Abstract
The herpes simplex virus (HSV) gH-gL complex is essential for virus infectivity and is a major antigen for the host immune system. The association of gH with gL is required for correct folding, cell surface trafficking, and membrane presentation of the complex. Previously, a mammalian cell line was constructed which produces a secreted form of gHt-gL complex lacking the transmembrane and cytoplasmic tail regions of gH. gHt-gL retains a conformation similar to that of its full-length counterpart in HSV-infected cells. Here, we examined the structural and antigenic properties of gHt-gL. We first determined its stoichiometry and carbohydrate composition. We found that the complex consists of one molecule each of gH and gL. The N-linked carbohydrate (N-CHO) site on gL and most of the N-CHO sites on gH are utilized, and both proteins also contain O-linked carbohydrate and sialic acid. These results suggest that the complex is processed to the mature form via the Golgi network prior to secretion. To determine the antigenically active sites of gH and gL, we mapped the epitopes of a panel of gH and gL monoclonal antibodies (MAbs), using a series of gH and gL C-terminal truncation variant proteins produced in transiently transfected mammalian cells. Sixteen gH MAbs (including H6 and 37S) reacted with the N-terminal portion of gH between amino acids 19 and 276. One of the gH MAbs, H12, reacted with the middle portion of gH (residues 476 to 678). Nine gL MAbs (including 8H4 and VIII 62) reacted with continuous epitopes within the C-terminal portion of gL, and this region was further mapped within amino acids 168 to 178 with overlapping synthetic peptides. Finally, plasmids expressing the gH and gL truncations were employed in cotransfection assays to define the minimal regions of both gH and gL required for complex formation and secretion. The first 323 amino acids of gH and the first 161 amino acids of gL can form a stable secreted hetero-oligomer with gL and gH792, respectively, while gH323-gL168 is the smallest secreted hetero-oligomer. The first 648 amino acids of gH are required for reactivity with MAbs LP11 and 53S, indicating that a complex of gH648-gL oligomerizes into the correct conformation. The data suggest that both antigenic activity and oligomeric structure require the amino-terminal portions of gH and gL.
Herpes simplex virus (HSV) is a double-stranded DNA virus which encodes information for at least 11 glycoproteins, 10 of which are found in the virion envelope as well as on the surfaces of infected mammalian cells. Because of their surface location, HSV glycoproteins act as major antigenic determinants for the cellular and immune responses of the host (33, 41, 42). Five of the glycoproteins are important for virus entry into mammalian cells. The initial interaction between virus and cell is through the binding of gC with cell surface heparan sulfate proteoglycans (17, 18, 50), which is followed by the specific binding of gD with a cellular receptor, termed HVEM (29, 47). Subsequently, in some undefined manner, gD in combination with a homodimeric form of gB and an oligomeric complex of gH and gL function together to carry out fusion of the virion envelope with the plasma membrane of the cell (43, 44).
In a previous study (35), we described the expression and initial characterization of a recombinant form of the gH-gL complex. We constructed a cell line (HL-7) which expresses and secretes a soluble complex consisting of gH truncated at residue 792 just prior to the transmembrance anchor (gHt) and full-length gL. The purified complex stimulated production of neutralizing antibodies and protected mice challenged with herpes simplex virus type 1 (HSV-1) against development of zosteriform lesions. Furthermore, the purified gHt-gL complex reacted with gH and gL monoclonal antibodies (MAbs), including the anti-gH MAb LP11, indicating that it retains its proper antigenic structure after secretion and purification. These findings suggest that the conformation of gHt-gL in the secreted complex was similar to that of its full-length counterpart produced in HSV-infected cells. This cell system allowed for production of sufficient quantities of conformationally correct purified gH-gL for biochemical and antigenic analysis.
HSV-1 gH contains 838 amino acids, the first 18 of which have been postulated to constitute a cleavable signal sequence (12, 27). The protein has seven consensus sites for N-linked oligosaccharides (N-CHO) (22) as well as 11 sites for O-linked glycosylation (O-CHO) (16). Until this study, it was not known how many of the CHO sites were actually utilized by mammalian cells. gH-1 and gH-2 (26) are 77% homologous, especially in the C-terminal one-fourth of the proteins. The spacing of six N-CHO sites is conserved in gH-1 and gH-2. gH-1 has eight cysteines (27), seven of which are conserved in gH-2 (26). Whereas the disulfide bond arrangements of gB (32), gC (39), and gD (25) have been solved, nothing is known about the disulfide bond formation of gH. Residues 804 to 824 constitute the transmembrane region (TMR), which is hydrophobic and presumably anchors the protein into membranes. However, a previous study showed that truncated forms of gH lacking the TMR are not secreted from cells (36). Proper transport of full-length gH from the endoplasmic reticulum to the infected cell surface requires cotransport of gL (19).
gL contains 224 amino acids, the first 19 of which have been postulated to constitute the signal sequence (26). The protein has one consensus site for N-CHO, which has been shown to be utilized (19). gL has three potential sites for the addition of O-CHO (16). The location of the N-CHO consensus site and the locations of the four cysteine residues are conserved between gL-1 and gL-2 (26). Lastly, gL does not have a TMR, and when it is expressed in the absence of gH, gL is secreted from transfected cells (5). Retention of gL on the surface of the cell therefore requires the coexpression of gH (5, 38).
The goal of the current study was to extend our understanding of native gH-gL structure and to further relate structure to function. We determined the stoichiometry of gHt-gL and analyzed its carbohydrate composition. The oligomer consists of 1 mol each of gH and gL, and most sites for N-CHO are utilized. Both proteins are also modified with O-CHO. Using a series of C-terminal truncation mutants of gH and gL as well as a panel of gH and gL MAbs, we localize neutralizing and nonneutralizing epitopes on each protein. With the truncations, we mapped the minimum length of each protein that was needed to form a complex that could be secreted from the cell. We found that the first 323 amino acids of gH and the first 168 amino acids of gL can form a stable, secreted complex which is reactive with MAb LP11. Based on these and other data, a model of gH-gL structure is proposed.
MATERIALS AND METHODS
Cells and virus.
African green monkey kidney (Vero) and mouse L cells were grown at 37°C in Dulbecco’s modified Eagle medium (DMEM) supplemented with 5% fetal bovine serum (FBS). Chinese hamster ovary (CHO-K1) cells were grown in Ham’s F-12 medium with 5% FBS. D14 cells (Vero derived), which express HSV-1 ICP-6 (51) were grown in DMEM with 5% FBS and G418 (25 μg/ml) at 37°C. HL-7 cells, which express gHt-gL, were grown in DMEM supplemented with 10% FBS and hygromycin B (50 μg/ml) (35). For protein production, hygromycin B was eliminated from medium. HSV-1(hrR3) (51) was propagated on D14 cells, and its titers on Vero cells were determined. The virus strain hrR3 and D14 cells were kindly provided by S. Weller.
Synthetic peptides and protein N-terminal sequencing.
Four synthetic peptides mimicking amino acid residues 168 to 208, 179 to 208, 184 to 208, and 194 to 208 of gL (HSV-1 NS) were synthesized by the Protein Chemistry Laboratory of the Medical School of the University of Pennsylvania. Peptides UL1-1 and UL1-2, representing residues 26 to 44 and 209 to 223 (HSV-1 NS), respectively, were kindly provided by D. Johnson (19). N-terminal sequencing of purified gHt-gL was carried out by the Protein Chemistry Laboratory of the Medical School of the University of Pennsylvania.
Antibodies used.
The anti-gL MAbs VIII 62, 82, 87, 200, 820, and rabbit polyclonal antibodies (PAbs) RS88 and RS89 were described previously (34). Hybridoma cell lines secreting anti-gH-1 MAbs 52S and 53S (40) were obtained from the American Type Culture Collection. Hybridoma cell lines secreting anti-gH-1 MAbs H1 to H13 and anti-gL MAbs L1 to L3 were obtained by immunizing mice with purified gHt-gL (35). Anti-gH MAb 37S was kindly provided by M. Zweig (40). Anti-gH MAb LP11 was the gift of A. Minson (2). Hybridomas secreting anti-gH MAbs MP6, MP7, and MP8 were generated based on previously described methods (7) with purified gH from HSV-1-infected cells (36). MAb 8H4, which recognizes a linear epitope on gL, was described previously (5). Rabbit antibodies αUL1-1 and αUL1-2, both of which were prepared against peptide sequences of gL, were kindly provided by D. Johnson (19). Rabbit antibody R83 (prepared against gH-1) was described previously (36). R137 is an anti-gHt-gL PAb which was described previously. Antihemagglutinin (anti-HA) MAb, 12CA5, was provided by R. Riccardi.
Plasmids.
The construction of plasmids pSR162, pSR124, pSR123, pCMV3gHTrunc (323), pSR125, and pCMV3gL-1 was described previously (5, 36). pSR162, pSR124, pSR123, and pSR125 encode gH truncated at residues 792, 648, 475, and 102, respectively. These pSR plasmids contain a promoter derived from the Rous sarcoma virus and were constructed by inserting SpeI oligonucleotide linkers containing termination codons into pSR92 (which contains the entire gH-1-coding region from HSV-1 NS) at PvuII, StuI, or NheI sites. pCMV3gHTrunc(323) is constructed by inserting an SpeI linker into the AvrII site of pCMV3gH-1 (5). pMN plasmids contain the cytomegalovirus promoter linked to various portions of the coding sequence for gL fused at the carboxy terminus to the Flu epitope as previously described (34).
gHt-gL purification from HL-7 cells.
For gHt-gL protein production, HL-7 cells were grown in roller bottles. The culture supernatant was collected after 3 days and replaced with fresh medium. Two harvests of culture supernatant were obtained from each roller and were clarified by low-speed centrifugation. The secreted gHt-gL complex was purified by chromatography on an immunoaffinity column of 53S, a gH-1-specific MAb, by a previously described method (35). Briefly, the clarified culture supernatant was passed over the column, and bound protein was eluted with 50 mM glycine buffer (pH 2.5) containing 0.5 M NaCl. The eluate was neutralized with 1 M Tris-base (pH 9.0) and concentrated. Protein was quantitated with the bicinchoninic acid kit (Pierce Chemical Co.).
SDS-PAGE, Western blot, and dot blot analysis.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) under denaturing or native conditions was done as previously described (4, 35), with Tris-glycine precast gels (Novex Experimental Technology). Silver staining was performed with a silver staining kit (Pharmacia Biotech). For dot blot analysis, peptides were dissolved in phosphate-buffered saline (PBS) at a concentration of 2 mg/ml, and 2 μl of each peptide was spotted onto a nitrocellulose membrane. For Western blot analysis, proteins were transferred to nitrocellulose as previously described (35). Blots were probed first with anti-gH or anti-gL antibodies, as specified in the experiment, and then with goat anti-rabbit or anti-mouse immunoglobulin G-peroxidase (Boehringer). Bands were visualized on X-ray film after the addition of enhanced chemiluminescence (ECL) substrate (Amersham).
Enzymatic treatment of gHt-gL.
Purified protein (2.5 μg) was treated with 80 mU of EndoF (Boehringer), 150 mU of neuraminidase (Sigma Chemical Co.), or 2 mU of EndoH (Boehringer) either alone or in various combinations at 37°C for 4 h in PBS. To remove O-linked carbohydrates, protein was first treated with neuraminidase for 4 h at 37°C, followed by a 2-h incubation with 0.5 mU of O-glycosidase (Boehringer) at 37°C.
Gel filtration and ELISA.
Fifty micrograms of purified gHt-gL complex was applied to a Superose 12 column (Pharmacia). The presence of gHt and gL in each column fraction was detected by enzyme-linked immunosorbent assay (ELISA) as follows. Equal amounts of each fraction were used to coat two plates overnight at 4°C. The plates were blocked with PBS containing 1% bovine serum albumin and 1% ovalbumin. MAb 8H4 diluted in PBS with 0.05% bovine serum albumin and 0.05% ovalbumin was added to one plate to detect gL, and MAb 37S was added to the second to detect gH. After 1 h at room temperature, the plates were washed three times with PBS–0.5% Tween 20, goat anti-mouse immunoglobulin G (IgG)-peroxidase (Boehringer) was added, and the plates were incubated at room temperature for 30 min. After a rinse with citrate buffer (20 mM citrate acid [pH 4.5]), ABTS substrate (2,2′-azino-di-3-ethylbenzthiozoline-6-sulfonic acid; Moss, Inc.) was added, and absorbance was read at 405 nm with a microtiter plate reader (BioTek Instruments).
Transfection.
The lipid transfection method was used to transfect CHO-K1 cells with plasmids. Basically, FuGene 6 (Boehringer) was mixed with plasmid DNA in Ham’s F-12 medium and added to 60% confluent CHO-K1 cells. At 72 h posttransfection, culture supernatants and cells were collected separately, and cells were washed twice with ice-cold PBS with 0.01 mM phenylmethylsulfonyl fluoride and lysed at 4°C with cell lysing buffer (0.02 M Tris-Cl, 0.05 M NaCl, 0.5% Nonidet P-40, 0.5% deoxycholate, 0.01 mM tolylsulfonyl phenylalanyl chloromethyl ketone [TPCK], and 0.01 mM Nα-p-tosyl-l-lysine chloromethyl ketone [TLCK]).
Metabolic labeling of gHt-gL.
HL-7 cells were grown in roller bottles to confluency, and the culture medium was replaced with cysteine-free medium for 2 h. Then [35S]cysteine (Amersham) was added (100 μCi/bottle), and the cells were incubated for a further 2 h and then overlaid with 45 ml of DMEM with 2% FBS. Incubation was continued for 16 h. Labeled gHt-gL was purified from the cell supernatant as described above. For phosphorimaging analysis, purified [35S]cysteine-labeled gHt-gL was applied to SDS-PAGE gel and the dried gel was scanned with a Storm 840 PhosphorImager (Molecular Dynamics).
RESULTS
The stoichiometry of gH and gL in the complex.
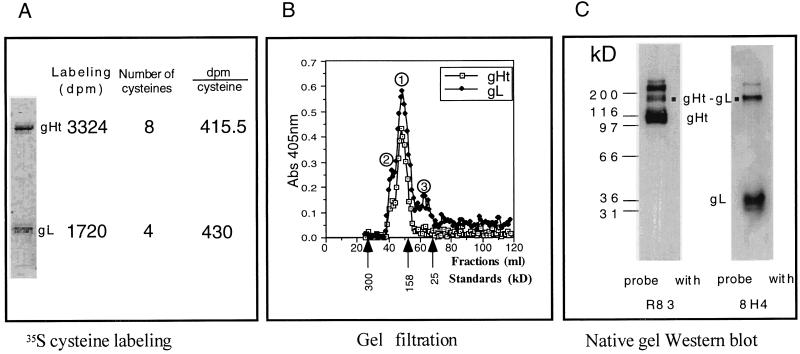
Previously, we showed that gHt-gL can be purified as a complex of the two proteins (35). To determine the stoichiometry of each protein in the complex, we took advantage of the 2:1 molar ratio of cysteine residues in gH and gL, respectively. If the two proteins were present in a 1:1 ratio in an oligomeric complex, then metabolic labeling should yield twice as many disintegrations per minute in gHt as in gL. HL-7 cells were grown in the presence of [35S]cysteine. The radiolabeled gHt-gL complex was purified, and the two proteins were separated on denaturing SDS-PAGE gel. The gel was dried, and the relative amount of label in each band was quantitated by phosphorimaging. In three separate experiments, one of which is shown (Fig. 1A), the gHt band contained twice as much radiolabel as the gL band. Since gHt contains eight cysteines per molecule and gL contains four, the disintegrations per minute per cysteine residue were equivalent. Thus, we conclude that the ratio of gHt to gL in the complex is 1:1. This result agrees with an estimate of the ratio, which was based on immunoprecipitation of [35S] cysteine-labeled full-length gH-gL from extracts of HSV-1-infected cells (19).
FIG. 1.
The stoichiometry of gHt and gL in the complex. (A) HL-7 cells were grown in medium containing [35S]cysteine. gHt-gL was purified and applied to SDS-PAGE gel. The gel was dried, and the disintegrations per minute (dpm) in each band were determined by phosphorimaging. (B) Fifty micrograms of purified gHt-gL was applied to a Superose 12 gel filtration column. One-milliliter fractions were collected and analyzed for gHt and gL by ELISA, with 37S MAb for gHt and 8H4 MAb for gL. The peaks are labeled ①, ②, and ③. Abs 405nm, absorbance at 405 nm. (C) Purified gHt-gL was mixed with sample buffer containing 0.1% SDS in the absence of reducing reagent. Samples were resolved on a 4 to 12% gradient SDS-PAGE gel and analyzed by Western blotting with R83 to detect gHt and 8H4 to detect gL. ▪ indicates the band detected by both R83 and 8H4.
As a second approach, and to determine the overall size of the complex, we subjected purified gHt-gL to Superose 12 gel filtration (Fig. 1B). gHt and gL were each detected in column fractions by ELISA. Ninety percent of gHt and gL eluted from the column in a single peak (Fig. 1B, peak 1), with a mass of approximately 180 kDa. A higher-mass shoulder (Fig. 1B, peak 2) also contained both proteins, suggesting that a small proportion of gHt and gL can also exist as higher-molecular-mass forms, i.e., greater than 180 kDa. A small amount of gL was found in the absence of gHt (Fig. 1B, peak 3), suggesting that the complex can dissociate under relatively mild elution conditions. As a third approach, gHt-gL was electrophoresed on a nondenaturing native gel, followed by Western blotting (Fig. 1C). Both anti-gH and anti-gL antisera reacted with a band migrating at 180 kDa as well as with a band migrating at >200 kDa. These results are in agreement with the gel filtration data. In addition, anti-gH serum reacted with a 110-kDa band, and anti- gL serum reacted with a 35-kDa band, corresponding to the monomeric sizes of the individual proteins. Since this represented the majority of the protein, we believe that much of the complex dissociated during electrophoresis, possibly due to the presence of 0.1% SDS in the native sample buffer. Hutchinson et al. (19) suggested that gH-gL is not disulfide bonded.
As a fourth approach, we attempted to determine the molar ratio of gL by performing N-terminal sequencing of the complex. However, the N terminus of gH was blocked. In contrast, we were able to determine the sequence of the first 20 amino acid residues of mature gL (which begin with GLPSTEYVIR) and found that glycine at amino acid 20 of the predicted sequence was the first amino acid of mature gL. Thus, we formally demonstrated that the predicted signal peptide of gL is cleaved at the predicted site (19).
We demonstrated that the mass of a gHt-gL complex is 180 kDa and that the ratio of gH and gL in the complex is 1:1. Since the molecular size of one gHt is 110 kDa and that of one gL is 35 kDa, there is only one gH in the gHt-gL complex. Thus, we conclude that the complex contains one gH and one gL.
Glycosylation of gHt and gL.
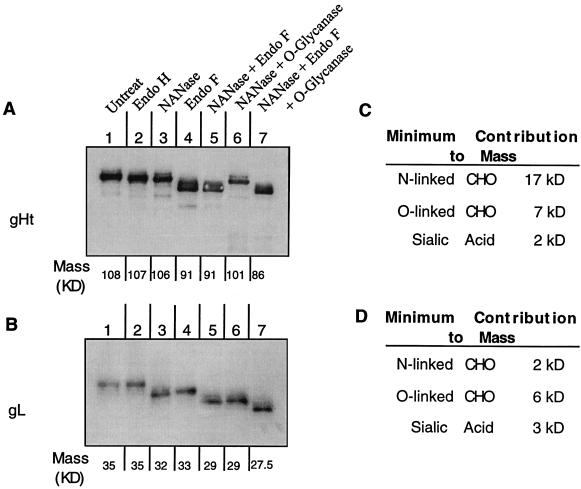
As the next step in structural analysis of gHt-gL, we determined the extent of glycosylation of the purified complex. The coding sequences for gH and gL contain predicted sites for N-CHO and O-CHO (Fig. 3). It was previously reported that both proteins within complexes obtained from infected-cell extracts or from baculovirus recombinants contain N-CHO (19, 46). Here we determined the type of glycosylation in each protein produced by HL-7 cells and estimated how many of the predicted sites were utilized. Purified gHt-gL was treated with glycosidases and with neuraminidase, either alone or in combination, and resolved by SDS- PAGE followed by Western blotting (Fig. 2A and B). Treatment with EndoH reduced the mass of gHt only slightly and had no effect on gL (Fig. 2A and B, lanes 2). In contrast, EndoF treatment had a more dramatic effect on both proteins, reducing the size of gHt from 108 to 91 kDa (Fig. 2A, compare lanes 1 and 4) and that of gL from 35 to 33 kDa (Fig. 2B, compare lanes 1 and 4). These data indicate that the majority of the N-CHO on gHt and the one N-CHO on gL were in the complex form. Neuraminidase treatment had a greater effect on the mobility of gL than on that of gHt (Fig. 2A and B, compare lanes 1 and 3), indicating that sialic acid was present, and combined EndoF and neuraminidase treatment increased the mobility of each protein even more, particularly that of gL (Fig. 2A and B, lanes 5). The latter results indicated that sialic acid was probably present on O-CHO, especially on gL. Treatment of gHt-gL with neuraminidase and O-glycanase also resulted in increased gHt mobility (Fig. 2A and B, lanes 6), another indication that both proteins contain O-CHO. When all three enzymes were used, gHt migrated to a molecular size of 86 kDa (Fig. 2A, lane 7), and gL migrated to a molecular size of 27.5 kDa (Fig. 2B, lane 7), values close to the predicted sizes of the unglycosylated molecules.
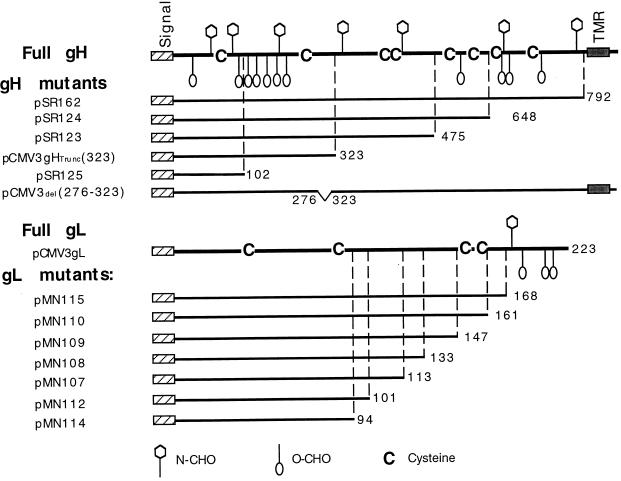
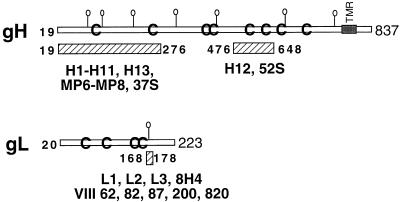
FIG. 3.
Schematic stick figures of full-length HSV-1 gH and gL and the C-terminal truncation mutants. Plasmids were constructed to express truncated forms of gH, a deletion mutant of gH pCMV3del(276–323), full-length gL (pCMV3gL), and truncated forms of gL. The signal peptides (signal), TMR, positions of the cysteine residues (C) and predicted N-CHO sites (open balloons) and predicted O-CHO sites (open hexagons), and the names of the plasmids are indicated.
FIG. 2.
Analysis of carbohydrates on gH and gL. Purified gHt-gL was incubated with no enzyme (untreated control) or with glycosidases and neuraminidase in the indicated combinations. The digests were resolved on a 10% denaturing polyacrylamide gel. Following transfer to a nitrocellulose membrane, one blot was probed with R83 anti-gH antibody (A). The bound antibody was detected with goat anti-rabbit IgG-peroxidase and chemiluminescent substrate. A second blot was probed with anti-gH MAb 8H4 (B) and then with goat anti-mouse IgG-peroxidase and chemiluminescent substrate. The molecular weight after each treatment of gH and gL was calculated according to molecular size markers on the gel (data not shown). The contributions of N-CHO, O-CHO, and sialic acid to the molecular weights of gH (C) and gL (D) were estimated from the difference between the untreated controls (A and B, lanes 1) and the Endo-F-treated (A and B, lanes 4), O-glycanase-treated (A and B, lanes 6), and neuraminidase-treated (A and B, lanes 7) samples.
We estimate that N-CHO contributes approximately 17 kDa to the mass of gHt; O-CHO contributes at least 7 kDa, and sialic acid contributes at least 2 kDa (Fig. 2C). In the case of gL, N-CHO contributes at least 2 kDa, O-CHO contributes 6 kDa, and sialic acid contributes 2 kDa in mass (Fig. 2D). Since there is a single N-CHO site on gL, which is used (mass, 2 kDa), we estimate that the mass of 17 kDa is sufficient to account for seven predicted N-CHO sites on gH. We conclude that most of the predicted N-linked sites on gHt are used and that they are present in the mature complex form. Our data show that both proteins also contain both O-CHO and sialic acid.
Antigenic analysis of gH-gL.
Thus far, only a limited number of anti-gH or gL MAbs have been available for the study of antigenic structure (2, 5, 34, 36, 40). In order to begin mapping antigenic domains on gHt-gL, we decided to generate more gH and gL MAbs. Mice were immunized with purified gHt-gL or full-length gH (36), and hybridoma supernatants were screened for production of gH- or gL-specific MAbs by an ELISA to detect antibodies which reacted against purified gHt-gL. To confirm their reactivity, we further screened positive clones by immunofluorescence, using cells transfected with gH and/or gL plasmids. Sixteen gH-specific and 4 gL-specific MAbs were obtained (Table 1). These 20 MAbs recognized gH or gL on Western blots of denaturing gels, indicating that all of them recognize linear epitopes.
TABLE 1.
gH and gL antibodies which react with linear epitopes
| Antibody | MAb or PAb | Glycogen specificity | Source of antigen |
|---|---|---|---|
| H1 to H13 | MAb | gH | Purified gHt-gL from HL-7 cells |
| MP6, MP7, MP8 | MAb | gH | Purified gH from HSV-1-infected cells |
| 37Sa | MAb | gH | HSV-1 virion |
| L1, L2, L3 | MAb | gL | Purified gHt-gL from HL-7 cells |
| 8H4b,c | MAb | gL | Purified gHt-gL from HL-7 cells |
| VIII 62, 82, 87, 200, 820d | MAb | gL | gL produced and purified from Escherichia coli |
| RS 88d | PAb | gL | gL produced and purified from Escherichia coli |
| RS 89d | PAb | gL | Rabbit immunized with gL plasmid DNA |
| R137c | PAb | gH-gL | Purified gHt-gL from HL-7 cells |
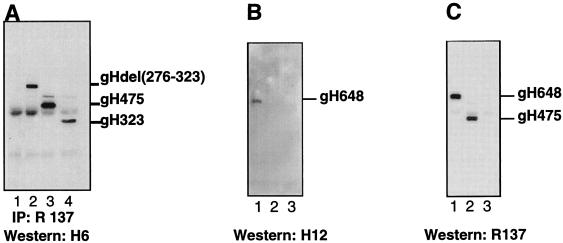
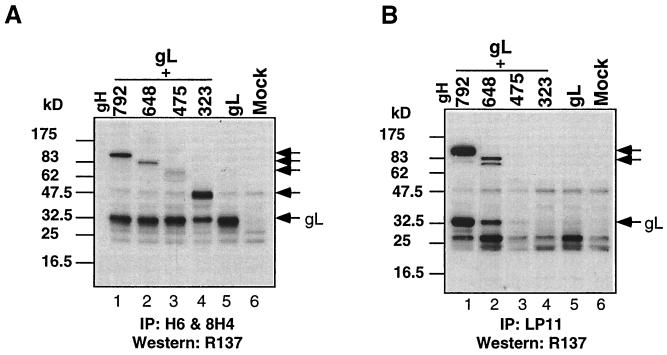
A series of gH and gL plasmids containing the gH and gL genes encoding C-terminal truncations of decreasing length (Fig. 3) was used to map these MAbs. In the first set of experiments, CHO-K1 cells were transiently transfected with plasmids expressing truncation or deletion mutants of gH, cell extracts were prepared and separated by SDS-PAGE, and Western blots were probed with gH MAbs. Surprisingly, 16 of the gH MAbs (represented in Fig. 4A by H6) reacted with the longer forms of gH (data not shown) as well as with gH475 (Fig. 4A, lane 3) and gH323 (lane 4). As a control, cells were mock transfected (Fig. 4A, lane 1). The band which reacts with the antibody in all four lanes, including lane 1, is antibody heavy chain (50 kDa). These data suggested that the epitopes for all 16 MAbs are located within residues 19 (at the end of the signal peptide) to 323. These MAbs also reacted with a gH mutant with residues 276 to 323 deleted (Fig. 4A, lane 2). Together, these results suggest that the epitope for H6 and those for 15 other gH-specific MAbs are located between residues 19 and 276. In contrast, MAb H12 reacted with gH648 but not with gH475 (Fig. 4B, compare lanes 1 and 2), indicating that its epitope is located between amino acids 475 and 648. As a control, a duplicate blot of Fig. 4B was probed with R137, a PAb prepared to purified gHt-gL (35). As expected, R137 reacted with both gH648 and gH475 (Fig. 4C, lanes 1 and 2).
FIG. 4.
Epitope mapping of anti-gH antibodies. CHO cells were transfected with gH truncation or deletion mutants. (A) Cell extracts were immunoprecipitated with R137, electrophoresed on a 12% denaturing polyacrylamide gel, transferred to nitrocellulose, and probed with MAb H6. Cell extracts from cells mock transfected (lane 1) or transfected with pCMV3gHdel(276–323) (lane 2), pSR123 (lane 3), or pCMV3gHtrunc(323) (lane 4) are shown. (B) Cell extracts were electrophoresed on a 12% denaturing polyacrylamide gel, transferred to nitrocellulose, and probed with MAb H12. (C) Cell extracts were electrophoresed on a 12% denaturing polyacrylamide gel, transferred to nitrocellulose, and probed with R137. Secondary antibodies were then added, and the blots were visualized by ECL. Lanes for panels B and C are cell extracts from pSR124- (lane 1), pSR123- (lane 2), and mock- (lane 3) transfected cells.
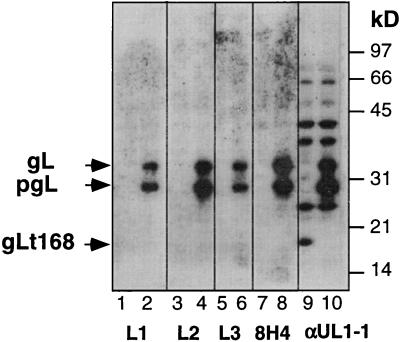
Similar studies were also done for the four newly prepared gL MAbs (L1, L2, L3, and 8H4). Cytoplasmic extracts prepared from pMN115 (encoding gL168)- or pCMV3gL (encoding full-length gL)-transfected CHO-K1 cells were separated by SDS-PAGE followed by Western blotting (Fig. 5). None of the MAbs reacted with gL168 (Fig. 5, lanes 1, 3, 5, and 7), although all of them reacted against precursor and product forms of full-length gL (Fig. 5, lanes 2, 4, 6, and 8). A control, αUL1-1, an antibody prepared against a synthetic peptide mimicking gL residues 26 to 44 (19), reacted with gL168 (Fig. 5, lane 9) and with full-length gL (Fig. 5, lane 10). These data suggest that the four gL MAbs recognize epitopes located between gL residues 169 and 224. Interestingly, Novotny and associates (34) generated five anti-gL MAbs with recombinant gL derived from bacteria as the immunogen, and the epitopes for these MAbs (called VIII 62, 82, 87, 200, and 820) were also located between gL residues 169 and 224.
FIG. 5.
Reactivity of anti-gL antibodies with gL168 or with full-length gL. CHO-K1 cells were transfected with pMN115 (gL168) (lanes 1, 3, 5, 7, and 9) or pCMV3gL (gL) (lanes 2, 4, 6, 8, and 10). Cell extracts were prepared and resolved on a 12% polyacrylamide denaturing gel. After Western blotting, separate strips of the membrane were probed with antibodies L1, L2, L3, 8H4, or αUL1-1, as indicated below the gel. Secondary antibodies were then added, and ECL was used to visualize the bands.
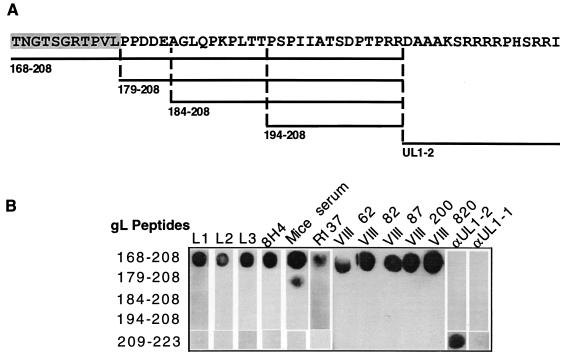
To fine map the gL epitope(s), a set of nested synthetic peptides which mimic amino acids 168 through 208 (peptide 168–208) was prepared (Fig. 6A). We also included the synthetic peptide mimicking C-terminal residues 209 to 223 (peptide 209–223) in this study. This peptide was originally used to prepare the rabbit polyclonal antibody αUL1-2 (19). As expected, αUL1-2 reacted only against the immunizing peptide 209–223, and αUL1-1, prepared against a synthetic peptide mimicking the amino terminus of gL, did not react with any peptides tested. We tested the reactivities of the four gL MAbs prepared in our lab and the five MAbs prepared by Novotny et al. against these peptides by immuno-dot blot analysis (Fig. 6B). All nine gL MAbs reacted with the peptide 168–208 but not with any of the other peptides. This result suggests that all of these anti-gL MAb epitopes are mainly contributed by amino acids 168 to 178 of gL. A pooled serum obtained from 10 mice immunized with purified gHt-gL as well as R137 was also tested in this assay. Both of these antisera reacted with peptide 168–208. The pooled mouse serum also reacted against peptide 179–208. Neither antiserum reacted with any other peptide. These data suggest that amino acids 168 to 178 of gL constitute a highly antigenic and immunogenic region of the protein. The mapping data for the gH- and gL-specific MAbs are summarized in Fig. 7.
FIG. 6.
Mapping of anti-gL antibody epitopes with synthetic peptides. (A) Diagram depicting the sequences of the set of overlapping synthetic peptides mimicking the gL sequence. The location of each peptide within the gL sequence is indicated. (B) Dot blot analysis of anti-gL antibodies with the peptides. Two microliters of each peptide (4 μg/dot) was spotted onto nitrocellulose membrane strips. After blocking, antibodies were added to each strip, and the reactivity was detected by ECL with goat anti-mouse peroxidase or goat anti-rabbit peroxidase.
FIG. 7.
Positions of the epitopes for MAbs to gH and gL mapped in this study. Schematic figures depict the linear amino acid sequences of gH and gL. The hatched bars depict the locations of the epitopes of anti-gH and anti-gL antibodies. The position of MAb 52S is according to the amino acid change (residue 536) of two MAR mutants selected by 52S (13).
gH and gL domains required for complex formation.
Retention of gH and gL in a stable secreted oligomeric complex does not require the transmembrane domain or the cytoplasmic tail (35). We therefore used the series of gH and gL truncations to determine the shortest fragment of each protein that was necessary to form a complex with the other protein. We knew from previous studies that several truncated forms of gH synthesized in transfected cells in the absence of gL failed to be secreted (36); however, cells transfected with a plasmid containing only gL were able to secrete gL protein (5). Thus, we decided to reexamine the properties of truncated gH, using a transient cotransfection system. We hypothesized that only properly folded protein complexes containing gH should be secreted from cells cotransfected with the gH and gL plasmids. To verify conformation, we examined the secreted complexes with MAbs.
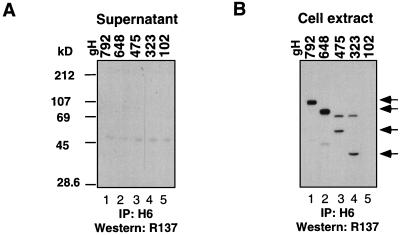
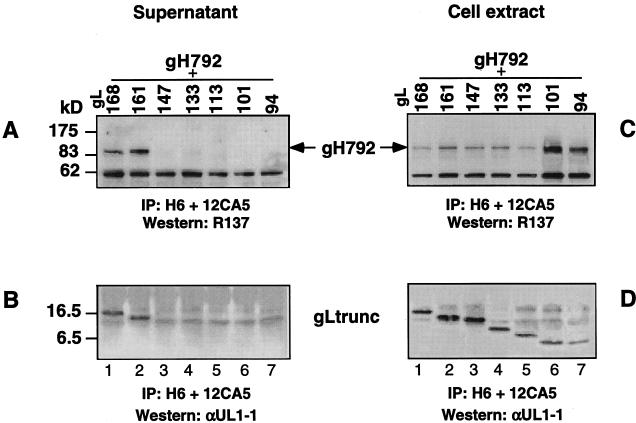
We first transfected CHO-K1 cells with the gH C-terminal plasmids (Fig. 3). Culture supernatants and cytoplasmic (cell) extracts obtained from each transfection were immunoprecipitated with anti-gH MAb H6, followed by Western blotting (Fig. 8). Both blots were then probed with polyclonal anti-gH serum R137. None of the truncated gH proteins were detected in the supernatant (Fig. 8A), despite the fact that the four longest truncations were found in the cell extract (Fig. 8B, lanes 1 to 4). Thus, as was shown before for gH792 (36), none of these gH truncations was secreted by itself. The shortest fragment, gH102, was not found in the cytoplasmic extract (Fig. 8B, lane 5), and therefore it was not used in any more studies. CHO-K1 cells were then cotransfected with pCMV3gL along with each one of the gH plasmids. Culture supernatants were immunoprecipitated with MAbs H6 (anti-gH) and 8H4 (anti-gL), and the precipitates were separated by SDS-PAGE, followed by Western blot analysis. Figure 9A shows that each of the four gH truncated proteins, terminating at residues 792, 648, 475, and 323, respectively, was found in the culture supernatant fluid (Fig. 9A, lanes 1 to 4). As a control, we transfected cells with pCMV3gL alone and found gL in the culture supernatant (Fig. 9A, lane 5). When the cotransfected culture supernatants were immunoprecipitated with anti-gH PAb (R83) alone, gL was found coimmunoprecipitated with gH truncations (data not shown). The results indicate that a fragment containing the first 323 amino acids of gH was sufficient for secretion when it was coexpressed with gL. Interestingly, this region is the same size as that reported to be necessary for complex formation of HHV6 gH with its cognate gL (1).
FIG. 8.
Expression of gH truncations by transfected CHO-K1 cells. (A) Culture supernatants were immunoprecipitated (IP) with gH-specific MAb H6, electrophoresed on a 12% denaturing polyacrylamide gel, transferred to nitrocellulose, and probed with R137. (B) Cell extracts were immunoprecipitated (IP) with gH-specific MAb H6, electrophoresed on a 12% denaturing polyacrylamide gel, transferred to nitrocellulose, and probed with R137. The bands corresponding to the various gH truncations are indicated with arrows. In both A and B, cells were transfected with pSR162 (lanes 1); pSR124 (lanes 2); pSR123 (lanes 3); pCMV3gH323 (lanes 4); and pSR125 (lanes 5).
FIG. 9.
Determination of the shortest C-terminal truncation of gH that forms a complex with full-length gL and is secreted. CHO-K1 cells were cotransfected with pCMV3gL, encoding full-length gL, and a plasmid encoding one of the five gH truncations. Culture supernatants were immunoprecipitated (IP) with MAbs H6 (anti-gH) and 8H4 (anti-gL) (A) or with LP11 (anti-gH-gL) (B). Proteins were resolved on a 12% denaturing polyacrylamide gel, transferred to nitrocellulose, and probed with R137. Lanes contain culture supernatants of cells cotransfected with pSR162 and pCMV3gL (lanes 1), pSR124 and pCMV3gL (lanes 2), pSR123 and pCMV3gL (lanes 3), pCMV3gH323 and pCMV3gL (lanes 4), or pCMV3gL (lanes 5) or of mock-transfected cells.
To determine the conformation of these truncated forms of gH-gL complexes, we tested their reactivities with LP11. Recognition by this MAb is particularly dependent on the correct conformation of the complex and is considered the “gold standard” for assessing proper folding of gH into its native biologically active form (13, 14, 40). CHO-K1 cells were cotransfected with pCMV3gL in combination with each of the plasmids expressing a gH truncation. Culture supernatants were immunoprecipitated with LP11, the precipitates were separated by SDS-PAGE, and Western blots were probed with R137 to detect gHt. gH792-gL and gH648-gL were immunoprecipitated with LP11 (Fig. 9B, lanes 1 and 2), but complexes containing the shorter truncations of gH were not (Fig. 9, lanes 3 to 5). This result suggests that (i) the LP11 epitope remains intact even in the absence of the last two cysteines of gH and that (ii) the LP11 epitope is located upstream of gH648. This result is consistent with the studies of HSV strains (MAb-resistant [MAR] mutants), which do not react with LP11 and contain point or insertion mutations within this region (10, 13). Similar results were also obtained for another conformation-dependent anti-gH MAb, 53S (data not shown).
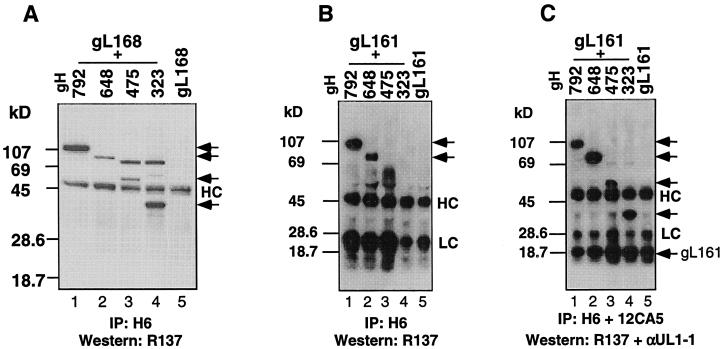
Next we determined the shortest truncation of gL which was sufficient for complex formation with gH. gL C-terminal truncations (Fig. 3) were constructed, with the Flu HA epitope attached to the C terminus of each protein (34) for the convenience of detection by anti-HA MAb 12CA5 (49). Each construct was transiently cotransfected into CHO-K1 cells along with plasmid pSR162, encoding amino acids 1 to 792 of gH. The assumption was that gH792 would be secreted only if the truncated gL protein formed a proper complex with it. Culture supernatants were immunoprecipitated with 12CA5. After Western blotting, R137 serum was used to detect secreted gH792 (Fig. 10A), and αUL1-1 antibody was used to detect each of the gL truncations (Fig. 10B). We found that gH792 was secreted when cells were cotransfected with pMN115, encoding gL168 (Fig. 10A, lane 1) or with pMN110, encoding gL161 (Fig. 10A, lane 2). gH792 was not secreted when cells were cotransfected with pSR162 and plasmids encoding shorter C-terminal truncations of gL (Fig. 10A, lanes 3 to 7). Interestingly, of all the gL plasmids tested by cotransfection, only gL168 and gL161 were detected in the culture supernatant (Fig. 10B, lanes 1 and 2). To show that each of the truncated gL proteins was actually synthesized, we carried out SDS-PAGE and Western blot analysis of the cytoplasmic extracts prepared from each transfection mixture (Fig. 10C and D). gH792 was present in each of the cell extracts (Fig. 10C), and each of the truncated forms of gL was also synthesized (Fig. 10D). We conclude that gL161 is the shortest gL truncation able to complex with gH792 and be secreted.
FIG. 10.
Determination of the minimal size of gL that forms a complex with gH792 and is secreted. CHO-K1 cells were cotransfected with pSR162, encoding gH792, and plasmids encoding one of the seven gL truncations. (A and B) Culture supernatants were immunoprecipitated (IP) with anti-gH MAb H6 and anti-HA MAb 12CA5 (directed at the HA epitope present in each of the gL truncations). The precipitated proteins were resolved on a 16% polyacrylamide denaturing gel and transferred to nitrocellulose. The blot was cut in half, and the top half was probed with R137 to detect gH (A); the bottom half was probed with αUL1-1 to detect gL (B). (C and D) Cell extracts were immunoprecipitated with H6 and 12CA5. The precipitated proteins were resolved on a 16% polyacrylamide denaturing gel and transferred to nitrocellulose. The blot was cut in half, the top half was probed with R137 to detect gH (C), and the bottom half was probed with αUL1-1 to detect gL (D). Lanes contain cells transfected with pSR 162 and pMN115 (lane 1); pMN110 (lane 2); pMN109 (lane 3); pMN108 (lane 4); pMN107 (lane 5); pMN112 (lane 6); and pMN114 (lane 7).
Having demonstrated that gH323 can be secreted when coexpressed with full-length gL and that gL161 is able to form a complex with gH792, we wondered whether a stable, secreted complex could form when these two short forms of each protein were coexpressed. Plasmids pMN115 and pMN110, expressing either gL168 or gL161, respectively, were cotransfected with each of the gH plasmids. Culture supernatants were immunoprecipitated with anti-gH MAb H6, separated by SDS-PAGE, transferred to nitrocellulose, and probed with R137 (Fig. 11). Truncations of gH ranging in length from 792 to 323 were present in the supernatant of cells cotransfected with pMN115, expressing gL168 (Fig. 11A, lanes 1 to 4). When a similar experiment was carried out with pMN110 (expressing gL161), we detected gH truncations ranging in length from 792 to 648 in the culture supernatant (Fig. 11B, lanes 1 and 2), whereas gH475 and gH323 were not detectably secreted. However, all of the truncated forms of gH and gL161 were readily detected in the cytoplasmic extracts from this set of transfections (Fig. 11C).
FIG. 11.
The smallest complexes formed and secreted by cells cotransfected with plasmids expressing truncated gH and truncated gL. CHO-K1 cells were cotransfected with gH truncation mutants and pMN115 (A) or pMN110 (B and C). (A) Culture supernatants were immunoprecipitated (IP) with anti-gH MAb H6, and the precipitates were resolved on 12% denaturing polyacrylamide gels, transferred to nitrocellulose, and probed with R137. Lane 1, pSR162 plus pMN115; lane 2, pSR124 plus pMN115; lane 3, pSR123 plus pMN115; lane 4, pCMV3gH323 plus pMN115; lane 5, pMN115. (B) Culture supernatants were immunoprecipitated (IP) with anti-gH MAb H6, and the precipitates were resolved on 12% denaturing polyacrylamide gels, transferred to nitrocellulose, and probed with R137. (C) Cell extracts were immunoprecipitated (IP) with anti-gH MAb H6 and anti-HA MAb 12CA5. Protein was resolved on 12% polyacrylamide denaturing gel, transferred to nitrocellulose, and probed with R137 and aUL1-1. Lanes in panels B and C are cells transfected with pSR162 plus pMN110 (lane 1); pSR124 plus pMN110 (lane 2); pSR123 plus pMN110 (lane 3); pCMV3gH323 plus pMN110 (lane 4); and pMN110 (lane 5). The positions of bands representing truncated gHs are indicated by arrows. The positions of IgG heavy chain (HC) and light chain (LC) are also indicated.
DISCUSSION
Four of the 11 HSV glycoproteins are essential for the entry of HSV into mammalian cells (3, 8, 24, 38, 44). Determination of the precise contribution of each virion glycoprotein to the entry process has been complicated by the sheer number of proteins involved as well as by the unknown relationship of one to another. It is now established that entry requires interaction between virion gD and one of several gD-specific cell surface receptors (11, 29, 30, 45, 47). This interaction coupled with the gC-glycosaminoglycan attraction may be viewed as a set of coordinated steps which lead to virus-cell fusion. The three remaining essential glycoproteins, gB and the gH-gL complex, are likely to be involved in the membrane fusion step (43). Turner et al. (44) recently showed that gD, gB, and gH-gL are necessary and sufficient for HSV-induced cell fusion.
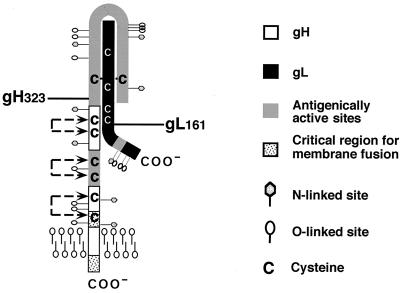
Which approaches can be used to shed light on this process and the molecules involved? We continued here by studying the properties of a secreted soluble form of the gH-gL complex. We previously showed that gHt-gL produced by HL-7 cells retains its native structure after purification as judged by its interaction with conformation-dependent MAbs and its ability to induce neutralizing antibodies and protect animals against viral challenge (35). Unlike soluble gD, gHt-gL did not block virus entry (35). Thus, although viral entry probably requires interaction of virion gH-gL with another virion glycoprotein or with a cell surface receptor, soluble gHt-gL failed to block this protein interaction. We decided that we lacked a good working model of gH-gL structure, and the goal here was to obtain data that would fill in details of antigenic structure and oligomerization. Based on the studies described in this article as well as on other known information about gH and gL, we propose a model (Fig. 12) to illustrate what we now know about this glycoprotein complex.
FIG. 12.
A proposed model for gH-gL structure. We speculate that the gH-gL complex contains one gH and one gL. The C termini of gH and gL are labeled coo−. The complex is anchored to the membrane through the TMR of gH. The gray area shows the antigenic active sites of gH and gL. Mutations affecting membrane fusion are located in the dot-shaded area of gH (10, 48). The positions of gH residue 323 and gL residue 161 are indicated as gH323 and gL161, respectively. These are the smallest regions required for complex formation and secretion. The positions of cysteines on both gH and gL are shown, and the possible disulfide bonds on gH are indicated by broken lines. The locations of N-linked and O-linked CHO sites are also shown.
Our first objective was to determine the stoichiometry of the complex. An estimate of 1:1 for the two proteins was made by Hutchinson et al. (19) by immunoprecipitation methods. Here we confirmed and extended these results by three different approaches to show that there is one molecule each of gH and gL in the truncated gH and gL complex. According to the size of the complex determined by gel filtration (180 kDa), it appears that the bulk of the complex exists in solution as a heterodimer. However, a small fraction of the protein eluted at a higher molecular weight, and higher-molecular-weight forms were seen by native SDS-PAGE. Analytical ultracentrifugation studies are now under way to examine this issue more precisely. Interestingly, higher oligomeric forms of gH-gL were not found in the virion by chemical cross-linking techniques (15). Thus, our working model (Fig. 12) contains one molecule each of gH and gL. Here, we show a full-length version of gH-gL which is anchored in the virion envelope via the TMR of gH.
Earlier, we detected complex oligosaccharides on gH during infection (36). In the present study, we showed that both proteins contained complex sugars and sialic acid. Our studies also suggest that most of the consensus sites for the addition of N-CHO in gH are utilized. These sites are distributed throughout the length of the protein (see model, Fig. 12). How much the N-CHO contributes to function is not known, but the most C-terminal N-CHO (NGT at residues 783, 784, and 785) is conserved among other gH homologs (12, 20, 21, 27, 28, 31, 37), although it is not required for infection (10). Furthermore, both gH and gL contain O-CHO and sialic acid. All of the predicted sites for O-glycosylation of gL are clustered near its C terminus. The locations of the oligosaccharides were taken into consideration when the model was being constructed.
We also incorporated published information about gH and gL MAbs as well as the new information from this study into the thinking behind this model. For example, LP11 recognizes a conformation-dependent epitope which requires proper oligomerization of gH and gL (14). This MAb protects animals by passive immunization (9). Because LP11 has a high level of virus-neutralizing activity, it has been used to isolate MAR mutant viruses (13). In addition, linker insertion mutants of gH have been examined for LP11 activity (10). These mutant sites have been mapped to gH residues 86 to 326 (10, 13). Most of the continuous epitopes of gH mapped in this study are located on gH323. Thus, the first 323 amino acids of gH contain a major antigenic site, with both continuous and discontinuous epitopes. A second antigenic site is located between amino acids 475 and 648. In this study, MAb H12 mapped to this site and a neutralizing antibody, 52S, originally described by Showalter et al. (40), also recognizes an epitope influenced by this site, based on isolation of a MAR mutant at residue 536 (13). Interestingly, from our studies, gH792-gL and gH648-gL were immunoprecipitated with LP11, but complexes containing the shorter truncations of gH were not, suggesting that residues between 326 and 648 are required to maintain the LP11 epitope.
Novotny et al. (34) found that five MAbs prepared to a bacterium-produced gL recognized linear epitopes in gL, blocked cell-to-cell spread, failed to neutralize virus, and were located in the C terminus of the molecule. The four anti-gL MAbs prepared against the gH-gL complex had the same properties (34a). Of particular interest for our model was the finding that the epitopes for all nine MAbs are located within amino acids 168 through 178 of gL. The structural reason why these MAbs all recognize the same stretch of amino acids remains to be determined. However, we propose that the N terminus of gL is hidden, possibly within the confines of gH (Fig. 12). According to hydropathy plots, the amino-terminal portion of gL is hydrophobic, consistent with our positioning of it within the gH polypeptide. Additionally, a PAb prepared against a peptide mimicking the N terminus of gL fails to detect the molecule within the complex but reacts readily with gL when it is separated from the complex (38). Moreover, R137, a polyclonal antibody against gHt-gL, reacted with full-length gL (Fig. 9A) but not with gL168 (Fig. 11A) or shorter gL forms. The N terminus of gL may serve as a scaffold for proper folding of gH around it. The positioning of the amino-terminal portion of gH on the outside is supported by the epitope mapping data as well as the location of nine N- and O-linked oligosaccharides. The C-terminal portion of gH after amino acid 648 may be partially buried in the membrane (not depicted in Fig. 12), as it appears to be critical for membrane fusion (10, 44).
Next, our model reflects the experiments carried out to determine the shortest portions of both gH and gL required for proper folding and secretion of the complex. Previous studies showed that in the absence of gL, neither full-length gH nor carboxyl terminal truncations of gH are secreted from cells (36), but gL is secreted from transfected cells that do not express gH (5). We found that the shortest fragment of gH that supported complex formation with gL and secretion is gH323. This estimate for gH must be tempered by the fact that the shortest fragment, gH102, was not detected in the transfected cell in the presence or absence of full-length gL. Interestingly, the shortest fragment necessary for gH-gL complex formation in HHV-6 requires the first 320 residues of gH (1). It is also of interest that this region of HSV-1 gH overlaps a major antigenic site.
Our studies showed that gL161 is the minimal portion of gL necessary for complex formation and secretion with gH792. However, gL168 was the shortest gL truncation which could fold and form a secreted complex with gH323. It should be noted that the major antigenic site on gL is downstream of the portion which interacts with gH. Thus, this portion of gL is probably exposed to the outside, as depicted in Fig. 12. Since gL161 and gL168 are truncated after the fourth cysteine, we argue that gL forms two disulfide bonds, both of which are necessary for proper folding and oligomerization with gH. A similar conclusion was drawn for varicella-zoster virus gL (6, 23).
The contribution of disulfide bonds to glycoprotein structure and function is critical. The disulfide bond arrangements of gD (25), gC (39), and gB (32) are known. What is the contribution of the cysteine residues to the structure of gHt-gL? It is relatively clear that gH is not disulfide bonded to gL, since the two molecules dissociate under nonreducing conditions (19). What do our data suggest about intramolecular disulfide bonding? First, gH792 contains eight cysteines in the full-length protein, while gH(648) contains six, gH(475) contains four, and gH (323) contains two. We hypothesize that since these four truncated forms of gH are able to complex with gL, there are no unpaired cysteines which could result in a malformed protein (25). According to this hypothesis, gH323 contains one disulfide bond (i.e., C1-C2), gH475 contains two (C1-C2 and C3-C4), and gH678 contains three (C1-C2, C3-C4, and C5-C6). Cysteines 7 and 8 are depicted as paired by default. We depicted the proposed disulfide bond arrangement as broken lines between the hypothetically paired cysteines in Fig. 12. Paired cysteines have also been found in the first 230 amino acids of HHV-6 (1). Clearly, this hypothesis should be tested by biochemical experiments.
Thus, our initial efforts have begun to probe the structure-functional relationships of the two proteins, and further biochemical and genetic studies are necessary.
ACKNOWLEDGMENTS
We thank S. Weller for strain hrR3 of HSV-1(KOS) as well as for D14 cells, D. Johnson for synthetic peptides UL1-1 and UL1-2 and antibodies αUL1-1 and αUL1-2, A. Minson for MAb LP11, R. Riccardi for MAb 12CA5, L. Spruce for peptide synthesis, and Y. Harrison-Shahan for N-terminal sequencing.
This study was supported by Public Health Service grants NS-30606 from the National Institute of Neurological Diseases and Stroke, AI-18289 from the National Institute of Allergy and Infectious Diseases, DE-08239 from the National Institute of Dental Research, CA 21776 from National Institutes of Health (P.G.S.), and CA 16520 and DK 19525 from Cancer and Diabetes Centers core support grants (J.D.L.). M.J.N. is supported by T32 GM08152.
REFERENCES
- 1.Anderson R A, Liu D X, Gompels U A. Definition of a human herpesvirus-6 betaherpesvirus-specific domain in glycoprotein gH that governs interaction with glycoprotein gL: substitution of human cytomegalovirus glycoproteins permits group-specific complex formation. Virology. 1996;217:517–526. doi: 10.1006/viro.1996.0146. [DOI] [PubMed] [Google Scholar]
- 2.Buckmaster E A, Gompels U, Minson A. Characterisation and physical mapping of an HSV-1 glycoprotein of approximately 115 × 103 molecular weight. Virology. 1984;139:408–413. doi: 10.1016/0042-6822(84)90387-8. [DOI] [PubMed] [Google Scholar]
- 3.Cai W, Gu B, Person S. Role of glycoprotein B of herpes simplex virus type 1 in viral entry and cell fusion. J Virol. 1988;62:2596–2604. doi: 10.1128/jvi.62.8.2596-2604.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cohen G H, Isola V J, Kuhns J, Berman P W, Eisenberg R J. Localization of discontinuous epitopes of herpes simplex virus glycoprotein D: use of a nondenaturing (“native” gel) system of polyacrylamide gel electrophoresis coupled with Western blotting. J Virol. 1986;60:157–166. doi: 10.1128/jvi.60.1.157-166.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dubin G, Jiang H. Expression of herpes simplex virus type 1 glycoprotein L (gL) in transfected mammalian cells: evidence that gL is not independently anchored to cell membranes. J Virol. 1995;69:4564–4568. doi: 10.1128/jvi.69.7.4564-4568.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duus K M, Hatfield C, Grose C. Cell surface expression and fusion by the varicella-zoster virus gH:gL glycoprotein complex: analysis by laser scanning confocal microscopy. Virology. 1995;210:429–440. doi: 10.1006/viro.1995.1359. [DOI] [PubMed] [Google Scholar]
- 7.Eisenberg R J, Long D, Ponce de Leon M, Matthews J T, Spear P G, Gibson M G, Lasky L A, Berman P, Golub E, Cohen G H. Localization of epitopes of herpes simplex virus type 1 glycoprotein D. J Virol. 1985;53:634–644. doi: 10.1128/jvi.53.2.634-644.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Forrester A, Farrell H, Wilkinson G, Kaye J, Davis-Poynter N, Minson T. Construction and properties of a mutant of herpes simplex virus type 1 with glycoprotein H coding sequences deleted. J Virol. 1992;66:341–348. doi: 10.1128/jvi.66.1.341-348.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Forrester A J, Sullivan V, Simmons A, Blacklaws B A, Smith G L, Nash A A, Minson A C. Induction of protective immunity with antibody to herpes simplex virus type 1 glycoprotein H (gH) and analysis of the immune response to gH expressed in recombinant vaccinia virus. J Gen Virol. 1991;72:369–375. doi: 10.1099/0022-1317-72-2-369. [DOI] [PubMed] [Google Scholar]
- 10.Galdiero M, Whiteley A, Bruun B, Bell S, Minson T, Browne H. Site-directed and linker insertion mutagenesis of herpes simplex virus type 1 glycoprotein H. J Virol. 1997;71:2163–2170. doi: 10.1128/jvi.71.3.2163-2170.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Geraghty, R. J., C. Krummenacher, R. J. Eisenberg, G. H. Cohen, and P. G. Spear. Entry of alphaherpesviruses mediated by poliovirus receptor related protein 1 and poliovirus receptor. Science, in press. [DOI] [PubMed]
- 12.Gompels U, Minson A. The properties and sequence of glycoprotein H of herpes simplex virus type 1. Virology. 1986;153:230–247. doi: 10.1016/0042-6822(86)90026-7. [DOI] [PubMed] [Google Scholar]
- 13.Gompels U A, Carss A L, Saxby C, Hancock D C, Forrester A, Minson A C. Characterization and sequence analyses of antibody-selected antigenic variants of herpes simplex virus show a conformationally complex epitope on glycoprotein H. J Virol. 1991;65:2393–2401. doi: 10.1128/jvi.65.5.2393-2401.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gompels U A, Minson A C. Antigenic properties and cellular localization of herpes simplex virus glycoprotein H synthesized in a mammalian cell expression system. J Virol. 1989;63:4744–4755. doi: 10.1128/jvi.63.11.4744-4755.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Handler C G, Eisenberg R J, Cohen G H. Oligomeric structure of glycoproteins in herpes simplex virus type 1. J Virol. 1996;70:6067–6075. doi: 10.1128/jvi.70.9.6067-6070.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hansen J E, Lund O, Engelbrecht J, Bohr H, Nielsen J O, Hansen J-E S, Brunak S. Prediction of O-glycosylation of mammalian proteins: specificity patterns of UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase. Biochem J. 1995;308:801–813. doi: 10.1042/bj3080801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Herold B C, Visalli R J, Sumarski N, Brandt C, Spear P G. Glycoprotein C-independent binding of herpes simplex virus to cells requires cell surface heparan sulfate and glycoprotein B. J Gen Virol. 1994;75:1211–1222. doi: 10.1099/0022-1317-75-6-1211. [DOI] [PubMed] [Google Scholar]
- 18.Herold B C, WuDunn D, Soltys N, Spear P G. Glycoprotein C of herpes simplex virus type 1 plays a principal role in the adsorption of virus to cells and in infectivity. J Virol. 1991;65:1090–1098. doi: 10.1128/jvi.65.3.1090-1098.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hutchinson L, Browne H, Wargent V, Davis-Poynter N, Primorac S, Goldsmith K, Minson A C, Johnson D C. A novel herpes simplex virus glycoprotein, gL, forms a complex with glycoprotein H (gH) and affects normal folding and surface expression of gH. J Virol. 1992;66:2240–2250. doi: 10.1128/jvi.66.4.2240-2250.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keller P M, Davison A J, Lowe R S, Riemen M W, Ellis R W. Identification and sequence of the gene encoding gpIII, a major glycoprotein of varicella-zoster virus. Virology. 1987;157:526–533. doi: 10.1016/0042-6822(87)90295-9. [DOI] [PubMed] [Google Scholar]
- 21.Klupp B G, Mettenleiter T C. Sequence and expression of the glycoprotein gH gene of pseudorabies virus. Virology. 1991;182:732–741. doi: 10.1016/0042-6822(91)90614-h. [DOI] [PubMed] [Google Scholar]
- 22.Kornfeld R, Kornfeld S. Assembly of asparagine-linked oligosaccharides. Annu Rev Biochem. 1985;54:631–664. doi: 10.1146/annurev.bi.54.070185.003215. [DOI] [PubMed] [Google Scholar]
- 23.Li Q, Buranathai C, Grose C, Hutt-Fletcher L M. Chaperone functions common to nonhomologous Epstein-Barr virus gL and varicella-zoster virus gL proteins. J Virol. 1997;71:1667–1670. doi: 10.1128/jvi.71.2.1667-1670.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ligas M W, Johnson D C. A herpes simplex virus mutant in which glycoprotein D sequences are replaced by β-galactosidase sequences binds to but is unable to penetrate into cells. J Virol. 1988;62:1486–1494. doi: 10.1128/jvi.62.5.1486-1494.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Long D, Wilcox W C, Abrams W R, Cohen G H, Eisenberg R J. Disulfide bond structure of glycoprotein D of herpes simplex virus types 1 and 2. J Virol. 1992;66:6668–6685. doi: 10.1128/jvi.66.11.6668-6685.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McGeoch D J, Cunningham C, Mcintyre G, Dolan A. Comparative sequence analysis of the long repeat regions and adjoining parts of the long unique regions in the genomes of herpes simplex viruses types 1 and 2. J Gen Virol. 1991;72:3057–3075. doi: 10.1099/0022-1317-72-12-3057. [DOI] [PubMed] [Google Scholar]
- 27.McGeoch D J, Davison A J. DNA sequence of the herpes simplex virus type 1 gene encoding glycoprotein gH, and identification of homologous in the genomes of varicella-zoster virus and Epstein-Barr virus. Nucleic Acids Res. 1986;14:4281–4292. doi: 10.1093/nar/14.10.4281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meyer A L, Petrovskis E A, Duffus W P, Thomsen D R, Post L E. Cloning and sequence of an infectious bovine rhinotracheitis virus (BHV-1) gene homologous to glycoprotein H of herpes simplex virus. Biochim Biophys Acta. 1991;1090:267–269. doi: 10.1016/0167-4781(91)90116-4. [DOI] [PubMed] [Google Scholar]
- 29.Montgomery R I, Warner M S, Lum B J, Spear P G. Herpes simplex virus-1 entry into cells mediated by a novel member of the TNF/NGF receptor family. Cell. 1996;87:427–436. doi: 10.1016/s0092-8674(00)81363-x. [DOI] [PubMed] [Google Scholar]
- 30.Nicola A V, Ponce de Leon M, Xu R, Hou W, Whitbeck J C, Krummenacher C, Montgomery R I, Spear P G, Eisenberg R J, Cohen G H. Monoclonal antibodies to distinct sites on the herpes simplex virus (HSV) glycoprotein D block HSV binding to HVEM. J Virol. 1998;72:3595–3601. doi: 10.1128/jvi.72.5.3595-3601.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nicolson L, Cullinane A A, Onions D E. The nucleotide sequence of an equine herpesvirus 4 gene homologue of the herpes simplex virus 1 glycoprotein H gene. J Gen Virol. 1990;71:1793–1800. doi: 10.1099/0022-1317-71-8-1793. [DOI] [PubMed] [Google Scholar]
- 32.Norais N, Tang D, Kaur S, Chamberlain S H, Masiarz F R, Burke R L, Marcus F. Disulfide bonds of herpes simplex virus type 2 glycoprotein gB. J Virol. 1996;70:7379–7387. doi: 10.1128/jvi.70.11.7379-7387.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Norrild B. Humoral response to herpes simplex virus infections. In: Roizman B, Lopez C, editors. The herpesviruses. Immunobiology and prophylaxis of human herpesvirus infections. Vol. 4. New York, N.Y: Plenum Press; 1985. pp. 69–86. [Google Scholar]
- 34.Novotny M, Parish M, Spear P. Variability of herpes simplex virus 1 gL and anti-gL antibodies that inhibit cell fusion but not viral infectivity. Virology. 1996;221:1–13. doi: 10.1006/viro.1996.0347. [DOI] [PubMed] [Google Scholar]
- 34a.Peng, T. Unpublished data.
- 35.Peng T, Ponce-de-Leon M, Jiang H, Dubin G, Lubinski J M, Eisenberg R J, Cohen G H. The gH-gL complex of herpes simplex virus (HSV) stimulates neutralizing antibody and protects mice against HSV type 1 challenge. J Virol. 1998;72:65–72. doi: 10.1128/jvi.72.1.65-72.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roberts S R, Ponce de Leon M, Cohen G H, Eisenberg R J. Analysis of the intracellular maturation of the herpes simplex virus type 1 glycoprotein gH in infected and transfected cells. Virology. 1991;184:609–624. doi: 10.1016/0042-6822(91)90431-a. [DOI] [PubMed] [Google Scholar]
- 37.Robertson G R, Scott N A, Miller J M, Sabine M, Zheng M, Bell C W, Whalley J M. Sequence characteristics of a gene in equine herpesvirus 1 homologous to glycoprotein H of herpes simplex virus. DNA Sequence. 1991;1:241–249. doi: 10.3109/10425179109020779. [DOI] [PubMed] [Google Scholar]
- 38.Roop C, Hutchinson L, Johnson D C. A mutant herpes simplex virus type 1 unable to express glycoprotein L cannot enter cells, and its particles lack glycoprotein H. J Virol. 1993;67:2285–2297. doi: 10.1128/jvi.67.4.2285-2297.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rux A H, Moore W T, Lambris J D, Abrams W R, Peng C, Friedman H M, Cohen G H, Eisenberg R J. Disulfide bond structure determination and biochemical analysis of glycoprotein C from herpes simplex virus. J Virol. 1996;70:5455–5465. doi: 10.1128/jvi.70.8.5455-5465.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Showalter S D, Zweig M, Hampar B. Monoclonal antibodies to herpes simplex virus type 1 proteins, including the immediate-early protein ICP 4. Infect Immun. 1981;34:684–692. doi: 10.1128/iai.34.3.684-692.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Spear P G. Antigenic structure of herpes simplex viruses. In: van Regenmortel M V H, Neurath A R, editors. Immunochemistry of viruses. The basis for serodiagnosis and vaccines. Amsterdam, The Netherlands: Elsevier Science Publishers B. V.; 1985. pp. 425–446. [Google Scholar]
- 42.Spear P G. Glycoproteins specified by herpes simplex viruses. In: Roizman B, editor. The herpesviruses. Vol. 3. New York, N.Y: Plenum Press; 1985. pp. 315–356. [Google Scholar]
- 43.Spear P G. Membrane fusion induced by herpes simplex virus. In: Bentz J, editor. Viral fusion mechanisms. Boca Raton, Fla: CRC Press, Inc.; 1993. pp. 201–232. [Google Scholar]
- 44.Turner A, Bruun B, Minson T, Browne H. Glycoproteins gB, gD, and gHgL of herpes simplex virus type 1 are necessary and sufficient to mediate membrane fusion in a Cos cell transfection system. J Virol. 1998;72:873–875. doi: 10.1128/jvi.72.1.873-875.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Warner, M. S., W. Martinez, R. J. Geraghty, R. I. Montgomery, J. C. Whitbeck, R. Xu, R. J. Eisenberg, G. H. Cohen, and P. G. Spear. A cell surface protein with herpesvirus entry activity (HveB) confers susceptibility to infection by herpes simplex virus type 2, mutants of herpes simplex virus type 1 and pseudorabies virus. Virology, in press. [DOI] [PubMed]
- 46.Westra D F, Glazenburg K L, Harmsen M C, Tiran A, Scheffer A J, Welling G W, The T H, Welling-Wester S. Glycoprotein H of herpes simplex virus type 1 requires glycoprotein L for transport to the surfaces of insect cells. J Virol. 1997;71:2285–2291. doi: 10.1128/jvi.71.3.2285-2291.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Whitbeck J C, Peng C, Lou H, Xu R, Willis S H, Ponce de Leon M, Peng T, Nicola A V, Montgomery R I, Warner M S, Soulika A M, Spruce L A, Moore W T, Lambris J D, Spear P G, Cohen G H, Eisenberg R J. Glycoprotein D of herpes simplex virus (HSV) binds directly to HVEM, a member of the tumor necrosis factor receptor superfamily and a mediator of HSV entry. J Virol. 1997;71:6083–6093. doi: 10.1128/jvi.71.8.6083-6093.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wilson D W, Davis-Poynter N, Minson A C. Mutations in the cytoplasmic tail of herpes simplex virus glycoprotein H suppress cell fusion by a syncytial strain. J Virol. 1994;68:6985–6993. doi: 10.1128/jvi.68.11.6985-6993.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wilson I A, Niman H L, Houghten R A, Cherenson A R, Connolly M L, Lerner R A. The structure of an antigenic determinant in a protein. Cell. 1984;37:767–778. doi: 10.1016/0092-8674(84)90412-4. [DOI] [PubMed] [Google Scholar]
- 50.WuDunn D, Spear P G. Initial interaction of herpes simplex virus with cells is binding to heparan sulfate. J Virol. 1989;63:52–58. doi: 10.1128/jvi.63.1.52-58.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yao F, Schaffer P A. An activity specified by the osteosarcoma line U2OS can substitute functionally for ICP0, a major regulatory protein of herpes simplex virus type 1. J Virol. 1995;69:6249–6258. doi: 10.1128/jvi.69.10.6249-6258.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]