Abstract
MetaCyc (http://metacyc.org) contains experimentally determined biochemical pathways to be used as a reference database for metabolism. In conjunction with the Pathway Tools software, MetaCyc can be used to computationally predict the metabolic pathway complement of an annotated genome. To increase the breadth of pathways and enzymes, more than 60 plant-specific pathways have been added or updated in MetaCyc recently. In contrast to MetaCyc, which contains metabolic data for a wide range of organisms, AraCyc is a species-specific database containing only enzymes and pathways found in the model plant Arabidopsis (Arabidopsis thaliana). AraCyc (http://arabidopsis.org/tools/aracyc/) was the first computationally predicted plant metabolism database derived from MetaCyc. Since its initial computational build, AraCyc has been under continued curation to enhance data quality and to increase breadth of pathway coverage. Twenty-eight pathways have been manually curated from the literature recently. Pathway predictions in AraCyc have also been recently updated with the latest functional annotations of Arabidopsis genes that use controlled vocabulary and literature evidence. AraCyc currently features 1,418 unique genes mapped onto 204 pathways with 1,156 literature citations. The Omics Viewer, a user data visualization and analysis tool, allows a list of genes, enzymes, or metabolites with experimental values to be painted on a diagram of the full pathway map of AraCyc. Other recent enhancements to both MetaCyc and AraCyc include implementation of an evidence ontology, which has been used to provide information on data quality, expansion of the secondary metabolism node of the pathway ontology to accommodate curation of secondary metabolic pathways, and enhancement of the cellular component ontology for storing and displaying enzyme and pathway locations within subcellular compartments.
The goal of the MetaCyc database is to catalog every experimentally determined biochemical pathway for small molecule metabolism (Krieger et al., 2004). MetaCyc had been initialized with all of the manually curated pathways in EcoCyc (Keseler et al., 2005), a model organism database for Escherichia coli. Pathways from more than 300 organisms have been subsequently added to MetaCyc, and more than 90% of its pathways are manually curated with literature citations and species information. The other 10% of the pathways, which were originally imported from the WIT database (http://www.cme.msu.edu/WIT/), are under manual curation. MetaCyc can be used as a reference database to create new Pathway Genome Databases (PGDB) from annotated genomes or genes, in conjunction with the Pathway Tools software (Paley and Karp, 2002). The Pathway Tools software contains three components: (1) PathoLogic, which matches the gene product names of an annotated genome against the enzymes and reactions in a reference database such as MetaCyc, and predicts the pathways for the organism using a scoring algorithm; (2) Pathway/Genome Editor, which allows manual updating of the derived database and supports data sharing among the derived databases; and (3) Pathway/Genome Navigator, which supports querying, browsing, visualization, as well as publishing of the database on the Web. AraCyc was the first plant metabolism database to be computationally predicted by PathoLogic using MetaCyc as the reference database (Mueller et al., 2003). With continued manual curation, AraCyc will eventually describe a complete set of metabolic pathways for Arabidopsis (Arabidopsis thaliana) and display genes and enzymes within their metabolic context. Though many pathways and enzymes in AraCyc have yet to be manually curated, AraCyc is currently the most comprehensive genome-wide metabolic database available for a single plant species. Features of MetaCyc and AraCyc are summarized in Table I. Both databases are readily accessible via the World Wide Web (http://metacyc.org and http://arabidopsis/tools/aracyc).
Table I.
Comparison of features in MetaCyc and AraCyc
| Features | MetaCyc (8.6) | AraCyc (2.0) |
|---|---|---|
| No. of pathways | 528 | 204 |
| No. of plant pathways | 77a | 204 |
| No. of plant species | 38 | 1 |
| No. of curated plant pathways | 77a | 51 |
| Can overlay user's data onto the metabolic map? | No | Yes |
| Supporting software | Pathway Tools | Pathway Tools |
The number includes pathways that are curated from microbes that also have a plant species included on the pathway species list.
With the release of the fully sequenced plant genomes of Arabidopsis (Arabidopsis Genome Initiative, 2000) and rice (Oryza sativa; International Rice Genome Sequencing Project, http://rgp.dna.affrc.go.jp/IRGSP; Goff et al., 2002; Yu et al., 2002), and the initiation of sequencing projects for many other plant species, there is a fast growing desire to place the sequenced and annotated genomes in a metabolic context. Indeed, the benefits of a species-specific metabolic pathway database are substantial: (1) it depicts the biochemical components of an organism; (2) it assists comparative studies of pathways across species and facilitates metabolic engineering to improve crop metabolism and traits; (3) it can be used as a platform to integrate and analyze data from large-scale experiments, such as gene expression, protein expression, or metabolite profiling; and (4) by presenting pathway steps lacking assigned genes or having genes assigned but solely based on computational prediction, we can discern what remains to be identified and experimentally characterized. Despite these advantages, the manual, de novo creation of a pathway database can be labor intensive and time consuming. SoyBase (http://soybase.agron.iastate.edu/), a metabolic pathway database specific to soybean (Glycine max), is the only other species-specific plant pathway database that has been created manually. Species-specific plant pathway databases can also be computationally predicted as a way to jump-start manual curation. For the predictions to be useful, an accurate and comprehensive reference database is key to the quality of the derived databases. Examples of comprehensive pathway databases include Kyoto Encyclopedia of Genes and Genomes (http://www.genome.jp/kegg/; Kanehisa and Goto, 2000) and Enzymes and Metabolic Pathways (http://www.empproject.com/). As they stand, their usefulness as reference databases for plant genomes is somewhat limited for one or more of the following reasons: (1) pathways are not associated with literature citations and, thus, it is hard to assess their accuracy; (2) individual pathway diagrams tend to be composites taken from several different species and are therefore not accurate for any single species; and (3) they include relatively few pathways specific to plants.
In this article, we describe how the MetaCyc and AraCyc databases are updated, including manual curation of new plant pathways and revision of predicted AraCyc pathways with information from the literature, updating the AraCyc pathway predictions using the latest genome annotations, recording evidence to pathways and enzymes, and enhancing data ontologies. We also describe general applications of the two databases to other plant genomes. Finally, we discuss the limitations and issues of these databases and future directions.
RESULTS
Manual Curation of Plant Pathways in MetaCyc and AraCyc
The number of manually curated plant pathways in MetaCyc and AraCyc have been expanded significantly in the last few years. To ensure that the newly added pathways benefit a broad user base, primary metabolic pathways universal to plants were given the highest priority. Pathways shared among a few species or those involving secondary metabolism were given a lower priority. Pathways are curated from literature following standard curation procedures developed by the curators at SRI International and the Carnegie Institution (http://bioinformatics.ai.sri.com/ptools/curatorsguide.pdf). In total, 63 plant pathways have been added to or updated in MetaCyc between release versions 6.5 (August 2002) and 8.6 (November 2004), and 28 pathways have been curated in AraCyc since last described (Mueller et al., 2003). Most notably, biosynthesis pathways of cellulose, pectin, suberin, lignin, wax, and cutin were added to the pathway class “Biosynthesis, Plant Cell Structure”; pathways describing biosynthesis and degradation of Suc and starch have been extensively updated. Within the “Fatty Acids and Lipids” class, biosynthesis of glycolipids, and desaturation pathways of glycolipids and phospholipids were added, and biosynthesis of phospholipids was significantly updated. To the category of “Generation of Precursor Metabolites and Energy” we added the cytosolic variant of glycolysis, photorespiration pathway, and the superpathway of glyoxylate cycle, which operates during seed germination to convert storage lipids to carbohydrates. Auxin biosynthesis was added to “Biosynthesis, Plant Hormones” to complete this class with the seven known plant hormones; biosynthetic pathways of vitamin C, vitamin E, and plastoquinone were added to “Biosynthesis, Cofactors, Prosthetic Groups and Electron Carriers.” Most of the curated pathways are shared between AraCyc and MetaCyc. A few pathways exist only in MetaCyc since there is no evidence that they operate in Arabidopsis. They include C4 photosynthesis, fructan metabolism, and the pathway variants of indole acetic acid metabolism.
Improvements and Changes in AraCyc 2.0
Prediction of Pathways Using PathoLogic
AraCyc was originally built (Mueller et al., 2003) using the PathoLogic software, which predicts pathways for an organism from a list of its annotated gene products by matching the annotations (names of the gene products) with enzyme names in MetaCyc and in turn assigning the genes to MetaCyc reactions and pathways (Paley and Karp, 2002). After a pathway has been initially predicted, where at least one of its reactions has a gene(s) assigned, PathoLogic evaluates whether to keep the pathway or prune it out based on how strong the evidence is for the pathway. The more reactions of a pathway having genes assigned, the stronger evidence that the pathway is present in the organism. Gene(s) assigned to a reaction that occurs only in one pathway, or a unique reaction, is also considered strong evidence. A pathway will be pruned out if the evidence is weak (for example, if the pathway consists of more than two reactions, but only one reaction, which is not a unique reaction, has a gene[s] assigned to it). The quality of the prediction depends on the annotations of the input file and the data in the reference database. The two components used in the automatic creation of AraCyc, the annotated Arabidopsis genome, and the reference database MetaCyc have been extensively updated since AraCyc's initial build in 2001. For example, At1g51110 was previously described as an anthranilate synthase, but that annotation no longer holds true in the most recent genome release (TIGR5.0, 2004). Accordingly, there was a need to update AraCyc to reflect the up-to-date knowledge of the metabolic context of the genome. In addition, the reference database MetaCyc had grown from 445 pathways and 1,115 enzymes in 2001 to 513 pathways and 1,840 enzymes in version 8.5 released in September 2004 (http://biocyc.org/metacyc/releasenotes.shtml). The expansion of MetaCyc over time will likely increase the number of pathways that can be predicted for Arabidopsis and for other genomes.
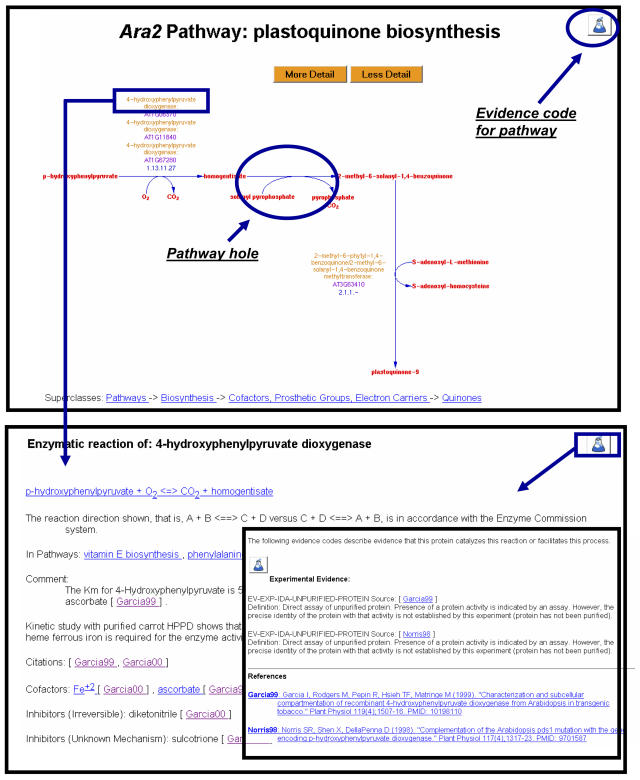
The starting point for building an updated version of AraCyc was making use of the increased quantity and quality of Arabidopsis genome annotation. The annotation file used in the original build of AraCyc was generated by manually extracting free-text gene descriptions of the The Institute for Genomic Research (TIGR) genome annotation (Mueller et al., 2003). Since that time, both The Arabidopsis Information Resource (TAIR) and TIGR have used controlled vocabularies for functional annotation of all the genes in Arabidopsis (Wortman et al., 2003; Berardini et al., 2004). The function of a gene is assigned to an unambiguously defined term, and the evidence of the assignment, either based on experimental data available from the literature or computational prediction based on sequence similarity, is also provided along with the annotation. The annotations obtained from the TAIR Web site (http://arabidopsis.org/servlets/Search?action=new_search&type=keyword) as of October 2004 included 7,900 loci that were annotated to catalytic activity (GO:0003824), a Gene Ontology (GO, http://www.geneontology.org/; Gene Ontology Consortium, 2004) term. Excluding those loci that are involved in macromolecule metabolism, such as peptidases, and loci whose catalytic activity is not involved in metabolism, such as transposases, 4,896 loci remained in the input file for building AraCyc 2.0. Using this list as the input, the PathoLogic program predicted 219 pathways and 1,102 unique reactions. Of the predicted reactions, 940 unique reactions were placed in the 219 pathways. Among the reactions mapped to the pathways, 586 unique reactions do not have Arabidopsis genes/enzymes mapped. These are referred to as pathway holes (Fig. 1). Nearly half (45%) of the pathway holes belong to EC1 (oxidoreductase, Enzyme Commission [EC] Nomenclature, http://www.chem.qmul.ac.uk/iubmb/enzyme/) or EC2 (transferase; Table II). After running the pathway-hole filler program (see “Materials and Methods”; Green and Karp, 2004), which BLASTs the 29,000 Arabidopsis protein sequences against sequences known to catalyze the same metabolic reactions in other species, 58 pathway holes (approximately 11%) were filled. As shown in Table II, among the six categories of enzymes from the EC, EC6 (ligase) had the highest rate (64%) of being filled by the hole-filler program, indicating that sequences of the Arabidopsis ligases share high similarity to sequences in other species. The filling rates of the other five categories of EC fall below 16%, suggesting low homology at the primary sequence level. Assuming that sequences of all of the pathway holes can be retrieved using BLAST and the current hole-filling rate holds true, no more than 21% (as the sum of the last column in Table II) of all the holes may be filled by the hole-filler program. This suggests that homology modeling or other ways of looking for more remote homologies should be considered to fill additional pathway holes.
Figure 1.
An example of an AraCyc pathway. A pathway evidence, which could be either computational (indicated by a computer icon) or experimental (indicated by a flask icon), provides assertion of the existence of the pathway in Arabidopsis. Similarly, evidence attached to an enzyme provides assertion of its catalytic activity involved in a specific reaction. Each piece of evidence is associated with a citation from where the source of the evidence can be found. Reactions without any Arabidopsis genes or enzymes assigned are called pathway holes. Pathways can be zoomed to show various levels of details. Compounds, reactions, enzymes, and genes on a pathway page are clickable for more information.
Table II.
Analysis of the results of the pathway hole-filler program in AraCyc 2.0
–, Data not available.
| Category of Holes | Holesa | Portion to All Holesb | Holes with Sequence Retrieveda | Holes Filleda | Hole-Filling Ratec | Maximum Percentile of Hole Fillingd |
|---|---|---|---|---|---|---|
| EC1 (oxidoreductase) | 114 | 21.6% | 74 | 12 | 16.2% | 3.5% |
| EC2 (transferase) | 128 | 24.2% | 98 | 15 | 15.3% | 3.7% |
| EC3 (hydrolase) | 41 | 7.8% | 32 | 2 | 6.3% | 0.5% |
| EC4 (lyase) | 40 | 7.6% | 29 | 2 | 6.9% | 0.5% |
| EC5 (isomerase) | 22 | 4.2% | 15 | 2 | 13.3% | 0.6% |
| EC6 (ligase) | 17 | 3.2% | 14 | 9 | 64.3% | 2.1% |
| No EC assignment | 166 | 31.4% | 50 | 16 | 32.0% | 10.1% |
| Sum | 528 | 100.0% | 312 | 58 | – | 20.9% |
Indicates number.
The number of holes divided by the total number of holes of all categories.
The number of holes filled divided by the number of holes where sequences can be retrieved from other organisms.
The number of holes multiplied by the hole-filling rate then divided by the total number of all holes.
Validation of Pathway Predictions
The 219 pathways of the AraCyc 2.0 initial build were validated manually by consulting the primary literature, review articles, and textbooks. A valid pathway is defined as a pathway whose existence in Arabidopsis is supported by experimental evidence described in the literature. If a pathway is not well known and a curator could not find it referenced in the literature, the following two criteria were considered for validation. First, do the reactions that are found only in this pathway (i.e. unique to this pathway) have any Arabidopsis genes assigned to them? When a gene could be associated to only one pathway, this pathway was kept. This criterion also applied even when the end product of the biosynthetic pathway had not been reported in Arabidopsis. For example, lipid A is a membrane component specifically found in gram-negative bacteria. It has not been described in plants. However, the Arabidopsis genome is predicted to include genes that catalyze at least three unique reactions of the lipid-A precursor biosynthesis pathway, suggesting plants may be able to synthesize the metabolite (http://www.biochem.duke.edu/Raetz/raetznew.html), or other similar compounds. The second criterion uses information about the existence of the metabolites. If none of the unique reactions of the pathway were associated with an Arabidopsis gene, we then looked for evidence of the existence of the metabolites (Robinson, 1983; Harborne and Baxter, 1993). For a biosynthetic pathway, we asked whether the end products and most of the intermediates are reported in Arabidopsis or in other plants. For a degradation pathway, we asked whether the starting compound and most of the catabolic intermediates are reported in Arabidopsis. For example, the glycerol teichoic acid biosynthesis pathway is not considered to be a valid pathway in Arabidopsis because none of the unique reactions in the pathway have Arabidopsis genes assigned, and the glycerol teichoic acid and most of the intermediate compounds in the pathway are reported to exist only in bacteria. In the course of the validation, 44 pathways were manually removed from the database. The removed pathways are tracked and documented on the AraCyc home page (http://arabidopsis.org/tools/aracyc/aracyc.deleted.pathways.jsp). The validated pathways were then assigned an evidence code (Fig. 1; see also below for further description of the evidence ontology used in AraCyc and MetaCyc; Karp et al., 2004). Evidence codes for the validated pathways were changed from EV-COMP-AINF (inferred from computational analysis without human review) to EV-COMP-HINF (inferred from computational analysis and reviewed by a curator). In ambiguous cases where further information was required, we kept the pathway in the database but left it with the evidence code EV-COMP-AINF.
Summary of AraCyc 2.0 Pathways
Previously, 173 pathways were predicted in the AraCyc 1.0 initial build (Mueller et al., 2003), whereas the new AraCyc 2.0 initial build predicted 219 pathways (Table III). Of these, 120 pathways were common to both versions (27 of them could not be validated and were subsequently removed from AraCyc 2.0). In the new build, 99 additional pathways not included in the first build were predicted (17 could not be validated and were removed). Excluding the 17 invalid pathways of these newly predicted 99 pathways, 27 represent plant pathways added to MetaCyc since it was used for the AraCyc 1.0 build. Another 17 pathways are variants of existing pathways that were predicted in AraCyc 1.0. For the remaining 38 pathways, we carefully examined the reasons for their identification in AraCyc 2.0 and not in AraCyc 1.0. Sixteen were due to gene annotation changes in Arabidopsis, which provided new supporting evidence for the pathways. Twenty-two pathways were updated in the reference database MetaCyc since 2001 and consequently contained enough supporting evidence to be predicted in the new run.
Table III.
Comparison of pathway data between the first and second AraCyc releases
–, Data not available.
| AraCyc 2.0 | AraCyc 2.0 Initial Build | AraCyc 1.0a | AraCyc 1.0 Initial Builda | |
|---|---|---|---|---|
| Pathways (excluding superpathways) | 204 | 219 | 174 | 173 |
| Pathways unique to the 1.0 initial build | – | – | – | 53 |
| Pathways unique to the 2.0 initial build | – | 99 | – | – |
| Pathways removed from initial build | 44 | – | 22 | – |
| Pathways added to initial build | 29 | – | 23 | – |
| Curated pathways | 51 | – | 23 | – |
| Unique reactions of pathways | 894 | 940 | 833 | 750 |
| Unique pathway holes | 401 (45%) | 528 (56%) | 403 (48%) | 408 (54%) |
| Unique genesb mapped onto pathways | 1,418 | 1,436 | 958 | 767 |
The numbers in these columns are according to Mueller et al. (2003).
Gene, or locus, is a mapped element that corresponds to a transcribed region in the Arabidopsis genome, or a genetic locus that segregates as a single genetic locus or quantitative trait.
On the other hand, 53 pathways that were previously predicted in AraCyc 1.0 were no longer called by the PathoLogic software in this run (Table III). Of these, 32 pathways either no longer exist in the reference database MetaCyc due to a lack of literature evidence, or were replaced by different pathway variants in AraCyc 2.0. Twelve pathways that were predicted in AraCyc 1.0 but not AraCyc 2.0 were due to changes in gene annotations that removed the supporting evidence for the pathways. One notable exception is the photosynthesis (light reaction) pathway. The two enzymes of the pathway in MetaCyc, PSI and PSII, are enzyme complexes. However, within the GO, which has been used in functional annotation of the Arabidopsis genes, PSI and PSII are categorized as children terms of cellular component, not catalytic activity. Therefore, the genes encoding their individual subunits were not included in our input file because the input file was restricted to genes annotated to the catalytic activity term and its children terms. The pathway was later manually added to AraCyc 2.0. The remaining nine pathways that were no longer predicted are due to a glitch in the PathoLogic algorithm. The bug was subsequently fixed in PathoLogic and the nine pathways were added to AraCyc2.0.
In addition, 19 plant pathways in MetaCyc were not predicted in AraCyc 2.0. There are several possible reasons for the missed predictions. First, slight name variations between the annotations in the input file and the enzymes in MetaCyc and poorly annotated names for the P450 cytochromes in MetaCyc made it difficult to match Arabidopsis genes to MetaCyc enzymes. The poorly annotated names were later fixed in MetaCyc. Second, in many cases lack of enough evidence from the Arabidopsis annotation input file made it impossible for the pathways to be predicted. For example, even though enzymes of the homogalacturonan biosynthesis pathway have been characterized from other plants (http://biocyc.org/META/NEW-IMAGE?type=PATHWAY&object=PWY-1061), no Arabidopsis genes could be assigned to the pathway based on the current genome annotation. Third, the same PathoLogic bug mentioned above failed to predict seven pathways for which supporting evidence exists. For example, within the lipoxygenase pathway, Arabidopsis genes can be assigned to at least one unique reaction of the pathway, which is one of the criteria for inclusion. Nevertheless, since all of the 19 pathways are either specific to Arabidopsis (e.g. glucosinolate biosynthesis) or universal to plants (e.g. homogalacturonan biosynthesis), they were imported into AraCyc 2.0 using the pathway import utility of Pathway Tools (Karp et al., 2002).
Overall, updates to the gene annotations resulted in prediction of 16 new pathways (or 7.8% of the 204 total pathways in AraCyc 2.0) since last described (Mueller et al., 2003) and pruned 12 previously predicted ones. Updates in MetaCyc predicted 66 new pathways (or 32.3% of the 204 total pathways in AraCyc 2.0) and pruned 32 previously predicted ones. In general, quality of the PathoLogic prediction remains comparable for the two AraCyc builds. The false-positive rate of PathoLogic prediction (invalid pathways/total predicted pathways) is 20% (44/219) and 13% (22/173) for AraCyc 2.0 and AraCyc 1.0, respectively. The false-negative rate (valid pathways that are not predicted/total valid pathways) is 7.8% (16/204) for AraCyc 2.0. Note, however, that PathoLogic is intentionally tuned to have a relatively high false-positive rate so that human curators have a chance to review all pathways for which there is even minor evidence for their presence.
After removal of the nonvalid pathways and addition of the missing pathways, AraCyc 2.0 contains 204 pathways with 1,418 unique genes assigned (Table III). The evidence and citations supporting the functional annotations of these genes were imported from TAIR and were associated to the corresponding enzymes (Fig. 1).The distribution of the 204 pathways according to the pathway ontology is summarized in Table IV. The three top categories, “Biosynthesis,” “Degradation/Utilization/Assimilation,” and “Generation of Precursor Metabolites and Energy,” contain 115, 74, and 26 pathway instances, respectively. Biosynthesis of all 20 protein amino acids, all DNA/RNA purine and pyrimidine nucleosides and nucleotides, commonly occurring sugars and polysaccharides, major fatty acid and lipid classes including triacylglycerol and phospho- and glyco-lipids, 15 cofactors, prosthetic groups and electron carries, and all seven known major plant hormones are represented in AraCyc 2.0. In addition, biosynthesis of the major molecules found in plant primary and secondary cell wall and plant epidermal structure, including cellulose, homogalacturonan (a pectin), lignin, suberin, wax, and cutin, are included. Pathways for central energy metabolism are well represented. It is not easy to assess the comprehensiveness of pathways under “Degradation/Utilization/Assimilation.” There is much less information available for catabolism than for biosynthesis in plants. Nonetheless, two known degradation pathways for odd chain and unusual fatty acids need to be added. Supplemental Table I to this manuscript provides a comprehensive list of all the pathways in AraCyc 2.0. For each pathway, it lists the number of reactions, the number of pathway holes, and the number of genes assigned to the pathway along with known genetic symbols. Pathways that have been curated are identified in the list.
Table IV.
Summary of pathways in AraCyc 2.0, grouped by category
| No. of Pathways | |
|---|---|
| Biosynthesis | 115 |
| Amino acids | 28 |
| Cell structure | 12 |
| Cofactors, prosthetic groups, electron donors | 22 |
| Fatty acids and lipids | 13 |
| Hormones | 6 |
| Nucleosides and nucleotides | 4 |
| Secondary metabolism | 10 |
| Sugars and polysaccharides | 9 |
| Others | 11 |
| Degradation | 74 |
| Amino acids | 29 |
| Fatty acids and lipids | 5 |
| Inorganic nutrients | 6 |
| Sugar derivatives | 6 |
| Sugars and polysaccharides | 12 |
| Others | 16 |
| Generation of precursor metabolites and energy | 26 |
Enhancement to the Database Ontology
Data objects in MetaCyc and AraCyc, including pathways, compounds, subcellular compartments, and evidence types, are structured in a hierarchical ontology (Gruber, 1993; Karp, 2000). The ontology describes concepts (terms) and relationships between them. Terms are organized into classes, subclasses, and instances according to the primary “is-a” relationship. The “is-a” relationship classifies what type of a concept a term is. The broader concepts, or parent terms such as classes, appear on the top level of the hierarchy tree. More specific concepts, or children terms such as subclasses and instances, are grouped under the broader concepts. Several improvements to the existing ontologies have made them more robust.
The secondary metabolism class of the pathway ontology has been significantly enhanced. Secondary (sometimes referred to as specialized) metabolites are widespread in higher plants. They contribute substantially to the reproduction, fitness, and adaptations that plants acquired throughout the course of evolution (Pichersky and Gang, 2000; Dixon, 2001; Verpoorte and Memelink, 2002; Singer et al., 2003; Wink, 2003). More than 100,000 of these low-molecular-weight organic compounds have been identified to date (Wink, 1988, 2003; Hadacek, 2002), revealing their complex composition and high structural diversity. Previous curation efforts for plant pathways have focused on primary metabolism, and only a few secondary metabolic pathways have been curated in MetaCyc and AraCyc to date. To accommodate a planned increase in curation for secondary metabolism, the secondary metabolism class of the pathway ontology was significantly expanded to include eight main subclasses, which, in turn, are composed of 26 subclasses, covering all of the major secondary metabolites reported in plants. The main classes include phenylpropanoid derivatives, xanthones, flavonoids, polyketides, fatty acid derivatives, isoprenoids, and nitrogen-containing secondary metabolites. The division of the main classes was structured strictly according to the biosynthetic origin of the metabolites, with the exception of phytoalexins. Although phytoalexins are derived from different precursors, the grouping of all phytoalexins together allows users to search at the functional level. Individual phytoalexins are also cross-listed under other main classes from which they arise. The separation of xanthones and flavonoids from phenylpropanoid derivatives reflects their hybrid biosynthetic origin.
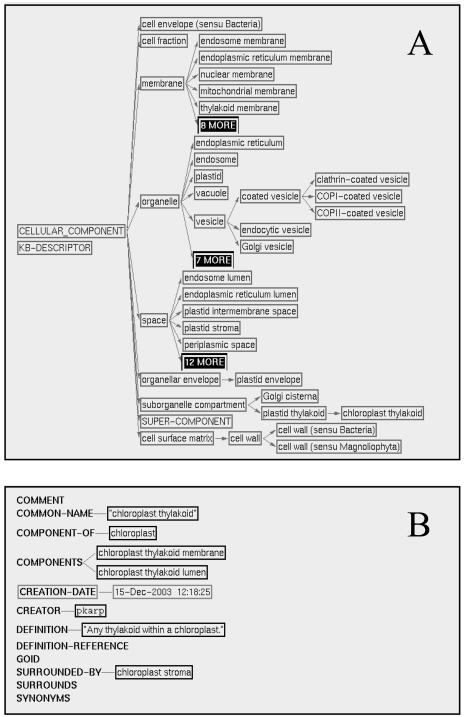
To represent pathways in a cellular context, MetaCyc and AraCyc also store protein subcellular location information. We have expanded the cellular component terms from 35 terms (Mueller et al., 2003) to more than 150. In addition, the terms that were stored as a flat list are now organized into a tree structure according to the “is-a” (this describes what kind of subcellular component a term is). The ontology shown in Figure 2 has nine top-level classes, including organelle, suborganelle compartment (defined as a membrane-enclosed compartment that is a part of an organelle, e.g. a chloroplast is a type of organelle and a chloroplast thylakoid is a type of suborganelle compartment), membrane, space (defined as a three-dimensional extent in which objects and events occur, such as the thylakoid lumen), organellar envelope, cell envelope, cell surface matrix, cell fraction, and supercomponent (a space plus other components that are located in the space). Cytoplasm cannot be classified in any of the other top classes. It refers to the cytosol plus an array of organelles within the cytosol. Therefore, it was placed under supercomponent. Each term is defined and, when applicable, has a database cross-link to the GO's cellular component ontology (Gene Ontology Consortium, 2004). Two additional relationships besides “is-a” are implemented in the ontology: “component-of” and “surrounded-by.” The “component-of” (or “part-of”) relationship describes whether one term is a physical constituent of another term. For example, chloroplast membrane is a component of the chloroplast. The “surrounded-by” relationship provides relative positional information of two terms within a cell. For example, the vacuolar lumen is surrounded by the vacuolar membrane. The ontology not only aids powerful querying such as “find all of the membrane-localized enzymes and their catalyzed reactions” or those enzymes whose locations are annotated to the term membrane and all of its children terms, but also could facilitate the development of graphical visualization software to display subcellular locations of enzymes and their catalyzed reactions and pathways. Using the “surrounded-by” relationship, a computer program can be guided to draw a chloroplast inner membrane encased within a chloroplast intermembrane space, which is in turn drawn encased within the chloroplast outer membrane. While there is essentially a one-to-one mapping between the terms in the GO component ontology and our component ontology, the “surrounds by” relationship that only exists in our ontology causes the structures of the two ontologies to be significantly different.
Figure 2.
A snapshot of the cellular component ontology in AraCyc and MetaCyc. The cellular component terms are hierarchically classified according to the “is-a” relationship. The nine top-level classes along with some of their subclasses are shown in A. The subclasses can be expanded to display all the instance terms. An example of a term detail page is shown in B. A term can have additional relationships such as “component-of” and “surrounded-by.”
There is an increasing need for attaching evidence to data objects to distinguish data that have been curated with experimental evidence from those that are computationally predicted (Gene Ontology Consortium, 2001). An evidence ontology has been recently developed and implemented to the Pathway Tools software (Karp et al., 2004), used by MetaCyc and AraCyc. Curators record evidence about the existence of an object in the database, for example, existence of a pathway in the species or existence of an activity for an enzyme, using an evidence code (see table I in Karp et al., 2004). There are four top-level evidence codes, EV-COMP (stands for inferred from computational analysis), EV-EXP (inferred from experiment), EV-AS (author statement), and EV-IC (inferred by curator). Lower-level evidence codes under each of the top-level nodes describe more specific types of evidence. For example, use of the “EV-EXP-IDA-purified-protein” evidence code asserts that the enzyme activity of the protein in catalyzing a certain reaction is based on direct assay using purified enzyme. Intuitive icons were created for the four top-level evidence codes and are displayed on the object pages regardless of whether the object is annotated directly with the top-level codes or annotated with more granular codes (Fig. 1). For example, a user will see a flask icon for “EV-EXP-IDA-purified-protein.” An evidence code is assigned to an object along with the citations from which the evidence came so that a user can follow up to a greater depth.
Applications for Arabidopsis and Other Plant Genomes
Although MetaCyc and AraCyc share a significant overlap in data and data access/analysis software, the two databases have different purposes and, thus, different applications. The goal of MetaCyc is to represent all experimentally verified metabolic pathway data from all organisms. On the other hand, the goal of AraCyc is to represent the complement of metabolism of one organism, including both experimentally determined and computationally predicted data. In general, MetaCyc is suitable for use as a reference database to predict the metabolism complement for any newly sequenced and annotated genome because it is designed to maximize sensitivity by including pathways from many organisms. On the other hand, AraCyc may be at least as good as or better than MetaCyc for predicting metabolic pathways for genomes that are evolutionarily closer to Arabidopsis (such as other flowering plants) because pathways that are predicted to exist in plants but have not been experimentally validated can exist in AraCyc but not in MetaCyc. Also, MetaCyc contains a number of pathway variants that are specific to different organisms, whereas AraCyc contains only the plant variants. Therefore, it may take more time to curate the results of the prediction program from MetaCyc. To maximize sensitivity and specificity of the program's results, it may be worthwhile to generate a new metabolism database using more than one reference database (e.g. using both MetaCyc and AraCyc). Regardless of which database is used as reference, the critical importance of validating and curating the outputs of the prediction cannot be overemphasized. Computational pathway prediction is meant to serve as a starting point for building the metabolic content of an organism. After the newly created database has undergone curation, comparison to another organism may shed light on the growth/development and physiology of each organism.
For individual species such as Arabidopsis, the Omics Viewer of Pathway Tools (the AraCyc version is at http://arabidopsis.org:1555/expression.html) can be used in data analysis. The tool paints data from gene expression, protein expression, gene family analysis, or metabolite profiling experiments onto a diagram of the full metabolic network of Arabidopsis. Each reaction (represented as a line connecting the compounds) can be color-coded according to the expression level of the gene or protein that catalyzes the reaction. Metabolite levels can also be depicted by color-coding the symbols for compounds (represented as squares or triangles connected by the reaction lines). Note that only those genes and compounds that are included in the pathways of AraCyc can be displayed on the metabolic map. However, it is possible to extrapolate from the Omics Viewer to identify additional components of a pathway. For example, if a set of genes from a gene expression microarray experiment appears to be involved in the same pathway and show similar changes in expression values, one could cluster the original dataset to identify other genes having a similar expression profile. These genes in turn may represent components of the pathway missing from AraCyc.
Database Access
MetaCyc and AraCyc can be accessed in a number of different ways: They can be queried and browsed using the Pathway Tools software through the Web (http://metacyc.org and http://arabidopsis.org/tools/aracyc). Datasets for AraCyc can be obtained as text files (ftp://ftp.arabidopsis.org/home/tair/Pathways/). The complete databases can also be downloaded (http://biocyc.org/download.shtml and http://arabidopsis.org/tools/aracyc). The first two options are freely accessible to anyone without a license, whereas the last option is available with a license (free to academic users). In addition, both databases can be queried and browsed using the desktop version of Pathway Tools, which provides more functionality than the Web version and is available for Windows/PC, Linux/PC, and SUN workstations (http://biocyc.org/download.shtml).
DISCUSSION
We have described recent updates of MetaCyc and AraCyc, which aimed to increase the breadth of data coverage and the accuracy of plant metabolism data. The remaining issues regarding quality of existing data, breadth of data curation, and limitations of data displaying are discussed below.
Currently, about 25% of the AraCyc pathways have been manually curated, meaning that we have verified and corrected the pathway diagrams according to literature information, and added pathway comments and literature citations. The noncurated pathway diagrams, which were curated from microorganisms in the reference database MetaCyc and were predicted to exist in Arabidopsis by PathoLogic, represent what is experimentally verified in other organisms. These pathways need to be further curated from literature to represent what is experimentally verified in Arabidopsis and other plants. For example, additional intermediate reactions may be required for plants to synthesize the same compound. Or, plants may use different cosubstrates in converting compound A to compound B.
The PathoLogic assignments of genes (and their encoded enzymes) to reactions and pathways are solely based on gene annotations. There are inevitably false-positive associations of genes with pathways. For example, isoenzymes localized in different subcellular compartments, though catalyzing the same reaction, may be involved in different pathways. These isoenzymes are not distinguished by PathoLogic and thus may be assigned to pathways in which they are not involved. For example, an isoenzyme catalyzing reaction X is localized in the cytoplasm. It may be incorrectly assigned to a pathway that contains reaction X but is located in the chloroplast. This kind of false-positive assignment needs to be removed during curation. False-positive assignments to reactions may also arise because of low-quality gene annotations. At present, functional annotations for the majority of the Arabidopsis genes lack experimental evidence. AraCyc users are advised to be cautious when using any of the noncurated data or data without experimental support.
Our immediate goal for enhancement of the database content for MetaCyc and AraCyc is to expand the breadth of existing coverage of plant secondary metabolism, i.e. curation of representative pathways for each of the main compound classes, followed by increasing the depth, i.e. curation of additional pathways representing each of the major subclasses. In addition, we plan to curate and integrate transporters into their relevant pathways in AraCyc and MetaCyc. Transporters will also be added to the Metabolic Overview Map. In addition to data curation, many enhancements to the data visualization capabilities of the Pathway Tools software are planned, such as the ability to overlay expression values of individual isoenzymes onto reactions (currently the highest value is overlaid), to zoom in from the Metabolic Overview Map overlaid with expression data to pathway detail pages color-coded in the same way, and to display pathways in the context of subcellular location information.
Pathway curation is a time-consuming process. One way to expedite the rate of curation and increase the quality of data is to encourage data submission by users, especially experts in a particular metabolism field. Submissions could include updates or corrections to an existing pathway or a new pathway to be added. An easy data submission form will be developed and available in the near future. Currently, users are encouraged to contact AraCyc curators if they notice errors or omissions in the data (curator@arabidopsis.org).
MATERIALS AND METHODS
PathoLogic Prediction of AraCyc 2.0
AraCyc 2.0 was built by running the PathoLogic (Pathway Tools 8.5) as described previously (Mueller et al., 2003). The input gene annotation file was extracted from TAIR (http://arabidopsis.org/servlets/Search?action=new_search&type=keyword), which contains genes that are annotated to the GO keyword or term “catalytic activity” (GO:0003824) or children terms of “catalytic activity” as of October 22, 2004. GO organizes terms such that the broader concepts, or parent terms such as catalytic activity, appear on the top level of the hierarchy tree. The more specific concepts, or children terms such as phytoene synthase activity, are grouped under catalytic activity. The input file was manually checked to exclude genes whose functions are apparently not related to small-molecule metabolism and contained 4,896 genes.
Running the Pathway Hole Filler
The amino acid sequence of the Arabidopsis genome annotation ATH1_pep_cm_20040228 (ftp://ftp.arabidopsis.org/home/tair/Sequences/blast_datasets/) was used as the input sequence file for the PathoLogic hole-filler program. EcoCyc was chosen as the training data for the hole filler. The probability cutoff was set to 0.9.
Validation and Manual Curation of Pathways
Curators follow standard procedures and guides to collect and enter information into the databases (http://bioinformatics.ai.sri.com/ptools/curatorsguide.pdf). Information is collected from major textbooks describing general plant biochemistry or specific areas of plant biochemistry, and from primary literature searched at databases including PubMed (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=PubMed) and Scirus (http://www.scirus.com/srsapp/). Curated pathway diagrams are entered with evidence codes and literature citations. Curators also write a summary describing the pathway's role and significance. Reactions are curated with EC numbers or EC classes and subclasses. Chemical structures are entered for compounds. Enzymes are curated with physical and catalytic properties and their coding genes. Evidence codes along with literature citations are assigned to enzyme activities.
Ontology Development
To enhance the secondary metabolic pathway ontology and develop the subcellular component ontology, existing terms are collected from textbooks or other resources, including GO (http://www.genetonology.org). Additional terms are created when necessary to meet the database needs. Each term is defined and classified according to the “is-a” relationship. Synonyms andadditional relationships to other terms such as “surrounded-by” and “component-of” are added if they exist.
Acknowledgments
We thank Aleksey Kleytman and Shijun Li for technical assistance, and Tanya Berardini, Leonore Reiser, and Wolf Frommer for reviewing the cellular component ontology of AraCyc and MetaCyc. We are grateful to Leonore Reiser and Eva Huala for critical reading the manuscript.
This work was supported by the National Science Foundation (grant no. DBI–9978564) and by the National Institutes of Health (National Institute of General Medical Sciences; grant no. 1–R01–GM65466–01).
The online version of this article contains Web-only data.
References
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Berardini TZ, Mundodi S, Reiser R, Huala E, Garcia-Hernandez M, Zhang P, Mueller LA, Yoon J, Doyle A, Lander G, et al (2004) Functional annotation of the Arabidopsis genome using controlled vocabularies. Plant Physiol 135: 745–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon RA (2001) Natural products and plant disease resistance. Nature 411: 843–847 [DOI] [PubMed] [Google Scholar]
- Gene Ontology Consortium (2001) Creating the gene ontology resource: design and implementation. Genome Res 11: 1425–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gene Ontology Consortium (2004) The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res 32: D258–D261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff SA, Ricke D, Lan TH, Presting G, Wang R, Dunn M, Glazebrook J, Sessions A, Oeller P, Varma H, et al (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 296: 92–100 [DOI] [PubMed] [Google Scholar]
- Green ML, Karp PD (2004) A Bayesian method for identifying missing enzymes in predicted metabolic pathway databases. BMC Bioinformatics 5: 76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber TR (1993) A translation approach to portable ontology specifications. Knowl Acquis 5: 199–220 [Google Scholar]
- Hadacek F (2002) Secondary metabolites as plant traits: current assessment and future perspectives. Crit Rev Plant Sci 21: 273–322 [Google Scholar]
- Harborne JP, Baxter H (1993) Phytochemical Dictionary: A Handbook of Bioactive Compounds from Plants. Taylor & Francis, London
- Kanehisa M, Goto S (2000) KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res 28: 27–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karp PD (2000) An ontology for biological function based on molecular interactions. Bioinformatics 16: 269–285 [DOI] [PubMed] [Google Scholar]
- Karp PD, Paley S, Krieger CJ, Zhang P (2004) An evidence ontology for use in pathway/genome databases. Pac Symp Biocomput 9: 190–201 [DOI] [PubMed] [Google Scholar]
- Karp PD, Paley S, Romero P (2002) The Pathway Tools software. Bioinformatics 18: S225–S232 [DOI] [PubMed] [Google Scholar]
- Keseler IM, Collado-vides J, Gama-Castro S, Ingraham J, Paley S, Paulsen IT, Peralta-Gil M, Karp PD (2005) EcoCyc: a comprehensive database resource for Escherichia coli. Nucleic Acids Res 33: D334–D337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieger CJ, Zhang P, Mueller LA, Wang A, Paley S, Arnaud M, Pick J, Rhee SY, Karp PD (2004) MetaCyc: a multiorganism database of metabolic pathways and enzymes. Nucleic Acids Res 32: D438–D442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller LA, Zhang P, Rhee SY (2003) AraCyc: a biochemical pathway database for Arabidopsis. Plant Physiol 132: 453–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paley S, Karp PD (2002) Evaluation of computational metabolic-pathway predictions for Helicobacter pylori. Bioinformatics 18: 715–724 [DOI] [PubMed] [Google Scholar]
- Pichersky E, Gang DR (2000) Genetics and biochemistry of secondary metabolites in plants: an evolutionary perspective. Trends Plant Sci 5: 439–445 [DOI] [PubMed] [Google Scholar]
- Robinson T (1983) The Organic Constituents of Higher Plants. Cordus Press, North Amherst, MA
- Singer AC, Crowley DE, Thompson IP (2003) Secondary plant metabolites in phytoremediation and biotransformation. Trends Biotechnol 21: 123–130 [DOI] [PubMed] [Google Scholar]
- Verpoorte R, Memelink J (2002) Engineering secondary metabolite production in plants. Curr Opin Biotechnol 13: 181–187 [DOI] [PubMed] [Google Scholar]
- Wink M (1988) Plant breeding: importance of plant secondary metabolites for protection against pathogens and herbivores. Theor Appl Genet 75: 225–233 [Google Scholar]
- Wink M (2003) Evolution of secondary metabolites from an ecological and molecular phylogenetic perspective. Phytochemistry 64: 3–19 [DOI] [PubMed] [Google Scholar]
- Wortman JR, Haas BJ, Hannick LI, Smith RK Jr, Maiti R, Ronning CM, Chan AP, Yu C, Ayele M, Whitelaw CA, et al (2003) Annotation of the Arabidopsis genome. Plant Physiol 132: 461–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Hu S, Wang J, Wong GK, Li S, Liu B, Deng Y, Dai L, Zhou Y, Zhang X, et al (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science 296: 79–92 [DOI] [PubMed] [Google Scholar]