Abstract
Bovine leukemia virus (BLV) is a member of the human T-cell leukemia virus (HTLV)/BLV group of retroviruses. These viruses regulate their own transcription by producing Tax, a protein which activates the virus promoter region, the long terminal repeat (LTR). To explore the molecular mechanisms involved in the transactivation, we identified protein binding elements by in vivo footprinting and analyzed their function by site- directed mutagenesis. We used in vivo dimethyl sulfate footprinting by ligation-mediated PCR to detect constitutive in vivo protein-DNA interactions in a BLV-producing cell line, Bat2Cl6. The U3 region and part of the R region of the LTR were footprinted. In addition to the cis-acting elements (three cyclic AMP-responsive elements [CREs] and two AP4 sites) reported by others to be important for Tax-mediated activation of the BLV LTR, we found footprints in regions flanking these elements and in the core promoter region. The importance of these sites for transcriptional activation was studied by site-directed mutagenesis followed by promoter function analysis of the mutants with a chloramphenicol acetyltransferase reporter system. Our data corroborate those of others showing that the CREs are necessary for transactivation of the LTR, and they identify two new functional sites not previously reported by others. We show that the middle region of the BLV U3 contains multiple dual-functioning cis-acting elements which act as either positive or negative regulatory elements depending on the cell type tested. This is the first report of a functional mapping of the cis-acting elements of a virus of the HTLV/BLV group.
Bovine leukemia virus (BLV) is the etiologic agent of enzootic bovine leukosis, a disease characterized by persistent lymphocytosis and development of B-cell lymphomas after a long latent period. It also causes chronic lymphocytic leukemia and lymphomas in experimental animals such as sheep and goats (reviewed in reference 24). BLV is a retrovirus closely related to human T-cell leukemia viruses (HTLV-1 and HTLV-2). Like HTLV, BLV contains a pX region between the env gene and its 3′ long terminal repeat (LTR), in addition to three genes common to all retroviruses: gag (group-specific antigen), pol (polymerase, endonuclease, and integrase) and env (envelope proteins). One of the proteins encoded in the pX region is p34 Tax (transactivator from the X region), which acts on the 5′ LTR to stimulate transcription of the BLV genome (9, 10, 25, 44). p34Tax can also activate some cellular promoters such as c-fos promoter (23). Expression of BLV in vivo is believed to be regulated by cellular trans-acting factors that bind to the cis-acting elements in the U3 region of the BLV LTR in the presence of p34 Tax (reviewed in reference 24).
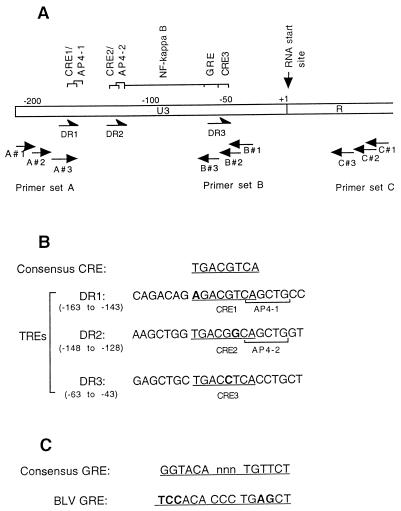
The LTR region of a retrovirus consists of U3, R, and U5 regions. Although the R and U5 regions are essential for virus replication, it is the U3 region that is critical for transcriptional control of a retrovirus (8). As with HTLV, the BLV U3 region contains three imperfect direct repeats (DRs) of 21 bp (DR1, bp −163 to −143; DR2, bp −138 to −118; DR3, bp −63 to −43), which are called tax-responsive elements (TREs) (9, 25, 44). In the middle of each 21-bp repeat is an octamer homologous to the core sequence of the cyclic AMP-response element (CRE), each with only a 1-bp difference from the consensus CRE sequence (Fig. 1). By analyzing the promoter function of deletion mutants, Derse (9) and Katoh et al. (23) found that the three CREs are cis-acting elements important for the responsiveness of the BLV LTR to p34Tax. A few other elements which may also be important in transcription regulation have been identified: an E-box sequence (CAGCTG), which is the binding site for the cellular transcription factor AP4 (the upstream AP4 site [−151 to −146] is hereafter designated AP4-1, and the downstream one [−124 to −119] is designated AP4-2) (42); a nuclear factor κB (NF-κB) binding site (6); and a glucocorticoid response element (GRE) (29, 45a) (Fig. 1).
FIG. 1.
cis-acting elements in the U3 region of BLV. (A) Schematic map of the BLV LTR showing the locations of the cis-acting elements in the U3 region and the binding sites for the primers used for in vivo footprinting. (B) Nucleotide sequence comparison of response elements in the BLV U3 region: the TREs, consensus CRE and CRE-like sequences including the AP4 sites, and the consensus GRE and GRE-like sequence. Bases different from the consensus sequences are shown in boldface type.
Although p34Tax does not bind DNA directly (1, 44), it has been shown, by in vitro studies of others, to interact directly with the proteins binding to some of the cis-acting sequences, e.g., members of the CREB/ATF family of bZip proteins which were shown to bind the CRE-like sequences in vitro (4, 25). p34Tax may also interact with other cellular transcription factors that recognize some of these sequences, such as AP4 and NF-κB (6, 42). For HTLV, it was shown that transactivation of the LTR involves interactions between p40Tax and multiple cellular transcription factors (reviewed in reference 1). Transcription factor CREB functions as either a positive or negative regulator depending on whether it binds to the CRE-like sequences in the U3 region or to a nonconsensus CRE sequence in the R region (46). It has been reported that both positive and negative regulatory elements are present in the LTR region of other retroviruses such as human immunodeficiency virus, mouse mammary tumor virus, and Rous sarcoma virus, and that cellular regulatory factors that bind to specific sequences in the LTR region of these viruses are responsible for the regulation of viral expression and tissue specificity of these viruses (5, 18, 21, 40, 47).
BLV is usually nondetectable in vivo in infected lymphocytes or tumor cells (reviewed in reference 24), but nothing is known about how BLV expression is suppressed in vivo. It is obvious that interactions between p34Tax and cellular proteins activate the viral promoter, but whether similar interactions are involved in repressing transcription has not been studied. We sought to clarify these interactions by determining the sites in the U3 region of the BLV at which LTR proteins are bound and whether these proteins function cooperatively with BLV p34Tax to control the expression of BLV by binding to negative regulatory elements (NREs) and/or positive regulatory elements (PREs). First we looked at constitutive in vivo protein-DNA interactions in the U3 region in a virus-producing cell line, Bat2Cl6. Then we applied site-directed mutagenesis to delete or mutate the individual protein-binding sites we detected (13 of the protein-binding sites and 1 site with no visible footprints were mutated or deleted, and 25 mutants were created). The wild-type and mutant LTRs were cloned into chloramphenicol acetyltransferase (CAT) reporter constructs, and promoter function was analyzed in six cell lines, representing different species and tissue types, by transient gene transfection assays in the presence or absence of Tax.
MATERIALS AND METHODS
Cell lines.
Tb1Lu, a bat lung epithelial cell line negative for BLV (16), was from the University of California San Francisco Cell Culture Facility (San Francisco, Calif.). Bat2Cl6, a BLV-producing cell line derived from Tb1Lu cells infected with the virus by cocultivation with lymphocytes from a BLV-infected cow (16), was obtained from K. Radke (University of California Davis, Davis, Calif.). FLK, a BLV-producing fetal lamb kidney fibroblast line (43), and MCF-7, a mammary epithelial cell line established from a human breast carcinoma (38), were obtained from the former Naval Biosciences Laboratory (Oakland, Calif.). GR, a cell line derived from a mouse mammary tumor (26), was obtained from G. Firestone (University of California Berkeley, Berkeley, Calif.). Raji, a human B-cell line derived from a Burkitt’s lymphoma (12, 32), was obtained from R. Emmons (California Department of Health Services, Berkeley, Calif.). BL3, a BLV-infected bovine B-lymphocyte line, was obtained from Harris Lewin (University of Illinois, Champaign, Ill.). Cell lines were maintained in Dulbecco’s modified Eagle’s medium (DMEM) (GIBCO, Grand Island, N.Y.) or, for BL3, Liebovitz medium (GIBCO) supplemented with 5 to 10% heat-inactivated fetal bovine serum (FBS) (Sigma, St. Louis, Mo.), 10 μg of insulin (Sigma) per ml, and antimicrobial agents and incubated at 37°C in a humidified atmosphere of 5% CO2.
In vivo footprinting with DMS. (i) Primers and linkers used for footprinting.
The primers and linkers used are described in Table 1 and are based on the BLV provirus sequence reported by Sagata et al. (35). Oligonucleotides were synthesized either by the Microchemical Facility, Department of Molecular and Cell Biology, University of California, Berkeley, or by Operon Biotechnologies, Alameda, Calif. A common linker was prepared by annealing equimolar amounts of each linker in 250 mM Tris-HCl by the method of Pfeifer and Riggs (31).
TABLE 1.
Sequences of primers and linkers used in this study
| Name | Location | Sequence |
|---|---|---|
| Gene-specific primers used for in vivo footprinting | ||
| Primer set A (noncoding strand) | ||
| A1 | (bp −211 to −191) | 5′-TGTATGAAAGATCATGCCGA-3′ |
| A2 | (bp −200 to −180) | 5′-ATCATGCCGACCTAGGAGCCG-3′ |
| A3 | (bp −181 to −156) | 5′-CCACCGCCCCGTAAACCAGACAGA-3′ |
| Primer set B (coding strand) | ||
| B1 | (bp 8 to −16) | 5′-AGCTCAATCGCCGTGGTCTTCGCAA-3′ |
| B2 | (bp −6 to −30) | 5′-CCGCTAACTCGACAGGGCCGGCATT-3′ |
| B3 | (bp −30 to −57) | 5′-TTATTAATTTATCAGCAGGTGAGGTCAG-3′ |
| Primer set C (coding strand) | ||
| C1 | (bp 97 to 74) | 5′-TACCTGACCGCTGCCGGATAGCCG-3′ |
| C2 | (bp 84 to 60) | 5′-CCGGATAGCCGACCAGAAGGTCTCG-3′ |
| C3 | (bp 60 to 34) | 5′-GGGAGCAAGAGAGCTCAGGACCGA-3′ |
| Linkers | ||
| 1 | 5′-GCGGTGACCCGGGAGATCTGAATTC-3′ | |
| 2 | 5′-GAATTCAGATC-3′ | |
| Primers used for mutagenesis | ||
| Vector primers | ||
| 1 | 5′-ACGGUUAUCCACAGAAUCA-3′ | |
| 2 | 5′-ACUGGAACAACACUCAACC-3′ | |
| Universal primers | ||
| 1 | 5′-AUUCUGUGGAUAACCGUA-3′ | |
| 2 | 5′-AGUGUUGUUCCAGUTTGG-3′ | |
| Primers for CAT plasmid constructs | ||
| 1 | LTR 5′ primer | 5′-TGTATGAAAGATCATGCCGAC-3′ |
| 2 | LTR 3′ primer | 5′-ATTGTTTGCCGGTCTCTCCT-3′ |
(ii) DNA preparation, primer extension, and ligation.
DNA preparation, primer extension, and ligation were done by the method of Pfeifer and Riggs (31). Briefly, DNA was extracted by standard phenol-chloroform procedures (36) from Bat2Cl6 cells with or without DMS treatment. DNA from cells not treated with DMS was treated with DMS after purification. DNA methylated with DMS was then cut with hot piperidine (90°C), precipitated, dried, and redissolved in distilled water at 0.5 to 1.0 μg/μl. Primer extension and ligation were done exactly as described by Pfeifer and Riggs (31) with 1 μg of each cleaved template for each reaction. Primers A1, B1, and C1 (Table 1) were used for primer extension to make double-stranded DNA fragments, which were ligated to the common linker (prepared by annealing linkers 1 and 2) (Table 1).
(iii) PCR.
A two-round PCR was used to amplify the U3 region of BLV provirus DNA (from Bat2Cl6 cells) in a thermal cycler (Perkin-Elmer Cetus, Norwalk, Conn.). Ligated DNA (50 μl) was mixed with 50 μl of freshly made 2× PCR reaction mix (10 μl of 10× Taq polymerase buffer [Mg2+ free; Promega, Madison, Wis.]; 10 μl of 25 mM MgCl2; 1 μl each [20 pmol] of primer A2, B2, or C2; 1 μl [20 pmol] of linker 1; 0.8 μl of 10 mM deoxynucleoside triphosphates [dNTPs; Promega], 3 U of Taq polymerase; 27 μl of water). The reaction mix was heated to 95°C for 5 min to denature the DNA and then run with the following cycle profile: 94°C for 1 min, 60°C for 2 min, and 75°C for 3 min for 35 cycles. To extend all DNA fragments completely, an additional incubation at 72°C was done for 20 min. The PCR products were extracted by the standard phenol-chloroform procedure, precipitated, dried, and dissolved in 20 μl of distilled water. The second-round PCR was run in a 10-μl volume containing 1× Taq polymerase buffer (Promega), 2 μl of the PCR-amplified DNA (see above), 0.5 U of Taq polymerase, 2.5 mM MgCl2, 200 μM each dNTP, and about 100 fmol (25,000 cpm) of primer A3, B3, or C3, which was 5′-end labeled with 32P by using T4 DNA kinase (Promega) and [γ-32P]ATP (Amersham, Arlington Heights, Ill.) and gel purified by standard methods (36). The samples were heated at 95°C for 2.5 min, and then 25 to 30 cycles of 95°C for 30 s, 66°C for 1 min, and 75°C for 2 min were run with a 20-min incubation at 72°C at the end. A 5-μl volume of stop buffer (95% formamide, 1 mM EDTA, 0.5% xylene cyanol, 0.5% bromphenol blue) was then added to each reaction mixture. The PCR products were analyzed by a 6% polyacrylamide–urea sequencing gel run as suggested by the manufacturer (Bio-Rad Laboratories, Hercules, Calif.). The DNA concentration of the samples was standardized by loading equal counts per minute. After electrophoresis, the gel was dried and exposed to Fuji X-ray film for 12 to 96 h. The film was developed manually.
Site-directed mutagenesis of the BLV LTR. (i) Preparation of plasmid DNA.
The pGEM-T plasmid used to clone the BLV LTR was purchased from Promega. pBL CAT3 plasmid, which contains only the basic chloramphenicol acetyltransferase (CAT) gene without any eukaryotic promoter (27), was kindly provided by Isabelle Cludts. Escherichia coli TG1 cells were transformed with the appropriate plasmid as described by Sambrook et al. (36). Large-scale plasmid DNA was prepared as described by Sambrook et al. (36) by the alkali lysis and CsCl-ethidium bromide banding methods. Plasmid minipreps were prepared by Qiagen Quick Spin plasmid purification kits as specified by the manufacturer.
(ii) DNA sequencing.
DNA sequencing was done with the Silver Sequence DNA-sequencing system (Promega), which uses the enzymatic method of Sanger et al. (37). The sequencing reaction was done in a thermal cycler (Perkin-Elmer Cetus). The sequencing gel was stained with silver nitrate, and permanent records were made by exposing the gel (for 10 to 15 s) to Silver Sequence film (Promega), which was developed manually.
(iii) Restriction enzyme digestion and ligation reaction.
All the restriction enzymes and T4 DNA ligase were purchased from Promega. For restriction enzyme digestion, 1 to 20 μg of plasmid DNA was digested at 37°C for 2 h to overnight in the buffer provided by the manufacturer. DNA fragments of interest were purified with a GeneClean kit (Bio 101, La Jolla, Calif.). All the ligation reactions were performed as described by Sambrook et al. (36). Briefly, 50 to 100 ng of each vector and insert DNA (equimolar) was mixed with 0.2 U of T4 DNA ligase in 10 μl of 1× ligation buffer (20 mM Tris HCl [pH 7.7], 5 mM MgCl2, 5 mM dithiothreitol, 1 mM ATP) and incubated at 16°C for 3 h. Then 1 to 2 μl of the ligation mix was used to transform competent TG1 cells as described above.
(iv) Construction of CAT plasmids.
Construction of CAT plasmids was generally done by standard procedures (36). The whole LTR region of BLV was amplified by PCR with Bat2Cl6 DNA as the template and BLV LTR specific oligonucleotides as primers (Table 1). The LTR PCR product was first cloned into the pGEM-T vector (Promega) in the 5′-to-3′ direction (confirmed by sequencing) as specified by the manufacturer. The LTR DNA was then subcloned into plasmid pBL CAT3 upstream of the CAT gene. Clones containing inserts (which have a higher molecular weight than the parental plasmid) were selected, and the cloned DNA was further amplified in TG1 cells. The orientations of the LTR inserts were determined by restriction enzyme digestion, and the U3 region of BLV LTR in the constructs was sequenced to confirm that the sequence was correct. The same method was used to construct mutant LTR-CAT plasmids after the mutations were made in plasmid pGEM-T/LTR (described below).
(v) PCR amplification.
The reaction was done in 50 μl of 1× PCR buffer (10 mMTris-HCl [pH 8.3], 50 mM KCl, 2 mM MgCl2, 5 mM 2-mercaptoethanol, 0.005% gelatin) containing 10 pmol of each primer, 100 to 500 ng of template DNA, 2.5 U of Taq polymerase, and 0.2 mM each dNTP. The reaction was performed in a thermal cycler with the following cycling profile: 94°C for 1 min, 50 to 56°C for 1 min, and 72°C for 2 min for 30 cycles followed by an extension cycle at 72°C for 10 min. The PCR products were stored at −20°C until use.
(vi) Mutagenesis of cloned BLV LTR.
The method of Rashtchian et al. (33) was modified to create mutations or deletions in the cloned BLV LTR. Vector and universal primers used for mutagenesis are listed in Table 1. In addition, one pair of mutagenic primers was synthesized for each mutant to introduce a small deletion or base substitution. Mutagenic primers are not included in Table 1, but the mutations introduced are summarized in Table 2. Each mutagenic primer was 20 bases long and corresponded to the wild-type LTR sequence except for the introduced mutation approximately in the center of the sequence. Three PCRs were run separately with three pairs of primers: vector primers were used to amplify most of the pGEMT vector DNA (bp 1000 to 3000), universal primer 1 coupled with a mutagenic primer 1, and universal primer 2 with a mutagenic primer 2 were used to amplify part of the plasmid and part of the cloned LTR. Three PCRs amplified the complete sequence of the vector containing the cloned LTR. Since the universal primers overlapped the vector primers, since mutagenic primer 1 overlapped mutagenic primer 2, and since the thymidines in the overlapping region were replaced by uridines, amplification of the template plasmid by PCR resulted in the incorporation of the primers and the desired mutation into the PCR products. Excision of the deoxyuracil residues in the PCR products by uracil deoxynucleotide glycosylase (UDG) destabilized base pairing at the ends of DNA molecules and thus generated 3′ protruding ends. Due to the overlapping nature of the primers, the resulting 3′ protruding ends were complementary and could anneal after treatment with UDG.
TABLE 2.
Sequence changes of 25 BLV LTR mutants made by site-directed mutagenesis
| Namea | Site of mutationb | Original sequence | Mutated sequence |
|---|---|---|---|
| d-CRE1 | −157 to −153 | AGACG | Deleted |
| d-CRE2 | −131 to −129 | GAC | Deleted |
| d-CRE3 | −57 to −54 | TGAC | Deleted |
| s-CRE3 | −56 | G | T |
| d-CRE 1/2 | −157 to −153 | AGACG | Deleted |
| −131 to −129 | GAC | Deleted | |
| d-CRE 1/3 | −157 to −153 | AGACG | Deleted |
| −57 to −54 | TGAC | Deleted | |
| dCRE 1/s-GRE | −157 to −153 | AGACG | Deleted |
| −71 to −60 | ACA---TG-CT | TAT---AC--TA | |
| dCRE2/s-GRE | −131 to −129 | GAC | Deleted |
| −71 to −60 | ACA---TG--CT | TAT---AC--TA | |
| d-CRE2/3 | −131 to −129 | GAC | Deleted |
| −57 to −54 | TGAC | Deleted | |
| d-CRE3/a-GRE | −57 to −54 | TGAC | Deleted |
| −71 to −72 | Wild type | TATCC added | |
| d-CRE1/2/3 | −157 to −153 | AGACG | Deleted |
| −131 to −129 | GAC | Deleted | |
| −57 to −54 | TGAC | Deleted | |
| d-CRE1/2/s-GRE | −157 to −153 | AGACG | Deleted |
| −131 to −129 | GAC | Deleted | |
| −71 to −60 | ACA---TG--CT | TAT---AC--TA | |
| d-CRE2/3/a-GRE | −131 to −129 | GAC | Deleted |
| −57 to −54 | TGAC | Deleted | |
| −71 to −72 | Wild type | TATCC added | |
| d-CRE1/2/3/a-GRE | −157 to −153 | AGACG | Deleted |
| −131 to −129 | GAC | Deleted | |
| −57 to −54 | TGAC | Deleted | |
| −71 to −72 | Wild type | TATCC added | |
| s-GRE | −71 to −60 | ACA---TG--CT | TAT---AC--TA |
| s-167 | −167 | A | G |
| s-AP4-1 | −151 | G | T |
| s-142,3 | −142 to −143 | AG | CT |
| s-140 | −140 | A | G |
| s-AP4-2 | −124 | G | T |
| s-116 | −116 | C | A |
| s-107,8 | −107 to −108 | CC | AT |
| s-99 | −99 | C | T |
| s-83,4 | −83 to −84 | TC | CT |
| s,a-TATA | −41 to −40 | TA | AT |
| −37 to −38 | Wild type | T added |
Abbreviations: a, addition; d, deletion; s, substitution; AP4, activating protein 4.
Positions of sequence changes are indicated by the numbers which represent the distance (in base pairs) relative to the RNA start site (+1).
After the PCRs, 5- to 10-μl volumes of each of the three PCR products were combined and 1 U of UDG was added to the mixture, which was then incubated at 37°C for 30 min, heated at 98°C for 15 min to cleave the abasic nucleotides, and reincubated at 37°C for 30 min (annealing). The UDG-treated PCR products were then ethanol precipitated and dissolved in 5 to 10 μl of water. A 2-μl volume of this DNA solution was used to transform competent TG1 cells. The template DNA was treated so that no intact circular plasmid DNA was left; i.e., no transforming colony could be found after transforming the cells by using the PCR products without UDG treatment. Only the mutant plasmid DNA formed by annealing of the UDG-treated PCR DNA could transform the bacteria. For reactions with universal and mutagenic primers, the plasmid was linearized with SspI, which cuts the vector three times at positions 2199, 2384, and 2408. For reaction with the vector primer, the template was treated with SacI and SacII, which cut the vector at bp 94 and 46, respectively. Miniprep plasmid DNA was prepared from the transformed colonies, and sequencing analysis was done to screen the mutants containing the desired sequence changes.
Gene transfer.
Tb1Lu cells were used to detect the promoter function of the BLV LTRs (wild type and mutants) which were cloned upstream of the CAT gene. For each transfection, 4 × 105 to 5 × 105 cells were seeded into a 35-mm petri dish and cultured for 24 h, and then the cells were transfected with plasmid DNA by using Lipofectamine (GIBCO) as specified by protocol provided by the manufacturer. Serum-free DMEM was the transfection medium, and 6 μl of Lipofectamine was used. Cells were incubated with the DNA-Lipofectamine mixture for 20 h before DMEM with FBS was added to a final concentration of 10% FBS. The cells (∼106/dish) were harvested for CAT assay 48 h after the transfection was started. To detect basal promoter function, 1 μg of LTR-CAT plasmid DNA was used for each 35-mm dish. To detect promoter function in response to p34Tax, 0.5 μg of each LTR CAT DNA and 0.5 μg of pSG-tax plasmid DNA (a kind gift from Luc Willems), which contains the BLV tax gene under the control of the simian virus 40 early promoter, were used for each 35-mm plate.
CAT ELISA.
The promoter function of the BLV LTR was measured as the amount of CAT protein produced by the transfected cells. CAT was detected by an enzyme-linked immunosorbent assay (ELISA) with the CAT ELISA kit (Boehringer-Mannheim, Indianapolis, Ind.) as specified by the manufacturer. Briefly, 48 h after transfection, cells were lysed with 0.5 to 1 ml of kit lysis buffer. Cell lysates were pelleted, and 50 to 100 μl of the supernatant was used for the ELISA. The absorbance readings were translated into CAT protein concentrations from a standard curve established simultaneously in each ELISA experiment, and the amount of CAT protein in each sample was adjusted according to the concentration of total proteins in the cell lysate. The protein concentration was measured with the bicinchoninic protein assay system as specified by the manufacturer (Pierce, Rockford, Ill.).
EMSA.
Oligonucleotides representing wild-type or mutated protein binding sites were used as probes (Table 1). Double-stranded oligonucleotides were obtained by mixing the complementary strands in a 1:1 ratio, heating to 80°C for 5 min, and cooling at room temperature for 1 h to anneal; they were stored at 4°C. For each electrophoretic mobility shift assay (EMSA) experiment, 1 pmol of each oligonucleotide was 5′-end labeled with [γ-32P]ATP (Amersham) and gel purified (36). Nuclear protein extracts were prepared from Bat2Cl6 and Tb1Lu cells essentially as described by Adam et al. (1). Extracts were stored at −70°C until use.
A 1-μl volume of cell extract (≈2 μg of total protein) was mixed with 1 μg of sheared calf thymus DNA and 5 μl of 32P-labeled oligonucleotide probe (10 fmol; 30,000 cpm) in 13 μl of binding buffer (40 mM NaCl, 20 mM Tris-HCl [pH 7.5], 4 mM DTT, 1 mM EDTA, 0.002% Triton X-100, 750 μg of bovine serum albumin per ml, 10% glycerol). The mixture was incubated for 1 h at room temperature and then separated on a 5% polyacrylamide gel (10 V/cm) in 90 mM Tris-borate buffer with 2 mM EDTA. The gels were dried and subjected to autoradiography at −70°C with intensifying screens.
Statistical analysis.
For CAT assays, differences between the various mutants and the wild type were analyzed by the Dunnett t test. This is a multiple-comparison method which uses a pooled standard deviation and allows a comparison of multiple values to a single control. The minimum significance level was P < 0.05 (two-sided test).
RESULTS
In vivo footprinting reveals multiple protein-binding sites in the U3 region of BLV.
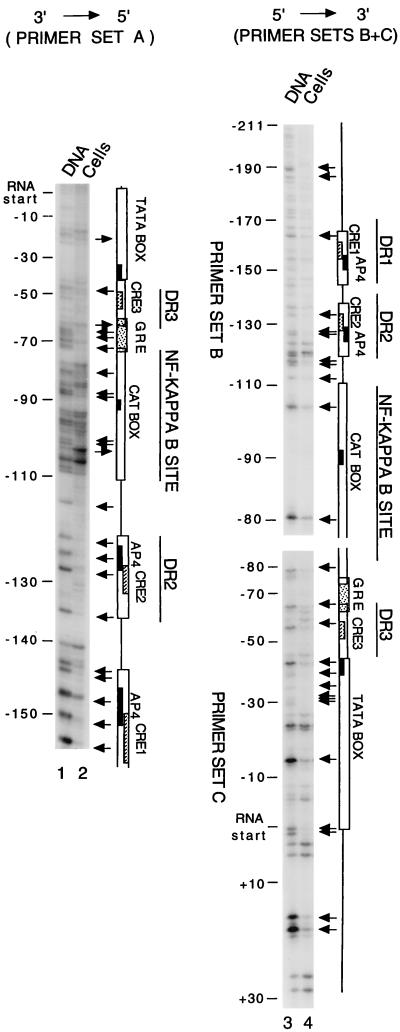
Figure 2 shows the DMS methylation patterns of both the noncoding strand (lanes 1 and 2, primer set A) and the coding strand (lanes 3 and 4, primer sets B and C) of the U3 region of BLV. At three CRE sites, two AP4 sites, the TATA box, and the putative GRE site, the guanidines were protected from DMS methylation on at least one strand, suggesting that these areas are protein binding sites. In the NF-κB site (bp −70 to −118), which contains a CAT box in the middle, the guanidines were protected at several sites on both strands, but one hypersensitive sites (bp −106 to −107) was found on the noncoding strand. This suggests that the NF-κB site is bound by multiple proteins or a large protein complex. In addition, protein contacts were found in sequences on both sides of the DR1/DR2 region, in the core promoter region (bp −40 to +1), and in the R region shown on this gel (from RNA start site to bp +30) and showed decreased sensitivity to DMS for most of the residues, with a few hypersensitive ones.
FIG. 2.
In vivo DMS footprinting of the BLV U3 region in the presence of Tax. BLV-infected Bat2Cl6 cells were used as the source of DNA. Lanes: 1 and 2, noncoding strand with primer set A; 3 and 4, coding strand with primer sets B and C. In vitro (purified DNA) DMS-treated samples are in lanes 1 and 3. In vivo (living cells) DMS-treated samples are in lanes 2 and 4. The locations of the bands relative to the RNA start site are labeled at the left of each gel (negative numbers for sequences upstream and positive numbers for the downstream of the start site). Increased sensitivity to DMS (band in lane 2 darker than in lane 1, and band in lane 4 darker than in lane 3) is indicated by arrows pointing away from the gel, and decreased sensitivity to DMS (lighter or no bands in lanes 2 and 4) is indicated by arrows pointing toward the gel. cis-acting elements reported in the literature are indicated by open boxes on the right of the gel, the consensus sequences of the AP4 sites are indicated by solid boxes, the core sequences of three CRE-like elements are indicated by the hatched boxes, and the GREs are indicated by dotted boxes.
Site-directed mutagenesis, EMSA (gel shift), and transient gene transfection studies were used to further evaluate the in vivo protein binding sites. The sequences of 13 protein binding sites and 1 site with no detectable protein contacts were mutated by site-specific mutagenesis, resulting in a total of 25 mutants (Table 2). Sequences of these mutants were confirmed by sequencing both before and after subcloning into pBLCAT3 plasmid (data not shown). The promoter function of these mutant LTRs was determined by a CAT ELISA after the plasmid DNA was transfected into Tb1Lu cells (a cell line identical to Bat2Cl6, except that it is not infected with BLV). The basal promoter activity of the wild-type and mutant LTRs (in the absence of p34Tax) was detected by transfecting Tb1Lu cells only with the LTR-CAT plasmid. To determine the function of the wild-type or a mutant LTR in response to p34Tax, we cotransfected the LTR-CAT plasmid DNA into Tb1Lu cells with plasmid pSGtax, which contains the BLV tax gene driven by the simian virus 40 early promoter.
Mutations in protein binding sites decrease or abolish protein binding.
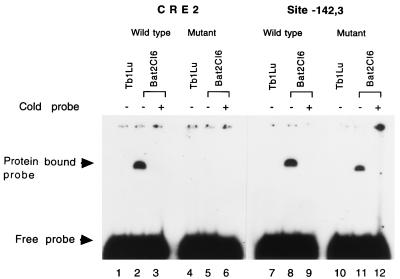
Three of the protein binding sites detected by in vivo footprinting were further tested for protein binding by EMSAs. Figure 3 shows that wild-type sequences corresponding to CRE2 and site −142 to −143 bound to nuclear proteins from BLV-producing Bat2Cl6 cells but not from BLV-negative Tb1Lu cells. In vitro protein binding at CRE2 was abolished by a 3-bp deletion; protein binding at site −142 to −143 was diminished by a 2-bp substitution. As reported in a separate communication, the putative GRE site demonstrated binding to purified glucocorticoid receptor protein, and this binding was abrogated by each of six different types of mutations (45a).
FIG. 3.
Gel shift assay to detect in vitro protein binding to wild-type and some mutant cis-acting elements in the BLV LTR. Synthetic double-stranded oligonucleotides were end labeled with [γ-32P]ATP and used as probes. Nuclear protein extracts were prepared from Bat2Cl6 (BLV-producing cell line) and Tb1Lu (BLV-free parental cell line of Bat2Cl6). Labeled probes were incubated with nuclear extracts in the presence (+) or absence (−) of unlabeled probe (100 times the concentration of the labeled probe) and separated on a 5% nondenaturing polyacrylamide gel. Protein-bound and free probes are indicated by the arrowheads. Tb1Lu cell extracts were used for lanes 1, 4, 7, and 10 and Bat2Cl6 cell extracts were used for the remaining lanes.
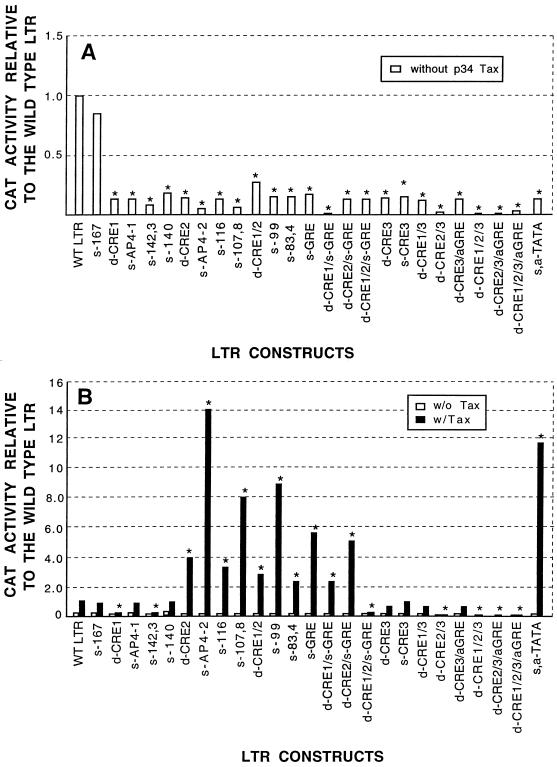
Basal LTR promoter activity (in the absence of p34Tax) is abolished by any mutation at any of the protein binding sites.
In the absence of p34Tax, all mutants except the negative control mutant (s-167) showed three- to sixfold lower activity than the wild-type LTR (Fig. 4A); the difference was statistically significant by the Dunnett t test. The negative-control sequence was from a site where no footprints were found; this mutant (s-167) was the only one to show a similar function to the wild-type LTR in both the absence and presence of p34Tax (Fig. 4A and B).
FIG. 4.
Basal promoter function of the wild-type BLV U3 and 25 LTR mutants in the absence (A) and presence (B) of p34Tax (solid bars) with values in the absence of p34Tax (open bars) shown for comparison. LTRs were inserted into a CAT reporter construct and transfected into BLV-negative Tb1Lu cells with or without cotransfection with a tax-containing plasmid. Mutants are arranged according to their location on the U3 map (Fig. 1). Promoter activity was measured as nanograms of CAT protein per 10 μg of total protein, and values were standardized by scaling the activity of the wild-type LTR to 1.0. Each value is the mean of five (A) or two (B) trials in separate experiments. Asterisks indicate that the CAT activity of the mutant is significantly different from the wild-type LTR (P ≤ 0.05 [Dunnett t test]). Abbreviations: a, addition; d, deletion; s, substitution; WT, wild type. Acronyms in capital letters refer to sites illustrated in Fig. 1.
The LTR promoter activity of mutants in the presence of p34Tax is decreased, increased, or unchanged depending on the location of the mutated protein binding site. (i) Mutants with activity similar to the wild type.
In addition to the negative control (s-167), six mutants showed activity similar to the wild-type LTR. These six involved deletions or mutations in the DR1 region (s-AP4-1 and s-140), the DR3 region (d-CRE3, s-CRE3, and d-CRE3/aGRE), or both (d-CRE1/3) (Fig. 4B).
(ii) Mutants with decreased promoter activity.
Seven mutants showed only 0 to 20% of the wild-type LTR promoter activity. Two of these mutants involved a mutation or deletion of a single protein binding site in the DR1 region (s-142-3 and d-CRE1) (Fig. 4B). The other five mutants all had deletions or mutations in two or all the CREs with or without mutations in the GRE region.
(iii) Mutants with increased promoter activity.
Eleven of the mutants had significantly stronger responsiveness to Tax than did the wild-type LTR (Fig. 4B; from left to right, d-CRE2, s- AP4-2, s-116, s-107,8, d-CRE1/2, s-99, s-83,4, s-GRE, d-CRE1/s-GRE, d-CRE2/s-GRE, and s,a-TATA). All these mutants involve sequence changes in the middle region of U3, including the DR2, GRE, CAT box, and its flanking sequences (NF-κB site). Deletion in both CRE1 and CRE2 (d-CRE1/2) resulted in significantly higher CAT activity than that of the wild type. A 3-nucleotide deletion in CRE2 (Fig. 4B, d-CRE2) increased the promoter function threefold, and the mutant with both a deletion and a GRE mutation (d-CRE2/s-GRE) showed a further increase in promoter activity.
Of these 11 mutants, 3 contained only a single mutation (Table 2) (s-AP4-2, s-116, and s-99). It is interesting that a single-base-pair mutation in AP4-1 (Table 2, s-AP4-1) did not change the promoter function significantly but the same mutation in AP4-2 (s-AP4-2) gave rise to the strongest promoter function of all mutants, 14 times that of the wild-type LTR. Point mutations at −116, (s-116), −107 to −108 (s-107,8), −99 (s-99), and −83 to −84 (s-83,4) increased promoter function three- to fivefold, respectively (Fig. 4B). The TATA box (−43 to −36) was mutated so that it contained an AT-to-TA change at the beginning and a T addition at the 3′ end (Table 2). This dramatically increased the CAT activity (≈ 12-fold).
The effect of mutations on LTR promoter activity varies in different cell types.
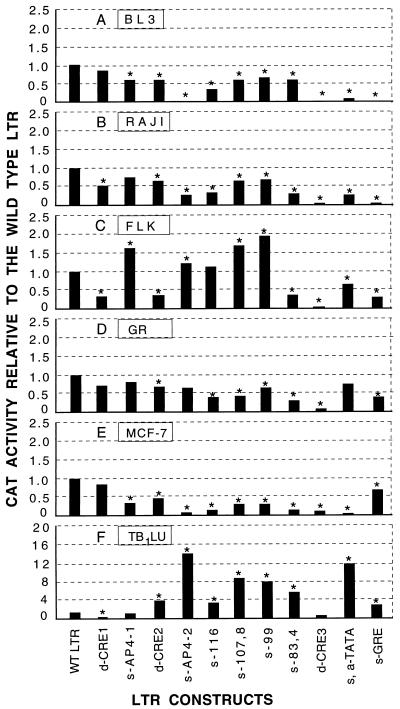
To examine the role of the host cell in the function of BLV cis-acting elements, we determined the CAT activity of 11 LTR mutants in five other cell lines besides Tb1Lu: BL3, a BLV-infected bovine B-lymphocyte cell line; Raji, a human B-lymphocyte cell line; FLK, a BLV-producing fibroblast cell line from fetal lamb kidney; GR, a mouse mammary epithelial cell line; and MCF-7, a human mammary epithelial cell line. These cell lines were chosen because they originated from different cell types and species and also because they are convenient for transfection studies. The constructs used were the ones that contained a mutation or deletion at only one protein binding site, so that the functions of individual protein-binding sites could be analyzed. The effects of mutations on promoter function are shown as increased or decreased CAT activity of the mutants relative to the wild-type LTR. The CAT activity of the LTR mutants in the six different cell lines is shown in Fig. 5. Three types of responses of the mutants to p34Tax were found: (i) mutations at CRE1 and CRE3 are the only ones that showed decreased CAT activity in all the cell lines (some decreases were not statistically significant); (ii) the CAT activity of some mutants was similar to that of the wild-type LTR in some cell lines but increased or decreased in other cell lines; and (iii) FLK and Tb1Lu were the only cell lines to show any increased activity with mutations at any site.
FIG. 5.
Promoter function of the wild-type BLV LTR and 11 LTR mutants in six different cell lines: BL3 (bovine B lymphocytes) (A), Raji (human B lymphocytes) (B), FLK (sheep fibroblasts) (C), GR (mouse mammary epithelial cells) (D), MCF-7 cells (human mammary epithelial cells) (E), and Tb1Lu (bat lung epithelium) (F). For BL3 and FLK cells, only the plasmid with the wild-type or mutant LTR sequences was transfected, since these cell lines contain endogenous p34Tax. For the other four lines, wild-type or mutant LTR was cotransfected into cells with the pSGtax plasmid. Bars represent the mean values (nanograms of CAT protein per 10 μg of total protein) from two to four independent experiments. Values were standardized by scaling the activity of the wild-type LTR to 1.0. Asterisks represent statistically significant differences (P ≤ 0.05 [Dunnett t test]) relative to the wild-type LTR.
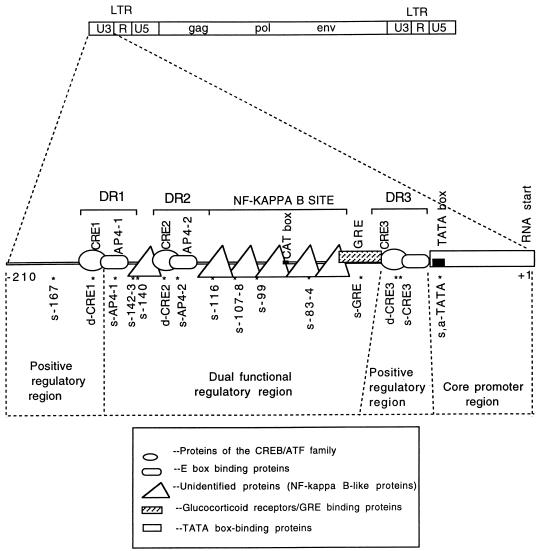
The BLV U3 region of BLV can be largely divided into four functional units.
Figure 6 summarizes the data from in vivo footprinting of the BLV U3 region in the virus-producing Bat2Cl6 cells and the CAT assays to detect promoter function of the mutants in Tb1Lu cells. According to CAT activities of mutant LTRs, the U3 region was shown to contain four functional units: a core promoter region, from the RNA start site to the 5′ of the TATA box; a dual regulatory region, from the 5′ end of DR3 to the 5′ end of AP4 (including CRE2, AP4, the NF-κB site, and GRE); and two positive regulatory regions, CRE1 and DR3.
FIG. 6.
Functional regions of the BLV U3. The genome structure of the whole BLV provirus is shown at the top. A schematic map of the U3 region is shown in the middle of the figure. The locations of three DRs are indicated by brackets. Possible protein binding sites are marked with boxes of different shapes (indicating different types of proteins). The locations of sequence changes in 15 mutants are indicated by asterisks (14 of the sites had altered in vivo footprints; no footprints were detected at bp −167). Mapping of the U3 region was based on the promoter activity of the mutants. Mutants with increased promoter activity were used to define an NRE, and the ones with decreased activity were used to define a PRE. The core promoter region was defined as reported in the literature (6, 9, 10). Regulatory regions were defined according to the reactions represented in Fig. 4B. Elements within the dually functional region function as NREs in some cell lines and as PREs in other cell lines.
DISCUSSION
Using in vivo footprinting and EMSA, we demonstrated multiple protein binding sites in the U3 region of the BLV LTR, and by site-directed mutagenesis and transient gene transfection studies, we found that protein-DNA interactions at these sites are important for the transactivation of the BLV LTR. In the absence of p34Tax (Fig. 4A), all except one of the mutants showed lower CAT activity than the wild-type LTR, indicating that basal promoter activity of the BLV LTR requires an intact U3 region in which none of the protein binding elements are dispensable. The only mutant (s-167) that showed basal CAT activity similar to the wild-type LTR was the one at a position where no footprints were found (Fig. 2). It was used as a negative control. These results suggest that binding of cellular proteins to viral cis-acting elements is necessary for the basal-level promoter function of the LTR and may explain how BLV transcription is launched in the initial absence of p34Tax.
In the presence of p34Tax, however, the promoter function of the mutants varied with each mutated protein binding site and with the cell type used. Footprints were obvious at three CRE-like cis-acting elements shown by others to be recognized by cellular proteins in EMSAs (1, 44). Partial deletion of the core sequences of CRE1 and CRE3 reduced the CAT activity in all cell lines tested (Fig. 5), suggesting that these elements may be the binding sites for ubiquitous positive regulatory proteins. Since CRE3 is immediately upstream of the TATA box, the transcription factor specific for this element may help stabilize the binding of the TATA factor (TFIID) to the TATA box. Hirano and Wong (20) demonstrated that such interaction is possible and that proteins binding to the promoter containing the TATA box and the upstream enhancer elements are cooperative in their function. This may explain why a deletion in CRE3 had a greater effect than the one in CRE1. The cooperative effects of the three CREs on the promoter function of BLV LTR were studied in the Tb1Lu line. Various mutants were made to contain a deletion of 3 to 5 bp in one, two, or all three of the CRE core sequences. Our findings that CRE2 alone (when both CRE1 and CRE3 were partially deleted) was able to maintain some normal function and that the CRE2 sequence was able to be bound by proteins in vitro (Fig. 3) are consistent with the results of previous studies by Katoh et al. (23), who reported that a synthetic promoter containing only CRE2 was responsive to BLV p34Tax. We found that CRE1 had no function in the absence of intact downstream CREs, although it may function as an upstream enhancer in p34Tax-mediated transactivation of the BLV LTR (9). In our study, the mutant containing CRE3 alone (both CRE1 and CRE2 were partially deleted) had significantly increased function. In contrast, Derse (9) found that CRE3 was almost nonfunctional in the absence of the upstream cis-acting elements. In his study, large deletions created by restriction enzymes, rather than small deletions made by site-directed mutagenesis, were used and all the sequences upstream of CRE3 were deleted. These technical differences may explain the discrepant results. Truncation of the upstream sequence may affect protein binding at downstream cis-acting elements close to the core promoter. Parekh and Hatfield (30) reported that proteins binding to an upstream activating sequence can activate transcription from a downstream promoter by DNA bending.
We found footprints at two AP4 sites shown by others to be recognized by proteins in gel shift assays (42) and in in vitro (DNase I) footprinting assays (6). Like Unk et al. (42), we found that transcription factor AP4 may be involved in the transactivation of BLV LTR, but according to our data, the effect may vary with the cell line and is affected by sequences flanking the E-box, since the same point mutation in AP4-1 and AP4-2 affected the function of the LTR differently in different cell lines. Since the AP4 sites overlap those of the CREs, binding of AP4 to its consensus sequence may also be affected by other regulatory proteins such as CREBs. AP4 may either enhance or suppress the function of the LTR depending on which of the two sites it binds.
Mutations in the NF-κB site (Fig. 6; Table 2, s-83, 4, s-99, and s-116) also showed opposite effects in different cell types. Our finding are consistent with previous reports that the NF-κB site is a dual-function element. In addition to functioning as a transcriptional enhancer element for a variety of genes (reviewed in reference 6), it can exert a negative effect on some cellular genes (13–15). The negative effect of the NF-κB site can also result from binding of other repressors that compete with NF-κB for the DNA sequence (15).
It is surprising that mutation of the TATA box also had different effects in different cell lines. Possible explanations come from previous observations that the function of the transcription initiation complex is affected by the upstream enhancer binding transcription factors (17, 20, 22). Grayson et al. (17) also showed that subtle sequence changes in the TATA box affected the tissue-specific function of the myoglobin gene promoter. It is likewise possible that the presence or absence of tissue-specific transcriptional factors determines whether the BLV TATA box is functional.
Our data indicated that novel cis-acting elements other than CREs, AP4 and NF-κB sites may play a role in the BLV LTR transactivation. One such element, as yet unnamed, is presumably located at the 3′ end of DR1, since a 2-base mutation at −142 and −143 significantly reduced the promoter activity of the LTR (Fig. 4B, s-142,3, P < 0.005) and greatly reduced protein binding in the gel shift assay (Fig. 3). Another such element is the GRE-like sequence just upstream of the CRE3, which we have previously shown to be a key element in the responsiveness of BLV to glucocorticoids such as dexamethasone (29, 45a).
Both positive and negative regulation may be involved in the transcriptional activation of the BLV LTR. If a deletion or mutation results in decreased activity, it indicates that the cis-acting element is necessary for promoter activation and therefore is a PRE. On the other hand, if a deletion or mutation leads to increased promoter function, it suggests that this sequence is an NRE normally bound by a protein which suppresses the transcription activation of the LTR. In the presence of p34Tax, two AP4 sites, the second 21-bp repeat (DR2), the NF-κB site, and the GRE function as either PREs or NREs. However, in the absence of p34Tax, all sites function as PREs. There are at least two possible explanations for this: (i) some cellular proteins may function as transcription factors to maintain low-level transcriptional activity in the absence of p34Tax, but they can function as repressors in the presence of p34Tax to prevent overactivation of the LTR; and (ii) p34Tax may activate its own promoter both by recruiting critical transcription factors and by releasing transcription repressors. In the absence of p34Tax, no such transcription factors are available for transcription activation and the LTR cannot be activated even in the absence of repressors. Therefore deletion or mutation of a repressor sequence does not increase the promoter activity of the LTR in the absence of p34Tax.
cis-acting elements in the U3 region of proviruses are the major determinants for retrovirus tissue tropism at the replication level (8), although sequences outside the LTR may also affect the tissue tropism of some retroviruses (45). Our results suggest that BLV transcription varies in different cell types and raise the possibility that host regulatory factors play an important role. This is particularly intriguing, since the tissue tropism of BLV in vivo is now known to extend beyond bovine B lymphocytes to mammary epithelial cells (7) and possibly to T lymphocytes (2, 28) and endothelial cells (34). Tissue-specific and/or differentiation-specific proteins may be required for the activation of BLV transcription, as is the case for the human papillomaviruses (39). This is supported by previous observations that BLV expression (virus particles, RNA, or viral proteins) was not detected in freshly isolated peripheral blood lymphocytes of animals infected with the virus or in BLV-positive tumor cells, but these cells were able to produce virus after 24 to 48 h in culture (reviewed in reference 24). Virus production was further increased with mitogens such as concanavalin A, lipopolysaccharide, and pokeweed mitogen (3, 11, 41). In vivo, positive lymphocytes were detected only in lymph nodes, where B cells would be undergoing differentiation to plasma cells (19).
Expression of BLV appears to be regulated by soluble host factors (41, 48, 49). Zandomeni et al. reported that both stimulatory and inhibitory factors are present in plasma and lymphatic fluid of infected cows. After removal of the stimulatory factor, the inhibitory activity can consistently be detected. Although these investigators identified the host inhibitory factor as a molecule that binds to immunoglobulin G, the nature of the inhibitory factor is still unclear and even less is known about the molecular mechanisms involved in this phenomenon. It is possible that NREs in the U3 region of the BLV LTR are the target for this host inhibitory factor, which may be responsible for BLV latency. A possible advantage of an NRE is to keep BLV expression at a low level in vivo and hence to reduce the host immune response to the virus.
In this research, we have for the first time delineated in vivo protein-DNA interactions in the entire U3 region of BLV and systematically analyzed the interaction sites by site-directed mutagenesis. Similar work has not been reported for any member of the HTLV/BLV group of retroviruses. Based on our data, we propose that the U3 region of BLV LTR consists of four functional units (Fig. 6): the core promoter region; two positive regulatory elements (the 5′ region of the U3 region containing the DR1 and part of DR3 containing CRE3), and a dually functional region containing the cis-acting elements CRE2, AP4-2, and the NF-κB site. Although site-directed mutagenesis was not performed in the first 60 nucleotides of the U3 5′ region, previous studies by Derse showed that this region is an enhancer element. Further experiments testing the expression pattern of recombinant viruses containing mutations in these regions in different cell types are necessary to determine if this dually functional region is responsible for the latent status of BLV in vivo in infected peripheral B lymphocytes and tumor cells.
ACKNOWLEDGMENTS
We gratefully acknowledge Caroline Kane and George Sensabaugh for helpful advice, and we thank numerous colleagues (named in the text) for gifts of cell lines and plasmids.
This study was partially supported by the Vice Chancellor’s Research Fund from the University of California, Berkeley; the Grossman Endowment from the School of Public Health, University of California, Berkeley; and a grant-in-aid award from Sigma Xi. J.X. was supported by Graduate Fellowships from the Graduate Division, University of California, Berkeley, and by a Levine Fellowship from the School of Public Health, University of California, Berkeley.
REFERENCES
- 1.Adam E, Kerkhofs P, Mammerickx M, Kettmann R, Burny A, Droogmans L, Willems L. Involvement of the cyclic AMP-responsive element binding protein in bovine leukemia virus expression in vivo. J Virol. 1994;68:5845–5853. doi: 10.1128/jvi.68.9.5845-5853.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allen L J, Kabbur M B, Cullor J S, Gardner I A, Stott J L, George L W. Alterations in blood lymphocyte subpopulations and hematologic values in neonatal calves after administration of a combination of multiple-antigen vaccines. J Am Vet Med Assoc. 1996;209:638–642. [PubMed] [Google Scholar]
- 3.Baliga V, Ferrer J. Expression of the bovine leukemia virus and its internal antigen in blood lymphocytes. Proc Soc Exp Biol Med. 1977;156:388. doi: 10.3181/00379727-156-39942. [DOI] [PubMed] [Google Scholar]
- 4.Boros I M, Tie F, Giam C Z. Interaction of bovine leukemia virus transactivator Tax with bZip proteins. Virology. 1995;214:207–214. doi: 10.1006/viro.1995.9939. [DOI] [PubMed] [Google Scholar]
- 5.Bramblett D, Hsu C L, Lozano M, Earnest K, Fabritius C, Dudley J. A redundant nuclear protein binding site contributes to negative regulation of the mouse mammary tumor virus long terminal repeat. J Virol. 1995;69:7868–7876. doi: 10.1128/jvi.69.12.7868-7876.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brooks P A, Nyborg J K, Cockrell G L. Identification of an NF-κB binding site in the bovine leukemia virus promoter. J Virol. 1995;69:6005–6009. doi: 10.1128/jvi.69.10.6005-6009.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buehring G C, Kramme P M, Schultz R D. Evidence for bovine leukemia virus in mammary epithelial cells of infected cows. Lab Invest. 1994;71:359–365. [PubMed] [Google Scholar]
- 8.Coffin J M. Retroviridae and their replication. 2nd ed. Vol. 1. New York, N.Y: Raven Press; 1990. [Google Scholar]
- 9.Derse D. Bovine leukemia virus transcription is controlled by a virus-encoded trans-acting factor and by cis-acting response elements. J Virol. 1987;61:2462–2471. doi: 10.1128/jvi.61.8.2462-2471.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Derse D, Casey J W. Two elements in the bovine leukemia virus long terminal repeat that regulate gene expression. Science. 1986;231:1437–1440. doi: 10.1126/science.3006241. [DOI] [PubMed] [Google Scholar]
- 11.Djilali S, Parodi A, Levy D. Bovine leukemia virus replicates in sheep B-lymphocytes under a T-cell released factor. Eur J Cancer Clin Oncol. 1987;23:81. doi: 10.1016/0277-5379(87)90423-8. [DOI] [PubMed] [Google Scholar]
- 12.Epstein M A, Barr Y M. Characterization and mode of growth of a tissue culture strain (EB1) of human lymphoblasts from Burkitt’s lymphoma. J Natl Cancer Inst. 1965;34:231–240. doi: 10.1093/jnci/34.2.231. [DOI] [PubMed] [Google Scholar]
- 13.Fong A M, Santoro S A. Transcriptional regulation of alpha IIb integrin gene expression during megakaryocytic differentiation of K562 cells: role of a silencer element. J Biol Chem. 1994;269:18441–18447. [PubMed] [Google Scholar]
- 14.Fong C L, Mark D F. The NF-kappa B-like site in the TNF-alpha repressor element is essential for its repressor function. Biochem Biophys Res Commun. 1995;212:879–886. doi: 10.1006/bbrc.1995.2051. [DOI] [PubMed] [Google Scholar]
- 15.Goldring C E, Narayanan R, Lagadec P, Jeannin J F. Transcriptional inhibition of the inducible nitric oxide synthase gene by competitive binding of NF-kappa B/Rel proteins. Biochem Biophys Res Commun. 1995;209:73–79. doi: 10.1006/bbrc.1995.1472. [DOI] [PubMed] [Google Scholar]
- 16.Graves D C, Ferrer J F. In vitro transmission and propagation of the bovine leukemia virus in monolayer cell cultures. Cancer Res. 1976;36:4152–4159. [PubMed] [Google Scholar]
- 17.Grayson J, Williams R S, Yu Y T, Bassel-Duby R. Synergistic interactions between heterologous upstream activation elements and specific TATA sequences in a muscle-specific promoter. Mol Cell Biol. 1995;15:1870–1878. doi: 10.1128/mcb.15.4.1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hartig E, Nierlich B, Mink S, Nebl G, Cato A C. Regulation of expression of mouse mammary tumor virus through sequences located in the hormone response element: involvement of cell-cell contact and a negative regulatory factor. J Virol. 1993;67:813–821. doi: 10.1128/jvi.67.2.813-821.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heeney J L, Valli P J, Jacobs R M, Valli V E. Evidence for bovine leukemia virus infection of peripheral blood monocytes and limited antigen expression in bovine lymphoid tissue. Lab Invest. 1992;66:608–617. [PubMed] [Google Scholar]
- 20.Hirano A, Wong T. Functional interaction between transcriptional elements in the long terminal repeat of reticuloendotheliosis virus: cooperative DNA binding of promoter- and enhancer-specific factors. Mol Cell Biol. 1988;8:5232–5244. doi: 10.1128/mcb.8.12.5232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoover T, Mikovits J, Court D, Liu Y L, Kung H F, Raziuddin A nuclear matrix-specific factor that binds a specific segment of the negative regulatory element (NRE) of HIV-1 LTR and inhibits NF-kappa(B) activity. Nucleic Acids Res. 1996;24:1895–1900. doi: 10.1093/nar/24.10.1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Horikoshi M, Hai T, Lin Y S, Green M R, Roeder R G. Transcription factor ATF interacts with the TATA factor to facilitate establishment of a preinitiation complex. Cell. 1988;54:1033–1042. doi: 10.1016/0092-8674(88)90118-3. [DOI] [PubMed] [Google Scholar]
- 23.Katoh I, Yoshinaka Y, Ikawa Y. Bovine leukemia virus transactivator p34Tax activates heterologous promoters with a common sequence known as a cAMP-responsive element or the binding site of a cellular transcription factor ATF. EMBO J. 1989;8:497–503. doi: 10.1002/j.1460-2075.1989.tb03403.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kettmann R, Burny A, Callebaut I, Droogmans L, Mammerickx M, Willems L, Portetelle D. Bovine leukemia virus. 3rd ed. Vol. 3. New York, N.Y: Plenum Press; 1994. [Google Scholar]
- 25.Kiss-Toth E, Paca-uccaralertkun S, Unk I, Boros I. Member of the CREB/ATF protein family, but not CREB alpha plays an active role in BLV tax transactivation in vivo. Nucleic Acids Res. 1993;21:3677–3682. doi: 10.1093/nar/21.16.3677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lasfargues E Y, Kramarsky B, Sarkar N H, Lasfargues J C, Moore D H. An established RIII mouse mammary tumor cell line: kinetics of mouse mammary tumor virus (MTV) production. Proc Soc Exp Biol Med. 1972;139:242–247. doi: 10.3181/00379727-139-36119. [DOI] [PubMed] [Google Scholar]
- 27.Luckow B, Schutz G. CAT constructions with multiple unique restriction sites for the functional analysis of eukaryotic promoters and regulatory elements. Nucleic Acids Res. 1987;15:5490. doi: 10.1093/nar/15.13.5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murakami K, Okada K, Amanuma H, Aida Y. The gamma delta T cell population in sheep experimentally infected with bovine leukemia virus. Vet Pathol. 1994;31:103–105. doi: 10.1177/030098589403100113. [DOI] [PubMed] [Google Scholar]
- 29.Niermman G, Buehring G C. Hormone regulation of bovine leukemia virus. Virology. 1997;239:249–258. doi: 10.1006/viro.1997.8868. [DOI] [PubMed] [Google Scholar]
- 30.Parekh B S, Hatfield G W. Transcriptional activation by protein-induced DNA bending: evidence for a DNA structural transmission model. Proc Natl Acad Sci USA. 1996;93:1173–1177. doi: 10.1073/pnas.93.3.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pfeifer G P, Riggs A D. Genomic footprinting by ligation mediated polymerase chain reaction. Methods Mol Biol. 1993;15:153–168. doi: 10.1385/0-89603-244-2:153. [DOI] [PubMed] [Google Scholar]
- 32.Pulvertaft J V. Cytology of Burkitt’s tumor (African lymphoma) Lancet. 1964;i:238–240. doi: 10.1016/s0140-6736(64)92345-1. [DOI] [PubMed] [Google Scholar]
- 33.Rashtchian A, Thornton C G, Heidecker G. A novel method for site-directed mutagenesis using PCR and uracil DNA glycosylase. PCR Methods Appl. 1992;2:124–130. doi: 10.1101/gr.2.2.124. [DOI] [PubMed] [Google Scholar]
- 34.Rovnak J, Casey J W, Boyd A L, Gonda M A, Cockerell G L. Isolation of bovine leukemia virus infected endothelial cells from cattle with persistent lymphocytosis. Lab Invest. 1992;65:192–202. [PubMed] [Google Scholar]
- 35.Sagata N, Yuanaga T, Tsuzuku-Kawamura J, Phishi K, Igawa Y. Complete nucleotide sequence of the genome of bovine leukemia virus: its evolutionary relationship to other retroviruses. Proc Natl Acad Sci USA. 1985;82:677–681. doi: 10.1073/pnas.82.3.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 37.Sanger F, Nicklen S, Coulson A R. A rapid method for determining sequences in DNA by primed synthesis with DNA polymerase. J Mol Biol. 1977;94:441. doi: 10.1016/0022-2836(75)90213-2. [DOI] [PubMed] [Google Scholar]
- 38.Soule H D, Vazquez J, Long A, Albert S, Brennan M. A human cell line from a pleural effusion derived from breast carcinoma. J Natl Cancer Inst. 1973;51:1409–1413. doi: 10.1093/jnci/51.5.1409. [DOI] [PubMed] [Google Scholar]
- 39.Stoler M H, Wolinsky S M, Whitbeck A, Broker T R, Chow L T. Differentiation-linked human papilloma virus types 6 and 11 transcription in genital condylomata revealed by in situ hybridization with message-specific RNA probes. Virology. 1989;172:331–340. doi: 10.1016/0042-6822(89)90135-9. [DOI] [PubMed] [Google Scholar]
- 40.Swingler S, Morris A, Easton A. Tumour necrosis factor alpha and interleukin-1 beta induce specific subunits of NFKB to bind the HIV-1 enhancer: characterization of transcription factors controlling human immunodeficiency virus type 1 gene expression in neural cells. Biochem Biophys Res Commun. 1994;203:623–630. doi: 10.1006/bbrc.1994.2228. [DOI] [PubMed] [Google Scholar]
- 41.Taylor J A, Jacobs R M. Effects of plasma and serum on the in vitro expression of bovine leukemia virus. Lab Invest. 1993;69:340–346. [PubMed] [Google Scholar]
- 42.Unk I, Kiss-Toth E, Boros I. Transcription factor AP-4 participates in activation of bovine leukemia virus long terminal repeat by p34 Tax. Nucleic Acids Res. 1994;22:4872–4875. doi: 10.1093/nar/22.23.4872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Van der Maaten M J, Miller J M. Replication of bovine leukemia virus in monolayer cell cultures. Int J Cancer. 1976;31:791. doi: 10.1159/000399166. [DOI] [PubMed] [Google Scholar]
- 44.Willems L, Kettmann R, Chen G, Portetelle D, Burny A, Derse D. A cyclic AMP-responsive DNA-binding protein (CREB2) is a cellular transactivator of the bovine leukemia virus long terminal repeat. J Virol. 1992;66:766–772. doi: 10.1128/jvi.66.2.766-772.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wolff L, Ruscetti S. Tissue tropism of a leukemogenic murine retrovirus is determined by sequences outside of the long terminal repeats. Proc Natl Acad Sci USA. 1986;83:3376–3380. doi: 10.1073/pnas.83.10.3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45a.Xiao, J., and G. C. Buehring. Submitted for publication.
- 46.Xu X, Brown D A, Kitajima I, Bilakovics J, Fey L W, Nerenberg M I. Transcriptional suppression of the human T-cell leukemia virus type I long terminal repeat occurs by an unconventional interaction of a CREB factor with the R region. Mol Cell Biol. 1994;14:5371–5383. doi: 10.1128/mcb.14.8.5371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yeh C H, Shatkin A J. Down-regulation of Rous sarcoma virus long terminal repeat promoter activity by a HeLa cell basic protein. Proc Natl Acad Sci USA. 1994;91:11002–11006. doi: 10.1073/pnas.91.23.11002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zandomeni R O, Carrera-Zandomeni M, Esteban E, Donawick W, Ferrer J F. Induction and inhibition of bovine leukaemia virus expression in naturally infected cells. J Gen Virol. 1992;73:1915–1924. doi: 10.1099/0022-1317-73-8-1915. [DOI] [PubMed] [Google Scholar]
- 49.Zandomeni R O, Esteban E, Carrera-Zandomeni M, Ferrer J F. Host soluble factors with blocking and stimulating activity on the expression of the bovine leukemia virus. J Infect Dis. 1994;170:787–794. doi: 10.1093/infdis/170.4.787. [DOI] [PubMed] [Google Scholar]