters, AR2 does not functionally substitute for AR1 at its natural position (AR1mut).
The number of EP repeats in AR1 is critical for E1A transactivation.
To define the significant functional feature of AR1 for E1A transcriptional activation, mutant plasmids expressing E1A proteins with reduced or increased EP repeat length were constructed (Fig. 2). The abilities of these mutant proteins to activate transcription of the E4-CAT reporter plasmid were determined in transiently transfected HeLa cells. As shown in Fig. 2, reducing the number of EP repeats from six to four resulted in an approximately 50% reduction of the E1A transactivation capacity. Further reduction of EP repeats, from four to two or zero, essentially abolished the effect of AR1 on E1A transactivation. This suggests that the number of EP repeats in AR1 is of critical importance for E1A transactivation. Interestingly, increasing the EP repeat length from 6 to 8 or 10 did not result in superactivation of E4 transcription; rather, we observed a reduction of the stimulatory effect of AR1.
Collectively, these results suggest that the EP repeat length is critical for AR1 function, with the wild-type number of six EP repeats being optimal.
The acidic nature of AR1 is required for its function.
Although AR1 is essential for efficient transactivation by the Ad2 E1A protein, it is not well conserved among E1A proteins from other adenovirus serotypes (see Discussion). Also, AR2 substitutes for AR1 when artificially juxtaposed to CR3 (Fig. 1B). It is noteworthy that AR1 and AR2 do not show primary sequence homology. However, both regions are predicted to expose an acidic surface (5). Thus, the negative charge in AR1 might be the sequence feature required for enhancement of CR3 transactivation.
To test this hypothesis, plasmids expressing E1A mutant proteins with conservative or nonconservative amino acid substitutions in AR1 were constructed. As shown in Fig. 3, replacing four of the six glutamic acid residues in the EP repeat unit with aspartic acid (4D/P) resulted in a functional E1A transactivator protein. In contrast, substituting alanine (4A/P) or the positively charged amino acid lysine (4K/P) for glutamic acid either annulled or repressed E1A transactivation below the basal level of promoter activity (4A/P and 4K/P, respectively). Interestingly, replacement of four of the six prolines in the EP repeat unit with glycine was found to generate a functional E1A transactivator protein.
Taken together, these results suggest that neither the glutamic acids nor the prolines in the EP repeat unit are essential for AR1 function. Since aspartic acid functionally substitutes for glutamic acid, the results strengthen the hypothesis that the AR1 element needs a negative surface to enhance E1A transactivation.
AR1 is not part of the CR3 promoter-targeting signal.
Although all promoters tested in this study require AR1 for efficient E1A activation, they do not share a binding site for a common upstream-binding transcription factor. The promoters tested contain binding sites for multiple unrelated transcription factors that are E1A responsive (29).
One mechanism by which E1A has been shown to activate transcription is via recruitment to responsive promoters through an association with sequence-specific upstream-binding transcription factors (7, 13, 22, 23, 28). Several factors, such as c-Jun, ATF-2, and Sp1, have been shown to specifically interact with E1A via the promoter-targeting signal, located at the C terminus of CR3 (22). It is noteworthy that AR1 is located immediately downstream of the CR3 promoter-targeting signal. To investigate whether AR1 helps to direct the binding of transcription factors to CR3, a series of in vitro protein binding experiments was performed. For these experiments, GST, GST/c-Jun, GST/ATF-2, and GST/Sp1(DBD) fusion proteins were expressed in E. coli. Their abilities to bind in vitro-translated 35S-labelled E1A proteins (Fig. 4A) were tested in a standard GST pull-down assay. As shown in Fig. 4B, E1A (Δ1–120) (wild type) and 6E/P (Δ1–120) (AR1+) bound strongly to Sp1 and weakly to ATF-2 and c-Jun. The GST fusion proteins also bound efficiently to E/P− (Δ1–120) (AR−), suggesting that the interactions were AR independent. Importantly, none of the transcription factors bound to the 243R (Δ1–120) protein, demonstrating that the interactions were CR3 dependent. Moreover, Sp1, ATF-2, and c-Jun interactions with the CR3pt− (Δ1–120) protein were drastically reduced, suggesting that the CR3 promoter-targeting signal (20, 35) is required for a stable interaction with E1A. Taken together, these results suggest that, at least in vitro, AR1 is dispensable for interaction of E1A with upstream-binding transcription factors and, thus, does not appear to be part of the E1A promoter-targeting signal.
FIG. 4.
AR1 is not part of the CR3 promoter-targeting signal. (A) A schematic representation of the N-terminally truncated E1A mutant proteins used for in vitro binding. (B) In vitro-transcribed and -translated, 35S-labelled deletion mutants of E1A, illustrated in panel A, were tested for binding to GST, GST/c-Jun, GST/ATF-2, and GST/Sp1(DBD). The positions of protein molecular size standards are marked on the right side (in kilodaltons). The first panel (in vitro translation products [IVT]) was run with 1/10 of the E1A protein input used in the GST fusion protein binding assay.
DISCUSSION
We have previously shown that two interchangeable elements in the nonconserved Ad2 E1A C-terminal exon (AR1 and AR2) are required for CR3-mediated transactivation of the viral E4 promoter (5). Here we extended this study by showing that AR1 is an essential element required for efficient E1A activation of all viral early promoters and the cellular HSP70 promoter (Fig. 1B). These results suggest that AR1 may be of global importance for CR3-mediated transactivation. Since the six EP repeats constituting AR1 are of general importance for E1A activation of transcription, we propose that the CR3 transactivation domain should be extended to include AR1.
We also showed that AR2 functionally imitates AR1 only when artificially juxtaposed to CR3 (Fig. 1B). Thus, in the wild-type E1A-289R protein, AR2 is nonfunctional as a CR3 enhancer domain, suggesting that the stimulatory effect of AR2 on CR3-mediated transactivation is artifactual. In contrast, AR2 appears to serve a biologically significant function in E1A transformation. Thus, AR2 has been shown to suppress the invasive propensity of E1A-transformed cells by inhibiting expression of metalloproteases (21). Consequently, AR1 and AR2 may both be biologically important elements, promoting different aspects of E1A biology.
On the basis of the great significance of Ad2 AR1 for CR3 transactivation, it appears likely that E1A proteins from other adenovirus serotypes also contain AR1-like elements. Since the primary sequence of AR1 is not well conserved among E1A proteins from other serotypes (Fig. 5), we designed experiments to identify the attributes of AR1 that are important for its function. In summary, these experiments suggest that the primary sequence of AR1 is not essential for its function. More likely, the net negative charge in AR1 is critical for its function as a CR3 enhancer. The number of EP repeats appears to be important, with six negative amino acids being optimal (Fig. 2). However, glutamic acid does not appear to be essential, since four of the six glutamic acids could be replaced by aspartic acids and still generate a functional E1A protein (Fig. 3). Also, four of the six prolines could be replaced with glycine without having much of an effect on AR1 function. Interestingly, replacing glutamic acid in AR1 with the basic amino acid lysine converted CR3 from a transcriptional activator to a transcriptional repressor domain. Additional support for our hypothesis that the net negative charge of AR1 is critical for function comes from an independent study by Webster and Ricciardi (35). They showed that the glutamic acid and proline of the first EP repeat could be individually replaced with aspartic acid and glycine, respectively, without CR3 transactivation being negatively affected. Also, AR2, which exhibits no primary sequence homology with AR1, substitutes for AR1 with a high efficiency when artificially juxtaposed to CR3 (Fig. 1B). AR2 has been mapped to a 23-amino-acid serine-threonine-rich region (5), of which many residues appear to be phosphorylated in vivo (34). Thus, the common denominator in AR1 and AR2 appears to be their acidic character. Collectively, these results strongly suggest that negative charges positioned next to CR3 are a prerequisite for efficient E1A activation of transcription. Although acidic regions are reported to function as transcriptional activation domains, AR1 did not function as such in a Gal4-E1A fusion protein assay (5).
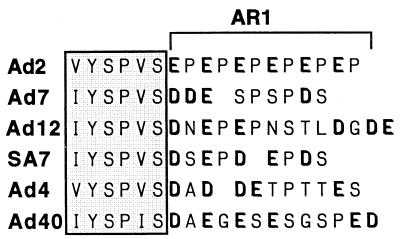
FIG. 5.
E1A proteins from six adenovirus serotypes have an excess of negatively charged amino acid residues at the hypothetical AR1 position. The boxed region indicates the highly conserved CR3 promoter-targeting domain. Negatively charged residues are indicated in boldface type.
With this limited genetic dissection of the Ad2 AR1 element, we searched for potentially homologous elements in E1A proteins from other serotypes. As shown in Fig. 5, all E1A proteins appear to have an excess of negatively charged amino acids positioned immediately downstream of the highly conserved C-terminal end of CR3. In some cases, the potential AR1 region is rich in serine residues, which theoretically may be phosphorylated in vivo. Our analysis suggests that a minimum of four negatively charged amino acids are required to create an E1A protein with a reasonable transactivation capacity (Fig. 2). Such a criterion appears to be fulfilled in all serotypes. Thus, it is possible that AR1-homologous elements exist in other E1A proteins.
Little is known about the mechanism by which AR1 enhances CR3-mediated transactivation. Our results showed that AR1 is essential for efficient CR3 transactivation in the context of the E1A wild-type protein. However, artificial tethering of CR3 to a promoter via the Gal4 DNA binding domain makes CR3 transactivation essentially AR independent (6). These results are consistent with the hypothesis that AR1 is required for recruitment of E1A to upstream-binding transcription factors. It is noteworthy that AR1 is positioned immediately downstream of the highly conserved promoter-targeting signal at the C-terminal end of CR3 (20, 35). This region has previously been shown to bind several transcription factors, such as c-Jun, ATF-2, Sp1, and USF (22). Thus, the most obvious hypothesis is that AR1 is required for interaction of CR3 with upstream-binding transcription factors. As shown in Fig. 4B, interaction of E1A with Sp1, ATF-2, or c-Jun required CR3 (243R) and was dependent on the CR3 promoter-targeting signal (CR3pt−). It is also noteworthy that these interactions were not enhanced by AR1 (compare the results for E/P+ and E/P−). Our results, therefore, are compatible with the hypothesis that AR1 is not part of the CR3 promoter-targeting signal. However, we cannot exclude the possibility that AR1 may help recruit transcription factors to CR3 in vivo, a stimulatory activity that is not reproduced in vitro in GST pull-down assays.
We have also considered the possibility that AR1 is selectively required for E1A activation through induced transcription factor phosphorylation. Thus, we previously showed that E1A induction of E4 promoter activity is primarily mediated by the cellular E4F transcription factor. Since E1A activation of E4F appears to involve an E1A-induced phosphorylation of E4F (10, 27), we previously hypothesized that the AR elements might be selectively required for E1A transactivation through transcription factor phosphorylation (5). However, our demonstration that AR1 is required for E1A activation of the HSP70 promoter and all adenovirus early promoters, most which are not believed to be regulated through transcription factor phosphorylation, argues against this hypothesis. E1A does not have an intrinsic protein kinase activity, but the CR3 domain has recently been found to associate with a cellular CTD (C-terminal domain of the largest subunit of RNA polymerase II) protein kinase (14, 16). We have therefore tested whether AR1 enhances CTD phosphorylation. Although we detected an E1A-associated CTD kinase activity, its binding to E1A was not dependent on the presence of AR1 (data not shown).
In summary, our results suggest that AR1 is an integral part of the CR3 transactivation domain, essential for Ad2 E1A transactivation of all tested promoters. However, the function of AR1 in E1A transactivation remains elusive.
ACKNOWLEDGMENTS
We are grateful to C. Svensson, K. Sollerbrant, and M. Green for kind gifts of plasmids. We also thank M. Mannervik for helpful comments on the manuscript.
This work was supported by the Swedish Cancer Society.
REFERENCES
- 1.Agoff S N, Wu B. CBF mediates adenovirus E1a trans-activation by interaction at the C-terminal promoter targeting domain of conserved region 3. Oncogene. 1994;9:3707–3711. [PubMed] [Google Scholar]
- 2.Akusjärvi G. Proteins with transcription regulatory properties encoded by human adenoviruses. Trends Microbiol. 1993;1:163–170. doi: 10.1016/0966-842x(93)90085-6. [DOI] [PubMed] [Google Scholar]
- 3.Bayley S T, Mymryk J S. Adenovirus E1A proteins and transformation. Int J Oncol. 1994;5:425–444. doi: 10.3892/ijo.5.3.425. [DOI] [PubMed] [Google Scholar]
- 4.Bondesson M, Mannervik M, Akusjärvi G, Svensson C. An adenovirus E1A transcriptional repressor domain functions as an activator when tethered to a promoter. Nucleic Acids Res. 1994;22:3053–3060. doi: 10.1093/nar/22.15.3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bondesson M, Svensson C, Linder S, Akusjärvi G. The carboxy-terminal exon of the adenovirus E1A protein is required for E4F-dependent transcription activation. EMBO J. 1992;11:3347–3354. doi: 10.1002/j.1460-2075.1992.tb05413.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bondesson M, Öhman K, Mannervik M, Fan S, Akusjärvi G. Adenovirus E4 open reading frame 4 protein autoregulates E4 transcription by inhibiting E1A transactivation of the E4 promoter. J Virol. 1996;70:3844–3851. doi: 10.1128/jvi.70.6.3844-3851.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chatton B, Bocco J L, Gaire M, Hauss C, Reimund B, Goetz J, Kedinger C. Transcriptional activation by the adenovirus larger E1a product is mediated by members of the cellular transcription factor ATF family which can directly associate with E1a. Mol Cell Biol. 1993;13:561–570. doi: 10.1128/mcb.13.1.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chiang C M, Roeder R G. Cloning of an intrinsic human TFIID subunit that interacts with multiple transcriptional activators. Science. 1995;267:531–536. doi: 10.1126/science.7824954. [DOI] [PubMed] [Google Scholar]
- 9.Dery C V, Herrmann C H, Mathews M B. Response of individual adenovirus promoters to the products of the E1A gene. Oncogene. 1987;2:15–23. [PubMed] [Google Scholar]
- 10.Fernandes E R, Rooney R J. The adenovirus E1A-regulated transcription factor E4F is generated from the human homolog of nuclear factor φAP3. Mol Cell Biol. 1997;17:1890–1903. doi: 10.1128/mcb.17.4.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Flint J, Shenk T. Adenovirus E1A protein paradigm viral transactivator. Annu Rev Genet. 1989;23:141–161. doi: 10.1146/annurev.ge.23.120189.001041. [DOI] [PubMed] [Google Scholar]
- 12.Geisberg J V, Chen J-L, Ricciardi R P. Subregions of the adenovirus E1A transactivation domain target multiple components of the TFIID complex. Mol Cell Biol. 1995;15:6283–6290. doi: 10.1128/mcb.15.11.6283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Geisberg J V, Lee W S, Berk A J, Ricciardi R P. The zinc finger region of the adenovirus E1A transactivating domain complexes with the TATA box binding protein. Proc Natl Acad Sci USA. 1994;91:2488–2492. doi: 10.1073/pnas.91.7.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gold M O, Tassan J P, Nigg E A, Rice A P, Herrmann C H. Viral transactivators E1A and VP16 interact with a large complex that is associated with CTD kinase activity and contains CDK8. Nucleic Acids Res. 1996;24:3771–3777. doi: 10.1093/nar/24.19.3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Herrmann C H, Dery C V, Mathews M B. Transactivation of host and viral genes by the adenovirus E1B 19K tumor antigen. Oncogene. 1987;2:25–35. [PubMed] [Google Scholar]
- 16.Herrmann C H, Gold M O, Rice A P. Viral transactivators specifically target distinct cellular protein kinases that phosphorylate the RNA polymerase II C-terminal domain. Nucleic Acids Res. 1996;24:501–508. doi: 10.1093/nar/24.3.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoeffler W K, Kovelman R, Roeder R G. Activation of transcription factor IIIC by the adenovirus E1A protein. Cell. 1988;53:907–920. doi: 10.1016/s0092-8674(88)90409-6. [DOI] [PubMed] [Google Scholar]
- 18.Kimelman D, Miller J S, Porter D, Roberts B E. E1a regions of the human adenoviruses and of the highly oncogenic simian adenovirus 7 are closely related. J Virol. 1985;53:399–409. doi: 10.1128/jvi.53.2.399-409.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kraus V B, Moran E, Nevins J R. Promoter-specific trans-activation by the adenovirus E1A12S product involves separate E1A domains. Mol Cell Biol. 1992;12:4391–4399. doi: 10.1128/mcb.12.10.4391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lillie J W, Green M R. Transcription activation by the adenovirus E1a protein. Nature. 1989;338:39–44. doi: 10.1038/338039a0. [DOI] [PubMed] [Google Scholar]
- 21.Linder S, Popowicz P, Svensson C, Marshall H, Bondesson M, Akusjärvi G. Enhanced invasive properties of rat embryo fibroblasts transformed by adenovirus E1A mutants with deletions in the carboxy-terminal exon. Oncogene. 1992;7:439–443. [PubMed] [Google Scholar]
- 22.Liu F, Green M R. Promoter targeting by adenovirus E1a through interaction with different cellular DNA-binding domains. Nature. 1994;368:520–525. doi: 10.1038/368520a0. [DOI] [PubMed] [Google Scholar]
- 23.Liu F, Green M R. A specific member of the ATF transcription factor family can mediate transcription activation by the adenovirus E1a protein. Cell. 1990;61:1217–1224. doi: 10.1016/0092-8674(90)90686-9. [DOI] [PubMed] [Google Scholar]
- 24.Mazzarelli J M, Atkins G B, Geisberg J V, Ricciardi R P. The viral oncoproteins Ad5 E1A, HPV16 E7 and SV40 TAg bind a common region of the TBP-associated factor-110. Oncogene. 1995;11:1859–1864. [PubMed] [Google Scholar]
- 25.Mazzarelli J M, Mengus G, Davidson I, Ricciardi R P. The transactivation domain of adenovirus E1A interacts with the C terminus of human TAFII135. J Virol. 1997;71:7978–7983. doi: 10.1128/jvi.71.10.7978-7983.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Raychaudhuri P, Bagchi S, Nevins J R. DNA-binding activity of the adenovirus-induced E4F transcription factor is regulated by phosphorylation. Genes Dev. 1989;3:620–627. doi: 10.1101/gad.3.5.620. [DOI] [PubMed] [Google Scholar]
- 27.Rooney R J, Raychaudhuri P, Nevins J R. E4F and ATF, two transcription factors that recognize the same site, can be distinguished both physically and functionally: a role for E4F in E1A trans activation. Mol Cell Biol. 1990;10:5138–5149. doi: 10.1128/mcb.10.10.5138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scholer H R, Ciesiolka T, Gruss P. A nexus between Oct-4 and E1A: implications for gene regulation in embryonic stem cells. Cell. 1991;66:291–304. doi: 10.1016/0092-8674(91)90619-a. [DOI] [PubMed] [Google Scholar]
- 29.Shenk T, Flint J. Transcriptional and transforming activities of the adenovirus E1A proteins. Adv Cancer Res. 1991;57:47–85. doi: 10.1016/s0065-230x(08)60995-1. [DOI] [PubMed] [Google Scholar]
- 30.Sollerbrant K, Akusjärvi G, Svensson C. Repression of RNA polymerase III transcription by adenovirus E1A. J Virol. 1993;67:4195–4204. doi: 10.1128/jvi.67.7.4195-4204.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ström A C, Forsberg M, Lillhager P, Westin G. The transcription factors Sp1 and Oct-1 interact physically to regulate human U2 snRNA gene expression. Nucleic Acids Res. 1996;24:1981–1986. doi: 10.1093/nar/24.11.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Svensson C, Akusjärvi G. A novel effect of adenovirus VA RNA1 on cytoplasmic mRNA abundance. Virology. 1990;174:613–617. doi: 10.1016/0042-6822(90)90116-9. [DOI] [PubMed] [Google Scholar]
- 33.Svensson C, Pettersson U, Akusjärvi G. Splicing of adenovirus 2 early region 1A mRNAs is non-sequential. J Mol Biol. 1983;165:475–495. doi: 10.1016/s0022-2836(83)80214-9. [DOI] [PubMed] [Google Scholar]
- 34.Tremblay M L, McGlade C J, Gerber G E, Branton P E. Identification of the phosphorylation sites in early region 1A proteins of adenovirus type 5 by amino acid sequencing of peptide fragments. J Biol Chem. 1988;263:6375–6383. [PubMed] [Google Scholar]
- 35.Webster L C, Ricciardi R P. trans-Dominant mutants of E1A provide genetic evidence that the zinc finger of the trans-activating domain binds a transcription factor. Mol Cell Biol. 1991;11:4287–4296. doi: 10.1128/mcb.11.9.4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weeks D L, Jones N C. E1A control of gene expression is mediated by sequences 5′ to the transcriptional starts of the early viral genes. Mol Cell Biol. 1983;3:1222–1234. doi: 10.1128/mcb.3.7.1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wigler M, Pellicer A, Silverstein S, Axel R. Biochemical transfer of single-copy eucaryotic genes using total cellular DNA as donor. Cell. 1978;14:725–731. doi: 10.1016/0092-8674(78)90254-4. [DOI] [PubMed] [Google Scholar]
- 38.Wu B J, Hurst H C, Jones N C, Morimoto R I. The E1A 13S product of adenovirus 5 activates transcription of the cellular human HSP70 gene. Mol Cell Biol. 1986;6:2994–2999. doi: 10.1128/mcb.6.8.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]