Abstract
We have developed a rice (Oryza sativa) genome annotation database (Osa1) that provides structural and functional annotation for this emerging model species. Using the sequence of O. sativa subsp. japonica cv Nipponbare from the International Rice Genome Sequencing Project, pseudomolecules, or virtual contigs, of the 12 rice chromosomes were constructed. Our most recent release, version 3, represents our third build of the pseudomolecules and is composed of 98% finished sequence. Genes were identified using a series of computational methods developed for Arabidopsis (Arabidopsis thaliana) that were modified for use with the rice genome. In release 3 of our annotation, we identified 57,915 genes, of which 14,196 are related to transposable elements. Of these 43,719 nontransposable element-related genes, 18,545 (42.4%) were annotated with a putative function, 5,777 (13.2%) were annotated as encoding an expressed protein with no known function, and the remaining 19,397 (44.4%) were annotated as encoding a hypothetical protein. Multiple splice forms (5,873) were detected for 2,538 genes, resulting in a total of 61,250 gene models in the rice genome. We incorporated experimental evidence into 18,252 gene models to improve the quality of the structural annotation. A series of functional data types has been annotated for the rice genome that includes alignment with genetic markers, assignment of gene ontologies, identification of flanking sequence tags, alignment with homologs from related species, and syntenic mapping with other cereal species. All structural and functional annotation data are available through interactive search and display windows as well as through download of flat files. To integrate the data with other genome projects, the annotation data are available through a Distributed Annotation System and a Genome Browser. All data can be obtained through the project Web pages at http://rice.tigr.org.
Rice (Oryza sativa) has emerged as a model species for the cereals, a group of grass species that includes not only rice but also the major crop species maize (Zea mays), wheat (Triticum aestivum), barley (Hordeum vulgare), sorghum (Sorghum bicolor), oats (Avena sativa), and millet (Eleusine coracana). Features of rice that have contributed to its utility as a model species include its small stature in comparison to other cereals, transformability, dense genetic map, well-developed genomic resources, and small genome (430 Mb; Arumuganathan and Earle, 1991) that was the target of four genome sequencing projects (Sasaki and Burr, 2000; Barry, 2001; Goff et al., 2002; Yu et al., 2002). In addition, rice is collinear with other larger genome cereals (Gale and Devos, 1998), serving as the central species for comparative studies in the cereals.
Of the four rice genome sequencing projects, the public effort of the International Rice Genome Sequencing Project (IRGSP) has generated the highest quality and most complete genome sequence, that of the japonica subspecies, cultivar Nipponbare. However, although the IRGSP has generated a near-complete finished sequence for rice (http://rgp.dna.affrc.go.jp/IRGSP/), the annotation of the rice genome is still ongoing. Annotation, in which features are noted on the genome sequence, is a dynamic, iterative process. The primary component of any genome annotation effort is identifying the genes, also termed structural annotation. This is a challenging task as it relies heavily on computational methods with less than optimal sensitivity and specificity. Structural annotation can be improved dramatically by access to experimental evidence, such as transcripts and protein sequences. However, the strength of experimental evidence is variable. The most powerful evidence is that of full-length cDNAs (FL-cDNAs). For rice, a collection of approximately 32,000 FL-cDNA sequences are available (The Rice Full-Length cDNA Consortium, 2003). Second in value for structural annotation are expressed sequence tags (ESTs), with 298,857 ESTs available for rice (dbEST Release 121004; http://www.ncbi.nlm.nih.gov/dbEST/dbEST_summary.html). Last, alignment to similar protein structures provides powerful, although not high resolution, evidence for gene structure.
Layered on top of structural annotation is functional annotation. This involves identifying the function of the genes, as well as associated genome sequences, with biologically relevant features. Assigning gene function is perhaps the most subjective aspect of functional annotation. This is typically done based on transitive annotation as only a small portion of genes within any genome have been verified for function at the experimental level. Gene function can be assigned based on sequence similarity with known proteins or through the presence of protein domains with known function. Gene ontologies have been developed to provide controlled vocabularies to annotate gene function, thereby allowing for cross-kingdom querying of genes (The Gene Ontology Consortium, 2000). Other types of functional annotation include integration of the genetic map with physical and sequence maps, thereby creating a unified map resource. It also can include the identification of the target sequence (and gene) of tagged insertion lines to provide researchers with catalogs of mutant lines. Another level of annotation is the identification of related sequences within the genome and between the target genome and related species. Identification of paralogous genes within the genome provides a resource for annotation as well as for evolutionary studies to examine gene and genome duplication events within rice. Alignment of the genome with sequences from related species provides for identification of orthologs and paralogs, which can be powerful resources for accelerating research in other species.
We have generated a public annotation database for the rice genome based on the near-complete sequence generated by the IRGSP. This database, termed Osa1 for Oryza sativa 1, is a Sybase relational database that stores and tracks rice genome sequence and annotation. Annotation data are generated using a series of bioinformatic processes initially developed for annotating the Arabidopsis (Arabidopsis thaliana) genome (Wortman et al., 2003) that have been modified for use with the rice genome. These processes result in the identification of genes, determination of gene structure, identification of domain and motif composition, construction of paralogous families, and assignment of gene function, all of which are stored in Osa1. Additional functional annotation data types are generated through other bioinformatic processes to further improve the depth of annotation as well as the gene structure. In this article, we describe the content and features of the Osa1 database along with the bioinformatic processes utilized to annotate features of the rice genome. From these activities, we can report on basic features of the rice genome and predicted rice proteome. We also provide information on our curation efforts, data access, and scheduled updates for our annotation.
OSA1 DATABASE STRUCTURE AND CONTENT
The goal of The Institute for Genomic Research (TIGR) rice annotation database is to provide high-quality, uniform structural and functional annotation of the rice genome. This involves identifying all the genes, constructing gene models (including alternative splice forms), and identifying putative function for these genes. In addition, the rice genome is annotated with functional annotation data types to provide biologists with the highest quality of content as possible. The bulk of the data is stored in the Osa1 database. This database is similar in structure to other eukaryotic annotation databases at TIGR, such as the Ath1 database utilized in the reannotation of Arabidopsis (Wortman et al., 2003).
ANNOTATION PROCESS
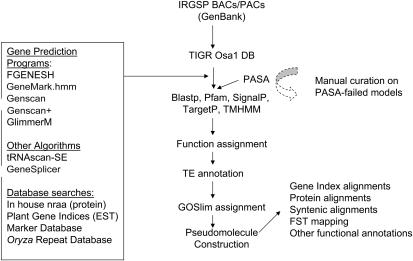
The basic sequence unit of our rice genome annotation pipeline is the bacterial artificial chromosome (BAC)/P1 artificial chromosome (PAC) clones generated by the IRGSP. The BAC/PACs are processed using the Eukaryotic Genome Control (EGC) pipeline (Wortman et al., 2003) to identify genes and construct gene models in which the gene structure is resolved and alternative splice forms are identified (Fig. 1). The EGC pipeline employs ab initio gene finders and database searches to generate evidence for gene model construction. The ab initio gene finders used in the rice EGC pipeline include FGENESH (monocot matrix; Salamov and Solovyev, 2000), GeneMark.hmm (rice matrix; Lukashin and Borodovsky, 1998), Genscan (maize matrix) and Genscan+ (Arabidopsis matrix; Burge and Karlin, 1997), as well as GlimmerM (rice matrix; Yuan et al., 2001). Splice sites are identified using GeneSplicer (Pertea et al., 2001). Sequences are searched against protein and nucleotide sequence databases using the Analysis and Annotation Tool (AAT) package (Huang et al., 1997). An in-house plant nonredundant amino acid database that contains all publicly available plant protein sequences (e.g. GenBank, SwissProt, etc.) is searched using the dps and nap programs of AAT. Searches of expressed transcript sequences are performed by aligning the BAC/PAC sequences against the TIGR plant gene indices (Quackenbush et al., 2001) using the dds and gap2 programs of AAT (Huang et al., 1997). Repetitive elements are identified by searching the TIGR Oryza Repeat Database (Ouyang and Buell, 2004). Transfer-RNAs are identified using tRNAScan-SE (Lowe and Eddy, 1997).
Figure 1.
Diagram of rice genome annotation pipeline. BAC/PAC sequences were obtained from GenBank, loaded into the Osa1 database, and processed through the EGC pipeline. Currently, multiple gene prediction programs are run on the sequence (FGENESH, GeneMark.hmm, Genscan, Genscan+, and GlimmerM). Algorithms are run to identify tRNAs and splice donor/acceptor sites. The sequences are also searched against a series of databases to identify regions of sequence similarity. The output of the FGENESH program is used to make the initial model for each gene. The gene structure is improved using the PASA program, in which EST and FL-cDNA evidence is incorporated resulting in the final gene model(s) for that locus. Protein similarity and domains are identified through BLASTP searches against amino acid databases and domain/motif-finding algorithms. Putative annotation (gene function) is assigned to the genes, including identification of TE-related genes. GOSlim assignments are made prior to construction of the pseudomolecules. Following construction of the pseudomolecules, other types of functional annotation are performed.
Comparison of sensitivity and specificity of the five ab initio gene finders listed above revealed FGENESH as superior in predicting rice genes (Q. Yuan and C.R. Buell, unpublished data). Thus, for all of our automated annotation, we utilized the output of the FGENESH program and not the other ab initio gene finders to generate gene models that were improved through use of the Program to Assemble Spliced Alignments (PASA; Haas et al., 2003), in which FL-cDNA and EST evidence are used to update current gene models and create new models based on spliced alignments. As rice has ample FL-cDNA and EST data, alternative splice forms of the genes can be detected. Thus, a single gene can have multiple gene models if there is experimental evidence of alternative splicing. For domain and motif searches, the deduced protein sequence of the gene model is searched against the Pfam database (Bateman et al., 2002) using Hmmpfam (http://bio.ifom-firc.it/docs/Software/hmmer-html/node25.html) and against the other InterPro databases (Mulder et al., 2003) using InterProScan (Zdobnov and Apweiler, 2001). For sequence similarity, the protein sequence is searched against a nonredundant amino acid database using BLASTP. The protein and Pfam domain evidence is used to assign putative function to the gene, which is further refined through a series of simple scripts to remove transitive annotation errors. One critical component in our annotation process is robust identification of genes that are transposable element (TE)-related as these are captured in the EGC pipeline. To do this, we search the gene models using the TIGR Oryza Repeat Database (Ouyang and Buell, 2004) as well as identify any gene models that contain TE-related Pfam domains. These genes are then transitively annotated using the TIGR Oryza Repeat Database nomenclature or the Pfam domain name. The gene, its underlying models, the evidence used in its construction, and other functional annotation data are viewable through a Web interface termed the Manatee page (Wortman et al., 2003).
CONSTRUCTING PSEUDOMOLECULES
Pseudomolecules (virtual contigs) of the 12 rice chromosomes are constructed to remove the overlapping sequences between the BAC/PAC clones. The BAC/PAC clones are aligned on the chromosome based on the clone order as reported by the IRGSP, and overlaps between the BAC/PAC clones are confirmed (http://rgp.dna.affrc.go.jp/cgi-bin/statusdb/irgsp-status.cgi). An overwhelming majority of the BAC/PAC clones match perfectly with most discrepancies involving polymorphisms of copy number of mono- and dinucleotide simple sequence repeats. The pseudomolecule DNA sequence is constructed by trimming the overlap region at junction points in which the genes are least disrupted and the annotation data are transferred from the BAC/PAC clones to the pseudomolecules. As a consequence of identifying junction points within the overlap region based on gene location, minimal resolution of incongruent gene models must be made. Statistics on our current pseudomolecules can be found in Table I and through the project Web pages (http://rice.tigr.org/tdb/e2k1/osa1/pseudomolecules/info.shtml).
Table I.
Features of the Osa1 release 3 pseudomolecules
| Chromosome | Length | No. Genesa | No. Gene Modelsb |
|---|---|---|---|
| bp | |||
| 1c | 43,249,587 | 6,905 (1,256) | 7,378 (1,266) |
| 2 | 35,876,369 | 5,422 (1,032) | 5,770 (1,042) |
| 3 | 36,347,804 | 5,986 (1,026) | 6,544 (1,039) |
| 4 | 35,023,746 | 5,534 (1,622) | 5,831 (1,633) |
| 5 | 29,695,855 | 4,662 (1,272) | 4,977 (1,279) |
| 6 | 31,198,625 | 4,837 (1,239) | 5,071 (1,242) |
| 7 | 29,688,601 | 4,635 (1,137) | 4,858 (1,143) |
| 8 | 28,309,183 | 4,327 (1,198) | 4,536 (1,205) |
| 9 | 22,680,691 | 3,409 (950) | 3,561 (952) |
| 10c | 22,698,374 | 3,743 (1,002) | 3,933 (1,007) |
| 11 | 28,369,397 | 4,286 (1,138) | 4,436 (1,139) |
| 12 | 27,492,551 | 4,169 (1,324) | 4,355 (1,327) |
| Total | 370,630,783 | 57,915 (14,196) | 61,250 (14,274) |
Number of genes reported is the total number of genes with the number of TE-related genes in parentheses.
Number of gene models reported is the total number of gene models with the number of TE-related gene models in parentheses.
Pack-MULEs were only identified on chromosomes 1 and 10 using data available from Jiang et al. (2004).
The total length of the 12 pseudomolecules is 370.6 Mb, smaller than the 430 Mb genome size reported for rice (Arumuganathan and Earle, 1991). Some of this discrepancy can be attributed to physical gaps within the IRGSP sequence and, consequently, our pseudomolecules. Release 3 of the pseudomolecules contains 45 physical gaps (excluding 10 centromeric and 23 telomeric gaps) that are due to either a physical gap in the BAC/PAC tiling path or a lack of availability of finished sequence for that segment. A surprisingly large number of BAC/PAC clones (494) available in GenBank/DDBJ/EMBL are not incorporated into the 12 pseudomolecules. These primarily represent redundant sequences and thus are already represented within the pseudomolecules (429 BAC/PACs). However, some clones could not be mapped to our pseudomolecules (65 BAC/PACs) and represent unanchored sequences. Sequence, annotation, and position on the pseudomolecules of these clones are available through the project Web pages (http://rice.tigr.org/tdb/e2k1/osa1/unusedBAC/index.shtml).
Recently, another public version of the rice pseudomolecules was released by the IRGSP (http://rgp.dna.affrc.go.jp/IRGSP/Build3/build3.html). While the foundation of both pseudomolecule sets is the same, our pseudomolecules and those of the IRGSP differ in several ways. First, the IRGSP molecules were constructed in July 2004 and thus represent a slightly older build with a higher percentage of unfinished sequence. Second, the IRGSP utilized a “left greedy” approach in constructing the pseudomolecules, and, thus, sequence in the overlap regions between the two builds will differ on occasion. Third, to date, no annotation of the genes is available from the IRGSP pseudomolecules, although the IRGSP pseudomolecules were the substrate used in the Rice Annotation Project 1 annotation effort in which FL-cDNAs were annotated (http://rgp.dna.affrc.go.jp/IRGSP/Build3/build3.html) by a community of rice and bioinformatics experts.
In release 3 of our pseudomolecules, we identified a total of 57,915 genes with 14,196 related to TE, leaving 43,719 non-TE-related genes (Table I). Of these non-TE-related genes, we were able to assign a putative function to 18,545 genes (42.4%), while 5,777 (13.2%) were annotated as encoding an expressed protein due to the presence of EST and/or FL-cDNA support. The remaining 19,397 genes (44.4%) lacked experimental evidence or sequence similarity with known proteins or domains and were annotated as encoding hypothetical proteins. Pack-MULEs, which are chimeric Mutator-like elements that have assimilated host sequences, were reported in the rice genome by Jiang et al. (2004). Although Jiang et al. (2004) identified more than 3,000 putative Pack-MULEs in the rice genome, their approach utilized automated methods, which when manually inspected revealed unsatisfactory accuracy. Consequently, their automated Pack-MULE annotation cannot be readily incorporated into our annotation. However, Pack-MULEs on chromosomes 1 and 10 were manually annotated by Jiang et al. (2004), and using their manual annotation, we have been able to identify 238 and 145 genes on our chromosome 1 and 10 pseudomolecules, respectively, as Pack-MULE related. As this represents such a small fraction of the entire set of Pack-MULEs, we have not incorporated this into our annotation and instead provide a list of chromosome 1 and 10 genes that we have identified as Pack-MULE related based on manual curation by Jiang et al. (2004; http://rice.tigr.org/tdb/e2k1/osa1/pack_mule.shtml). As more TEs are described and annotated in the rice genome, such as those of Juretic et al. (2004), we will continue to identify these within our database to provide robust annotation of this type of gene.
Abundant transcript evidence (approximately 300,000 ESTs and approximately 32,000 FL-cDNAs) is available for rice and provides evidence for gene expression as well as experimental data to refine gene model structure. Using cutoff criteria of 95% identity over 50% of the length of the EST/FL-cDNA sequence, 25,410 genes could be aligned with either a rice EST and/or FL-cDNA, suggesting that at least 43.9% of the genes are expressed. To provide users with access to the pattern and frequency of gene expression, we developed the Expression Viewer Tool (http://rice.tigr.org/tdb/e2k1/osa1/expression/expression.info.shtml) for gene models in the Osa1 database. Alternative splicing does occur in rice, and we identified 2,538 genes representing a total of 5,873 alternative splice forms in the rice genome using both EST and FL-cDNA evidence. These can be viewed in our Alternative Splice Form Viewer Tool (http://rice.tigr.org/tdb/e2k1/osa1/expression/alt_spliced.info.shtml). To improve the gene structure in an automated manner, we used the PASA program, which employs more stringent criteria than a simple alignment of EST and FL-cDNA sequence to the genome (Haas et al., 2003). Due to problems associated with a subset of the FL-cDNAs and ESTs, such as genomic contamination or incomplete/aberrant splicing, automated annotation updates can only occur when the modified gene structure passes a series of stringent validation tests, such as length and homology comparisons with the current annotation. In release 3 of our annotation using the PASA program, the structures of 15,165 genes (18,252 gene models) have been updated based on EST/FL-cDNA evidence. Since release 3, we have been able to update additional gene models, such that 19,419 genes (22,651 gene models) have been updated and will be included in release 4. However, 3,948 genes aligned with FL-cDNAs and ESTs using PASA but failed the validation process, and, thus, their structure cannot be updated. These genes will be the target of manual curation efforts in the near future.
LOCUS IDENTIFIERS
In previous releases of our pseudomolecules, we referred to the genes and gene models using an internal identifier (feat_name) that was cumbersome for the user and not readily convertible through releases of the pseudomolecules. However, implementation of a more stable identifier prior to release 3 was not feasible due to the instability of the unfinished genome sequence. In release 3, 98% of the underlying sequence is finished with the remaining 2% sequence derived from phase II HGS BAC/PAC clones, which although unfinished are in ordered and oriented contigs. Thus, for release 3, we have implemented locus identifiers for the genes. The convention we have implemented is similar to the nomenclature used in Arabidopsis with adaptations made for the larger size of the rice genome and to the nomenclature currently under discussion by the rice community (http://www.gramene.org/documentation/nomenclature/). However, as the nomenclature has not been finalized and alternative builds of the pseudomolecules are publicly available (http://rgp.dna.affrc.go.jp/IRGSP/Build3/build3.html), we have chosen to use locus nomenclature that clearly denotes the TIGR loci. Each gene is labeled LOC_OsXXg#####, with LOC referring to locus, Os referring to rice, XX referring to chromosome (01–12), g referring to gene, and a 5-digit number referring to the gene order on the chromosome. We have sequentially numbered the genes (loci) on each of the chromosomes in increments of 10 to allow for insertion of future loci. To accommodate additional sequence that may be identified in the physical gaps, we have provided larger spacing in the locus numbering at the physical gaps. As locus identifiers are associated with release 3 and not our previous releases, we have developed a Version Converter (http://rice.tigr.org/tdb/e2k1/osa1/v_converter/index.shtml) to allow users to find locus identifiers for genes and models from releases 1 and 2, which had been identified solely with feat_names. A tab-delimited flat file is also available at the project FTP site. As with the addition of newly identified loci, we have scripts in place to handle merging, splitting, and retirement of locus identifiers as the annotation improves over the course of our project.
QUALITY OF STRUCTURAL ANNOTATION
The rice EGC annotation pipeline is highly automated. Manual curation, in which a trained annotator inspects the gene model and evidence(s) supporting the model and then creates the most congruent model possible, is considered the highest level of annotation possible. While this is clearly desirable at the whole-genome level, this is labor intensive and not feasible with a genome the size of rice or with the iterative updates of the annotation that are required with incremental releases of new experimental evidence. However, a portion of the release 3 gene model set is derived from manually curated genes (282 BACs, 5,420 genes). This provided us the opportunity to assess the accuracy of our automated annotation pipeline to determine the qualitative impact of automation on annotation. As shown in Table II, our automated annotation pipeline captures similar gene structure as manually curated genes. One clear difference is the slightly higher density of genes in manually curated annotation versus automated annotation, suggesting lack of capture of all genes in the automated method. This can be explained by the use of a sole ab initio gene finder (FGENESH) for the identification of genes in the automated pipeline. With manual curation, an annotator would examine all ab initio gene finder output (five programs in total) as well as experimental evidence and create a gene if the evidence warrants. Another feature that differs in manual versus automated annotation is the assignment of putative function, a subjective process at best. This is clearly an aspect of annotation that can be highly benefited by manual curation, and we will be addressing this in future curation activities through annotation of paralogous families. Another aspect that we feel warrants manual inspection is curation of gene model structure of genes with EST and/or FL-cDNA evidence that failed our PASA validation tests (3,948 genes in release 3).
Table II.
Statistics of manual versus automated rice genome annotation
| Statistic | Manual | Automated |
|---|---|---|
| Number of BACs | 286 | 3,617 |
| Total length (bp)a | 38,489,150 | 476,318,197 |
| Average BAC GC content (%) | 43.5 | 43.5 |
| Average intergenic GC content (%) | 40.9 | 41.4 |
| Average exon GC content (%) | 54.3 | 53.1 |
| Average intron GC content (%) | 38.7 | 38.7 |
| No. genesb | 6,717 (1,311) | 78,950 (17,665) |
| Average gene sizec | 2,411 (3,111) | 2,519 (3,383) |
| Total gene length (bp) | 16,197,893 (42.1%) | 198,925,545 (41.8%) |
| Gene density (kb/gene) | 5.7 | 6.2 |
| Known/putative genes | 3,668 (54.6%) | 44,287 (56.1%) |
| Expressed genes | 702 (10.5%) | 6,468 (8.2%) |
| Hypothetical genes | 2,347 (34.9%) | 28,195 (35.7%) |
| Total no. of gene modelsd | 7,232 (1,315) | 82,921 (17,730) |
| Average exon no. per model | 4.2 | 4.2 |
| Average exon size (bp) | 289 | 312 |
| Average intron size (bp) | 375 | 364 |
Total length is that for all BAC/PAC clones, including the overlapping regions between clones.
Genes annotated are at the BAC/PAC level, and the numbers include the duplicated genes in the overlap regions. The numbers in parentheses are the numbers of TE-related genes.
Gene size is reported for all genes; within the parentheses is the size of TE-related genes.
Total number of gene models is at the BAC/PAC level and includes the duplicated models in the overlap region. The number of TE-related gene models is within the parentheses.
FUNCTIONAL ANNOTATION DATA TYPES
In addition to structural annotation of the genes, we annotated a number of other data types within the rice genome. At the sequence level, we have mapped >10,000 sequence-based genetic markers derived primarily from rice to our pseudomolecules (http://rice.tigr.org/tdb/e2k1/osa1/BACmapping/markers_physical_map.shtml). This collection of markers (13,900) includes RFLP-based (Causse et al., 1994; Harushima et al., 1998) markers as well as transcript-derived markers that were positioned on the genetic map through anchoring to the yeast artificial chromosome physical map and integration with the existing RFLP-based genetic map (Wu et al., 2002). Using cutoff criteria of ≥97% identity over ≥90% length of the marker, 10,279 markers could be aligned to 10,695 locations within the rice genome. Reducing this stringency to ≥95% identity over ≥90% length of the marker resulted in 10,945 markers aligning to 11,630 locations within the genome. Markers with multiple alignments within the genome are denoted on our Rice Genetic Marker search page.
Multiple efforts are under way to generate a large collection of tagged insertion mutants in rice using T-DNA tagging (Sallaud et al., 2004), Ac/Ds (Greco et al., 2001; Kim et al., 2004), and Tos17 (Miyao et al., 2003). We downloaded from the dbGSS division of GenBank a total of 26,725 sequences reported to be flanking sequence tags (FSTs) from various projects. Using 95% identity over 80% of the FST length, a total of 23,140 (86.6%) FSTs could be aligned to the rice genome (41,378 total alignments). With respect to genic insertions, 17,929 FSTs could be mapped into or within 500 bp of 11,094 genes, providing a ready set of putative mutants for functional genomic research.
To provide resources for other plant biologists, we have aligned the rice genome with sequences from other plant species. These alignments are displayed through several Web interfaces: through a single interface aligning the pseudomolecules with 20 of the TIGR Plant Gene Indices (http://rice.tigr.org/tigr-scripts/alignTC/db/chrs.pl?taxon=16), through the evidence window on the Manatee page for each gene, and through a genome browser (see below). For the alignments with the Gene Indices, we used the BLAT program (Kent, 2002) and displayed matches with minimum cutoff criteria of ≥75% identity over ≥25% length (monocot sequence) and ≥10% length (dicot sequence). For the evidence displayed in the Manatee page, we used a dps final chain cutoff score of ≥100 and a dds final chain score of ≥100. In addition, we have aligned sequence-based genetic markers from maize (4,906 markers) available from the IBM2 Neighbors map at Maize GDB (http://www.maizegdb.org/) and wheat (Qi et al., 2004; 5,996 bin-mapped markers) available from the GrainGenes Database (http://wheat.pw.usda.gov/cgi-bin/westsql/map_locus.cgi) with the rice BAC/PAC clones to provide syntenic maps between rice and these cereal species. Displays, as well as downloadable tab-delimited files, of the alignments between the maize and wheat genetic markers with the rice genome are available from the project Web pages.
At the proteome level, we provide several layers of annotation. A catalog of domains and motifs for the predicted rice proteome are available, including Pfam domains (Bateman et al., 2002), transmembrane domains (Krogh et al., 2001), signal peptide motifs (Nielsen et al., 1997), and signal anchor motifs. A total of 29,570 predicted proteins within the predicted rice proteome contained at least one Pfam domain above the trusted cutoff, with 2,219 Pfam domains represented in total. Excluding TEs, the most frequent Pfam domain was PF00069 (protein kinase domain). A total of 8,205 proteins contained a putative transmembrane domain, and 12,750 proteins contained a putative signal peptide. All predicted domain and motif data are available through the project Web pages using a series of search tools (http://rice.tigr.org/tdb/e2k1/osa1/irgsp.shtml). We also have assigned gene ontology molecular function terms to the predicted rice proteome. This was done using BLASTP searches of the predicted rice proteome against the TIGR Arabidopsis GO-curated proteins (release 5). We excluded proteins that are related to TEs, proteins with no known function (hypothetical and expressed proteins), and GO terms with no known function. To reduce errors in our transitive GO annotation, the GO assignments were mapped to the GOSlim/Plant ontologies (www.geneontology.org). In total, 69,215 GO assignments were made to 17,169 rice proteins. As with our other annotation data, the GO assignments are accessible through search tools using the GO tree display on the project Web pages (http://rice.tigr.org/tdb/e2k1/osa1/GO.retrieval.shtml).
DATA ACCESS
Multiple access modes are available for the sequence and annotation data associated with the current release. Perhaps the most user-friendly access is a set of Web pages with text and search tools for searching the sequence and annotation data. These can all be accessed through the project homepage (http://rice.tigr.org). In addition, all of the sequence and a majority of the annotation data are available through anonymous FTP download in XML and GFF3 format (ftp://ftp.tigr.org/pub/data/Eukaryotic_Projects/o_sativa/annotation_dbs/). A subset of the sequence and annotation data can be accessed through the Data Extractor tool, which generates flat files of user-selected datasets (http://rice.tigr.org/tdb/e2k1/osa1/data_download.shtml). In addition, the current sequence and annotation are available through a Genome Browser (Fig. 2; Stein et al., 2002; http://rice.tigr.org/tigr-scripts/osa1/gbrowse/ricegenomebrowser). A total of 22 tracks of annotation are available in the Osa1 Genome Browser, including the BAC/PAC tiling path, TIGR annotations, GenBank annotations, ab initio gene prediction output, rice EST evidence, rice FL-cDNA evidence, protein evidence, alignment with genetic markers (rice, maize, wheat), alignment with the TIGR Plant Gene Indices, and FSTs. A description of the tracks as well as the alignment programs and cutoff criteria employed are provided on the Track Description and Citation page (http://rice.tigr.org/tigr-scripts/osa1/gbrowse/ricegenomebrowser?help=citations). A search function is available on the browser in which chromosomes and coordinates can be selected or the user can search based on gene name, marker name, or locus identifier. We also make our annotation data available through a Distributed Annotation System (Dowell et al., 2001; http://rice.tigr.org/tdb/e2k1/osa1/irgsp/DAS.shtml). Sequence and annotation data for previous releases are archived on the project FTP site (ftp://ftp.tigr.org/pub/data/Eukaryotic_Projects/o_sativa/annotation_dbs/).
Figure 2.
Display view available from the Osa1 Genome Browser. In A, the search and navigation functions of the browser are shown. Users can select chromosome, coordinates, marker name, locus identifier, feat_name, or putative annotation. B depicts output from the browser for three genes (loci), along with annotation data from the selected tracks. The gene on the far left (LOC_Os01g51840) encodes a putative IQ calmodulin-binding motif protein. There are two gene models (LOC_Os01g51840.1, LOC_Os01g51840.2) for this gene representing two splice isoforms. The middle gene encodes a hypothetical protein (LOC_Os01g51850), while the far right gene encodes an expressed protein with EST and FL-cDNA support. C displays the key to the selected tracks. D shows the tracks available on the browser that are hyperlinked to a descriptive page and the options available for viewing and displaying the tracks.
DATA UPDATE SCHEDULE
The size of the rice genome sequence and the volume of accompanying annotation data limit our ability to rapidly provide and track updates in these two data sets. Thus, we schedule updates to the sequence (i.e. the pseudomolecules) and the accompanying annotation on a biannual basis. Release 1 of the pseudomolecules was in September 2003, followed by release 2 in April 2004. The third release was made in late December 2004. Each release has been accompanied with a major improvement in sequence and annotation quality. With each release, we have been able to expand the annotation data types to augment the data available to the community. In the near future, we will be providing updates quarterly through addition of new tracks in the Genome Browser. With the release of the IRGSP pseudomolecules, we will provide users alignments between our pseudomolecules and those of the IRGSP to provide a cross-reference of these two datasets. We will also provide any annotation made available by the IRGSP or other entities as separate tracks in our Genome Browser, which will allow the users of the Osa1 database the ability to see alternative annotations of the rice genome and select their preferred annotation.
Acknowledgments
The efforts of the TIGR Bioinformatics department in generating a suite of tools and resources for eukaryotic sequence and annotation are appreciated.
This work (on rice genome annotation) was supported by the National Science Foundation (grant no. DBI–0321538 to C.R.B.) and the U.S. Department of Agriculture (grant no. 2003–35317–13173 to C.R.B.).
References
- Arumuganathan K, Earle ED (1991) Nuclear DNA content of some important plant species. Plant Mol Biol Rep 9: 208–219 [Google Scholar]
- Barry GF (2001) The use of the Monsanto draft rice genome sequence in research. Plant Physiol 125: 1164–1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman A, Birney E, Cerruti L, Durbin R, Etwiller L, Eddy SR, Griffiths-Jones S, Howe KL, Marshall M, Sonnhammer EL (2002) The Pfam protein families database. Nucleic Acids Res 30: 276–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge C, Karlin S (1997) Prediction of complete gene structures in human genomic DNA. J Mol Biol 268: 78–94 [DOI] [PubMed] [Google Scholar]
- Causse MA, Fulton TM, Cho YG, Ahn SN, Chunwongse J, Wu K, Xiao J, Yu Z, Ronald PC, Harrington SE, et al (1994) Saturated molecular map of the rice genome based on an interspecific backcross population. Genetics 138: 1251–1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowell RD, Jokerst RM, Day A, Eddy SR, Stein L (2001) The Distributed Annotation System. BMC Bioinformatics 2: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale MD, Devos KM (1998) Comparative genetics in the grasses. Proc Natl Acad Sci USA 95: 1971–1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Gene Ontology Consortium (2000) Gene ontology: tool for the unification of biology. Nat Genet 1: 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff SA, Ricke D, Lan TH, Presting G, Wang R, Dunn M, Glazebrook J, Sessions A, Oeller P, Varma H, et al (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 296: 92–100 [DOI] [PubMed] [Google Scholar]
- Greco R, Ouwerkerk PB, Taal AJ, Favalli C, Beguiristain T, Puigdomenech P, Colombo L, Hoge JH, Pereira A (2001) Early and multiple Ac transpositions in rice suitable for efficient insertional mutagenesis. Plant Mol Biol 46: 215–227 [DOI] [PubMed] [Google Scholar]
- Haas BJ, Delcher AL, Mount SM, Wortman JR, Smith RK Jr, Hannick LI, Maiti R, Ronning CM, Rusch DB, Town CD, et al (2003) Improving the Arabidopsis genome annotation using maximal transcript alignment assemblies. Nucleic Acids Res 31: 5654–5666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harushima Y, Yano M, Shomura A, Sato M, Shimano T, Kuboki Y, Yamamoto T, Lin S, Antonio BA, Parco A, et al (1998) A high-density rice genetic linkage map with 2275 markers using a single F2 population. Genetics 148: 479–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Adams MD, Zhou H, Kerlavage AR (1997) A tool for analyzing and annotating genomic sequences. Genomics 46: 37–45 [DOI] [PubMed] [Google Scholar]
- Jiang N, Bao Z, Zhang X, Eddy SR, Wessler SR (2004) Pack-MULE transposable elements mediate gene evolution in plants. Nature 431: 569–573 [DOI] [PubMed] [Google Scholar]
- Juretic N, Bureau TE, Bruskiewich RM (2004) Transposable element annotation of the rice genome. Bioinformatics 20: 155–160 [DOI] [PubMed] [Google Scholar]
- Kent WJ (2002) BLAT—the BLAST-like alignment tool. Genome Res 12: 656–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim CM, Piao HL, Park SJ, Chon NS, Je BI, Sun B, Park SH, Park JY, Lee EJ, Kim MJ, et al (2004) Rapid, large-scale generation of Ds transposant lines and analysis of the Ds insertion sites in rice. Plant J 39: 252–263 [DOI] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer ELL (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305: 567–580 [DOI] [PubMed] [Google Scholar]
- Lowe TM, Eddy SR (1997) tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25: 955–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukashin AV, Borodovsky M (1998) GeneMark.hmm: new solutions for gene finding. Nucleic Acids Res 26: 1107–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyao A, Tanaka K, Murata K, Sawaki H, Takeda S, Abe K, Shinozuka Y, Onosato K, Hirochika H (2003) Target site specificity of the Tos17 retrotransposon shows a preference for insertion within genes and against insertion in retrotransposon-rich regions of the genome. Plant Cell 15: 1771–1780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder NJ, Apweiler R, Attwood TK, Bairoch A, Barrell D, Bateman A, Binns D, Biswas M, Bradley P, Bork P, et al (2003) The InterPro Database, 2003 brings increased coverage and new features. Nucleic Acids Res 31: 315–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen H, Engelbrecht J, Brunak S, von Heijne G (1997) Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng 10: 1–6 [DOI] [PubMed] [Google Scholar]
- Ouyang S, Buell CR (2004) The TIGR Plant Repeat Databases: a collective resource for identification of repetitive sequences in plants. Nucleic Acids Res (Database Issue) 32: D360–D363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea M, Lin X, Salzberg SL (2001) GeneSplicer: a new computational method for splice site prediction. Nucleic Acids Res 29: 1185–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi LL, Echalier B, Chao S, Lazo GR, Butler GE, Anderson OD, Akhunov ED, Dvorak J, Linkiewicz AM, Ratnasiri A, et al (2004) A chromosome bin map of 16,000 expressed sequence tag loci and distribution of genes among the three genomes of polyploid wheat. Genetics 168: 701–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quackenbush J, Cho J, Lee D, Liang F, Holt I, Karamycheva S, Parvizi B, Pertea G, Sultana R, White J (2001) The TIGR Gene Indices: analysis of gene transcript sequences in highly sampled eukaryotic species. Nucleic Acids Res 29: 159–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Rice Full-Length cDNA Consortium (2003) Collection, mapping, and annotation of over 28,000 cDNA clones from japonica rice. Science 301: 376–379 [DOI] [PubMed] [Google Scholar]
- Salamov AA, Solovyev VV (2000) Ab initio gene finding in Drosophila genomic DNA. Genome Res 10: 516–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallaud C, Gay C, Larmande P, Bes M, Piffanelli P, Piegu B, Droc G, Regad F, Bourgeois E, Meynard D (2004) High throughput T-DNA insertion mutagenesis in rice: a first step towards in silico reverse genetics. Plant J 39: 450–464 [DOI] [PubMed] [Google Scholar]
- Sasaki T, Burr B (2000) International Rice Genome Sequencing Project: the effort to completely sequence the rice genome. Curr Opin Plant Biol 3: 138–141 [DOI] [PubMed] [Google Scholar]
- Stein LD, Mungall C, Shu S, Caudy M, Mangone M, Day A, Nickerson E, Stajich JE, Harris TW, Arva A, et al (2002) The generic genome browser: a building block for a model organism system database. Genome Res 10: 1599–1610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wortman JR, Haas BJ, Hannick LI, Smith RK, Maiti R, Ronning CM, Chan AP, Yu C, Ayele M, Whitelaw CA, et al (2003) Annotation of the Arabidopsis genome. Plant Physiol 132: 461–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Maehara T, Shimokawa T, Yamamoto S, Harada C, Takazaki Y, Ono N, Mukai Y, Koike K, Yazaki J, et al (2002) A comprehensive rice transcript map containing 6591 expressed sequence tag sites. Plant Cell 14: 525–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Hu S, Wang J, Wong GK, Li S, Liu B, Deng Y, Dai L, Zhou Y, Zhang X, et al (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science 296: 79–92 [DOI] [PubMed] [Google Scholar]
- Yuan Q, Ouyang S, Liu J, Suh B, Cheung F, Sultana R, Lee D, Quackenbush J, Buell CR (2001) The TIGR Rice Genome Annotation Resource: annotating the rice genome and creating resources for plant biologists. Nucleic Acids Res 31: 229–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zdobnov EM, Apweiler R (2001) InterProScan—an integration platform for the signature-recognition methods in InterPro. Bioinformatics 9: 847–848 [DOI] [PubMed] [Google Scholar]