Abstract
The capsid of flock house virus is composed of 180 copies of a single type of coat protein which forms a T=3 icosahedral shell. High-resolution structural analysis has shown that the protein subunits, although chemically identical, form different contacts across the twofold axes of the virus particle. Subunits that are related by icosahedral twofold symmetry form flat contacts, whereas subunits that are related by quasi-twofold symmetry form bent contacts. The flat contacts are due to the presence of ordered genomic RNA and an ordered peptide arm which is inserted in the groove between the subunits and prevents them from forming the dihedral angle observed at the bent quasi-twofold contacts. We hypothesized that by deleting the residues that constitute the ordered peptide arm, formation of flat contacts should be impossible and therefore result in assembly of particles with only bent contacts. Such particles would have T=1 symmetry. To test this hypothesis we generated two deletion mutants in which either 50 or 31 residues were eliminated from the N terminus of the coat protein. We found that in the absence of residues 1 to 50, assembly was completely inhibited, presumably because the mutation removed a cluster of positively charged amino acids required for neutralization of encapsidated RNA. When the deletion was restricted to residues 1 to 31, assembly occurred, but the products were highly heterogeneous. Small bacilliform-like structures and irregular structures as well as wild-type-like T=3 particles were detected. The anticipated T=1 particles, on the other hand, were not observed. We conclude that residues 20 to 30 are not critical for formation of flat protein contacts and formation of T=3 particles. However, the N terminus of the coat protein appears to play an essential role in regulating assembly such that only one product, T=3 particles, is synthesized.
Assembly of viral coat protein and nucleic acid into infectious virions is a highly regulated process that rarely leads to formation of aberrant particles in vivo. The precision with which assembly proceeds is particularly remarkable with icosahedral capsids whose triangulation number exceeds 1. In these capsids the coat protein subunits must be able to adopt several different conformations and the various conformers must be positioned at precise locations on the icosahedral surface lattice. How protein subunits “know” which conformation to adopt at what point during assembly is unknown. High-resolution structural analyses of icosahedral virus capsids have shown that the variations in protein conformation are quite subtle (e.g., see references 13, 17, 19, 27, and 29). Usually they involve an alteration between order and disorder of flexible regions in the protein, often located near the N and C termini. These regions are also referred to as molecular switches since their settings determine the conformational status of a particular protein subunit. Elimination of the molecular switching region from a coat protein subunit is expected to result in loss of precision in viral assembly and conceivably in the appearance of nonnative structures. In this report we describe the effects of deleting the molecular switch from the coat protein of flock house virus (FHV), a member of the family Nodaviridae.
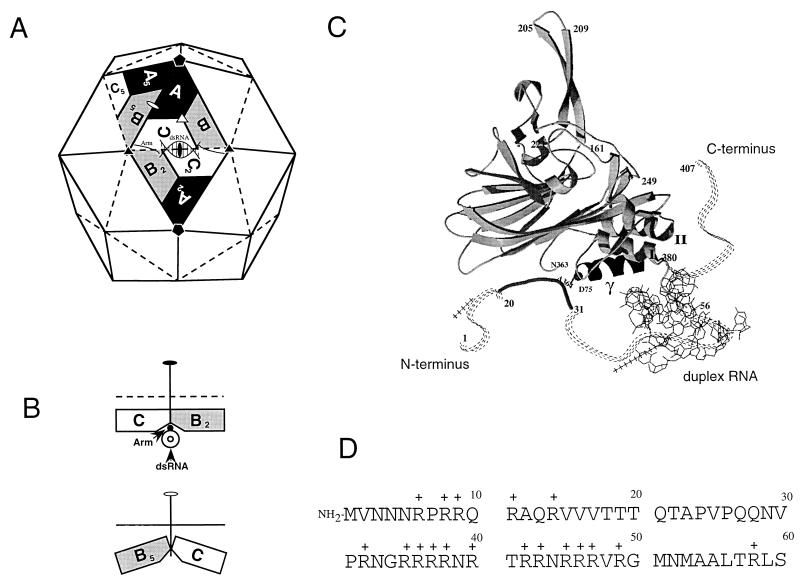
FHV is a nonenveloped, icosahedral virus with a positive-sense, bipartite RNA genome (for a review, see reference 24). RNA 1 (3.1 kb) encodes replication functions, and RNA 2 (1.4 kb) encodes the precursor of the coat protein, protein alpha (43 kDa) (1, 8). In infected cells, protein alpha rapidly assembles into noninfectious precursor particles, called provirions (9, 25). Provirions contain 180 alpha protein subunits and one copy each of the two genomic RNAs. The assembly process triggers an autocatalytic maturation cleavage in which the 407-amino-acid alpha chains are cleaved between residues Asn363 and Ala364 to yield two polypeptides, beta (38 kDa) and gamma (5 kDa) (9, 11). Both beta and gamma remain associated with the mature, infectious virion. The structure of the mature virion was solved to near atomic resolution by X-ray crystallography (5). The 180 protein subunits form a T=3 capsid which is composed of 60 triangular units (Fig. 1A). Each triangular unit contains three chemically identical but conformationally slightly different protein subunits, A, B, and C, that are related by icosahedral symmetry. In the A and B subunits, amino acid residues 1 to 54 are disordered and not visible in the electron density, whereas a subset of the same residues, amino acids 20 to 30, forms an ordered peptide “arm” in the C subunits (Fig. 1C). This subtle variation in tertiary structure, which is similar to that originally described for the capsid protein of tomato bushy stunt virus (10), contributes to a significant difference in the nature of the contacts between the triangular units across the icosahedral twofold and quasi-twofold axes (Fig. 1B). While the interactions between the protein subunits related by the quasi-twofold axes are bent, the interactions between subunits related by the icosahedral twofold axes are flat because the ordered peptide arm in the C subunits folds into the groove between the subunits, preventing them from forming the dihedral angle seen at the quasi-twofold axes. This flat joint is further stabilized by an ordered segment of encapsidated duplex RNA that wedges under each of the 30 twofold axes of the particle. The precise alternation between flat and bent contacts is crucial for formation of a properly dimensioned virus particle.
FIG. 1.
Subunit interactions as seen in the structure of FHV and the N-terminal sequence of the coat protein. (A) Schematic representation of the FHV capsid as a rhombic triacontahedron. Each trapezoid represents a protein subunit which consists of 407 amino acids. The labels A, B, and C represent the three subunits in each of the 60 icosahedral asymmetric units in the T=3 particle. Although A, B, and C represent identical gene products, they are not related by strict symmetry and they are structurally slightly different. The icosahedral twofold, threefold, and fivefold axes are represented by the filled oval, triangle, and pentagon, respectively. The quasi-twofold and quasi-threefold axes are represented by the open oval and triangle. Quasisymmetry axes relate subunits only locally outside the context of the entire shell. For example, the quasi-twofold axis shown between the central triangle and its left neighbor relates subunits A, B, and C to B5, C5, and A5, respectively, thus bringing subunits clustered around a fivefold axis into coincidence with those clustered around a threefold axis. dsRNA, double-stranded RNA. (B) Side view of the joints between asymmetric units. The top diagram shows a joint at the base of the central triangle viewed along the line connecting the two threefold-symmetry axes. The dihedral angle between these two triangular surfaces is 180°C, i.e., they are coplanar. The bottom diagram shows a joint at the left of the central triangle viewed along the line connecting a threefold axis to a fivefold axis. The dihedral angle between these two surfaces is 144°. For proper assembly to occur, the trimer interfaces must be able to “hinge” with a molecular switch that determines the dihedral angle between asymmetric units. In FHV, the switch is a 10-bp RNA duplex and an 11-amino-acid peptide (residues 20 to 30) that are ordered only in the C subunits. The RNA and peptide arm form a wedge that prevents the bending of the joint at the bottom of the central triangle. (C) Ribbon diagram showing the tertiary structure of the C subunit of FHV. The subunit is oriented such that the virus exterior is at the top and the virus interior is at the bottom. The ordered peptide arm (residues 20 to 30) is represented as a solid line, whereas the disordered portions are represented as dashed lines. The duplex RNA is shown as a line diagram. (D) Amino-terminal sequence of the FHV coat protein. Note the presence of 17 positively charged amino acids, all arginines, within the first 50 residues.
The ordered peptide arm of the coat protein and ordered portions of the genomic RNA represent the molecular switches that direct the T=3 quaternary organization of the FHV virion. In theory, deletion of amino acids 20 to 30 from the N terminus of the coat protein should prevent formation of flat contacts unless the encapsidated RNA alone is sufficient for its formation and maintenance in the particle. Assembly of a particle lacking flat contacts and containing only bent contacts, however, may result in a T=1 particle, since all of the subunit interactions would be conformationally equivalent. We tested this hypothesis by expressing two N-terminal deletion mutants of the FHV coat protein subunit in insect cells with recombinant baculoviruses. In this expression system, wild-type FHV coat protein assembles into virus-like particles (VLPs) that are structurally indistinguishable from native virions, even at a 3-Å resolution (unpublished data). Our new results show that deletion of amino acids 1 to 50 from coat precursor protein alpha completely inhibits assembly of particles but that deletion of residues 1 to 31 results in formation of variously shaped particles, including wild-type-like T=3 particles. Surprisingly, the expected T=1 particles were not detected. Our results demonstrate that while formation of T=3 particles does not depend on the presence of the ordered peptide arm, the absence of residues 1 to 31 leads to accumulation of defective structures. The N-terminal region of the coat protein is thus required for maintaining the precision with which particles are normally assembled in infected cells.
MATERIALS AND METHODS
Cells and infection.
Trichoplusia ni cells were propagated at 27°C in EX-CELL 401 serum-free medium (JRH Biosciences) supplemented with 2 mM l-glutamine, 100 U of penicillin per ml, and 100 μg of streptomycin per ml. Stock cultures (50 ml) were maintained in suspension on a gyratory shaker at 100 rpm and subpassaged when the cell density reached 4 × 106 cells per ml. For large-scale infections, cultures were expanded to 1 liter by successively diluting cell suspensions with fresh medium when the density reached 4 × 106 cells per ml. Virus inoculum for a 1-liter culture was prepared by infecting 15 × 106 Spodoptera frugiperda cells at a multiplicity of 0.5 to 2 PFU per cell and incubating the infected cells at 27°C until they showed cytopathic effect, usually 4 to 5 days later. The cell supernatant (30 ml) was harvested and used directly to infect a 1-liter culture of T. ni cells at a density of 2 × 106 cells per ml.
S. frugiperda cells (line IPLB-Sf21) were propagated and infected as described previously (23).
Site-directed mutagenesis of FHV coat protein alpha.
Plasmid p2BS(+)-wt (25) containing a cDNA copy of the wild-type FHV coat protein gene was used to generate two deletion mutants in which the coding sequences for amino acid residues 1 to 31 and 1 to 50 were removed. The deletions were created by inverse PCR (12), i.e., oligonucleotide primers were designed in inverted tail-to-tail directions to amplify the cloning vector as well as the target sequence except for the area to be deleted. Specifically, primers used to create p2BS(+)-Δ31 had the sequences 5′ CAT TTT GGA ACT TGG AAT TG 3′ and 5′ CGT AAT GGT AGA CGC CGA CG 3′; primers used to create p2BS(+)-Δ50 had the sequences 5′ CAT TTT GGA ACT TGG AAT TG 3′ and 5′ ATG AAC ATG GCG GCG CTA AC 3′. PCR samples (100 μl) contained 1× Pfu buffer provided as a 10× stock from the manufacturer (Stratagene), 2.5 U of Pfu polymerase (Stratagene), and 2 ng of plasmid DNA. Primer concentrations were 2 mM, and deoxynucleoside triphosphate concentrations were 200 mM. Cycle conditions were as follows: 95°C for 5 min followed by 55°C for 5 min and then 25 cycles at 72°C for 4 min, 94°C for 2 min, and 55°C for 2 min. Products were purified by agarose gel electrophoresis and with a GeneClean purification kit (Bio 101), religated with T4 DNA ligase (New England Biolabs), and transformed into DH5α competent cells. Plasmid DNAs were isolated from three to five independent clones, and the sequence across the mutated sites was verified by the dideoxy sequencing method of Sanger et al. (22).
Construction of recombinant baculoviruses ΔNterm-31(bac) and ΔNterm-50(bac).
Recombinant baculoviruses containing the gene for FHV coat protein deletion mutants lacking residues 1 to 31 and 1 to 50 were generated with a BacPAK baculovirus expression system kit (Clontech). The genes for both mutants were first amplified by PCR with plasmids p2BS(+)-Δ31 and p2BS(+)-Δ50. Amplification primers matched the sequences at the 5′ and 3′ ends of the coat protein genes and in addition contained restriction sites used for subcloning into the transfer vector pBacPAK9. Specifically, primers had the sequences 5′ ATG CAG GAT CCG TAA ACA ATT CCA AGT TC 3′ (BamHI-5′RNA2) and 5′ TAC GTT CTA GAA CCT TAG TCT GTT GAC TT 3′ (XbaI-3′RNA2). PCR mixture samples (100 μl) contained 1× PFU buffer (Stratagene), 2.5 U of Pfu polymerase (Stratagene), and 0.5 ng of plasmid DNA. Primer concentrations were 0.3 mM, and deoxynucleoside triphosphate concentrations were 200 mM. Cycle conditions were as follows: 95°C for 5 min followed by 55°C for 5 min and then 30 cycles at 72°C for 2 min, 95°C for 1.5 min, and 55°C for 1 min. Amplification products were purified as described above, digested with BamHI and XbaI (New England Biolabs), purified, and ligated into BamHI- and XbaI-digested pBacPAK9. Following transformation, plasmid DNA was isolated from several clones and the presence of the inserted DNA was verified by PCR. To generate recombinant baculoviruses, protocols provided by the manufacturer were used. Briefly, the transfer vector pBacPAK9 containing the gene for either the deletion mutant lacking residues 1 to 31 or that lacking residues 1 to 50 was mixed with Bsu36I-linearized BacPAK6 viral DNA and transfected into Sf21 cells. Three days after transfection, cell supernatants were harvested and putative recombinant viruses were isolated by plaquing the supernatants once on Sf21 cell monolayers. Individual plaque isolates were amplified following confirmation of the presence and expression of the FHV coat protein gene.
Electrophoretic analysis of proteins and immunoblot analysis.
Electrophoresis was performed with discontinuous sodium dodecyl sulfate (SDS)-polyacrylamide gels according to the procedure of Laemmli (15). Samples were mixed with an equal volume of 2× electrophoresis buffer (125 mM Tris-HCl [pH 6.8], 4.6% SDS, 10% 2-mercaptoethanol [2-ME], 20% glycerol) and heated to 100°C for 10 min. Slab gels (8 by 6 by 0.75 cm) contained 12% (wt/vol) acrylamide in the resolving gel (5-cm length) and 5% (wt/vol) acrylamide in the stacking gel (1-cm length). Electrophoresis was at 200 V for 45 min. Gels were fixed and stained with Coomassie brilliant blue R-250 by following standard procedures. For immunoblot analysis, proteins were electrophoretically separated and electroblotted in 10 mM CAPS (3-[cyclohexylamino]-1-propane-sulfonic acid [pH 11]) containing 10% methanol onto nitrocellulose (Nitrobind; Micron Separations Inc.) at 200 mA for 1 h. Following transfer, the membrane was incubated in blot buffer (5% [wt/vol] Carnation nonfat dry milk in phosphate-buffered saline [PBS]) for 1 h at room temperature. Rabbit anti-FHV serum was diluted 1,000-fold in the blot buffer, and incubation was continued for another hour. The membrane was then washed three times for 10 min with PBS-Tween buffer (0.05% [vol/vol] Tween 20 in PBS) and transferred to fresh blot buffer containing goat anti-rabbit horseradish peroxidase-conjugated antibodies (Pierce) at a 2,500-fold dilution. The membrane was incubated for 1 h at room temperature and then washed again three times for 10 min with PBS-Tween buffer. Immune complexes were visualized by enhanced chemiluminescence (Amersham) and exposure to X-ray film (Kodak Biomax MR).
Purification of VLPs.
For biochemical characterization, VLPs were purified from Sf21 cells 4 days after infection. To this end, cell monolayers were dislodged into the growth medium and the resulting suspension was made 0.5% in Nonidet P-40 and 0.1% in 2-ME. After incubation on ice for 15 min, cell debris was pelleted in a Beckman JA17 rotor at 10,000 rpm for 10 min at 4°C. The supernatant was treated with RNase A at a final concentration of 10 μg/ml at 27°C for 30 min. VLPs were then pelleted through a 1-ml 30% (wt/wt) sucrose cushion in 50 mM HEPES (pH 7)–0.1% 2-ME–0.1% bovine serum albumin–5 mM CaCl2 at 41,000 rpm in an SW41 rotor for 3.5 h at 4°C. The pellet was resuspended in 50 mM HEPES (pH 7)–0.1% 2-ME–5 mM CaCl2 and layered on a 10-ml 10 to 40% (wt/wt) sucrose gradient in the same buffer. VLPs were sedimented at 40,000 rpm for 1.5 h at 11°C. The gradient was fractionated on an ISCO gradient fractionator at 0.75 ml/min and 0.5 min per fraction.
For crystallization experiments, VLPs were purified from 1-liter suspension cultures of T. ni cells 3 to 4 days after infection. Nonidet P-40 was added directly to the cell suspension to a final concentration of 0.5% (vol/vol). After 15 min on ice, cell debris was removed by centrifugation in a Beckman JA17 rotor at 10,000 rpm for 10 min at 4°C. Polyethylene glycol 8000 (Fisher) and NaCl were added to the supernatant to final concentrations of 8% (wt/vol) and 0.2 M, respectively. The sample was stirred at 4°C for 1 h, and the precipitate was pelleted by centrifugation in a Beckman JA17 rotor at 10,000 rpm for 10 min at 4°C. Pellets containing the VLPs were resuspended in HEPES buffer (50 mM HEPES [pH 7], 5 mM CaCl2, 0.1% 2-ME), clarified by low-speed centrifugation, transferred to 26-ml ultracentrifuge tubes, and underlaid with 3 ml of 30% (wt/wt) sucrose in HEPES buffer containing 0.1% (wt/vol) bovine serum albumin. After centrifugation in a Beckman 50.2 Ti rotor at 50,000 rpm for 3 h at 4°C, the VLP pellets were resuspended in HEPES buffer and 2 ml was loaded onto a 40-ml 10 to 40% (wt/wt) sucrose gradient in HEPES buffer. The gradients were centrifuged in a Beckman SW28 rotor at 28,000 rpm for 5 h at 4°C. Following centrifugation, the gradients were fractionated on an ISCO gradient fractionator at 1.5 ml/min and 1 min per fraction. Fractions containing putative T=3 particles were collected and dialyzed against 25 mM HEPES (pH 7). Following dialysis, protein was concentrated to 15 to 20 mg/ml in a microconcentrator (Filtron) that has a molecular mass cutoff of 100,000 Da.
RNA extraction and Northern (RNA) blot analysis.
RNA was extracted from sucrose gradient fractions with acidic phenol-chloroform by standard procedures. Electrophoresis of RNA in agarose-formaldehyde gels and Northern blot analysis were performed as described previously (26). The probe used for hybridization was a digoxigenin-UTP-labeled antisense RNA complementary to nucleotides 600 to 888 of FHV RNA 2. To create this probe, a cDNA fragment spanning positions 600 to 888 of RNA 2 was generated by PCR with plasmid p2BS(+)-wt as the template, a sense primer of the sequence 5′ CCC TGT AAA GCT GAG TAC TG 3′ and an antisense primer of the sequence 5′ CTA CTC CAC TGG TGG CTT CT 3′. PCR conditions were as described above under “Construction of recombinant baculoviruses” except that the annealing temperature was reduced to 50°C. The PCR product was purified and cloned into pBluescript KSII(+) (Stratagene) prepared with 3′ T overhangs (18). Following transformation of DH5α competent cells, plasmids containing inserts were selected and the orientation of the insert relative to those of the T7 and T3 promoters was determined. The plasmid chosen for RNA probe synthesis was linearized with XbaI and transcribed with T3 RNA polymerase. Transcription of digoxigenin-UTP-labeled antisense RNA was performed according to the protocols of the manufacturer (Boehringer Mannheim).
Electron microscopy.
Samples of gradient-purified VLPs were negatively stained with 1% (wt/vol) uranyl acetate (Ted Pella, Tustin, Calif.). To this end, a drop of each sample was applied to a glow-discharged collodion-covered copper grid (400 mesh) and allowed to adsorb for 1 to 2 min. Excess solution was removed with filter paper, and the grids were washed and blotted with filter paper three times by floating them on droplets of 50 mM HEPES, pH 7. Each grid was then treated three times with a drop of 1% uranyl acetate solution (filtered through a 2-μm-pore-size filter), and the third drop was left on for 1 to 2 min before the grid was blotted and air dried. The samples were viewed in a Phillips CM 100 transmission electron microscope at 100 kV.
Crystallization and preliminary X-ray analysis.
Crystallization was performed by the hanging-drop vapor-diffusion method. Crystals were obtained at room temperature (20 to 22°C) with a reservoir solution containing 5.4% (wt/vol) polyethylene glycol 8000, 10 mM HEPES (pH 7.0), 150 mM Li2SO4, 10 mM NaCl, and 2.6% isopropanol, and a drop was prepared by mixing equal volumes (1 to 2 μl) of reservoir solution with gradient-purified VLPs at 15 to 20 mg/ml in 25 mM HEPES (pH 7.0). Crystals ranging in size from 0.4 to 0.8 mm (body diagonal) were mounted and drained in quartz capillaries for X-ray-diffraction experiments. Oscillation data were collected at the Cornell High Energy Synchrotron Source (F1 station) at ambient temperature with a Fuji image plate detector.
RESULTS
Expression of ΔNterm-50 and ΔNterm-31 in S. frugiperda cells.
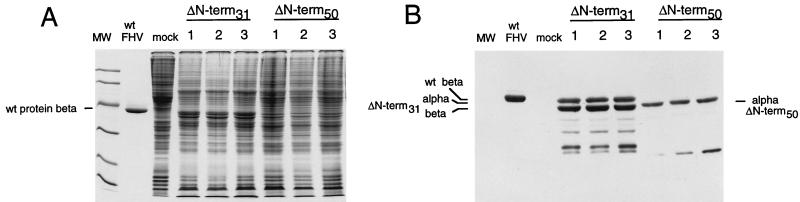
The amino acid sequence at the N terminus of the FHV coat protein is shown in Fig. 1D. Like capsid proteins of other viruses, it is very basic, containing 17 arginines within the first 50 residues. Amino acids 1 to 54 are completely invisible in the electron density of the A and B subunits of crystalline virus particles, whereas residues 20 to 30 form an ordered peptide arm in the C subunits (5). We constructed two N-terminal deletion mutants. In the mutant ΔNterm-50, residues 1 to 50 were deleted from coat precursor protein alpha, which eliminated all of the positive charges as well as the portion that constitutes the ordered peptide arm in C. In the mutant ΔNterm-31, the deletion was restricted to residues 1 to 31. This construct lacked the portion required for formation of the ordered peptide arm as well as a proline residue at position 31, but it retained 12 of the 17 arginine residues which we suspected might be critical for assembly of ribonucleoprotein particles. The construction of recombinant baculoviruses expressing ΔNterm-31 and ΔNterm-50 was by standard procedures. Three independent plaque isolates for each construct were initially tested for expression of the mutant coat protein. To this end, lysates from infected Sf21 cells were electrophoresed through a polyacrylamide gel and stained with Coomassie brilliant blue (Fig. 2A). With ΔNterm-31, the lysates contained two new bands that were not present in mock-infected cells. These two proteins migrated slightly faster than the alpha and beta proteins of native FHV, as was expected for a deletion mutant. In the case of ΔNterm-50, there also appeared to be at least one additional band located at a position between the putative alpha and beta bands of the ΔNterm-31 mutant.
FIG. 2.
Protein gel electrophoresis and immunoblot analysis of Sf21 cell lysates infected with three independent plaque isolates of the recombinant baculoviruses ΔNterm-31(bac) and ΔNterm-50(bac). (A) Monolayers (5 × 105 cells) of Sf21 cells were infected with 100 μl of a 0.5-ml solution containing individual plaque isolates of ΔNterm-31(bac) and ΔNterm-50(bac). Five days after infection, medium was removed from the dishes and cells were suspended in 1 ml of PBS, collected by centrifugation, and washed once with 1 ml of PBS. The final pellet was resuspended in 100 μl of PBS, diluted with an equal volume of electrophoresis buffer, and heated in a boiling water bath for 10 min. Aliquots of 10 μl were electrophoresed through an SDS–12% polyacrylamide gel, followed by staining with Coomassie brilliant blue. Molecular weight (MW) markers from top to bottom were 97,000, 68,000, 43,000, 29,000, 18,000, and 14,000. wt, wild-type. (B) Proteins were subjected to electrophoresis as described for panel A and transferred to a nitrocellulose membrane. The membrane was incubated with polyclonal FHV antiserum and horseradish peroxidase- conjugated secondary antibodies as described in Materials and Methods. Immune complexes were visualized by enhanced chemiluminescence and exposure to X-ray film.
To determine whether the newly detected proteins indeed represented FHV coat protein, Western blot analysis was performed with anti-FHV antibodies and the same cell lysates. The results (Fig. 2B) confirmed the presence of two major bands for ΔNterm-31 plus several minor smaller products. The presence of the two major proteins indicated that the ΔNterm-31 coat protein was not only synthesized in Sf21 cells but was probably also assembled into particles, since cleavage of precursor protein alpha into beta and gamma is not observed in monomeric alpha protein (9). In addition, the pattern of the smaller products was reminiscent of that often detected for gradient-purified authentic FHV. For ΔNterm-50, on the other hand, only a single major band was detected, providing the first indication that this protein was not able to form particles.
Sucrose gradient analysis of putative VLPs.
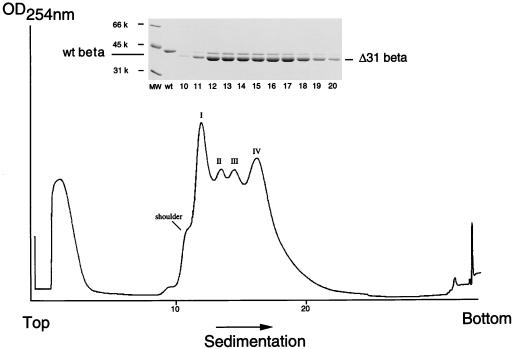
To establish conclusively whether ΔNterm-31 and ΔNterm-50 proteins assembled into VLPs, infected Sf21 cell lysates were subjected to a virus purification protocol normally used for purification of wild-type VLPs. The protocol was slightly modified, however, in anticipation of the presence of particles that were smaller than the native T=3 structure. Briefly, putative particles were released from Sf21 cells on day 4 after infection and pelleted through a 30% (wt/wt) sucrose cushion. The resuspended pellets were then sedimented through a 10 to 40% (wt/wt) sucrose gradient, and the gradients were fractionated, with continuous absorbance at 254 nm. From previous experiments (23) it was known that particles assembled from full-length coat protein in the same system yield a single peak near the center of the gradient after centrifugation. However, the sedimentation profile of the ΔNterm-50 mutant did not contain any peaks (data not shown), confirming that this protein was not able to assemble into particles. The profile of the ΔNterm-31 mutant, on the other hand, showed an unexpected pattern of four peaks in addition to a peak of soluble protein normally detected on top of the gradient (Fig. 3). This four-peak pattern was exceptionally reproducible: the first peak routinely had the highest absorbance and contained a small shoulder, while the fourth peak was located at a position normally observed for wild-type T=3 particles. The two intermediate peaks were always smaller than the two flanking peaks. Electrophoretic analysis of the fractions comprising the four peaks showed that they all contained a major and a minor protein, both of which migrated slightly faster than the beta protein of wild-type FHV (Fig. 3, inset). The minor protein presumably was residual mutant alpha protein, most of which appeared to have efficiently cleaved into the corresponding beta and gamma proteins. The gamma protein, which represents the 44 carboxy-terminal amino acids of precursor alpha, is usually not visible on Tris-glycine SDS-polyacrylamide gels. However, analysis of material in the respective fractions by mass spectroscopy confirmed that all contained the gamma peptide (data not shown). Optical density measurements of purified particles indicated that the ratio of values at 260 and 280 nm was 1.68. This ratio was very similar to that of native FHV (1.60), indicating that the particles maintained a protein-to-RNA ratio comparable to that of authentic virus.
FIG. 3.
Sedimentation profile of VLPs synthesized in ΔNterm-31(bac)-infected Sf21 cells. Monolayers of Sf21 cells (approximately 8 × 106 cells per 100-mm-diameter tissue culture plate) were infected with ΔNterm-31(bac) at a multiplicity of one PFU per cell. Dishes were incubated for 4 days, followed by purification of VLPs as described in Materials and Methods. VLPs from three to four plates were pooled and sedimented through an 11-ml 10 to 40% (wt/wt) sucrose gradient. The gradient was fractionated on an ISCO gradient fractionator, with continuous absorbance at 254 nm. (Inset) Electrophoretic analysis of gradient fractions 10 through 20. An aliquot (5 μl) of each fraction (approximately 375 μl) was mixed with an equal volume of 2× electrophoresis buffer, heated to 95°C for 10 min, and electrophoresed through an SDS–12% polyacrylamide gel. The gel was fixed and stained with Coomassie brilliant blue. wt, wild type; MW, molecular weight.
Electron microscopic analysis of ΔNterm-31 VLPs.
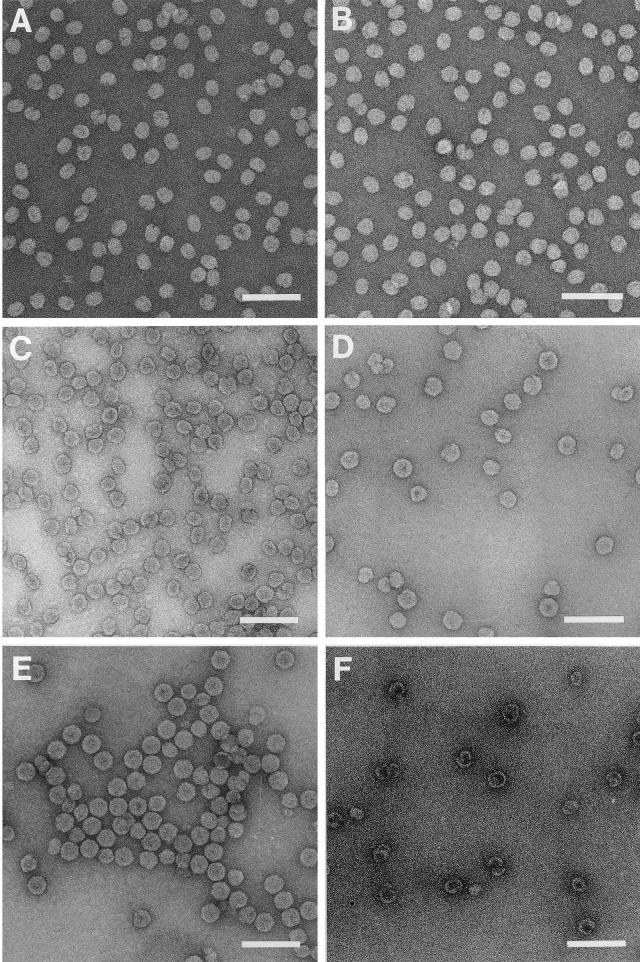
Sucrose gradient fractions representing the four peaks of a profile similar to that shown in Fig. 3 were used to record electron micrographs of negatively stained specimen (Fig. 4). Surprisingly, none of the peaks contained the anticipated T=1 particles. Instead, the first peak (peak I) and its shoulder (Fig. 4A and B) were formed by particles that had a round to ellipsoidal appearance. This appearance was not due to distortions caused by the staining protocol, because cryo-electron microscopy images verified the oval appearance of these structures (data not shown). The length of the smaller axis of the particles was approximately 20 nm, while the length of the longer axis varied between 23, 27, and 30 nm. Peaks II and III (Fig. 4C and D) contained particles whose shapes were more irregular than those in peak I, and they were generally more spheroidal than elongated. Last, peak IV (Fig. 4E) contained mostly hexagonal particles with diameters of 33 nm. These particles were virtually identical to native T=3 FHV, as was already suggested by their sedimentation rate on the sucrose gradient.
FIG. 4.
Electron micrographs of negatively stained, gradient-purified ΔNterm-31 VLPs. (A) Particles recovered from the shoulder of peak I; (B) particles representing peak I; (C) particles representing peak II; (D) particles representing peak III; (E) particles representing peak IV; (F) assembly products observed only in T. ni cells following infection with ΔNterm-31(bac). These products sedimented more slowly than all other VLPs and were usually detected midway between the top of the gradient and peak I. Bars, 100 nm.
RNA contents of ΔNterm-31 VLPs.
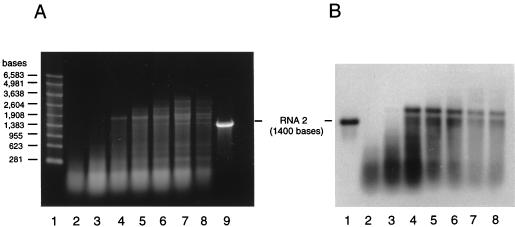
The ratio of optical densities at 260 and 280 nm of the ΔNterm-31 VLPs and their sedimentation rates on sucrose gradients suggested that they contained nucleic acids. In previous experiments (23) we had shown that VLPs assembled from wild-type coat protein in Sf21 cells package RNA, and we therefore assumed that the nucleic acids in ΔNterm-31 particles likewise represented RNA and not DNA. This assumption was also justified by the fact that assembly occurs in the cytoplasms and not in the nuclei of infected cells. To determine the nature of the encapsidated RNAs, sucrose gradient fractions were extracted with phenol and chloroform and the purified RNAs were subjected to denaturing agarose gel electrophoresis and ethidium bromide staining. The results (Fig. 5A) showed that the particles contained a heterogeneous mixture of RNA species whose lengths increased with the sedimentation rate and size of the assembled particles. For example, the slowest-sedimenting particles contained RNAs of 100 to 300 bases, while RNAs of up to 3,600 bases were observed in the fastest-sedimenting peak IV particles. Although the particles typically contained a broad collection of RNA species, a few individual bands could be discerned within the nucleic acid smear of the faster-sedimenting VLPs.
FIG. 5.
Agarose gel electrophoresis and Northern blot analysis of RNAs encapsidated by ΔNterm-31 VLPs. (A) RNA was phenol-chloroform extracted from successive sucrose gradient fractions spanning the four peaks of a sedimentation profile similar to that shown in Fig. 3. An aliquot (4 μg) of each sample was electrophoresed through a 1% agarose-formaldehyde gel, and RNAs were visualized by staining with ethidium bromide. Lane 1, RNA size markers; lanes 2 to 8, RNAs extracted from successive gradient fractions spanning peaks I to IV (left to right); lane 9, in vitro-synthesized full-length transcript of FHV RNA 2 (1,400 bases). (B) Northern blot analysis of RNA samples shown in panel A. RNA (65 ng) was size fractionated on a 1% agarose-formaldehyde gel and transferred to a nylon membrane. Following UV cross-linking, the immobilized RNAs were hybridized to a digoxigenin-labeled probe complementary to bases 600 to 888 of FHV RNA 2. Lane 1, full-length in vitro-synthesized RNA 2 transcript (1,400 bases); lanes 2 to 8, same as lanes 2 to 8 in panel A.
The RNAs were further examined by Northern blot analysis to determine whether they included the mRNA of the FHV coat protein. The results obtained with an antisense RNA probe complementary to bases 600 to 888 of FHV RNA 2 (1,400 bases in total) is shown in Fig. 5B. Similar results were obtained with probes complementary to the 5′ end and the 3′ end of RNA 2 (data not shown). The probes hybridized with virtually the entire collection of RNAs in each fraction, except those that were longer than approximately 1,900 to 2,000 bases. Overexposure, however, revealed that even with these longer RNAs, some hybridization occurred (data not shown). The pattern of bands discernible by ethidium bromide staining was not observed by Northern blot analysis. The 1,900- to 2,000-base RNA presumably represented the full-length message of the FHV coat protein. It is longer than native RNA 2 due to the presence of heterologous sequences at both the 5′ and 3′ ends of the transcript and the presence of a poly(A) tail not observed for native RNA 2. The longer transcripts which we detected upon overexposure might have been due to the use of secondary transcription termination sites downstream of the major polyadenylation site of the polyhedrin gene (23).
Crystallization and preliminary X-ray analysis of wild-type-like ΔNterm-31 VLPs.
The results described in the previous sections indicated that even in the absence of the ordered peptide arm, particles that appeared identical to native T=3 virions were synthesized. To understand how protein-protein contacts, in particular the flat contacts across the icosahedral twofold axes, are formed in the absence of residues 20 to 30, high-resolution structural analysis of the putative T=3 ΔNterm-31 particles is required. To this end, we expressed the ΔNterm-31 coat protein in T. ni cells, which are capable of producing higher levels of recombinant protein than Sf21 cells (2, 30). Obtaining large quantities of the VLPs was required for crystallization and X-ray analysis. From a 1-liter T. ni culture, 5 to 10 mg of peak IV particles were typically obtained. Interestingly, the gradient sedimentation profile of the ΔNterm- 31 VLPs obtained from T. ni cells contained an additional peak not observed during expression in Sf21 cells (data not shown). This peak was located midway between the top of the gradient and peak I, and the size of peak I was usually reduced (data not shown). Analysis of material in this new peak by electron microscopy of negatively stained samples revealed hollow shells, often incompletely closed or wound upon themselves (Fig. 4F). These structures were somewhat reminiscent of phage P22 coat protein assembled in the absence of scaffolding protein (3).
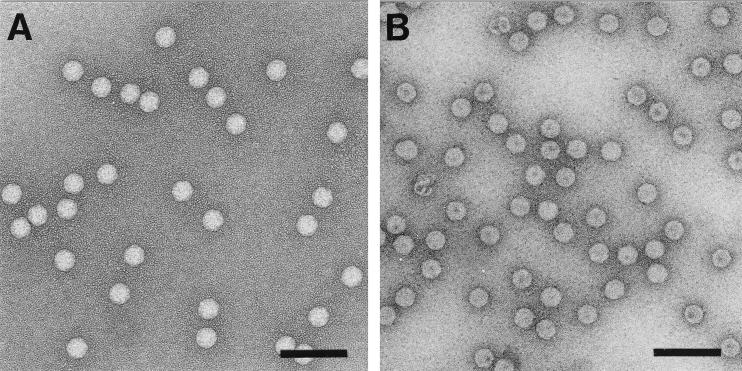
Figure 6 shows an electron micrograph of a typical preparation of peak IV particles. In contrast to native virions and particles assembled from full-length coat protein in Sf21 cells (23) they appeared somewhat more stain penetrable. However, they could be crystallized by the hanging-drop vapor-diffusion method and gave rise to rhombohedral crystals that were isomorphous with crystals generated from native FHV. Specifically, the crystals appeared within 1 week and grew to maximum size within 2 weeks. Crystals ranging in size from 0.4 to 0.8 mm along the body diagonal diffracted X rays to a 2.4-Å resolution (data not shown). Auto indexing of the diffraction patterns with the program DENZO (20) identified the crystal space group as R3, with unit cell dimensions very similar to those of native virus (a = 325.4 Å, α = 61.6°). High-resolution structural analysis of the peak IV particles is under way and should reveal how the putative T=3 symmetry is maintained in the absence of the ordered peptide arm.
FIG. 6.
Electron micrographs of negatively stained FHV particles. (A) Native FHV particles; (B) ΔNterm-31 (peak IV) particles used for crystallization and X-ray analysis. Bars, 100 nm.
DISCUSSION
We generated two N-terminal deletion mutants of the FHV coat protein to examine the role of the N terminus in virus assembly and more specifically the function of residues 20 to 30 in regulating T=3 symmetry of the final particle. Deletion of amino acids 1 to 50 blocked particle formation completely, suggesting that these amino acids include essential elements required for assembly of a ribonucleoprotein particle. Alternatively, it is possible that deletion of residues 1 to 50 resulted in misfolding of the remaining peptide chain such that assembly was inhibited. We believe that the latter possibility is unlikely because amino acids 1 to 50 are naturally flexible (Fig. 1C) and it is doubtful that their absence would affect the structure of the beta barrel or the loops that connect its individual strands. It is more likely that assembly was inhibited because deletion of residues 1 to 50 eliminated a cluster of positively charged amino acids that are normally present at the N terminus of the protein. Seventeen arginine residues are found within the first 50 residues of wild-type coat protein, and these arginines are presumably required to condense and neutralize the negatively charged sugar-phosphate backbone of the viral RNA when it is packaged into virions. Lack of the N terminus probably arrests assembly at an intermediate stage, prior to which condensation of the RNA is not required. Computational chemistry analysis suggests that this assembly intermediate might be a trimer of coat protein subunits (21), and we are in the process of purifying the mutant protein to determine its oligomeric state experimentally.
N-terminal deletion mutants analogous to FHV ΔNterm-50 have previously been generated for a number of T=3 plant viruses, including southern bean mosaic virus (4) and turnip crinkle virus (16, 28). Proteolytic digestion of the N-terminal portions of the coat proteins of these viruses led to assembly of a nucleic acid-free T=1 particle. Neither full nor empty T=1 particles, however, were detected for FHV ΔNterm-50. A possible explanation for this observation is based on recent data from our laboratory (unpublished) indicating that the C terminus of the coat protein, which is cleaved to form protein gamma in mature particles, interacts with viral RNA and is in fact required for specific recognition of FHV RNAs during assembly. It is reasonable to assume that this recognition event occurs early in assembly and that it results in formation of a nucleoprotein complex. Completion of a closed shell after formation of this complex presumably requires the neutralizing capacity of the N terminus such that in its absence assembly stalls. If this assumption is correct, we should be able to detect the putative nucleoprotein complex in infected cell lysates. Also, the model predicts that in the absence of both the N and C termini, empty T=1 particles might form because interactions between coat protein and RNA would have been eliminated. However, our preliminary results indicate that a double deletion mutant lacking 50 amino acids at the N terminus and 26 amino acids at the C terminus does not form particles in Sf21 cells.
The second mutant, ΔNterm-31, lacked amino acids 1 to 31. Unlike with ΔNterm-50, 12 of the 17 arginine residues at the N terminus were retained in this mutant. Unexpectedly, the ΔNterm-31 protein gave rise to multiple types of particles that could be separated by sucrose gradient sedimentation into at least four groups. Electron microscopic analyses of negatively stained samples revealed that the particles in each group differed in size as well as in shape. Moreover, a marked difference was detected with regard to the encapsidated RNAs, whose lengths increased with the sedimentation rate, i.e., size, of the assembly products. For example, particles in peak I contained RNAs no longer than 300 nucleotides (nt) whereas those representing peak IV contained RNAs ranging from approximately 150 to 3,600 nt. Northern blot analysis indicated that each fraction contained RNAs representing the full-length message of the coat protein or smaller breakdown products, but ethidium bromide staining of the entire sample suggested that other, presumably cellular, RNAs were encapsidated as well. These results were in line with observations made previously for expression of wild-type FHV coat protein (23). We did not attempt to determine the identities of the other encapsidated RNAs, but it is reasonable to assume that they were cellular mRNAs and rRNAs. The bright signal obtained for RNAs smaller than the 281-nt marker on the ethidium bromide-stained gel is most likely due to tRNAs.
Despite the differences in shape, size, and nucleic acid contents, coat protein maturation cleavage occurred with wild-type efficiency in all types of particles, suggesting that the individual protein subunits retained the critical quaternary interactions required for cleavage, as was observed in native virions (31). Indeed, the fastest-sedimenting particles (peak IV) were detected at a position on the gradient normally observed for wild-type FHV, and electron microscopic analysis confirmed that these assembly products had the same overall shapes and dimensions as native virions although they were somewhat more stain penetrable. X-ray structural analysis in progress has confirmed that these particles indeed have T=3 symmetry. Clearly, the presence of residues 20 to 30 is not essential for formation of flat protein contacts across the twofold axes. Either encapsidated RNA alone is sufficient for their formation, or the protein subunits adjust in such a way that other residues fill the space normally occupied by the ordered peptide arm. The high-resolution structure of the peak IV particles is expected to provide detailed insights into the organization of the protein subunits and RNA at this location. The fact that the particles crystallized isomorphously with wild-type FHV indicates that there are no changes at the surfaces of the mutant particles, an observation that is in agreement with the fact that the deletion was made at a site located in the interior of the capsid. The high-resolution structure may also provide insights into why the mutant particles take up stain more readily than authentic virions.
The most striking observation was the presence of multiple types of assembly products, all of them smaller than native T=3 particles but larger than the anticipated T=1 particles. The assembly products in peak I and its shoulder, for example, were oval or ellipsoidal and resembled short bacilliform structures similar to those observed for alfalfa mosaic virus (AMV). AMV is a multipartite RNA virus that forms particles, whose shapes and sizes are dictated by the size of the encapsidated RNA (6). For example, the small subgenomic RNA4 is packaged into particles that have T=1 symmetry while RNAs 1, 2, and 3, which range in size from 2 to 3.6 kb, are packaged into cylindrical, rod-like structures whose lengths are proportional to the length of the encapsidated nucleic acid. The rod-like particles are composed of multiple rings of coat protein hexamers capped at each end by one-half of a T=1 icosahedral shell such that the cylinders have threefold rotational symmetry along their long axis (Fig. 7).
FIG. 7.
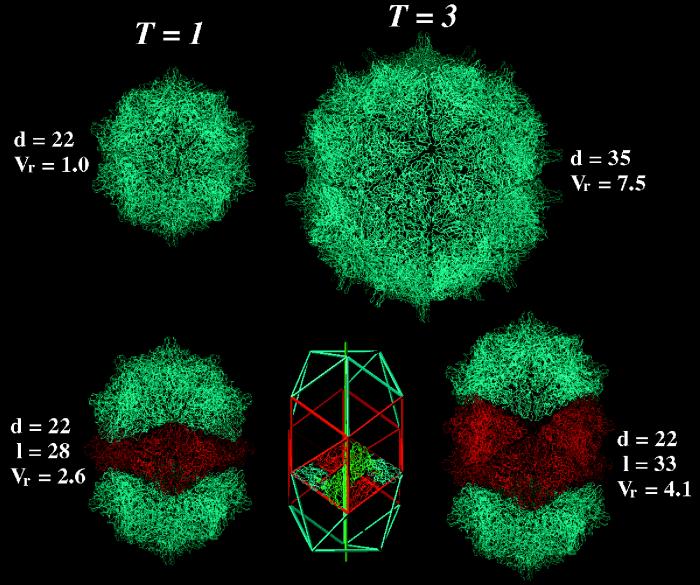
A comparison of the native FHV capsid with hypothetical structures built with Cα coordinates of the coat protein subunit by applying suitable symmetry operations. In the top row at the left is a T=1 particle built from 60 subunits that form 20 trimeric units. Each trimer interacts with its three neighbors via bent contacts. Note that a T=1 particle is significantly smaller than the native T=3 particle shown to the right. Its interior volume (4.35 × 105 Å3) can accommodate approximately 660 nt. The maximal diameter (d) of the particle is given in nanometers. Vr represents the relative interior volumes of the various particles. It was set to 1 for the T=1 particle. In the top row at right is the Cα representation of the native T=3 structure of FHV as determined by X-ray crystallography. This particle, which is shown diagramatically in Fig. 1A, contains 180 protein subunits. In the bottom row at left is the smallest possible bacilliform particle, which consists of one ring of hexameric coat protein subunits (shown in red) and two caps, each representing one-half of a T=1 particle (shown in blue). Note that the line of separation between the two halves of the T=1 particle zigzags and that the caps are rotated relative to each other by 120°. The smaller axis of this particle (d) is identical to the diameter of the T=1 particle shown in the top row, whereas the maximal length (l) of the particle depends on the number of hexameric coat protein rings inserted between the two caps. In the bottom row in the center is a schematic representation of a bacilliform particle containing two rings of hexamers and T=1 icosahedral caps. The organization of the subunits in the hexamer is indicated by the Cα chain trace of the FHV coat protein. Note that the cylindrical portion of the bacilliform particle has threefold rotational symmetry, as is indicated by the green vertical line. In the bottom row at right is a particle built from FHV coat protein subunits and representing the diagram shown in the center of the row.
Our results show that the FHV ΔNterm-31 coat protein, similar to the AMV capsid protein, has the ability to form particles of various sizes and shapes, and we hypothesize that the morphology of the final product is at least in part controlled by the size of the nucleic acid around which the coat protein initially nucleates. Theoretically, the oval particles present in peak I and its shoulder may have an AMV-like architecture. This is shown in Fig. 7, in which the high-resolution structure of the FHV coat protein was modeled into bacilliform particles by using the appropriate symmetry operations. Although no adjustments needed to be made in the conformation of the subunits to avoid an overlap of protein domains, some gaps remain between the icosahedral caps and the cylindrical portion at this point. It is likely that in an actual particle, some adjustments in the subunits occur. A comparison of the predicted dimensions of the models shown in Fig. 7 with the dimensions of the particles shown in the electron micrograph in Fig. 4 indicates that the particles we isolated are smaller in length and diameter than the models. However, negative staining procedures are known to result in some shrinkage of the specimen such that the measured values are artifactually smaller. For comparative purposes, it is thus more appropriate to use the ratio of the values obtained for diameter and length. Our experimentally obtained particles (constant diameter of 20 nm; lengths of 23, 27, and 30 nm) exhibited ratios of 1.15, 1.35, and 1.50. Two of these ratios, 1.35 and 1.50, closely resemble or match the values of 1.27 and 1.50, which correspond to the structures containing either one or two rings of hexamers between the icosahedral caps (Fig. 7). Particles whose dimensions correspond to structures containing three or more rings of hexamers were not detected, perhaps because the protein switches to T=3 icosahedral symmetry when it nucleates around a large-enough RNA.
The internal volumes of the T=1 particle, the bacilliform particles, and the T=3 particle (Fig. 7) may be used to estimate the approximate number of ribonucleotides that can be accommodated within each structure. From the crystal structure of duplex RNA it is known that the volume occupied by one hydrated nucleotide is approximately 655 Å3 (14). Based on this number, a T=3 particle could theoretically encapsidate 4,980 nt. The experimentally observed value for the native T=3 FHV particle, however, is only 4,500 nt (the sum of nucleotides in FHV RNAs 1 and 2). A T=1 particle could theoretically accommodate 660 nt, a bacilliform particle with one ring of protein hexamers could accommodate 1,740 nt, and a bacilliform particle with two rings of coat protein hexamers could accommodate 2,700 nt. These numbers are larger than what we observed experimentally for the particles in peak I, its shoulder, and peak IV (Fig. 5A). However, we may not have been able to detect some of the larger RNAs by ethidium bromide staining if they were present in low amounts. Also, as demonstrated for the native T=3 particle, the experimentally observed values can be significantly smaller than those based on theoretical calculations, indicating that much smaller RNAs allow formation of a closed protein shell.
Obviously, further experiments are required to confirm that the oval particles have the general architecture of bacilliform particles. Crystallization and X-ray analysis are a possible strategy, but they have not been successful for AMV. Other procedures such as light scattering and X-ray solution scattering might provide insights into the molecular properties of the particles such as their exact mass and dimensions. It may also be possible to use cryo-electron microscopy and image reconstruction to generate a low-resolution map of the particle into which the known structure of the FHV coat protein could then be modeled.
In addition to the oval particles in peak I and the wild-type-like particles in peak IV, we observed other types of assembly products whose appearance did not fit the predicted models in Fig. 7 or the structure of native FHV. The architecture of these particles, in particular those present in peaks II and III, appeared ill defined and possibly represented intermediates between particles with quasi-icosahedral symmetry, i.e., bacilliform architecture, and true T=3 icosahedral symmetry. Such types of particles have been observed for tobacco streak virus (7), which belongs to the same family as AMV. Tobacco streak virus particles lack a well-defined surface lattice and are thought to contain pentameric and hexameric assembly units at random positions on their capsids. Similarly, FHV ΔNterm-31 coat protein may form particles whose capsids lack a precise geometric definition in addition to the cylindrical and isometric forms described above.
Last, during overexpression of the protein in T. ni cells, we observed for the first time assembly products that did not contain nucleic acid and that appeared to be partial shells and shells that were incorrectly closed. These products were reminiscent of the coat protein spirals observed for bacteriophage P22 capsid protein assembled in the absence of scaffolding protein (3). Perhaps in FHV the RNA plays a role similar to that of scaffolding protein in P22.
In summary, our results show that the N terminus of the FHV coat protein plays a critical role in viral assembly. Its positively charged character is almost certainly required for neutralizing the negatively charged RNA as it is encapsidated into virions. Residues 20 to 30, on the other hand, do not appear to be required for formation of flat contacts across the icosahedral twofold axes of the T=3 particles. Nonetheless, when residues 1 to 31 are absent, the assembly process loses its precision and large amounts of aberrant particles are synthesized. These particles are dead-end products, since most of them are too small to encapsidate the normal complement of FHV RNAs. How the N terminus regulates assembly such that only T=3 particles are formed is currently not understood, but the high-resolution structure of the ΔNterm-31 T=3 particles may provide a possible explanation.
ACKNOWLEDGMENTS
We thank Dawn Marshall for excellent technical assistance and Vijay Reddy for help in generating Fig. 1.
This work was supported by NIH grants GM53491 (A.S.), GM34220 (J.E.J.), and GM54076 (J.E.J.).
Footnotes
Manuscript no. 11439MB from The Scripps Research Institute.
REFERENCES
- 1.Dasmahapatra B, Dasgupta R, Ghosh A, Kaesberg P. Structure of the black beetle virus genome and its functional implications. J Mol Biol. 1985;182:183–189. doi: 10.1016/0022-2836(85)90337-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davis T R, Wickham T J, McKenna K A, Granados R R, Shuler M L, Wood H A. Comparative recombinant protein production of eight insect cell lines. In Vitro Cell Dev Biol Anim. 1993;29A:388–390. doi: 10.1007/BF02633986. [DOI] [PubMed] [Google Scholar]
- 3.Earnshaw W, King J. Structure of phage P22 coat protein aggregates formed in the absence of the scaffolding protein. J Mol Biol. 1978;126:721–747. doi: 10.1016/0022-2836(78)90017-7. [DOI] [PubMed] [Google Scholar]
- 4.Erickson J W, Rossmann M G. Assembly and crystallization of a T = 1 icosahedral particle from trypsinized southern bean mosaic virus coat protein. Virology. 1982;116:128–136. doi: 10.1016/0042-6822(82)90408-1. [DOI] [PubMed] [Google Scholar]
- 5.Fisher A J, Johnson J E. Ordered duplex RNA controls capsid architecture in an icosahedral animal virus. Nature. 1993;361:176–179. doi: 10.1038/361176a0. [DOI] [PubMed] [Google Scholar]
- 6.Francki R I B, Milne R G, Hatta T. Atlas of plant viruses. Vol. 2. Boca Raton, Fla: CRC Press; 1985. pp. 93–102. [Google Scholar]
- 7.Francki R I B, Milne R G, Hatta T. Atlas of plant viruses. Vol. 2. Boca Raton, Fla: CRC Press; 1985. pp. 81–91. [Google Scholar]
- 8.Friesen P D, Rueckert R R. Synthesis of black beetle virus proteins in cultured Drosophila cells: differential expression of RNAs 1 and 2. J Virol. 1981;37:876–886. doi: 10.1128/jvi.37.3.876-886.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gallagher T, Rueckert R R. Assembly-dependent maturation cleavage in provirions of a small icosahedral insect ribovirus. J Virol. 1988;62:3399–3406. doi: 10.1128/jvi.62.9.3399-3406.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harrison S C. Protein interfaces and intersubunit bonding. The case of tomato bushy stunt virus. Biophys J. 1980;32:139–153. doi: 10.1016/S0006-3495(80)84930-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hosur M V, Schmidt T, Tucker R C, Johnson J E, Gallagher T M, Selling B H, Rueckert R R. Structure of an insect virus at 3.0 Å resolution. Protein Struct Funct Genet. 1987;2:167–176. doi: 10.1002/prot.340020302. [DOI] [PubMed] [Google Scholar]
- 12.Imai Y, Matsushima Y, Sugimura T, Terada M. A simple and rapid method for generating a deletion by PCR. Nucleic Acids Res. 1991;19:2785. doi: 10.1093/nar/19.10.2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson J E. Functional implications of protein-protein interactions in icosahedral viruses. Proc Natl Acad Sci USA. 1996;93:27–33. doi: 10.1073/pnas.93.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson J E, Rueckert R R. Packaging and release of the viral genome. In: Chiu W, Burnett R M, Garcea R L, editors. Structural biology of viruses. New York, N.Y: Oxford University Press; 1997. pp. 269–287. [Google Scholar]
- 15.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 16.Leberman R, Finch J T. The structure of turnip crinkle and tomato bushy stunt viruses. I. A small protein particle derived from turnip crinkle virus. J Mol Biol. 1970;50:209–213. doi: 10.1016/0022-2836(70)90186-5. [DOI] [PubMed] [Google Scholar]
- 17.Liddington R C, Yan Y, Moulai J, Sahli R, Benjamin T L, Harrison S C. Structure of simian virus 40 at 3.8Å resolution. Nature. 1991;354:278–284. doi: 10.1038/354278a0. [DOI] [PubMed] [Google Scholar]
- 18.Marchuk D, Drumm M, Saulino A, Collins F S. Construction of T-vector, a rapid and general system for direct cloning of unmodified PCR products. Nucleic Acids Res. 1990;19:1154. doi: 10.1093/nar/19.5.1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Olson A J, Bricogne G, Harrison S C. Structure of tomato bushy stunt virus IV. The virus particle at 2.9Å resolution. J Mol Biol. 1983;171:61–93. doi: 10.1016/s0022-2836(83)80314-3. [DOI] [PubMed] [Google Scholar]
- 20.Otwinowski Z, Minor W. Processing of X-ray diffraction in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 21.Reddy V S, Giesing H A, Morton R T, Kumar A, Post C B, Brooks III C L, Johnson J E. Energetics of quasiequivalence: computational analysis of protein-protein interactions in icosahedral viruses. Biophys J. 1998;74:546–558. doi: 10.1016/S0006-3495(98)77813-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schneemann A, Dasgupta R, Johnson J E, Rueckert R. Use of recombinant baculovirus in synthesis of morphologically distinct viruslike particles of flock house virus, a nodavirus. J Virol. 1993;67:2756–2763. doi: 10.1128/jvi.67.5.2756-2763.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schneemann A, Reddy V, Johnson J E. The structure and function of nodavirus particles: a paradigm for understanding chemical biology. Adv Virus Res. 1998;50:381–446. doi: 10.1016/s0065-3527(08)60812-x. [DOI] [PubMed] [Google Scholar]
- 25.Schneemann A, Zhong W, Gallagher T M, Rueckert R R. Maturation cleavage required for infectivity of a nodavirus. J Virol. 1992;66:6728–6734. doi: 10.1128/jvi.66.11.6728-6734.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schneider P A, Schneemann A, Lipkin W I. RNA splicing in Borna disease virus, a nonsegmented, negative-strand RNA virus. J Virol. 1994;68:5007–5012. doi: 10.1128/jvi.68.8.5007-5012.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Silva A M, Rossmann M G. Refined structure of southern bean mosaic virus at 2.9 Å resolution. J Mol Biol. 1987;197:69–87. doi: 10.1016/0022-2836(87)90610-3. [DOI] [PubMed] [Google Scholar]
- 28.Sorger P K, Stockley P G, Harrison S C. Structure and assembly of turnip crinkle virus II. Mechanism of reassembly in vitro. J Mol Biol. 1986;191:639–658. doi: 10.1016/0022-2836(86)90451-1. [DOI] [PubMed] [Google Scholar]
- 29.Speir J A, Munshi S, Wang G, Baker T S, Johnson J E. Structures of the native and swollen forms of cowpea chlorotic mottle virus determined by X-ray crystallography and cryo electron microscopy. Structure. 1995;3:63–78. doi: 10.1016/s0969-2126(01)00135-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wickham T J, Davis T, Granados R R, Shuler M L, Wood H A. Screening of insect cell lines for the production of recombinant proteins and infectious virus in the baculovirus expression system. Biotechnol Prog. 1992;8:391–396. doi: 10.1021/bp00017a003. [DOI] [PubMed] [Google Scholar]
- 31.Zlotnick A, Reddy V S, Dasgupta R, Schneemann A, Ray W J, Rueckert R R, Johnson J E. Capsid assembly in a family of animal viruses primes an autoproteolytic maturation that depends on a single aspartic acid residue. J Biol Chem. 1994;269:13680–13684. [PubMed] [Google Scholar]