Abstract
The maturation and subcellular localization of hepatitis C virus (HCV) core protein were investigated with both a vaccinia virus expression system and CHO cell lines stably transformed with HCV cDNA. Two HCV core proteins, with molecular sizes of 21 kDa (p21) and 23 kDa (p23), were identified. The C-terminal end of p23 is amino acid 191 of the HCV polyprotein, and p21 is produced as a result of processing between amino acids 174 and 191. The subcellular localization of the HCV core protein was examined by confocal laser scanning microscopy. Although HCV core protein resided predominantly in the cytoplasm, it was also found in the nucleus and had the same molecular size as p21 in both locations, as determined by subcellular fractionation. The HCV core proteins had different immunoreactivities to a panel of monoclonal antibodies. Antibody 5E3 stained core protein in both the cytoplasm and the nucleus, C7-50 stained core protein only in the cytoplasm, and 499S stained core protein only in the nucleus. These results clearly indicate that the p23 form of HCV core protein is processed to p21 in the cytoplasm and that the core protein in the nucleus has a higher-order structure different from that of p21 in the cytoplasm. HCV core protein in sera of patients with HCV infection was analyzed in order to determine the molecular size of genuinely processed HCV core protein. HCV core protein in sera was found to have exactly the same molecular weight as the p21 protein. These results suggest that p21 core protein is a component of native viral particles.
Hepatitis C virus (HCV) is the major causative agent of posttransfusion and sporadic non-A, non-B hepatitis (3, 18). Persistent HCV infection often progresses to chronic hepatitis, cirrhosis, and hepatocellular carcinoma (5, 16, 32). HCV contains a positive-strand RNA genome that consists of about 9,600 nucleotides that encode a single polyprotein composed of 3,010 amino acid residues. The similarity of the genomic organization of HCV to that of flaviviruses (4, 13, 41) led to the assumption that the precursor polyprotein is processed into structural and nonstructural proteins by a cellular signal peptidase and viral proteases. This assumption has subsequently been proven (6–8, 34, 45). Morphologically, HCV particles are spherical, measuring 55 to 65 nm in diameter, with fine spike-like surface projections and an inner core measuring 30 to 35 nm in diameter (11). Putative core protein is located at the N terminus of the open reading frame and is considered to form the viral capsid, as core protein can be detected in viral particles in patient serum by immunoelectron microscopy (11, 12, 40, 43). Different forms of core protein have been identified by both sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blot analysis (22, 26, 33, 39), and subcellular localization correlates with these different forms (1, 22, 36). Furthermore, core protein has been shown to have multiple functions, within the cell, such as transactivation of suppression of cell growth (29, 31). Immunologically, core protein presents several epitopes for both T and B cells (15, 38). The HCV core gene contains the most conserved sequence in the coding region of most HCV genotypes, which implies an important biological function. Since suitable viral culture systems are not generally available (10, 14, 21, 37), analysis of HCV genome organization and viral-product function is important to understand the viral life cycle and the pathogenesis of HCV infection.
The objectives of the present study were to clarify the maturation process of HCV core protein, the molecular species involved, and their subcellular localization. We found two species of HCV core protein, p21 and p23, in mammalian cells and demonstrated that HCV core protein exists in both the cytoplasm and the nucleus, primarily as p21. We also determined that native viral particles in patient sera are composed of p21 core protein.
MATERIALS AND METHODS
Cells and viruses.
Rabbit kidney cells (RK13) were maintained in Dulbecco’s modified Eagle’s minimal essential medium (DMEM) supplemented with 5% newborn calf serum. HPB-Ma cells (37), generously provided by Y. Shimizu and H. Yoshikura (University of Tokyo, Tokyo, Japan), were maintained in RPMI 1640 medium supplemented with 8% heat-inactivated fetal calf serum. CHO cells were maintained in F12 medium supplemented with 10% fetal calf serum.
Recombinant vaccinia viruses were grown in RK13 cells, and titers of infectious progeny were determined by plaque assay in these cells.
HCV cDNA constructs.
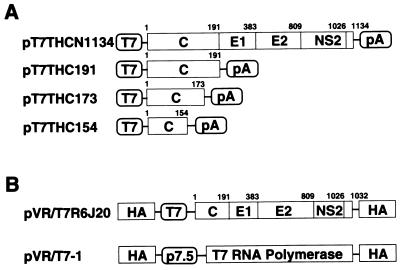
HCV cDNA (pHCR6; genotype 1b, nucleotide positions 1 to 9610) was cloned by reverse transcription-PCR using Superscript II reverse transcriptase (GIBCO-BRL, Rockville, Md.), Taq DNA polymerase (Perkin-Elmer, Branchburg, N.J.), Vent DNA polymerase (New England Biolabs, Beverly, Mass.), and specific primers from chronic hepatitis C patient plasma (R6) that was designated “plasma K” by Shimizu et al. (37). Another genotype 1b HCV cDNA was also cloned from chronic hepatitis C patient serum (TH) (46). For the recloning of desired regions into expression vectors, PCR amplification of the areas of interest was performed. Plasmid pT7THC154 contains an insert consisting of HCV cDNA nucleotides 342 to 800. pT7THC173, pT7THC191, and pT7THCN1134 contain HCV cDNA nucleotides 342 to 870, 342 to 914, and 342 to 3745, respectively.
RVV construction.
R6 HCV cDNA between nucleotides 274 and 3495 (CNS2) was cloned into the pVR vector (35, 45) downstream of the T7 promoter sequence and within the hemagglutinin (HA) gene of the vaccinia virus (Fig. 1B). The plasmid thus obtained was designated pVR/T7R6J20. RK13 cells were infected with the Lister strain of vaccinia virus at a multiplicity of infection (MOI) of 10, followed by electroporation of plasmid pVR/T7R6J20 using a Gene pulsar (Bio-Rad, Hercules, Calif.). Recombinant viruses (HA negative) were selected by HA assay and screened by gene hybridization using an HCV cDNA probe as previously described (17). The recombinant vaccinia virus (RVV) strain thus constructed was named LO-R6J20 and used in the following analyses. An RVV named LO-T7-1, which expresses the T7 RNA polymerase under the control of the P7.5 vaccinia virus early promoter (Fig. 1B), was also established by the same procedure as that used for LO-R6J20 construction (12).
FIG. 1.
Construction of HCV cDNA expression vectors. (A) Structure of the HCV genome. The coding frame of the polyprotein is represented by an open box. C, core; E1 and E2, envelope 1 and 2; NS2, nonstructural protein 2. Genes were subcloned under control of the T7 promoter. (B) RVV genomes.
Continuous expression of HCV core protein.
CHO cells were transfected with the vector pChmBpl CNS2, which was under the control of the SRα promoter (42). This expression vector contained an insert consisting of HCV cDNA encoding core, E1, E2, and NS2 regions. Transformants were selected with hygromycin B.
Transient expression of HCV core protein by cDNA transfection and RVV infection.
Confluent RK13 cells in 60-mm plastic dishes were doubly infected with LO-R6J20 and LO-T7-1 RVVs at an MOI of 10 for each virus. Infected cells were cultured at 37°C and harvested at 3, 6, 12, 24, 48, and 72 h after RVV inoculation.
Subconfluent RK13 cells in 60-mm plastic dishes were transfected with 10-μg HCV expression vectors by a modified calcium phosphate precipitation method (2). The transfected cells were infected 6 h after transfection with LO-T7-1 RVV at an MOI of 10. Transfected and infected cells were cultured at 37°C and harvested 12 h after RVV inoculation. Collected cells were lysed with RIPA buffer (10 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1% Triton X-100, 0.1% SDS, 1% sodium deoxycholate, and 1 mM phenylmethylsulfonyl fluoride) for Western blot analysis or fixed for immunostaining in a 1:1 solution of acetone-methanol at −20°C for 10 min.
Preparations of HCV core protein by in vitro translation.
Plasmids pT7THC154, T7THC173, and pT7THC191 were transcribed and translated simultaneously by using the TNT coupled in vitro translation system (Promega, Madison, Wis.) according to the manufacturer’s instructions. Aliquots of the reaction mixture were separated by SDS-PAGE and analyzed by Western blot using anticore monoclonal antibodies.
Subcellular fractionation of RK13 cells infected with LO-R6J20 and LO-T7-1 RVVs.
Subconfluent RK13 cells grown in 175-cm2 tissue culture flasks were double infected with LO-R6J20 and LO-T7-1 RVVs (MOI, 10). After a 12-h incubation at 37°C, cells were harvested by trypsinization. The cells were centrifuged at 800 × g and resuspended in homogenization buffer (250 mM sucrose in 10 mM Tris-HCl [pH 7.4], 0.5% Nonidet P-40, 1 mM MgCl2, and 1 mM phenylmethylsulfonyl fluoride). This cell suspension was then homogenized on ice by 30 strokes in a Dounce homogenizer (Wheaton; Milliville, N.J.). The homogenate was centrifuged at 800 × g for 5 min at 4°C to obtain the crude-nucleus (N1) pellet. The supernatant was removed and used as the soluble cytoplasmic fraction. The N1 crude nuclei were further purified by ultracentrifugation at 50,000 × g for 45 min at 4°C on a 2.3 M sucrose cushion. The pellet obtained after ultracentrifugation was used as the pure nuclear fraction (N2).
Indirect immunofluorescence microscopy.
RK13 cells cultured on glass slides were infected with LO-R6J20 and LO-T7-1 RVVs and fixed after designated culture periods. The fixed cells were first incubated with antibody dilution buffer (1% bovine serum albumin [BSA], 2.5 mM EDTA, phosphate-buffered saline) for 10 min at room temperature. The cells were then incubated for 60 min at room temperature with one of the following anti-HCV core monoclonal antibodies as the primary antibody at 100 μg/ml: 515S (12), 5E3 (12), C7-50 (25), and 499S. The epitope of 5E3, 515S, and C7-50 consists of amino acids 21 to 40. That of 499S is unknown. Binding was detected at room temperature for 60 min with fluorescein isothiocyanate (FITC)-conjugated anti-mouse immunoglobulin G (IgG), A, M (Cappel, Durham, N.C.) diluted 1:100 with antibody dilution buffer. The cells were then mounted with PermaFluor aqueous mounting medium (Immunon, Pittsburgh, Pa.) and observed under a fluorescence microscope (Carl Zeiss, Oberkochen, Germany).
Detailed analysis of the subcellular localization of HCV core protein was performed by confocal laser scanning microscopy (Carl Zeiss). For this study, cells were treated with RNase A at 1 mg/ml for 60 min at room temperature and nuclei were counterstained with propidium iodide (Molecular Probe, Eugene, Oreg.) at 5 μg/ml for 15 min.
Western blot analysis of HCV core protein.
The protein samples prepared from RVV-infected and DNA-transfected RK13 cells and in vitro translation reaction were separated by 10 to 20% gradient and SDS–15% PAGE, respectively. The low-molecular-weight calibration kit (Pharmacia LKB, Piscataway, N.J.) was used for molecular weight markers. After electrophoresis, proteins were electrotransferred to Immobilon-P membranes (Millipore, Bedford, Mass.) with a semidry blotting apparatus. After transfer, membranes were washed and incubated with blocking buffer (5% nonfat dry milk, 2.5% BSA, 20 mM Tris-HCl [pH 7.6], 0.5 M NaCl, 0.05% Tween 20). The primary antibody was 10 μg of the anti-HCV core monoclonal antibody 515S per ml diluted in blocking buffer containing 1% nonfat dry milk and 0.5% BSA. Membranes were subsequently treated with horseradish peroxidase-conjugated donkey anti-mouse Ig (Amersham, Arlington Heights, Ill.) at a 1:2,000 dilution for 60 min at room temperature. Protein binding was detected with the ECL system (Amersham).
Sucrose density gradient centrifugation of patient sera.
Four milliliters of each chronic hepatitis C patient serum was layered onto a 10, 20, 30, 40, 50, and 60% discontinuous sucrose density gradient in TNE buffer (50 mM Tris-HCl, 100 mM NaCl, 1 mM EDTA [pH 7.5]) and centrifuged at 40,000 rpm for 16 h at 4°C in a Beckman SW41 rotor (Beckman Instruments, Palo Alto, Calif.). A 500-μl aliquot of each fraction was collected from the bottom of the tube. The HCV core protein titer of each fraction was quantified by fluorescent enzyme immunoassay (FEIA) (12, 43). The HCV RNA titer of each fraction was quantified by real-time detection reverse transcription-PCR (42a).
Western blot analysis of native core protein.
In order to detect HCV core protein, sera of patients with HCV infection were screened by FEIA (12, 43). Two sera that showed high titers (19 and 27 ng/ml) were chosen for the following analysis. One milliliter of the selected serum or normal control human serum was precipitated with polyethylene glycol 4000 at a final concentration of 4% and incubated for 3 h at 4°C. The mixture was then centrifuged at 4,000 × g for 60 min at 4°C. The pellet was dissolved in 80 μl of 8 M urea, mixed with the same volume of 2× Laemmli sample buffer (19), and then boiled for 5 min. Proteins in the supernatant were separated by SDS-PAGE and analyzed by Western blot as described above, except that the membrane was incubated with 0.5 μg of biotinylated anti-HCV core protein monoclonal antibody (5E3) per ml for 60 min at room temperature and then treated with horseradish peroxidase-labeled streptavidine (Pierce, Rockford, Ill.) at a 1:2,000 dilution in blocking buffer containing 5% nonfat dry milk and 1% BSA.
RESULTS
Kinetics of HCV core protein expressed in RK13 cells.
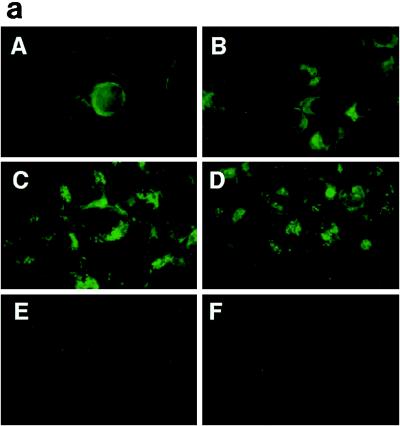
RK13 cells were simultaneously infected with LO-R6J20 and LO-T7-1 RVVs and harvested 3, 6, 12, 24, 48, and 72 h later. Expression of HCV core protein at each time point was analyzed by indirect immunofluorescence microscopy (Fig. 2a) and Western blot (Fig. 2b). By 3 h after RVV infection, core protein was observed around the nuclear membrane. After 6 h, core protein was distributed diffusely in the cytoplasm. The core protein distribution pattern between 3 and 6 h represents localization in the endoplasmic reticulum (ER) (33, 34). After 12 h, HCV core protein formed granules which appeared to be localized in both the cytoplasm and the nucleus. At 24 h, the formation of granules was more prominent.
FIG. 2.
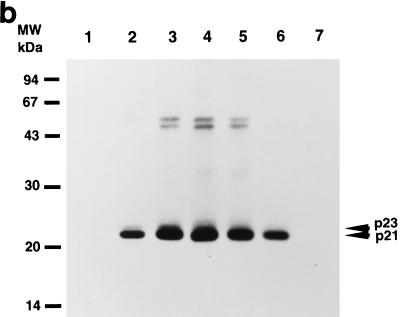
Time course of changes in expression of HCV core protein. (a) Indirect immunofluorescence microscopy. RK13 cells were infected with both RVVs, LO-R6J20 and LO-T7-1, fixed with acetone and methanol at −20°C for 10 min, and stained 3 (A), 6 (B), 12 (C), and 24 (D) h after infection. E, 24 h after infection with LO-T7-1; F, negative serum. Immunostaining was performed with monoclonal antibody 515S. (b and c) Western blot analysis. RK13 cells were infected with both LO-R6J20 RVV and LO-T7-1 RVV and harvested at various time points after infection: 3 h (lane 1), 6 h (lane 2), 12 h (lane 3), 24 h (lane 4), 48 h (lane 5), and 72 h (lane 6). As a negative control, RK13 cells were infected with only LO-T7-1 RVV and harvested at 24 h after infection (lane 7). Western blotting was performed with monoclonal antibody 515S. In panel b, the same amount (15 μg/lane) of infected RK13 cell lysate was loaded in all lanes. In panel c, he total loaded protein was 54, 6, 3, 3, 3, 6, and 54 μg in lanes 1 to 7, respectively. The two HCV core proteins, p21 and p23, are indicated on the right, and the sizes of protein molecular weight (MW) markers are indicated on the left.
Western blot analysis with the 515S anti-HCV core monoclonal antibody revealed two discrete bands 3 and 6 h after RVV infection (Fig. 2b, lanes 1 and 2, respectively) but not 12, 24, 48, and 72 h after RVV infection (Fig. 2b, lanes 3 to 6, respectively). Based on the molecular weight, the upper molecule was designated the p23 core protein and the lower molecule was designated the p21 core protein. p23 was detected between 3 and 6 h after inoculation of the RVVs and disappeared thereafter, while expression of p21 was detected at 3 h reached a maximum after 24 h, and remained at that level until 48 h.
Analysis of p23 and p21 HCV core proteins.
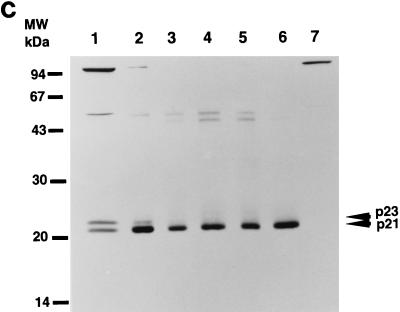
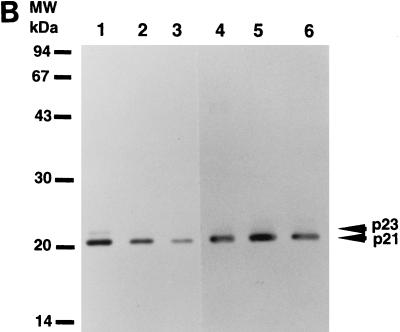
To determine the origin of the two forms of HCV core protein, we transfected plasmids pT7THC154, pT7THC173, pT7THC191, and pT7THCN1134 into RK13 cells and then infected the cells with LO-T7-1 RVV. The cells were harvested at 12 h after infection. We also performed in vitro translation experiments using the same plasmid DNA as the source of the RNA template. Both p23 and p21 were detected in the cell lysate of cells transfected with pT7THC191, but only p23 was detected in the in vitro-translated product of pT7THC191 (Fig. 3). In the cell lysate of cells transfected with pT7THC191 or RK13 cells infected with both LO-T7-1 and LO-R6J20 RVVs, p21 and p23 were detected at the same position on the gel. Furthermore, with pT7THC173, a slightly smaller core protein, p20.5, was detected at the same position on the gel for both the cell lysate and the in vitro-translated product. Similarly, from pT7THC154, only p18 was detected. These results indicate that p23 is the 191-amino-acid translation product and that p23 is further processed to p21 by cleavage at its C terminus between amino acids 174 and 191.
FIG. 3.
Determination of the origins of the two forms of HCV core proteins, p21 and p23 by Western blot analysis. Plasmids pT7THC154, pT7THC173, and pT7THC191 were transiently expressed in an in vitro translation system (lanes 1, 3, and 5, respectively) and were also transfected into RK13 cells, which were then infected with LO-T7-1 RVV and harvested 12 h after infection (lanes 2, 4, and 6, respectively). Plasmid pT7THCN1134 was also transfected into RK13 cells, which were then infected with LO-T7-1 RVV and harvested 12 h after infection (lanes 7). RK13 cells infected with both LO-T7-1 RVV and LO-R6J20 RVV were then harvested 3 h after infection (lane 8). The same sample was loaded in lanes 6 and 9.
Subcellular localization of HCV core protein.
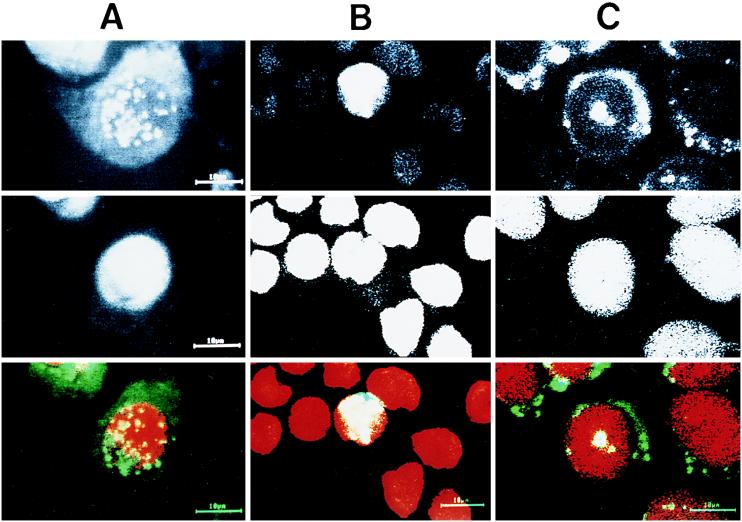
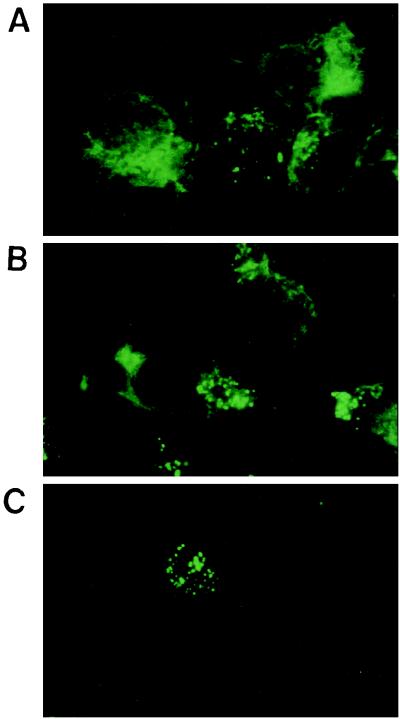
Laser scanning confocal microscopy was used to examine the granular staining of HCV core in the nucleus in greater detail. By using horizontal sections, 1 μm thin, of RK13 cells infected with LO- R6J20 and LO-T7-1 RVVs, HCV core protein was stained with monoclonal antibody 515S (Fig. 4A). HCV core protein was found predominantly in the cytoplasm. In addition, merging of both the core protein (FITC; green) and nucleus DNA (propidium iodide; red) images produced a yellow area that indicates the presence of HCV core protein inside the nucleus or on the nuclear membrane (1). Similar findings were observed with HPB-Ma cells infected with LO-R6J20 and LO-T7- 1 RVVs (Fig. 4B) and CHO cells stably expressing HCV core protein (Fig. 4C).
FIG. 4.
Subcellular localization of HCV core protein. RK13 cells (A) and HPB-Ma cells (B) were infected with both the LO-R6J20 and the LO-T7-1 RVVs. Cells were fixed with acetone and methanol at −20°C for 10 min 12 h after infection, counterstained with FITC-conjugated anti-mouse Ig (for HCV core protein) and propidium iodide (for nuclear DNA), and viewed with a confocal laser scanning microscope. (C) CHO cells stably transformed with an HCV core protein expression vector. Upper panels, HCV core protein; middle panels, nuclear DNA; lower panels, merged image of HCV core protein (green) and nucleus (red).
Subcellular fractionation of HCV core protein in cells.
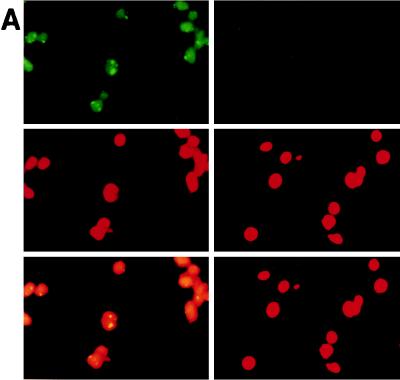
RK13 cells infected with LO-R6J20 and LO-T7-1 RVVs were homogenized and fractionated as described in Materials and Methods. Pure nuclear fraction N2 was stained with anticore monoclonal antibody and counterstained with propidium iodide. Core protein was clearly detected inside the nucleus and was found only within the nucleus (Fig. 5A). Aliquots of the cytoplasmic, crude nuclear, and pure nuclear fractions were separated by SDS-PAGE followed by Western blot analysis. HCV core protein was detected by the 515S monoclonal antibody as shown in Fig. 5B. All fractions clearly showed bands of the same molecular size, namely 21 kDa. These data suggest that the sizes of the core proteins localized in the cytoplasm and nucleus were similar to that of the 21 kDa protein detected by Western blot analysis.
FIG. 5.
Subcellular fractionation of HCV core protein. (A) Immunostaining of pure nuclear fraction N2. N2 fractions from RK13 cells infected with LO-R6J20 and LO-T7-1 RVVs for 12 h were separated as described in Materials and Methods, stained with FITC-conjugated anti-mouse Ig (for HCV core protein) (upper panels), and counterstained with propidium iodide (for nucleus DNA) (middle panels), and the resultant images were merged (lower panels). Fraction N2 was stained with anticore monoclonal antibody 515S (upper left) or anti-poliovirus receptor monoclonal antibody as a negative control (upper right). (B) The cytoplasmic fraction (Cy), crude nuclear fraction (N1), and pure nuclear fraction (N2) from RK13 cells infected with LO-R6J20 RVV and LO-T7-1 RVV for 6 h (lanes 1 to 3) or 12 h (lanes 4 to 6) were separated as described in Materials and Methods. The HCV core protein in each fraction was analyzed by Western blotting. Lanes 1 and 4, Cy; lanes 2 and 5, N1; lanes 3 and 6, N2.
Conformational analysis of HCV core protein.
For further analysis of localization, immunostaining of HCV core protein was performed with the anti-HCV core protein monoclonal antibodies 5E3, 515S, C7-50, and 499S. HCV core protein was stained in both the cytoplasm and the nucleus with 5E3 and 515S, only in the cytoplasm with C7-50, and only in the nucleus with 499S (Fig. 6). These findings suggest that the HCV core proteins in the cytoplasm and nucleus have different higher-order structures.
FIG. 6.
Subcellular localization of HCV core protein determined by immunostaining with different antibodies. RK13 cells infected with both LO-R6J20 RVV and LO-T7-1 RVV were fixed with acetone and methanol at −20°C for 10 min 12 h after infection and stained with the anti-HCV core protein monoclonal antibodies 5E3 (A), C7-50 (B), and 499 (C).
Correlation of HCV RNA with core protein in the sucrose density gradient centrifugation fraction.
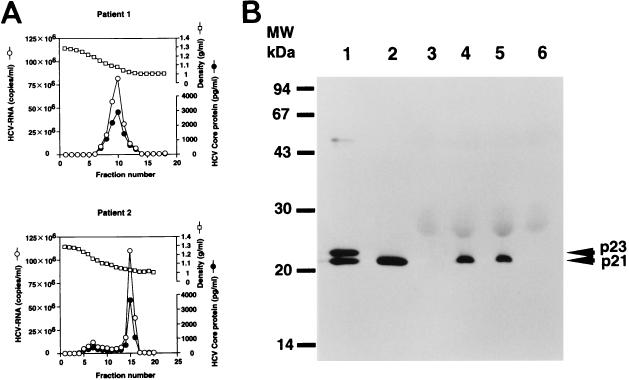
The quantities of HCV core protein and HCV RNA in each fraction obtained from sucrose density gradient centrifugation of the patient sera were examined (Fig. 7A). The fraction numbers of peak HCV core protein and peak RNA were identical for these sera. This finding indicates that native HCV viral particles consist of HCV core protein and HCV RNA.
FIG. 7.
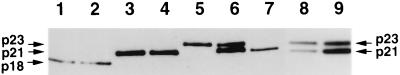
(A) Quantification of HCV core protein and HCV RNA in fractions from sucrose density gradient centrifugation of the patient sera. (B) Western blot analysis of HCV native core protein in HCV-infected-patient sera or HCV-negative sera. Lanes 1 and 2, HCV core protein in RK13 cells harvested 3 and 24 h, respectively, after infection with both the LO-R6J20 and the LO-T7-1 RVVs; lanes 4 and 5, HCV core protein from HCV-infected-patient sera; lanes 3 and 6, HCV-negative sera. MW, molecular weight markers.
Western blot analysis to determine the size of HCV core protein found in viral particles in patients’ sera.
In order to determine the molecular weight of core protein in natural viral particles, core protein was detected by Western blot analysis in the sera of two patients who showed high titers of HCV core protein upon screening by FEIA. These serum samples were examined by Western blotting using the high-affinity monoclonal antibody 5E3 (12, 43), which was one of the two antibodies used in FEIA. The molecular weight of the HCV core protein in both patient’s sera was the same as that of the p21 HCV core protein expressed in RK13 cells by using the RVV LO-R6J20 (Fig. 7B).
DISCUSSION
The maturation process of HCV core protein in mammalian cells was characterized by using an RVV system. We found two molecular species of HCV core protein, p21 and p23, by Western blot analysis (Fig. 2b and c). The time course study showed that p23 was detected transiently only in the early phase (between 3 and 6 h after infection with LO-R6J20 and LO-T7-1 RVVs) but disappeared thereafter, while p21 was detected in all phases. Therefore, p21 may be the mature and stable form of core protein that accumulates in the cell and which eventually constitutes the viral capsid. In support of this, the core protein that formed viral particles in the patient sera was determined to be p21 by Western blot analysis.
Previous studies have indicated that the HCV polyprotein is processed by cellular and viral proteinases. The host signal peptidase on the ER catalyzes cleavages within the structural region (core/E1, E1/E2, and E2/NS2) (7). In the present study, the staining pattern of HCV core protein at 3 and 6 h after infection with the RVVs seems to represent ER localization (Fig. 2a) (33, 34). Based on this localization pattern, p23 is believed to be an ER-related form of HCV core protein. The time course of HCV core protein expression suggests that p23 is associated with the ER and is further processed to yield p21. The 50-kDa band in Fig. 2b and c may represent unprocessed polyprotein of core to NS2. The 100-kDa bands most likely reflect cross-reactivity of the 515S antibody to proteins other than HCV, as indicated by the presence of these 100-kDa bands in the negative-control sample (Fig. 2c, lane 7).
HCV core protein is located in the N-terminal portion of the HCV polyprotein and is thought to consist of 191 amino acids. However, few details are known about the C-terminal site of mature core protein. In the present study, the origin of p21 and p23 was examined by expression of a series of C-terminally truncated core proteins. We constructed three core protein expression vectors, pT7THC154, pT7THC173, and pT7THC191, which correspond to amino acids 1 to 154, 1 to 173, and 1 to 191, respectively. By using pT7THC191, only p23 was detected in a cell-free translation study, while both p21 and p23 were expressed in a cell culture study (Fig. 3). The sizes of the protein products of pT7THC154 and pT7THC173 were 18 and 20.5 kDa, respectively (Fig. 3). These results show that p23 is composed of 191 amino acids and p21 is a truncated form of p23, with further processing occurring between amino acids 174 and 191, which may be just before the signal sequence (9, 33). On the other hand, the possibility that it was the N-terminal region that was cleaved certainly exists. Amino acid sequence analysis of the C-terminal region of p21 will be required to clarify this issue.
The subcellular localization of HCV core protein is controversial (1, 7, 20, 22, 23, 27, 33, 34). We examined the localization of HCV core protein expressed in RK13 cells and HPB- Ma cells using LO-R6J20 and LO-T7-1 RVVs and in stably transformed CHO cells. A confocal laser scanning microscope was used to obtain high-resolution microtomographic images of the subcellular localization of HCV core protein in these cells. Our findings demonstrate that while HCV core protein exists predominantly in the cytoplasm, it also appears to localize in the nucleus (Fig. 4). On the other hand, these confocal microscopic images may actually represent core protein localized on the cytoplasmic side of the nuclear membrane (1). Similar results were obtained with two expression systems and three cell lines, including HPB-Ma cells, which are reported to support HCV replication (37). Therefore, this localization pattern is not a function of vaccinia virus infection, the level of expression, or the cellular species. Although contamination by the ER fraction cannot be completely excluded, the subcellular fractionation study strongly suggests the presence of p21 in both cytoplasmic and nuclear fractions (Fig. 5). The molecular weights of the two fractions were similar by Western blot analysis.
The different localization patterns of HCV core protein led us to analyze whether cytoplasmic and nuclear core proteins have different properties. Immunoreactivities of both forms were first studied with a panel of well-characterized monoclonal antibodies. As shown in Fig. 6, immunostaining with different monoclonal antibodies showed different localization of HCV core protein. This finding and the truncated protein expression pattern in Fig. 3 suggest that the HCV core protein in the cytoplasm is cleaved between amino acids 174 and 191, after which it folds into a different conformation and moves into the nucleus. A previous study (22) of HCV protein using the C7-50 monoclonal anticore antibody found that core proteins of 173 amino acids and 191 amino acids exist. They suggested that the 173-amino-acid core protein moves into the nucleus but that the coexistence of the 191-amino-acid core protein within the cytoplasm causes some of the 173-amino-acid core protein to remain in the cytoplasm. We also detected core protein in the cytoplasm using the same monoclonal antibody, C7-50, in the CNS2 expression system. When we stained these cells with the 5E3 and 515S monoclonal antibodies, core protein was detected in both the cytoplasm and the nucleus. Furthermore, core protein was detected primarily in the nucleus when the 499S antibody was used (Fig. 4). These results strongly suggest that p21 folds and changes its conformation prior to entering the nucleus. It is also possible that p21 interacts with the nuclear transporter, changes its conformation, and then enters the nucleus. However, further studies are necessary to clarify the reason or function of the different distribution patterns of core protein in a cell.
To examine the properties of native HCV core protein, we determined the molecular weight of HCV core protein found in viral particles in sera of HCV-infected patients. The recently developed FEIA enables measurement of a small amount of HCV core protein in the sera of patients with HCV infection (12, 43). Using FEIA, we screened and selected two patients’ sera that had a high titer of HCV core protein. These sera were fractionated by sucrose density gradient centrifugation. The fraction that contained peak HCV RNA was identical to that containing peak core protein (Fig. 7A). The buoyant density of the HCV RNA and core protein peak fractions was consistent with previously published results (11). The HCV core protein derived from these sera was the same 21-kDa protein as the p21 protein expressed in cells of the RVV system (Fig. 7B). This finding clearly indicates that p21, which has been characterized in this study, is similar to the native HCV core protein.
HCV core protein has been reported to bind to the lymphotoxin β receptor (24), or viral and cellular promoters (28, 31), and thus may influence cellular growth control (29, 30). Several truncated forms of core protein are expressed in in vitro-cultured cells, and only a short form (<p16) has been detected in the nucleus (27, 38); however, this form is usually not detected in vivo. We have demonstrated that the HCV core protein, p23, is processed at the C-terminal end to yield p21. Although p21 exists primarily in the cytoplasm, some molecules fold and move into the nucleus of the cell, either actively or passively. More importantly, the size of core protein in patient sera is the same as that of p21, suggesting that p21 is the mature form of the HCV core protein. Thus, the results of the present study on HCV core protein maturation shed light not only on the replication of HCV but also on the pathogenesis of HCV infection.
ACKNOWLEDGMENTS
We express gratitude to Youko Shimizu, Hiroshi Yoshikura, and Akira Hasegawa for generously providing the HPB-Ma cells and 5E3 antibody, and Atsuko Fujisawa for creating the figures.
This study was supported in part by a grant-in-aid for Specially Promoted Research on Viral Diseases from the Tokyo metropolitan government; a grant from the Institute for Adult Diseases Asahi Life Foundation; a grant from the Ministry of Education, Science, and Culture of Japan; and a grant from the Ministry of Health and Welfare of Japan.
REFERENCES
- 1.Barba B, Harper F, Harada T, Kohara M, Goulinet S, Matsuura Y, Eder G, Schaff Z, Chapman M J, Miyamura T, Brechot C. Hepatitis C virus core protein shows a cytoplasmic localization and associates to cellular lipid storage droplets. Proc Natl Acad Sci USA. 1997;94:1200–1205. doi: 10.1073/pnas.94.4.1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen C, Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol Cell Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Choo Q-L, Kuo G, Weiner A J, Overby L R, Bradley D W, Houghton M. Isolation of cDNA derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989;244:359–362. doi: 10.1126/science.2523562. [DOI] [PubMed] [Google Scholar]
- 4.Choo Q-L, Richman K, Han J H, Berger K, Lee C, Dong C, Gallegos C, Coit D, Medina-Selby A, Barr P, Weiner A J, Bradley D W, Kuo G, Houghton M. Genetic organization and diversity of the hepatitis C virus. Proc Natl Acad Sci USA. 1991;88:2451–2455. doi: 10.1073/pnas.88.6.2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Esteban J I, Esteban R, Viladomiu L, Lopez-Talavera J C, Gonzalez A, Hernandez J M, Roget M, Vargas V, Genesca J, Buti M, Guardia J. Hepatitis C virus antibodies among risk groups in Spain. Lancet. 1989;ii:294–296. doi: 10.1016/s0140-6736(89)90485-6. [DOI] [PubMed] [Google Scholar]
- 6.Grakoui A, Wychowski C, Lin C, Feinstone S M, Rice C M. Expression and identification of hepatitis C virus polyprotein cleavage products. J Virol. 1993;67:1385–1395. doi: 10.1128/jvi.67.3.1385-1395.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hijikata M, Kato N, Ootsuyama Y, Nakagawa M, Shimotohno K. Gene mapping of the putative structural region of the hepatitis C virus genome by in vitro processing analysis. Proc Natl Acad Sci USA. 1991;88:5547–5551. doi: 10.1073/pnas.88.13.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hijikata M, Mizushima H, Tanji Y, Komoda Y, Hirowatari Y, Akagi T, Kato N, Kimura K, Shimotono K. Proteolytic processing and membrane association of putative nonstructural proteins of hepatitis C virus. Proc Natl Acad Sci USA. 1993;90:10773–10777. doi: 10.1073/pnas.90.22.10773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hussy P, Langen H, Mous J, Jacobsen H. Hepatitis C virus core protein: carboxy-terminal boundaries of two processed species suggest cleavage by a signal peptide peptidase. Virology. 1996;224:93–104. doi: 10.1006/viro.1996.0510. [DOI] [PubMed] [Google Scholar]
- 10.Ito T, Mukaigawa J, Zou J, Hirabayashi Y, Mitamura H, Yasui K. Cultivation of hepatitis C virus in primary hepatocyte culture from patients with chronic hepatitis C results in release of high titre infectious virus. J Gen Virol. 1996;77:1043–1054. doi: 10.1099/0022-1317-77-5-1043. [DOI] [PubMed] [Google Scholar]
- 11.Kaito M, Watanabe S, Tsukiyama-Kohara K, Yamaguchi K, Kobayashi Y, Konishi M, Yokoi M, Ishida S, Suzuki S, Kohara M. Hepatitis C virus particle detected by immunoelectron microscopic study. J Gen Virol. 1994;75:1755–1760. doi: 10.1099/0022-1317-75-7-1755. [DOI] [PubMed] [Google Scholar]
- 12.Kashiwakuma T, Hasegawa A, Kajita T, Takata A, Mori H, Ohta Y, Tanaka E, Kiyosawa K, Tanaka T, Tanaka S, Hattori N, Kohara M. Detection of hepatitis C virus specific core protein in serum of patients by a sensitive fluorescence enzyme immunoassay (FEIA) J Immunol Methods. 1996;190:79–89. doi: 10.1016/0022-1759(95)00261-8. [DOI] [PubMed] [Google Scholar]
- 13.Kato N, Hijikata M, Ootsuyama Y, Nakagawa M, Ohkoshi S, Sugimura T, Shimotohno K. Molecular cloning of the human hepatitis C virus genome from Japanese patients with non A, non B hepatitis. Proc Natl Acad Sci USA. 1990;87:9524–9528. doi: 10.1073/pnas.87.24.9524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kato N, Nakazawa T, Mizutani T, Shimotono K. Susceptibility of human T-lymphotropic virus type I infected cell line MT-2 to hepatitis C virus infection. Biochem Biophys Res Commun. 1995;206:863–869. doi: 10.1006/bbrc.1995.1123. [DOI] [PubMed] [Google Scholar]
- 15.Kita H, Hiroishi K, Moriyama T, Okamoto H, Kaneko T, Ohnishi S, Yazaki Y, Imawari M. A minimal and optimal cytotoxic T cell epitope within hepatitis C virus nucleoprotein. J Gen Virol. 1995;76:3189–3193. doi: 10.1099/0022-1317-76-12-3189. [DOI] [PubMed] [Google Scholar]
- 16.Kiyosawa K, Sodeyama T, Tanaka E, Gibo Y, Yoshizawa K, Nakano Y, Furuta S, Akahane Y, Nishioka K, Purcell R H, Alter H J. Interrelationship of blood transfusion, non-A, non-B hepatitis and hepatocellular carcinoma: analysis by detection of antibody to hepatitis C virus. Hepatology. 1990;12:671–675. doi: 10.1002/hep.1840120409. [DOI] [PubMed] [Google Scholar]
- 17.Kohara M, Tsumiyama-Kohara K, Maki N, Asano K, Yamaguchi K, Miki K, Tanaka S, Hattori N, Matsuura Y, Saito I, Miyamura T, Nomoto A. Expression and characterization of glycoprotein gp35 of hepatitis C virus using recombinant vaccinia virus. J Gen Virol. 1992;73:2313–2318. doi: 10.1099/0022-1317-73-9-2313. [DOI] [PubMed] [Google Scholar]
- 18.Kuo G, Choo Q-L, Alter H J, Gitnick G L, Redeker A G, Purcell R H, Miyamura T, Dienstag J L, Alter M J, Stevens C E, Tegtmeier G E, Bonino F, Colombo M, Lee W-S, Kuo C, Berger K, Shuster J R, Overby L R, Bradley D W, Houghton M. An assay for circulating antibodies to a major etiologic virus of human non-A, non-B hepatitis. Science. 1989;244:362–364. doi: 10.1126/science.2496467. [DOI] [PubMed] [Google Scholar]
- 19.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 20.Lanford R E, Notvall L, Chavez D, White R, Frenzel G, Simonsen C, Kim J. Analysis of hepatitis C virus capsid, E1, and E2/NS1 proteins expressed in insect cells. Virology. 1993;197:225–235. doi: 10.1006/viro.1993.1583. [DOI] [PubMed] [Google Scholar]
- 21.Lanford R E, Sureau C, Jacob J R, White R, Fuerst T R. Demonstration of in vitro infection of chimpanzee hepatocytes with hepatitis C virus using strand specific RT/PCR. Virology. 1994;202:606–614. doi: 10.1006/viro.1994.1381. [DOI] [PubMed] [Google Scholar]
- 22.Liu Q, Tackney C, Bhat R A, Prince A M, Zhang P. Regulated processing of hepatitis C virus core protein is linked to subcellular localization. J Virol. 1997;71:657–662. doi: 10.1128/jvi.71.1.657-662.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lo S-Y, Masiarz F, Hwang S B, Lai M M M, Ou J-H. Differential subcellular localization of hepatitis C virus core gene products. Virology. 1995;213:455–461. doi: 10.1006/viro.1995.0018. [DOI] [PubMed] [Google Scholar]
- 24.Matsumoto M, Hsieh T-Y, Zhu N, VanArsdale T, Hwang S B, Jeng K-S, Gorbalenya A E, Lo S-Y, Ou J-H, Ware C F, Lai M M C. Hepatitis C virus core protein interacts with the cytoplasmic tail of lymphotoxin-β receptor. J Virol. 1997;71:1301–1309. doi: 10.1128/jvi.71.2.1301-1309.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moradpour D, Wakita T, Tokushige K, Carlson R I, Krawczynski K, Wands J R. Characterization of three novel monoclonal antibodies against hepatitis C virus core protein. J Med Virol. 1996;48:234–241. doi: 10.1002/(SICI)1096-9071(199603)48:3<234::AID-JMV4>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 26.Moradpour D, Englert C, Wakita T, Wands J R. Characterization of cell lines allowing tightly regulated expression of hepatitis C virus core protein. Virology. 1996;222:51–63. doi: 10.1006/viro.1996.0397. [DOI] [PubMed] [Google Scholar]
- 27.Ravaggi A, Natoli G, Primi D, Albertini A, Levrero M, Cariani E. Intracellular localization of full-length and truncated hepatitis C virus core protein expressed in mammalian cells. J Hepatol. 1994;20:833–836. doi: 10.1016/s0168-8278(05)80157-6. [DOI] [PubMed] [Google Scholar]
- 28.Ray R B, Lagging L M, Meyer K, Steele R, Ray R. Transcriptional regulation of cellular and viral promoters by the hepatitis C virus core protein. Virus Res. 1995;37:209–220. doi: 10.1016/0168-1702(95)00034-n. [DOI] [PubMed] [Google Scholar]
- 29.Ray R B, Lagging L M, Meyer K, Ray R. Hepatitis C virus core protein cooperates with ras and transforms primary rat embryo fibroblasts to tumorigenic phenotype. J Virol. 1996;70:4438–4443. doi: 10.1128/jvi.70.7.4438-4443.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ray R B, Meyer K, Ray R. Supression of apoptotic cell death by hepatitis C virus core protein. Virology. 1996;226:176–182. doi: 10.1006/viro.1996.0644. [DOI] [PubMed] [Google Scholar]
- 31.Ray R B, Steele R, Meyer K, Ray R. Transcriptional repression of p53 promoter by hepatitis C virus core protein. J Biol Chem. 1997;272:10983–10986. doi: 10.1074/jbc.272.17.10983. [DOI] [PubMed] [Google Scholar]
- 32.Saito I, Miyamura T, Ohbayashi A, Harada H, Katayama T, Kikuchi S, Watanabe Y, Koi S, Onji M, Ohta Y, Choo Q-L, Houghton M, Kuo G. Hepatitis C virus infection is associated with the development of hepatocellular carcinoma. Proc Natl Acad Sci USA. 1990;87:6547–6549. doi: 10.1073/pnas.87.17.6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Santolini E, Migliaccio G, La Monica N. Biosynthesis and biochemical properties of the hepatitis C virus core protein. J Virol. 1994;68:3631–3641. doi: 10.1128/jvi.68.6.3631-3641.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Selby M J, Choo Q-L, Berger K, Kuo G, Glazer E, Eckart M, Lee C, Chien D, Kuo C, Houghton M. Expression, identification and subcellular localization of the proteins encoded by the hepatitis C viral genome. J Gen Virol. 1993;74:1103–1113. doi: 10.1099/0022-1317-74-6-1103. [DOI] [PubMed] [Google Scholar]
- 35.Shida H, Tochikura T, Sato T, Konno T, Hirayoshi K, Seki M, Ito Y, Hatanaka M, Hinuma Y, Sugimoto M, Takahashi-Nishimaki F, Maruyama T, Miki K, Suzuki K, Morita M, Sashiyama H, Hayami M. Effect of the recombinant vaccinia viruses that express HTLV-I envelope gene on HTLV-I infection. EMBO J. 1987;6:3379–3384. doi: 10.1002/j.1460-2075.1987.tb02660.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shih C M, Lo S J, Miyamura T, Chen S Y, Lee Y-H W. Suppression of hepatitis B virus expression and replication by hepatitis C virus core protein in Huh-7 cells. J Virol. 1993;67:5823–5832. doi: 10.1128/jvi.67.10.5823-5832.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shimizu Y K, Purcell R H, Yoshikura H. Correlation between the infectivity of hepatitis C virus in vivo and its infectivity in vitro. Proc Natl Acad Sci USA. 1993;90:6037–6041. doi: 10.1073/pnas.90.13.6037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shirai M, Okada H, Nishioka M, Akatuka T, Wychowski C, Houghten R, Pendleton C D, Feinstone S M, Berzofsky J A. An epitope in hepatitis C virus core region recognized by cytotoxic T cells in mice and humans. J Virol. 1994;68:3334–3342. doi: 10.1128/jvi.68.5.3334-3342.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Suzuki R, Matsuura Y, Suzuki T, Ando A, Chiba J, Harada S, Saito I, Miyamura T. Nuclear localization of the truncated hepatitis C virus core protein with its hydrophobic C-terminus deleted. J Gen Virol. 1995;76:53–61. doi: 10.1099/0022-1317-76-1-53. [DOI] [PubMed] [Google Scholar]
- 40.Takahashi K, Kishimoto S, Yoshizawa H, Okamoto H, Yoshikawa A, Mishiro S. p26 protein and 33-nm particle associated with nucleocapsid of hepatitis C virus recovered from the circulation of infected hosts. Virology. 1992;191:431–434. doi: 10.1016/0042-6822(92)90204-3. [DOI] [PubMed] [Google Scholar]
- 41.Takamizawa A, Mori C, Fuke I, Manabe S, Fujuta J, Onishi E, Andoh T, Yoshida I, Okayama H. Structure and organization of the hepatitis C virus genome isolated from human carriers. J Virol. 1991;65:1105–1113. doi: 10.1128/jvi.65.3.1105-1113.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takebe Y, Seiki M, Fujisawa J, Hoy P, Yokota K, Arai K, Yoshida M, Arai N. SRα promoter: an efficient and versatile mammalian cDNA expression system composed of the simian virus 40 early promoter and the R-U5 segment of human T-cell leukemia virus type 1 long terminal repeat. Mol Cell Biol. 1988;8:466–472. doi: 10.1128/mcb.8.1.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42a.Takeuchi, T., et al. Unpublished data.
- 43.Tanaka T, Lau J Y N, Mizokami M, Orito E, Tanaka E, Kiyosawa K, Yasui K, Ohta Y, Hasegawa A, Tanaka S, Kohara M. Simple fluorescent enzyme immunoassay for detection and quantification of hepatitis C viremia. J Hepatol. 1995;23:742–745. doi: 10.1016/0168-8278(95)80043-3. [DOI] [PubMed] [Google Scholar]
- 44.Tomei L, Failla C, Santolini E, De Francesco R, La Monica N. NS3 is a serine protease required for processing of hepatitis C virus polyprotein. J Virol. 1993;67:4017–4026. doi: 10.1128/jvi.67.7.4017-4026.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tsukiyama K, Yoshikawa Y, Kamata H, Imaoka K, Asano K, Funahashi S, Maruyama T, Shida H, Sugimoto M, Yamanouchi K. Development of heat-stable recombinant rinderpest vaccine. Arch Virol. 1989;107:225–235. doi: 10.1007/BF01317919. [DOI] [PubMed] [Google Scholar]
- 46.Wakita T, Wands J R. Specific inhibition of hepatitis C virus expression by antisense oligodeoxynucleotides. J Biol Chem. 1994;269:14205–14210. [PubMed] [Google Scholar]