Abstract
We have used mass spectrometry (MS) to characterize protein signaling in lipopolysaccharide (LPS)-stimulated macrophages from human blood, human THP1 cells, mouse bone marrow, and mouse Raw264.7 cells. Protein ADP-ribosylation was truncated down to phosphoribose, allowing for enrichment and identification of the resulting phosphoribosylated peptides alongside phosphopeptides. Size exclusion chromatography-MS (SEC-MS) was used to separate proteoforms by size; protein complexes were then identified by weighted correlation network analysis (WGCNA) based on their correlated movement into or out of SEC fractions following stimulation, presenting an analysis method for SEC-MS that does not rely on established databases. We highlight two modules of interest: one linked to the apoptosis signal-regulating kinase (ASK) signalosome and the other containing poly(ADP-ribose) polymerase 9 (PARP9). Finally, PARP inhibition was used to perturb the characterized systems, demonstrating the importance of ADP-ribosylation for the global interactome. All post-translational modification (PTM) and interactome data have been aggregated into a meta-database of 6729 proteins, with ADP-ribosylation characterized on 2905 proteins and phosphorylation characterized on 2669 proteins. This database—titled MAPCD, for Macrophage ADP-ribosylation, Phosphorylation, and Complex Dynamics—serves as an invaluable resource for studying crosstalk between the ADP-ribosylome, phosphoproteome, and interactome.
Keywords: ADP-ribosylation, post-translational modifications, phosphorylation, phosphoproteomics, macrophage, ASK signalosome, PARP9
Graphical Abstract

INTRODUCTION
Lipopolysaccharide (LPS), a component of Gram-negative cell walls, is a highly immunogenic molecule capable of activating Toll-like receptor 4 (TLR4), thereby triggering cellular signaling pathways responsible for cytokine release.1,2 As understimulation of these pathways can lead to an insufficient immune response,3 and overstimulation can lead to the deadly cytokine storm associated with septic shock,4,5 fast and reliable regulation is necessary. Protein-level regulation, in the form of protein–protein interactions and post-translational modifications (PTMs), provides the necessary speed and precision for this purpose and is arguably best studied through the use of unbiased mass spectrometry (MS)-based methods.6 In this study, we have broadly characterized protein complex dynamics and PTMs downstream of TLR4 activation and examined changes in this system after treatment with PARP inhibitors, a chemotherapeutic class of drugs that block the formation of ADP-ribosylation by poly(ADP-ribose) polymerases (PARPs).
Protein ADP-ribosylation is a PTM composed of ADP-ribose (ADPr) subunits derived from the precursor nicotinamide adenine dinucleotide (NAD+). The PTMs can be monomeric (referred to as MAR or mono[ADP-ribose]) or polymeric (PAR or poly[ADP-ribose]); PAR can be linear or branched and up to 200 subunits long in vivo.7 As a heterogeneous, potentially quite large (up to 100 kDa, due to the 0.5 kDa mass of a single subunit), and negatively charged PTM, ADPr can affect cellular signaling in multiple ways. First, the use of NAD+ can lead to a potentially lethal drop in energy levels, resulting in cell death in the form of necroptosis8 and/or parthanatos, a necrotic form of cell death dependent on PARP-1.9 Even depletion of NAD+ at levels insufficient for heralding death has the potential to modulate a wide variety of cellular signaling paradigms, including calcium signaling, histone deacetylation, mitochondrial biogenesis, and a range of other processes regulated by free NAD+ and/or the Sirtuin deacetylases and Nudix hydro-lases.10,11 Second, ADP-ribosylation may act as a “traditional” PTM, affecting cellular signaling by directly blocking other events—such as ubiquitin conjugation,12 STAT1α phosphorylation,13 p53 interaction with Crm1,14 or p65 phosphorylation/translocation15—or turning on downstream events, such as PAR-dependent ubiquitination (PARdU) by the E3 ligase Iduna.16 Third, PARylation may specifically interfere with charged interactions, such as those that mediate chromatin remodeling17 and/or RNA regulation.18 Finally, PARylation may act as a protein scaffold, responsible for recruiting proteins for the purpose of sequestration/silencing (e.g., in the stress granule 19) or to orchestrate multistep processes such as DNA repair17 and formation of the IκB kinase (IKK) complex:.20 This “scaffolding” role is made possible by the wide variety of ADPr binding domains that target distinct subunits of the PAR polymer; there are at least 12 ADPr binding domains known today.21 With so many ways to affect cellular signaling, the study of protein ADP-ribosylation is both important and challenging, and the field is still benefitting greatly from new methodologies and basic insights into fundamental protein ADP-ribosylation.
This study employed a systems biology approach to discover new aspects of ADP-ribosylation-mediated control of cellular signaling by combining in-depth study of the coordinated changes in protein ADP-ribosylation, protein phosphorylation, and protein complexes following LPS stimulation and/or PARP inhibition. Enzymatic digestion of mono- and poly(ADP-ribose) to a phosphoribose (pR) “tag” allowed for phosphoenrichment and characterization of phosphoribosylated peptides alongside phosphopeptides, a relatively new approach22 that is presented here at an unprecedented scale. We used this strategy to characterize PTM changes in response to LPS stimulation on over 1000 proteins, in four different cellular macrophage models (human and mouse primary cells, human and mouse cell lines), to generate Database #1. To generate Database #2, we combined relative quantification of changing ADP-ribosylation and phosphorylation sites on over 2000 proteins in LPS-stimulated primary human monocyte-derived macrophages (hMDMs) through application of dimethyl labeling (DML), a peptide-level isotopic labeling technique. We then utilized size exclusion chromatography-MS (SEC-MS) to measure changes in protein complex dynamics by comparing SEC elution profiles of lysates from the human THP1 macrophage-like cell line with and without cellular LPS stimulation (Database #3). Using machine learning to search the data for proteins that coordinately move between fractions, we were able to identify known and potentially novel protein complexes responding to LPS stimulation. Finally, we perturbed the carefully characterized system by pretreating human macrophage-like U937 cells with a PARP inhibitor (PARPi) before stimulating with LPS over a 30 minute time course to generate Database #4. The treated cells were lysed in a native lysis buffer and SEC-fractionated to separate high molecular weight (HMW) from low molecular weight (LMW) complexes/proteins. The HMW and LMW fractions were treated with snake venom phosphodiesterase (SVP) and immobilized metal affinity chromatography (IMAC) enriched to allow for characterization of protein ADP-ribosylation and phosphorylation in these SEC-separated samples. All four databases are presented here in a curated “meta-database” of 6729 proteins, which reveals known and novel changes in protein signaling dynamics downstream of TLR4 activation in macrophages.
Two intriguing hits from these analyses were further characterized following immunoprecipitation of key complex components: (1) the ASK signalosome, containing ASK1, ASK2, and ASK3, which appeared to dissociate and also be ADP-ribosylated in response to LPS stimulation, and (2) a PARP9-centric complex that appeared to form upon LPS stimulation. The ASK2 and PARP9 interactomes, including protein ADP-ribosylation sites and protein phosphorylation sites, are included in the meta-database for study alongside global protein changes.
EXPERIMENTAL PROCEDURES
Murine Sepsis Model
129SJ mice were purchased from Jackson Laboratories and maintained in specific pathogen-free conditions at an American Association for Laboratory Animal Care-accredited animal facility at the National Institute of Allergy and Infectious Diseases (NIAID, NIH) and were used under study protocol LISB4E approved by the NIAID Animal Care and Use Committee (National Institutes of Health, NIH). Eight month old female mice were injected intraperitoneally with vehicle (phosphate-buffered saline (PBS)) or PJ34 (10 mg/kg) 1 h before challenge with LPS from Salmonella minnesota R595 (20 mg/kg intraperitoneal), and then every 12 h following until the onset of moribundity when the mice were immediately euthanized.
Cloning
Lenti-MAP3K6-TAP was generated by amplifying the coding amplicon from MAP3K6-TAP, a gift from Daniel Liebler (Addgene plasmid # 69727), with primers F:CGGGTTTGCCGCCAGAACACAGGACCGGTCGACTCACTATAGGGAGACCCAAGCTGG, R:CAGCAGAGAGAAGTTTGTTGCGCCGGATCCGGTGCTATCCAGGCCCAGCAGC followed by Gibson assembly into pLenti Mod1.1 EF1a-BFP 2A Blast where the BFP cassette was removed by AgeI BamHI.
Virus Production and Stable Line Generation
Lentivirus was produced by transfecting a 10 cm plate of 3 × 106 293T17 cells with a 3:1:0.2 ratio of Lenti-MAP3K6-TAP:pDR8.91:VSV-G using TransIT-Lenti via the manufacturer’s protocols. After 48 h, the supernatant was harvested and centrifuged at 2000g to remove the cellular debris. The supernatant was then concentrated with a LentiX-Concentrator, and 1/4 total volume was added to 5 × 105 U937 cells in one well of a six-well plate. After 48 h, 25 μg/mL blasticidin Dulbecco’s modified Eagle’s medium (DMEM) was added to the transduced cells in a 1:1 volume (12.5 μg/mL final concentration). The cells were allowed to select for 10 days. The selected cells were assessed or pooled expression via Western blot, and a limiting dilution of 0.7 cells/well in a 384-well plate was performed to generate clonal lines. Individual lines were then screened via immunofluorescence and Western blotting to assess the level of expression.
Note: U937 cells were amenable to transfection and were therefore used here instead of THP1 cells.
Cell Culture
Primary Murine Cells.
C57Bl/6 mice were maintained in specific pathogen-free conditions, and all procedures were approved by the NIAID Animal Care and Use Committee (NIH). Bone marrow progenitors were flushed from femurs and tibias and differentiated into bone-marrow-derived macrophage (BMDM) during a 6 day culture in complete Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 60 ng/mL recombinant mouse M-CSF (R&D Systems).
Primary Human Cells.
Elutriated monocytes from de-identified, screened, healthy donors were obtained under the NIH Clinical Center IRB-approved protocol 99-CC-0168 from the NIH Department of Transfusion Medicine. The cells were cultured in an X-VIVO 15 serum-free hematopoietic cell medium (Lonza) supplemented with 10 ng/mL human M-CSF (Peprotech) for 6 days.
Mouse and Human Cell Lines.
Raw264.7 cells were obtained from American Type Culture Collection (ATCC) and cultured in DMEM supplemented with 10% fetal bovine serum (FBS) (Gemini). THP1 (ATCC) cells were cultured in Roswell Park Memorial Institute (RPMI) 1640 with 10% FBS and differentiated with 100 ng/mL phorbol 12-myristate 13-acetate (PMA) for 3 days, and U937 cells (ATCC) were cultured in RPMI 1650 with 10% FBS and differentiated with 5 ng/mL PMA for 3 days. The PMA dose for both THP1 and U937 cell lines was optimized to determine a dose that was effective (resulted in cells attaching to the plate and adopting a macrophage-like phenotype, as determined by cell shape) and minimally stressful (did not result in cell clumping); see representative images of cell differentiation states in Figure S1. Both cell lines were differentiated in all cases, unless otherwise noted.
PARPi Treatment.
The PARP inhibitors PJ34 and olaparib (both from Selleck Chemicals) were titrated to determine the effective and nontoxic dose as 1 μM. PJ34 stock was made in water and olaparib in dimethyl sulfoxide (DMSO). The cells were treated with PARPi for 48 h, unless otherwise indicated.
LPS Stimulation.
The cells were stimulated with 100 ng/mL LPS from S. minnesota, R595 (Enzo), unless otherwise indicated, for the reported times.
Nuclear-Cytoplasmic Fractionation
THP1 or Raw264.7 cells were stimulated with 100 ng/mL LPS for the indicated times and then scraped into cold PBS and lysed as specified in the NE-PER kit (Thermo).
Cytokine Measurement
Cytometric bead array (CBA) kits were obtained from BD for human tumor necrosis factor (TNF) protein, and the manufacturer’s instructions were followed to measure protein levels in cell media taken from primary human monocyte-derived macrophages treated with 10 ng/mL LPS (S. minnesota R595, Enzo) for 4 h. Data were collected on a BD Fortessa and analyzed in FlowJo.
Immunoprecipitation
U937 cells expressing recombinant ASK2-HA-V5 were differentiated into macrophages and then treated with PARPi and/or LPS, as indicated. The cells were then washed in cold PBS and lysed in an MS-compatible, modified radioimmunoprecipitation assay (RIPA) buffer (50 mM N-(2-hydroxyethyl)piperazine-N′-ethanesulfonic acid (HEPES) pH 7, 150 mM NaCl, 1% N-octylgluconate, 0.1% sodium deoxycholate, 2 μM ADP-HPD [Enzo], 40 μM PJ34, 1× Phos-STOP [Sigma-Aldrich], 1× cOmplete protease inhibitor cocktail with ethylenediaminetetraacetic acid (EDTA) [Sigma-Aldrich]) with bath sonication at 4 °C. The samples were cleared by centrifugation and then quantified and aliquoted into 500 μg samples. The samples were combined with 2 μg of antibody (rabbit αV5 [Cell Signaling Technology (CST)], αPARP9 [Abcam], or immunoglobulin G (IgG) [Cell Signaling Technology]) and incubated overnight, rotating, at 4 °C. Twenty-five microliters of A/G magnetic beads (Pierce) were added to the lysate and rotated for 1 h at 4 °C. The samples were washed in a wash buffer (50 mM HEPES pH 7, 150 mM NaCl) and eluted into either 8 M urea containing 1 mM tris(2-carboxyethyl)phosphine (TCEP) and 50 mM Tris, pH 7.3, or 6 M guanidinium HCl, pH 4, as indicated.
Sodium Dodecyl Sulfate (SDS)-Polyacrylamide Gel Electrophoresis (PAGE) and Western Blots
Samples were either eluted or lysed into 1× Laemmli buffer (Bio-Rad), incubated at 95 °C for 5 min, and then quantified using the 660 nm protein assay (Thermo). Ten micrograms of protein was loaded per lane in a NuPAGE bis–Tris, 4–12%, 1 mm thick gel (Thermo). Gels were transferred to nitrocellulose, and total protein was visualized using Ponceau S stain (Sigma-Aldrich). Membranes were blocked in 1% fish gelatin (VWR) for 1 h at room temperature and then probed with primary antibody (1:1000 MαJNK [CST 3708], 1:1000 Rαp-JnK [CST 4668S], 1:2000 MαERK1/2 [CST 4696], Rαp-ERK1/2 [CST 4370S], or αPAR cocktail [Trevigen 4335-MC-100 and 4336-BPC-100], where CST = Cell Signaling Technologies) overnight at 4 °C. Membranes were then washed in TBS containing 0.01% Tween (TBS-T) and probed with secondary antibody (Sigma NA9340 [αRabbit] or NA9310 [αMouse]) for 1 h at room temperature. Finally, membranes were washed in TBS-T, developed with SuperSignal West Pico PLUS (Thermo), and imaged using the ChemiDoc MP Imaging System (Bio-Rad).
Liquid Chromatography-Tandem Mass Spectrometry (LC-MS/MS)
LC-MS/MS was performed on one of two systems.
“Velos Pro” (DB1, DB2, DB3, DB4A, Select AP-MS).
A Nano-LC ultra-two-dimensional (2D) high-performance liquid chromatography (HPLC) system (Eksigent Technologies [acquired by AB Sciex Pte. Ltd.]) was coupled via electrospray to an LTQ Orbitrap Velos Pro (Thermo Fisher Scientific Inc.). Chromatography was performed using custom-made columns (15 cm length, 50 μm i.d.) packed with a Halo ES C18 resin (Michrom Bioresources, Inc. [acquired by Bruker Corp., Billerica, MA]) (2.7 μm diameter; flow = 200 nL/min; 0.1% v/v formic acid; 6–32% v/v acetonitrile; gradient: 90′ [DB1, DB3], 120′ [DB4A], 180′ [DB2]). Mass spectrometry used data-dependent acquisition (scan range: 300–1700 [DB1], 400–1600 [DB2], 300–1600 [DB3], 350–1400 [DB4A]) of the selected precursors (top 5 → collision-induced dissociation (CID)/high-energy collision dissociation (HCD) [DB1], top 18 → CID [DB2,3,4A]; CID scan range: 100–2000, HCD scan range: 100–1500).
“QE-HF” (DB4B, Select AP-MS).
An UltiMate 3000 HPLC system (Thermo Fisher Scientific Inc.) was coupled via electrospray to a Q Exactive HF (Thermo Fisher Scientific Inc.). Chromatography was performed using custom-made columns (as above, except a 0–30% gradient over 180′). Mass spectrometry used data-dependent acquisition (scan range: 350–1400) of the selected precursors (top 20 → HCD, scan range: 200–2000).
Note: instrumentation details per experiment are available in the first tab of each table in Supporting Information.
Data Availability.
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the data set identifier PXD016469.
ANALYSIS OF MASS SPECTROMETRY DATA
All proteomic experiments were performed, at minimum, in duplicate. MaxQuant 1.5.3.8 was used to analyze all RAW files, with default settings and the following settings, as appropriate: allowing for phosphorylation on S, T, and Y residues; allowing for phosphoribosylation on D, E, K, R, and C residues; and searching for dimethylation (light, medium, heavy) on the peptide N-terminus as well as K. For Database #2, the “main search maximum combinations” were raised to 1000 from 400, to allow for more PTM combinations to be considered (dimethyl labeling is searched as a post-translational modification). Match between runs was turned on as appropriate; “requantify’ was never turned on. FASTA databases were downloaded from UniProt for the reviewed proteomes of Mus musculus and Homo sapiens. An in-house contaminant database was used. Specific settings for the generation of each database can be found in the first tab of the tables in the Supporting Information.
A note on amino acids selected as acceptors of ADP-ribose: D, E, K, R, and C represent a subset of the 11 known amino acids, which can carry this PTM since not all amino acids are included in this search; it is likely that there are instances of mislocalization by the search algorithm of a phosphoribose group, which is on an excluded acceptor residue (e.g., serine or phosphoserine) to a neighboring amino acid that is included in the search. For this reason, site localization confidence is always reported in search results, manual validation is performed to confirm PTM localization on peptides of interest, and, of course, all RAW data is provided for readers to search with their own amino acid combinations if desired. We have performed parallel analyses of a subset of our data using six different amino acid combinations (DRC ± E, K, S); the detailed results are reported in Note S1 and Table S12.
ISOTOPIC LABELING FOR MASS SPECTROMETRY
Stable Isotope Labeling by Amino Acids in Cell Culture (SILAC)
U937 cells were grown in SILAC RPMI 1640 (Gibco) supplemented with 5% FBS, 0.4 mM lysine, and 0.8 mM arginine (light = K0, R0; medium = K4, R6; heavy = K8, R10; all from Cambridge Isotope Laboratories) for at least five doublings. Amino acid incorporation was measured as described previously23 and confirmed to be greater than 95%. The labeled cells were treated separately, lysed, and quantified. Equal amounts of the labeled protein were combined for downstream analysis.
Dimethyl Labeling
As DML is typically performed at pH 8.5 and with ammonia-based quenching of labeling reagents (see Boersema et al.24), this protocol was adjusted to be compatible with protein ADP-ribosylation, which is alkaline-sensitive.25 Specific changes include the use of formic acid to lower the triethylammonium bicarbonate (TEAB) buffer pH to 7 and quenching with neutral Tris–HCl instead of the standard ammonia treatment. Briefly: following cell lysis and protein quantification by 660 nm absorption analysis (Thermo kit, catalog # 226600), 250 μg of protein was denatured in 8 M urea, reduced in 1 mM TCEP, alkylated in 2 mM 2-chloroacetamide (CAM), and digested by trypsin and Lys-C; the peptides were then brought to 1 mL in 100 mM TEAB, pH 7, and 40 μL of 4% formaldehyde was added (light: CH2O, medium: HD2O, heavy: 13CD2O); the samples were briefly mixed and spun down, and 40 μL of 0.6 M sodium cyanoborohydride was added (light and medium: NaBH3CN, heavy: NaBD3CN) and the samples were agitated for 1 h at room temperature. The samples were quenched with 160 μL of 1 M Tris pH 7 followed by 80 μL of 5% formic acid. Light, medium, and heavy samples were mixed together and concentrated to approximately 1 M urea. Snake venom phosphodiesterase was then added, and the peptides were desalted (see the next section).
ADP-RIBOSYLATION AND PHOSPHORYLATION SITE IDENTIFICATION
SVP Purification
Snake venom phosphodiesterase I (Worthington) was purified on a 1 mL blue sepharose column as described26 without glycerol in the buffers and with 1 mM TCEP added to the elution and dialysis buffers. The eluted enzyme was dialyzed against salt-free buffer in a 10 kDa molecular-weight-cutoff (MWCO) snakeskin tubing (Thermo).
Protein Digestion
Proteins were digested overnight at neutral pH in Tris buffer by Lys-C and trypsin, as described.26
ADPr Digestion
Five micrograms (approximately 50 μL of the 1 mL elution) of SVP was added to the protein digestion mixture following overnight incubation (after proteins have been digested to peptides and, for DB#2, dimethyl-labeled) and incubated for 2 h at 37 °C. For DB#1, SVP was added to 500 μg of peptides, and for DB#2, SVP was added to 750 μg of peptides.
Phosphoenrichment
The fully digested peptides were desalted on C18 tips (Omix, from Agilent), speed-vacuumed to dryness, and phosphoenriched on PHOS-Select IMAC beads (Sigma-Aldrich), as described.26 The beads were eluted twice, consecutively, and then desalted again before analysis by LC-MS/MS.
SIZE EXCLUSION CHROMATOGRAPHY
Sample Preparation
Differentiated U937 cells expressing recombinant ASK2-HA-V5 were scraped into a native lysis buffer (50 mM Hepes pH 7, 150 mM NaCl, 1 mM dithiothreitol (DTT), 1× cOmplete protease inhibitor cocktail with EDTA [Sigma-Aldrich], 1× Phos-STOP phosphatase inhibitor [Sigma-Aldrich], 1 μM ADP-HPD [Enzo], 80 μM PJ34 [Selleck]) and subjected to three rounds of rapid freezing and thawing. The lysates were cleared by centrifuging at 16 000 rpm for 10 min at 4 °C and then running the supernatant through a 0.22 μm filter. Protein concentration was quantified, and lysates were adjusted to a final concentration of 5 mg/mL.
Chromatography
A Superose 6 increase 10/300 GL prepacked glass column (GE) was equilibrated on an AKTA purifier system (GE) in SEC running buffer (50 mM HEPES pH 7, 150 mM NaCl, 1 mM DTT), and protein standards (high molecular weight calibration kit from GE) or up to 500 μL/10 mg of protein was loaded onto the column. Forty-eight milliliters of buffer was run through the column, and 0.5 mL fractions were collected.
Visualization of Elution Profiles
Unicorn Software (GE) was used to control the instrumentation and export result files, which were then processed with PyCORN (https://github.com/pyahmed/PyCORN).
Analysis
The fractions were lyophilized and resuspended in 8 M urea, and a 10 kDa MWCO spin column was used to exchange the buffer for a final sample mixture of 0.25 M urea, 150 mM NaCl, 50 mM HEPES pH 7. The protein concentration was then measured, and the samples were analyzed by SDS-PAGE/Western blot and/or LC-MS/MS.
WEIGHTED CORRELATION NETWORK ANALYSIS (WGCNA)
Data Preparation
Raw intensity files (as reported in Table S3) were smoothed using the exponential smoothing function from excel with an α value of 0.5 and normalized as follows: the highest values in each data set were set to 1, and all other values were divided by this max value resulting in data ranging from 0 to 1. Values of less than 0.1 were adjusted to 0.05 to minimize noise. The matched fractions from untreated and LPS-treated samples were then used to generate fold changes due to treatment; the resulting values are in the “ratios” tab of Table S4. Values greater than 1 represent an increase of protein in that fraction under LPS, and values less than 1 signify loss of that protein from that fraction after LPS. The data was then analyzed in two ways: the first specifically looking for proteins coming together into fractions (ignoring loss from other fractions) and the second looking specifically for proteins coordinately leaving the fractions (regardless of which they end up going into). To create the “up in LPS” data set, values <1 were converted to 1. To create the “down in LPS” data set, values >1 were converted to 1. To minimize the effects of the monomers on the final clustering, fractions 17–25 were removed from further analysis.
Analysis
WGCNA is an R package and is publicly available.27 Briefly, the network was created by calculating the component-wise minimum values for topologic overlap (TO). Using 1 – TO (dissTOM) as the distance measure, genes were hierarchically clustered. Initial module assignments were determined by using a dynamic tree-cutting algorithm. The resulting modules or clusters of co-eluting genes were used to calculate module eigengenes (MEs or the first principal component of the module). The core WGCNA function is netblockwise. The parameters for the complex formation are as follows: power = 12, mergeCutHeight = 0.2, corType = ″bicor″, networkType = ″signed″, pamStage = TRUE, pamRespectsDendro = TRUE, deepSplit = 2, TOMDenom = ″mean″, verbose = 3, saveTOMs = FALSE, minModuleSize = 20, minKMEtoStay = 0.3, maxBlockSize = 10 000, and reassignThreshold = 0.05. The parameter for the complex disassociation was as follows: power = 9, mergeCutHeight = 0.2, corType = ″bicor″, networkType = ″signed″, pamStage = TRUE, pamRespectsDendro = TRUE, deepSplit = 2, TOMDenom = ″mean″, verbose = 3, saveTOMs = FALSE, minModuleSize = 10, minKMEtoStay = 0.3, maxBlockSize = 10 000, and reassignThreshold = 0.05.
GO-ELITE GENE ONTOLOGY ANALYSIS
The functional enrichment of proteins within the WGCNA modules was determined using the GO-Elite (v1.2.5) python package.28 GO-Elite Hs (human) databases were downloaded on or after June 2016. The set of all proteins identified and considered in the network was used as the background. The Z score determines the over-representation of ontologies in a module, and a one-tailed Fisher’s exact test (Benjamini–Hochberg false discovery rate (FDR) corrected) was used to assess the significance of the Z score. A Z score cutoff of 1.96, a P value cutoff of 0.01, and a minimum of five genes per ontology were used as filters prior to pruning the ontologies.
PRINCIPAL COMPONENT ANALYSIS (PCA): DATABASE #4
Data Preparation
Due to the distinct protein composition of the HMW and LMW fractions, they were analyzed separately. Protein intensities were log 2-transformed. The median of the nonzero numerical values was calculated for each fraction. A correction value was calculated by dividing the median of each fraction by the average of medians. To normalize, the log 2 intensity of each protein in each fraction was multiplied by the correction value. Protein rows with non-numeric or zero values were removed. The resulting data was then used for principal component analysis.
Analysis
The principal component analysis was performed in the software package Perseus v1.6.7.0 using the default parameters for the PCA function.
RESULTS
Protein ADP-Ribosylation and Phosphorylation in LPS-Stimulated Macrophages
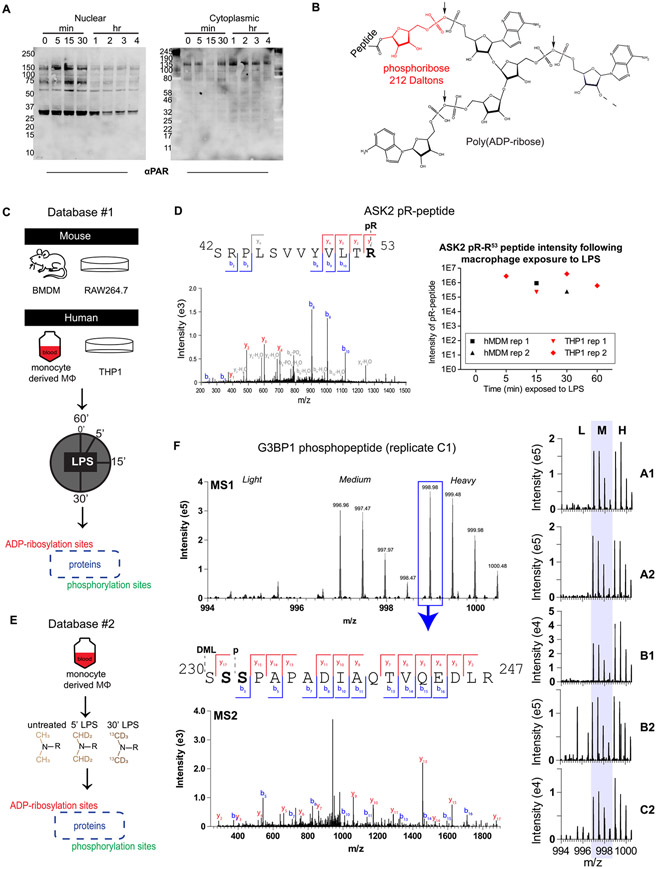
Macrophages contain protein ADP-ribosylation that changes upon LPS stimulation, as evidenced by Western blots detecting PARylation in the nuclear and cytoplasmic fractions of macrophages stimulated over a time course with LPS (Figures 1A and S1). To explore this phenomenon at the molecular level, cells from two macrophage-like cell lines (murine Raw264.7 and human THP1) and two primary sources (murine bone marrow and human blood) were treated with LPS over a time course, and then proteins were digested to peptides and treated with snake venom phosphodiesterase (SVP), an enzyme that cleaves ADP-ribose down to a phosphoribose “tag”, allowing for phosphoenrichment and subsequent MS-aided identification (Figure 1B,C). The resulting “Database #1” contained 959 ADP-ribosylated peptides from 882 proteins and 1008 phosphopeptides from 899 proteins. The overlap between mouse and human proteomes was quite low, with only 3% of proteins with ADP-ribosylation site identifications overlapping and 14% overlapping in the phosphoproteome (Figure S3A). The overlap was higher between primary cells and cell lines within the same species (16–19% for pR, 28–33% for phospho) and between biological replicates within the same cell type (7–23% for pR, 12–37% for phospho). As this ADPr site identification method relies on complete enzymatic degradation of polymers, it is expected that the efficiency of site coverage is lower for phosphoribosylation sites (which require full digestion to the pR tag) than for phosphorylation sites (which are identified intact). Finally, the proteins identified in this data set were analyzed to determine the enrichment of biological processes, which are reported as a tree map in Figure S3B, and selected ADP-ribosylation sites of interest from the top biological properties are shown. As expected, a large number of proteins involved in DNA repair were confidently identified as ADP-ribosylated, with novel sites such as D90 on XRCC3, E31 on BARD1, and K416 on AIFM3 being reported for the first time (based on a comparison with ADPriboDB29). In addition, proteins known to be important in regulating immune signaling were also identified with novel ADP-ribosylation sites, for example, K203 on STAT1, K136 on TRAF4, D980/R982 on NLRC3, R423 on CARD11 (CARMA1), and R616/E617 on PRKCQ (PKCΘ). Additionally, we show that ASK2 and ASK3 (gene symbols MAP3K6 and MAP3K15, respectively) are site-specifically ADP-ribosylated (ASK2 is shown in Figure 1D). Both proteins are core components of the ASK signalosome,30 which is emerging as an important promoter of late-phase transcriptional responses following LPS stimulation.31 In the case of R53 on ASK2, ADP-ribosylation is only observed following LPS stimulation and is found in all four human sample sets (Figure 1D, right). All phosphorylation and phosphoribosylation sites identified and quantified in Database #1 can be found in Tables S1 and S2, and 289 selected peptide-spectrum matches (PSMs) are shown with annotation in the Supporting Spectra document.
Figure 1.
Assessment of the ADP-ribosylated and phosphorylated proteomes in LPS-stimulated macrophages and generation of Database #1 and Database #2. Nuclear-cytoplasmic fractionation of LPS-stimulated Raw264.7 cells (A) reveals changes in global PARylation patterns during LPS stimulation (100 ng/mL; LPS from S. minnesota). To detect site-specific changes in ADP-ribosylation, snake venom phosphodiesterase was used to cleave the monomer or polymer down to its phosphoribose (pR) attachment site (B) following 100 ng/mL LPS stimulation of human or mouse macrophages for various amounts of time (C). Site identification was achieved through localization of the 212 Da pR group, and PTM dynamics was measured from extracted ion chromatograms (XICs) of modified peptides (example from the ASK2 protein shown in (D), z = 2, m/z = 801.42, source = hMDM rep 1). Primary human monocyte-derived macrophages (hMDMs) were further characterized using dimethyl labeling (DML) to achieve robust relative quantification (E). DML-enabled quantification of an example peptide from replicate C1 of Database #2 is shown in (F); the base peak from the heavy labeled form was selected for MS/MS, and fragmentation reveals that this isolated peak is either a combination of S231 and S232, or a single form that cannot be distinguished. The summed MS1 spectra from other replicates are shown on the right, indicating that this peptide is identified exclusively or primarily in the medium and heavy (LPS-treated) forms. A/B/C indicates a biological replicate (unique human donor), and 1/2 indicates technical replicate (unique derivation of macrophages from blood monocytes). BMDM, bone-marrow-derived macrophages from mice; hMDM, human monocyte-derived macrophages from healthy human donor blood.
In an effort to more fully characterize the human ADP-ribosylated and phosphorylated proteomes in primary human macrophages, a second database was generated using human monocyte-derived macrophages (hMDMs) from three healthy donors; these hMDMs were generated twice from each set of donor cells, resulting in six total replicates that were stimulated with LPS for either 0, 5, or 30 min. The differentially stimulated samples were processed as in Database #1, with the addition of dimethyl labeling (DML) at the peptide level to allow for robust, relative quantification following combined SVP digestion and phosphoenrichment (Figure 1E). Exemplary data is shown in Figure 1F, where a peptide from G3BP1—a protein important in the innate immune response32 and stress granule formation33—is shown to be phosphorylated at either S231 or S232 (or, potentially, both proteoforms are present) following LPS stimulation in all six replicates. Additional examples are shown in Figure S4, demonstrating that phospho-S252 on LSP1 is observed consistently during LPS stimulation and phosphoribose-C194 on ZAR1L disappears upon LPS stimulation. Database #2 yielded 1564 phosphoribosylated peptides from 1407 proteins and 1410 phosphopeptides from 1211 proteins, with an 8–12% overlap between technical replicates for phosphoribosylated proteins and 12–22% for phosphoproteins (Figure S5). Taken together, Databases #1 and #2 provide a broad yet detailed view of the macrophage ADP-ribosylated proteome.
Protein Complex Dynamics in LPS-Stimulated Macrophages
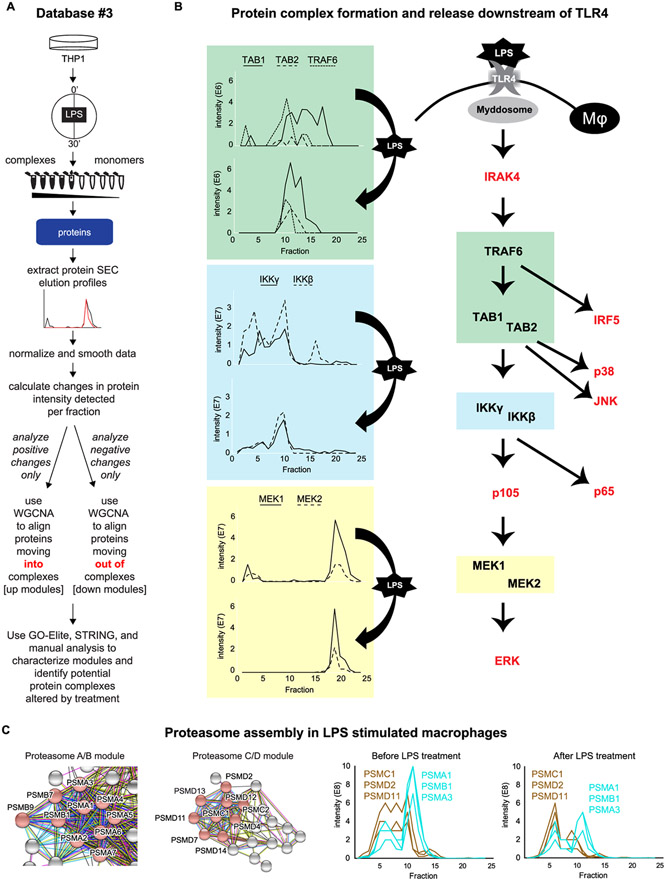
Size exclusion chromatography (SEC) separates proteins and protein complexes based on their size, as shown by protein standards in Figure S6A. To retain protein complexes from untreated and LPS-treated (100 ng/mL, 30 min) THP1 cells, a native lysis was performed and protein samples were SEC-fractionated for downstream analysis by UV–vis, Western blot, and LC-MS/MS (Figure 2A, top). UV–vis and total protein blots reveal fractionation of protein complexes and monomers by size (Figure S6B-E), and LC-MS/MS analysis of fractions confirms that proteins tend to be identified consistently in the same or tightly neighboring fractions, and most proteins do not shift into new fractions following LPS stimulation (Figure S6F; all protein IDs and quantification can be found in Table S3). This consistent background allows for the identification of LPS-responsive protein shifts from the 2723 proteins analyzed in this data set; noteworthy examples of protein SEC elution profiles shifting in response to stimulus are highlighted in Figure 2B. TRAF6, TAB1, and TAB2 are shown together (green rectangles) as they are known to interact in response to stimulus34 and appear to come together into fraction #11 after LPS stimulation. IKKγ (gene symbol IKBKG, also known as NEMO) and IKKβ (gene symbol IKBKB) are shown coming together in fraction #10 in the blue rectangles, representing the canonical IKK complex, which is known to phosphorylate inhibitors of NF-κB signaling, a signal that initiates their degradation by the proteome.35 In these examples, protein elution profiles simplify following a stimulus, suggesting that multiple proteoforms exist for each protein at a steady state, but a dominant proteoform emerges following cellular activation. A simpler example is shown in the yellow rectangles, where the MEK1–MEK2 (gene symbols MAP2K1 and MAP2K2, respectively) dimer appears to dissociate following LPS treatment since MEK1–MEK2 dimerization is critical for the negative regulation of ERK1/2 signaling;36 this may contribute to ERK activation following LPS stimulus. SEC elution profiles for proteins shown in red in Figure 2B can be found in Figure S7; it is notable that these elution profiles tend to be fairly simple, suggesting that few proteoforms exist for these proteins.
Figure 2.
Database #3: fractionation of whole-cell lysate by size exclusion chromatography for the study of protein complex dynamics. Whole-cell lysate from LPS- or vehicle-treated THP1 cells was fractioned by size exclusion chromatography, and proteins were identified by mass spectrometry ((A) top) to generate Database #3. The elution profiles of proteins were grouped into modules based on their movement into or out of a fraction/complex ((A) bottom). The associated dendrograms are displayed in Figure S7. Exemplary data shows SEC elution profiles from proteins downstream of TLR4/myddosome activation that are identified in DB#3; proteins in red are shown in Figure S7. (B) Related protein modules are shown in (C), where the brown “down” module consists primarily of proteins known to form the C and D subunits of the proteasome, and the turquoise “up” module contains proteins known to form the A and B subunits of the proteasome. Full interactomes for these modules can be found in Figure S9. WGCNA, weighted correlation network analysis.
To globally identify proteins that coordinately move between SEC fractions in response to LPS, protein elution profiles were analyzed by weighted correlation network analysis (WGCNA), an unsupervised clustering algorithm that groups proteins based on correlations in relative abundance across samples, in this case, SEC fractions. Proteins that have similar changes in their elution profiles following stimulation are clustered together into groups referred to as modules, with each module carrying a color designation. We hypothesized that these modules represent protein complexes. As we were searching for proteins that coordinately move both into a fraction (complex) and out of a fraction (complex), we analyzed the elution profiles in two ways: in the first, we considered only positive fold changes—or an accumulation of proteins in fractions following LPS stimulation; we referred to these protein modules as up modules. In the second analysis, we considered only proteins that coordinately left fractions (complexes), referring to these modules as down modules. To assess whether known or novel complexes may be associated with each module, the modules were then analyzed using GO-Elite28 and STRING37 (Figure 2A, bottom). Modules are depicted by colors in Figure S8, along with “nicknames” for each module based on functional enrichments derived from GO-Elite and STRING analyses. Proteins included in each module, along with the kME values depicting association or near-association with each module, can be found in Table S4.
The proteasome A/B and proteasome C/D modules are shown in Figure 2C, and full interactome maps for these modules are shown in Figure S9. The LC-MS/MS elution profiles of three representative proteins from each module are shown at the bottom of Figure 2C, revealing that prior to LPS stimulation, the A/B components are eluting together and the C/D components are eluting together, but the groups have distinct elution profiles between them. Following LPS stimulation, the proteins come together in fraction 6, indicating that the proteasome is fully formed following 30 min of LPS treatment, consistent with the known critical role for proteasome engagement to activate NF-κB and MAPK pathways downstream of TLR4.38 As the proteasome A/B module contains both the Lmp9 (PSMB7) and Lmp2 (PSMB9) proteins, this complex appears to be an intermediary between the constitutive- and immuno-proteasome.39 The PA28 proteasome regulator module contains PSME1 and PSME2, which appear to complex in fraction 11, suggesting that multiple forms of the proteasome are dynamically regulated in response to LPS stimulation.
PARP Inhibition Alters Protein Signaling in Macrophages
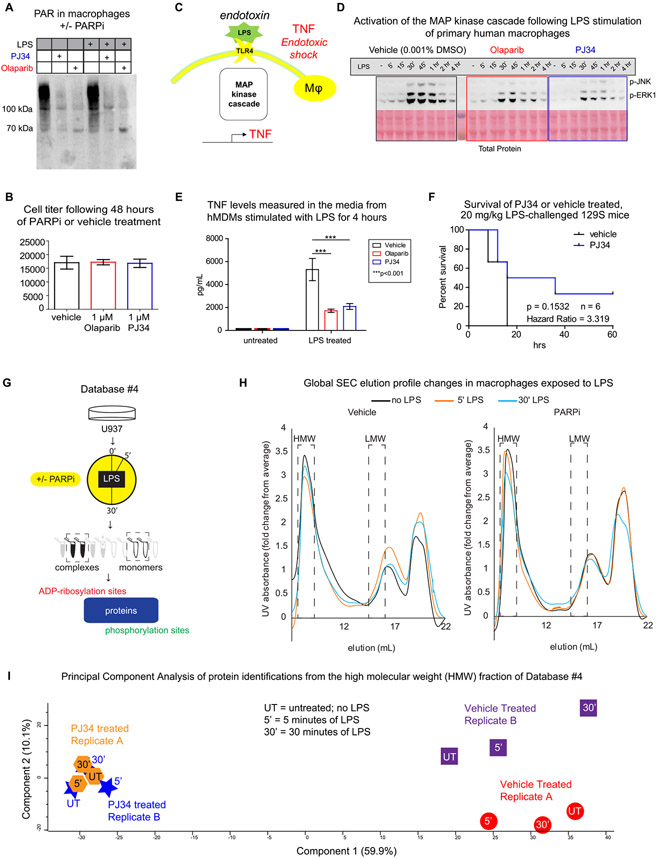
PARP inhibition by PJ34 and/or olaparib is an effective and nontoxic way to block the formation of protein ADP-ribosylation in macrophages (Figures 3A,B and S10). Here we have characterized the impact of PARP inhibition on events following LPS activation of TLR4, a process that is known to lead to, among other pathways, the activation of MAP kinase cascades and subsequent TNF production, which is part of the cytokine storm associated with septic/endotoxic shock (Figure 3C). Figure 3D-F reveals that PARP inhibition decreases this response at all three discussed stages: in Figure 3D, it is evident that there are less JNK and ERK phosphorylation in response to LPS in cells treated by PARP inhibitors. In Figure 3E, TNF levels from the media of LPS-stimulated hMDMs are lower in PARP inhibitor-treated cells. Finally, in Figure 3F, the PARP inhibitor PJ34 appears to confer resistance to endotoxic shock in mice, as modeled by the intraperitoneal injection of LPS, a finding that reproduces the original experiment.40 These data provided the rationale to delve into detailed experiments with PARP inhibitors.
Figure 3.
PARP inhibition and generation of Database #4. Protein ADP-ribosylation was inhibited in macrophages using a 1 μM, 48 h treatment, which was both effective (A), as shown by a Western blot revealing levels of protein ADP-ribosylation, and nontoxic (B), as measured by the commercial CellTiter-Glow kit; full titration data is shown in Figure S10. The physiological impact of PARP inhibition on the LPS activation of TLR4 is measured here at the signaling, secretion, and whole animal level (C). By Western blot, both olaparib and PJ34 decreased the amount of phosphorylation detected on JNK and ERK1/2 during LPS stimulation of primary human macrophages (D) and decreased the cytokine release from these cells (E). The PARP inhibitor PJ34 effect on mice, as modeled by the intraperitoneal injection of LPS (F). PJ34 response was measured in unstimulated and LPS-treated U937 cells, which were subjected to native lysis and SEC separation of high molecular weight (HMW) from low molecular weight (LMW) protein complexes. These fractions were analyzed by LC-MS/MS to determine the identity of proteins and their phosphorylation and ADP-ribosylation sites (G). The elution profiles, as measured by UV absorption, are shown in (H), with a separate replicate shown in Figure S12. Principal component analysis (PCA) of the proteins identified by MS in the HMW fractions is shown in (I), and the PCA analysis for the LMW fractions is shown in Figure S13.
To determine whether PARP inhibition is altering the PTMs and/or interactomes of cellular processes downstream of TLR4 signaling, the human macrophage cell line U937 was treated with vehicle or PJ34 prior to LPS stimulation for 0, 5, or 30 min. These stimulated cells were then lysed and fractionated using our established native lysis and SEC protocols (as described for Database #3), and two fractions—either high molecular weight (HMW, approximately 1.4–3.1 MD) or low molecular weight (LMW, approximately 140–300 kDa)—were analyzed to determine their ADP-ribosylation, phosphorylation, and protein identification profiles (Figure 3G; the exemplary peptide data is shown in Figure S11; the standards are shown in Figure S12). The results of this analysis make up Database #4 and can be found in Tables S5-S7. The UV–vis elution profiles of the cellular protein complexes are shown in Figure 3H (replicate A; replicate B is shown in Figure S12); these profiles appear to shift in response to LPS (black vs orange vs blue lines), and this shift is altered between the vehicle-treated (left) and PARP inhibitor-treated (right) cells, suggesting that PARP inhibition affects the composition of protein complexes in response to LPS. This hypothesis is supported by analysis of the proteins identified in the HMW and LMW fractions (boxed out in Figure 3H): a principal component analysis of protein identifications from the HMW fractions revealed that the proteomic profile of the HMW fraction is highly affected by PARP inhibition and to a lesser extent affected by LPS stimulation (Figure 3I). Furthermore, the proteomic profile of the HMW fractions from PARPi-treated cells did not change as much in response to LPS stimulation as those from the vehicle-treated cells (component 2 of Figure 3I), suggesting that the overall interactome is less affected by LPS when ADP-ribosylation is blocked. While the separation between PARPi- and vehicle-treated cells on the first component is visible in the LMW fraction (Figure S13), the impact on response to LPS by PARPi is less clear in these fractions containing mostly monomers and small complexes. In summary, Database #4 demonstrates the impact of PARP inhibition on the macrophage interactome and provides a detailed view of macromolecular structures (from the HMW fraction) changing in response to LPS and PARPi.
ASK Complex
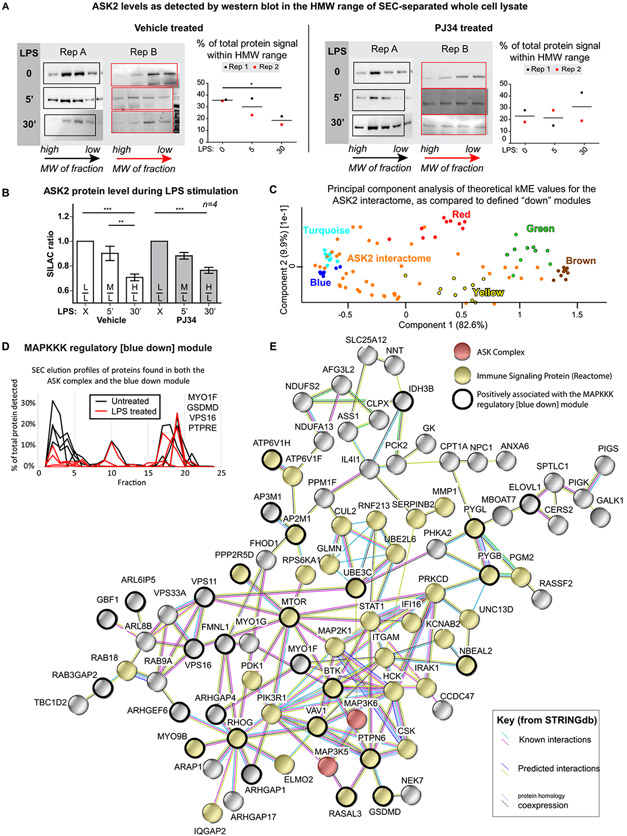
Our initial observation that all three ASK proteins (ASK1, ASK2, ASK3) that make up the core of the ASK signalosome are ADP-ribosylated in Database #1 or #2 (see Figure 1D, Table S2, and Supporting Spectra) led us to investigate the dynamics of this complex more directly. Since we did not detect native ASK proteins in the SEC-separated fractions of Database #3, we generated a macrophage-like cell line stably expressing ASK2-HA-V5 by subcloning this TAP-tagged protein fusion from Federspiel et al.30 into U937 cells, which was used for the generation of Database #4 and all following experiments. Subsequent Western blot analysis of SEC fractions from this cell line detected the recombinant protein, allowing for quantification of ASK2 in the HMW and LMW fractions (Figure S14). In the HMW fraction, ASK2 appears to transiently shift into a higher molecular weight range after 5 min of LPS stimulation, before leaving the HMW range after 30 min of LPS stimulation (Figure 4A, vehicle-treated samples), a response that appears to be blocked by PARP inhibition (Figure 4A, PJ34-treated samples). To better understand this response, ASK2 was immunoprecipitated using the V5 tag and visualized by Western blot using the HA tag (Figure S16); subsequent stable isotope labeling of amino acids in cell culture (SILAC41)-based MS quantification of ASK2 from four independently performed pulldowns is shown in Figure 4B. The global ASK2 protein level decreases in response to LPS but is unaffected by PARP inhibition. Taken together, this analysis of global and complexed ASK2 shows that ASK2 is both (1) degraded and (2) diminished in an HMW protein complex following 30 min of LPS stimulation. As PARP inhibition has no effect on the LPS-induced ASK2 degradation, but blocks ASK2 departure from a protein complex, it can be hypothesized that (1) ADP-ribosylation plays a role in ASK2 complex dynamics, specifically in breaking ASK2 away from a protein complex, and (2) it is likely the noncomplexed form of ASK2 is degraded, as this degradation is independent of PARP inhibition.
Figure 4.
ASK2 protein is degraded in response to LPS stimulation of macrophages and is shown to interact with proteins from the blue down module of Database #3. Western blots visualizing the presence of recombinant ASK2-HA-V5 protein in the fractions that were combined to make the HMW samples (0.5 mL fractions from the 8 to 10 mL elution range) analyzed in Database #4 are shown in (A), along with the changes in relative quantification as compared to the total intensity from all fractions; unpaired t-test results are shown. Full elution profiles and quantification of the LMW range are shown in Figure S14, and full blots are shown in Figure S15. SILAC quantification of ASK2 protein levels following PARPi treatment, varying amounts of LPS stimulation, lysis, and immunoprecipitation are shown in (B), revealing that the global ASK2 protein level decreases in response to LPS, and this response is unaffected by PARP inhibition. These SILAC ratios are based on data from four independent replicates that were analyzed with a one-way ANOVA with Bonferroni post-testing. A PCA-based comparison of the ASK2 interactome with the top 10 proteins from each of the known down modules from Database #3 reveals similarity between the ASK2 interactome and the MAPKKK regulatory [blue down] module, suggesting that the ASK interactome may be represented by this module (C). The SEC elution profiles of four proteins that are included in both the MAPKKK regulatory module and the ASK interactome are shown in (D), both before (black) and after (red) a 30 min LPS stimulation. A STRINGdb37 diagram of the overall ASK2 interactome is shown in (E); inclusion in the interactome was based on the proteins being identified by at least two peptides in at least three of eight biological replicates, in the bait (αV5) but not the isotype control pulldown, and appearing in the CRAPome42 25 times or less. *p = <0.05, **p = <0.01, and ***p = <0.001.
The ASK2 interactome was determined by consensus from eight independently performed biological replicates; the reported proteins had (1) at least two peptides for each identification, (2) were identified in at least three of the eight biological replicates, (3) were never seen in the isotype control, and (4) were present in the CRAPome42 less than 25 times. This final interactome list of 129 proteins (Table S8) was compared with the identified modules from Database #3, revealing an overlap with the MAPKKK regulatory [blue down] module (Figure 4C), which contained four of the ASK2 interactome members (components of the blue down module are depicted as a STRING map in Figure S17 and listed in Table S4). Furthermore, when the ASK2 interactome proteins that appeared in Database #3 were examined, they had an average kME value of 0.47 for the blue module, suggesting that many of the interactome proteins have a similar LPS-induced change in their SEC elution profile to that of the blue module. This change is depicted in Figure 4D, using the four ASK2 interactome members to represent the module. As with the Western blot data showing ASK2 leaving the HMW fraction upon LPS stimulation, the module proteins appear to be coordinately leaving the HMW range in response to LPS, suggesting that this module/complex falls apart upon stimulation. Finally, the ASK interactome is shown in Figure 4E, with those proteins that have positive kME values for the blue module circled in black. Taken together, this data suggests that the ASK signalosome dissociates upon LPS stimulation, and complex dynamics is regulated by protein ADP-ribosylation.
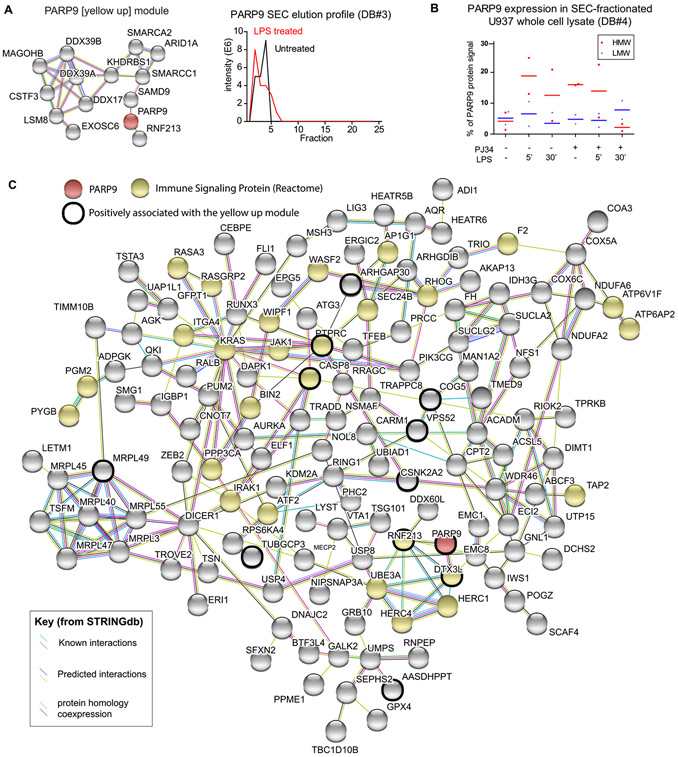
PARP9 as a Hub Protein of an LPS-Responsive Protein Module
The “yellow-up” module from Database #3 appears to form in response to LPS signaling and has PARP9 as a hub protein—i.e., a protein with a very high kME value for the module, indicating that it links tightly to all other proteins in the PARP9 module (Table S4). The PARP9 module is shown as a STRING map in Figure 5A, with the elution profile of PARP9 representing the module’s shift into a higher molecular weight fraction upon LPS stimulation. This pattern was also seen in Database #4, where PARP9 was more abundant in the HMW fraction following LPS stimulation, a response that was not seen in the PJ34-treated samples (Figure 5B). The PARP9 interactome was directly assessed by immunoprecipitation of PARP9 from U937 cells expressing ASK2-HA-V5. Western blots show efficient pulldown and potential interaction between PARP9 and ASK2, as ASK2 was detected in the PARP9 IP, though the reverse could not be confirmed (see Figure S18). The PARP9 interactome is shown in Figure 5C, with proteins that have kME values positively associated with the yellow module circled in black. The core of this interactome contains, in addition to PARP9, five E3 ubiquitin ligases: DTX3L (PARP9’s heterodimeric binding partner43), RNF213, UBE3A, HERC1, and HERC4, reflecting the importance of PARP9 in protein ubiquitination. The presence of IRAK1, a core component of the myddosome that gets ubiquitylated in response to LPS stimulation,44 suggests a link to the TLR4 response. Taken together, this evidence suggests that a PARP9-centric protein complex is forming in response to LPS stimulation, and this complex may be disturbed by PARP inhibition.
Figure 5.
PARP9 interactome overlaps with the yellow-up modules from Database #3. The yellow-up module (A) contains PARP9 (red) as a hub protein, indicating that it is a central component in this complex, which is forming in response to LPS stimulation. The PARP9 elution profile before and after LPS stimulation is shown. Quantification of the PARP9 levels in the HMW or LMW fractions of Database #4 is shown in (B), revealing that PARP9 movement into the HMW fraction in response to LPS is affected by PJ34. The PARP9 interactome is shown as a STRING diagram, with nodes colored to reflect the bait protein (PARP9, red), proteins known to be involved in immune signaling, as annotated in the Reactome59 database (green), and those that have a positive correlation with the yellow-up modules from Database #3 (black outline). Inclusion in the interactome was based on proteins being identified by at least two peptides in both replicates, at an intensity at least 3× that of the IgG control, and appearing in the CRAPome42 25 times or less.
DISCUSSION
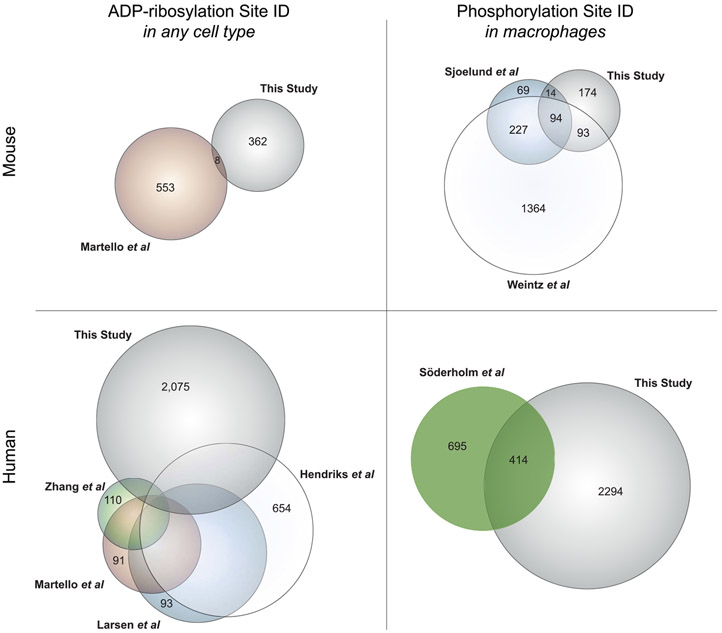
This study has produced a meta-database of six unique data sets—Databases #1–4, as described, and the ASK2 and PARP9 interactomes, including protein ADP-ribosylation and phosphorylation sites identified from these immunoprecipitation experiments. This meta-database includes ADP-ribosylation sites on 2905 proteins, expanding the number of proteins with the characterized endogenous ADP-ribosylation sites (current large-scale studies45-48 are compared in Figure 6, left side), and represents the first data set of ADP-ribosylation sites from primary human cells. A detailed overlap of human ADP-ribosylation studies is shown in Figure S19, as visualized on an UpSet plot49 and a Venn diagram, and in Table S11. This meta-database also includes phosphorylation sites on 2669 proteins, of which 2294 are from human cells, providing the first look at LPS-responsive phosphorylation in human macrophages (current studies50-52 are compared in Figure 6, right side). All databases have been brought together in Table S10 and can be searched from a single search line at mapcd.niaid.nih.gov (see Note S2); Macrophage ADP-ribosylation, Phosphorylation, and Complex Dynamics (MAPCD) allows users to quickly see whether their protein of interest has been identified in any databases presented in this paper. As all experiments were designed, executed, and analyzed by the same scientist, using the same stock of LPS, in the same laboratory, these databases are well suited for combined interpretation. Our hope is that MAPCD users will be enabled to further study crosstalk between protein ADP-ribosylation, protein phosphorylation, and protein complex dynamics by way of this unique and powerful resource.
Figure 6.
Overlap between protein ADP-ribosylation and phosphorylation databases presented in this study and other available databases. Protein lists, as designated by their universal gene symbols, were generated from PTM databases presented in this study or other studies, as noted: Martello et al.,45 wherein ADP-ribosylation sites were identified from mouse liver; Zhang et al.,46 in which human HCT116 cells were analyzed for protein ADP-ribosylation following PARG knockdown; Martello et al.,45 Larsen et al.,47 and Hendriks et al.,48 in which human HeLa cells were analyzed for ADP-ribosylation sites; Sjoelund et al.,51 wherein phosphorylation sites were quantified in murine immortalized macrophages; Weintz et al.,50 in which phosphorylation was measured in murine bone-marrow-derived macrophages; and Söderholm et al.,52 wherein phosphorylation was analyzed in human blood monocyte-derived macrophages infected with influenza A. Area-proportional Euler diagrams were generated using VennMaster.60 Full overlap data for the human ADP-ribosylome is visualized in Figure S19.
In analyzing Database #3, we have used an unbiased, machine-learning-based method to identify known and novel protein complexes from SEC-MS data. WGCNA successfully grouped together proteins with similar changes in SEC elution profiles following LPS stimulation, without reliance on established databases. Upon characterization of these protein modules by gene ontology and STRING interaction networks, it was evident that the proteins grouping together were very likely to be working together in a complex, validating the approach. With the exception of a single study,53 which determined protein complex assignments from SEC-MS data by comparing protein elution profiles from evolutionarily distant species, SEC-MS analysis has historically relied on established databases to interpret protein–protein interactions and thus identify protein complexes.54 As our approach tracks shifts in elution profiles following stimulation, focusing either on shifts into a complex or on shifts out of a complex (reducing confusion from uncoordinated protein movements in either one direction or the other), a database is not required for high-confidence assignments, allowing for unbiased and unrestricted analysis. We believe this approach has great potential to discover stimulus-responsive complexes from any and all cell types and assist in both isolating and characterizing known and novel protein complexes.
The ASK signalosome has already been linked to TLR4 signaling, most notably in 2005 by Matsuzawa et al., who showed that ASK1-deficient mice are protected from LPS-induced endotoxic shock due to a lack of intracellular reactive oxygen species formation and subsequent cytokine release.31 In 2007, it was shown that ASK2 forms a functional heteromeric complex with ASK1,55 and in 2016, the highly precise stoichiometry of the ASK signalosome proteins revealed ASK1/ASK2/ASK3 interactions of 1:1:0.1.30 Here we have shown that this complex appears to dissociate in response to LPS signaling, and this may be affected by ADP-ribosylation based on deregulation in the presence of PARP inhibition. Further studies are needed to determine the precise role of protein ADP-ribosylation in the ASK signalosome.
PARP9 and its heterodimeric partner DTX3L are known regulators of interferon signaling56 and have been reported to block ubiquitination.12 PARP9, working with PARP14, has also been shown to regulate STAT1 activation.13 However, there is generally very little known about this particular PARP, which historically has been reported to lack ADP-ribosylation activity.57,58 In this study, we have identified PARP9 as a central (hub) protein in a complex that forms following LPS stimulation, suggesting a new role for this protein in the macrophage TLR4 response to LPS.
The databases presented here form a broad picture of the changing proteomic landscape in macrophages and provide an unprecedented view of the ADP-ribosylated proteome and its possible regulatory effects on the innate immune response.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Dr. Rachel Gottschalk for her advice and insightful comments during the early days of this project. This research was supported by the Intramural Research Program of NIAID, NIH, and BCBB Support Services Contract HHSN316201300006W/HHSN27200002.
Footnotes
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.jproteome.0c00261.
Phosphorylation sites from Database #1 and Database #2 (Table S1); phosphoribosylation sites from Database #1 and Database #2 (Table S2); protein group identifications from Database #3 (Table S3); WGCNA output from Database #3 (Table S4); protein groups from Database #4 (Table S5); phosphorylation sites from Database #4 (Table S6); phosphoribosylation sites from Database #4 (Table S7); the ASK2 interactome (Table S8); the PARP9 interactome (Table S9); curated master database summarizing Databases #1–4 and the interactomes (protein IDs only) (Table S10); comparative protein list, showing gene names for ADP-ribosylated proteins characterized in the discussed studies (Table S11); a comparison of phosphoribose site identifications from parallel analyses of five RAW files from Database #1 (Table S12); the full list of gene ontology biological process (GOBP) terms and p-values used to generate the tree maps in Figure S3B (Table S13) (ZIP)
291 annotated MS2 spectra, manually exported from MaxQuant, showing phosphoribosylation sites on representative proteins from Database #1 (Supporting Spectra) (PDF)
Optimization of PMA differentiation of monocytic-like human cell lines THP1 and U937 to macrophage-like cells (Figure S1); nuclear-cytoplasmic fractionation of human and mouse macrophage cells following LPS stimulation shows a changing ADP-ribosylated proteome (Figure S2); overlap between human and mouse subsets of Database #1, and the presentation of selected pR sites (Figure S3); examples of dimethyl-labeled peptides (Figure S4); overlap between replicates in Database #2 (Figure S5); development of the SEC-based method for separating proteoforms (Figure S6); SEC elution profiles of selected TLR4-responsive proteins (Figure S7); WGCNA analysis of SEC elution profiles from Database #3 yields protein modules coordinately moving into and out of fractions (Figure S8); full interactomes of proteasome C/D and proteasome A/B modules (DB#3) (Figure S9); macrophage cell viability following 48 h of PARP inhibitor treatment (Figure S10); examples of phosphopeptide data from DB#4 (Figure S11); SEC-UV elution profiles of standards and samples, DB#4 (Figure S12); principal component analysis of protein identifications from the low molecular weight (LMW) fraction of Database #4, replicates A and B (Figure S13); ASK2-HA-V5 SEC elution profile from Database #4 (Figure S14); uncropped images of the Western blots used in Figures 4 and S14 (Figure S15); purification of ASK2-HA-V5 and a human macrophage cell line (Figure S16); depiction of the interactome identified in the MAPKKK regulatory [blue down] module (DB#3) (Figure S17); purification of ASK2-HA-V5 and PARP9 from human macrophage cell lines (Figure S18); and overlap between the major studies profiling protein ADP-ribosylation in human cells (Figure S19); discussion on the various amino acid residues that are known to carry mono- and/or poly(ADP-ribose), and the rationale for choosing D, E, K, R, and C as residues for the MaxQuant search in this project (Note S1); overview of MAPCD, the website associated with this project that allows readers to easily search all supplemental documents for their protein of interest (Note S2) (PDF)
The authors declare no competing financial interest.
Contributor Information
Casey M. Daniels, Functional Cellular Networks Section, Laboratory of Immune System Biology, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, Maryland 20892-6612, United States
Pauline R. Kaplan, Functional Cellular Networks Section, Laboratory of Immune System Biology, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, Maryland 20892-6612, United States
Isaac Bishof, Bioinformatics and Computational Biosciences Branch, Office of Cyber Infrastructure and Computational Biology, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, Maryland 20892-6612, United States.
Clinton Bradfield, Signaling Systems Section, Laboratory of Immune System Biology, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, Maryland 20892-6612, United States.
Trisha Tucholski, Functional Cellular Networks Section, Laboratory of Immune System Biology, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, Maryland 20892-6612, United States.
Arthur G. Nuccio, Functional Cellular Networks Section, Laboratory of Immune System Biology, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, Maryland 20892-6612, United States
Nathan P. Manes, Functional Cellular Networks Section, Laboratory of Immune System Biology, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, Maryland 20892-6612, United States
Samuel Katz, Signaling Systems Section, Laboratory of Immune System Biology, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, Maryland 20892-6612, United State.
Iain D. C. Fraser, Signaling Systems Section, Laboratory of Immune System Biology, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, Maryland 20892-6612, United States
Aleksandra Nita-Lazar, Functional Cellular Networks Section, Laboratory of Immune System Biology, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, Maryland 20892-6612, United States.
REFERENCES
- (1).Poltorak A; He X; Smirnova I; Liu MY; Van Huffel C; Du X; Birdwell D; Alejos E; Silva M; Galanos C; Freudenberg M; Ricciardi-Castagnoli P; Layton B; Beutler B Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science 1998, 282, 2085–2088. [DOI] [PubMed] [Google Scholar]
- (2).Tan Y; Kagan JC A cross-disciplinary perspective on the innate immune responses to bacterial lipopolysaccharide. Mol. Cell 2014, 54, 212–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Hoshino K; Takeuchi O; Kawai T; Sanjo H; Ogawa T; Takeda Y; Takeda K; Akira S Cutting edge: Toll-like receptor 4 (TLR4)-deficient mice are hyporesponsive to lipopolysaccharide: evidence for TLR4 as the Lps gene product. J. Immunol 1999, 162, 3749–3752. [PubMed] [Google Scholar]
- (4).Roger T; Froidevaux C; Le Roy D; Reymond MK; Chanson AL; Mauri D; Burns K; Riederer BM; Akira S; Calandra T Protection from lethal gram-negative bacterial sepsis by targeting Toll-like receptor 4. Proc. Natl. Acad. Sci. U.S.A 2009, 106, 2348–2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Cohen J The immunopathogenesis of sepsis. Nature 2002, 420, 885–891. [DOI] [PubMed] [Google Scholar]
- (6).Aebersold R; Mann M Mass-spectrometric exploration of proteome structure and function. Nature 2016, 537, 347–355. [DOI] [PubMed] [Google Scholar]
- (7).Hassa PO; Haenni SS; Elser M; Hottiger MO Nuclear ADP-ribosylation reactions in mammalian cells: where are we today and where are we going? Microbiol. Mol. Biol. Rev 2006, 70, 789–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Pajuelo D; Gonzalez-Juarbe N; Tak U; Sun J; Orihuela CJ; Niederweis M NAD(+) Depletion Triggers Macrophage Necroptosis, a Cell Death Pathway Exploited by Mycobacterium tuberculosis. Cell Rep. 2018, 24, 429–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Robinson N; Ganesan R; Hegedus C; Kovacs K; Kufer TA; Virag L Programmed necrotic cell death of macrophages: Focus on pyroptosis, necroptosis, and parthanatos. Redox Biol. 2019, 26, No. 101239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Yang Y; Sauve AA NAD(+) metabolism: Bioenergetics, signaling and manipulation for therapy. Biochim. Biophys. Acta, Proteins Proteomics 2016, 1864, 1787–1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Nikiforov A; Kulikova V; Ziegler M The human NAD metabolome: Functions, metabolism and compartmentalization. Crit. Rev. Biochem. Mol. Biol 2015, 50, 284–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Yang CS; Jividen K; Spencer A; Dworak N; Ni L; Oostdyk LT; Chatterjee M; Kusmider B; Reon B; Parlak M; Gorbunova V; Abbas T; Jeffery E; Sherman NE; Paschal BM Ubiquitin Modification by the E3 Ligase/ADP-Ribosyltransferase Dtx3L/Parp9. Mol. Cell 2017, 66, 503–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Iwata H; Goettsch C; Sharma A; Ricchiuto P; Goh WW; Halu A; Yamada I; Yoshida H; Hara T; Wei M; Inoue N; Fukuda D; Mojcher A; Mattson PC; Barabasi AL; Boothby M; Aikawa E; Singh SA; Aikawa M PARP9 and PARP14 cross-regulate macrophage activation via STAT1 ADP-ribosylation. Nat. Commun 2016, 7, No. 12849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Kanai M; Hanashiro K; Kim SH; Hanai S; Boulares AH; Miwa M; Fukasawa K Inhibition of Crm1-p53 interaction and nuclear export of p53 by poly(ADP-ribosyl)ation. Nat. Cell Biol 2007, 9, 1175–1183. [DOI] [PubMed] [Google Scholar]
- (15).Zerfaoui M; Errami Y; Naura AS; Suzuki Y; Kim H; Ju J; Liu T; Hans CP; Kim JG; Abd Elmageed ZY; Koochekpour S; Catling A; Boulares AH Poly(ADP-ribose) polymerase-1 is a determining factor in Crm1-mediated nuclear export and retention of p65 NF-kappa B upon TLR4 stimulation. J. Immunol 2010, 185, 1894–1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Kang HC; Lee YI; Shin JH; Andrabi SA; Chi Z; Gagne JP; Lee Y; Ko HS; Lee BD; Poirier GG; Dawson VL; Dawson TM Iduna is a poly(ADP-ribose) (PAR)-dependent E3 ubiquitin ligase that regulates DNA damage. Proc. Natl. Acad. Sci. U.S.A 2011, 108, 14103–14108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Ray Chaudhuri A; Nussenzweig A The multifaceted roles of PARP1 in DNA repair and chromatin remodelling. Nat. Rev. Mol. Cell Biol 2017, 18, 610–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Ji Y; Tulin AV Post-transcriptional regulation by poly(ADP-ribosyl)ation of the RNA-binding proteins. Int. J. Mol. Sci 2013, 14, 16168–16183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Leung AK; Vyas S; Rood JE; Bhutkar A; Sharp PA; Chang P Poly(ADP-ribose) regulates stress responses and microRNA activity in the cytoplasm. Mol. Cell 2011, 42, 489–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Stilmann M; Hinz M; Arslan SC; Zimmer A; Schreiber V; Scheidereit C A nuclear poly(ADP-ribose)-dependent signalosome confers DNA damage-induced IkappaB kinase activation. Mol. Cell 2009, 36, 365–378. [DOI] [PubMed] [Google Scholar]
- (21).Teloni F; Altmeyer M Readers of poly(ADP-ribose): designed to be fit for purpose. Nucleic Acids Res. 2016, 44, 993–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Daniels CM; Ong SE; Leung AK A phosphoproteomic approach to characterize protein mono and poly(ADP-ribosyl)ation sites from whole cell lysate. J. Proteome Res 2014, 13, 3510–3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Ong SE; Mann M A practical recipe for stable isotope labeling by amino acids in cell culture (SILAC). Nat. Protoc 2006, 1, 2650–2660. [DOI] [PubMed] [Google Scholar]
- (24).Boersema PJ; Aye TT; van Veen TA; Heck AJ; Mohammed S Triplex protein quantification based on stable isotope labeling by peptide dimethylation applied to cell and tissue lysates. Proteomics 2008, 8, 4624–4632. [DOI] [PubMed] [Google Scholar]
- (25).Cervantes-Laurean D; Jacobson EL; Jacobson MK Preparation of low molecular weight model conjugates for ADP-ribose linkages to protein. Methods Enzymol. 1997, 280, 275–287. [DOI] [PubMed] [Google Scholar]
- (26).Daniels CM; Ong SE; Leung AKL ADP-Ribosylated Peptide Enrichment and Site Identification: The Phosphodiesterase-Based Method. Methods Mol. Biol 2017, 1608, 79–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Langfelder P; Horvath S WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 2008, 9, No. 559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Zambon AC; Gaj S; Ho I; Hanspers K; Vranizan K; Evelo CT; Conklin BR; Pico AR; Salomonis N GO-Elite: a flexible solution for pathway and ontology over-representation. Bioinformatics 2012, 28, 2209–2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Vivelo CA; Wat R; Agrawal C; Tee HY; Leung AK ADPriboDB: The database of ADP-ribosylated proteins. Nucleic Acids Res. 2017, 45, D204–D209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Federspiel JD; Codreanu SG; Palubinsky AM; Winland AJ; Betanzos CM; McLaughlin B; Liebler DC Assembly Dynamics and Stoichiometry of the Apoptosis Signal-regulating Kinase (ASK) Signalosome in Response to Electrophile Stress. Mol. Cell. Proteomics 2016, 15, 1947–1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Matsuzawa A; Saegusa K; Noguchi T; Sadamitsu C; Nishitoh H; Nagai S; Koyasu S; Matsumoto K; Takeda K; Ichijo H ROS-dependent activation of the TRAF6-ASK1-p38 pathway is selectively required for TLR4-mediated innate immunity. Nat. Immunol 2005, 6, 587–592. [DOI] [PubMed] [Google Scholar]
- (32).Liu ZS; Cai H; Xue W; Wang M; Xia T; Li WJ; Xing JQ; Zhao M; Huang YJ; Chen S; Wu SM; Wang X; Liu X; Pang X; Zhang ZY; Li T; Dai J; Dong F; Xia Q; Li AL; Zhou T; Liu ZG; Zhang XM; Li T G3BP1 promotes DNA binding and activation of cGAS. Nat. Immunol 2019, 20, 18–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Tourriere H; Chebli K; Zekri L; Courselaud B; Blanchard JM; Bertrand E; Tazi J The RasGAP-associated endoribonuclease G3BP assembles stress granules. J. Cell Biol 2003, 160, 823–831. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- (34).Takaesu G; Kishida S; Hiyama A; Yamaguchi K; Shibuya H; Irie K; Ninomiya-Tsuji J; Matsumoto K TAB2, a novel adaptor protein, mediates activation of TAK1 MAPKKK by linking TAK1 to TRAF6 in the IL-1 signal transduction pathway. Mol. Cell 2000, 5, 649–658. [DOI] [PubMed] [Google Scholar]
- (35).Israel A The IKK complex, a central regulator of NF-kappaB activation. Cold Spring Harbor Perspect. Biol 2010, 2, No. a000158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Catalanotti F; Reyes G; Jesenberger V; Galabova-Kovacs G; de Matos Simoes R; Carugo O; Baccarini M A Mek1-Mek2 heterodimer determines the strength and duration of the Erk signal. Nat. Struct. Mol. Biol 2009, 16, 294–303. [DOI] [PubMed] [Google Scholar]
- (37).Snel B; Lehmann G; Bork P; Huynen MA STRING: a web-server to retrieve and display the repeatedly occurring neighbourhood of a gene. Nucleic Acids Res. 2000, 28, 3442–3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Qureshi N; Vogel SN; Van Way C 3rd; Papasian CJ; Qureshi AA; Morrison DC The proteasome: a central regulator of inflammation and macrophage function. Immunol. Res 2005, 31, 243–260. [DOI] [PubMed] [Google Scholar]
- (39).Ferrington DA; Gregerson DS Immunoproteasomes: structure, function, and antigen presentation. Prog. Mol. Biol. Transl Sci 2012, 109, 75–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Pacher P; Cziraki A; Mabley JG; Liaudet L; Papp L; Szabo C Role of poly(ADP-ribose) polymerase activation in endotoxin-induced cardiac collapse in rodents. Biochem. Pharmacol 2002, 64, 1785–1791. [DOI] [PubMed] [Google Scholar]
- (41).Ong SE; Blagoev B; Kratchmarova I; Kristensen DB; Steen H; Pandey A; Mann M Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics 2002, 1, 376–386. [DOI] [PubMed] [Google Scholar]
- (42).Mellacheruvu D; Wright Z; Couzens AL; Lambert JP; St-Denis NA; Li T; Miteva YV; Hauri S; Sardiu ME; Low TY; Halim VA; Bagshaw RD; Hubner NC; Al-Hakim A; Bouchard A; Faubert D; Fermin D; Dunham WH; Goudreault M; Lin ZY; Badillo BG; Pawson T; Durocher D; Coulombe B; Aebersold R; Superti-Furga G; Colinge J; Heck AJ; Choi H; Gstaiger M; Mohammed S; Cristea IM; Bennett KL; Washburn MP; Raught B; Ewing RM; Gingras AC; Nesvizhskii AI The CRAPome: a contaminant repository for affinity purification-mass spectrometry data. Nat. Methods 2013, 10, 730–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Takeyama K; Aguiar RC; Gu L; He C; Freeman GJ; Kutok JL; Aster JC; Shipp MA The BAL-binding protein BBAP and related Deltex family members exhibit ubiquitin-protein isopeptide ligase activity. J. Biol. Chem 2003, 278, 21930–21937. [DOI] [PubMed] [Google Scholar]
- (44).Balka KR; De Nardo D Understanding early TLR signaling through the Myddosome. J. Leukocyte Biol 2019, 105, 339–351. [DOI] [PubMed] [Google Scholar]
- (45).Martello R; Leutert M; Jungmichel S; Bilan V; Larsen SC; Young C; Hottiger MO; Nielsen ML Proteome-wide identification of the endogenous ADP-ribosylome of mammalian cells and tissue. Nat. Commun 2016, 7, No. 12917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Zhang Y; Wang J; Ding M; Yu Y Site-specific characterization of the Asp- and Glu-ADP-ribosylated proteome. Nat. Methods 2013, 10, 981–984. [DOI] [PubMed] [Google Scholar]
- (47).Larsen SC; Hendriks IA; Lyon D; Jensen LJ; Nielsen ML Systems-wide Analysis of Serine ADP-Ribosylation Reveals Widespread Occurrence and Site-Specific Overlap with Phosphorylation. Cell Rep. 2018, 24, 2493.e4–2505.e4. [DOI] [PubMed] [Google Scholar]
- (48).Hendriks IA; Larsen SC; Nielsen ML An Advanced Strategy for Comprehensive Profiling of ADP-ribosylation Sites Using Mass Spectrometry-based Proteomics. Mol. Cell. Proteomics 2019, 18, 1010–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Lex A; Gehlenborg N; Strobelt H; Vuillemot R; Pfister H UpSet: Visualization of Intersecting Sets. IEEE Trans. Visualization Comput. Graphics 2014, 20, 1983–1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Weintz G; Olsen JV; Fruhauf K; Niedzielska M; Amit I; Jantsch J; Mages J; Frech C; Dolken L; Mann M; Lang R The phosphoproteome of toll-like receptor-activated macrophages. Mol. Syst. Biol 2010, 6, No. 371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Sjoelund V; Smelkinson M; Nita-Lazar A Phosphoproteome profiling of the macrophage response to different toll-like receptor ligands identifies differences in global phosphorylation dynamics. J. Proteome Res 2014, 13, 5185–5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Söderholm S; Kainov DE; Öhman T; Denisova OV; Schepens B; Kulesskiy E; Imanishi SY; Corthals G; Hintsanen P; Aittokallio T; Saelens X; Matikainen S; Nyman TA Phosphoproteomics to Characterize Host Response During Influenza A Virus Infection of Human Macrophages. Mol. Cell. Proteomics 2016, 15, 3203–3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Wan C; Borgeson B; Phanse S; Tu F; Drew K; Clark G; Xiong X; Kagan O; Kwan J; Bezginov A; Chessman K; Pal S; Cromar G; Papoulas O; Ni Z; Boutz DR; Stoilova S; Havugimana PC; Guo X; Malty RH; Sarov M; Greenblatt J; Babu M; Derry WB; Tillier ER; Wallingford JB; Parkinson J; Marcotte EM; Emili A Panorama of ancient metazoan macro-molecular complexes. Nature 2015, 525, 339–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Salas D; Stacey RG; Akinlaja M; Foster LJ Next-generation Interactomics: Considerations for the Use of Co-elution to Measure Protein Interaction Networks. Mol. Cell. Proteomics 2020, 19, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Takeda K; Shimozono R; Noguchi T; Umeda T; Morimoto Y; Naguro I; Tobiume K; Saitoh M; Matsuzawa A; Ichijo H Apoptosis signal-regulating kinase (ASK) 2 functions as a mitogen-activated protein kinase kinase kinase in a heteromeric complex with ASK1. J. Biol. Chem 2007, 282, 7522–7531. [DOI] [PubMed] [Google Scholar]
- (56).Zhang Y; Mao D; Roswit WT; Jin X; Patel AC; Patel DA; Agapov E; Wang Z; Tidwell RM; Atkinson JJ; Huang G; McCarthy R; Yu J; Yun NE; Paessler S; Lawson TG; Omattage NS; Brett TJ; Holtzman MJ PARP9-DTX3L ubiquitin ligase targets host histone H2BJ and viral 3C protease to enhance interferon signaling and control viral infection. Nat. Immunol 2015, 16, 1215–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Vyas S; Matic I; Uchima L; Rood J; Zaja R; Hay RT; Ahel I; Chang P Family-wide analysis of poly(ADP-ribose) polymerase activity. Nat. Commun 2014, 5, No. 4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Aguiar RC; Takeyama K; He C; Kreinbrink K; Shipp MA B-aggressive lymphoma family proteins have unique domains that modulate transcription and exhibit poly(ADP-ribose) polymerase activity. J. Biol. Chem 2005, 280, 33756–33765. [DOI] [PubMed] [Google Scholar]
- (59).Fabregat A; Jupe S; Matthews L; Sidiropoulos K; Gillespie M; Garapati P; Haw R; Jassal B; Korninger F; May B; Milacic M; Roca CD; Rothfels K; Sevilla C; Shamovsky V; Shorser S; Varusai T; Viteri G; Weiser J; Wu G; Stein L; Hermjakob H; D’Eustachio P The Reactome Pathway Knowledgebase. Nucleic Acids Res. 2018, 46, D649–D655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Kestler HA; Muller A; Kraus JM; Buchholz M; Gress TM; Liu H; Kane DW; Zeeberg BR; Weinstein JN VennMaster: area-proportional Euler diagrams for functional GO analysis of microarrays. BMC Bioinformatics 2008, 9, No. 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the data set identifier PXD016469.