This cohort study examines the frequency of germline CDH1 sequence variations in women with the hereditary lobular breast cancer phenotype and the association of genetic profiles with clinical-pathological data and survival.
Key Points
Question
What is the frequency of germline CDH1 variants in hereditary lobular breast cancer (HLBC) predisposition?
Findings
In this cohort study of 394 women with LBC, 15 germline CDH1 variants were identified in 15 families with HLBC; 40.0% were pathogenic or likely pathogenic (P/LP). The overall frequency of P/LP CDH1 variants was 1.5% and was associated with age of 45 years or younger at LBC diagnosis and positive family history of BC.
Meaning
The identification of P/LP germline CDH1 variants in young women with LBC with (or without) family history of BC, not fulfilling the classic CDH1 genetic screening criteria, may provide an indication to test for CDH1 gene.
Abstract
Importance
Pathogenic or likely pathogenic (P/LP) germline CDH1 variants are associated with risk for diffuse gastric cancer and lobular breast cancer (LBC) in the so-called hereditary diffuse gastric cancer (HDGC) syndrome. However, in some circumstances, LBC can be the first manifestation of this syndrome in the absence of diffuse gastric cancer manifestation.
Objectives
To evaluate the frequency of germline CDH1 variants in women with the hereditary LBC (HLBC) phenotype, somatic CDH1 gene inactivation in germline CDH1 variant carriers’ tumor samples, and the association of genetic profiles with clinical-pathological data and survival.
Design, Setting, and Participants
This single-center, longitudinal, prospective cohort study was conducted from January 1, 1997, to December 31, 2021, with follow-up until January 31, 2023. Women with LBC seen at the European Institute of Oncology were included. Testing for germline CDH1, BRCA1, and BRCA2 genes was performed. Somatic profiling was assessed for germline CDH1 carriers.
Main Outcomes and Measures
Accurate estimates of prevalence of germline CDH1 variants among patients with HLBC and the association of somatic sequence alteration with HLBC syndrome. The Kaplan-Meier method and a multivariable Cox proportional hazards regression model were applied for overall and disease-free survival analysis.
Results
Of 5429 cases of primary LBC, familial LBC phenotype accounted for 1867 (34.4%). A total of 394 women with LBC were tested, among whom 15 germline CDH1 variants in 15 unrelated families were identified. Among these variants, 6 (40.0%) were P/LP, with an overall frequency of 1.5% (6 of 394). Of the 6 probands with P/LP CDH1 LBC, 5 (83.3%) had a positive family history of BC and only 1 (16.7%) had sporadic juvenile early-onset LBC. No germline BRCA1 and BRCA2 variants were identified in CDH1 carriers. An inactivating CDH1 mechanism (second hit) was identified in 4 of 6 explored matched tumor samples (66.7%) in P/LP germline carriers. The P/LP CDH1 LBC variant carriers had a significantly lower age at diagnosis compared with the group carrying CDH1 variants of unknown significance or likely benign (42.5 [IQR, 38.3-43.0] vs 51.0 [IQR, 45.0-53.0] years; P = .03).
Conclusions and Relevance
In this cohort study, P/LP germline CDH1 variants were identified in individuals not fulfilling the classic clinical criteria for HDGC screening, suggesting that identification of these variants may provide a novel method to test women with LBC with early age at diagnosis and/or positive family history of BC.
Introduction
Hereditary lobular breast cancer (HLBC) is a rare inherited cancer predisposition syndrome associated with germline pathogenic or likely pathogenic (P/LP) variants in the CDH1 gene, in which LBC is the first manifestation without a clear family history of diffuse gastric cancer (DGC).1 In 1999, a germline CDH1 variant was defined as the hallmark of hereditary DGC (HDGC)2; further research identified P/LP germline CDH1 variants also in women with LBC but without clinical evidence of DGC.3,4 Initially excluding LBC, the International Gastric Cancer Linkage Consortium (IGCLC) suggested 4 revisions of the clinical criteria for CDH1 genetic testing,2,5,6,7 and at the 2020 IGCLC meeting, LBC was considered a pivotal cancer index for CDH1 genetic testing independently from DGC manifestation. The IGCLC LBC-oriented criteria recommend to test for CDH1 in patients with (1) 2 or more independent cases of LBC in the same family at younger than 50 years and (2) bilateral LBC, diagnosed at younger than 70 years (both with no cases of DGC).7 In 2018, some of our group established a panel of experts to provide recommendations for HLBC management, and after revisiting retrospective literature data, additional clinical criteria were proposed for CDH1 genetic testing in women with LBC.1 In sporadic LBC, the frequency of germline CDH1 variant detection is approximately 0.2% to 0.5%,8 but in the setting of the HLBC phenotype, the overall frequency is not well established due to a lack of large, prospective cohort studies.9
In the current study, we tested a large series of women with diagnosed LBC fulfilling the HLBC clinical criteria, aiming to assess the frequency of germline variants in the CDH1 gene, genomic inactivation in matched tumor samples, and disease-free and overall survival. The BRCA1 and BRCA2 genes were also tested to verify a possible association (or exclusion) between CDH1 HLBC and the hereditary breast-ovarian cancer syndromes in these families.
Methods
Ethics Statement
This cohort study was approved by the European Institute of Oncology ethical committee, and all available participants gave their written consent to be included in the study. The study was conducted in accordance with the Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) reporting guideline.
Study Participants
For this longitudinal, prospective cohort study, a genetic analysis of the CDH1, BRCA1, and BRCA2 genes was conducted from January 1, 1997, to December 31, 2021, on blood samples from selected women with LBC at the European Institute of Oncology. Clinical criteria for genetic testing are described in eTable 1 in Supplement 1. Personal and family history along with clinical and histopathological data were collected in a dedicated institutional database. Available biologic samples (whole blood and tumor specimens) were stored in our institutional biobank. Follow-up was conducted using medical records, pathology reports, and telephone consultations conducted by dedicated personnel. The end of the follow-up period for outcome ascertainment was January 31, 2023. Genetic and psychological counseling was offered to all patients with variants in the BRCA1, BRCA2, or CDH1 gene.
Library Preparation and Next-Generation Sequencing
Genomic DNA was extracted from peripheral blood samples using a MagCore Super Automated Nucleic Acid Extractor (Diatech), and for next-generation sequencing library preparation, DNA was quantified using the Qubit dsDNA HS Assay Kit with the Qubit 3.0 Fluorometer (Life Technologies) following the manufacturer’s instructions. Available samples were analyzed with the Hereditary Cancer Solution CE-IVD multigene panel by SOPHiA Genetics. Library preparation was optimized for 200 ng of total genomic DNA (Qubit quantification) using an enrichment protocol (version PM_T1_5.1.5_r2en July 2017). Libraries were quantified through the 4200 TapeStation (Agilent) and Qubit 3.0 Fluorometer (Life Technologies) and diluted to 4 nM. Following denaturation, a 10-pM dilution was loaded on the Illumina MiSeq System with 3% PhiX Control using the MiSeq, version 2 standard reagent kit and 2 × 250 cycles. The results were retrieved and analyzed in the dedicated platform SOPHiA DDM, which allows for the accurate detection of single nucleotide variations, insertions or deletions, and copy number variations.
Germline Variant Classification
The identified genetic variants were divided into 5 classes according to the International Agency for Research on Cancer recommendations.10 Variant pathogenicity was assessed using the ClinVar database,11 the Leiden Open Variation Database,12 the Clinical Genome Resource CDH1 Variant Curation Expert Panel,13 and the BRCA Exchange14 for BRCA1 and BRCA2 genes following the American College of Medical Genetics and Genomics guidelines.15 In an attempt to assess the pathogenicity of germline CDH1 missense variants identified in this study, we also used predictive in silico models, such as PROVEAN, SIFT, PolyPhen, and FoldX (details are provided in eTable 2 in Supplement 1).16,17,18
Tumor Genomic Profiling
Next-generation sequencing analysis of primary LBC tumor samples from CDH1 germline variant carriers was performed to investigate the potential genetic-based mechanism of E-cadherin inactivation (ie, second hit), including second CDH1 gene somatic variants and/or loss of heterozygosity (LOH), and to explore the genomic landscape of these tumors. In brief, 10 unstained sections were cut from representative formalin-fixed, paraffin-embedded tumor tissue blocks retrieved from the archives of the Division of Pathology of the European Institute of Oncology and submitted to the FoundationOne CDx assay (Foundation Medicine) according to FoundationOne CDx specimen instructions.
Intragenic Loss of Heterozygosity and Somatic CDH1 Promoter Methylation Analyses
For intragenic LOH analysis, we used the following intragenic markers: promoter c.-161C>A transversion (rs16260), silent substitution c.2076T>C at exon 13 (rs1801552), and c.*54C>T polymorphism (rs1801026). CDH1 promoter methylation analysis was carried out 160 base pairs upstream of the translation start site, encompassing 17 CpG sites. Both protocols were described in detail previously.19
E-Cadherin Immunohistochemistry
E-cadherin immunoreactivity was evaluated using the standard protocol on tumor and normal tissues. We considered the predominant expression pattern as normal (complete membrane staining), aberrant (cytoplasmic and heterogeneous staining), or absent (no staining).
Statistical Analysis
For continuous variables, the median and IQR were reported, and absolute and relative frequencies were assessed as summary measures of categorical variables. According to the nature of variables, χ2 tests and Kruskal-Wallis tests were performed to investigate the associations between germinal variant status and characteristics of patients and tumors. The Mann-Whitney U test was used to compare ages between probands and family members. Disease-free survival was estimated using the Kaplan-Meier method, and survival distributions were compared using the log-rank test. A multivariable Cox proportional hazards regression model was used to determine the independent association of germinal variant status with cancer progression or death, adjusting for age. The significance level was set at a global 2-tailed P < .05 for all analyses. The statistical analyses were performed with RStudio, version 4.2.3 (RStudio).
Results
Germline Genetic Testing Results
From an initial consecutive population of 5429 cases of primary LBC, we selected 1867 patients with LBC using the HLBC clinical criteria (eTable 1 in Supplement 1). We were able to enroll 421 women with LBC, of whom 394 (93.6%) were actually tested. Twenty-seven individuals (6.4%) eventually refused to participate or were not available to provide informed consent (eFigure 1 in Supplement 1). All women with LBC were White and from the Italian geographic area. Familial LBC phenotype accounted for 1867 of the 5429 cases (34.4%) (eFigure 1 in Supplement 1). For CDH1, 15 of the 394 index cases (3.8%) that met previous1 and new, expanded criteria (including early-onset sporadic LBC at age 45 years or younger) were found to have a germline heterozygous CDH1 variant. Thirteen distinct variants were found (Figure 1); c.1003C>T and c.1633C>G variants were identified in 2 unrelated index cases. Six of the 15 variants (40.0%) were classified as P/LP. The overall frequency of identified P/LP CDH1 variants was 1.5% (6 of 394) (Table 1), and the variants were identified only in the invasive LBC histotype. Pathogenic or likely pathogenic BRCA1 and BRCA2 occurred with a frequency of 0.2% and 1.2%, respectively. Missense variants were also evaluated using in silico tools, and the results are reported in eTable 2 in Supplement 1. Lastly, no co-occurrence of germline BRCA1 and BRCA2 variants (eTables 3 and 4 in Supplement 1) was observed in any germline CDH1 variant carriers (Table 1).
Figure 1. Localization of CDH1 Variants Identified in Our Study.
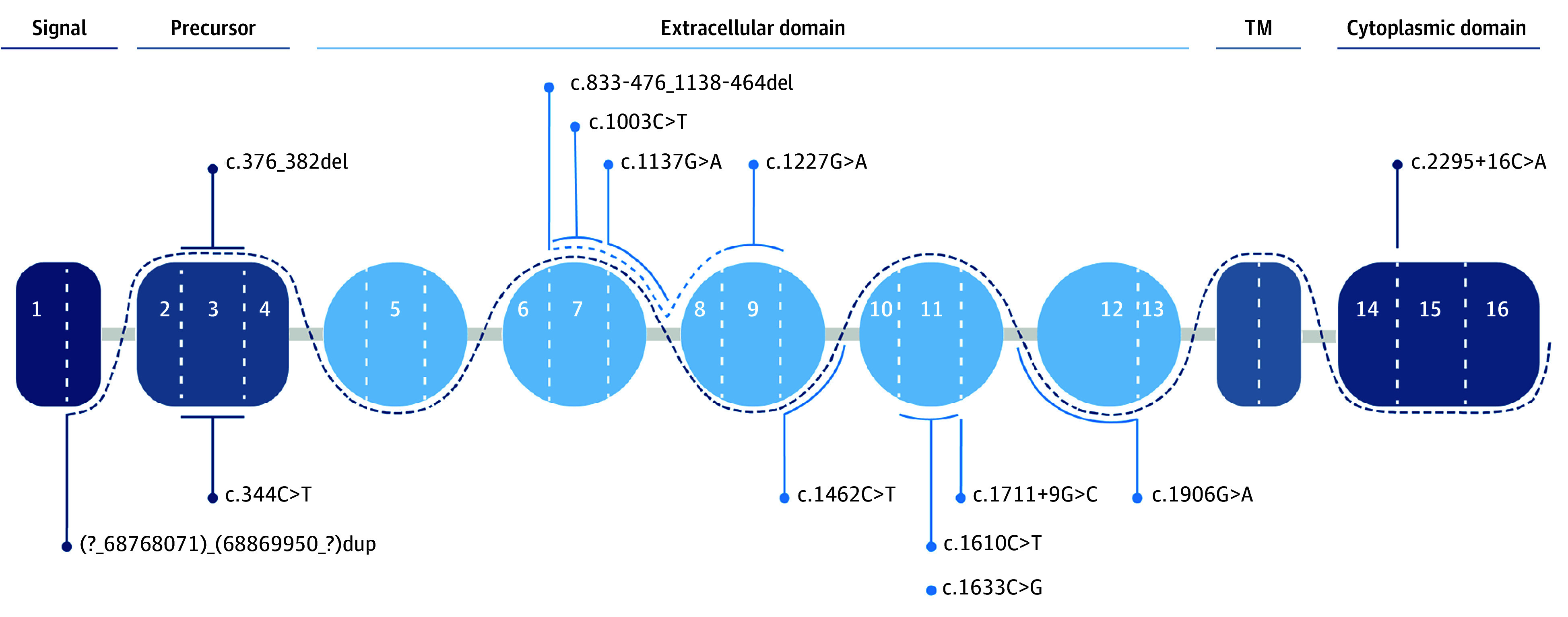
Germline pathogenic or likely pathogenic variants are given below the dashed line, and variants of unknown significance or benign or likely benign variants are given above the dashed line. TM indicates transmembrane.
Table 1. Germline CDH1 Variants Identified in 15 Patients With LBC.
| Proband identification No. | Age, y | Sequence variationa | Protein change | Type | Localization | Interpretation |
|---|---|---|---|---|---|---|
| 834-001 | 52 | c.1906G>A | p.(Ala636Thr) | Missense | Exon 12 | VUS |
| 834-003 | 43 | c.1003C>T | p.(Arg335Ter) | Nonsense | Exon 7 | P/LP |
| 834-016 | 43 | c.2295+16C>A | (p.?) | Intronic | Intron 14 | B/LB |
| 834-042 | 63 | c.1462C>T | p.(Pro488Ser) | Missense | Exon 10 | VUS |
| 834-059 | 44 | c.376_382del | p.(Pro126IlefsTer87) | Frameshift | Exon 3 | P/LP |
| 834-096 | 37 | c.1137G>A | p.(Thr379 = ) | Synonymous | Exon 8 | P/LP |
| 834-155 | 54 | c.1610C>T | p.(Pro537Leu) | Missense | Exon 11 | VUS |
| 834-163 | 53 | c.1711+9G>C | (p.?) | Intronic | Intron 11 | B/LB |
| 834-173 | 36 | c.833-476_1138-464del | p.(Gly278ValfsTer7) | Deletion | Exon 7-8 | P/LP |
| 834-183 | 39 | c.1633C>G | p.(Arg545Gly) | Missense | Exon 11 | VUS |
| 834-190 | 45 | c.344C>T | p.(Thr115Met) | Missense | Exon 3 | VUS |
| 834-263 | 43 | c.1227G>A | p.(Trp409Ter) | Nonsense | Exon 9 | P/LP |
| 834-269 | 50 | (?_68768071)_(68869950_?)dup | NA | CNV | Exon 1-16 | VUS |
| 834-323 | 41 | c.1633C>G | p.(Arg545Gly) | Missense | Exon 11 | VUS |
| 834-369 | 43 | c.1003C>T | p.(Arg335Ter) | Nonsense | Exon 7 | P/LP |
Abbreviations: B/LB, benign or likely benign; CNV, copy number variation; LBC, lobular breast cancer; NA, not applicable; P/LP, pathogenic or likely pathogenic; VUS, variant of unknown significance.
Human Genome Variation Society nomenclature (reference sequence [Human Feb. 2009 - GRCh37/hg19 Assembly]: NM_004360.51).
Pedigree Features of Germline CDH1 Variant Carriers
The reported germline CDH1 variants were identified in 15 unrelated families. Excluding c.1137G>A and c.833-476_1138-463del alterations,20 the remaining identified CDH1 variants in this study were not previously reported in patients with LBC, to our knowledge.21 The identified P/LP CDH1 variants were associated with a positive family history for BC in 5 cases (83.3%; 4 early-onset LBC at age <45 years at diagnosis, 1 also with bilateral LBC manifestation), and 1 (16.7%) case was a sporadic juvenile early-onset LBC (eFigure 2 in Supplement 1).
Somatic CDH1 Genetic and Epigenetic Alterations
Data from tumor genomic testing were obtained for 12 of the 15 CDH1 germline variant carriers (80.0%). No tumor samples were available for 2 cases (13.3%) (1 benign or likely benign [B/LB] [6.7%] and 1 variant of unknown significance [VUS] [6.7%]); 1 tumor specimen (P/LP) did not yield sufficient DNA for the analysis (6.7%). Eleven of 12 explored samples (91.7%) manifested at least a somatic alteration (7 of 12 [58.3%], sequence variations; 6 of 12 [50.0%], intragenic LOH; 3 of 12 [25.0%], promoter methylation) (Table 2 and Table 3). In 5 of the 12 samples (41.7%), we aimed to explore the second-hit mechanism and, in 7 samples (58.3%), somatic inactivation. In P/LP germline CDH1 carriers, an inactivating CDH1 mechanism (second hit) was identified in 4 of 6 explored matched tumor samples (66.7%). In this group, we also identified the germline variants in matched tumor specimens but not additional CDH1 somatic variants. Intragenic LOH was the most common inactivating second-hit mechanism (3 of 5 cases [60.0%]), and methylation was identified in only 1 of 5 cases (20.0%) (Table 2).
Table 2. Identified Somatic Alterations, or Second Hit, in 6 Patients With HLBC With Germline P/LP CDH1 Variant.
| Proband identification No. | HLBC CDH1 variant | Tumor | Structural | Epigenetic, methylation | E-cadherin IHC expression | Second-hit CDH1 | Other P/LP variants | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Germline CDH1 variant detected | VAF, % | Somatic CDH1 variant detected | VAF, % | Variant | iLOH | ||||||
| 834-003 | c.1003C>T | Yes | 82.4 | No | No | No | Yes | No | Negative | iLOH | AKT3, MDM4, IKBKE, PARP1, PIK3C2B |
| 834-059 | c.376_382del | Yes | 77.2 | No | No | No | Yes | No | Negative | iLOH | CCND1, C11ORF30, FGF19, FGF3, FGF4 |
| 834-096 | c.1137G>A | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| 834-173 | c.833-476_1138-464del | Yes | CNV | No | No | No | Yes | No | Negative | iLOH | PIK3CA, MDM4, GATA3, PIK3C2B |
| 834-263 | c.1227G>A | Yes | 70.6 | No | No | No | No | No | Negative | Not found | PIK3CA, CCND1, C11ORF30 |
| 834-369 | c.1003C>T | Yes | 58.5 | No | No | No | No | Yes | Negative | Methylation | ERBB2, PTEN, BRD4, NOTCH3, PRKAR1A, SMARCA4 |
Abbreviations: CNV, copy number variation; HLBC, hereditary lobular breast cancer; IHC, immunohistochemistry; iLOH, intragenic loss of heterozygosity; NA, unavailable tumor sample; P/LP, pathogenic or likely pathogenic; VAF, variant allele frequency.
Table 3. Overall Somatic Alteration in 9 Sporadic LBCs With Germline VUS or LB CDH1 Germline Variant Carrier.
| Proband identification No. | LBC CDH1 variant | Tumor | Structural | Epigenetic, methylation | E-cadherin IHC expression | Other P/LP variants | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Germline CDH1 variant detected | VAF, % | Somatic CDH1 variant detected | VAF, % | Variation | iLOH | |||||
| 834-001 | c.1906G>A | Yes | NA | c.163+1G>C | 72 | Yes | Yes | No | Negative | PIK3CA, ACVR1B, CBFB |
| 834-016 | c.2295+16C>A | No | NA | c.511_518delTTTCCTAA | 26 | Yes | ND | Yes | Negative | ERBB2 |
| 834-042 | c.1462C>T | Yes | NA | c.67C>T | 75.3 | Yes | Yes | No | Negative | EPHA3 |
| 834-155 | c.1610C>T | Yes | NA | c.532-1G>A | 51.2 | Yes | ND | No | Negative | ERBB2, ERBB3 |
| 834-163 | c.1711+9G>C | NA | NA | NA | NA | NA | NA | NA | Negative | NA |
| 834-183 | c.1633C>G | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| 834-190 | c.344C>T | Yes | NA | c.2095C>T | 46.6 | Yes | No | No | Negative | CBFB, GATA3, NSD3, TP53, ZNF703 |
| 834-269 | (?_68768071)_(68869950_?)dup | No | NA | c.367C>T | 2.4 | Yes | No | Yes | Positive | CCND1, FGF19, FGF3, FGF4, TP53 |
| 834-323 | c.1633C>G | Yes | NA | c.67C>T | 51.8 | Yes | Yes | No | Negative | AKT1 |
Abbreviations: IHC, immunohistochemistry; iLOH, intragenic loss of heterozygosity; LB, likely benign; LBC, lobular breast cancer; NA, unavailable tumor sample; ND, not defined (intragenic allelic analysis was not resolutive for iLOH definition); P/LP, pathogenic or likely pathogenic; VAF, variant allele frequency; VUS, variant of unknown significance.
In sporadic LBC (germline CDH1 VUS or LB), discordances between germline and tumor testing were observed in 2 of 7 cases (28.6%), including 1 germline intronic CDH1 variant (c.2295+16C>A) and 1 germline CDH1 copy number variant ([?_68768071]_[68869950_?]dup) missed by tumor sequencing. However, in these 2 cases, somatic CDH1 variants were detected in tumor samples. A second somatic CDH1 variant was detected in all LBC samples. Intragenic LOH was detected in 3 of 7 LBCs (42.9%), and promoter methylation was detected in 2 (28.6%) (Table 3) (Figure 2). We noted a high accumulation of genomic aberrations in HLBC samples (Table 2) compared with sporadic LBCs (Table 3): 23 in 5 HLBC groups and 18 in 7 sporadic LBC groups. E-cadherin immunohistochemistry revealed an absent pattern in 12 of 13 analyzed available sample tumors (92.3%) (eFigure 3 in Supplement 1). We observed that epigenetic and genetic CDH1 somatic alterations occurred only alone in HLBC (Table 2) but simultaneously in sporadic LBC neoplastic lesions (Table 3).
Figure 2. Recurrent Somatic Genomic Alterations in Lobular Breast Cancer Samples From Germline CDH1 Variant Carriers.
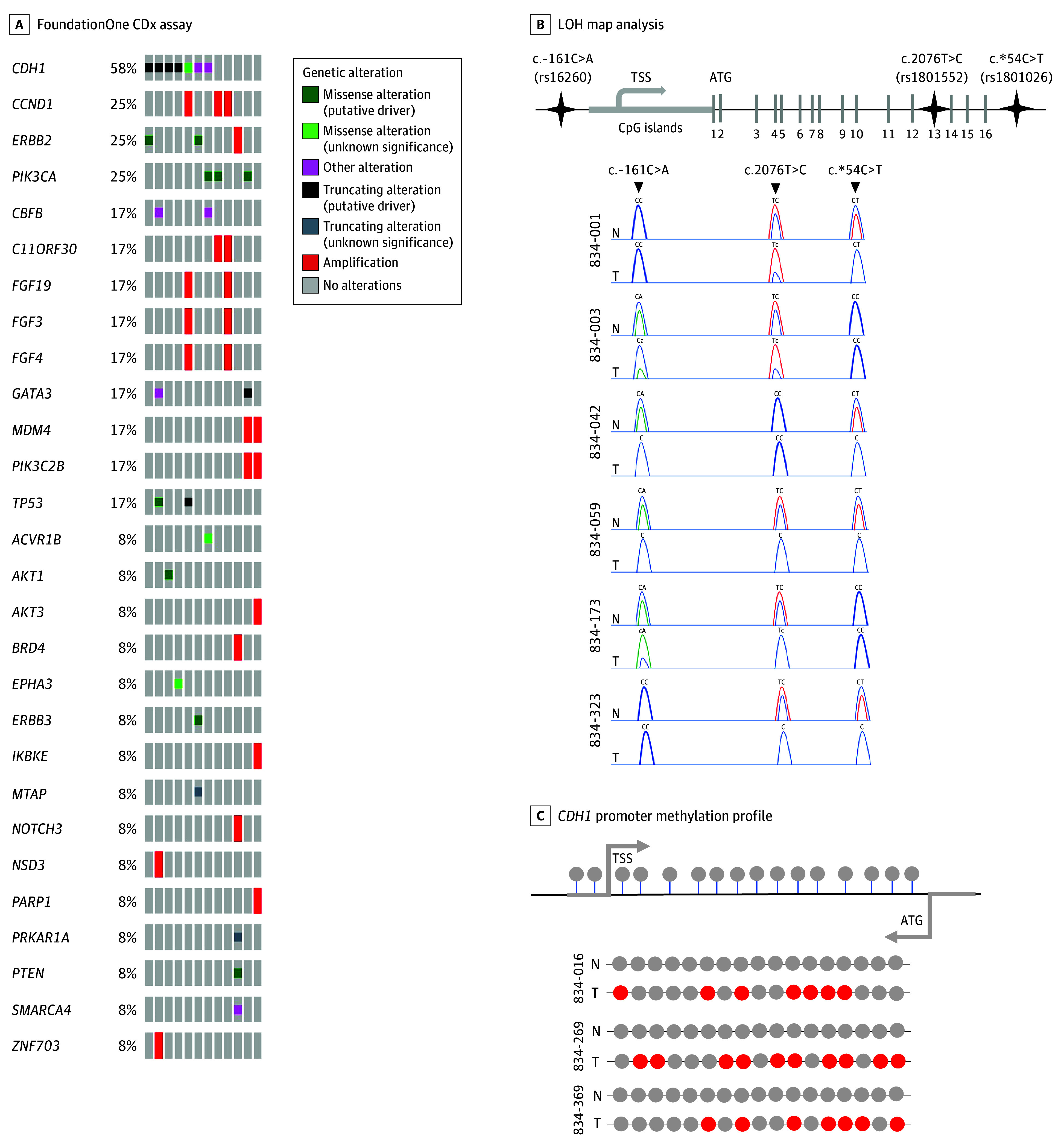
LOH indicates loss of heterozygosity.
Clinical Follow-Up
None of the 15 families with LBC who were CDH1 variant carriers showed a positive first-degree familial history of DGC. Due to the lack of strong evidence in terms of the benefits of prophylactic total gastrectomy (PTG) in the absence of a clear potential gastric cancer (GC) risk, these patients were followed up in accordance with the latest IGCLC recommendations.7 For breast surveillance, we suggested annual breast magnetic resonance imaging and mammography, 6-month breast ultrasonography, and annual physical examination for all 15 CDH1 variant carriers.1 With 1 exception, no patients developed DGC during a median of 5.24 years (IQR, 4.93-5.67 years) of follow-up. Conversely, a CDH1 c.833-476_1138-464del carrier, who was previously tested following a diagnosis of bilateral LBC and then enrolled in this study, had already undergone PTG at enrollment, and gastric histopathological analysis revealed 3 foci of invasive DGC (pT1aN0). Subsequently, this patient also developed a Krukenberg ovary tumor from primary LBC.
Clinical-Pathological Data
A total of 377 invasive and 17 in situ LBCs were tested for genetic screening. No P/LP variants were detected in the in situ LBCs. With a median 5.24 years of follow-up (IQR, 4.93-5.67 years), no associations were observed among germline CDH1 variant carriers, BRCA1 and BRCA2 genes, wild-type groups, and the clinical-pathological features of LBC (eTables 5 and 6 in Supplement 1). Furthermore, variant carrier or wild-type status was not associated with disease-free survival (eFigure 4 in Supplement 1). Using an age-adjusted Cox proportional hazards regression model, no associations were detected among the explored groups. Exploring the 15 CDH1 pedigrees, the median age of P/LP CDH1 variant carriers at LBC diagnosis was significantly lower (42.5 years [IQR, 38.3-43.0 years]) compared with the group carrying VUS plus LB CDH1 variants (51.0 years [IQR, 45.0-53.0 years]; P = .03) and also with the CDH1 wild-type group (47.0 years [IQR, 43.0-53.0 years]; P = .009) (eFigure 1 and eTable 5 in Supplement 1). A lower age at diagnosis was identified in CDH1 carriers compared with BRCA1 and BRCA2 carriers and the wild-type group, but the differences were not significant (eTable 6 in Supplement 1).
Discussion
CDH1 Genetic Testing in HLBC
To our knowledge, this is the largest single-center longitudinal study to report results from germline CDH1 genetic testing in families with suspected HLBC. In this series, the HLBC phenotype occurred in 34.4% of patients with LBC, whereas a US study reported a lower incidence (15%).22 There are some substantial differences between these studies (eTable 8 in Supplement 1). Our study was a European, longitudinal, prospective cohort analysis, and we tested for women with CDH1 LBC prospectively from phenotype (HLBC) to genotype (CDH1). The US study reported a retrospective frequency of approximately 60% of germline CDH1 variants in its HLBC series. Our frequency of germline CDH1 variants was substantially lower (3.8%) and seems to be more in accordance with the rarity of this syndrome.
The identification of P/LP germline CDH1 variants in women with early-onset LBC suggests new criteria for genetic screening selection and recommendations to test this group of patients with LBC. Positive BC family history and juvenile age at LBC diagnosis should be considered to be pivotal criteria for CDH1 genetic testing in women with LBC. We identified that 83.3% of germline CDH1 variant carriers had a diagnosis at 45 years or younger and a positive BC family history.
BRCA vs CDH1
We demonstrated the mutual exclusion of BRCA and CDH1 genes in the pathway of families with HLBC. A German study suspected this evidence in just 1 pedigree analysis.23 In the present HLBC series, we identified that P/LP BRCA1, BRCA2, and CDH1 variants occurred with a frequency of 0.2%, 1.2%, and 1.5%, respectively. In cases of sporadic LBC, Yadav et al8 reported a frequency of P/LP BRCA1, BRCA2, and CDH1 variants of 0.3%, 2.2%, and 0.5%. In an ongoing independent study by our team testing only sporadic LBC (LobularCard Breast trial), the frequency of P/LP variants was 0.9% in BRCA1 and 2.2% in BRCA2 genes, and no P/LP CDH1 variants were identified (eTable 9 in Supplement 1). It seems that there is an association between CDH1 and HLBC (more than BRCA1 and BRCA2), and this mutual exclusion suggests that CDH1 HLBC is an inherited cancer predisposition syndrome unrelated to BRCA.
Somatic CDH1 Alterations, Second Hit, and E-Cadherin Inactivation
In our study of LBC cases associated with germline CDH1 variants, 11 of 12 (91.7%) manifested a somatic alteration (7 of 12 [58.3%], sequence variations; 6 of 12 [50.0%], intragenic LOH; 3 of 12 [25.0%], promoter methylation)(Table 2 and Table 3). All CDH1 variant carriers presented with an invasive LBC histotype, thus demonstrating a pivotal role of E-cadherin protein inactivation in the process of lobular breast tumorigenesis. Loss of E-cadherin expression is an early gatekeeper event in in situ LBC and a precursor of invasive LBC.24,25
Inactivated sequence variations in the CDH1 gene have been frequently described as the cause of E-cadherin protein deregulation in invasive LBC.26,27,28,29 In this study, all sporadic LBCs had a second structural CDH1 somatic sequence variation (Table 3); intragenic LOH and methylation were detected in 42.9% and 28.6% of LBCs, respectively. In HLBC series, we were able to identify a second-hit mechanism in 66.7% of analyzed samples matched with identified germline P/LP CDH1 variants. Different from the HDGC tumors in which CDH1 promoter methylation appears to be the predominant inactivating mechanism (around 32%),30 we rarely observed methylation phenomena in HLBC tumors (Table 2 and eTable 7 in Supplement 1). The difference in genetic and epigenetic mechanisms described between HDGC and HLBC tumors could be a possible explanation for different risk for lobular and gastric tumors in these 2 syndromes.
By exploring FoundationOne data (Table 2 and Table 3), we noted the absence of additional somatic CDH1 variations, and respective germline variations were confirmed in all HLBC tumors. We noted also that in HLBC tumors, there was a higher accumulation of aberrant variations (Table 2) compared with sporadic LBC (Table 3). We could suppose that this phenomenon is associated with a long progression of CDH1 HLBC tumorigenesis resulting in more aggressive and undifferentiated tumors.
BC Risk in Families With HLBC
A recent European study20 demonstrated an association of P/LP germline CDH1 truncating alterations with LBCs, not fulfilling the 2020 HDGC criteria.7 However, the exact LBC risk in this nonclassic context is under evaluation; it seems variable in accordance with some different clinical phenotypes.31,32,33 Our study demonstrated a significantly earlier age at diagnosis of LBC manifestation in P/LP CDH1 variant carriers compared with other groups (wild-type, VUS plus B/LB) and also BRCA1 and BRCA2 genes. This evidence supports the hypothesis that germline P/LP CDH1 sequence variations may determine the earliest LBC manifestation in the HLBC phenotype.
GC Risk in Families With HLBC
The GC risk in asymptomatic P/LP germline CDH1 variant carriers fluctuates depending on the family history, the age at GC diagnosis, and the number of GC cases in the same family.31,32,33 In the context of CDH1 HLBC, the exact GC risk is unknown. Considering the absence or unclear diagnosis of a positive GC family history, we could consider that the GC risk is presumably lower, but it should not be ignored.22 The 2020 IGCLC guidelines reduce the emphasis on PTG if GC family history is weak,7 as in HLBC syndrome. In patients not undergoing PTG, endoscopic surveillance remains the unique option.34 In our analysis of 15 pedigrees, only 1 documented DGC was reported and none of the remaining 14 women with LBC developed GC within a median 5 years of follow-up. In accordance with the latest IGCLC indications,7 the benefits and risks of different preventive options (including PTG) were discussed in a multidisciplinary evaluation.
Limitations
The following limitations must be considered. First, due to the low frequency of identified germline CDH1 variants, we were unable to estimate the exact risk for LBC predisposition in families with HLBC. We could assume that the association between the early diagnosis of LBC (age ≤45 years) and P/LP CDH1 variants correlates with relevant risk for developing LBC. Second, the identified frequency of CDH1 variants in this study may have been underestimated because we were able to test only 394 women with LBC due to the lack of availability of contacts, patient refusal, and unavailability of informed consent or stored biologic samples. Third, 46.7% of identified germline variants were classified as VUS, and we were not able to assess definitively their pathogenicity. Further studies should explore new predictive tools to solve this relevant issue since the current models are not sufficient to solve this point.
Conclusions
The data from this cohort study provide a detailed analysis of genotype-phenotype associations in women with HLBC phenotype who have been tested for germline CDH1 variants. We defined a group of patients (women with LBC with an early age at diagnosis and/or a positive family history for BC) with P/LP germline CDH1 variants not fulfilling the classic HDGC criteria and with an uncertain risk of developing GC. These data may assist in the genetic counseling on individual risk for families with HLBC carrying P/LP CDH1 variants, particularly in the absence of germline BRCA1 and BRCA2 variants. In addition, our data provide new evidence of CDH1 second-hit alterations as a main mechanism of HLBC tumorigenesis, suggesting future investigations for the definition of new theranostic biomarkers.
eTable 1. Clinical Criteria Adopted in the Study for Germline CDH1 Genetic Testing in LBC Women
eTable 2. In Silico Predictions for CDH1 Missense Variants
eTable 3. Germline BRCA1 Variants Identified in Patients With LBC Enrolled in the Study
eTable 4. Germline BRCA2 Variants Identified in Patients With LBC Enrolled in the Study
eTable 5. Median Age at Diagnosis Between Pathogenic vs Other Identified CDH1 Variants or Wild-Type
eTable 6. Comparison of Clinical-Pathological Variables in Relation to the Genetic Profile of Patients With LBC
eTable 7. Overall Frequency of CDH1 Structural and Epigenetic Alterations Described in Literature and in This Study’s HLBC Tumors
eTable 8. Differences Between the 2 Studies That Explored CDH1 Gene Testing in HLBC
eTable 9. Identified Germline P/LP BRCA1, BRCA2, and CDH1 Variants
eFigure 1. Flow Diagram of the Study Organization, Germline Genetic Testing Results, and Clinical Features of Families With Germline CDH1 Variant
eFigure 2. Pedigree With Germline Pathogenic CDH1 Variation Carriers
eFigure 3. Loss of E-Cadherin Expression in Invasive Lobular Breast Carcinoma From CDH1 Germline Variant Carrier
eFigure 4. Disease-Free Survival Results Between Different Variant Statuses, Including Wild-Type Population
Data Sharing Statement
References
- 1.Corso G, Figueiredo J, La Vecchia C, et al. Hereditary lobular breast cancer with an emphasis on E-cadherin genetic defect. J Med Genet. 2018;55(7):431-441. doi: 10.1136/jmedgenet-2018-105337 [DOI] [PubMed] [Google Scholar]
- 2.Caldas C, Carneiro F, Lynch HT, et al. Familial gastric cancer: overview and guidelines for management. J Med Genet. 1999;36(12):873-880. [PMC free article] [PubMed] [Google Scholar]
- 3.Benusiglio PR, Malka D, Rouleau E, et al. CDH1 germline mutations and the hereditary diffuse gastric and lobular breast cancer syndrome: a multicentre study. J Med Genet. 2013;50(7):486-489. doi: 10.1136/jmedgenet-2012-101472 [DOI] [PubMed] [Google Scholar]
- 4.Petridis C, Shinomiya I, Kohut K, et al. Germline CDH1 mutations in bilateral lobular carcinoma in situ. Br J Cancer. 2014;110(4):1053-1057. doi: 10.1038/bjc.2013.792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fitzgerald RC, Hardwick R, Huntsman D, et al. ; International Gastric Cancer Linkage Consortium . Hereditary diffuse gastric cancer: updated consensus guidelines for clinical management and directions for future research. J Med Genet. 2010;47(7):436-444. doi: 10.1136/jmg.2009.074237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van der Post RS, Vogelaar IP, Carneiro F, et al. Hereditary diffuse gastric cancer: updated clinical guidelines with an emphasis on germline CDH1 mutation carriers. J Med Genet. 2015;52(6):361-374. doi: 10.1136/jmedgenet-2015-103094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blair VR, McLeod M, Carneiro F, et al. Hereditary diffuse gastric cancer: updated clinical practice guidelines. Lancet Oncol. 2020;21(8):e386-e397. doi: 10.1016/S1470-2045(20)30219-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yadav S, Hu C, Nathanson KL, et al. Germline pathogenic variants in cancer predisposition genes among women with invasive lobular carcinoma of the breast. J Clin Oncol. 2021;39(35):3918-3926. doi: 10.1200/JCO.21.00640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Girardi A, Magnoni F, Vicini E, et al. CDH1 germline mutations in families with hereditary lobular breast cancer. Eur J Cancer Prev. 2022;31(3):274-278. doi: 10.1097/CEJ.0000000000000688 [DOI] [PubMed] [Google Scholar]
- 10.Plon SE, Eccles DM, Easton D, et al. ; IARC Unclassified Genetic Variants Working Group . Sequence variant classification and reporting: recommendations for improving the interpretation of cancer susceptibility genetic test results. Hum Mutat. 2008;29(11):1282-1291. doi: 10.1002/humu.20880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.National Center for Biotechnology Information, National Institutes of Health. ClinVar. Accessed October 24, 2023. https://www.ncbi.nlm.nih.gov/clinvar/
- 12.Leiden Open Variation Database. Home page. Accessed October 24, 2023. https://www.lovd.nl/
- 13.Luo X, Maciaszek JL, Thompson BA, et al. ; ClinGen CDH1 Variant Curation Expert Panel . Optimising clinical care through CDH1-specific germline variant curation: improvement of clinical assertions and updated curation guidelines. J Med Genet. 2023;60(6):568-575. doi: 10.1136/jmg-2022-108807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.BRCA Exchange. Home page. Accessed October 24, 2023. https://brcaexchange.org/
- 15.Richards S, Aziz N, Bale S, et al. ; ACMG Laboratory Quality Assurance Committee . Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405-424. doi: 10.1038/gim.2015.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Simões-Correia J, Figueiredo J, Lopes R, et al. E-cadherin destabilization accounts for the pathogenicity of missense mutations in hereditary diffuse gastric cancer. PLoS One. 2012;7(3):e33783. doi: 10.1371/journal.pone.0033783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4(7):1073-1081. doi: 10.1038/nprot.2009.86 [DOI] [PubMed] [Google Scholar]
- 18.Van Durme J, Delgado J, Stricher F, Serrano L, Schymkowitz J, Rousseau F. A graphical interface for the FoldX forcefield. Bioinformatics. 2011;27(12):1711-1712. doi: 10.1093/bioinformatics/btr254 [DOI] [PubMed] [Google Scholar]
- 19.Corso G, Carvalho J, Marrelli D, et al. Somatic mutations and deletions of the E-cadherin gene predict poor survival of patients with gastric cancer. J Clin Oncol. 2013;31(7):868-875. doi: 10.1200/JCO.2012.44.4612 [DOI] [PubMed] [Google Scholar]
- 20.Garcia-Pelaez J, Barbosa-Matos R, Lobo S, et al. Genotype-first approach to identify associations between CDH1 germline variants and cancer phenotypes: a multicentre study by the European Reference Network on Genetic Tumour Risk Syndromes. Lancet Oncol. 2023;24(1):91-106. doi: 10.1016/S1470-2045(22)00643-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Corso G, Corso F, Bellerba F, et al. Geographical distribution of E-cadherin germline mutations in the context of diffuse gastric cancer: a systematic review. Cancers (Basel). 2021;13(6):1269. doi: 10.3390/cancers13061269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gamble LA, Rossi A, Fasaye GA, et al. Association between hereditary lobular breast cancer due to CDH1 variants and gastric cancer risk. JAMA Surg. 2022;157(1):18-22. doi: 10.1001/jamasurg.2021.5118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stuebs F, Heidemann S, Caliebe A, Mundhenke C, Arnold N. CDH1 mutation screen in a BRCA1/2-negative familial breast-/ovarian cancer cohort. Arch Gynecol Obstet. 2018;297(1):147-152. doi: 10.1007/s00404-017-4551-1 [DOI] [PubMed] [Google Scholar]
- 24.Oka H, Shiozaki H, Kobayashi K, et al. Expression of E-cadherin cell adhesion molecules in human breast cancer tissues and its relationship to metastasis. Cancer Res. 1993;53(7):1696-1701. [PubMed] [Google Scholar]
- 25.Hunt NC, Douglas-Jones AG, Jasani B, Morgan JM, Pignatelli M. Loss of E-cadherin expression associated with lymph node metastases in small breast carcinomas. Virchows Arch. 1997;430(4):285-289. doi: 10.1007/BF01092751 [DOI] [PubMed] [Google Scholar]
- 26.De Leeuw WJ, Berx G, Vos CB, et al. Simultaneous loss of E-cadherin and catenins in invasive lobular breast cancer and lobular carcinoma in situ. J Pathol. 1997;183(4):404-411. doi: [DOI] [PubMed] [Google Scholar]
- 27.Berx G, Cleton-Jansen AM, Nollet F, et al. E-cadherin is a tumour/invasion suppressor gene mutated in human lobular breast cancers. EMBO J. 1995;14(24):6107-6115. doi: 10.1002/j.1460-2075.1995.tb00301.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huiping C, Sigurgeirsdottir JR, Jonasson JG, et al. Chromosome alterations and E-cadherin gene mutations in human lobular breast cancer. Br J Cancer. 1999;81(7):1103-1110. doi: 10.1038/sj.bjc.6690815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Droufakou S, Deshmane V, Roylance R, Hanby A, Tomlinson I, Hart IR. Multiple ways of silencing E-cadherin gene expression in lobular carcinoma of the breast. Int J Cancer. 2001;92(3):404-408. doi: 10.1002/ijc.1208 [DOI] [PubMed] [Google Scholar]
- 30.Oliveira C, Sousa S, Pinheiro H, et al. Quantification of epigenetic and genetic 2nd hits in CDH1 during hereditary diffuse gastric cancer syndrome progression. Gastroenterology. 2009;136(7):2137-2148. doi: 10.1053/j.gastro.2009.02.065 [DOI] [PubMed] [Google Scholar]
- 31.Hansford S, Kaurah P, Li-Chang H, et al. Hereditary diffuse gastric cancer syndrome: CDH1 mutations and beyond. JAMA Oncol. 2015;1(1):23-32. doi: 10.1001/jamaoncol.2014.168 [DOI] [PubMed] [Google Scholar]
- 32.Roberts ME, Ranola JMO, Marshall ML, et al. Comparison of CDH1 penetrance estimates in clinically ascertained families vs families ascertained for multiple gastric cancers. JAMA Oncol. 2019;5(9):1325-1331. doi: 10.1001/jamaoncol.2019.1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xicola RM, Li S, Rodriguez N, et al. Clinical features and cancer risk in families with pathogenic CDH1 variants irrespective of clinical criteria. J Med Genet. 2019;56(12):838-843. doi: 10.1136/jmedgenet-2019-105991 [DOI] [PubMed] [Google Scholar]
- 34.Lee CYC, Olivier A, Honing J, et al. Endoscopic surveillance with systematic random biopsy for the early diagnosis of hereditary diffuse gastric cancer: a prospective 16-year longitudinal cohort study. Lancet Oncol. 2023;24(1):107-116. doi: 10.1016/S1470-2045(22)00700-8 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eTable 1. Clinical Criteria Adopted in the Study for Germline CDH1 Genetic Testing in LBC Women
eTable 2. In Silico Predictions for CDH1 Missense Variants
eTable 3. Germline BRCA1 Variants Identified in Patients With LBC Enrolled in the Study
eTable 4. Germline BRCA2 Variants Identified in Patients With LBC Enrolled in the Study
eTable 5. Median Age at Diagnosis Between Pathogenic vs Other Identified CDH1 Variants or Wild-Type
eTable 6. Comparison of Clinical-Pathological Variables in Relation to the Genetic Profile of Patients With LBC
eTable 7. Overall Frequency of CDH1 Structural and Epigenetic Alterations Described in Literature and in This Study’s HLBC Tumors
eTable 8. Differences Between the 2 Studies That Explored CDH1 Gene Testing in HLBC
eTable 9. Identified Germline P/LP BRCA1, BRCA2, and CDH1 Variants
eFigure 1. Flow Diagram of the Study Organization, Germline Genetic Testing Results, and Clinical Features of Families With Germline CDH1 Variant
eFigure 2. Pedigree With Germline Pathogenic CDH1 Variation Carriers
eFigure 3. Loss of E-Cadherin Expression in Invasive Lobular Breast Carcinoma From CDH1 Germline Variant Carrier
eFigure 4. Disease-Free Survival Results Between Different Variant Statuses, Including Wild-Type Population
Data Sharing Statement


