Abstract
An antibody made against the herpes simplex virus 1 US5 gene predicted to encode glycoprotein J was found to react strongly with two proteins, one with an apparent Mr of 23,000 and mapping in the S component and one with a herpes simplex virus protein with an apparent Mr of 43,000. The antibody also reacted with herpes simplex virus type 2 proteins forming several bands with apparent Mrs ranging from 43,000 to 50,000. Mapping studies based on intertypic recombinants, analyses of deletion mutants, and ultimately, reaction of the antibody with a chimeric protein expressed by in-frame fusion of the glutathione S-transferase gene to an open reading frame antisense to the gene encoding glycoprotein B led to the definitive identification of the new open reading frame, designated UL27.5. Sequence analyses indicate the conservation of a short amino acid sequence common to US5 and UL27.5. The coding sequence of the herpes simplex virus UL27.5 open reading frame is strongly homologous to the sequence encoding the carboxyl terminus of the herpes simplex virus 2 UL27.5 sequence. However, both open reading frames could encode proteins predicted to be significantly larger than the mature UL27.5 proteins accumulating in the infected cells, indicating that these are either processed posttranslationally or synthesized from alternate, nonmethionine-initiating codons. The UL27.5 gene expression is blocked by phosphonoacetate, indicating that it is a γ2 gene. The product accumulated predominantly in the cytoplasm. UL27.5 is the third open reading frame found to map totally antisense to another gene and suggests that additional genes mapping antisense to known genes may exist.
In this report, we describe the identification of a new open reading frame (ORF) in the genomes of herpes simplex virus types 1 and 2 (HSV-1 and HSV-2). What makes this ORF particularly interesting is its location antisense to UL27, the gene encoding glycoprotein B (gB) (5, 22). The initial objectives of this study were quite different from its outcome.
The sequence of the HSV-1 genome (22) corroborated the existence of several glycoproteins and led to the discovery of others, bringing the total to 11 (gB, gC, gD, gE, gG, gH, gI, gJ, gK, gL, and gM) (reviewed in references 28, 30, and 32). Of these, gJ was reported to be dispensable for viral replication in cell culture and was the least well understood (4, 21, 33). To initiate these studies, we immunized rabbits with a fusion protein consisting of maltose binding protein fused in frame to the entire sequence of gJ. The resulting antibody reacted with a protein (presumed to be gJ) whose gene maps in the S component and with higher-Mr proteins in lysates of HSV-1- and HSV-2-infected cells. Extensive mapping studies led ultimately to the conclusion that these protein bands are encoded by a gene mapping antisense to gB. A possible explanation for the reactivity of the polyclonal antibody with both proteins rests on the observation that HSV-1 and HSV-2 UL27.5 proteins have a short amino acid sequence common to the predicted amino acid sequence of gJ.
Transcripts antisense to known ORFs were first reported by Stevens et al. (31). The latency-associated transcript, however, does not appear to affect the expression of ICP0, the gene to which it is partially antisense. In the past several years, this laboratory reported two sets of antisense genes. Thus, ORFs O and P map antisense to the γ134.5 gene, and the UL43.5 gene maps antisense to the UL43 gene (6, 20, 26). The striking feature of the antisense genes is that their expression is mutually exclusive. Thus, derepression of ORF P leads to its expression early in infection and grossly reduces the normal expression of the γ134.5 gene (27). UL43 and UL43.5 are also expressed at different times after infection (6). The gB/UL27.5 genes are the third set of genes located antisense to each other. gB is expressed relatively early in infection, whereas the UL27.5 gene belongs to the γ2 group in that its expression is totally dependent on viral DNA synthesis (14, 15).
MATERIALS AND METHODS
Cells and viruses.
Rabbit skin cells and Vero cells were obtained from John McLaren and American Type Culture Collection, respectively, and were maintained in Dulbecco’s modified Eagle medium supplemented with 5% newborn calf serum. BHK(TK+) cells (American Type Culture Collection) and 143TK− cells (obtained from Carlo Croce) were maintained in the same medium supplemented with 5% fetal bovine serum. Infected cells were maintained in mixture 199 supplemented with 1% calf serum (199V) unless indicated otherwise. HSV-1(F) and HSV-2(G) are prototypes of HSV-1 and HSV-2 strains, respectively, used in this laboratory (10). Intertypic recombinants HSV-1(F) × HSV-2(G) were described previously (1, 8). In R7015 the HSV-1 S component was replaced by the homologous HSV-2(G) sequences. HSV-1(F)Δ305 was derived from HSV-1(F) and has a 501-bp deletion in UL23 (thymidine kinase) and UL24 genes (25). The HSV-1(KOS) mutant, lacking the UL26 gene (11), was the generous gift of Steven Weinheimer (Bristol-Myers Squibb).
Antibodies.
Mouse monoclonal antibody CH28 to the human cytomegalovirus (HCMV) gB epitope or G1102 to ICP35 was purchased from Goodwin Institute (Plantation, Fla.). ICP0 polyclonal antibody was described previously (18). Goat anti-mouse or anti-rabbit immunoglobulin G alkaline phosphatase-conjugated secondary antibodies were purchased from Bio-Rad (Hercules, Calif.).
Generation of US5 polyclonal antibody.
The US5 maltose binding protein expression plasmid pRB5154, containing the entire coding region of US5, was transfected into Escherichia coli. The induction and purification procedure of maltose binding fusion protein was done as recommended by the manufacturer (New England BioLabs, Beverly, Mass.). A New Zealand White female rabbit was injected subcutaneously five times, each time with 250 to 400 μg of fusion protein emulsified with Freund adjuvant.
Detection of viral proteins in lysates electrophoretically separated in a denaturing polyacrylamide gel.
Mock-infected or HSV-1-infected Vero or BHK (TK+) cells were harvested in disruption buffer (50 mM Tris-HCl [pH 7.0], 2% sodium dodecyl sulfate (SDS), 0.7 M β-mercaptoethanol, 2.75% sucrose), electrophoretically separated in a denaturing polyacrylamide gel cross-linked with N,N′-diallyltartardiamide, electrophoretically transferred to a nitrocellulose membrane (Schleicher & Schuell), and probed with appropriate antibodies. The immunoblotting procedure was as described elsewhere (7). The molecular weight marker was obtained with the LMW electrophoresis calibration kit (Pharmacia Biotech, Uppsala, Sweden).
Generation of plasmids and recombinant viruses.
All molecular cloning was done by standard techniques as described elsewhere (29). The maltose binding protein-US5 chimeric construct was cloned as pRB5154 and contained the entire US5 coding sequence. The US5 coding sequence with an insertion at the EcoO109 site of an oligomer encoding the HCMV gB epitope (5′ AAAAGGG ACAGAAGCCCAACCTGCTAGACCGACTGCGACACCGCAAAAACGG GTACCGACACC 3′) was ligated to the UL26.5 promoter sequence (−599 to +44, relative to the transcription start site of the UL26.5 gene) to create pRB4152. The UL26.5 promoter-US5 construct was cloned into the HSV-1(F) thymidine kinase (tk) gene in place of the BglII/EcoNI sequence of the coding domain of the tk gene to create pRB5175. In turn, the HSV sequence in pRB5175 was recombined into HSV-1(F) as previously described to generate the recombinant virus R5175.
pRB4351 contained an α27-tk construct cloned into the NcoI site between the UL26 and UL27 genes (Fig. 1C, line 6). pRB4492 contained an α27-tk construct between the UL25 and UL26 genes. In order to insert the α27-tk construct and at the same time provide a promoter for the UL26 gene, a series of clones were generated. A BglII/EcoRI fragment from pRB4454 containing the α27-tk cassette was cloned into the BamHI/EcoRI sites of pRB4428 to create pRB4463. pRB4463 contains the 3′ terminus of the UL25 gene and the inserted α27-tk construct. Subsequently, a NarI fragment containing an α4 promoter (−604 to +25 relative to the transcription start site of the α4 gene) UL26 5′ terminal sequence from pRB4060 was cloned into the NarI site of pRB4463 to create pRB4492. pRB4351 and pRB4492 were recombined into the HSV-1(F)Δ305 genome to create recombinant viruses R4351 and R4492, respectively.
FIG. 1.
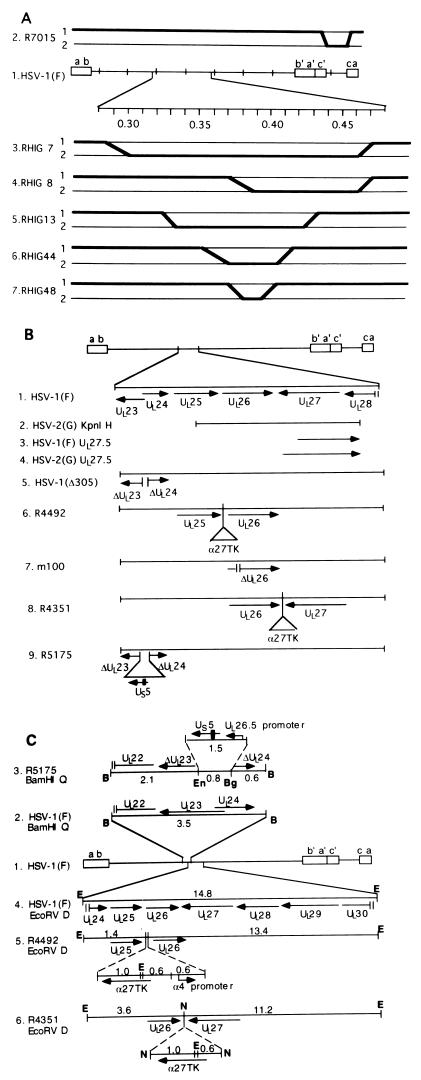
(A) Schematic diagram of genome structures of HSV-1(F) and HSV-1(F) × HSV-2(G) intertypic recombinants. Line 1, HSV-1(F) genome arrangement. Open rectangles represent inverted repeats, and the single lines in between represent unique long (UL) and unique short (US) regions of the genome. Lines 2 to 7, schematic diagram of genome arrangement of intertypic recombinants used in this study. Lines labeled 1 represent HSV-1(F) sequences, and lines labeled 2 represent HSV-2(G) sequences. Crossovers are indicated by bold-faced lines. R7015 (line 2) has the HSV-1(F) unique short region replaced with the HSV-2(G) unique short sequence. Lines 3 to 7 are a set of intertypic recombinants that are basically HSV-1(F), with various portions between map units 0.3 and 0.45 replaced with the HSV-2(G) sequence. (B) Schematic diagram of HSV-1(F) and various recombinants used in this study. Line 1, genome arrangement of HSV-1(F) and location of ORFs UL23 to UL28. Arrow indicates the polarity and position of each ORF. Line 2, position of the HSV-2(G) KpnI H fragment relative to the location of ORFs. Lines 3 and 4, positions of proposed new ORF UL27.5 in the HSV-1(F) and HSV-2(G) genomes, respectively. Lines 5 to 9, genome organization of various recombinants. Short vertical lines represent deletions (lines 5, 7, and 9). Triangles represent insertions (lines 6, 8, and 9). Filled rectangle represents HCMV epitope tag (line 9). (C) Line 1, sequence arrangement of HSV-1(F). Line 2, ORFs in HSV-1(F) BamHI Q fragment. Line 3, in R5175, an 0.8-kb EcoNI/BglII fragment within the UL23-to-UL24 region is replaced by a 1.5-kb UL26.5 promoter-US5 construct. Line 4, ORF arrangement in HSV-1(F) EcoRV D fragment. Line 5, in R4492, a 2.2-kb α27tk-α4 promoter construct is inserted between the UL25 and UL26 genes. Line 6, in R4351, a 1.6-kb α27tk construct is inserted between the UL26 and UL27 genes. B, BamHI; Bg, BglII; E, EcoRV; En, EcoNI; N, NcoI.
The HSV-2(G) KpnI H fragment subcloned from pAV6 (a kind gift from Aviron, Mountain View, Calif.), containing HSV-2 genes UL25 to UL28, was cloned into pGEM3Zf(+) as pRB812. pRB812 was recombined into the R4492 viral genome to create a series of intertypic recombinant viruses (K-2 to K-7, K-9, and K-10).
All recombinant viruses made for these studies were generated by homologous recombination. Rabbit skin cells were transfected with viral DNA and cotransfected with appropriate plasmid DNA as previously described (7). The transfection was plated on 143TK− cells and selected either for tk+ progeny viruses, e.g., R4351 and R4492 in HAT medium (15 μg of hypoxanthine, 0.2 μg of aminopterin, and 5 μg of thymidine per ml) or tk mutant progeny viruses (e.g., R5175) in medium containing 40 μg of bromodeoxyuridine per ml. Individual isolates were plaque purified on Vero cells, and their sequences and gene expression were verified by Southern blot and immunoblot analyses, respectively.
Analyses of viral DNA by hybridization.
Cytoplasmic DNAs were purified from infected cells as described elsewhere (16), digested with either BamHI or EcoRV, separated by agarose gel electrophoresis, and transferred to a nylon membrane (Bio-Rad). The hybridization procedure was done according to the manufacturer’s recommendations. Either the HSV-1(F) BamHI Q fragment cloned as pRB165 or the gel-purified HSV-1(F) EcoRV D fragment was used as a probe. All probes were internally labeled with [α32P]dCTP (800 Ci/mmol; DuPont, NEN, Boston, Mass.) with a nick translation kit (DuPont, NEN).
Generation of GST-UL27.5 fusion protein.
The sequence encoding UL27.5 codons 299 to 412 was amplified by PCR and cloned into glutathione S-transferase (GST) expression vector pGEX-KG as pRB5132 (12). The induction and purification of a GST fusion protein were done as recommended by the manufacturer (Promega, Madison, Wis.). Affinity-purified fusion protein was loaded onto an SDS–15% polyacrylamide gel and stained with Coomassie blue or electrophoretically transferred to a nitrocellulose membrane and subjected to immunoblot analysis.
In vitro transcription and translation.
The UL27.5 ORF amplified by PCR was cloned into pGEM3Zf(+) as pRB5169 and used as a template for in vitro transcription and translation. The reaction was carried out in a TNT coupled transcription/translation system (Promega) with T7 RNA polymerase in the presence of [14C]leucine (0.25 μCi, 342 mCi/mmol; DuPont, NEN). The final product was denatured in disruption buffer and subjected to electrophoresis on an SDS–15% polyacrylamide gel and electrophoretically transferred to a nitrocellulose sheet. The membrane was treated with En3Hance spray (Dupont, NEN) and subjected to autoradiography.
Cytoplasmic and nuclear fractionation.
BHK(TK+) cells were mock infected or exposed to 10 PFU of HSV-1(F) or HSV-2(G) per cell and harvested at 18 h after infection. Infected cells were collected by low-speed centrifugation (1,000 × g for 5 min), resuspended in buffer A (10 mM HEPES buffer [pH 7.4], 10 mM NaCl, 1.5 mM MgCl2), and stored on ice for 10 min. The cells were subjected to five strokes of Dounce homogenization and incubated on ice for 10 min. The cytoplasmic fraction was collected after centrifugation (12,000 × g at 4°C for 20 min). The pellet was washed once with buffer A and resuspended in buffer B (10 mM HEPES buffer [pH 7.4], 420 mM NaCl, 1.5 mM MgCl2). The pellet was briefly sonicated to facilitate resuspension, and the nuclear fraction was collected after the removal of any insoluble material by centrifugation (12,000 × g at 4°C for 5 min). The proteins in the cytoplasmic and nuclear fractions were denatured by the addition of disruption buffer, boiled for 5 min, and subjected to SDS-polyacrylamide gel electrophoresis.
RESULTS
Genomic structure of recombinant viruses R4351, R4492, and R5175.
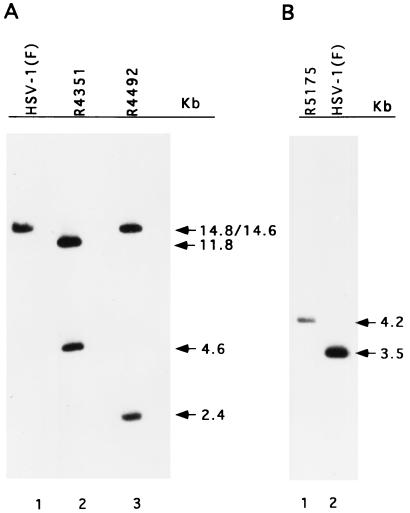
Recombinant viruses were constructed by double homologous recombination as described in Materials and Methods, and their structures were verified by Southern blot analyses. Cytoplasmic DNAs from infected cells were digested with either the BamHI or the EcoRV enzyme, separated on an agarose gel, transferred to a nylon membrane, and hybridized with appropriate probes. As illustrated in Fig. 1C, lines 2 and 3, the replacement of the BglII/EcoNI fragment of the tk gene with a UL26.5 promoter-US5-HCMV tag construct increased the size of HSV-1(F) BamHI-Q from 3.5 to 4.2 kb, as detected when 32P-labeled pRB165 was used as the probe (Fig. 2B). To verify the predicted genomic structure of R4351 and R4492, electrophoretically separated EcoRV digests of the recombinant virus DNAs were probed with a 32P-labeled HSV-1(F) EcoRV D fragment. There are two closely positioned EcoRV sites in the α27-tk construct. The EcoRV-D probe showed, as predicted, that insertion of the α27-tk construct and the α4 promoter into R4492 (Fig. 1C, lines 4 and 5) replaced the original EcoRV D fragment (14.8 kb) with two fragments of 2.4 and 14.6 kb in length (Fig. 2A, lanes 1 and 3). In the same fashion, the insertion of the α27-tk cassette into the R4351 viral genome (Fig. 1C, line 6) resulted in the replacement of the EcoRV D fragment with two fragments of 4.6 and 11.8 kb in length (Fig. 2A, lane 2). We conclude that the three recombinant viruses made in these studies show the expected genotype.
FIG. 2.
Autoradiography of electrophoretically separated DNA hybridized with 32P-labeled probe. DNA of HSV-1(F) or recombinant viruses was digested with EcoRV (A) or BamHI (B), electrophoretically separated in agarose gel, transferred to a nylon membrane, and hybridized with labeled EcoRV D fragment (A) or BamHI Q fragment (B).
US5 rabbit polyclonal antibody reacted with two viral proteins by immunoblot analyses.
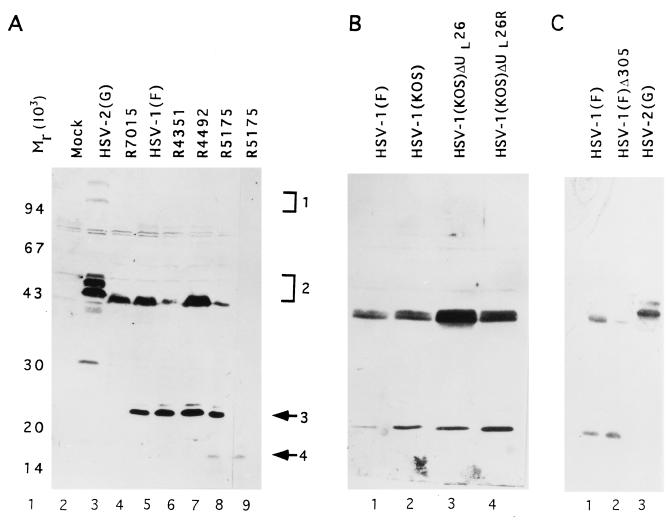
Although the rabbit polyclonal antibody was made against the US5 protein, in immunoblot assays the anti-US5 polyclonal antibody reacted with two bands containing different proteins. Mock-infected or HSV-1(F)-infected BHK(TK+) cells were harvested in disruption buffer at 18 h after infection, electrophoretically separated in an SDS–15% polyacrylamide gel, electrophoretically transferred to a nitrocellulose sheet, and probed with the US5 polyclonal antiserum. The results (Fig. 3A, lane 5) reproducibly showed that the serum reacted with two virus-specific HSV-1 proteins with apparent Mrs of 43,000 and 23,000, respectively (designated band 2 and band 3, respectively). The antibody did not react with lysates of mock-infected cells (Fig. 3A, lane 1). In HSV-2(G)-infected cell lysate, only one virus-specific signal, with a mobility on a denaturing gel equivalent to that of HSV-1(F) band 2, was detected as a heterogeneous group of bands with apparent Mrs ranging from 43,000 to 50,000. The HSV-2 equivalent of HSV-1(F) band 3 was not detected (Fig. 3A, lane 3). Overdeveloped immunoblots occasionally showed another faint band with an apparent Mr of greater than 100,000 (designated band 1).
FIG. 3.
Photograph of immunoblot of electrophoretically separated proteins reacted with proper antibodies. Vero cells mock infected or infected with HSV were harvested at 18 h postinfection, electrophoretically separated onto an SDS–15% polyacrylamide gel, electrically transferred to a nitrocellulose membrane, and reacted with US5 polyclonal antibody (panel A, lanes 2 to 8, and panels B and C) or monoclonal antibody recognizing the HCMV epitope (CH28) (panel A, lane 9). (B) Lane 3, HSV-1(KOS)ΔUL26 (UL26 deletion virus m100); lane 4, HSV-1(KOS)ΔUL26R (UL26 repaired virus). Molecular weight markers are shown in lane 1, and bands 1 to 4 are indicated at the right of panel A.
The reactive band 3 was encoded by a gene residing in the US sequence of HSV-1(F) inasmuch as the signal was missing in the intertypic recombinant virus R7015 (Fig. 3, lane 4). As noted in Materials and Methods, in this recombinant, the US sequence of HSV-1(F) was replaced with the corresponding sequence of HSV-2(G). The hypothesis that the protein contained in band 3 is encoded by the US5 gene was supported by studies of recombinant R5175. In this recombinant, a second copy of US5 coding sequence tagged with an HCMV gB epitope was inserted into the tk gene. The polyclonal rabbit serum made against the US5 protein reacted with a fourth band in electrophoretically separated lysates of R5175 virus-infected cell lysates (Fig. 3A, lane 8, band 4). Band 4 comigrated with the signal detected in R5175 reacted only with the monoclonal antibody to the HCMV gB epitope (Fig. 3, compare lanes 8 and 9). The CMV epitope-tagged second copy US5 protein migrated with an apparent Mr of about 18,000, which is faster than the authentic US5 protein. One explanation for the discrepancy in the electrophoretic mobilities is that insertion of the epitope blocks the glycosylation of the protein. The studies with the gJ protein will be dealt with elsewhere.
The gene encoding protein in band 2 maps in UL.
In this section, we describe three series of experiments designed to map the gene encoding band 2 protein. We took advantage of the difference in the electrophoretic mobilities of the HSV-1 and HSV-2 homologs of band 2 protein.
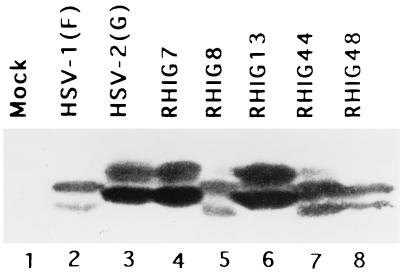
The first series of experiments was done with a series of HSV-1 × HSV-2 recombinants shown in Fig. 1A. These recombinants have been extensively studied for mapping studies, and the crossover sites are well known. Replicate Vero cell cultures were exposed to 10 PFU of a wild-type or recombinant virus, incubated at 37°C, harvested at 18 h after infection, solubilized in disruption buffer, electrophoretically separated in a denaturing polyacrylamide gel, transferred to a nitrocellulose sheet, and reacted with the anti-US5 antibody. The results shown in Fig. 4 indicate that the recombinant viruses RHIG7 and RHIG13 encoded the HSV-2(G) form of band 2 protein (Fig. 4, lanes 3, 4, and 6), whereas the recombinants RHIG8, RHIG44, and RHIG48 yielded the HSV-1(F) form of band 2 protein (Fig. 4, lanes 2, 5, 7, and 8). These results indicated that the sequence encoding the band 2 protein maps between map units 0.33 and 0.37, that is, within the genome domain encoding the UL23 to UL27 genes (Fig. 1A) (22).
FIG. 4.
Photograph of immunoblot of electrophoretically separated proteins reacted with US5 polyclonal antibody. Vero cells mock infected or infected with wild-type HSV or intertypic recombinants, harvested at 18 h postinfection, electrophoretically separated in a denaturing polyacrylamide gel, transferred to a nitrocellular membrane, and reacted with US5 polyclonal antibody.
The second series of studies was designed to determine whether the gene encoding band 2 protein maps to UL23 to UL26 on the basis of the synthesis of band 2 protein in cells infected with mutants with changes in these genes. Thus, several recombinant viruses with mutations in this region, including deletions in the domains of UL23 and UL24 [HSV-1(F)Δ305 and R5175] and UL26 [HSV-1(KOS)m100] (11) and disruption of the UL25-to-UL26 (R4492) and UL26-to-UL27 interfaces (R4351), were tested for alteration in the mobility of band 2 protein. In this series of experiments, Vero cells were infected and processed as described above. The results were that the electrophoretic mobility of band 2 protein specified by the recombinants tested in these studies was no different from that predicted (9, 17) or specified by HSV-1(F) (Fig. 3A, lanes 5 to 7, Fig. 3B, lanes 1 to 4, and Fig. 3C, lanes 1 to 3). Band 3 protein was detected in all of the recombinant viruses tested in this series of experiments.
We conclude from this series of experiments that the band 2 protein was not encoded by UL23, UL24, or UL26 ORFs inasmuch as deletions in these genes had no effect on the mobility of band 2. Furthermore, the mobility of band 2 on denaturing polyacrylamide gel was different from what would be expected for UL25 or UL27 gene products (2, 3, 13, 22–24). Therefore, we conclude that band 2 protein is derived from a previously unidentified viral ORF between map units 0.33 and 0.37.
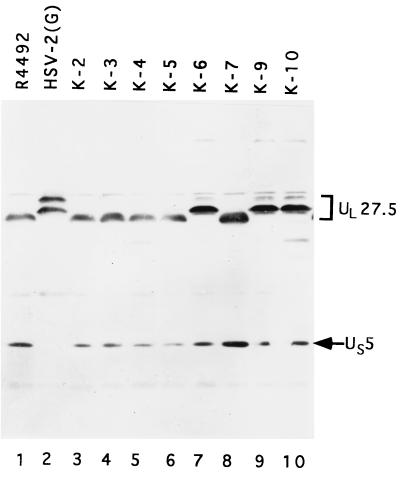
In the third series of experiments, we generated a series of recombinant viruses in which the HSV-1 ORFs UL25 to UL28 were replaced with the equivalent HSV-2(G) sequences. The strategy was to cotransfect the HSV-2 sequences in pRB812 with intact R4492 viral DNA and select for the TK− phenotype. In the process, the α27-tk chimeric gene inserted into the R4492 genome in the intergenic domain between UL25 and UL26 was replaced with the HSV-2(G) DNA sequences cloned in pRB812. Replicate Vero cell cultures, each exposed to 10 PFU of intertypic recombinant per cell, were then analyzed for the presence of the HSV-2(G) band 2 protein in the HSV-1(F) background. As shown in Fig. 5, the lysates of cells infected with isolates K-6, K-9, and K-10 exhibited band 2 proteins which comigrated with band 2 of HSV-2(G), that is, migrated more slowly than the HSV-1 band 2 protein present in the lysate from parent virus R4492 (Fig. 5, lane 1). The slower-migrating species, designated UL27.5 in this figure for reasons detailed in the next section, appeared as a doublet similar to the band 2 in HSV-2(G)-infected lysate, although the upper band was not as prominent as in HSV-2(G)-infected lysate. Since the crossover could have occurred proximal to the location of the ORF encoding band 2 protein, not all of the recombinants in this series exhibited or were predicted to exhibit an HSV-2(G) band 2 phenotype. In no instance was the electrophoretic mobility of the band 3 (US5) protein affected. Based on the observation that an insertion between UL26 and UL27 (R4351) or between UL25 and UL26 (R4492) and a truncation in the amino terminus of the UL26 gene (m100) had no effect on the mobility of band 2, we conclude that the new ORF could reside completely within the UL25 ORF, within the carboxyl-terminal portion of the UL26 ORF, or within the region between UL27 and UL28 (Fig. 1B).
FIG. 5.
Photograph of immunoblot of electrophoretically separated proteins reacted with US5 polyclonal antibody. Vero cells were infected with various intertypic recombinants and harvested at 18 h postinfection. Proteins were electrophoretically separated on an SDS–15% polyacrylamide gel, transferred to a nitrocellulose membrane, and reacted with US5 polyclonal antibody. The bands formed by the UL27.5 and US5 proteins are identified at the right.
Sequence analysis predicts new ORF antisense to UL27.
We next searched for potential ORFs in the target regions stated above. We focused on new ORFs conserved in HSV-1 and HSV-2 with the capacity to encode at least 200 amino acids. As shown in Fig. 1B, lines 3 and 4, only one ORF, designated UL27.5 and predicted to encode an HSV-1 protein of 575 amino acids, and an HSV-2 protein of 985 amino acids met these criteria. Sequence comparison showed that, except for the amino-terminal region of the predicted HSV-2 UL27.5 ORF, the HSV-1 and HSV-2 UL27.5 ORFs were homologous (Fig. 6). Since the US5 polyclonal antibody cross-reacted with the denatured band 2 protein, the cross-reacting epitope is predicted to be linear and could potentially be deduced by comparing primary amino acid sequences. Amino acid sequence comparison of HSV-1(F) and HSV-2(G) UL27.5 and US5 revealed very limited sequence homology (Fig. 7). The sequence of the US5 gene predicts a hydrophobic protein. Therefore, the observation that the limited homology resided in a rather hydrophobic stretch of amino acids was not unexpected.
FIG. 6.
Amino acid sequence alignment of the proposed new ORF UL27.5 from HSV-1(F) (top line) and HSV-2(G) (middle line). The consensus sequence is shown on the bottom line. Numbers indicate the amino acid number based on the sequence of the HSV-2(G) ORF. Note the slight sequence variation between HSV-1(F) and HSV-2(G) from 855 to 934, which gives the HSV-2(G) UL27.5 ORF a total amino acid residue count of 985.
FIG. 7.
Amino acid sequence alignment between HSV-2(G) UL27.5 amino acids 701 to 847 (the second line), the equivalent sequence in HSV-1(F) UL27.5 (top line), and the entire coding sequence of HSV-1(F) US5 (the third line). The consensus sequence is shown at the bottom.
A GST-UL27.5 fusion protein expressing the US5 homologous region of UL27.5 reacted with the US5 polyclonal antibody by immunoblot analysis.
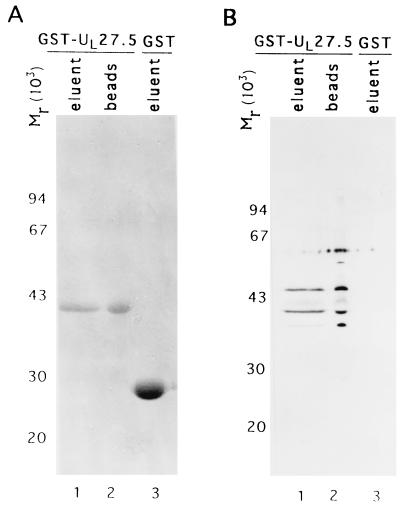
A GST-UL27.5 fusion protein expressing the homologous region of the HSV-1 UL27.5 coding sequence (Fig. 6, HSV-1 amino acids 299 to 412) was constructed, purified on affinity columns, and subjected to electrophoresis in denaturing polyacrylamide gels. The eluted GST-UL27.5 chimeric protein, the chimeric protein remaining bound to the affinity resin and eluted by solubilization in disruption buffer, and the eluted GST protein were each separated on a 15% denaturing gel and stained with Coomassie blue (Fig. 8A, lanes 1, 2, and 3, respectively). Portions of the same preparation were electrophoretically separated in the same gel, electrically transferred to a nitrocellulose membrane, and probed with US5 polyclonal antibody (Fig. 8B). The results were that the GST-UL27.5 fusion protein migrated at an expected Mr of 42,000 on denaturing gel (panel A, lanes 1 and 2) (2). Both eluted GST-UL27.5 and resin-associated GST-UL27.5 reacted with the US5 polyclonal antibody on an immunoblot, whereas GST alone did not (Fig. 8B, lanes 1, 2, and 3). These results are consistent with the results of the mapping studies and indicate that UL27.5 encodes a protein containing an epitope common to the US5 protein.
FIG. 8.
Photograph of gel stained with Coomassie blue (A) or immunoblot of proteins electrophoretically separated in an SDS-polyacrylamide gel and reacted with US5 polyclonal antibody (B). GST-UL27.5 fusion protein was induced as described in Materials and Methods. The eluent (lane 1), the remaining fusion protein bound to the affinity resin (lane 2), and the eluent of GST protein (lane 3) were electrophoretically separated in an SDS–15% polyacrylamide gel and stained with Coomassie blue (A). Equal amounts of each sample were separated in the same fashion, electrically transferred to a nitrocellulose membrane, and reacted with US5 polyclonal antibody (B). Molecular weight marker is shown at the left.
In vitro-translated product of the UL27.5 coding sequence migrated as a protein with a molecular weight of 65,000.
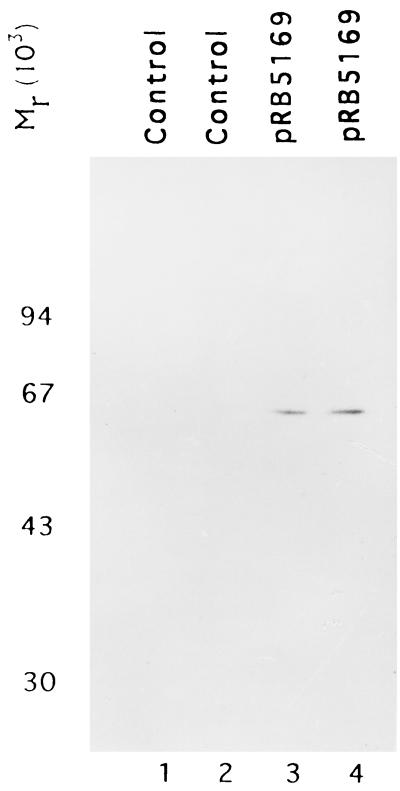
The primary sequence of the UL27.5 ORF has the capacity to encode a protein of 575 amino acids. The apparent molecular weight of band 2 on denaturing polyacrylamide gel was 43,000. To test if the discrepancy between the predicted and observed size is due to the nature of the protein or to posttranslational modification, we translated the UL27.5 open reading frame with a coupled in vitro transcription translation system and analyzed the protein in a denaturing gel. As shown in Fig. 9, lanes 3 and 4, the only [14C]leucine-labeled species, supposedly the full-length product, migrated with an apparent Mr of 65,000. A minor protein band with an apparent Mr of 65,000 was also observed in immunoblots reacted with the US5 antibody (Fig. 4).
FIG. 9.
Autoradiography of [14C]leucine-labeled in vitro-translated UL27.5 protein immobilized on a nitrocellulose membrane. Vector pGEM3Zf(+) (lanes 1 and 2) or pRB5169 containing the proposed HSV-1(F) UL27.5 coding sequence (lanes 3 and 4) was in vitro translated in a programmed rabbit reticulocyte lysate, labeled with [14C]leucine, electrophoretically separated in an SDS–15% polyacrylamide gel, electrically transferred onto a nitrocellulose membrane, and subjected to autoradiography. Lanes 1, 2, 3, and 4 represent four independent experiments.
As a general rule, HSV proteins migrate with an apparent Mr larger than that predicted on the basis of their amino acid sequences. The apparent Mr obtained for the in vitro transcription-translation product is consistent with an HSV protein of 575 amino acids and suggests that either the domain translated is smaller than the ORF or that the protein is processed by cleavage in the environment of the infected cell.
UL27.5 is a γ2 gene.
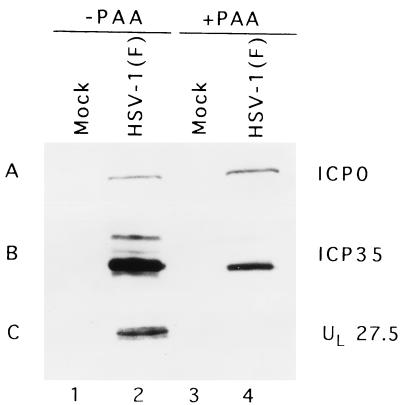
In this series of experiments, replicate Vero cultures were either mock infected or infected with HSV-1(F) and either left untreated or incubated in medium containing phosphonoacetic acid (PAA) (300 μg/ml of medium; a gift of Abbott Laboratories) throughout the course of infection. The cells were harvested at 18 h after infection, solubilized in disruption buffer, subjected to electrophoresis, electrically transferred onto a membrane, and probed with appropriate antibodies to viral proteins. As shown in Fig. 10B, lanes 2 and 4, the treatment of PAA was effective, as evidenced by the reduced accumulation of ICP35 (a γ1 gene). In contrast to ICP0 and the ICP35 protein, UL27.5 was not detected in infected cells treated with PAA (Fig. 10C). We conclude therefore that UL27.5 is a γ2 gene totally dependent on viral DNA synthesis for its expression.
FIG. 10.
Photograph of immunoblot of electrophoretically separated infected-cell proteins reacted with indicated antibodies. Vero cells were mock infected or infected with HSV-1(F) in the presence or absence of PAA and harvested at 18 h postinfection. Lysates were electrophoretically separated in a denaturing polyacrylamide gel, transferred to a nitrocellulose membrane, and reacted with antibodies to ICP0 (A) and ICP35 (B) and the US5 polyclonal antibody (C).
UL27.5 protein accumulates in the cytoplasm.
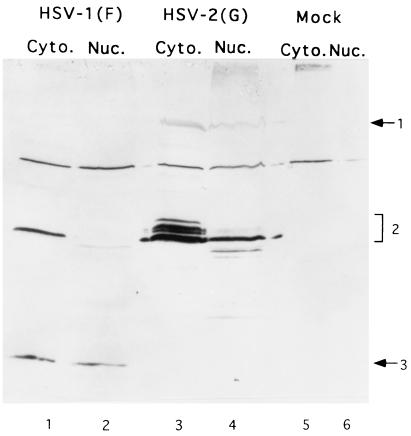
BHK(TK+) cells were either mock infected or exposed to 10 PFU of HSV-1(F) or of HSV-2(G) per cell and incubated at 37°C. The cells were harvested at 18 h after infection. The cytoplasmic and nuclear fractions were collected as described in Materials and Methods, solubilized, electrophoretically separated onto a denaturing polyacrylamide gel, electrically transferred to a membrane, and reacted with the anti-US5 antibody. As shown in Fig. 11, lanes 1 and 2, the UL27.5 protein accumulated exclusively in the cytoplasmic fraction. In HSV-2(G)-infected cell lysates, the majority of the signal was detected in the cytoplasmic fraction. A slightly more rapidly migrating protein band was detected in both the nuclear and cytoplasmic fractions whereas a less abundant and more rapidly migrating protein band was detected only in the nuclear fraction (Fig. 11, lanes 3 and 4).
FIG. 11.
Photograph of immunoblot of electrophoretically separated proteins reacted with US5 polyclonal antibody. BHK(TK+) cells were either mock infected or infected with HSV for 18 h. The cytoplasmic and nuclear fractions were collected as described in Materials and Methods, electrophoretically separated onto an SDS–15% polyacrylamide gel, electrically transferred to a nitrocellulose membrane, and reacted with US5 polyclonal antibody. Band 1, 2, and 3 proteins are indicated at the right.
DISCUSSION
A polyclonal rabbit antibody made against gJ reacted with a protein expressed by both HSV-1 and HSV-2. The gene encoding this protein has been mapped to an ORF antisense to the gene encoding gB. In this article, we report on the mapping of the gene and preliminary characterization of the product of the ORF. The salient features of this report are as follows.
(i) The protein is encoded by the UL27.5 gene on the basis of three kinds of data, i.e., mapping of two independently derived intertypic recombinants, analyses of insertion and deletion mutants, and direct reaction of the antibody with a chimeric protein containing the relevant domain of the ORF expressed in E. coli.
(ii) The UL27.5 ORFs predict an HSV-1 protein of 575 amino acids and an HSV-2 protein of 985 amino acids. The HSV-1 sequence predicts a single methionine codon, whereas the HSV- 2 sequence predicts two methionines at positions 1 and 669. Translation in vitro of the entire HSV-1 ORF yielded a protein with an apparent Mr of 65,000. However, this observation does not prove that translation initiation occurs at the initiator methionine of the ORF, and there is no substantive evidence that the accumulating HSV-2 proteins are derived from a high-molecular-weight precursor. Initiation at the second methionine codon of the HSV-2 UL27.5 open reading frame would predict a protein much smaller than the HSV-2 protein accumulating in the infected cell. We cannot at this time exclude the possibility that translation initiation begins internally from an alternate translation initiation codon (19).
(iii) Most of the HSV-1 and HSV-2 UL27.5 accumulated in the cytoplasm. A small amount of HSV-2 UL27.5 protein migrating faster than the cytoplasmic protein partitioned in the nucleus. We should note that the amount of HSV-2 protein accumulating in infected cells is higher or reacts better with the antibody than the corresponding HSV-1 protein. It is conceivable that a small amount of HSV-1 UL27.5 also accumulates in the nucleus.
Because UL27.5 maps antisense to a gene essential for viral replication, it has not been possible at this time to determine whether the UL27.5 gene product is also essential. Experiments now in progress should allow us to assess the role of this gene in cell culture and in an experimental animal system.
(iv) The discovery of a gene antisense to gB (UL27) was totally unexpected. Most analytical tools used to analyze nucleotide sequences are based on the assumption that a coding domain is present in the form of a linear array of nucleotides on one strand only. The gB-UL27.5 pair of antisense genes is the third set discovered within the HSV-1 genome (6, 20, 26). The ease with which they have been discovered in the past few years suggests that there may be more such pairs. Given the fact that the size of the capsids is conserved and virtually identical for all herpesviruses and that the capsids could package >240 kbp of DNA (e.g., the HCMV genome), the question arises as to why HSV encodes genes antisense to each other rather than stringing these ORFs in linear arrays. Among the many possible explanations, three are worthy of further discussion.
The first, less-interesting hypothesis is that even the large herpesviruses contain genes arranged antisense to each other and that the actual number of genes in HCMV is grossly underestimated. It is conceivable that the antisense arrangements antedate the divergence of the primordial herpesvirus into the various subfamilies now in existence.
The second, more-attractive hypothesis is that the antisense arrangement is a form of regulation of gene expression that determines both the timing of synthesis and the abundance of the gene product. In the two preceding cases, that is, γ134.5/ORF P and ORF O and UL43/UL43.5, we have found that the expression of the genes situated antisense to each other was sequential or even mutually exclusive (6, 20, 26). In this instance, gB is expressed very early in infection in abundant amounts, whereas the UL27.5 gene expression appears to be a late event, dependent on viral DNA synthesis. One test of the hypothesis would be to reverse the timing of the expression of the two genes to determine whether early expression of the UL27.5 is deleterious.
Lastly, the possibility that the sequences of two ORFs fit such that both proteins contain only the amino acid sequences essential for their function is probably remote. It is more likely that key amino acid domains of one protein correspond to neutral or linker domains in the product of the antisense ORF. Once the precise sequence encoding the UL27.5 protein is elucidated, it may be possible to probe more accurately the corresponding domains of the gB gene.
ACKNOWLEDGMENTS
We thank Min Gao for HSV-1(KOS)ΔUL26 virus and A. Louise McCormick for the HSV-1(KOS)ΔUL26R virus.
These studies were aided by grants from the National Cancer Institute (CA47451 and CA71933) to the University of Chicago laboratory and by AIDS Project from Istituto Superiore di Sanita BIOMED2 BMH4 CT95 1016 from the European Community Target Project in Biotechnology to the University of Bologna laboratory.
REFERENCES
- 1.Ackermann M, Longnecker R, Roizman B, Pereira L. Identification, properties, and gene location of a novel glycoprotein specified by herpes simplex virus 1. Virology. 1986;150:207–220. doi: 10.1016/0042-6822(86)90280-1. [DOI] [PubMed] [Google Scholar]
- 2.Addison C, Rixon F J, Palfreyman J W, O’Hara M, Preston V G. Characterisation of a herpes simplex virus type 1 mutant which has a temperature-sensitive defect in penetration of cells and assembly of capsids. Virology. 1984;138:246–259. doi: 10.1016/0042-6822(84)90349-0. [DOI] [PubMed] [Google Scholar]
- 3.Ali M A, Forghani B, Cantin E M. Characterization of an essential HSV-1 protein encoded by the UL25 gene reported to be involved in virus penetration and capsid assembly. Virology. 1996;216:278–283. doi: 10.1006/viro.1996.0061. [DOI] [PubMed] [Google Scholar]
- 4.Balan P, Davis-Poynter N, Bell S, Atkinson H, Browne H, Minson T. An analysis of the in vitro and in vivo phenotypes of mutants of herpes simplex virus type 1 lacking glycoproteins gG, gE, gI or the putative gJ. J Gen Virol. 1994;75:1245–1258. doi: 10.1099/0022-1317-75-6-1245. [DOI] [PubMed] [Google Scholar]
- 5.Bzik D J, Fox B A, Deluca N A, Person S. Nucleotide sequence specifying the glycoprotein gene gB of herpes simplex virus type 1. Virology. 1984;133:301–314. doi: 10.1016/0042-6822(84)90397-0. [DOI] [PubMed] [Google Scholar]
- 6.Carter K L, Ward P L, Roizman B. Characterization of the products of the UL43 gene of herpes simplex virus 1: potential implications for regulation of gene expression by antisense transcription. J Virol. 1996;70:7663–7668. doi: 10.1128/jvi.70.11.7663-7668.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang Y E, Roizman B. The product of the UL31 gene of herpes simplex virus 1 is a nuclear phosphoprotein which partitions with the nuclear matrix. J Virol. 1993;67:6348–6356. doi: 10.1128/jvi.67.11.6348-6356.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Conley A J, Knipe D M, Jones P C, Roizman B. Molecular genetics of herpes simplex virus. VII. Characterization of a temperature-sensitive mutant produced by in vitro mutagenesis and defective in DNA synthesis and accumulation of γ polypeptides. J Virol. 1981;37:191–206. doi: 10.1128/jvi.37.1.191-206.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cook W J, Coen D M. Temporal regulation of herpes simplex virus type 1 UL24 mRNA expression via differential polyadenylation. Virology. 1996;218:204–213. doi: 10.1006/viro.1996.0180. [DOI] [PubMed] [Google Scholar]
- 10.Ejercito P M, Kieff E D, Roizman B. Characterization of herpes simplex virus strains differing in their effects on social behaviour of infected cells. J Gen Virol. 1968;2:357–364. doi: 10.1099/0022-1317-2-3-357. [DOI] [PubMed] [Google Scholar]
- 11.Gao M, Matusick-Kumar L, Hurlburt W, Ditusa S F, Newcomb W W, Brown J C, McCann III P J, Deckman I, Colonno R J. The protease of herpes simplex virus type 1 is essential for functional capsid formation and viral growth. J Virol. 1994;68:3702–3712. doi: 10.1128/jvi.68.6.3702-3712.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guan K, Dixon J E. Eukaryotic protein expression in Escherichia coli: an improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal Biochem. 1991;192:262–267. doi: 10.1016/0003-2697(91)90534-z. [DOI] [PubMed] [Google Scholar]
- 13.Holland L E, Sandri-Goldin R M, Goldin A L, Glorioso J C, Levine M. Transcriptional and genetic analysis of the herpes simplex virus type 1 genome: coordinates 0.29 to 0.45. J Virol. 1984;49:947–959. doi: 10.1128/jvi.49.3.947-959.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Honess R W, Roizman B. Regulation of herpes virus macromolecular synthesis. I. Cascade regulation of the synthesis of three groups of viral proteins. J Virol. 1974;14:8–19. doi: 10.1128/jvi.14.1.8-19.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Honess R W, Roizman B. Regulation of herpesvirus macromolecular synthesis: sequential transition of polypeptide synthesis requires functional viral polypeptides. Proc Natl Acad Sci USA. 1975;72:1276–1280. doi: 10.1073/pnas.72.4.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Igarashi K, Fawl R, Roller R J, Roizman B. Construction and properties of a recombinant herpes simplex virus 1 lacking both S-component origins of DNA synthesis. J Virol. 1993;67:2123–2132. doi: 10.1128/jvi.67.4.2123-2132.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jacobson J G, Martin S L, Coen D M. A conserved open reading frame that overlaps the herpes simplex virus thymidine kinase gene is important for viral growth in cell culture. J Virol. 1989;63:1839–1843. doi: 10.1128/jvi.63.4.1839-1843.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kawaguchi Y, Van Sant C, Roizman B. Herpes simplex virus 1 alpha regulatory protein ICP0 interacts with and stabilizes the cell cycle regulator cyclin D3. J Virol. 1997;71:7328–7336. doi: 10.1128/jvi.71.10.7328-7336.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kozak M. Recognition of AUG and alternative initiator codons is augmented by G in position +4 but is not generally affected by the nucleotides in positions +5 and +6. EMBO J. 1997;16:2482–2492. doi: 10.1093/emboj/16.9.2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lagunoff M, Roizman B. The regulation of synthesis and properties of the protein product of open reading frame P of the herpes simplex virus 1 genome. J Virol. 1995;69:3615–3623. doi: 10.1128/jvi.69.6.3615-3623.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Longnecker R, Roizman B. Clustering of genes dispensable for growth in culture in the S component of the HSV-1 genome. Science. 1987;236:573–576. doi: 10.1126/science.3033823. [DOI] [PubMed] [Google Scholar]
- 22.McGeoch D J, Dalrymple M R, Davison A J, Dolan A, Frame M C, McNab D, Perry L, Scott J E, Taylor P. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J Gen Virol. 1988;69:1531–1574. doi: 10.1099/0022-1317-69-7-1531. [DOI] [PubMed] [Google Scholar]
- 23.McNab A R, Desai P, Person S, Roof L L, Thomsen D R, Newcomb W W, Brown J C, Homa F L. The product of the herpes simplex virus type 1 UL25 gene is required for encapsidation but not for cleavage of replicated viral DNA. J Virol. 1998;72:1060–1070. doi: 10.1128/jvi.72.2.1060-1070.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pellett P E, Kousoulas K G, Pereira L, Roizman B. Anatomy of the herpes simplex virus 1 strain F glycoprotein B gene: primary sequence and predicted protein structure of the wild type and of monoclonal antibody-resistant mutants. J Virol. 1985;53:243–253. doi: 10.1128/jvi.53.1.243-253.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Post L E, Roizman B. A generalized technique for deletion of specific genes in large genomes: α gene 22 of herpes simplex virus 1 is not essential for growth. Cell. 1981;25:227–232. doi: 10.1016/0092-8674(81)90247-6. [DOI] [PubMed] [Google Scholar]
- 26.Randall G, Lagunoff M, Roizman B. The product of ORF O located within the domain of herpes simplex virus 1 genome transcribed during latent infection binds to and inhibits in vitro binding of infected cell protein 4 to its cognate DNA site. Proc Natl Acad Sci USA. 1997;94:10379–10384. doi: 10.1073/pnas.94.19.10379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Randall G, Roizman B. Transcription of the derepressed open reading frame P of herpes simplex virus 1 precludes the expression of the antisense γ134.5 gene and may account for the attenuation of the mutant virus. J Virol. 1997;71:7750–7757. doi: 10.1128/jvi.71.10.7750-7757.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roizman B, Sears A E. The replication of herpes simplex viruses. In: Fields B N, Knipe D M, Howley P, Chanock R M, Hirsch M S, Melnick J L, Monath T P, Roizman B, editors. Virology. 3rd ed. New York, N.Y: Raven Press; 1995. pp. 2231–2295. [Google Scholar]
- 29.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 30.Spear P G. Membrane fusion induced by herpes simplex virus. In: Bentz J, editor. Viral fusion mechanisms. Boca Raton, Fla: CRC Press Inc.; 1993. pp. 201–232. [Google Scholar]
- 31.Stevens J G, Wagner E K, Devi-Rao G B, Cook M L, Feldman L T. RNA complementary to a herpesvirus α gene mRNA is prominent in latently infected neurons. Science. 1987;235:1056–1059. doi: 10.1126/science.2434993. [DOI] [PubMed] [Google Scholar]
- 32.Ward P L, Roizman B. Herpes simplex genes: the blueprint of a successful human pathogen. Trends Genet. 1994;10:267–274. doi: 10.1016/0168-9525(90)90009-u. [DOI] [PubMed] [Google Scholar]
- 33.Weber P C, Levine M, Glorioso J C. Rapid identification of nonessential genes of herpes simplex virus type 1 by Tn5 mutagenesis. Science. 1987;236:576–579. doi: 10.1126/science.3033824. [DOI] [PubMed] [Google Scholar]