Abstract
Background
Antimicrobial resistance (AMR) is a major health concern with high rates in low-income countries. Bacteriology laboratories sustain the fight against AMR by providing antibiotic susceptibility testing (AST) results to ensure appropriate therapies. These laboratories generate a lot of data, which are usually used for prospective interventions. Our study conducted in a lower-middle-income hospital setting aimed to describe the profile of bacteria isolated from the specimens received over 3 years, assess their susceptibility profile and identify potential gaps or area of improvement from the analysis of our data.
Methods
Monthly data were retrieved from registers for all specimens received between January 2020 until December 2022. Data were compiled and analysed using the R and WHONET software.
Results
Out of 3582 specimens received, 797 were culture positive (22.3%). Escherichia coli and Klebsiella pneumoniae were frequently isolated (30.5% and 24.2%, respectively). AST results analysis showed high resistance of Gram-negative bacteria to penams and cephems, whereas low resistance was observed to carbapenems. Susceptibility to antibiotics based on the AWaRe antibiotic classification was variable. The bacteriological profile in the various types of specimen was established and rational information to design a therapeutic protocol adapted to our hospital setting was obtained.
Conclusions
AST results may not only be used for prospective guidance for treatment, but rather cumulative data analysis can contribute to design effective antibiotic prescriptions and improve general practices at the laboratory. This is, however, dependent on a good record-keeping, standardization of practices and collaboration between clinicians and laboratory scientists.
Introduction
Antimicrobial resistance (AMR) is a global health concern considered as the next pandemic and threat worldwide.1 It is estimated that by 2050 up to 10 million deaths could occur annually due to AMR.2 Although the development of antibiotic resistance (ABR) can arise from pathogens’ natural defence mechanisms, it is enhanced by inappropriate behaviours, overuse or misuse of antibiotics in humans, animals and the environment.3,4,5 In parallel, there has not been much discovery of new antibiotics over the past two decades. Even if intensive research is ongoing, leading to promising future discovery,6 it is urgent to target other effective areas of interventions, which could quickly help in the control of ABR. Such areas of interventions include antimicrobial stewardship. Antimicrobial stewardship is an integrated and multidisciplinary approach to select appropriate drugs for appropriate patients for a proper duration to minimize the risk of developing AMR. This approach also promotes implementation of guidelines, and continuous medical education.7 The impact of antimicrobial stewardship in reducing AMR was clearly demonstrated by several studies;8,9 however, it is also advisable that such interventions should be designed and implemented based on available resources and expertise.10 The selection of the appropriate drug for the appropriate patient mainly relies on the use of the antibiotic susceptibility testing (AST) results, which definitely provide the insight of which antibiotic to use based on the pathogen isolated. Bacteriology laboratories performing AST may therefore generate a lot of data, which are frequently used for prospective interventions, either readjustment of a patient’s treatment or surveillance. This paper describes how the analysis of retrospective data generated from AST could be used as part of the antibiotic stewardship process at the level of the health facility. Specifically, the study aimed to describe the profile of bacteria isolated from the specimens received over 3 years, assess their antibiotic susceptibility profile and identify potential gaps or areas of improvement from the analysis of our data.
Methods
Setting
The study was conducted at the bacteriology laboratory of the Yaoundé General Hospital. This is a first-category health facility in Yaoundé, the capital city of Cameroon, with a general capacity of 200 beds and up to 21 medical and subsurgical specialties. The health facility has many reference pools such as the oncology management centre. The bacteriology unit receives specimen from inpatients and outpatients.
Data entry system
Data retrieved from the registers were entered into the WHONET® software. This is a free desktop Windows application for the management and analysis of microbiology laboratory data with a particular focus on AMR surveillance. It has been developed and supported by the WHO Collaborating Centre for Surveillance of Antimicrobial Resistance at the Brigham and Women’s Hospital in Boston, MA, USA. WHONET supports local, national, regional and global surveillance efforts in over 2300 hospitals, public health, animal health and food laboratories in over 130 countries worldwide.11 Aerobic bacteria isolated from specimens received from January 2020 until December 2022 were recorded and their susceptibility profiles were analysed. The specimens considered were urine, stool, blood, catheters, body fluids and pus (from wounds, swabs or surgery). Genital samples were not included. Anaerobic bacteria were not included in the study.
Laboratory methods
Culture and identification methods
The general culturing method was an incubation at 36°C ± 5°C in aerobic conditions or in a 5% CO2-enriched atmosphere in a jar, for blood-enriched culture media.
Urine samples were plated on cystine lactose electrolyte-deficient (CLED) agar. Stool samples were both simultaneously plated on Hektoen and Salmonella Shigella agar, whereas another part was enriched in Muller–Kauffmann or selenite-F broth before plating on to agar after 24 h incubation. Blood samples were collected on diphasic culture bottles, which were incubated at aerobic conditions for at least seven days, and crosschecked every 48 h. Pus specimens were directly plated on blood-enriched based media and incubated in a jar under a 5% CO2-enriched environment.
Identification was performed using Gram staining, and biochemical orientation tests such as catalase and oxidase. API 20E®mini galleries were used for identification of Enterobacteriaceae isolates. For Staphylococcus species, identification was confirmed after coagulase and DNase tests, whereas for Streptococcus species, blood agar haemolytic patterns, Gram staining and Streptokit® were used for general group identification.
AST
The Kirby–Bauer disc diffusion method was the antibiotic susceptibility technique performed at the laboratory over the 3 years surveyed. The main brands supplied were Oxoid® and Bio-Analyze®. The disc concentrations were those recommended by the EUCAST guidelines of the corresponding year. Discs of antibiotics were placed on the top of Mueller–Hinton agar, inoculated with a 0.5 McFarland suspension of the isolate. After a minimum of 16 h incubation, diameters of the inhibitory zones were measured manually using an electronic sliding caliper. Susceptibility results were interpreted based on the EUCAST guidelines.
Ethics
We received administrative authorization to perform data analysis. Ethical clearance was delivered by the Institutional Research Ethics Committee, with the reference number 121/UY1/FMSB/VDRC/DAASR/CSO.
Results
Statistics of samples received from 2020 to 2022
In total, 3582 samples were received for culture between 2020 and 2022. The most frequent samples were urine, followed by blood and stool samples. The number of samples increased over the years for urine, blood, stool, pus and body fluids. The proportions of positive samples per type of specimen in 2020 and 2022 are presented in Table 1. The overall percentage positivity of specimens cultured decreased between 2020 and 2022. The proportion of positive urine samples also considerably reduced, from 33.7% to 11.4%. The highest positivity rates were observed for catheters and suppurated specimens.
Table 1.
Proportion of positive samples per type of specimen
| Type of samples | Samples received/positive (+) samples | 2020 | 2021 | 2022 | |
|---|---|---|---|---|---|
| Samples from sterile sites | Blood | Samples received, n | 265 | 297 | 244 |
| + samples, n (%) | 68 (25.7) | 60 (20.2) | 78 (32) | ||
| Body fluid | Samples received, n | 34 | 56 | 122 | |
| + abdominal fluid, n (%) | 5 (14.7) | 2 (3.6) | 0 | ||
| + CSF, n (%) | 1 (3) | 2 (3.6) | 3 | ||
| + knee fluid, n (%) | 1 (3) | 0 | 0 | ||
| + others: biopsy, cyst, pleural, n (%) | 2 (6) | 2 (3.6) | 2 | ||
| Catheters | Samples received, n | 22 | 14 | 62 | |
| + urine catheters, n (%) | 8 (36) | 8 (57) | 39 (63) | ||
| + venous catheters, n (%) | 6 (27) | 3 (21.4) | 5 (8) | ||
| Urine | Samples received, n | 347 | 587 | 709 | |
| + samples, n (%) | 117 (33.7) | 160 (27.3) | 81 (11.4) | ||
| Total | Samples received, n | 668 | 954 | 1137 | |
| + samples, n (%) | 208 (31.1) | 237 (24.8) | 208 (18.3) | ||
| Samples from non- sterile sites | Pus | Samples received, n | 53 | 106 | 118 |
| + samples, n (%) | 24 (45.3) | 59 (55.7) | 56 (47.5) | ||
| Stool | Samples received, n | 147 | 289 | 100 | |
| + samples, n (%) | 3 (2) | 1 (0.3) | 1 (1) | ||
| Total | Samples received, n | 200 | 395 | 218 | |
| + samples, n (%) | 27 (13.5) | 60 (15.2) | 57 (26.1) | ||
| Total samples from sterile and non- sterile sites | Samples received, n | 868 | 1349 | 1355 | |
| + samples, n (%) | 235 (27) | 297 (22) | 265 (19.6) | ||
Age group and sex distribution of patients with positive cultured samples
The age group and sex demographics of patients with positive cultures are presented in Table 2. The mean age of patients with positive culture samples was 45 years. We observed a slight male predominance in 2021 and 2022.
Table 2.
Sex and age group distribution of patients with positive samples
| 2020 | 2021 | 2022 | ||
|---|---|---|---|---|
| Sex, n (%) | Female | 127 (54) | 141 (47.6) | 122 (46.2) |
| Male | 108 (46) | 156 (52.4) | 143 (53.8) | |
| Age (years) | Mean | 45 | 46 | 46 |
| Min | 0 | 1 | 1 | |
| Max | 91 | 98 | 93 | |
| Age group (years), n (%) | 0–4 | 16 (6.8) | 18 (6) | 24 (9) |
| 5–14 | 9 (3.7) | 15(5) | 7 (3) | |
| 15–24 | 29 (12.3) | 25 (8.4) | 12 (5) | |
| 25–34 | 33 (14.1) | 57 (19.1) | 13 (5) | |
| 35–44 | 22 (9.2) | 31 (10.4) | 55 (21) | |
| 45–54 | 25 (10.4) | 21 (7) | 65 (24) | |
| 55–64 | 49 (20.9) | 47 (16) | 42 (16) | |
| 65–74 | 32 (13.5) | 47 (16) | 30 (11) | |
| 75–84 | 14 (6.1) | 25 (8.4) | 14(5) | |
| >85 | 6 (3) | 11 (3.7) | 3 (1) | |
| Total | 235 | 297 | 265 |
Pathogens isolated from specimens
The most frequent bacteria isolated from the various types of specimen are presented in Table 3.
Table 3.
Frequency of bacteria isolated from targeted specimens
| Bacteria | Overall, n (%) | Blood, n (%) | Body fluid, n (%) | Catheter, n (%) | Pus, n (%) | Urine, n (%) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2020 | 2021 | 2022 | 2020 | 2021 | 2022 | 2020 | 2021 | 2022 | 2020 | 2021 | 2022 | 2020 | 2021 | 2022 | ||
| Gram-negative isolates | ||||||||||||||||
| Acinetobacter baumannii | 1 (0.1) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (0.85) | 0 (0.0) | 0 (0.0) |
| Citrobacter freundii | 18 (2.4) | 2 (2.9) | 1 (1.7) | 2 (2.6) | 0 (0.0) | 1 (16.7) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 2 (4.5) | 0 (0.0) | 4 (6.9) | 0 (0.0) | 1 (0.85) | 4 (2.5) | 1 (1.2) |
| Citrobacter koseri | 1 (0.1) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (20.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Enterobacter cloacae | 28 (3.6) | 6 (8.8) | 2 (3.4) | 2 (2.6) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 3 (6.8) | 0 (0.0) | 5 (8.6) | 8 (14) | 0 (0.0) | 5 (3.1) | 3 (3.7) |
| E. coli | 241 (30.5) | 5 (7.4) | 1 (1.7) | 10 (12.8) | 4 (40.0) | 1 (16.7) | 2 (40.0) | 4 (26.7) | 4 (40.0) | 16 (36.4) | 2 (8.3) | 8 (13.8) | 8 (14) | 63 (53.8) | 77 (48.1) | 32 (39.5) |
| Klebsiella aerogenes | 9 (1.1) | 0 (0.0) | 1 (1.7) | 3 (3.8) | 1 (10.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (0.9) | 0 (0.0) | 3 (3.7) |
| Klebsiella oxytoca | 15 (1.9) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 5 (8.6) | 2 (3.5) | 1 (0.9) | 4 (2.5) | 3 (3.7) |
| K. pneumoniae subsp. pneumoniae | 191 (24.2) | 11 (16.2) | 8 (13.6) | 14 (17.9) | 2 (20.0) | 2 (33.2) | 1 (20.0) | 2 (13.3) | 4 (40.0) | 10 (22.7) | 4 (16.7) | 17 (29.3) | 13 (22.8) | 24 (20.5) | 53 (33.1) | 26 (32.1) |
| Morganella morganii | 2 (0.2) | 0 (0.0) | 1 (1.7) | 1 (1.3) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| P. mirabilis | 8 (1.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 3 (5.2) | 2 (3.5) | 0 (0.0) | 1 (0.6) | 2 (2.5) |
| P. aeruginosa | 50 (6.4) | 1 (1.5) | 7 (11.9) | 4 (5.1) | 0 (0.0) | 1 (16.7) | 0 (0.0) | 1 (6.7) | 0 (0.0) | 3 (6.8) | 10 (41.7) | 7 (12.1) | 7 (12.3) | 2 (1.7) | 2 (1.3) | 5 (6.2) |
| Salmonella spp. | 8 (1.0) | 5 (7.4) | 2 (3.4) | 1 (1.3) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Serratia liquefaciens | 1 (0.1) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (20.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Serratia marcescens | 32 (4.0) | 2 (2.9) | 3 (5.1) | 2 (2.6) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (6.7) | 1 (10.0) | 3 (6.8) | 0 (0.0) | 1 (1.7) | 6 (10.6) | 6 (5.1) | 5 (3.1) | 2 (2.5) |
| Gram-positive isolates | ||||||||||||||||
| Enterococcus spp. | 1 (0.1) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (1.8) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| S. aureus subsp. aureus | 157 (19.9) | 27 (39.7) | 28 (47.5) | 34 (43.6) | 3 (30.0) | 1 (16.7) | 0 (0.0) | 7 (46.6) | 1 (10.0) | 6 (13.6) | 6 (25) | 8 (13.8) | 10 (17.5) | 18 (15.4) | 4 (2.5) | 4 (4.9) |
| CoNS | 21 (2.7) | 8 (11.8) | 4 (6.8) | 4 (5.1) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 5 (3.2) | 0 (0.0) |
| Streptococcus spp. | 6 (0.7) | 1 (1.4) | 1 (1.5) | 1 (1.3) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (2.4) | 2 (8.3) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
Negative samples were considered only for growth under aerobic conditions.
Over the 3 years’ time frame, Escherichia coli, Klebsiella pneumoniae and Staphylococcus aureus remained the most frequent bacteria isolated in urine. Other bacteria such as Citrobacter, Enterobacter and Proteus mirabilis were isolated with variable frequency over the years. Staphylococcus saprophyticus, considered for urine samples, was isolated only in the year 2022.
Staphylococcus aureus was the most prevalent bacterium isolated from blood cultures, followed by K. pneumoniae. CoNS species were also isolated with decreasing occurrence from 11.8% in 2020 to 5% in 2022.
Apart from a high prevalence of Pseudomonas aeruginosa observed in suppurated specimens collected in 2020, the main bacterium isolated in these specimens was K. pneumoniae followed by S. aureus. Streptococcus species were isolated only in 2020.
S. aureus was the most prevalent bacterium isolated from catheters in 2020 then the prevalence decreased by 10%–15% in 2021 and 2022. Globally, E. coli and K. pneumoniae tend to be the most frequent bacteria isolated from catheters. The frequency of bacteria isolated from body fluids over the years was variable.
For stool culture, Salmonella typhi was the main bacterium isolated in 2020 (2%) and 2022 (1%). Only one species of Shigella was isolated from stool in 2021.
Antibiotic susceptibility patterns of priority pathogens
The susceptibility patterns for priority pathogens according to the WHO GLASS tool12 are presented in Figures 1–3. Antibiotics are classified according to the WHO Access, Watch, Reserve (AWaRe) classification.13
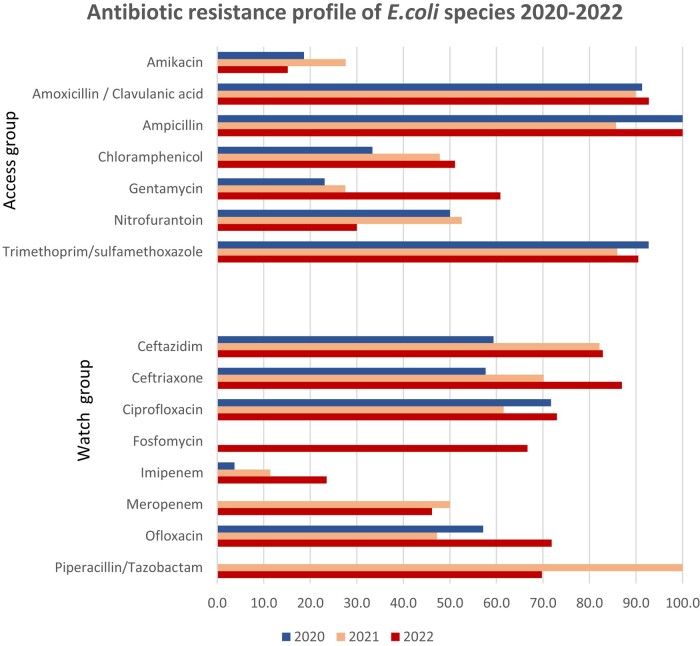
Figure 1.
Antibiotic resistance profile of E. coli 2020–22.
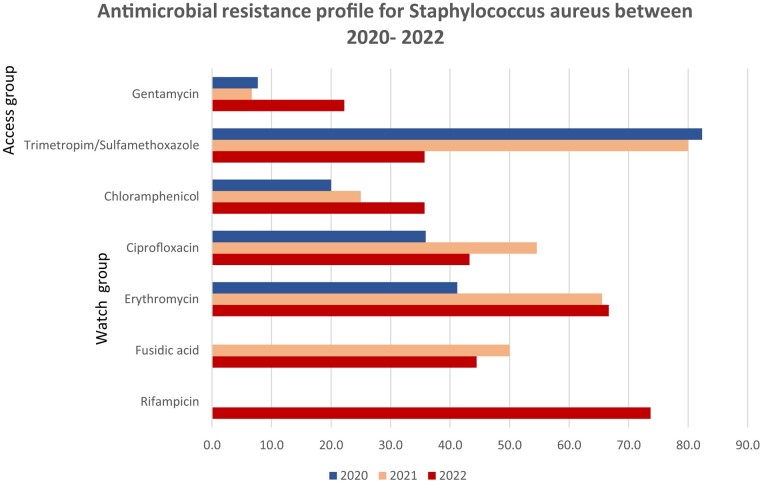
Figure 3.
Antimicrobial resistance profile of S. aureus 2020–22.
Among the Access group of antibiotics, ampicillin and amoxicillin/clavulanic acid had the lowest susceptibility rates. The trimethoprim/sulfamethoxazole combination also showed low rates of susceptibility. The highest rates were found for nitrofurantoin and amikacin. For species isolated in urine, more than 50% of isolates were susceptible to nitrofurantoin and a slightly lower rate was observed for fosfomycin.
Within the Watch group, the lowest susceptibility rates were observed with third-generation cephalosporins. The highest susceptibility was observed with imipenem.
According to the 2022 GLASS report, surveillance of resistance for E. coli species focuses on third- and fourth-generation cephalosporins, quinolones, carbapenems, sulphonamides and polymyxins, especially colistin. Colistin is also classified within the Reserve group of the AWaRe classification, but it was not part of the antibiotics tested within the laboratory, as the recommended testing method is MIC determination.
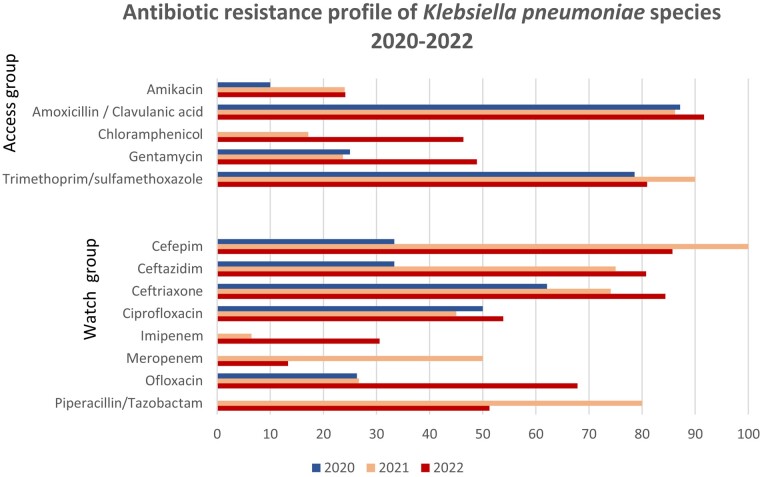
E. coli showed high resistance rates to most of the antibiotics under surveillance, according to the GLASS report. Antibiotic resistance of K. pneumoniae are presented in Figure 2. The resistance profile for K. pneumoniae followed the same patterns as E. coli. Resistance patterns in Staphylococcus species were as follows: gentamicin showed the highest susceptibility rates for S. aureus, followed by fusidic acid. Trimethoprim/sulfamethoxazole showed the lowest susceptibility rates.
Figure 2.
Antibiotic resistance profile of K. pneumoniae 2020–22.
Discussion
Samples received and bacterial ecology
Although the total number of samples received increased from 2020 to 2022, concordant with a parallel increase in the frequency of patients attending the health facility, data showed a progressive decrease in the proportion of positive samples over the years. This was particularly noticeable for urine samples, which dropped from 34.6% in 2020 to 11.4% in 2022. This global decrease may have been impacted by two key factors. From 2020, the laboratory was selected as part of the national network for surveillance of AMR. Being part of this network, it involved retraining of laboratory staff and capacity building for improvement of procedures and AST. It also involved more rigorous screening of sample collection procedures, which would have decreased the number of samples contaminated. The decrease may have also been influenced by an improved collaboration between clinicians and biologists, as cases with less significant bacteriuria were discussed before taking the decision to pursue with AST, depending on the patient’s condition and symptoms.14
Stool culture presented the lowest percentages of positivity. As litterature suggests that the presence of Salmonella and Shigella species is associated with age, especially children.15,16 The low positivity rates observed may partially be linked to our observed population which were mainly adults. The level of the health facility, which is a reference hospital, could also explain these findings, as people with acute diarrhoea would preferably consult in peripheral health structures rather than reference hospitals or would have performed self-medication.
Catheters had the highest percentages of positivity but the yearly number of samples tested remained quite low. This suggests that the indication for such cultures was guided by strong evidence of these catheters being the source of the infection.
E. coli, K. pneumoniae and S. aureus were globally the most frequent bacteria isolated from urine in our study. If the first two are clearly described as main causes of urinary tract infections, S. aureus has been reported in lower frequencies.17 The relatively higher frequency of S. aureus was observed in patients with cancer and this may probably be the same in our setting,18,19,20 the Yaoundé General Hospital being a reference centre for cancer treatment in the country. Globally, authors have acknowledged great variations in the bacterial ecology in urine, depending on age, patient’s status and region, thus highlighting the importance of having local epidemiology data available.17
For blood cultures, the patterns observed were as described in several other studies.21, 22 Bacteria isolated throughout the years were also described among those who can be responsible for sepsis.23
Besides knowing the bacteria’s profile in our hospital setting, the data provided over these 3 years will also support accurate procurement for reagents and consumables.
AST
The global patterns of susceptibility for E. coli were comparable to data observed in other studies. This involved low susceptibility to aminopenicillins, third-generation cephalosporins and sulphonamides, medium susceptibility to quinolones and high susceptibility to aminoglycosides, especially amikacin, carbapenems and nitrofurantoin.24,25 However, the proportions observed from our data were consistently lower. Lower susceptibility of the amoxicillin/clavulanic acid combination may have been influenced by batch-to-batch variations and storage conditions. This observation has led to improvement of quality assurance for AST, which will be described in the upcoming sections. All this information guided specific improvement interventions at the level of the laboratory. When considering imipenem, we observed about 10% phenotypic resistance. Although some authors described the presence of phenotypic resistance patterns without the presence of resistance genes,26 others near us have found the presence of resistance genes even though in a lower proportion compared with phenotypic resistance patterns.27 This finding raised our concern to quickly set up additional tests for phenotypic carbapenemase detection, and also storage of strains for further genotypic assays. We also found low susceptibility to fosfomycin compared with other studies,28 as the disc diffusion method for fosfomycin implies some additional specificity such as adding G6PD solution, which is ideal and more accurate in the determination of MIC. For nitrofurantoin, we recorded more than 60% susceptibility of the strains, rendering this antibiotic a potential good option for the treatment of uncomplicated urinary tract infections.
Improvement areas and ways forward
From the analysis of our data, we found that there were some gaps in terms of quality assurance, especially for AST and specifically preservation of the quality of the discs. If we confirmed the quality of the discs upon reception and before use, we observed that there was potentially a decrease of potency over time. Thus, we performed a global check of all the discs that were still in stock and those that presented with inappropriate results were discarded. We also systematically adopted keeping discs frozen, for those for whom the temperature storage ranged between 2°C and −20°C. Furthermore, rather than purchasing extremely high quantities of antibiotics to be stocked in order to avoid shortages, we decided to perform quarterly procurement based on our statistics from the past 3 years. Also, we systematically introduced quality control of discs every month.
As for some of the antibiotics tested, determination of MIC is reported to provide more effective results, considering the fact that manual measurement of inhibitory diameters was not repeatedly performed to minimize operator errors at the laboratory. Based on our findings, we got engaged in an advocacy with the managers to quickly implement such methods in our laboratory.
We have also integrated procurement of reagents that will help in the systematic research of resistance patterns, ESBLs, MRSA or carbapenem-resistant strains.
Our data showed variable susceptibility to Access group antibiotics, which are considered as the first choice for antibiotic prescription. Molecules such as nitrofurantoin could be recommended for uncomplicated urinary tract infections based on our results. Based on the profile described per specimen and the antibiotic susceptibility profile, working groups between biologists and clinicians have been launched to define strategic antibiotic prescription guidelines for the health facility.
Improvement can also be planned for data collection, as for example, systematic recording of the specimen origin could lead to additional description of bacterial epidemiology depending on the units.
Conclusions
The review of the data from the bacteriology laboratory over the past 3 years provided a lot of information, from the trends in the number of samples received to the epidemiology of bacteria isolated and their susceptibility patterns. Use of such data is strongly dependent on good record-keeping, and software such as the WHONET® is helpful both for recording and data analysis. Analysis of results and comparison with other findings helped to detect potential gaps in quality control for AST and also raised several areas of improvement (internal control quality at reduced periodicity for AST, appropriate methods for AST). Furthermore, the process of exploitation of the data for general improvement of antibiotic prescription and use within the hospital enhanced more effective collaboration between clinicians and laboratory scientists. Overall, despite the numerous challenges that may be encountered in laboratories in low and middle-income settings, there is great interest to use AST data as part of the implementation of antimicrobial stewardship.
Contributor Information
Marie Paule Ngogang, Faculty of Medicine and Biomedical Sciences, Yaoundé I University, Yaounde, Cameroon; Yaoundé General Hospital, Yaounde, Cameroon.
Abel fils Nkoth, Yaoundé General Hospital, Yaounde, Cameroon.
Welysiane Ngaleu, Yaoundé General Hospital, Yaounde, Cameroon.
Heroine Mfouapon, Yaoundé General Hospital, Yaounde, Cameroon.
Priscille Ekoume, Yaoundé General Hospital, Yaounde, Cameroon.
Yannick Nibeye, Faculty of Medicine and Biomedical Sciences, Yaoundé I University, Yaounde, Cameroon.
Christiane Medi Sike, Faculty of Medicine and Biomedical Sciences, Yaoundé I University, Yaounde, Cameroon.
Esther Voundi Voundi, Faculty of Medicine and Biomedical Sciences, Yaoundé I University, Yaounde, Cameroon.
Mohammed Moctar Mouliom Mouiche, Infectious Diseases Detection and Surveillance, USAID, Yaounde, Cameroon.
Marie Christine Fonkoua, Infectious Diseases Detection and Surveillance, USAID, Yaounde, Cameroon.
Michel Toukam, Faculty of Medicine and Biomedical Sciences, Yaoundé I University, Yaounde, Cameroon.
Francois-Xavier Mbopi-Keou, Faculty of Medicine and Biomedical Sciences, Yaoundé I University, Yaounde, Cameroon.
Funding
This study was carried out as part of our routine work.
Transparency declarations
None to declare.
References
- 1. Gautam A. Antimicrobial resistance: the next probable pandemic. JNMA J Nepal Med Assoc 2022; 60: 225–8. 10.31729/jnma.7174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. UN Environment Programme . Antimicrobial resistance: a global threat. https://www.unep.org/explore-topics/chemicals-waste/what-we-do/emerging-issues/antimicrobial-resistance-global-threat.
- 3. Aljeldah MM. Antimicrobial resistance and its spread is a global threat. Antibiotics (Basel) 2022; 11: 1082. 10.3390/antibiotics11081082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ngogang MP, Ernest T, Kariuki J et al. Microbial contamination of chicken litter manure and antimicrobial resistance threat in an urban area setting in Cameroon. Antibiotics 2021; 10: 20. 10.3390/antibiotics10010020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Polianciuc SI, Gurzău AE, Kiss B et al. Antibiotics in the environment: causes and consequences. Med Pharm Rep 2020; 93: 231–40. 10.15386/mpr-1742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hutchings MI, Truman AW, Wilkinson B. Antibiotics: past, present and future. Curr Opin Microbiol 2019; 51: 72–80. 10.1016/j.mib.2019.10.008 [DOI] [PubMed] [Google Scholar]
- 7. Razzaque MS. Implementation of antimicrobial stewardship to reduce antimicrobial drug resistance. Expert Rev Anti Infect Ther 2021; 19: 559–62. 10.1080/14787210.2021.1840977 [DOI] [PubMed] [Google Scholar]
- 8. Huang L-J, Chen S-J, Hu Y-W et al. The impact of antimicrobial stewardship program designed to shorten antibiotics use on the incidence of resistant bacterial infections and mortality. Sci Rep 2022; 12: 913. 10.1038/s41598-022-04819-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Neels AJ, Bloch AE, Gwini SM et al. The effectiveness of a simple antimicrobial stewardship intervention in general practice in Australia: a pilot study. BMC Infect Dis 2020; 20: 586. 10.1186/s12879-020-05309-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Garwan YM, Alsalloum MA, Thabit AK et al. Effectiveness of antimicrobial stewardship interventions on early switch from intravenous-to-oral antimicrobials in hospitalized adults: a systematic review. Am J Infect Control 2023; 51: 89–98. 10.1016/j.ajic.2022.05.017 [DOI] [PubMed] [Google Scholar]
- 11. Agarwal A, Kapila K, Kumar S. WHONET software for the surveillance of antimicrobial susceptibility. Med J Armed Forces India 2009; 65: 264–6. 10.1016/S0377-1237(09)80020-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. WHO . Global Antimicrobial Resistance and Use Surveillance System (GLASS) Report 2022. 2022. https://www.who.int/publications/i/item/9789240062702.
- 13. WHO . 2021 AWaRe Classification. 2021. https://www.who.int/publications/i/item/2021-aware-classification.
- 14. Daley P, Garcia D, Inayatullah R et al. Modified reporting of positive urine cultures to reduce inappropriate treatment of asymptomatic bacteriuria among nonpregnant, noncatheterized inpatients: a randomized controlled trial. Infect Control Hosp Epidemiol 2018; 39: 814–9. 10.1017/ice.2018.100 [DOI] [PubMed] [Google Scholar]
- 15. Teshome B, Teklemariam Z, Admassu Ayana D et al. Salmonella and Shigella among patients with diarrhea at public health facilities in Adama, Ethiopia: prevalence, antimicrobial susceptibility pattern, and associated factors. SAGE Open Med 2019; 7: 2050312119846041. 10.1177/2050312119846041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tosisa W, Mihret A, Ararsa A et al. Prevalence and antimicrobial susceptibility of Salmonella and Shigella species isolated from diarrheic children in Ambo town. BMC Pediatr 2020; 20: 91. 10.1186/s12887-020-1970-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Huang L, Huang C, Yan Y et al. Urinary tract infection etiological profiles and antibiotic resistance patterns varied among different age categories: a retrospective study from a tertiary general hospital during a 12-year period. Front Microbiol 2022; 12: 813145. 10.3389/fmicb.2021.813145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shrestha G, Wei X, Hann K et al. Bacterial profile and antibiotic resistance among cancer patients with urinary tract infection in a national tertiary cancer hospital of Nepal. Trop Med Infect Dis 2021; 6: 49. 10.3390/tropicalmed6020049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. AbuSara A, Tayyeb N, Matalka L et al. Prevalence and predictors of multi-drug resistant organisms among ambulatory cancer patients with urinary tract infections. IDR 2023; 16: 747–53. 10.2147/IDR.S388680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sime WT, Biazin H, Zeleke TA et al. Urinary tract infection in cancer patients and antimicrobial susceptibility of isolates in Tikur Anbessa Specialized Hospital, Addis Ababa, Ethiopia. PLoS One 2020; 15: e0243474. 10.1371/journal.pone.0243474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Birru M, Woldemariam M, Manilal A et al. Bacterial profile, antimicrobial susceptibility patterns, and associated factors among bloodstream infection suspected patients attending Arba Minch General Hospital, Ethiopia. Sci Rep 2021; 11: 15882. 10.1038/s41598-021-95314-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Duan N, Sun L, Huang C, et al. Microbial distribution and antibiotic susceptibility of bloodstream infections in different intensive care units. Front Microbiol 2021; 12: 792282. 10.3389/fmicb.2021.792282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Minasyan H. Sepsis: mechanisms of bacterial injury to the patient. Scand J Trauma Resusc Emerg Med 2019; 27: 19. 10.1186/s13049-019-0596-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Naqid IA, Balatay AA, Hussein NR et al. Antibiotic susceptibility pattern of Escherichia coli isolated from various clinical samples in Duhok City, Kurdistan Region of Iraq. Int J Infect 2020; 7: e103740. 10.5812/iji.103740 [DOI] [Google Scholar]
- 25. Ait-Mimoune N, Hassaine H, Boulanoir M. Bacteriological profile of urinary tract infections and antibiotic susceptibility of Escherichia coli in Algeria. Iran J Microbiol 2022; 14: 156–60. 10.18502/ijm.v14i2.9180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Khavandi S, Arzanlou M, Teimourpour R et al. Phenotypic and molecular characterization of carbapenems resistant Escherichia coli isolated from patients with urinary tract infections in Ardabil province, Iran. Iran J Pathol 2022; 17: 261–7. 10.30699/ijp.2022.538613.2716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dwomoh FP, Kotey FCN, Dayie NTKD et al. Phenotypic and genotypic detection of carbapenemase-producing Escherichia coli and Klebsiella pneumoniae in Accra, Ghana. PLoS One 2022; 17: e0279715. 10.1371/journal.pone.0279715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tutone M, Bjerklund Johansen TE, Cai T et al. SUsceptibility and Resistance to Fosfomycin and other antimicrobial agents among pathogens causing lower urinary tract infections: findings of the SURF study. Int J Antimicrob Agents 2022; 59: 106574. 10.1016/j.ijantimicag.2022.106574 [DOI] [PubMed] [Google Scholar]