Abstract
The intracellular region (RAMIC) of the mouse Notch1 receptor interacts with RBP-J/CBF-1, which binds to the DNA sequence CGTGGGAA and suppresses differentiation by transcriptional activation of genes regulated by RBP-J. Epstein-Barr virus nuclear antigen 2 (EBNA2) is essential for immortalization of human B cells by the virus. EBNA2 is a pleiotropic activator of viral and cellular genes and is targeted to DNA at least in part by interacting with RBP-J. We found that EBNA2 and the Notch1 RAMIC compete for binding to RBP-J, indicating that their interaction sites on RBP-J overlap at least partially. EBNA2 and Notch1 RAMIC transactivated the same set of viral and host promoters, i.e., the EBNA2 response element of the Epstein-Barr virus TP1 and the HES-1 promoter. Furthermore, EBNA2 functionally replaced the Notch1 RAMIC by suppressing differentiation of C2C12 myoblast progenitor cells.
RBP-J/CBF-1 is a ubiquitous DNA binding protein which recognizes the sequence CGTGGGAA (9, 17, 18). Suppressor of Hairless, the Drosophila homolog of RBP-J, is involved in signal transduction of the receptor Notch, which plays a pivotal role in cell fate determination of various lineages including nerve, muscle, and germ cells (1, 20). Interaction of Notch with the ligand Delta results in differentiation suppression of progenitor cells of various lineages. The intracellular region (RAMIC) of Notch has been shown to have transactivation activity when overexpressed in various cell lines (10–14). RAMIC directly binds the nuclear protein RBP-J at two regions: the RAM domain located immediately C terminus to the transmembrane region (22) and the CDC 10/ankyrin repeats (2, 14). The RAM domain appears to compete with an endogenous corepressor of RBP-J (14), while the role of the ankyrin repeats is not clear. The transactivation activity of RAMIC is responsible for suppression activity of muscle differentiation because two activities of RAMIC and its mutants are correlated (14). Mammalian Notch has four family members, all of which interact with RBP-J through the RAM domain (11, 13). Chromosomal translocations that cause expression of the truncated form of human Notch (TAN-1) are found in a subset of acute human T-cell lymphoblastic lymphoma (5).
RBP-J is also involved in transcriptional regulation by Epstein-Barr virus nuclear antigen 2 (EBNA2) which is essential for transformation of human B and occasionally T cells by the virus. EBNA2, a pleiotropic activator of viral and cellular genes, is unable to bind to DNA by itself but is targeted to DNA at least in part by interacting with RBP-J (6, 9, 10, 16, 25, 29). RBP-J also contains a family member called RBP-L, which is almost exclusively expressed in the lung and is marginally expressed in the brain and spleen (18). Although RBP-L does not have a strong interaction with Notch1, -2, -3, or -4, RBP-L showed functional interaction with EBNA2. Although EBNA2 and RAMIC are structurally distinct, both interact with the same DNA binding protein (RBP-J) and can stimulate proliferation of cells. However, it is not known whether EBNA2 and RAMIC share other functions such as suppression of differentiation.
We compared RAMIC and EBNA2 for activities of RBP-J binding, transactivation, and myogenic suppression. We found that RAMIC and EBNA2 that can transactivate promoters carrying RBP-J binding motifs in COS cells suppress myogenesis, whereas mutants of EBNA2 that are transactivation incompetent cannot block myogenesis. The mutagenesis experiments indicate that the transactivation activities of EBNA2 are mediated by RBP-J. The results indicate that RAMIC and EBNA2 have similar biological functions.
MATERIALS AND METHODS
DNA binding assay.
We examined the DNA binding activity of each RBP-J mutant by electrophoresis mobility shift assay (EMSA) essentially as described previously (3) except that in vitro-translated mutant proteins rather than in vivo transfection products were used. Although we essentially confirmed the previous conclusion that the N region (residues 212 to 227) and C region (residues 275 to 323) are important for DNA binding, the relative DNA binding activity of each mutant was considerably different from that previously reported. The difference is likely due to small amounts of endogenous RBP-J protein and quantitation errors for the RBP-J mutant proteins in the COS7 transfection system, as the present method allowed for more accurate quantitation of the input protein.
Construction of plasmids. (i) Plasmids used for yeast two-hybrid assay.
After digestion of the CDM8-RBP-J deletion constructs (23) with EcoRI and BamHI, the fragments were isolated and inserted into the EcoRI-BamHI sites of the pGBT9 vector (CLONTECH Laboratories, Inc.) and the pBluescript II vector. To make the construct with the GAL4 activation domain (ad) fused to EBNA2, the RBP-J interacting region of EBNA2 was excised from pU294-6 (28) by digestion with BamHI and the fragment was inserted in frame into the pGAD424 vector (CLONTECH Laboratories, Inc.). The GAL4(ad)-RAM23 construct and glutathione S-transferase (GST) fusion plasmids of RAM23 and EBNA2 were previously described (22, 28).
(ii) Plasmids used for transactivation and differentiation assay.
pEF-BOSneo-RAMIC was constructed from pCS2+MTmNotchIC (15) and RAM23 (22). RAM23 fragments synthesized by PCR were flanked by a c-Myc tag (22). All of these fragments were cloned into pEF-BOSneo vector (19). pEF-BOSneo-RBP-J or pEF-BOSneo-RBP(R218H) was constructed from CDM8-RBP-J or CDM8-RBP(R218H), respectively (3), in the pEF-BOSneo vector. EBNA2 expression plasmid was constructed in the SG5 vector (Stratagene) (16). pGa981-6 contains the hexamerized 50-bp EBNA2 response element of the TP-1 promoter in front of the minimal β-globin promoter driving the luciferase gene. HES-1-luc contains the −194 to +60 promoter fragment of the HES-1 gene (21) cloned upstream of the luciferase gene in the pGV-B basic vector (TOYO-INKI Co Ltd., Tokyo, Japan). pSG5-EBNA2 WW323SS was described previously (18). Original EBNA2 WW323SS was obtained from E. Kieff (27). For pSG5-EBNA2 ΔHincII-StuI, a 657-bp fragment between the HincII and StuI sites (residues 19 to 237) within the coding region of EBNA2 was removed.
Protein-protein interaction assay.
The yeast two-hybrid assay was done according to standard procedures (24). Coprecipitation experiments using GST-fusion proteins were carried out as described previously (22). The amounts of GST-fusion proteins were monitored by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis of Sepharose-GST-fusion protein complexes, and equal amounts were used for each set of experiments. Bound proteins were released by boiling in sample loading buffer at 95°C for 5 min. The samples were run on an SDS-polyacrylamide gel. The gel was dried with a gel drier, and the signals on the gel were analyzed using an Imaging Analyzer BAS2000 (Fuji Film Co. Ltd.).
Transactivation assay.
COS7 cells were transfected in six-well plates by the lipofection method with Lipofectamine (GIBCO BRL) and 0.5 μg of luciferase vector alone or in combination with 1.0 μg of pSG5-EBNA2 or pEF-BOSneo-RAMIC and various amounts (1.0 or 2.0 μg) of pEF-BOSneo-RBP(R218H) or pEF-BOSneo-RAM23 construct; 250 ng of SV40-lacZ construct was included in each transfection as an internal control. Three independent experiments were carried out. The luciferase activity was measured 48 h after transfection by using a LumatLB9501 luminometer (Berthold, Wildbach) and was normalized according to the β-galactosidase activity; the induction of luciferase activity was calculated as the ratio of the luciferase activity of the reporter plasmid containing the RBP-J binding site to that of a negative control plasmid which has no RBP-J binding site.
C2C12 cells were transfected with expression plasmids for RAMIC and EBNA2 and its derivatives. Differentiation induction and differentiation suppresion assays were carried out as described previously (14). EBNA2-related proteins and RAMIC were stained with the anti-EBNA2 monoclonal antibody PE2 (YLEM, Rome, Italy) and anti-Myc monoclonal antibody, respectively, and the antibodies were detected by DTAF-labeled anti-mouse immunoglobulin G antibody (Chemicon International, Inc.). Myoglobin expression was monitored by staining the cells with anti-myoglobin polyclonal antibody (Nichirei) and Texas Red-labeled anti-rabbit immunoglobulin G antibodies (Organon Teknika N.V.-Capped Products).
RESULTS AND DISCUSSION
Notch1 and EBNA2 interact with similar but not identical regions of RBP-J.
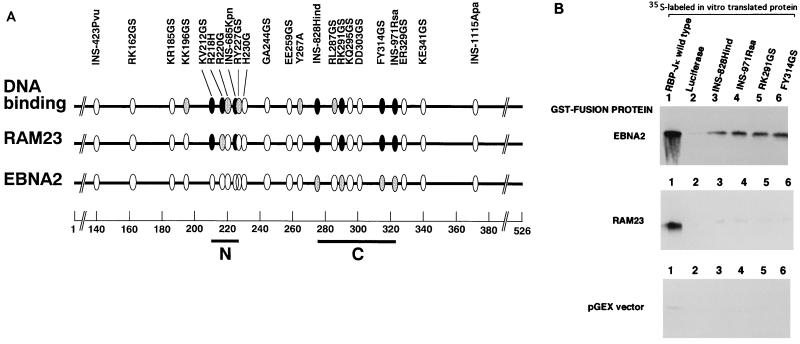
To identify regions of RBP-J that interact with EBNA2 and RAMIC of mouse Notch 1, a series of RBP-J mutants was tested in the yeast two-hybrid system for interaction with wild-type EBNA2 and the RAM domain (RAM23) which is the major RBP-J binding site of Notch (22). Experiments using RBP-J deletion mutants indicated that both EBNA2 and RAM23 interacted with the central portion (amino acids 196 to 372) of RBP-J (data not shown) that contains the N and C regions previously shown to be essential for DNA binding (3). These results are in general agreement with reports by others (10, 11). Subsequent experiments using replacement mutants in the N and C regions of RBP-J showed that most of the replacement mutations that lost DNA binding activity either lost or had a markedly reduced ability to interact with RAM23 (Fig. 1A). The same mutations in the C region also reduced EBNA2 binding activity but to a less extent. On the other hand, the mutations in the N region did not affect EBNA2 binding activity.
FIG. 1.
Interaction regions of RBP-J with EBNA2 and RAM23. (A) Positions of the point and insertional mutations in RBP-J (3) are shown by ellipses. N (residues 212 to 227) and C (residues 275 to 323) regions shown by bars are DNA binding regions and are slightly enlarged compared with those from a previous study (3). Residue numbers are shown at the bottom. The top line shows DNA binding activity relative to that of the wild type. The second and third lines indicate the relative interacting abilities of the mutants with RAM23 and EBNA2, respectively. Both yeast two-hybrid assays and coprecipitation experiments using GST-fusion proteins were carried out to measure interaction activities. Closed ellipse, less than 5% of that of the wild type; shaded ellipse, less than 15% of that of the wild type; open ellipse, more than 50% of that of the wild type. (B) In vitro interactions of 35S-labeled products of RBP-J wild type (lane 1), luciferase (lane 2, negative control), or RBP-J mutants (lanes 3 to 6) with GST-RAM23, GST-EBNA2, or GST (pGEX) vector. The same amounts of the wild type and mutant forms of RBP-J were used.
To confirm the results of the yeast two-hybrid assay, four mutants (INS-828Hind, INS-971Rsa, RK291GS, and FY314GS), which had partly or completely lost the ability to interact with EBNA2 and RAM23, were also tested in coprecipitation experiments using GST-fusion proteins of RAM23 and EBNA2 (Fig. 1B). GST-fusion proteins were immobilized on glutathione-Sepharose beads and tested for their ability to bind 35S-labeled RBP-J mutants. None of the four mutants coprecipitated with GST-RAM23, and all bound to GST-EBNA2 albeit weakly (about 10% of that of the wild type), in agreement with the yeast two-hybrid assay. The results of the yeast two-hybrid assay of the N region mutants were also confirmed by the GST pull down assay: these mutants lost or had markedly reduced binding to RAM23 but not to EBNA2 (data not shown). These results suggest that the C region of RBP-J appears to be involved in the interaction with both EBNA2 and RAM23 whereas the N region may be involved in RAM23 interaction only. Differences in the interaction sites of RBP-J to Notch and EBNA2 were also reported for a mutant with mutations at residues 275 to 277 of human RBP-J (11).
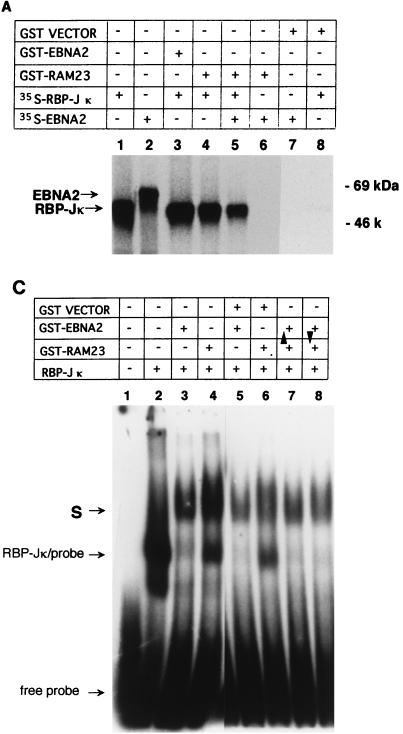
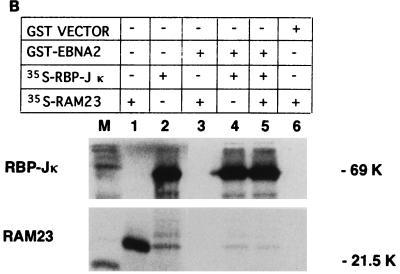
Since the interaction regions of RBP-J with RAM23 and EBNA2 overlap but are not identical, we tested whether these three molecules were able to form a ternary complex in two in vitro systems. First, a coprecipitation assay using 35S-labeled in vitro translation products and GST-fusion proteins was performed. When 35S-labeled products of RBP-J and EBNA2 were mixed with GST-RAM23, only 35S-RBP-J and not 35S-EBNA2 was coprecipitated with RAM23 (Fig. 2A, lane 5). Similarly, when 35S-RBP-J, 35S-RAM23, and GST-EBNA2 were mixed, GST-EBNA2 formed a complex with RBP-J only (Fig. 2B, lane 5). We were also unable to see evidence of a ternary complex among RBP-J, RAM23, and EBNA2 by EMSA. Either GST-EBNA2 (Fig. 2C, lane 3) or GST-RAM23 (lane 4) supershifted the RBP-J-DNA complex. However, the addition of both GST-EBNA2 and GST-RAM23 did not further supershift the EBNA2 (or RAM23)-RBP-J-DNA complex (Fig. 2C, lanes 7 and 8). These results exclude the formation of a ternary complex among RBP-J, RAM23, and EBNA2. Such a complex does not form probably because EBNA2 and RAM23 share at least some of the binding sites of RBP-J and compete with each other for binding to RBP-J.
FIG. 2.
Competition of RAM23 and EBNA2 for their interaction with RBP-J. (A) GST-RAM23 coprecipitates 35S-labeled RBP-J but not 35S-labeled EBNA2. Added samples in each lane are shown above. 35S-labeled in vitro-translated RBP-J (lane 1) or EBNA2 (lane 2) was clearly distinguished by size. Samples in lanes 3 to 8 were coprecipitated as described previously (22). (B) GST-EBNA2 coprecipitates 35S-RBP-J but not 35S-RAM23. Added samples in each lane are shown above. Samples in lanes 3 to 6 were coprecipitated. (C) EMSA for ternary complex formation among EBNA2, RAM23, and RBP-J. EMSA was carried out as previously described (7) with 2 ng of 32P-labeled Epstein-Barr virus Cp promoter probe (16). Added samples other than DNA probe are listed above. Arrowheads indicate the sample added at the end. Sequential addition of both GST-EBNA2 and GST-RAM23 did not change the mobility of the supershifted band with either GST-RAM23 or GST-EBNA2 alone (lanes 7 and 8). Neither GST-RAM23 nor GST-EBNA2 formed a complex with the probe (data not shown). Note that the complex of RBP-J and GST-EBNA2 migrates slightly faster than that of RBP-J and GST-RAM23. S, supershifted band.
EBNA2 and RAMIC transactivate the same set of promoters.
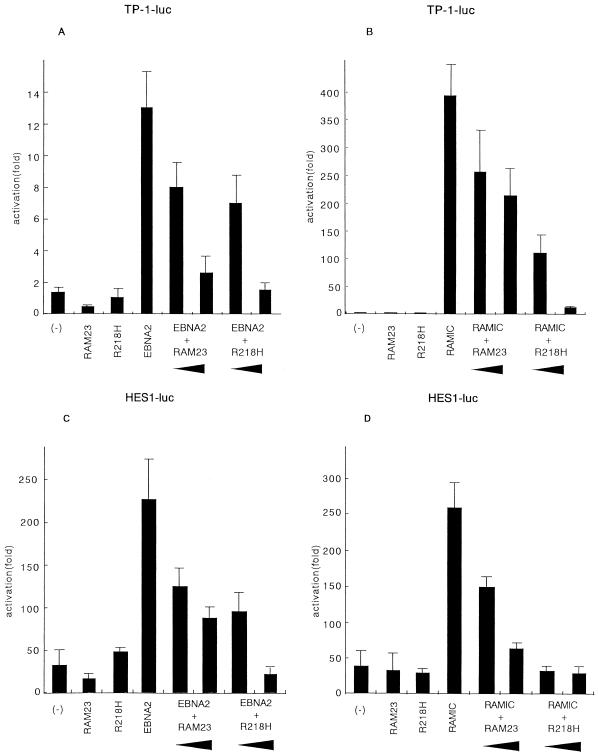
Since both EBNA2 and mouse Notch1 RAMIC require RBP-J for binding to DNA and interact with somewhat similar regions of RBP-J, EBNA2 and RAMIC may transactivate similar target genes. To test this possibility, we used two typical promoters, the EBNA2 response element of the Epstein-Barr virus TP1 (ERE-TP1) promoter and the HES-1 promoter, which have been shown to be regulated by EBNA2 (29) and Notch1 (12), respectively. RAMIC as well as EBNA2 transactivated the ERE-TP1 promoter by cotransfection of COS7 cells (Fig. 3A and B) in agreement with our previous report (13). Similarly, we found that both EBNA2 and RAMIC transactivated the HES-1 promoter (Fig. 3A and B). In both cases endogenous RBP-J appears to be involved in transactivation by EBNA2 and RAMIC because the EBNA2 activities were blocked by competition with either RAM23 or a mutant of RBP-J (R218H) that cannot bind to DNA. The RAM domain would compete for binding to endogenous RBP-J while the R218H mutant would compete for binding to RAMIC, EBNA2, or unknown cofactors with endogenous RBP-J. The transactivation activity of RAMIC was also blocked by RAM23.
FIG. 3.
Transactivation of the viral and host genes by EBNA2 or RAMIC. COS7 cells were transfected with a luciferase reporter plasmid alone (-) or together with various expression vectors [pEF-BOSneo-RAM23, pEF-BOSneoRBP(R218H), pSG5-EBNA2, pEF-BOSneo-RAMIC] as indicated. A total of 1.0 μg of pSG5-EBNA2 (A and C) or pEF-BOSneo-RAMIC (B and D) per 3.5-cm-diameter dish was introduced into COS7 cells together with a reporter plasmid ERE-TP1-luc (pGa981-6) (A and B) or HES1-luc (C and D) and increasing amounts (1 or 2 μg/dish) of pEF-BOSneo-RAM23 or pEF-BOSneoRBP(R218H) DNA as indicated by bottoms of arrowheads. The luciferase reporter plasmid/pSG5-EBNA2/pEF-BOSneo-RAMIC ratio was 1/2/2 for all cotransfection experiments. Each transfection assay was carried out in comparison with a negative control reporter, ptk-luc177 (28) against pGa981-6 or pGV-B vector (TOYO-INKI Inc.) against HES1-luc. Error bars indicate standard deviations.
Previously, EBNA2 mutants which lost association activity with RBP-J were shown to have no transactivation activity (6, 9, 10, 25, 29). One such mutant, WW323SS, was confirmed to have little, if any, transactivation activity with either promoter used in the present study (data not shown). We also found no transactivation activity in the EBNA2 ΔHincII-StuII mutant, which has a deletion from residues 19 to 237.
EBNA2 and RAMIC suppress differentiation of muscle progenitor cells.
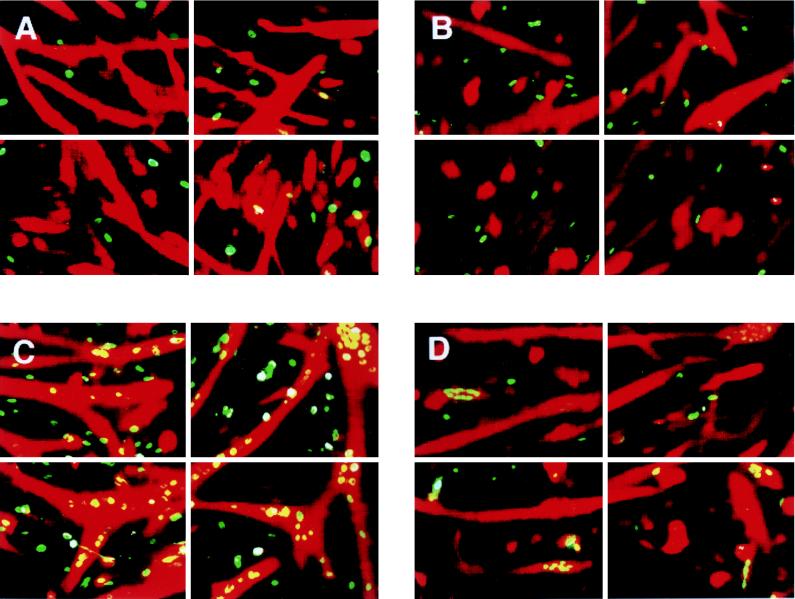
We then investigated whether EBNA2 can replace the biological function of RAMIC in suppressing differentiation of the myogenic cell line C2C12 into myotubes in a differentiation induction medium (15). C2C12 cells were transfected by RAMIC, EBNA2, or EBNA2 mutant constructs. To quantitate differentiation suppression activity, we carried out two color-staining experiments that detect expression of both transgene and myoglobin as differentiation markers. Such experiments clearly showed that EBNA2 could suppress the muscle cell differentiation of C2C12 cells as strongly as RAMIC (Fig. 4A and B). Examination of 277 EBNA2-expressing nuclei showed that the level of C2C12 cell differentiation was suppressed to 17% of that of nontransfected cells. Previously, we showed that RAMIC expression suppressed C2C12 differentiation to 28% of that of the control (14). However, the EBNA2 WW323SS mutant (27), which cannot bind RBP-J, showed very weak, if any, suppressive effect on C2C12 cell differentiation (Fig. 4C). Another EBNA2 mutant (EBNA2 ΔHincII-StuI) which has no transactivation activity also showed no differentiation suppression activity (Fig. 4D). Again, quantitation of differentiation suppression was carried out by examining 452 or 392 nuclei expressing EBNA2 ΔHincII-StuI or EBNA2 WW323SS, respectively, and comparing the number of myoglobin-expressing cells with that in nontransfected cells. Differentiation of C2C12 cells expressing EBNA2 ΔHincII-StuI or EBNA2 WW323SS was 100 or 90.6%, respectively, of that of the control cells. These results indicate that EBNA2 also suppresses myogenic differentiation as effectively as the active form of Notch1.
FIG. 4.
Suppression of myogenic differentiation of C2C12 cells by RAMIC and EBNA2 mutants. RAMIC (A), EBNA2 (B), and EBNA2 mutant proteins (C and D) are stained green and differentiated cells, judged by myoglobin expression, are stained red. Note that nuclei of transfected cells that underwent differentiation look yellow because of overlap with the red staining of myoglobin. Green nuclei indicate expression of RAMIC or EBNA2 or EBNA 2 mutants without differentiation into myotube. Differentiation of C2C12 cells is suppressed strongly with RAMIC (A) and EBNA2 (B). No or weak, if any, differentiation suppression was observed for EBNA2 WW323SS (C) and EBNA2 ΔHincII-StuI (D).
Our experiments have shown that EBNA2 and RAMIC are biologically equivalent in transactivation of viral and cellular genes as well as suppression of myogenic cell differentiation. Since differentiation suppression by Notch is likely to be due to gene activation, the two proteins may transactivate a similar, if not identical, set of genes by interaction with the RBP-J DNA binding protein. In addition, both EBNA2 and the truncated form of Notch are involved in transformation of lymphocytes (4, 5, 8). Is EBNA2 the functional homolog of Notch? Functional homologs of biologically important host molecules are encoded by several viral genomes; proteins involved in cell death regulation are good examples (26). Obviously, EBNA2 can mimic only a part of the Notch function because EBNA2 lacks the extracellular region. In addition, the two proteins may not always transactivate identical genes because (i) different accessory proteins may be involved in their transactivation, (ii) the interaction sites on RBP-J are similar but not identical, and (iii) EBNA2 and Notch1 may be engaged in other pathways unrelated to RBP-J. With all these differences in mind, comparison of various aspects of RAMIC and EBNA2 is still important and informative for the elucidation of mechanisms of transformation associated with EBNA2 as well as for understanding the role of Notch in stem cell maintenance.
ACKNOWLEDGMENTS
We thank E. Kieff for providing the EBNA2 WW323SS mutant, S. D. Hayward for providing the GST-fusion EBNA2 expression plasmid, R. Kopan for providing the mNotch1 cDNA, and R. Kageyama for providing the HES-1 promoter DNA.
This investigation was supported by grants for the COE program from the Ministry of Education, Science, Sports and Culture of Japan and from the Deutsch Forschungsgemeinshaft (Str 461/1-1).
REFERENCES
- 1.Artavanis-Tsakonas S, Matsuno K, Fortini M E. Notch signaling. Science. 1995;268:225–232. doi: 10.1126/science.7716513. [DOI] [PubMed] [Google Scholar]
- 2.Aster J C, Robertson E S, Hasserjian R B, Turner J R, Kieff E, Sklar J. Oncogenic forms of NOTCH1 lacking either the primary binding site for RBP-Jκ or nuclear localization sequences retain the ability to associate with RBP-Jκ and activate transcription. J Biol Chem. 1997;272:11336–11343. doi: 10.1074/jbc.272.17.11336. [DOI] [PubMed] [Google Scholar]
- 3.Chung C N, Hamaguchi Y, Honjo T, Kawaichi M. Site-directed mutagenesis study on DNA binding regions of the mouse homologue of Suppressor of Hairless, RBP-Jκ. Nucleic Acids Res. 1994;22:2938–2944. doi: 10.1093/nar/22.15.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cohen J I, Wang F, Mannick J, Kieff E. Epstein-Barr virus nuclear protein 2 is a key determinant of lymphocyte transformation. Proc Natl Acad Sci USA. 1989;86:9558–9562. doi: 10.1073/pnas.86.23.9558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ellisen L W, Bird J, West D C, Soreng A L, Reynolds T C, Smith S D, Sklar J. TAN-1, the human homolog of the Drosophila Notch gene, is broken by chromosomal translocations in T lymphoblastic neoplasms. Cell. 1991;66:649–661. doi: 10.1016/0092-8674(91)90111-b. [DOI] [PubMed] [Google Scholar]
- 6.Grossman S R, Johannsen E, Tong X, Yalamanchili R, Kieff E. The Epstein-Barr virus nuclear antigen 2 transactivator is directed to response elements by the Jκ recombination signal binding protein. Proc Natl Acad Sci USA. 1994;91:7568–7572. doi: 10.1073/pnas.91.16.7568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hamaguchi Y, Matsunami N, Yamamoto Y, Honjo T. Purification and characterization of a protein that binds to the recombination signal sequence of the immunoglobulin Jκ segment. Nucleic Acids Res. 1989;17:9015–9026. doi: 10.1093/nar/17.22.9015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hammerschmidt W, Sugden B. Genetic analysis of immortalizing functions of Epstein-Barr virus in human B lymphocytes. Nature. 1989;340:393–397. doi: 10.1038/340393a0. [DOI] [PubMed] [Google Scholar]
- 9.Henkel T, Ling P D, Hayward S D, Peterson M G. Mediation of Epstein-Barr virus EBNA2 transactivation by recombination signal-binding protein Jκ. Science. 1994;265:92–95. doi: 10.1126/science.8016657. [DOI] [PubMed] [Google Scholar]
- 10.Hsieh J J, Hayward S D. Masking of the CBF1/RBPJκ transcriptional repression domain by Epstein-Barr virus EBNA2. Science. 1995;268:560–563. doi: 10.1126/science.7725102. [DOI] [PubMed] [Google Scholar]
- 11.Hsieh J J, Nofziger D E, Weinmaster G, Hayward S D. Epstein-Barr virus immortalization: Notch2 interacts with CBF1 and blocks differentiation. J Virol. 1997;71:1938–1945. doi: 10.1128/jvi.71.3.1938-1945.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jarriault S, Brou C, Logeat F, Schroeter E H, Kopan R, Israel A. Signalling downstream of activated mammalian Notch. Nature. 1995;377:355–358. doi: 10.1038/377355a0. [DOI] [PubMed] [Google Scholar]
- 13.Kato H, Sakai T, Tamura K, Minoguchi S, Shirayoshi Y, Hamada Y, Tsujimoto Y, Honjo T. Functional conservation of mouse Notch receptor family members. FEBS Lett. 1996;395:221–224. doi: 10.1016/0014-5793(96)01046-0. [DOI] [PubMed] [Google Scholar]
- 14.Kato H, Taniguchi Y, Kurooka H, Minoguchi S, Sakai T, Nomura-Okazaki S, Tamura K, Honjo T. Involvement of RBP-J in biological functions of mouse Notch1 and its derivatives. Development. 1997;124:4133–4141. doi: 10.1242/dev.124.20.4133. [DOI] [PubMed] [Google Scholar]
- 15.Kopan R, Nye J S, Weintraub H. The intracellular domain of mouse Notch: a constitutively activated repressor of myogenesis directed at the basic helix-loop-helix region of MyoD. Development. 1994;120:2385–2396. doi: 10.1242/dev.120.9.2385. [DOI] [PubMed] [Google Scholar]
- 16.Ling P D, Rawlins D R, Hayward S D. The Epstein-Barr virus immortalizing protein EBNA-2 is targeted to DNA by a cellular enhancer-binding protein. Proc Natl Acad Sci USA. 1993;90:9237–9241. doi: 10.1073/pnas.90.20.9237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matsunami N, Hamaguchi Y, Yamamoto Y, Honjo T. A protein binding to the Jκ recombination sequence of immunoglobulin genes contains a sequence related to the integrase motif. Nature. 1989;342:934–937. doi: 10.1038/342934a0. [DOI] [PubMed] [Google Scholar]
- 18.Minoguchi S, Taniguchi Y, Kato H, Okazaki T, Strobl L J, Zimber-Strobl U, Bornkamm G W, Honjo T. RBP-L, a transcription factor related to RBP-Jκ. Mol Cell Biol. 1997;17:2679–2687. doi: 10.1128/mcb.17.5.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mizushima S, Nagata S. pEF-BOS, a powerful mammalian expression vector. Nucleic Acids Res. 1990;18:5322. doi: 10.1093/nar/18.17.5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Muskavitch M A. Delta-notch signaling and Drosophila cell fate choice. Dev Biol. 1994;166:415–430. doi: 10.1006/dbio.1994.1326. [DOI] [PubMed] [Google Scholar]
- 21.Takebayashi K, Sasai Y, Sakai Y, Watanabe T, Nakanishi S, Kageyama R. Structure, chromosomal locus, and promoter analysis of the gene encoding the mouse helix-loop-helix factor HES-1. Negative autoregulation through the multiple N box elements. J Biol Chem. 1994;269:5150–5156. [PubMed] [Google Scholar]
- 22.Tamura K, Taniguchi Y, Minoguchi S, Sakai T, Tun T, Furukawa T, Honjo T. Physical interaction between a novel domain of the receptor Notch and the transcription factor RBP-Jκ/Su(H) Curr Biol. 1995;5:1416–1423. doi: 10.1016/s0960-9822(95)00279-x. [DOI] [PubMed] [Google Scholar]
- 23.Tun T, Hamaguchi Y, Matsunami N, Furukawa T, Honjo T, Kawaichi M. Recognition sequence of a highly conserved DNA binding protein RBP-Jκ. Nucleic Acids Res. 1994;22:965–971. doi: 10.1093/nar/22.6.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vojtek A B, Hollenberg S M, Cooper J A. Mammalian Ras interacts directly with the serine/threonine kinase Raf. Cell. 1993;74:205–214. doi: 10.1016/0092-8674(93)90307-c. [DOI] [PubMed] [Google Scholar]
- 25.Waltzer L, Logeat F, Brou C, Israel A, Sergeant A, Manet E. The human Jκ recombination signal sequence binding protein (RBP-Jκ) targets the Epstein-Barr virus EBNA2 protein to its DNA responsive elements. EMBO J. 1994;13:5633–5638. doi: 10.1002/j.1460-2075.1994.tb06901.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.White E. Life, death, and the pursuit of apoptosis. Genes Dev. 1996;10:1–15. doi: 10.1101/gad.10.1.1. [DOI] [PubMed] [Google Scholar]
- 27.Yalamanchili R, Tong X, Grossman S, Johannsen E E, Mosialos G, Kieff E. Genetic and biochemical evidence that EBNA2 interaction with a 63-kDa cellular GTG-binding protein is essential for B lymphocyte growth transformation by EBV. Virology. 1994;204:634–641. doi: 10.1006/viro.1994.1578. [DOI] [PubMed] [Google Scholar]
- 28.Zimber-Strobl U, Kremmer E, Grasser F, Marschall G, Laux G, Bornkamm G W. The Epstein-Barr virus nuclear antigen 2 interacts with an EBNA2 responsive cis-element of the terminal protein 1 gene promoter. EMBO J. 1993;12:167–175. doi: 10.1002/j.1460-2075.1993.tb05642.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zimber-Strobl U, Strobl L J, Meitinger C, Hinrichs R, Sakai T, Furukawa T, Honjo T, Bornkamm G W. Epstein-Barr virus nuclear antigen 2 exerts its transactivating function through interaction with recombination signal binding protein RBP-Jκ, the homologue of Drosophila Suppressor of Hairless. EMBO J. 1994;13:4973–4982. doi: 10.1002/j.1460-2075.1994.tb06824.x. [DOI] [PMC free article] [PubMed] [Google Scholar]