Abstract
Morphological comparisons and multi locus phylogenetic analyses (base on the combined genes of ITS, LSU, rpb2 and tub) demonstrated that three new saprobic taxa isolated from bamboo belong to Cainiaceae. These taxa comprise a novel genus Paramphibambusa (P.bambusicolasp. nov.) and two new species, Arecophilaxishuangbannaensis and A.zhaotongensis. The three new taxa belong to Cainiaceae (Xylariales, Sordariomycetes) a poorly studied family, which now comprises eight genera. Paramphibambusa can be distinguished from other Cainiaceae genera in having ascomata with a neck and ascospores lacking longitudinal striation, germ slits or germ pores. The two new Arecophila species clustered in a clade with Arecophila sp. and A.bambusae. Detailed morphological descriptions, illustrations, and an updated phylogenetic tree are provided for the new taxa.
Key words: Bambusicolous fungi, multilocus phylogeny, taxonomy, Xylariales
Introduction
During our continuous investigation of bambusicolous fungi in Yunnan, China, we have collected one new genus and two new Arecophila K.D. Hyde species in Cainiaceae J.C. Krug. The family Cainiaceae (Xylariales, Sordariomycetes) was established by Krug (1978), with Cainia Arx & E. Müll as the type genus. Hongsanan et al. (2017) and Wijayawardene et al. (2020) accommodated Cainiaceae in Xylariomycetidae family incertae sedis. However, Hyde et al. (2020), Samarakoon et al. (2021), and Wijayawardene et al. (2022a) accepted Cainiaceae in Xylariales.
Maharachchikumbura et al. (2015, 2016) accepted five genera (viz., Amphibambusa D.Q. Dai & K.D. Hyde, Arecophila, Atrotorquata Kohlm. & Volkm.-Kohlm, Cainia, and Seynesia Sacc.) in Cainiaceae based on morphology and phylogeny. Subsequently, Mapook et al. (2020) introduced Longiappendispora Mapook & K.D. Hyde as a new member of Cainiaceae. Konta et al. (2021) transferred Endocalyx Berk. & Broome from Apiosporaceae to Cainiaceae. Li et al. (2022) revisited the monospecific genus Alishanica Karun et al. and synonymized it under Arecophila. Hence, seven genera (Amphibambusa, Arecophila, Atrotorquata, Cainia, Endocalyx, Longiappendispora, Seynesia) are accepted in Cainiaceae according to Hyde et al. (2020), Mapook et al. (2020), Jiang et al. (2021), and Konta et al. (2021).
Members of Cainiaceae are often found in tropical and temperate regions as saprobic fungi, which are usually associated with monocotyledons (mainly grasses) and fabaceous dicotyledons. Some Cainia species have been reported as causative agents of plant diseases, e.g., C.desmazieri C. Moreau & E. Müll (Krug 1978). Cainiaceae is morphologically characterized by immersed ascomata with a papillate ostiole, unitunicate asci, with a unique J+, apical ring or series of rings, and hyaline to pigmented, 1-septate ascospores with longitudinal striations or germ slits or germ pores, and usually surrounded by a sheath or appendages (Maharachchikumbura et al. 2016; Hyde et al. 2020). Asexual morphs of this family were reported as coelomycetous taxa, viz., Cainia and Endocalyx, that are characterized by black, pycnidial conidiomata, denticulate, sympodially proliferating conidiophores, branched or simple, septate, and phialidic conidiogenous cells, and hyaline, fusiform, or falcate to lunate conidia (Maharachchikumbura et al. 2016; Hyde et al. 2020; Konta et al. 2021; Wijayawardene et al. 2021a).
Arecophila was introduced by Hyde (1996) with A.gulubiicola K.D. Hyde as the type species. The genus Arecophila was initially regarded as a member of Amphisphaeriaceae G. Winter based on the morphology. Subsequently, Kang et al. (1999) accepted Arecophila as a member of Cainiaceae. Afterwards, the placement of Arecophila within the Cainiaceae has been confirmed based on analyses of partial LSU gene sequences (Jeewon et al. 2003; Senanayake et al. 2015; Li et al. 2022). Currently, 18 epithets are listed under Arecophila based on morpho-molecular study (Li et al. 2022; Index Fungorum 2023), and 15 epithets are listed under Arecophila in Species Fungorum (2023).
According to Jiang et al. (2022) and previous studies (Eriksson and Yue 1998; Hyde et al. 2002a, b; Zhou and Hyde 2002; Cai et al. 2003), only four Cainiaceae species are associated with bamboo (Amphibambusahongheensis H.B. Jiang & Phookamsak, Arecophilabambusae Umali & K.D. Hyde, A.coronata (Rehm) Umali & K.D. Hyde and A.nypae K.D. Hyde) in China. In this study, we aim to collect bamboo samples in Yunnan, China, describe and introduce a new genus Paramphibambusa to accommodate P.bambusicola, and two new species Arecophilaxishuangbannaensis and A.zhaotongensis in the family of Cainiaceae. This study enriches the species diversity of bambusicolous Cainiaceae species in China.
Materials and methods
Sample collection, single spore isolation and morphological study
Bamboo culms were collected in northeastern (Zhaotong), northwestern (Shangri-La), and southwestern (Xishuangbanna) Yunnan Province, China, stored in disposable plastic Ziplock bags and brought back to the laboratory for examination and study. Morphological observation and single spore isolation were followed as described in Dai et al. (2017). The ascomata on the host surface were observed by Leica using a S8AP0 microscope and photographed by HDMI 200C. Micro-morphological features were observed using an Olympus BX53 compound microscope and captured with an Olympus DP74 camera (Olympus SZ61; Olympus Corporation, Tokyo, Japan). The asci were stained by Meltzer’s reagent to examine the J-/J+ ring at the tip of the asci. India ink was used to stain the ascospores for checking the mucilaginous sheath. The micro-morphological features and fruiting bodies were measured by Tarosoft (R) Image FrameWork (IFW). The photo plates were created by Adobe Photoshop CS6 software (Adobe Systems Inc., San Jose, CA, USA). Herbarium material and living cultures were deposited at the Herbarium of Guizhou Medical University (GMB), Guizhou Medical University Culture Collection (GMBCC) Guiyang, Zhongkai University of Agriculture and Engineering (ZHKU), Zhongkai University of Agriculture and Engineering Culture Collection (ZHKUCC) Guangdong, China, and the Guizhou Culture Collection (GZCC), Guiyang, China. MycoBank numbers were obtained from MycoBank database (https://www.mycobank.org/; accessed on 23 January 2024) to register the newly described taxa (MycoBank 2024).
DNA extraction, PCR amplification and sequencing
Fungal genomic DNA was extracted from fresh mycelium using the Biospin Fungus Genomic DNA Extraction Kit (BioFlux) according to the manufacturer’s instructions. When culture could not be obtained, fruiting bodies were used to extract genomic DNA by using E.Z.N.A. Forensic DNA Kit (BIO-TEK) followed the protocols. Genomic DNA was conducted by polymerase chain reaction (PCR). Four phylogenetic markers, internal transcribed spacer (ITS), large-subunit ribosomal RNA (LSU), RNA polymerase II (rpb2), and tub, were amplified using primer pairs ITS4/ITS5 (White et al. 1990), LR5/LR0R (Vilgalys and Hester 1990), RPB2-5F/RPB2-7cR (Liu et al. 1999), Bt2a/Bt2b (Hsieh et al. 2005), respectively. Amplification conditions were performed according to Dai et al. (2022) and Li et al. (2022). The purified PCR fragments were sequenced at Shanghai Myobio Biomedical Technology Co. and China UW Genetics Solutions (BGI-Tech), in Shanghai, China. The newly obtained sequence data were deposited in GenBank (https://www.ncbi.nlm.nih.gov).
Sequence alignment and phylogenetic analyses
The newly generated reverse and forward sequences were assembled with Geneious (Restricted) 9.1.2 (https://www.geneious.com, accessed on 20 May 2023) and subjected to BLAST searches in GenBank (https://blast.ncbi.nlm.nih.gov/, accessed on 20 May 2023) for revealing closely matched strains (Table 1). The related sequences of families in the order Xylariales were downloaded based on the latest article Li et al. (2022). The single gene matrix was aligned via the server version of MAFFT v. 7 (Katoh and Standley 2013) (https://mafft.cbrc.jp/alignment/server). The aligned sequence datasets were trimmed by trimAl.v1.2rev59. The alignments were combined via SequenceMatrix 1.9 (Vaidya et al. 2011). The AliView 1.26 (Larsson 2014) was used to obtain phylip and nexus format files for RAxML analysis and Bayesian analysis, respectively.
Table 1.
Sequences used for phylogenetic analyses in this study. The newly generated sequences are in bold. Type strains or type specimens are labelled with HT (holotype), ET (epitype), IT (isotype), and PT (paratype), T (Type), “N/A” indicates no available sequences.
| Species | Strain/voucher No. | Status | GenBank accession numbers | |||
|---|---|---|---|---|---|---|
| ITS | LSU | rpb2 | tub | |||
| Amphibambusabambusicola | MFLUCC 11-0617 | HT | KP744433 | KP744474 | NA | NA |
| Amphibambusahongheensis | KUN-HKAS 112723 | HT | MW892971 | MW892969 | NA | NA |
| Amphibambusahongheensis | KUMCC 20-0334 | HT | MW892972 | MW892970 | NA | NA |
| Amphiroselliniafushanensis | HAST 91111209 | HT | GU339496 | NA | GQ848339 | GQ495950 |
| Amphirosellinianigrospora | HAST 91092308 | HT | GU322457 | NA | GQ848340 | GQ495951 |
| Annulohypoxylonatroroseum | ATCC 76081 | – | AJ390397 | KY610422 | KY624233 | DQ840083 |
| Annulohypoxylonstygium | MUCL 54601 | – | KY610409 | KY610475 | KY624292 | KX271263 |
| Apiosporaarundinis | CBS 464.83 | – | KF144888 | KF144933 | NA | KF144979 |
| Apiosporahysteriana | ICMP 6889 | – | NA | DQ368630 | DQ368649 | DQ368621 |
| Apiosporakogelbergense | CBS 117206 | – | KF144895 | KF144941 | NA | KF144987 |
| Apiosporasetosa | ATCC 58184 | – | NA | AY346259 | NA | NA |
| Arecophilaaustralis | GZUCC0112 | HT | MT742126 | MT742133 | NA | MT741734 |
| Arecophilaaustralis | GZUCC0124 | PT | MT742125 | MT742132 | NA | NA |
| Arecophilabambusae | HKUCC 4794 | – | NA | AF452038 | NA | NA |
| Arecophilaclypeata | GZUCC0110 | HT | MT742129 | MT742136 | MT741732 | NA |
| Arecophilaclypeata | GZUCC0127 | PT | MT742128 | MT742135 | NA | NA |
| Arecophilamiscanthi | GZUCC0122 | – | MT742127 | MT742134 | NA | NA |
| Arecophilamiscanthi | MFLU 19-2333 | HT | NR_171235 | MK503827 | NA | NA |
| Arecophila sp. | HKUCC 6487 | – | NA | AF452039 | NA | NA |
| Arecophilaxishuangbannaensis | ZHKU 23-0280 | – | OR995737 | OR995744 | NA | NA |
| Arecophilaxishuangbannaensis | GMB-W1283 | HT | OR995736 | OR995743 | NA | NA |
| Arecophilazhaotongensis | GMBCC1145 | HT | OR995740 | OR995747 | OR995741 | NA |
| Arecophilazhaotongensis | ZHKU 23-0260 | – | OR995738 | OR995745 | NA | NA |
| Arecophilazhaotongensis | ZHKU 23-0259 | IT | OR995735 | OR995742 | NA | NA |
| Astrocystisconcavispora | MFLUCC 14-0174 | HT | KP297404 | KP340545 | KP340532 | KP406615 |
| Atrotorquatalineata | HKUCC 3263 | – | AF009807 | NA | NA | NA |
| Atrotorquataspartii | MFLUCC 13-0444 | HT | NA | KP325443 | NA | NA |
| Barrmaeliarappazii | CBS 142771 | HT | MF488989 | MF488989 | MF488998 | MF489017 |
| Barrmaeliarhamnicola | CBS 142772 | ET | MF488990 | MF488990 | MF488999 | MF489018 |
| Cainiaanthoxanthis | MFLUCC 15-0539 | HT | NR_138407 | KR092777 | NA | NA |
| Cainiadesmazieri | CAI | – | KT949896 | KT949896 | NA | NA |
| Cainiadesmazieri | CBS 137.62 | – | MH858124 | MH869702 | NA | NA |
| Cainiaglobosa | MFLUCC 13-0663 | HT | NR_171724 | KX822123 | NA | NA |
| Cainiagraminis | CBS 136.62 | – | MH858123 | AF431949 | NA | NA |
| Cainiagraminis | MFLUCC 15-0540 | – | KR092793 | KR092781 | NA | NA |
| Cainia sp. | LSU0560 | – | MT000421 | MT000513 | NA | NA |
| Camilleaobularia | ATCC 28093 | – | KY610384 | KY610429 | KY624238 | KX271243 |
| Camilleatinctor | YMJ 363 | – | JX507806 | NA | JX507790 | JX507795 |
| Collodisculabambusae | GZ 62 | – | KP054279 | KP054280 | KP276675 | KP276674 |
| Collodisculafangjingshanensis | GZUH 0109 | HT | KR002590 | KR002591 | KR002592 | KR002589 |
| Coniocessiamaxima | CBS 593.74 | HT | NR_137751 | MH878275 | NA | NA |
| Coniocessianodulisporioides | CBS 281.77 | IT | MH861061 | AJ875224 | NA | NA |
| Creosphaeriasassafras | STMA 14087 | – | KY610411 | KY610468 | KY624265 | KX271258 |
| Daldiniabambusicola | CBS 122872 | HT | KY610385 | KY610431 | KY624241 | AY951688 |
| Daldiniaconcentrica | CBS 113277 | – | AY616683 | KT281895 | KY624243 | KC977274 |
| Endocalyxcinctus | NBRC 31306 | – | MZ313191 | MZ313152 | NA | NA |
| Endocalyxcinctus | JCM 7946 | – | LC228648 | LC228704 | NA | NA |
| Endocalyxgrossus | JCM 5164 | HT | MZ313160 | MZ313138 | NA | NA |
| Endocalyxgrossus | JCM 5165 | – | MZ313159 | MZ313158 | NA | NA |
| Endocalyxgrossus | JCM 5166 | – | MZ313179 | MZ313171 | NA | NA |
| Endocalyxindumentum | JCM 5171 | HT | MZ313153 | MZ313161 | NA | NA |
| Endocalyxindumentum | JCM 8042 | – | MZ313162 | MZ313157 | NA | NA |
| Endocalyxmelanoxanthus | CBS147393 | – | MW718204 | MW718204 | NA | NA |
| Endocalyxmelanoxanthus | CBS147394 | – | MW718203 | MW718203 | NA | NA |
| Endocalyxptychospermatis | ZHKUCC 21-0008 | HT | MZ493352 | OK513439 | NA | NA |
| Endocalyxptychospermatis | ZHKUCC 21-0009 | HT | MZ493353 | OK513440 | NA | NA |
| Endocalyxptychospermatis | ZHKUCC 21-0010 | HT | MZ493354 | OK513441 | NA | NA |
| Entoleucamammata | JDR 100 | – | GU300072 | NA | GQ844782 | GQ470230 |
| Entonaemaliquescens | ATCC 46302 | – | KY610389 | KY610443 | KY624253 | KX271248 |
| Entosordariaperfidiosa | CBS 142773 | ET | MF488993 | MF488993 | MF489003 | MF489021 |
| Entosordariaquercina | RQ/CBS 142774 | HT | MF488994 | MF488994 | MF489004 | MF489022 |
| Graphostromaplatystomum | CBS 270.87 | HT | JX658535 | AY083827 | KY624296 | HG934108 |
| Hypocoprarostrata | NRRL 66178 | – | KM067909 | KM067909 | NA | NA |
| Hypocreagelatinosa | NBRC 104900 | ET | JN943358 | JN941453 | NA | NA |
| Hypomontagnellabarbarensis | STMA 14081 | HT | MK131720 | MK131718 | MK135891 | MK135893 |
| Hypomontagnellamonticulosa | MUCL 54604 | ET | KY610404 | KY610487 | KY624305 | KX271273 |
| Hypoxylonfragiforme | MUCL51264 | ET | KM186294 | KM186295 | KM186296 | KX271282 |
| Hypoxyloninvestiens | CBS 118185 | – | KC968924 | KY610451 | KY624260 | KC977269 |
| Jackrogersellamultiformis | CBS 119016 | ET | KC477234 | KT281893 | KY624290 | KX271262 |
| Kretzschmariadeusta | CBS 163.93 | – | KC477237 | KY610458 | KY624227 | KX271251 |
| Leiosphaerellachromolaenae | CBS 125586 | – | JF440976 | JF440976 | ||
| Longiappendisporachromolaenae | MFLUCC 17-1485 | HT | NR_169723 | NG_068714 | NA | NA |
| Lopadostomaamericanum | LG8 | HT | KC774568 | KC774568 | KC774525 | NA |
| Lopadostomadryophilum | LG21 | ET | KC774570 | KC774570 | KC774526 | MF489023 |
| Lopadostomafagi | LF1 | HT | KC774575 | KC774574 | KC774531 | NA |
| Lopadostomaquercicola | LG27 | HT | KC774610 | KC774610 | KC774558 | NA |
| Lopadostomaturgidum | LT2 | ET | KC774618 | KC774618 | KC774563 | MF489024 |
| Monographellanivalis | UPSC 3273 | – | NA | AF452030 | NA | NA |
| Nemaniaabortiva | BISH 467 | HT | GU292816 | NA | GQ844768 | GQ470219 |
| Nemaniabipapillata | HAST 90080610 | – | GU292818 | NA | GQ844771 | GQ470221 |
| Nemaniamaritima | HAST 89120401 | ET | GU292822 | NA | GQ844775 | GQ470225 |
| Nemaniaprimolutea | HAST 91102001 | HT | EF026121 | NA | GQ844767 | EF025607 |
| Obolarinadryophila | MUCL 49882 | – | GQ428316 | GQ428316 | KY624284 | GQ428322 |
| Oxydothisfrondicola | HKUCC 1001 | – | NA | AY083835 | NA | NA |
| Paramphibambusabambusicola | GMBCC1142 | HT | OR995739 | OR995746 | OR995740 | NA |
| Paramphibambusabambusicola | ZHKUCC 23-0976 | – | OR995741 | OR995748 | OR995739 | NA |
| Paraxylariaxylostei | MFLU 17-1636 | – | MW240640 | MW240570 | NA | MW820914 |
| Paraxylariaxylostei | MFLU 17-1645 | – | MW240641 | MW240571 | NA | MW820915 |
| Phylaciasagrana | CBS 119992 | – | AM749919 | NA | NA | NA |
| Podosordariamexicana | WSP 176 | – | GU324762 | NA | GQ853039 | GQ844840 |
| Podosordariamuli | WSP 167 | HT | GU324761 | NA | GQ853038 | GQ844839 |
| Poroniapileiformis | WSP 88113001 | ET | GU324760 | NA | GQ853037 | GQ502720 |
| Poroniapunctata | CBS 656.78 | HT | KT281904 | KY610496 | KY624278 | KX271281 |
| Pyrenopolyporusnicaraguensis | CBS 117739 | – | AM749922 | KY610489 | KY624307 | KC977272 |
| Rhopalostromaangolense | CBS 126414 | – | KY610420 | KY610459 | KY624228 | KX271277 |
| Roselliniaaquila | MUCL 51703 | – | KY610392 | KY610460 | KY624285 | KX271253 |
| Roselliniacorticium | MUCL 51693 | – | KY610393 | KY610461 | KY624229 | KX271254 |
| Rostrohypoxylonterebratum | CBS 119137 | HT | DQ631943 | DQ840069 | DQ631954 | DQ840097 |
| Ruwenzoriapseudoannulata | MUCL 51394 | HT | KY610406 | KY610494 | KY624286 | KX271278 |
| Sarcoxyloncompunctum | CBS 359.61 | – | KT281903 | KY610462 | KY624230 | KX271255 |
| Seynesiaerumpens | SMH 1291 | – | NA | AF279410 | AY641073 | NA |
| Stilbohypoxylonquisquiliarum | YMJ 172 | – | EF026119 | NA | GQ853020 | EF025605 |
| Thamnomycesdendroideus | CBS 123578 | – | FN428831 | KY610467 | KY624232 | KY624313 |
| Vialaeamangiferae | MFLUCC 12-0808 | HT | KF724974 | KF724975 | NA | NA |
| Vialaeaminutella | BRIP 56959 | – | KC181926 | KC181924 | NA | NA |
| Xylariahypoxylon | CBS 122620 | ET | KY610407 | KY610495 | KY624231 | KX271279 |
| Zygosporiumoscheoides | MFLUCC 14-0402 | – | MF621585 | MF621589 | NA | NA |
Maximum likelihood (ML) analysis was performed by RAxML-HPC2 on XSEDE (8.2.12) (Stamatakis et al. 2008; Stamatakis 2014) via the CIPRES Science Gateway V.3.3 web server (https://www.phylo.org/portal2/login!input. action) (Miller et al. 2010). The best model was GTRGAMMA, with 1000 replicates rapid bootstrapping. Bayesian inference (BI) analysis was performed by MrBayes on XSEDE (3.2.7a) in the website CIPRES Science Gateway (Ronquist et al. 2012). Markov Chain Monte Carlo (MCMC) was used to evaluate posterior probabilities (PP) (Rannala and Yang 1996; Zhaxybayeva and Gogarten 2002). The best model test for each gene was performed via MrMTgui (Ma 2016). Six simultaneous Markov chains were run for 1000000 generations, and trees were sampled every 100th generation (resulting in 10,000 total trees). The phylogenetic trees were visualized with FigTree v. 1.4.2 (http://tree.bio.ed.ac.uk software/figtree/) (Rambaut 2012), and edited by Adobe Illustrator CS v. 5.
Abbreviations
ATCC: American Type Culture Collection; BISH: Bishop Museum, Department of Natural Sciences; CAI: Cairo University, Botany Department; CBS: Culture Collection of the Westerdijk Fungal Biodiversity Institute, Utrecht, Netherlands; GMBCC: Guizhou Medical University Culture Collection, Guiyang, China; GZU: Karl-Franzens-Universitat Graz; GZUCC: Guizhou University Culture Collection, Guiyang, Guizhou, China; HAST: Research Center for Biodiversity, Academia Sinica; HKUCC: The University of Hong Kong Culture Collection, Hong Kong, P.R. China; JCM: Japan Collection of Microorganisms, Japan; JDR: J.D. Rogers; KUMCC: Kunming Institute of Botany Culture Collection; KUN-HKAS: Herbarium of Cryptogams Kunming Institute of Botany Academia Sinica; LF: LopadostomafagiL; LT: Lopadostomaturgidum; MFLU: Mae Fah Luang University Herbarium; MFLUCC: Mae Fah Luang University Culture Collection; MUCL: Agro-food & Environmental Fungal Collection; NBRC: Biological Resource Center IFO; NRRL: Agricultural Research Service Culture Collection; SMH: Sabine M. Huhndorf; KUN-HKAS: Herbarium of Cryptogams Kunming Institute of Botany Academia Sinica; STMA: HZI culture collection, Helmholtz Centre for Infection Research, Braunschweig, Germany; WSP: Washington State University, Plant Pathology Department; YMJ: YuMing, Ju; ZHKUCC: Zhongkai University of Agriculture and Engineering.
Results
Phylogenetic results
The combined dataset comprised 107 strains (Table 1). Hypocreagelatinosa (NBRC 104900) was selected as the outgroup taxon. The alignment comprised 4195 bp in total (ITS 580 bp, LSU 736 bp, rpb2 1197 bp, and tub 1682 bp). The final ML optimization likelihood value of -68750.486429 and the matrix had 2603 bp distinct alignment patterns, with 45.50% of undetermined characters or gaps. Estimated base frequencies were as follows: A = 0.240607, C = 0.260776, G = 0.259542, T = 0.239075, AC = 1.358257, AG = 3.703167, AT = 1.354909, CG = 1.087664, CT = 6.069506, GT = 1.000000; proportion of invariable sites I = 0.378984; and gamma distribution shape parameter α = 0.817253.
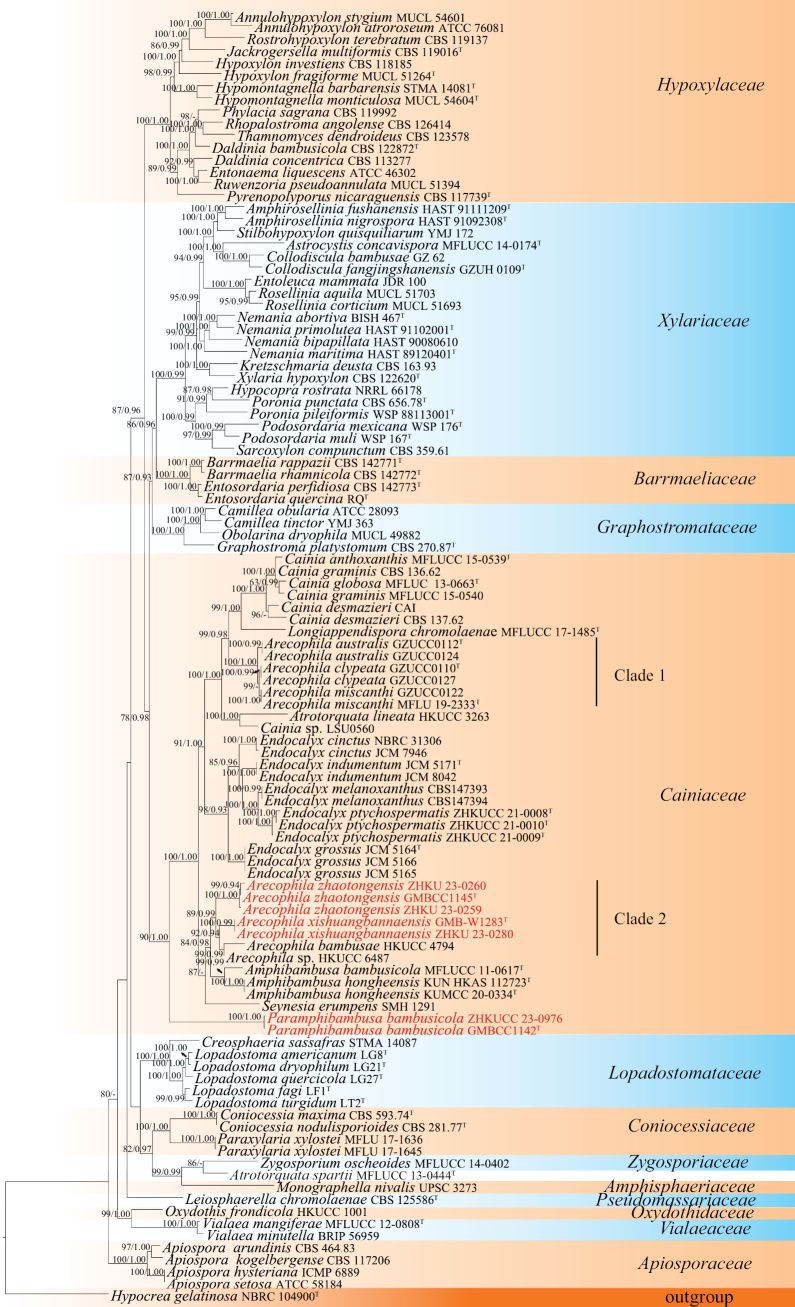
The final RAxML tree (Fig. 1) is based on maximum likelihood (ML), and Bayesian inference analyses with similar topology. The RAxML tree showed that Paramphibambusabambusicola (GMBCC1142, ZHKUCC 23-0976) formed a distinct, stable clade basal to the other genera of Cainiaceae with high statistical support (90% ML, 1.00 PP). Moreover, Arecophila strains form two clades (Fig. 1), which coincide with Li et al. (2022). Our new collections cluster with A.bambusae Umali & K.D. Hyde (HKUCC 4794) and Arecophila sp. (HKUCC 6487) forming a sister branch clustered in Clade 2 (Fig. 1).
Figure 1.
The RAxML tree was generated based on the combined ITS, LSU, rpb2, and tub sequence data. Bootstrap support values for ML equal to or greater than 60%, and Bayesian posterior probabilities (BYPP) equal to or higher than 0.90 are indicated above the nodes as ML/PP. Type materials are indicated by superscript “T”, while the newly generated sequences are shown in red.
Taxonomy
. Paramphibambusa
L.S. Han & D.Q. Dai gen. nov.
CD63B0CA-BDB9-5768-94F0-2CD297BA0F6A
MB851854
Etymology.
In reference to a new genus is morphologically similar to Amphibambusa, but phylogenetically distinct.
Description.
Saprobic on bamboo culms. Sexual morph: Ascomata deeply immersed beneath poorly developed clypeus, solitary, scattered, black, globose to subglobose, ostiolate, with a long neck. Peridium composed of several layers, thick-walled, hyaline to pale brown cells of textura angularis. Paraphyses hyaline, numerous, filiform to cylindrical, guttulate, branched, septate, tapering towards the apex. Asci 8-spored, rarely 6-spored, unitunicate, cylindrical, short pedicellate, straight or slightly curved, rounded at the apex, with an elliptical to trapezoidal, J+ sub-apical ring. Ascospores uniseriate or overlapping uniseriate, hyaline to golden brown, ellipsoidal, guttulate, 2–3-celled, tapering at the ends, slightly constricted at the septum, smooth-walled, surrounded by a mucilaginous sheath. Asexual morph: Undetermined.
Type species.
Paramphibambusabambusicola L.S. Han & D.Q. Dai
Notes.
A monotypic genus Paramphibambusa is introduced based on its different morphological characteristics and the support of phylogenetic affinity with the other members in Cainiaceae. The morphological characteristics of Paramphibambusa resemble Amphibambusa in having dark clypeus, immersed, globose to subglobose ascomata, unitunicate, short pedicellate asci with a J+, and sub-apical ring, and 1-septate ascospores, surrounded by a thick mucilaginous sheath (Liu et al. 2015; Jiang et al. 2021). Paramphibambusa can be easily distinguished from Amphibambusa in having an ostiole, with a long neck, and ascospores lacking longitudinal wall ornamentations. In addition, Paramphibambusa forms a well-separated branch basal to other cainiaceous genera with 90% ML, and 1.00 PP statistical supports (Fig. 1). Paramphibambusa differs from the sexual members of Cainiaceae in ascomata with a long neck leading up to the ostiole, and in that the ascospores lack longitudinal striations or germ slits or germ pores Endocalyx is an asexually typified genus and lacks a sexual morph to compare its morphology with Paramphibambusa. However, in the phylogenetic analyses, Paramphibambusa resides in a distinct phylogenetic lineage to Endocalyx (Fig. 1). Therefore, we consider Paramphibambusa as a distinct genus.
. Paramphibambusa bambusicola
L.S. Han & D.Q. Dai sp. nov.
9B44C681-26B4-5A60-B4FF-8DC259F346B3
MB851857
Figure 2.
Paramphibambusabambusicola (GMB-W1350, holotype) a bamboo specimen b black ostioles at the host surface c transverse section of ascomata d, e vertical section of ascomata with long necks and black clypeus f cells of peridium g paraphyses h–k asci l asci with J+, elliptical to trapezoidal, subapical ring (stained in Melzer’s reagent) m–s ascospores (s ascospore stained in Indian ink showing mucilaginous sheath) t a germinating ascospore u, v cultures on PDA after 20 days (u upper, v reverse). Scale bars: 300 µm (d, e); 15 µm (f, l–t); 30 µm (g); 50 µm (h–k).
Etymology.
With reference to its occurrence on host bamboo.
Holotype.
GMB-W1350.
Description.
Saprobic on dead culms of bamboo. Sexual morph: Ascomata 430–580 × 500–550 µm (x– = 474 × 519 µm, n = 20), deeply immersed beneath blackened poorly developed clypeus, solitary, scattered, black, globose to subglobose, ostiolate, with a long neck, 50–125 µm diam., 240–260 µm long. Peridium 15–25 µm thick, composed of several layers, thick-walled, hyaline to pale brown cells of textura angularis. Paraphyses 2–5.5 µm wide, hyaline, numerous, filiform to cylindrical, guttulate, branched, septate, tapering towards the apex. Asci 200–240 × 10–13.5 µm (x– = 215 × 11.5 µm, n = 20), 8-spored, rarely 6-spored, unitunicate, cylindrical, short pedicellate, straight or slightly curved, rounded at the apex, with a 3–4 µm wide, 1.5–2 µm high (x– = 3.6 × 1.7 µm, n = 20), elliptical to trapezoidal, J+, sub-apical ring. Ascospores 24–35 × 6–7.5 µm (x– = 27 × 6.6 µm, n = 20), uniseriate or overlapping uniseriate, hyaline to golden brown, ellipsoidal, 2–3-celled, tapering at the ends, slightly constricted at the septum, smooth-walled, surrounded by a 9–12 µm mucilaginous sheath. Asexual morph: Undetermined.
Culture characters.
Ascospores germinating within 24 h. Colonies reaching 45 mm diam. in 20 days under dark and at 28 °C conditions, circular, flocculent, yellowish from above and below.
Materials examined.
China, Yunnan Province, Zhaotong, Zhenxiong town, 27°36′8"N, 104°56′34"E, 1673.07 m, on dead culms of bamboo, 29 July 2021, Dong-Qin Dai, Li-Su Han, DDQ02077, (GMB-W1350, holotype), GMBCC1142, ex-type; ibid. (ZHKU 23-0256, isotype), GZCC 23-0629, ex-isotype; Zhaotong, Zhenxiong town, Shanzhai, 27°62′52"N, 104°81′98"E, 1666.10 m, on dead culms of bamboo, 4 August 2023, Dong-Qin Dai, Li-Su Han, HLS0114 (ZHKU 23-0257), living culture ZHKUCC 23-0976.
Notes.
In the phylogenetic tree, Paramphibambusabambusicola formed a stable clade basal to the other species of Cainiaceae with 90% ML, and 1.00 PP statistical supports (Fig. 1). In morphology, Paramphibambusabambusicola has Cainiaceae species typical characteristics that are cylindrical asci, with a J+, apical ring, and ellipsoidal ascospores surrounded by a mucilaginous sheath. However, the spores of Cainiaceae species have the ornamented walls with longitudinal striations or germ slits or germ pores. Paramphibambusabambusicola differs from the current Cainiaceae species by having smooth-walled ascospores. Therefore, based on morphological and phylogenetic studies, P.bambusicola is introduced hereby as a new species occurring on bamboo in Yunnan, China.
. Arecophila
K.D. Hyde, Nova Hedwigia 63(1-2): 82 (1996)
0742922E-C1E2-5321-AB86-754C598280DC
MB27653
Notes.
The genus Arecophila is characterized by immersed ascomata, usually with a clypeus, unitunicate, cylindrical asci, commonly producing an apical ring, and ascospores with longitudinal striation or a verrucose wall, and surrounded by a mucilaginous sheath (Hyde 1996; Li et al. 2022). Li et al. (2022) provided a morphological comparison of the main characters of Arecophila species. The asexual morph of Arecophila has not been reported. According to Li et al. (2022), this genus is distributed across 12 countries and is reported from 16 host species.
. Arecophila xishuangbannaensis
L.S. Han & D.Q. Dai sp. nov.
59991F35-DF11-50B4-8D83-E6C9350C81E9
MB851853
Figure 3.
Arecophilaxishuangbannaensis (GMB-W1283, holotype) a bamboo specimen b, c appearance of ostioles on host surface d–f vertical sections of ascomata g peridium h paraphyses i–m asci n asci with J+, wedge-shaped rings (Stained in Melzer’s reagent) o–t ascospores (s showing ascospore with longitudinal striations t ascospore stained in Indian ink showing mucilaginous sheath). Scale bars: 300 μm (d–f); 20 μm (g); 30 μm (h); 50 μm (i–m); 15 μm (n–t).
Etymology.
Named after the location “Xishuangbanna” where the new taxon was discovered.
Holotype.
GMB-W1283.
Description.
Saprobic on dead culms of bamboo. Sexual morph: Ascomata 540–700 × 320–450 µm (x– = 586 × 389 µm, n = 20), immersed beneath a black clypeus, forming white ring surrounding ostioles of ascomata, solitary or scattered, sometimes gregarious, globose to subglobose, dark brown to black. Ostioles papillate, central, black. Peridium 15–25 µm thick, comprised of several layers, thick-walled, dense, brown to hyaline, cells of textura angularis. Paraphyses 2.5–6 μm wide, hyaline, numerous, cylindrical, unbranched, septate. Asci 180–270 × 12–14 μm (x– = 213 × 12.8 μm, n = 20), 8-spored, unitunicate, cylindrical, pedicellate, straight or slightly curved, apically rounded, with a 3.7–4.7 μm wide, 2.5–3 μm high (x– = 4.3 × 2.7 μm, n = 20), wedge-shaped, J+, apical ring. Ascospores 23–27 × 8.5–9.5 μm (x– = 24.5 × 8.8 μm, n = 20), overlapping, uniseriate, initially hyaline, pale brown to dark brown when mature, ellipsoidal, medianly 1-septate, tapering towards both ends, slightly constricted at the septum, with longitudinal striation along entire length of the ascospore, surrounded by a 3.5–5 µm thick, distinct, globose to subglobose, mucilaginous sheath. Asexual morph: Undetermined.
Materials examined.
China, Yunnan Province, Xishuangbanna, Jinghong, Manzhang, Mengla, 21°91′97"N, 101°20′42"E, 617.14 m, on dead culms of bamboo, 16 August 2020, Dong-Qin Dai, Li-Su Han, DDQ00993, (GMB-W1283 holotype), ibid. (ZHKU 23-0258, isotype), ibid. DDQ00993-1 (ZHKU 23-0280).
Notes.
In the phylogenetic tree, our new collections of Arecophilaxishuangbannaensis (GMB-W1283, ZHKU 23-0280) formed a well-separated sister branch with A.bambusae (HKUCC 4794) and Arecophila sp. (HKUCC 6487) with 92% ML, 0.94 PP statistical supports (Fig. 1). Based on a nucleotide base pair comparison, A.xishuangbannaensis differs from A.bambusae (HKUCC 4794) in LSU gene (15/736 bp, 2%). Morphologically, A.xishuangbannaensis is similar to A.bambusae, in having cylindrical asci and ellispoidal ascospores. However, our new taxon differs A.bambusae by forming a white ring surrounding ostioles of ascomata and having larger asci (180–270 × 12–14 μm vs. 132.5–140 × 7.5–8 µm) and larger ascospores (23–27 × 8.5–9.5 μm vs. 19–22.5 × 5.5–7 µm) (Umali et al. 1999; Li et al. 2022). Arecophilaxishuangbannaensis also resembles A.notabilis K.D. Hyde, but it has larger ascomata (586 × 389 µm vs. 400 × 360 µm) (Hyde 1996). The spores of this species did not germinate on PDA or malt extract agar (MEA) media, thus no culture is available.
. Arecophila zhaotongensis
L.S. Han & D.Q. Dai sp. nov.
6E796CA0-45F3-5D29-907D-CA2900B11090
MB851836
Figure 4.
Arecophilazhaotongensis (GMB-W1353, holotype) a bamboo specimen b, c appearance of ostioles at the host surface d, e vertical sections of ascomata with ostioles and black clypei f peridium g paraphyses h–m asci n, o asci with a J+ trapezoidal ring (stained in Melzer’s reagent) p–t ascospores surrounded by mucilaginous sheath (t ascospore with longitudinal striations) u a germinating ascospore v, w cultures on PDA after 15 days (v upper, w reverse). Scale bars: 300 µm (d, e); 30 µm (f, g); 50 µm (h–m); 15 µm (n–u).
Etymology.
Named after the location “Zhaotong” where the new taxon was discovered.
Holotype.
GMB-W1353.
Description.
Saprobic on dead culms of bamboo. Sexual morph: Ascomata 600–960 × 450–550 µm (x– = 710 × 500 µm, n = 20), immersed beneath blackened clypeus, clypeus well-developed, darkened raised discs, or as tiny ostiolar dots, solitary, scattered, sometimes gregarious, dark brown to black, globose to subglobose, papillate, with a central ostiole. Peridium 15–25 µm thick, comprising several layers, thick-walled, brown cells of textura angularis. Paraphyses 1–3 µm wide, hyaline, numerous, filiform, branched. Asci 190–240 × 10.5–14 µm (x– = 215 × 11.6 µm, n = 20), 4- or 8-spored, rarely 6-spored, cylindrical, unitunicate, short pedicellate, straight or slightly curved, rounded at the apex, with a 4–4.5 μm wide, 2–2.5 µm high (x– = 4.2 × 2.2 µm, n = 20), trapezoidal, J+, apical ring. Ascospores 21–30 × 6–8 µm (x– = 25.5 × 7 µm, n = 20), uniseriate or overlapping uniseriate, brown, ellipsoidal, 1-septate, septate at the centre, slightly tapering at the ends, with longitudinal and sulcate striations, surrounded by a 5–10.5 µm wide, distinct, oval to spherical, mucilaginous sheath. Asexual morph: Undetermined.
Culture characters.
Ascospores germinating within 24 h. Colonies reach 20 mm diam. in 15 days under dark and at 28 °C conditions, circular, hairy, white from above, and yellow to yellowish from below.
Materials examined.
China, Yunnan Province, Diqin, Shangri-La, Bigu Mountain, on dead culms of bamboo, 22 July 2020, 27°36′56.9"N, 99°42′6.4"E, 3460 m, Dong-Qin Dai DDQ00740 (ZHKU 23-0261); Zhaotong, Zhenxiong S302, 27°36′8"N, 104°56′34"E, 1673.07 m, on dead culms of bamboo, 29 July 2021, Dong-Qin Dai, Li-Su Han, DDQ02079, (GMB-W1353, holotype), GMBCC1145, ex-type; ibid. (ZHKU 23-0259, isotype), ZHKUCC 23-0975, ex-isotype; ibid. DDQ02105 (ZHKU 23-0260).
Notes.
In the phylogenetic tree, the new species A.zhaotongensis (GMBCC 1145, ZHKU 23-0259, ZHKU 23-0260) formed a separated sister branch to A.bambusae (HKUCC 4794), Arecophila sp. (HKUCC 6487) and A.xishuangbannaensis (GMB-W1283, ZHKU 23-0280) with 89% ML, 0.99 PP statistical supports (Fig. 1). Based on a nucleotide pairwise comparison, A.zhaotongensis differs from A.bambusae (HKUCC 4794) in 26/736 bp of LSU (3.5%), and differs from A.xishuangbannaensis (GMB-W1283, ZHKU 22-0280) in 56/563 bp of ITS (9.9%), 18/736 bp of LSU (2.4%). Arecophilazhaotongensis has larger asci than A.bambusae (190–240 × 10.5–14 µm vs. 132.5–140 × 7.5–8 µm) and larger ascospores (21–30 × 6–8 µm vs. 19–22.5 × 5.5–7 µm) (Umali et al. 1999). Arecophilazhaotongensis differs from A.xishuangbannaensis (GMB-W1283, ZHKU 23-0280) in having narrower ascospores (21–30 × 6–8 µm vs. 23–27 × 8.5–9.5 µm). The new species also resembles A.muroiana (I. Hino & Katum.) You Z. Wang et al. (Wang et al. 2004). However, A.muroiana lacks a clypeus absent, while a blackened clypeus was observed in A.zhaotongensis.
Discussion
Paramphibambusa forms deeply immersed, dark ascomata, with a long neck, J+ asci and smooth-walled ascospores. Interestingly, genera in Cainiaceae usually form ascospores with longitudinal striations or germ slits or germ pores, however, these characters were not observed in our new collection (GMB-W1350). Hence, we introduced the new genus Paramphibambusa in Cainiaceae based on morphological characteristics and phylogenetic analyses (Fig. 1). Moreover, we introduced two new Arecophila species in Cainiaceae. The establishment of Paramphibambusa and the introduction of two new Arecophila species enriches the species diversity of the family Cainiaceae and the diversity of bambusicolous fungi.
Currently, some species in the Cainiaceae are monospecific, such as Longiappendispora (Mapook et al. 2020), and Paramphibambusa (this study), while Amphibambusa, and Atrotorquata each contain only two species (Kohlmeyer and Volkmann-Kohlmeyer 1993; Liu et al. 2015; Jiang et al. 2021). Hence, more samples are needed to better understand each genus. Wijayawardene et al. (2022b) mentioned that it is essential to carry out more studies on host plants (that have been extensively studied for fungi, such as bamboo) in biodiversity-rich regions to reveal more novel species. Yunnan is exceedingly rich in fungal diversity, especially in higher level taxa, such as ascomycetes and basidiomycetes (Wijayawardene et al. 2021b; Dai et al. 2022). Hence, we believe that future studies on bamboo-associated fungi in Yunnan Province would disclose more novel taxa.
Atrotorquata was introduced as a monotypic genus by Kohlmeyer and Volkmann-Kohlmeyer (1993) to accommodate A.lineata Kohlm. & Volkm.-Kohlm. Subsequently, Liu et al. (2015) introduced A.spartii Thambug et al. as the second species. These two species share similar morphology, but their phylogenetic relationship was not well-resolved by Liu et al. (2015). Due to a lack of sequence data in GenBank, Atrotorquata clusters outside of Cainiaceae. More sequences especially protein genes loci are needed, to clarify its family placement.
Eighteen epithets were listed in Arecophila (Li et al. 2022), but only four taxa and a unnamed species have available molecular data, viz., A.australis Q.R. Li et al. (GZUCC0112, GZUCC0124), A.bambusae (HKUCC 4794), A.clypeata Q.R. Li et al. (GZUCC0110, GZUCC0127), A.miscanthi Q.R Li & J.C. Kang (GZUCC0122, MFLU 19-2333), and Arecophila sp. (HKUCC 6487). Thus, it is necessary to recollect fresh specimens and designate epitypes or reference specimens. Li et al. (2022) divided Arecophila into two clades based on phylogenetic analyses. We obtained the same results in our study, probably because most species of Arecophila lack protein genes regions in GenBank. We may need to design more suitable primers for sequencing protein genes fragments of Arecophila to support phylogenetic study.
Supplementary Material
Acknowledgments
The authors are grateful to High-Level Talent Recruitment Plan of Yunnan Provinces (“Young Talents” Program and “High-End Foreign Experts” Program), National Natural Science Foundation of China (Grant No.31760013, 32100010 and 32060710), Mee-mann Chang Academician Workstation in Yunnan Province, (Grant No. 202205AF150002), and Science and technology plan project of Science and Technology Department of Yunnan Province (Grant No. 202305AC350252, 20210BA070001-076), Key Laboratory of Yunnan Provincial Department of Education of the Deep-Time Evolution on Biodiversity from the Origin of the Pearl River for support. We are grateful to Dr. Shaun Pennycook for suggestions of Latin names for the new taxa. Li-Su Han would like to thank Tian-Ye Du for helping with phylogenetic analyses. The authors extend their appreciation to the Researchers supporting Project Number (RSP2024R120) King Saud University, Riyadh, Saudi Arabia. Kazuaki Tanaka would like to thank the Japan Society for the Promotion of Science (JSPS, 23K05900).
Citation
Han L-S, Wijayawardene NN, Liu C, Han L-H, Promputtha I, Li Q, Elgorban AM, Al-Rejaie S, Tanaka K, Dai D-Q (2024) Paramphibambusa bambusicola gen. et. sp. nov., Arecophila xishuangbannaensis and A. zhaotongensis spp. nov. in Cainiaceae from Yunnan, China. In: Wijayawardene N, Karunarathna S, Fan X-L, Li Q-R (Eds) Taxonomy and secondary metabolites of wood-associated fungi. MycoKeys 104: 113–132. https://doi.org/10.3897/mycokeys.104.117872
Contributor Information
Chao Liu, Email: liuchao_80@163.com.
Dong-Qin Dai, Email: cicidaidongqin@gmail.com.
Additional information
Conflict of interest
The authors have declared that no competing interests exist.
Ethical statement
No ethical statement was reported.
Funding
National Natural Science Foundation of China (Grant No.31760013, 32100010 and 32060710), Mee-mann Chang Academician Workstation in Yunnan Province, (Grant No. 202205AF150002), Science and technology plan project of Science and Technology Department of Yunnan Province (Grant No. 202305AC350252, 20210BA070001-076), and Researchers Supporting Project Number (RSP2024R120), King Saud University, Riyadh, Saudi Arabia.
Author contributions
Data curation: QL, NNW, CL. Formal analysis: AME. Methodology: KT, LHH. Software: SAR. Writing - original draft: LSH. Writing - review and editing: DQD, IP.
Author ORCIDs
Li-Su Han https://orcid.org/0000-0001-5380-9928
Nalin N. Wijayawardene https://orcid.org/0000-0003-0522-5498
Chao Liu https://orcid.org/0000-0001-6811-2218
Li-Hong Han https://orcid.org/0000-0002-6127-0915
Itthayakorn Promputtha https://orcid.org/0000-0003-3376-4376
Qiang Li https://orcid.org/0000-0002-9735-8214
Abdallah M. Elgorban https://orcid.org/0000-0003-3664-7853
Salim Al-Rejaie https://orcid.org/0000-0002-9254-1087
Kazuaki Tanaka https://orcid.org/0000-0002-7037-0774
Dong-Qin Dai https://orcid.org/0000-0001-8935-8807
Data availability
All of the data that support the findings of this study are available in the main text.
References
- Cai L, Zhang K, Mckenzie EH, Hyde KD. (2003) Freshwater fungi from bamboo and wood submerged in the Liput River in the Philippines. Fungal Diversity 13: 1–12. [Google Scholar]
- Dai DQ, Phookamsak R, Wijayawardene NN, Li WJ, Bhat DJ, Xu JC, Taylor JE, Hyde KD, Chukeatirote E. (2017) Bambusicolous fungi. Fungal Diversity 82(1): 1–105. 10.1007/s13225-016-0367-8 [DOI] [Google Scholar]
- Dai DQ, Wijayawardene NN, Dayarathne MC, Kumla J, Han LS, Zhang GQ, Zhang X, Zhang TT, Chen HH. (2022) Taxonomicand phylogenetic characterizations reveal four new species, two new asexual morph reports, and six new country records of bambusicolous Roussoella from China. J Fungi 8(5): 532. 10.3390/jof8050532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson OE, Yue JZ. (1998) Bambusicolous pyrenomycetes, an annotated checklist. Myconet 1(2): 25–78. [Google Scholar]
- Hongsanan S, Maharachchikumbura SSN, Hyde KD, Samarakoon MC, Jeewon R, Zhao Q, Al-Sadi AM, Bahkali AH. (2017) An updated phylogeny of Sordariomycetes based on phylogenetic and molecular clock evidence. Fungal Diversity 84(1): 25–41. 10.1007/s13225-017-0384-2 [DOI] [Google Scholar]
- Hsieh HM, Ju YM, Rogers JD. (2005) Molecular phylogeny of Hypoxylon and closely related genera. Mycologia 97(4): 914–923. 10.1080/15572536.2006.11832776 [DOI] [PubMed] [Google Scholar]
- Hyde KD. (1996) Fungi from palms. XXIX. Arecophila gen. nov. (Amphisphaeriaceae, Ascomycota), with five new species and two new combinations. Nova Hedwigia 63: 81–100. [Google Scholar]
- Hyde KD, Zhou D, McKenzie EHC, Ho WH, Dalisay T. (2002a) Vertical distribution of saprobic fungi on bamboo culms. Fungal Diversity 11: 109–118. [Google Scholar]
- Hyde KD, Zhou D, Dalisay T. (2002b) Bambusicolous fungi: A review. Fungal Diversity 9: 1–14. [Google Scholar]
- Hyde KD, Norphanphoun C, Maharachchikumbura SSN, Bhat DJ, Jones EBG, Bundhun D, Chen YJ, Bao DF, Boonmee S, Calabon MS, Chaiwan N, Chethana KWT, Dai DQ, Dayarathne MC, Devadatha B, Dissanayake AJ, Dissanayake LS, Doilom M, Dong W, Fan XL, Goonasekara ID, Hongsanan S, Huang SK, Jayawardena RS, Jeewon R, Karunarathna A, Konta S, Kumar V, Lin CG, Liu JK, Liu NG, Luangsa-ard J, Lumyong S, Luo ZL, Marasinghe DS, McKenzie EHC, Niego AG, Niranjan M, Perera RH, Phukhamsakda C, Rathnayaka AR, Samarakoon MC, Samarakoon SMBC, Sarma VV, Senanayake IC, Shang QJ, Stadler M, Tibpromma S, Wanasinghe DN, Wei DP, Wijayawardene NN, Xiao YP, Yang J, Zeng XY, Zhang SN, Xiang MM. (2020) Refined families of Sordariomycetes. Mycosphere 11(1): 305–1059. 10.5943/mycosphere/11/1/7 [DOI] [Google Scholar]
- Index Fungorum (2024) Index Fungorum. http://www.indexfungorum.org/names/Names.asp [Accessed on 10 December 2023]
- Jeewon R, Liew ECY, Hyde KD. (2003) Molecular systematics of the Amphisphaeriaceae based on cladistic analyses of partial LSU rDNA gene sequences. Mycological Research 107(12): 1392–1402. 10.1017/S095375620300875X [DOI] [PubMed] [Google Scholar]
- Jiang HB, Zhang SJ, Phookamsak R, Promputtha I, Kakumyan P, Xu JC. (2021) Amphibambusahongheensis sp. nov., a novel bambusicolous ascomycete from Yunnan, China. Phytotaxa 505(2): 201–212. 10.11646/phytotaxa.505.2.6 [DOI] [Google Scholar]
- Jiang HB, Phookamsak R, Hongsanan S, Bhat DJ, Mortimer PE, Suwannarach N, Kakumyan P, Xu JC. (2022) A review of bambusicolous Ascomycota in China with an emphasis on species richness in southwest China. Studies in Fungi 7(1): 1–33. 10.48130/SIF-2022-0020 [DOI] [Google Scholar]
- Kang JC, Hyde KD, Kong RYC. (1999) Studies on Amphisphaeriales: The Cainiaceae. Mycological Research 103(12): 1621–1627. 10.1017/S0953756299001264 [DOI] [Google Scholar]
- Katoh K, Standley DM. (2013) MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Molecular Biology and Evolution 30(4): 772–780. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlmeyer J, Volkmann-Kohlmeyer B. (1993) Atrotorquata and Loratospora: New ascomycete genera on Juncusroemerianus. Systema Ascomycetum 12: 7–22. [Google Scholar]
- Konta S, Hyde KD, Eungwanichayapant PD, Karunarathna SC, Samarakoon MC, Xu JC, Aluthwattha ST, Dauner LAP, Tibpromma S, Lumyong S. (2021) Morphology and multigene phylogeny reveal Haploanthostomella gen. et sp. nov. and familial placement of Endocalyx (Xylariales, Sordariomycetes, Ascomycota). Life (Chicago, Ill. ) 11(6): 486. 10.3390/life11060486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krug JC. (1978) The genus Cainia and a new family, Cainiaceae. Sydowia 30: 122–133. [Google Scholar]
- Larsson A. (2014) AliView: A fast and lightweight alignment viewer and editor for large datasets. Bioinformatics (Oxford, England) 30(22): 3276–3278. 10.1093/bioinformatics/btu531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li QR, Zhang X, Lin Y, Samarakoon MC, Hyde KD, Shen XC, Liao WQ, Karunarathna A, Long SH, Kang YQ, Kang JC. (2022) Morpho-molecular characterisation of Arecophila, with A.australis and A.clypeata sp. nov. and A.miscanthi comb. nov. MycoKeys 88: 123–149. 10.3897/mycokeys.88.79475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YL, Whelen S, Hall BD. (1999) Phylogenetic relationships among ascomycetes: Evidence from an RNA polymerase II subunit. Molecular Biology and Evolution 16(12): 1799–1808. 10.1093/oxfordjournals.molbev.a026092 [DOI] [PubMed] [Google Scholar]
- Liu JK, Hyde KD, Jones EBG, Buyck B, Chethana KWT, Dai DQ, Dai YC, Daranagama DA, Dissanayake AJ, Doilom M, D’souza MJ, Fan XL, Goonasekara ID, Hirayama K, Hongsanan S, Jayasiri SC, Jayawardena RS, Karunarathna SC, Li WJ, Mapook A, Norphanphoun C, Pang KL, Perera RH, Peršoh D, Pinruan U, Senanayake IC, Somrithipol S, Suetrong S, Tanaka K, Thambugala KM, Tian Q, Tibpromma S, Udayanga D, Wijayawardene NN, Wanasinghe D, Wisitrassameewong K, Zeng XY, Abdel-Aziz FA, Adamčík S, Bahkali AH, Boonyuen N, Bulgakov T, Callac P, Chomnunti P, Greiner K, Hashimoto A, Hofstetter V, Kang JC, Lewis D, Li XH, Liu XZ, Liu ZY, Matsumura M, Mortimer PE, Rambold G, Randrianjohany E, Sato G, Sri-Indrasutdhi V, Tian CM, Verbeken A, von Brackel W, Wang Y, Wen TC, Xu JC, Yan JY, Zhao RL, Camporesi E. (2015) Fungal diversity notes 1–110: Taxonomic and phylogenetic contributions to fungal species. Fungal Diversity 72(1): 1–197. 10.1007/s13225-015-0324-y [DOI] [Google Scholar]
- Ma XM. (2016) Study on complete mitochondrial genome of Cypridopsisvidua and molecular phylogeny of Ostracoda. Ph.D. Thesis, East China Normal University, Shanghai, China.
- Maharachchikumbura SSN, Hyde KD, Jones EBG, McKenzie EHC, Huang SK, Abdel-Wahab MA, Daranagama DA, Dayarathne M, D’souza MJ, Goonasekara ID, Hongsanan S, Jayawardena RS, Kirk PM, Konta S, Liu JK, Liu ZY, Norphanphoun C, Pang KL, Perera RH, Senanayake IC, Shang QJ, Shenoy BD, Xiao YP, Bahkali AH, Kang JC, Somrothipol S, Suetrong S, Wen TC, Xu JC. (2015) Towards a natural classification and backbone tree for Sordariomycetes. Fungal Diversity 72(1): 199–301. 10.1007/s13225-015-0331-z [DOI] [Google Scholar]
- Maharachchikumbura SSN, Hyde KD, Jones EBG, McKenzie EHC, Bhat DJ, Dayarathne MC, Huang SK, Norphanphoun C, Senanayake IC, Perera RH, Shang QJ, Xiao YP, D’souza MJ, Hongsanan S, Jayawardena RS, Daranagama DA, Konta S, Goonasekara ID, Zhuang WY, Jeewon R, Phillips AJL, Abdel-Wahab MA, Al-Sadi AM, Bahkali AH, Boonmee S, Boonyuen N, Cheewangkoon R, Dissanayake AJ, Kang JC, Li QR, Liu JK, Liu XZ, Liu ZY, Luangsa-ard JJ, Pang KL, Phookamsak R, Promputtha I, Suetrong S, Stadler M, Wen TC, Wijayawardene NN. (2016) Families of Sordariomycetes. Fungal Diversity 79(1): 1–317. 10.1007/s13225-016-0369-6 [DOI] [Google Scholar]
- Mapook A, Hyde KD, McKenzie EHC, Jones EBG, Bhat DJ, Jeewon R, Stadler M, Samarakoon MC, Malaithong M, Tanunchai B, Buscot F, Wubet T, Purahong W. (2020) Taxonomic and phylogenetic contributions to fungi associated with the invasive weed Chromolaenaodorata (Siam weed). Fungal Diversity 101(1): 1–175. 10.1007/s13225-020-00444-8 [DOI] [Google Scholar]
- Miller MA, Pfeiffer W, Schwartz T. (2010) Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Proceedings of the Gateway Computing Environments Workshop (GCE). New Orleans, Louisiana, 1–8. 10.1109/GCE.2010.5676129 [DOI]
- MycoBank (2024) MycoBank. https://www.mycobank.org/ [Accessed on 23 January 2024]
- Rambaut A. (2012) FigTree v1. 4.0. a Graphical viewer of phylogenetictrees. http://tree.bio.ed.ac.uk/software/figtree/ [Accessed on 3 January 2024]
- Rannala B, Yang Z. (1996) Probability distribution of molecular evolutionary trees, a new method of phylogenetic inference. Journal of Molecular Evolution 43(3): 304–311. 10.1007/BF02338839 [DOI] [PubMed] [Google Scholar]
- Ronquist F, Teslenko M, Van Der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. (2012) MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61(3): 539–542. 10.1093/sysbio/sys029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samarakoon MC, Hyde KD, Maharachchikumbura SSN, Stadler M, Jones EBG, Promputtha I, Suwannarach N, Camporesi E, Bulgakov TS, Liu JK. (2021) Taxonomy, phylogeny, molecular dating and ancestral state reconstruction of Xylariomycetidae (Sordariomycetes). Fungal Diversity 112(1): 1–88. 10.1007/s13225-021-00495-5 [DOI] [Google Scholar]
- Senanayake IC, Maharachchikumbura SSN, Hyde KD, Bhat JD, Jones EBG, McKenzie EHC, Phookamsak R, Phukhamsakda C, Shenoy BD. (2015) Towards unraveling relationships in Xylariomycetidae (Sordariomycetes). Fungal Diversity 73(1): 73–144. 10.1007/s13225-015-0340-y [DOI] [Google Scholar]
- Species Fungorum (2024) Species Fungorum. https://www.speciesfungorum.org/Names/Names.asp [Accessed on 10 October 2023]
- Stamatakis A. (2014) RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics (Oxford, England) 30(9): 1312–1313. 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A, Hoover P, Rougemont J. (2008) A rapid bootstrap algorithm for the ML web servers. Systematic Biology 57(5): 758–771. 10.1080/10635150802429642 [DOI] [PubMed] [Google Scholar]
- Umali TE, Hyde KD, Quimio TH. (1999) Arecophilabambusae sp. nov. and A.coronata comb. nov., from dead culms of bamboo. Mycoscience 40(2): 185–188. 10.1007/BF02464296 [DOI] [Google Scholar]
- Vaidya G, Lohman DJ, Meier R. (2011) Sequence Matrix: Concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics 27(2): 171–180. 10.1111/j.1096-0031.2010.00329.x [DOI] [PubMed] [Google Scholar]
- Vilgalys R, Hester M. (1990) Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. Journal of Bacteriology 172(8): 4238–4246. 10.1128/jb.172.8.4238-4246.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YZ, Aptroot A, Hyde KD. (2004) Revision of the genus Amphisphaeria. Hong Kong SAR, China. Fungal Diversity Research Series 13: 1–168. [Google Scholar]
- White TJ, Bruns T, Lee S, Taylor J. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR protocols: a guide to methods and applications 18: 315–322. 10.1016/B978-0-12-372180-8.50042-1 [DOI] [Google Scholar]
- Wijayawardene NN, Hyde KD, Al-Ani LKT, Tedersoo L, Haelewaters D, Rajeshkumar KC, Zhao RL, Aptroot A, Leontyev DV, Saxena RK, Tokarev YS, Dai DQ, Letcher PM, Stephenson SL, Ertz D, Lumbsch HT, Kukwa M, Issi IV, Madrid H, Phillips AJL, Selbmann L, Pfliegler WP, Horváth E, Bensch K, Kirk PM, Kolaříková K, Raja HA, Radek R, Papp V, Dima B, Ma J, Malosso E, Takamatsu S, Rambold G, Gannibal PB, Triebel D, Gautam AK, Avasthi S, Suetrong S, Timdal E, Fryar SC, Delgado G, Réblová M, Doilom M, Dolatabadi S, Pawłowska JZ, Humber RA, Kodsueb R, Sánchez-Castro I, Goto BT, Silva DKA, de Souza FA, Oehl F, da Silva GA, Silva IR, Błaszkowski J, Jobim K, Maia LC, Barbosa FR, Fiuza PO, Divakar PK, Shenoy BD, Castañeda-Ruiz RF, Somrithipol S, Lateef AA, Karunarathna SC, Tibpromma S, Mortimer PE, Wanasinghe DN, Phookamsak R, Wang Y, Tian F, Alvarado P, Li DW. (2020) Outline of Fungi and fungus-like taxa. Mycosphere: Journal of Fungal Biology 11(1): 1060–1456. 10.5943/mycosphere/11/1/8 [DOI] [Google Scholar]
- Wijayawardene NN, Hyde KD, Anand G, Dissanayake LS, Tang LZ, Dai DQ. (2021a) Towards incorporating asexually reproducing fungi in the natural classification and notes for pleomorphic genera. Mycosphere: Journal of Fungal Biology 12(1): 238–405. 10.5943/mycosphere/12/1/4 [DOI] [Google Scholar]
- Wijayawardene NN, Dissanayake LS, Li QR, Dai DQ, Xiao YP, Wen TC, Karunarathna SC, Wu HX, Zhang H, Tibpromma S. (2021b) Yunnan–Guizhou Plateau: A mycological hotspot. Phytotaxa 523(1): 1–31. 10.11646/phytotaxa.523.1.1 [DOI] [Google Scholar]
- Wijayawardene NN, Hyde KD, Dai DQ, Sanchez-Garcia M, Goto BT, Saxena RK, Erdogdu M, Selçuk F, Rajeshkumar KC, Aptroot A, Błaszkowski J, Boonyuen N, da Silva GA, de Souza FA, Dong W, Ertz D, Haelewaters D, Jones EBG, Karunarathna SC, Kirk PM, Kukwa M, Kumla J, Leontyev DV, Lumbsch HT, Maharachchikumbura SSN, Marguno F, Martínez-Rodríguez P, Mešić A, Monteiro JS, Oehl F, Pawłowska J, Pem D, Pfliegler WP, Phillips AJL, Pošta A, He MQ, Li JX, Raza M, Sruthi OP, Suetrong S, Suwannarach N, Tedersoo L, Thiyagaraja V, Tibpromma S, Tkalčec Z, Tokarev YS, Wanasinghe DN, Wijesundara DSA, Wimalaseana SDMK, Madrid H, Zhang GQ, Gao Y, Sánchez-Castro I, Tang LZ, Stadler M, Yurkov A, Thines M. (2022a) Outline of Fungi and fungus-like taxa. Mycosphere : Journal of Fungal Biology 13(1): 55–453. 10.5943/mycosphere/13/1/2 [DOI] [Google Scholar]
- Wijayawardene NN, Phillips AJL, Pereira DS, Dai DQ, Aptroot A, Monteiro JS, Druzhinina IS, Cai F, Fan XL, Selbmann L, Coleine C, Castañeda-Ruiz RF, Kukwa M, Flakus A, Fiuza PO, Kirk PM, Rajesh Kumar KC. (2022b) Forecasting the number of species of asexually reproducing fungi (Ascomycota and Basidiomycota). Fungal Diversity 114(1): 63–490. 10.1007/s13225-022-00500-5 [DOI] [Google Scholar]
- Zhaxybayeva O, Gogarten JP. (2002) Bootstrap bayesian probability and maximum likelihood mapping, exploring new tools for comparative genome analyses. MBC genomics 3: 1–15. 10.1186/1471-2164-3-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D, Hyde KD. (2002) Fungal succession on bamboo in Hong Kong. Fungal Diversity 10: 213–217. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All of the data that support the findings of this study are available in the main text.