Abstract
Expression of mouse mammary tumor virus (MMTV)-encoded superantigens in B lymphocytes is required for viral transmission and pathogenesis. The mechanism of superantigen expression from the viral sag gene in B cells is largely unknown, due to problems with detection and quantification of these low-abundance proteins. We have established a sensitive superantigen-luciferase reporter assay to study the expression and regulation of the MMTV sag gene in B-cell lymphomas. The regulatory elements for retroviral gene expression are generally located in the 5′ long terminal repeat (LTR) of the provirus. However, we found that neither promoters nor enhancers in the MMTV 5′ LTR play a significant role in superantigen expression in these cells. Instead, the essential regulatory regions are located in the pol and env genes of MMTV. We report here that maximal sag expression in B-cell lines depends on an enhancer within the viral pol gene which can be localized to a minimal 183-bp region. Regulation of sag gene expression differs between B-cell lymphomas and pro-B cells, where an enhancer within the viral LTRs is involved. Thus, MMTV sag expression during B-cell development is achieved through the use of two separate enhancer elements.
Retroviral transcription is generally initiated within the 5′ copy of the two long terminal repeats (LTRs) that flank the viral genes after integration into the host genome as a provirus (10). Promoter and enhancer elements within the LTR determine the transcription initiation rates, preferential transcription in certain cell types, and responses to external stimuli such as hormones.
Mouse mammary tumor virus (MMTV) is a murine B-type retrovirus and causes a high incidence of mammary gland carcinomas in infected females (43). MMTV-induced T-cell lymphomas are observed at a lower incidence. Infectious MMTV is transmitted from mother to offspring via milk (5, 20), and after passage through the gastrointestinal tract, infection of B lymphocytes plays an important role in the viral life cycle (4, 23).
MMTV encodes a superantigen (Sag) that, when expressed on the surface of B cells or other antigen-presenting cells, activates a large number of T cells by interaction with specific T-cell receptor β chains (1, 48). The resulting T-cell response in turn stimulates the infected B cells to proliferate (23) and thus amplifies the number of virus-infected cells (24) and potential target bystander cells. The subsequent clonal elimination of activated T cells results in a progressive depletion of a specific T-cell subset during the course of the infection (25). MMTV can also be genetically inherited in the form of endogenous proviruses (Mtv) in the host germ line. Sag expression from inherited proviruses usually leads to depletion of T-cell subsets expressing reactive T-cell receptor β chains (16), but it can have other consequences as well. In the SJL mouse model of follicular B-cell lymphoma, Sag expression from the endogenous provirus Mtv-RCS in major histocompatibility complex (MHC) class II-positive B-cell lymphomas provides the necessary T-cell stimulation in vivo to induce T-cell-dependent, interleukin-mediated proliferation of B lymphoma cells (54). A similar paracrine mechanism has been implicated in the generation of human follicular B-cell lymphoma (9, 17). Recently, human autoimmune type I diabetes mellitus has been linked to the superantigen expression from an MMTV-like endogenous retrovirus in the pancreas (11).
The viral sag gene encoding the MMTV superantigen is located within the 3′ viral LTR. Subgenomic MMTV sag mRNAs, initiated at four separate promoters and spliced to remove gag, pol, and env genes, have been described under different conditions. Candidate promoters are (i) the classical promoter at the U3-R border (P1196, also called P1) (7, 55), used for the expression of all retroviral genomes and structural genes; (ii) a second promoter within the U3 region (P698, also designated P2) (18); (iii) a T-cell-specific phorbol ester-inducible promoter in the env gene (P7246 or Penv) (15, 40, 58); and (iv) a second, LTR-proximal promoter (P8498 or Penv2) (2) within the env gene.
The lack of a sensitive and quantitative assay for MMTV superantigen expression has long prevented a detailed understanding of sag gene regulation. Superantigen activity determinations in mixed lymphocyte reactions require coexpression of MHC class II molecules on the same cell. Constant MHC class II antigen levels effectively limit the extent of T-cell stimulation in response to varying amounts of superantigen (36). Due to the very small amounts of Sag proteins that are available in B cells and sufficient for T-cell stimulation, Sag detection by monoclonal antibodies is problematic (57). Rigorous tests regarding the relative functional significance of putative MMTV promoters for sag gene expression and the identification of B-cell-specific regulatory elements have only recently become possible. We have described a Sag reporter assay on the basis of a recombinant Sag-placental alkaline phosphatase gene (50). This method combines high sensitivity with MHC class II-independent quantification and is thus superior to conventional Sag assays for the quantitative analysis of sag gene expression mechanisms.
MMTV gene expression has been extensively analyzed in mammary gland epithelial cells. In this cell type, MMTV transcription is constitutively activated through a mammary gland cell-specific enhancer element within the proviral LTRs (41). During pregnancy, MMTV gene transcription is further increased by glucocorticoid hormone interaction with steroid hormone-responsive elements near the major MMTV promoter. The second cell type exhibiting elevated levels of MMTV transcripts consists of cells of the B-lymphocyte lineage. Endogenous MMTV mRNAs are found in normal cells and cell lines representing pro-B, pre-B, and mature stages of B-cell development (35). MMTV transcription in mature B cells can be further increased in response to lipopolysaccharide or interleukin-5 (27, 49). We have recently described an enhancer within the MMTV LTRs that is required for efficient expression of the sag gene in a pro-B-cell line (50). However, the mechanisms that determine sag expression, putative enhancer elements, and interacting cellular transcription factors within the main Sag-presenting cell, the B lymphocyte, remain to be discovered.
In this study, we used an improved, firefly luciferase-based Sag reporter assay to extend our previous studies to B-cell lines. We found that the promoters and enhancers in the 5′ LTR are dispensable for sag expression. By contrast, the major determinants for sag gene expression in B lymphoma cells are found in the pol and env genes of the virus. We identify and map a novel enhancer element in the pol gene, present evidence for additional positive regulatory regions in the pol or env gene, and test the functional significance of these elements in the context of an intact MMTV provirus.
MATERIALS AND METHODS
Plasmids.
The Mtv-1/C3H-recombinant proviral clone, hybrid MMTV, in pBR322 (53) was a gift from H. Varmus. References to nucleotide positions are based on the Mtv-1 LTR (12) and C3H LTR (37) for the 5′ and 3′ LTR, respectively, and on MMTV(BR6) (42) for the internal region. The designation for MMTV promoters used throughout this paper identifies the promoters (P698, P1196, P7246, and P8498) by the position of the major transcription initiation site within the reference proviral sequence MMTV(BR6) (42). Plasmid CMV-luc was generated by introduction of the cytomegalovirus (CMV) immediate-early promoter from plasmid pRC/CMV (Invitrogen) as a BglII-BamHI fragment into vector pGL2-basic (Promega).
Recombinant viral constructs.
Plasmid pM including the hybrid MMTV provirus (53) has been described previously (50). Recombinant luciferase genes were generated by PCR with Pfu DNA polymerase (Stratagene) under standard conditions with primers luc5 (5′ TGACCTAGGGAAGACGCCAAA 3′) or luc5.2 (5′ CCAGATCTTAACGTGCTTCTTTTAAAAAAGAAAAAAGGGGGAAATGGAAGACGCC 3′) and luc3 (5′ CTAGCCTAGGCGGCCGCTTACAATTTGGA 3′) and template pGL2-basic (Promega). The product generated with primers luc5 and luc3 was digested with AvrII and inserted into the AvrII site within the 3′ LTR of the hybrid MMTV provirus (Msag17-luc). Products resulting from the amplification with the luc5.2-luc3 primer pair were cut with BglII and AvrII to replace the BglII-AvrII fragment at the junction of the env gene and 3′ LTR (Msag1-luc). Plasmid pMsag17-luc (EproB+/−) lacks the AvrII-AflII fragment of the 3′ LTR (C3H). Proviral 5′ truncations were generated with the EcoRI site in pGEM2 and MMTV sites StyI (position 1637), StyI (position 2412), StyI (position 3264), StyI (position 4251), StyI (position 5055), EcoRI (position 5803), ClaI (position 7478), and BglII (position 8532) or with the PstI site in pGEM2 and MMTV PstI (position 1459). The MMTV internal deletion mutants ΔE and ΔP were generated by PCR. For ΔE, two fragments surrounding the pol gene enhancer were amplified with primers FP3 (ACACACGCTACTCAGTGTCGACAAACATTTACAAAGGTTAATCC) and FMR5846 (AGATCTGTCGACTAGTGCGTGGCT) or FP1 (GGATGGGCTAGCATCATAATAGATCTACAAGA) and FP2 (ACACACGTCGACCTCCCTTCCTTGTATATA). For ΔP, two fragments flanking the CAAT box and TATA box were amplified with primers FP5 (ACACACGAATTCGCTAGTCGACGAAGTTGCCCCCCAAA) and FP6 (CTGCTTCATACCATCGAT) or FP3 and FP8 (GTACACGTCGACCCCTGAGTTCCCCAAG). The two flanking regions were combined by using the central SalI site and introduced into the Msag17-luc provirus to replace the wild-type sequence either singly (ΔE, ΔP) or in combination (ΔE+P). Mutant ΔE to P was generated by SalI digestion of ΔE+P. To generate the pGL2-pol plasmids 5+, 5−, 3+, and 3−, the MMTV pol gene region from nucleotides (nt) 5055 to 6255 was isolated from plasmid pMsag17-lucΔ5055 as a BamHI fragment and inserted into the BglII site or the BamHI site of plasmid pGL2.promoter (Promega). The orientation was determined by restriction analysis. Truncation mutants of the MMTV pol gene enhancer in pGL2-pol 3+ were generated by PCR with Pfu polymerase (Stratagene) and the following primers: FM5204 (GATGATGGATCCGACACCGC), FM5351 (GATGATGGATCCAAAAGACC), FM5499 (GATGATGGATCCTATAATTAAAG), FM5655 (GATGATGGATCCCAGAACAAA), FM5812 (GATGATGGATCCGCTTTAGAG), FM5952 (GATGATGGATCCGCAATTAGG), FM6109 (GATGATGGATCCAAGGATGTG), and FMR6252 (ACTACTAGATCTGTCGACTCCCCGTGAC) or FM5499 and FMR6017 (AGATCTGTCGACATGAGTAACATC), FMR5846 (AGATCTGTCGACTAGTGCGTGGCT), FMR5738 (AGATCTGTCGACGATATGACCAATG), FMR5681 (AGATCTGTCGACAGTTCTGTGTA), FMR5622 (AGATCTGTCGACACCCTGTCACAT), and FMR5559 (AGATCTGTCGACTAATGACTGCCA). PCR fragments were digested with BamHI and SalI and cloned into plasmid pGL2.promoter (Promega) cut with the same enzymes.
Cell lines and transfection procedures.
The interleukin-3-dependent mouse pro-B-cell line Ba/F3 (46), a gift from U. Klingmüller, was maintained as described previously. The mouse B-cell hybridoma line LBB-IIV.11 (44), abbreviated LBB.11 in this paper, and mouse B-cell lymphomas CH28 (22), A20 (26), and M12 (21) were obtained from B. Huber (Tufts University School of Medicine, Boston, Mass.). The mouse pre-B-cell lines 2M3/M (51) and 300-18 (31), immortalized by Abelson murine leukemia virus, were provided by N. Rosenberg (Tufts University School of Medicine). All lymphoid cell lines were maintained in RPMI 1640 medium supplemented with 10% fetal calf serum, 50 μM β-mercaptoethanol, penicillin (100 UI/ml), and streptomycin (100 μg/ml). DNA was introduced by electroporation with a Bio-Rad Gene Pulser. Equimolar amounts of test plasmids (2 pmol) were linearized outside the cloned provirus with a restriction enzyme. Carrier plasmid pGEM-2 and TE (10 mM Tris, 1 mM EDTA [pH 7.0]) were added to a constant DNA amount and volume. We used a single batch of cells for all the transfections of a particular cell line in a specific experiment to control for growth condition or density-dependent differences. Cells were harvested at the previously determined peak of luciferase activity, which may differ between cell lines.
Reporter gene assay.
Cells were harvested, washed twice in phosphate-buffered saline (137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4, 1.4 mM KH2PO4), and lysed in reporter lysis buffer (Promega). The luciferase assay system (Promega) was used for detection of firefly luciferase (duplicate assays). Light emission was determined for 10 s in an ILA911 luminometer (Tropix). The protein concentration in cellular extracts was measured by the Bio-Rad protein assay.
RESULTS
Sag-luciferase fusion protein as Sag reporter.
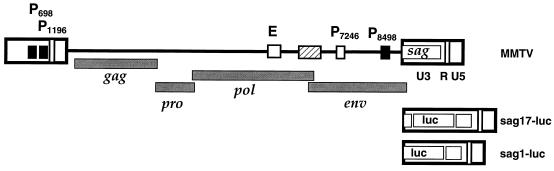
We have previously described a sensitive and quantitative assay for the MMTV sag gene product expressed from a cloned provirus. The assay is based on a recombinant Sag-human alkaline phosphatase fusion protein (50). This approach allowed us to characterize a region within the MMTV LTR that has an important function for sag gene expression as an enhancer element in a pro-B-cell line. However, analysis of sag gene expression in the main effector cell for Sag activity, B cells, was not possible due to high endogenous alkaline phosphatase levels in these cells (13; our unpublished results). To study the regulation of MMTV sag gene expression in B cells, we replaced the reporter gene in our MMTV constructs with the gene encoding firefly luciferase (luc). A luciferase gene lacking its original initiation codon was inserted into the 3′ LTR sag gene of the infectious and oncogenic provirus hybrid MMTV (53), a hybrid of the endogenous Mtv-1 and exogenous C3H proviruses. Insertion of the luciferase gene after sag gene codon 17 created a hybrid sag-luciferase gene (sag17-luc) (Fig. 1). All MMTV proviral sequences are present in this construct (pMsag17-luc). As previously demonstrated, expression of the reporter gene in this system depends on the cotranslation with the 17 N-terminal amino acids of the C3H strain Sag protein (50) and thus reflects functional sag expression at the major translational initiation site codon Met 1 (8, 29). This is also the case for our second MMTV proviral construct (pMsag1-luc), which contains a wild-type luc gene exactly in the position of the sag gene but lacks sag gene codons 2 to 17 (Fig. 1).
FIG. 1.
Schematic representation of the MMTV provirus, Sag-reporter proviruses, and regulatory elements for MMTV sag gene expression in B cells. The positions of viral genes (gag, pro, pol, env, and sag), promoters (P698, P1196, P7246, and P8498), newly characterized pol gene enhancer (E), and functional regions within the LTRs (U3, R, and U5) are indicated. An additional stimulatory element (hatched box) is located between E and P7246. The Sag-reporter proviruses Msag17-luc (sag17-luc) and Msag1-luc (sag1-luc) differ from the MMTV provirus in the 3′ LTRs only. The recombinant 3′ LTRs are depicted below the original LTR. The position of the reporter gene firefly luciferase (luc) in the recombinant constructs is shown. Regulatory elements with demonstrated relevance for sag gene expression are represented as open and hatched boxes.
Sag-reporter expression from newly introduced MMTV proviruses in cell lines representing pro-B, pre-B, and B-cell stages.
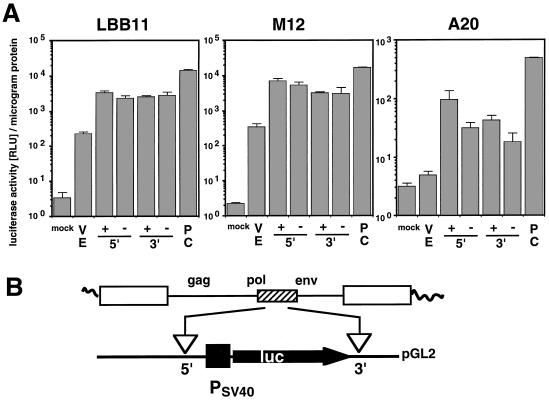
B cells are derived from hematopoietic stem cells and differentiate into plasma cells. Specific intermediate stages during this process can be identified based on characteristic events during immunoglobulin gene rearrangement and differences in gene expression: pro-B, pre-B, and B cells. Our previous experiments detected and analyzed MMTV sag expression in the pro-B-cell line Ba/F3 (50). In this study we extend our analysis to the pre-B and mature stages of B-cell development. As examples of pre-B cells, we selected the two Abelson murine leukemia virus-transformed lines 2M3/M and 300-18. The mature B-cell stage was represented by the murine B-cell lymphomas CH28, A20, and M12. We also tested LBB.11, an M12-derived B-cell hybridoma that is frequently used for Sag activity tests in mixed-lymphocyte stimulatory assays after the introduction of MMTV sag genes (3). All B-lymphoma and hybridoma lines are characterized by surface immunoglobulin (sIg+) and MHC class II antigen (Ia+) markers. The cell lines were tested for MMTV-dependent luciferase expression after transient transfection of sag reporter plasmids pMsag17-luc or pMsag1-luc and the corresponding negative control Δ8532, lacking the 5′ LTR and all but 43 nucleotides of the internal coding region of the provirus (see Fig. 3).
FIG. 3.
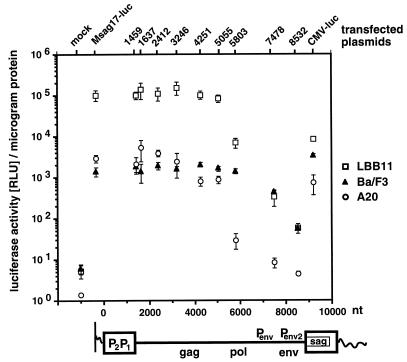
Functional analysis of MMTV proviral regions that are involved in sag gene expression by 5′ truncation analysis of MMTV sag-reporter provirus Msag17-luc. The sag-luciferase reporter provirus Msag17-luc, a series of 5′ truncation mutants (1459, 1637, 2412, 3246, 4251, 5055, 5803, 7478, and 8532), or positive control plasmid CMV-luc were transfected into Ba/F3 pro-B cells, A20 B-cell lymphoma, or LBB.11 B-cell hybridoma. We used 2 pmol of each test plasmid, except for CMV-luc, for which we used 0.5 pmol of plasmid. Cells were harvested after 24 h, and the lysate was tested for luciferase activity and protein concentration. The numbers represent the arithmetic mean ± SD of at least three separate experiments with two different DNA preparations. The MMTV proviral construct is depicted below the graph. The positions of the viral genes (gag, pol, env, and sag) and reported promoters (P) are indicated. The relative result for each truncation mutant is represented at the position of the truncation endpoint within the MMTV provirus.
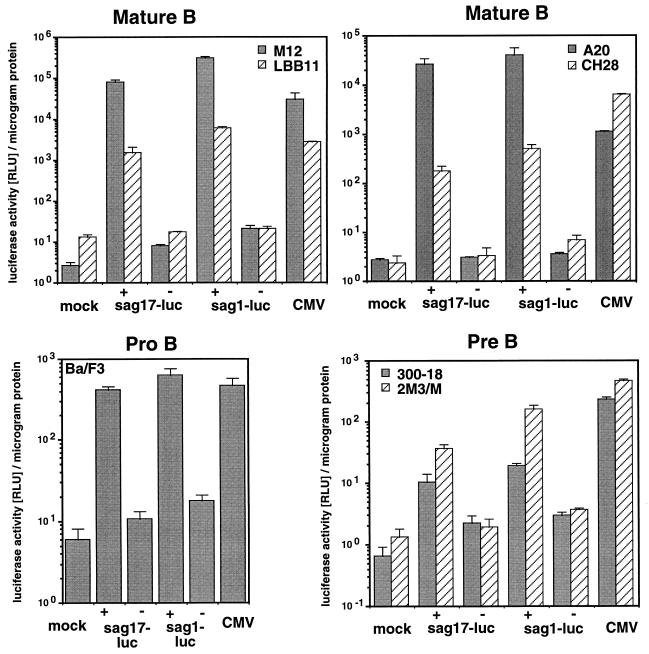
In all B-lymphoid cell lines tested, we were able to detect significantly higher sag-reporter expression with the intact MMTV proviral reporter constructs pMsag17-luc and pMsag1-luc than with the respective negative controls (Fig. 2). We found that the luciferase enzyme encoded by sag1-luc generally yielded larger absolute numbers than did the Sag-luciferase recombinant enzyme expressed from sag17-luc. The luciferase signal obtained with the two Sag-reporter plasmids (sag1-luc + sag17-luc) was on average 38-fold (in Ba/F3 cells), 8- and 31-fold (in pre-B cells 300-18 and 2M3/M), and 64, 188, 10,000, or 12,000-fold (in B cells CH28, LBB.11, A20, and M12) higher than that obtained with the negative controls.
FIG. 2.
MMTV sag-reporter activity in cell lines representing pro-B, pre-B, and mature stages of B-cell development. MMTV sag-reporter proviruses Msag17-luc (sag17-luc) and Msag1-luc (sag1-luc), and positive control plasmid CMV-luc were transfected into the pro-B-cell line Ba/F3, the pre-B-cell lines 300-18 and 2M3/M, the B-lymphoma lines A20, CH28, and M12, and the B-cell hybridoma LBB.11. We used 2 pmol of each test plasmid, except for CMV-luc, for which we used 0.5 pmol of plasmid. The full-length MMTV reporter constructs are denoted by plus signs, whereas the negative control constructs lacking the 5′ LTR and most of the internal region are represented by minus signs. Cells were harvested after 24 h and lysed. The relative luciferase activity (RLU) and protein concentration were determined with this extract. The numbers represent the arithmetic mean ± standard deviation (SD) of at least three separate experiments with two different DNA preparations.
We conclude that B-lymphoid cell lines representing the pro-B, pre-B, and B-cell stages allow expression of the MMTV sag gene from a newly introduced provirus. In all these cell types, sag gene expression can be sensitively monitored over a wide range through the relative activity of the Sag-luciferase reporter enzyme. The high background signals previously resulting from alkaline phosphatase-based negative control constructs (48) were not present in the luciferase reporter system. Thus, the sag-luc reporter proviruses provide excellent tools for the quantitative analysis of MMTV Sag expression at all stages of B-cell development.
MMTV sag gene expression in B-cell lymphomas is stimulated by a cell-type-specific regulatory element in the pol gene.
A number of MMTV-derived sag mRNAs, initiated at one of four different promoters and spliced to remove most of the gag, pol, and env genes, has been associated with Sag expression in B lymphoid cells. However, experimental evidence for the relative importance of specific mRNAs and promoters is not available, and B-cell-specific regulatory elements analogous to the mammary gland cell enhancer (41) have yet to be characterized.
To functionally define the regions within an MMTV provirus that are essential for expression of the sag gene in mature B-cell-derived lymphoma cell lines, we used our Sag-reporter system Msag17-luc. Plasmid pMsag17-luc carries a full-length MMTV provirus including the Sag-luciferase reporter gene sag17-luc. We generated a series of truncations from the 5′ end of the provirus progressively removing the 5′ LTR (Δ1463) and the internal region (Δ1637 to Δ8532). The respective numbers characterize the size of a specific truncation. The resulting mutants were tested in transient-transfection assays, and luciferase levels were determined.
Our initial experiments focused on the 5′ LTR. Partial truncation (data not shown) or complete deletion (Fig. 3) of the 5′ LTR did not significantly decrease the sag expression levels in B-cell hybridoma LBB.11. Subsequently, a wider panel of truncations spanning the 5′ LTR and the internal coding region were tested in both B-cell lymphoma A20 and B-cell hybridoma LBB.11 and compared to that in Ba/F3 pro-B cells (Fig. 3). In all three cell lines, sag reporter levels were not significantly affected by the removal of the 5′ LTR (Δ1459). In the B-cell lines A20 and LBB.11, truncations of the 5′ LTR and the internal coding region up to position 5055 in the pol gene affected sag reporter expression weakly or not at all. Further reduction of the MMTV provirus revealed that a region of the pol gene from nt 5055 to 5803 had a strong positive effect on sag expression (Fig. 3) in the more mature B-cell lines. Loss of this region (Δ5803) decreased sag expression to 1% (A20) and 7% (LBB.11) relative to the initial activity. The adjacent region from nt 5803 to 8532 also contributes to sag expression, as demonstrated by a further reduction to 0.3% (Δ7478) and 0.06% (Δ8532) in LBB.11 cells and 0.3% (Δ7478) and 0.2% (Δ8532) in A20 cells of initial sag gene expression.
In Ba/F3 pro-B cells, in agreement with our previous data (50), MMTV mutants lacking up to 5,803 nt (Δ5803) were still capable of generating approximately wild-type sag levels. Deletion of the pol gene region from nt 5055 to 5803 did not affect sag expression. Only upon further deletion of the internal region from positions 5803 to 7478 or 8532 was a reduction of sag-reporter activity to 30 and 4% found. Sag-reporter levels are maintained at these comparatively high levels due to the presence of a sag gene intragenic enhancer in the 3′ LTR (50).
We conclude that in B cells neither promoters nor enhancers in the 5′ LTR play a major role in MMTV Sag expression. However, due to the limited resolution of our assay, minor contributions from the 5′ LTR cannot be excluded. In B-cell lines but not pro-B cells, the pol gene region from nt 5055 to 5803 has a strong stimulatory effect on sag gene expression. Previously, no regulatory function has been assigned to this section of the MMTV pol gene.
MMTV sag gene expression in B-cell lymphomas does not involve the sag gene intragenic enhancer.
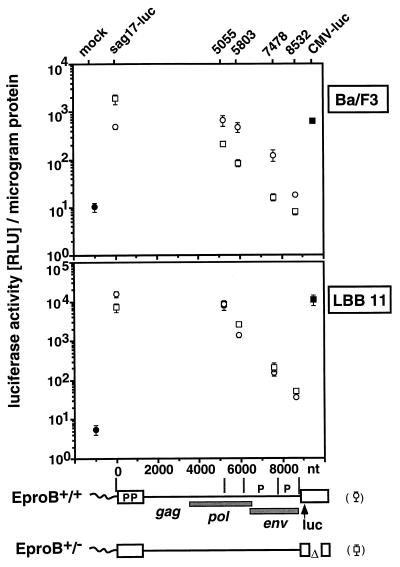
We have previously identified a sag gene intragenic enhancer element (EproB) that stimulates MMTV sag expression in a pro-B-cell line from both the 5′ and 3′ LTR (50). Its potential involvement in MMTV gene expression in the more mature stages of B-cell development has not yet been addressed. The 3′ LTR copy of this regulatory element was present in all the above-tested MMTV mutants and could potentially modulate sag reporter levels in our system. To test the effect of this pro-B-cell enhancer on sag expression in the more mature B cells, we deleted a 0.9-kb region, including EproB, from the 3′ LTR U3 region (nt 54 to 991) of provirus Msag17-luc. The resulting mutant provirus, Msag17-luc [EproB+/−], lacks the 3′ LTR copy of the EproB enhancer but retains the copy in the 5′ LTR, while the original Msag17-luc has an EproB+/+ enhancer status. Removal of the 5′ LTR renders these proviruses EproB−/− and EproB−/+, respectively. Proviruses Msag17-luc [EproB+/+], Msag17-luc [EproB+/−], and a number of 5′ truncation mutants (EproB−/+ or EproB−/−) were compared with regard to sag expression in LBB.11 and Ba/F3 cells following transient transfection.
In Ba/F3 pro-B cells (Fig. 4, top) all MMTV mutants lacking EproB in both the 5′ and 3′ LTR (EproB−/−), such as Msag17-luc [EproB+/−] Δ5055, Δ5803, Δ7478, and Δ8532, show clearly reduced sag expression compared to their EproB−/+ counterparts. This occurs even though for Msag17-luc [EproB+/+] the EproB deletion in the 3′ LTR results in an increased reporter expression, possibly due to a loss of negative regulatory elements (19). In LBB.11 cells, no significant differences in sag expression between EproB−/+ and EproB−/− reporter proviruses were found (Fig. 4, bottom). This indicates that the activity of the MMTV pro-B-cell enhancer EproB is limited to early B-lymphoid cells and has no effect in B cells. Consequently, for MMTV gene expression in B cells, a second enhancer must be postulated that is active predominantly in mature B cells and is located outside of the tested U3 LTR region of the MMTV provirus.
FIG. 4.
MMTV Sag expression in B cells is independent of the sag gene intragenic enhancer. Reporter plasmids pMsag17-luc(EproB+/+) (open circles) and pMsag17-luc(EproB+/−) (open squares), with their respective truncation mutants Δ5055, Δ5803, Δ7478, and Δ8532, and luciferase control plasmid CMV-luc (solid squares) were transfected into either Ba/F3 pro-B cells (top) or B-cell hybridoma LBB.11 (bottom). The mock transfection is represented by solid circles. We used 2 pmol of each test plasmid, except for CMV-luc, for which we used 0.5 pmol of plasmid. Cells were harvested after 24 h, and luciferase activity was determined. Results are expressed as relative luciferase units (RLU) per microgram of protein and represent the arithmetic mean ± SD of at least three separate experiments with two different DNA preparations. The viral constructs with nucleotide positions are depicted below the graphs. The approximate positions of coding regions (gag, pol, and env), promoters (P), and reporter gene luciferase (luc) are shown. The specific constructs transfected are identified above the graphs.
The MMTV pol gene contains an enhancer element for B cells.
We hypothesized that the stimulatory region that we had identified in the viral pol gene might be identical to the postulated B-cell enhancer. To test this hypothesis, we analyzed the effects of the corresponding pol region on gene expression from a heterologous promoter. A 1.2-kb region from the 3′ end of the MMTV pol gene (nt 5055 to 6255) was placed into the enhancer test plasmid pGL2, with a simian virus 40 (SV40) promoter regulating the expression of a luciferase reporter gene, to generate pGL2-pol. The MMTV test fragment was inserted in a position either 5′ or 3′ of the promoter/reporter unit in either the sense or antisense orientation. The effect on luciferase reporter levels was quantified after transient transfection into B-cell lymphomas M12 and A20 and B-cell hybridoma LBB.11.
All three B-cell lines, A20, M12, and LBB.11, showed increased reporter gene expression in response to the MMTV-derived test fragment (Fig. 5). Basal expression from the enhancerless SV40 promoter was stimulated up to 15-fold (LBB.11) or 20-fold (M12 and A20). The strongest effect was observed in all three cell lines, when the MMTV fragment was in the 5′ position and the sense orientation. However, alternative positions and orientations resulted in an increase of at least 11-fold (LBB.11), 9-fold (M12), or 4-fold (A20) over vector-induced reporter gene expression.
FIG. 5.
The MMTV pol gene contains an enhancer element for gene expression in B cells. (A) Stimulation of SV40 promoter-directed luciferase expression by an MMTV pol gene fragment (nt 5055 to 6255). (B) Schematic representation of enhancer test constructs pGL2-pol. Control plasmid pGL2-promoter (VE) contains an enhancerless SV40 promoter upstream of the firefly luciferase reporter gene. The MMTV pol gene fragment (nt 5055 to 6255) was introduced in a position either 5′ or 3′ from the SV40 promoter-luciferase reporter gene of enhancer test plasmid pGL2-promoter, in either the sense (+) or antisense (−) orientation, to generate pGL2-pol 5+, 5−, 3+, and 3−, respectively. These plasmids, pGL2.promoter (VE) and CMV.luc (PC), were transfected into B-cell lines LBB.11, M12, and A20. We used 2 pmol of each test plasmid, except for CMV-luc, for which we used 0.5 pmol of plasmid. The cells were harvested after 24 h (A20 and LBB.11) or 48 h (M12), and the luciferase activity and protein concentrations in the cell lysate were determined. Results are expressed as relative luciferase units (RLU) per microgram of protein and represent the arithmetic mean ± SD of at least three separate experiments with two different DNA preparations. As previously established (50a), introduction of an irrelevant piece of DNA into pGL2-promoter does not result in increased luciferase expression.
These results demonstrate a strong positive effect of the selected MMTV pol gene region on gene expression from a heterologous promoter. The ability of this fragment to stimulate gene expression from positions 5′ or 3′ of the reporter gene and in both orientations provides definite evidence for the presence of an enhancer element within the tested pol gene region. Although this enhancer is functionally comparable to the MMTV mammary gland enhancer or murine leukemia virus enhancer within the viral LTRs (30, 41), its location within the pol gene is highly unusual.
The MMTV B-cell enhancer is located within a 183-bp region in the RNase H domain of the pol gene.
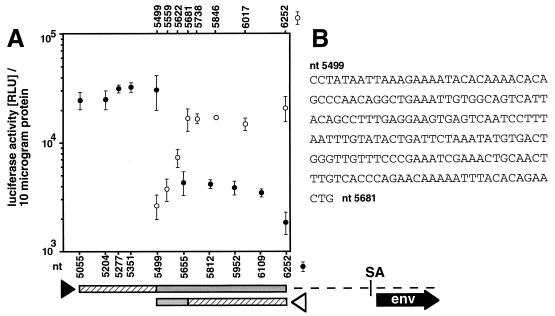
To define the size and position of the minimal functional enhancer element within the 1.2-kb pol gene region (nt 5055 to 6255), we generated a set of defined truncations of the MMTV pol gene enhancer fragment from either the 5′ or the 3′ terminus. The stimulatory activity of these fragments on SV40 promoter-driven luciferase gene expression was analyzed with the MMTV fragment in the 3′ position and sense orientation relative to the promoter and reporter gene so as to avoid interference with potential promoters in the tested sequence. Stimulation of reporter gene expression was assayed in the B-cell hybridoma line LBB.11 after transient transfection.
Truncation of the initial pol gene enhancer (nt 5055 to 6255) from the 5′ direction to position 5499 had no effect on enhancer activity in LBB.11 cells (Fig. 6). However, a sharp reduction was observed when an additional 156 bp (from positions 5499 to 5655) was eliminated. All further truncation mutants exhibited similar, slightly elevated expression levels compared to the vector control. With the 5′ extension of the functional enhancer thus identified between MMTV nt 5499 and 5655, the remaining functional region (MMTV nt 5499 to 6255) was progressively shortened from the 3′ terminus in several steps. We found that all truncations up to nt 5681 left the enhancer activity unchanged. Beyond this, more extensive truncations to nt 5622 and 5559 gradually reduced pol enhancer-induced reporter expression, suggesting the presence of several regulatory elements within this area. The 3′ boundary of the pol gene enhancer is located between nt 5622 and 5681.
FIG. 6.
Localization of the functional B-cell enhancer region within the MMTV pol gene. (A) Enhancer activity of MMTV pol gene regions. (B) Nucleotide sequence of the MMTV pol gene enhancer region. The MMTV pol region (nt 5055 to 6255) in pGL2-pol 3+ was successively truncated from the 5′ terminus (solid arrowhead) and from the 3′ terminus (open arrowhead). The truncation mutants were transfected into LBB.11 B cells. After 24 h, the cells were harvested and lysed and the luciferase activity and protein concentrations determined. The numbers indicate the position of the test fragment within the MMTV provirus. Specific truncation mutants are identified below the graph for the 5′ truncation series (solid circles) and above the graph for the 3′ truncations (open circles). The truncation mutant 6252 represents the vector control and is identical to the plasmid pGL2-Promoter (Promega). The bars below the graph represent the pol gene fragment analyzed, with the shaded portions indicating the parts required for enhancer activity in the respective 5′ and 3′ truncations and the hatched portions specifying the dispensable parts. Approximate positions of the env gene and splice acceptor (SA) site are indicated.
In our analysis, a 183-bp fragment from the MMTV pol gene (nt 5499 to 5681) was sufficient for full B-cell enhancer activity. The entire enhancer element is contained within the region encoding the RNase H domain of the viral enzyme reverse transcriptase. It covers the central portion of the RNase H domain and three of the seven conserved active-site amino acid residues (39).
In B cells the MMTV pol gene enhancer and a promoter within the env gene contribute to sag expression from an intact MMTV provirus.
The activity of the MMTV B-cell enhancer in the viral pol gene has been defined through reduction of the MMTV provirus to its essential functional regions and in the absence of dispensable parts of the provirus. To firmly establish the biological significance of this enhancer for sag gene expression, we tested its effect within a complete MMTV provirus. Since our data suggested the use of one or more non-LTR promoters in sag gene transcription, we also wished to functionally analyze the effect of the env gene promoter P7246 on this process.
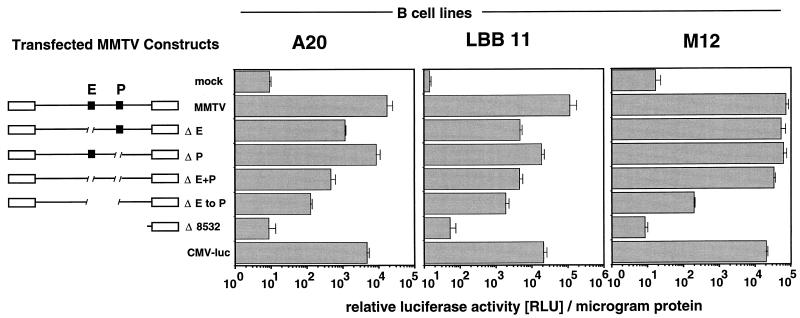
Specific, small deletions were introduced into the full-length MMTV provirus Msag17-luc to disable either the pol gene enhancer or the env gene promoter P7246 while preserving the overall architecture of the MMTV provirus. In particular, the 5′ LTR was present in all constructs and all deletions were generated at least 1.3 kb from the sag gene splice acceptor site. We removed either the complete 183-bp B-cell enhancer (ΔE) or a 29-bp fragment including the TATA box and putative CAT box of the P7246 env gene promoter (ΔP) from positions −52 to −24 with respect to the transcriptional start site. A third mutant combined these defects and lacked both the pol gene enhancer and the P7246 env promoter (ΔE+P). To test for potential functional elements between Epol and P7246, an additional mutant, lacking Epol, Penv, and the 1.3-kb intervening region (ΔE to P), was generated. The sag-luciferase expression from full-length MMTV provirus Msag17-luc and the mutant proviruses was compared after transfection into the B-cell lines A20, LBB.11, and M12.
In A20 and LBB.11 cells, specific excision of the pol gene enhancer reduced sag gene expression to 7% (A20) or 4% (LBB.11) of wild-type activity (Fig. 7). In contrast, removal of the conserved promoter elements from P7246 had weaker, but significant effects. Sag-reporter levels observed with these P7246 mutants in A20 or LBB.11 cells were 50 and 17%, respectively. This reduction, albeit moderate, represents a stronger effect on sag gene expression than the removal of the complete 5′ LTR, for which no significant difference was observed (Fig. 3). The combined Epol and Penv deletions resulted in only a minor further reduction (to 3% of wild-type activity in A20 and 4% in LBB.11) compared to the effect of the ΔE mutant alone. Similarly, in these two cell lines, removal of the proviral region between Epol and P7246 env had only a moderate effect on sag gene expression (reduction to 0.7% in A20 cells and 1.7% in LBB.11 cells), indicating the absence of major regulatory elements.
FIG. 7.
Effect of the MMTV pol gene enhancer and env gene promoter P7246 on sag gene expression in a complete provirus. An intact MMTV provirus (MMTV) and deletion mutants lacking the pol gene enhancer (ΔE), env gene promoter P7246 (ΔP), both elements (ΔE+P), or the pol gene enhancer, P7246 and the intervening region (ΔE to P), the negative control pMsag17-lucΔ8532 (Δ8532), or control plasmid CMV-luc were transfected into the B-cell lines A20, LBB.11, and M12. We used 2 pmol of each test plasmid, except for CMV-luc, for which we used 0.5 pmol of plasmid. After 24 h, the cells were harvested and lysed, and luciferase activity and protein concentrations were determined. Results are expressed as relative luciferase units per microgram of protein and represent the arithmetic mean ± SD of at least three separate experiments with two different DNA preparations.
The situation in M12 B-lymphoma cells was sharply different. In these cells, the Epol-mediated effects (30% of total sag expression [compare the second and third bars from the top in the right panel of Fig. 7]) as well as the combined effects of Epol and Penv (54% of total sag expression [compare the second and fifth bars from the top in the right panel of Fig. 7]) were much smaller than in the previous two cell lines. This result was surprising in view of the very similar activity of Epol in our previous heterologous enhancer assay in M12 and LBB.11 (Fig. 5). In M12 cells, it was only after further removal of the region between Epol and Penv that a drastic reduction of sag-luciferase expression to 0.3% of wild-type levels was observed. This result strongly argues for the presence of an auxiliary stimulatory element between the newly described pol gene enhancer and the P7246 env gene promoter. Thus, in M12 cells, sag gene expression is probably maintained at high levels through a second, parallel enhancer that is recognized by a different set of cellular proteins. In all three cell lines tested, the mutant provirus lacking Epol, P7246, and the intervening region (ΔE to P) exhibits only residual sag reporter expression of ≤2% of full activity.
In conclusion, the majority of MMTV sag expression (≥98%) in B-cell lines is controlled by stimulatory elements within a 1.7-kb region of the MMTV pol and env genes. This region includes the pol gene enhancer, env gene promoter P7246, and intervening sequence. The newly defined MMTV pol gene enhancer (Epol) contributes to sag gene expression to a large extent in at least two of three B-cell lines tested (93% in A20 and 96% in LBB.11 cells). In spite of the general activity of the pol gene enhancer in all three tested sIg+ Ia+ B-cell lines, in one of these lines an auxiliary stimulatory element, possibly associated with the env gene promoter P7246, ensures expression of the sag gene even after Epol has been removed. A potential candidate for this function is the MMTV env gene transcriptional activator, previously described as T-cell specific and activation dependent, located between Epol and P7246 (40, 52). It will be of great interest to determine whether this additional activity can be attributed to a specific physiological status or developmental stage of B lymphocytes.
DISCUSSION
The perception of MMTV as a predominantly mammary gland-tropic virus changed drastically after the discovery of the immuno-stimulatory Sags encoded in the LTR of this virus (16, 38). Subsequently, it was demonstrated that a critical part of the viral life cycle involves infection of lymphocytes and that the expression of the Sag-encoding sag gene in B lymphocytes is required for successful transmission of the infectious virus (4). However, the molecular mechanism of MMTV gene expression in B lymphoid cells remained virtually unknown. The clearly established function of the MMTV Sags in B cells made the sag gene a promising tool for the analysis of MMTV gene expression in B cells.
Conventional Sag assays, either by Sag activity measurements or by serological detection, have so far been unable to address the regulation of the MMTV sag gene within an intact provirus. We have established a sensitive, quantitative, and MHC class II-independent reporter assay for the expression of the MMTV sag gene in all cell types. This assay is based on the enzymatic activity of a Sag-firefly luciferase fusion protein and replaces an earlier alkaline phosphatase-based assay (48). The high background signals previously seen with alkaline phosphatase-based negative control constructs were not present in the luciferase reporter system. Luminometric analysis allows the detection and quantification of recombinant Sag-luciferase over a very wide dynamic range.
The analysis of MMTV sag gene expression in B cells revealed a number of unusual features (Fig. 1). (i) Promoters and enhancers within the 5′ LTR are not required for sag gene expression. The complete 5′ LTR can be deleted without any detectable effect on sag expression. These results argue against a major role of either of the two promoters in the U3 region of the LTR, P698 and P1196 (18, 55), in this process. However, minor contributions of these promoters to sag expression cannot be excluded.
(ii) A newly identified transcriptional enhancer (Epol) is present in the MMTV pol gene (Fig. 1). Its stimulatory activity has been demonstrated in all B-cell lines tested, and specific excision of Epol from an otherwise intact provirus strongly reduces (≥93%) sag expression in two of three cell lines analyzed. An alternative explanation, the reduction of sag expression through an effect on sag mRNA splicing, is considered unlikely due to the long distance between the pol gene enhancer and the LTR-proximal sag splice acceptor (2.8 kb), the sag splice donor in the env gene (1.7 kb), or the splice donor between 5′ LTR and gag gene (4.0 kb).
Retroviral enhancer elements are generally located within the U3 region of the LTRs. An additional internal enhancer within the viral coding region has been reported for the pol and vif genes of human immunodeficiency virus (56). Internal promoters with associated, conditional enhancers are known from the MMTV env gene (52) and the spumavirus env gene (6). Only for spumaviruses has the biological relevance of these findings been firmly established.
This is the first report to demonstrate an effect of an internal enhancer within the pol gene on retroviral gene expression. The entire B-cell enhancer element is located within the region encoding the RNase H domain of the viral enzyme reverse transcriptase. The enhancer covers the central portion of the RNase H domain and three of the seven conserved active-site amino acid residues (39). Within the retroviral genome, the nucleotide sequence of the pol gene shows the highest degree of conservation between different isolates of a virus or even between different viruses. As a consequence, the position of this enhancer within the pol gene strongly suggests that a functional B-cell enhancer is present in all MMTV strains and most endogenous Mtv proviruses. Nucleotide sequence comparison of our Mtv-1-derived pol gene enhancer with the corresponding region of proviruses Mtv-8/MMTV(GR40), MMTV(BR6), and MMTV(JYG) (14, 42, 45) identified no, two, and four differences, respectively within the 183-bp region. Comparison with the MMTV-like group of human endogenous retroviruses HERV-K revealed a 61% identity between the two sequences (47) in a 103-nt region. Genetically inherited Mtv proviruses within the germ line no longer require all the viral replication functions for their maintenance in the host. One notable example is the endogenous provirus Mtv-6. Sequence analysis of Mtv-6 revealed that most of the internal coding region including all of the pol gene and most of the env gene has been lost (7). Despite these limitations, Mtv-6 is able to express sufficient amounts of Sag to direct thymic deletion of a specific T-cell subset (16). The positive effect of a cellular enhancer in the vicinity of the Mtv-6 integration site may be responsible for the continued Mtv-6 sag expression in this case.
The MMTV pol gene enhancer for B cells is the second such element that we have identified for MMTV sag expression in the B-cell lineage. Previously, we have described the stimulatory function of an intragenic enhancer within the sag gene itself on sag gene expression in pro-B cells (50). Thus, MMTV sag expression during B-cell development is achieved through the use of two separate enhancer elements.
(iii) All major regulatory regions for MMTV sag expression in B cells are located within the pol and env genes. We found that in all B-cell lines tested, the internal region from the pol gene enhancer Epol to the P7246 env gene promoter directs 98 to 99.7% of sag gene expression (Fig. 7). Although the pol gene enhancer is responsible for most of this effect, excision of the P7246 TATA box and CAAT box causes a 30 and 50% reduction of sag expression, respectively, in two of the three cell lines tested. We would like to point out that the measured numbers are likely to be an underestimation of P7246 function, since promoter inactivation is frequently partially compensated by newly activated, cryptic promoters (34). In this particular case, compensation by one or more of the remaining three MMTV promoters appears possible. These results indicate that the overall contribution of the P7246 env promoter to sag gene expression is higher than the contribution of both LTR promoters (P698 and P1196), where no significant effect of their deletion was detected. In our system, the P8498 env gene promoter (2) alone is not sufficient to induce more then 0.3% of wild-type sag-reporter expression. However, despite our experimental support for involvement of the P7246 env gene promoter, contributions from other MMTV promoters are possible, in particular in response to external stimuli, cellular differentiation processes, or different host cell types.
Both MMTV and the spumaviruses use an internal promoter within the env gene for the regulation of viral gene expression (6, 33). In both MMTV and spumaviruses, the internal promoter is used to express accessory genes 5′ to or overlapping the 3′ LTR. In spumaviruses, the high basal activity of the internal promoter directs expression of the Bel 1 transactivator gene early after infection to initiate Bel 1-enhanced expression from the classical U3 and the internal promoters (32). Hypothesizing an analogous biphasic expression mechanism for MMTV, initial sag expression predominantly from the internal promoter is likely to induce or enhance T-cell-mediated B-cell activation and subsequent differentiation into antibody-secreting B cells. Concomitant induction of general MMTV gene expression from the 5′ LTR would stimulate the production of viral structural components to generate infectious virions. In support of this hypothesis are reports demonstrating induction of MMTV LTR-driven gene expression by agents such as lipopolysaccharide or interleukin-5 that activate B-cell differentiation (27, 28). These differentiating B cells increase immunoglobulin μ chain expression, J chain expression, and antibody secretion, can migrate to mucosal sites of secretory immunity, and are potential vehicles for the transmission of MMTV to the mammary gland.
ACKNOWLEDGMENTS
We are grateful to B. Huber, U. Klingmüller, N. Rosenberg, and H. Varmus for their generous gifts of cell lines or reagents and to S. Fenner for expert technical assistance.
J.M.C. is American Cancer Society Professor of Molecular Biology and Microbiology. This work was supported by National Cancer Institute award R35CA44385 to J.M.C. and a Leukemia Society of America Fellowship to F.U.R.
REFERENCES
- 1.Acha Orbea H, MacDonald H R. Superantigens of mouse mammary tumor virus. Annu Rev Immunol. 1995;13:459–486. doi: 10.1146/annurev.iy.13.040195.002331. [DOI] [PubMed] [Google Scholar]
- 2.Arroyo J, Winchester E, McLellan B, Huber B T. Shared promoter elements between a viral superantigen and the major histocompatibility complex class II-associated invariant chain. J Virol. 1997;71:1237–1245. doi: 10.1128/jvi.71.2.1237-1245.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beutner U, Frankel W N, Code M S, Coffin J M, Huber B T. Mls-1 is encoded by the long terminal repeat open reading frame of the mouse mammary tumor provirus Mtv-7. Proc Natl Acad Sci USA. 1992;89:4532–4536. doi: 10.1073/pnas.89.12.5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beutner U, Kraus E, Kitamura D, Rajewsky K, Huber B T. B cells are essential for murine mammary tumor virus transmission, but not for presentation of endogenous superantigens. J Exp Med. 1994;179:1457–1466. doi: 10.1084/jem.179.5.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bittner J J. Some possible effects of nursing on the mammary gland tumor incidence in mice. Science. 1936;84:162–169. doi: 10.1126/science.84.2172.162. [DOI] [PubMed] [Google Scholar]
- 6.Campbell M, Renshaw-Gegg L, Renne R, Luciw P A. Characterization of the internal promoter of simian foamy viruses. J Virol. 1994;68:4811–4820. doi: 10.1128/jvi.68.8.4811-4820.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cho K, Ferrick D A, Morris D W. Structure and biological activity of the subgenomic Mtv-6 endogenous provirus. Virology. 1995;206:395–402. doi: 10.1016/s0042-6822(95)80055-7. [DOI] [PubMed] [Google Scholar]
- 8.Choi Y, Marrack P, Kappler J W. Structural analysis of a mouse mammary tumor virus superantigen. J Exp Med. 1992;175:847–852. doi: 10.1084/jem.175.3.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clayberger C, Luna Fineman S, Lee J E, Pillai A, Campbell M, Levy R, Krensky A M. Interleukin 3 is a growth factor for human follicular B cell lymphoma. J Exp Med. 1992;175:371–376. doi: 10.1084/jem.175.2.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coffin J M. Retroviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, et al., editors. Virology. 3rd ed. Vol. 2. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 1767–1847. [Google Scholar]
- 11.Conrad B, Weissmahr R N, Bönl J, Arcari R, Schüpbach J, Mach B. A human endogenous retroviral superantigen as candidate autoimmune gene in type I diabetes. Cell. 1997;90:303–313. doi: 10.1016/s0092-8674(00)80338-4. [DOI] [PubMed] [Google Scholar]
- 12.Crouse C A, Pauley R J. Molecular cloning and sequencing of the MTV-1 LTR: evidence for a LTR sequence alteration. Virus Res. 1989;12:123–137. doi: 10.1016/0168-1702(89)90059-2. [DOI] [PubMed] [Google Scholar]
- 13.Culvenor J G, Harris A W, Mandel T E, Whitelaw A, Ferber E. Alkaline phosphatase in hematopoietic tumor cell lines of the mouse: high activity in cells of the B lymphoid lineage. J Immunol. 1981;126:1974–1977. [PubMed] [Google Scholar]
- 14.Deen K C, Sweet R W. Murine mammary tumor virus pol-related sequences in human DNA: characterization and sequence comparison with the complete murine mammary tumor virus pol gene. J Virol. 1986;57:422–432. doi: 10.1128/jvi.57.2.422-432.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elliott J F, Pohajdak B, Talbot D J, Shaw J, Paetkau V. Phorbol diester-inducible, cyclosporine-suppressible transcription from a novel promoter within the mouse mammary tumor virus env gene. J Virol. 1988;62:1373–1380. doi: 10.1128/jvi.62.4.1373-1380.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frankel W N, Rudy C, Coffin J M, Huber B T. Linkage of Mls genes to endogenous mammary tumor viruses of inbred mice. Nature. 1991;349:526–528. doi: 10.1038/349526a0. [DOI] [PubMed] [Google Scholar]
- 17.Gaidano G, Dalla Favera R. Biologic and molecular characterization of non-Hodgkin’s lymphoma. Curr Opin Oncol. 1993;5:776–784. doi: 10.1097/00001622-199309000-00002. [DOI] [PubMed] [Google Scholar]
- 18.Günzburg W H, Heinemann F, Wintersperger S, Miethke T, Wagner H, Erfle V, Salmons B. Endogenous superantigen expression controlled by a novel promoter in the MMTV long terminal repeat. Nature. 1993;364:154–158. doi: 10.1038/364154a0. [DOI] [PubMed] [Google Scholar]
- 19.Günzburg W H, Salmons B. Factors controlling the expression of mammary tumour virus. Biochem J. 1992;283:625–632. doi: 10.1042/bj2830625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hainaut P, Vaira D, Francois C, Calberg B C M, Osterrieth P M. Natural infection of Swiss mice with mouse mammary tumor virus (MMTV): viral expression in milk and transmission of infection. Arch Virol. 1985;83:195–206. doi: 10.1007/BF01309916. [DOI] [PubMed] [Google Scholar]
- 21.Hamano T, Kim K J, Leiserson W M, Asofsky R. Establishment of B cell hybridomas with B cell surface antigens. J Immunol. 1982;129:1403–1406. [PubMed] [Google Scholar]
- 22.Haughton G, Arnold L W, Bishop G A, Mercolino T J. The CH series of murine B cell lymphomas: neoplasmic analogues of Ly-1+ normal B cells. Immunol Rev. 1986;93:35–51. doi: 10.1111/j.1600-065x.1986.tb01501.x. [DOI] [PubMed] [Google Scholar]
- 23.Held W, Shakhov A N, Izui S, Waanders G A, Scarpellino L, MacDonald H R, Acha Orbea H. Superantigen-reactive CD4+ T cells are required to stimulate B cells after infection with mouse mammary tumor virus. J Exp Med. 1993;177:359–366. doi: 10.1084/jem.177.2.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Held W, Waanders G A, Shakhov A N, Scarpellino L, Acha Orbea H, MacDonald H R. Superantigen-induced immune stimulation amplifies mouse mammary tumor virus infection and allows virus transmission. Cell. 1993;74:529–540. doi: 10.1016/0092-8674(93)80054-i. [DOI] [PubMed] [Google Scholar]
- 25.Kappler J W, Staerz U, White J, Marrack P C. Self-tolerance eliminates T cells specific for Mls-modified products of the major histocompatibility complex. Nature. 1988;332:35–40. doi: 10.1038/332035a0. [DOI] [PubMed] [Google Scholar]
- 26.Kim K J, Kanellopoulos Langevin C, Merwin R M, Sachs D H, Asofsky R. Establishment and characterization of BALB/c lymphoma lines with B cell properties. J Immunol. 1979;122:549–554. [PubMed] [Google Scholar]
- 27.King L B, Corley R B. Lipopolysaccharide and dexamethasone induce mouse mammary tumor proviral gene expression and differentiation through distinct regulatory pathways. J Immunol. 1990;10:4211–4220. doi: 10.1128/mcb.10.8.4211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.King L B, Lund F E, White D A, Sharma S, Corley R B. Molecular events in B lymphocyte differentiation. Inducible expression of the endogenous mouse mammary tumor proviral gene, Mtv-9. J Immunol. 1990;144:3218–3227. [PubMed] [Google Scholar]
- 29.Korman A J, Bourgarel P, Meo T, Rieckhof G E. The mouse mammary tumour virus long terminal repeat encodes a type II transmembrane glycoprotein. EMBO J. 1992;11:1901–1905. doi: 10.1002/j.1460-2075.1992.tb05242.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laimins L A, Khoury G, Gorman C, Howard B, Gruss P. Host-specific activation of transcription by tandem repeats from simian virus 40 and Moloney murine sarcoma virus. Proc Natl Acad Sci USA. 1982;79:6453–6457. doi: 10.1073/pnas.79.21.6453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lewis S, Rosenberg N, Alt F, Baltimore D. Continuing k-gene rearrangement in a cell line transformed by Abelson murine leukemia virus. Cell. 1982;30:807–816. doi: 10.1016/0092-8674(82)90285-9. [DOI] [PubMed] [Google Scholar]
- 32.Löchelt M, Flügel R M, Aboud M. The human foamy virus internal promoter directs the expression of the Bel 1 transactivator and Bet protein early after infection. J Virol. 1994;68:638–645. doi: 10.1128/jvi.68.2.638-645.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Löchelt M, Muranyi W, Flügel R M. Human foamy virus genome possesses an internal, Bel-1-dependent and functional promoter. Proc Natl Acad Sci USA. 1993;90:7317–7321. doi: 10.1073/pnas.90.15.7317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Löchelt M, Yu S F, Linial M, Flügel R M. The human foamy virus internal promoter is required for efficient gene expression and infectivity. Virology. 1995;206:601–610. doi: 10.1016/s0042-6822(95)80077-8. [DOI] [PubMed] [Google Scholar]
- 35.Lund F E, Corley R B. Regulated expression of mouse mammary tumor proviral genes in cells of the B lineage. J Exp Med. 1991;174:1439–1450. doi: 10.1084/jem.174.6.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lund F E, Randall T D, Woodland D L, Corley R B. MHC class II limits the functional expression of endogenous superantigens in B cells. J Immunol. 1993;150:78–86. [PubMed] [Google Scholar]
- 37.Majors J E, Varmus H E. Nucleotide sequencing of an apparent proviral copy of env mRNA defines determinants of expression of the mouse mammary tumor virus env gene. J Virol. 1983;47:495–504. doi: 10.1128/jvi.47.3.495-504.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marrack P, Kushnir E, Kappler J. A maternally inherited superantigen encoded by a mammary tumour virus. Nature. 1991;349:524–526. doi: 10.1038/349524a0. [DOI] [PubMed] [Google Scholar]
- 39.McClure M A. Evolutionary history of reverse transcriptase. In: Skalka A M, Goff S, editors. Reverse transcriptase. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1993. pp. 425–444. [Google Scholar]
- 40.Miller C L, Garner R, Paetkau V. An activation-dependent, T-lymphocyte-specific transcriptional activator in the mouse mammary tumor virus env gene. Mol Cell Biol. 1992;12:3262–3272. doi: 10.1128/mcb.12.7.3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mink S, Hartig E, Jennewein P, Doppler W, Cato A C. A mammary cell-specific enhancer in mouse mammary tumor virus DNA is composed of multiple regulatory elements including binding sites for CTF/NFI and a novel transcription factor, mammary cell-activating factor. Mol Cell Biol. 1992;12:4906–4918. doi: 10.1128/mcb.12.11.4906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moore R, Dixon M, Smith R, Peters G, Dickson C. Complete nucleotide sequence of a milk-transmitted mouse mammary tumor virus: two frameshift suppression events are required for translation of gag and pol. J Virol. 1987;61:480–490. doi: 10.1128/jvi.61.2.480-490.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nandi S, McGrath C M. Mammary neoplasia in mice. Adv Cancer Res. 1973;17:353–414. [Google Scholar]
- 44.Nicolas J-F, Wegmann D, Lebrun P, Kaiserlian D, Tovey J, Glasebrook A L. Relationship of B cell Fc receptors to T cell recognition of Mls antigen. Eur J Immunol. 1987;17:1561–1565. doi: 10.1002/eji.1830171106. [DOI] [PubMed] [Google Scholar]
- 45.Nishio M, Xu L, Sasaki M, Haga S, Okumoto M, Mori N, Sarkar N H, Acha-Orbea H, Enami J, Imai S. Complete nucleotide sequence of mouse mammary tumor virus from JYG Chinese wild mice: absence of bacterial insertion sequences in the cloned viral gag gene. Breast Cancer. 1994;1:89–94. doi: 10.1007/BF02967037. [DOI] [PubMed] [Google Scholar]
- 46.Palacios R, Steinmetz M. Il-3-dependent mouse clones that express B-220 surface antigen, contain Ig genes in germ-line configuration, and generate B lymphocytes in vivo. Cell. 1985;41:727–734. doi: 10.1016/s0092-8674(85)80053-2. [DOI] [PubMed] [Google Scholar]
- 47.Patience C, Simpson G R, Coletta A A, Welch H M, Weiss R A. Human endogenous retrovirus expression and reverse transcriptase activity in the T47D mammary carcinoma cell line. J Virol. 1996;70:2654–2657. doi: 10.1128/jvi.70.4.2654-2657.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pullen A M, Wade T, Marrack P, Kappler J W. Identification of the region of T cell receptor beta chain that interacts with the self-superantigen MIs-1a. Cell. 1990;61:1365–1374. doi: 10.1016/0092-8674(90)90700-o. [DOI] [PubMed] [Google Scholar]
- 49.Randall T D, Lund F E, Brewer J W, Aldridge C, Wall R, Corley R B. Interleukin-5 (IL-5) and IL-6 define two molecularly distinct pathways of B-cell differentiation. Mol Cell Biol. 1993;13:3929–3936. doi: 10.1128/mcb.13.7.3929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reuss F U, Coffin J M. Stimulation of mouse mammary tumor virus superantigen expression by an intragenic enhancer. Proc Natl Acad Sci USA. 1995;92:9293–9297. doi: 10.1073/pnas.92.20.9293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50a.Reuss, F. U., and J. M. Coffin. Unpublished results.
- 51.Rosenberg N, Baltimore D, Scher D. In vitro transformation of lymphoid cells in Abelson murine leukemia virus. Proc Natl Acad Sci USA. 1975;72:1932–1936. doi: 10.1073/pnas.72.5.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sambasivarao D, Paetkau V. Interactions of a transcriptional activator in the env gene of the mouse mammary tumor virus with activation-dependent, T cell-specific transacting factors. J Biol Chem. 1996;271:8942–8950. doi: 10.1074/jbc.271.15.8942. [DOI] [PubMed] [Google Scholar]
- 53.Shackleford G M, Varmus H E. Construction of a clonable, infectious, and tumorigenic mouse mammary tumor virus provirus and a derivative genetic vector. Proc Natl Acad Sci USA. 1988;85:9655–9659. doi: 10.1073/pnas.85.24.9655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tsiagbe V K, Yoshimoto T, Asakawa J, Cho S Y, Meruelo D, Thorbecke G J. Linkage of superantigen-like stimulation of syngeneic T cells in a mouse model of follicular center B cell lymphoma to transcription of endogenous mammary tumor virus. EMBO J. 1993;12:2313–2320. doi: 10.1002/j.1460-2075.1993.tb05885.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.van Ooyen A, Michalides R J, Nusse R. Structural analysis of a 1.7-kilobase mouse mammary tumor virus-specific RNA. J Virol. 1983;46:362–370. doi: 10.1128/jvi.46.2.362-370.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Verdin E, Becker N, Bex F, Droogmans L, Burny A. Identification and characterization of an enhancer in the coding region of the genome of human immunodeficiency virus type 1. Proc Natl Acad Sci USA. 1990;87:4874–4878. doi: 10.1073/pnas.87.12.4874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Winslow G M, Scherer M T, Kappler J W, Marrack P. Detection and biochemical characterization of the mouse mammary tumor virus 7 superantigen (Mls-1a) Cell. 1992;71:719–730. doi: 10.1016/0092-8674(92)90549-r. [DOI] [PubMed] [Google Scholar]
- 58.Zhang D J, Tsiagbe V K, Huang C, Thorbecke G J. Control of endogenous mouse mammary tumor virus superantigen expression in SJL lymphomas by a promoter within the env region. J Immunol. 1996;157:3510–3517. [PubMed] [Google Scholar]