Abstract
The genome-wide protein sequences from Arabidopsis (Arabidopsis thaliana) and rice (Oryza sativa) spp. japonica were clustered into families using sequence similarity and domain-based clustering. The two fundamentally different methods resulted in separate cluster sets with complementary properties to compensate the limitations for accurate family analysis. Functional names for the identified families were assigned with an efficient computational approach that uses the description of the most common molecular function gene ontology node within each cluster. Subsequently, multiple alignments and phylogenetic trees were calculated for the assembled families. All clustering results and their underlying sequences were organized in the Web-accessible Genome Cluster Database (http://bioinfo.ucr.edu/projects/GCD) with rich interactive and user-friendly sequence family mining tools to facilitate the analysis of any given family of interest for the plant science community. An automated clustering pipeline ensures current information for future updates in the annotations of the two genomes and clustering improvements. The analysis allowed the first systematic identification of family and singlet proteins present in both organisms as well as those restricted to one of them. In addition, the established Web resources for mining these data provide a road map for future studies of the composition and structure of protein families between the two species.
Sequence similarity comparisons play an essential role in analyzing the phylogenetic and structure-function relationships of genes and proteins. They are critical for dissecting complex functional differences between the members of protein families to ultimately understand their full activity spectrum. Efficient tools for analyzing complex families are of particular importance to plant biology since the majority of the genome-encoded proteins from the model organisms Arabidopsis (Arabidopsis thaliana) and rice (Oryza sativa) are members of large sequence families. The number and sizes of these families frequently exceed the complexity in animal and other kingdoms (Riechmann and Ratcliffe, 2000; Wang et al., 2003; Nelson et al., 2004). The concomitant redundancy of genes causes often serious limitations for loss-of-function or knockout experiments due to unpredictable compensation effects of other family members with overlapping functionalities (paralogs). As a consequence, many plant researchers have to invest extensive time and experimental resources to dissect the specific sequence and structural features of each member for a family of interest. Unfortunately, most technical approaches for determining gene functions in vivo are limited to the analysis of one or a few candidate genes per experiment. This makes it very difficult and time consuming to examine the full functional diversity of families with frequently built-in redundancies. Moreover, the majority of the predicted proteins from sequenced plant genomes remain functionally unclassified (unknown proteins). Thus, many families are still lacking functional information for all of their members (unknown families). Since overlapping functions between entire families are less common, it can be expected that most of these unknown families contribute critical molecular baseline activities that are involved in many unexplored developmental and adaptation strategies in plants and other kingdoms.
Although computational approaches will not overcome these difficulties, the development of efficient bioinformatics tools for analyzing differences within and across families is critical for guiding future research in many plant science areas. Most in silico family analysis strategies are based on the simple but effective concept that sequences with a higher degree of similar residues share more structural and functional properties than those with weaker similarities. This guideline allows the assignment of putative functions to unidentified proteins sharing significant sequence similarities and, hence, functional properties with characterized candidates. Although weak similarities are less reliable indicators for predicting function, they commonly provide essential information for discovering proteins with novel properties. To perform the required comparisons, all related sequences of interest need to be identified by similarity searches, the retrieved candidates organized in multiple alignments, their conserved domains localized, and distance trees calculated. Those basic family analysis steps have guided the functional identification of countless uncharacterized genes in the past. Various software tools are available for grouping unclustered protein sets into families, largely by employing distance matrices derived from all-against-all comparisons (Enright and Ouzounis, 2000; Enright et al., 2002; Koonin et al., 2002; Pipenbacher et al., 2002). Furthermore, several Web resources with preclustered family information are available to simplify time-consuming family analysis steps for the user. Most of them are based on the clustering of well annotated sequences from all organisms contained in the major protein and structure databases (Tatusov et al., 1997, 2000, 2003; Kriventseva et al., 2001; Krause et al., 2002; Enright et al., 2003; Bateman et al., 2004; Leinonen et al., 2004). The corresponding Web interfaces of these databases allow the user to quickly identify orthologs for a sequence of interest across all available organisms. This global organism approach is ideally suited for comprehensive ortholog studies across a broad range of organisms. However, the present infrastructure is frequently insufficient for genome-wide studies of fully sequenced organisms since many of their predicted or weakly annotated proteins are often not included in the corresponding databases (Mohseni-Zadeh et al., 2004). In addition, more refined clustering and customized data structures are required to provide a complete inventory of family and singlet proteins for researchers focusing on specific organisms.
In this study, we have clustered all protein sequences from Arabidopsis and rice into similarity groups, calculated their corresponding alignments, localized their conserved domains, and generated distance trees. The resulting data sets provide comprehensive information about the similarities and dissimilarities between a monocotyledon and a dicotyledon representative with regard to the size, quantity, and composition of their family and singlet proteins. The provided data sets represent a foundation for future studies of the ortholog and paralog sequences of the two species. The user-friendly Genome Cluster Database (GCD; http://bioinfo.ucr.edu/projects/GCD) was designed to provide to the public an efficient cluster mining tool for Arabidopsis and rice to perform various intraspecies and interspecies comparisons, and also to retrieve related sequences from other organism groups.
RESULTS AND DISCUSSION
Protein Similarity Clustering
To identify and compare all family and singlet proteins from Arabidopsis and rice spp. japonica, their protein sequences from The Institute for Genomic Research (TIGR) were clustered into similarity groups. Two profoundly different approaches were chosen for this purpose to minimize the limitations inherent in most available methods for clustering large and diverse sequence sets with high sensitivity and low false-positive rates. To guide the reader through the following text, a summary of the two methods with regard to their relative performance for high-sensitivity clustering of remotely related sequences is provided in Table I. It is important to point out that the reported differences greatly depend on the parameter settings (see below) of the two methods.
Table I.
Relative performance differences of the two clustering methods
The table provides a comparison of the most important strengths and weaknesses (Performance Criteria) when the two approaches BCL and HCL are used with the clustering parameters of this study. The reported differences are based on benchmark comparisons with curated families. Altered settings can change the performance differences significantly. The “++” stands for better performance than “+” in this table.
| Performance Criteria | BCL | HCL |
|---|---|---|
| High sensitivitya | + | ++ |
| Low false-positive rate under high-sensitivity settings | + | ++ |
| Elimination of clusters with inconsistent domain composition | + | ++ |
| Rejection of clusters with unconnected domains | + | ++ |
| Control over length of similarity region relative to sequences | ++ | + |
| Not limited by domain knowledge | ++ | + |
References: Eddy (1996); Altschul et al. (1997).
The first approach (BCL) used the BLASTCLUST software from the National Center for Biotechnology Information (ftp://ftp.ncbi.nlm.nihi.gov/blast/executables) to automatically group the proteins based on BLASTP similarity scores and single-linkage clustering. Low-complexity regions had been masked in the sequences to avoid overclustering due to biased amino acid distributions in certain proteins (Promponas et al., 2000; Wootton and Federhen, 2003). While this masking step minimizes the false-positive rates in the overall clustering, it can prevent related sequences with repetitive elements from clustering into families (e.g. Pro-rich proteins; Fowler et al., 1999). The minimum criteria used for joining distantly related sequences into clusters were 50% overlap and 35% identity in their pairwise BLAST alignments. Prior test runs with the same data set had shown that BCL produces under the chosen parameters for well characterized families the most complete clusters with low false-positive rates (Table II). Lowering the identity value below 35% increased the number of false positives significantly (data not shown). In response to requests from the plant community, a transparent percentage value instead of a statistically more robust E value has been chosen here as a threshold. The relatively low overlap value of 50% between sequences was used to maximize the formation of complete families in this clustering. Increasing the overlap stringency results in far less complete clusters since members of the same family often show significant length differences due to variations in target sequences, terminal extensions, alternative splice events, truncated gene models, etc. (Girke et al., 2004). A disadvantage is that the relaxed overlap requirements can result in the contamination of clusters with unrelated proteins through indirect connectivity with multiple domain proteins. However, those events are relatively rare since the method only joins two proteins into a cluster when the alignment length coverage (overlap) of both members is 50%. To illustrate the effect of this restriction: The protein families cytochrome b5, nitrate reductase, and Δ8 sphingolipid desaturase form separate families in this clustering despite the fact that they all share a cytochrome b5 domain with sequence identities above the similarity threshold (Table II). They are not joined into a hybrid cluster due to the relatively short length of the shared domain. Nevertheless, very large gene families with extremely complex domain architectures can be contaminated with false-positive proteins. An example for this event is the kinase superfamily that contains unrelated sequences in this clustering. The following domain-based approach generates far more reliable results for subgroups of this extremely complex family with more than 2,000 members. Clustering all kinases into one superfamily with accurate separation of subgroups requires several manual curation steps and specialized clustering techniques as described by Wang et al. (2003) and the PlantsP project (Tchieu et al., 2003).
Table II.
Benchmarking of cluster qualities
The size and composition of expert-curated Arabidopsis families collected from literature (Reference column) is compared to results obtained with HCL and BCL. The last example represents a family of unknown function for which no Pfam domain or references are available. The provided numbers represent the family sizes, while several numbers in a field indicate the size of the obtained subfamilies in decreasing order. Only one gene/protein model was counted per gene. The number “1” stands for singlet (e.g. “5 × 1” stands for five singlets). Data sources are provided as footnotes. *, These counts do not include truncated genes or those that are absent in the latest genome annotation. n.a., Not available.
| Family Name | Reference | HCL | BCL |
|---|---|---|---|
| Cytochrome P450 familya | 244 | 239/5×1 | 151/31/28/19/4/3/8×1 |
| Acyl-group desaturasesb | 15 | 15 | 9/4/1/1 |
| Stearoyl-ACP desaturasesb | 7 | 7 | 7 |
| Xyloglucan xylosyltransferasesc | 7* | 7 | 7 |
| Xyloglucan fucosyltransferasesd | 9* | 9 | 9 |
| Phototropinse | 2 | 2 | 2 |
| Auxin response factorsf | 23 | 17/6 | 20/3×1 |
| Fatty acid multifunctional proteinsg | 2 | 2 | 2 |
| Phospholipase D familyh | 12* | 9/3×1 | 10/2 |
| Δ8 sphingolipid desaturasesi | 2 | 2 | 2 |
| Nitrate reductasesj | 2 | 2 | 2 |
| Expressed protein (BCL ID: 321) | n.a. | 10×1 | 10 |
The second clustering approach (hidden Markov model [HMM] domain-based clustering [HCL]) used the serial arrangement of Pfam domains in each protein to form families with the same order of known protein domains. The domains were identified in the two protein sets by HMMPFAM (http://hmmer.wustl.edu/) searches against the Pfam domain model database. A custom Perl script was developed to group the proteins according to their identified domain architecture. Similarly as above, the composition of known families was used as benchmark for parameter optimization. Our experience with more than 30 curated plant families from Girke et al. (2004) and other families (Table II) has shown that an HMM E value of ≤0.1 as cutoff allows clustering of nearly complete families with false positive rates close to zero. In addition, no constraints were set in this clustering regarding the domain coverage relative to the entire protein length. Those restrictions were avoided to favor the formation of complete families, even though limited coverage can result in false positives in which sequences share only short similarities. To further evaluate the cluster qualities by manual inspection of selected cases, multiple alignments and distance trees for all identified families of the two methods were calculated with the programs MultAlin and PHYLIP, respectively (Corpet, 1988; Felsenstein, 2004).
The outcome of the two clustering methods is summarized in Table III. The HCL data for Arabidopsis agree in large parts with the domain signature clustering results from Wortman et al. (2003). Within the provided cluster size intervals, BCL and HCL show a similar performance trend between the two organisms. Interestingly, both methods indicate that approximately 45% to 60% of the proteins from the two species belong to families with six or more members. This result clearly demonstrates the importance of protein families for plant biology. The number of singlets in the HCL data set is slightly higher than in the BCL results. This is expected since domain clustering is an empirical approach that is limited by the domain knowledge contained at present in the Pfam database. The higher degree of partial gene predictions in the less complete rice annotation results in a stronger relative performance difference between the two methods for identifying singlets since many protein fragments show no or poor coverage with known Pfam domains. Future improvements of the rice gene models will certainly improve this situation. In contrast to this, the HCL approach consistently assigns for both organisms more clusters with more members in the larger size intervals (cluster sizes above 10) than the BCL approach. This is also an anticipated trend due to the higher sensitivity of HMM searches in detecting weak similarities without incorporating too many false-positive search hits into the clusters. This robust performance of the HCL approach is demonstrated in Table II by comparisons with some expert curated families. The automated method assembles in most cases complete families without contaminations with unrelated candidates. Even in the case of the very large and diverse P450 family, HCL is able to identify almost all of its members. Only five (2%) members of this family are missed since their available sequences do not entirely cover the corresponding Pfam domain model. The auxin response factor family is annotated in the literature with 23 Arabidopsis members and contains three Pfam domains. Six members of this family form a separate HCL cluster due to deletions and truncations in the C-terminal Aux/IAA domain. In this case, the BCL approach forms a larger cluster since it is not affected by incomplete domain coverage. An additional important feature of the implemented HCL strategy is that it joins multiple domain proteins into families only if they share all domains in the same order. This prevents coclustering of proteins with inconsistent domain architectures or unrelated proteins through bridging effects of multiple domain proteins (Bolten et al., 2001). However, if no domains are available, then the approach fails to provide cluster information. An example of this situation is shown in the last row of Table II for a family of unknown function. In conclusion, the two clustering methods exhibit counterbalancing strengths for high-sensitivity clustering: HCL generates the most complete clusters for families with known domains, while BCL compensates its inability to cluster families with no or insufficient domain information. Since these complementary properties offer advanced query flexibilities for downstream analyses, both data sets were integrated as separate entities into the GCD database, and efficient query tools were developed to mine them in parallel (see below). Merging the two cluster sets would have unnecessarily complicated the evaluation of their qualities by reducing data transparency and preventing the public from choosing the better data set for a given scientific question.
Table III.
Amount and complexity of families in Arabidopsis and rice
The size (Members column) and number of clusters (Clusters column) within size intervals (Cluster Size column) are provided for both approaches (Method column): BCL and domain-based (HCL) clustering. The total number of proteins and clusters for each method is given in the same vertical arrangement. All percentage values are calculated relative to them. Readers should be aware that the provided information will be subject to future changes due to updates in the underlying sequence and domain databases. The most recent cluster statistics as well as version tracking data can be retrieved from http://bioinfo.ucr.edu/cgi-bin/clusterStats.pl.
| Cluster Size
|
Method
|
Arabidopsis
|
Rice
|
||
|---|---|---|---|---|---|
| Members | Clusters | Members | Clusters | ||
| 1 | BCL | 7,271 (25%) | 21,350 (36%) | ||
| HCL | 7,971 (28%) | 27,652 (46%) | |||
| 2–5 | BCL | 9,289 (32%) | 3,502 | 10,664 (18%) | 4,063 |
| HCL | 3,478 (12%) | 1,183 | 3,631 (6%) | 1,253 | |
| 6–10 | BCL | 3,513 (12%) | 477 | 3,667 (6%) | 487 |
| HCL | 2,764 (10%) | 370 | 2,710 (5%) | 360 | |
| 11–50 | BCL | 4,858 (17%) | 273 | 6,298 (11%) | 328 |
| HCL | 7,433 (26%) | 350 | 7,348 (12%) | 352 | |
| 51–100 | BCL | 1,650 (6%) | 25 | 1,885 (3%) | 29 |
| HCL | 2,709 (9%) | 39 | 3,889 (7%) | 56 | |
| ≥101 | BCL | 2,371 (8%) | 11 | 15,847 (27%) | 19 |
| HCL | 4,597 (16%) | 21 | 14,481 (24%) | 56 | |
| Total | BCL | 28,952 (100%) | 4,288 | 59,711 (100%) | 4,926 |
| HCL | 28,952 (100%) | 1,963 | 59,711 (100%) | 2,077 | |
The comparative proteome-wide clustering of this study allowed the systematic identification of most singlet and family proteins that are present only in one of the two organisms. Those organism-restricted clusters are a rich resource for studying the molecular and functional diversities between the two organisms. Table IV provides a summary of the statistics of these complex differences, and Table V shows several examples that are organism restricted according to both clustering methods and contain only one Pfam domain. The complete family information for all cluster intervals of Table IV can be retrieved through predefined queries from GCD's Advanced Search page. Overall, the relative abundance of organism-restricted singlet and family proteins is much higher in rice (45% and 57%) than in Arabidopsis (29% and 25%). This finding is in agreement with published search results between the two organisms (Kikuchi et al., 2003). With regard to functional diversity, this difference may not be as marked as it first appears since the dominance of transposon-related proteins in rice and its less completed genome annotation both result in an overestimation of the total amount of expressed full-length proteins in this organism. Interestingly, a large number of the families, which occur only in one organism, are families of unknown function. The lack of Pfam domains for many of these novel families explains why the HCL approach identifies a lower number of organism-restricted families than the BCL approach (Table IV).
Table IV.
Abundance of organism-restricted clusters and singlets
Family and singlet proteins with zero members in one of the two organisms are compared using the column arrangement and units of Table III. The percentage values are given in relation to the total number of proteins for each organism. The complete family information for the corresponding cluster intervals can be retrieved through predefined queries from the Advanced Search page of GCD.
| Cluster Size
|
Method
|
Arabidopsis Restricted
|
Rice Restricted
|
||
|---|---|---|---|---|---|
| Members | Clusters | Members | Clusters | ||
| 1 | BCL | 4,693 (16%) | 18,483 (31%) | ||
| HCL | 6,780 (23%) | 26,474 (44%) | |||
| 2–5 | BCL | 2,239 (8%) | 1,113 | 4,391 (7%) | 1,926 |
| HCL | 212 (0.7%) | 111 | 487 (0.8%) | 216 | |
| 6–10 | BCL | 429 (1%) | 60 | 911 (2%) | 123 |
| HCL | 51 (0.2%) | 8 | 258 (0.4%) | 35 | |
| 11–50 | BCL | 614 (2%) | 36 | 1,662 (3%) | 82 |
| HCL | 214 (0.7%) | 11 | 507 (0.9%) | 25 | |
| 51–100 | BCL | 85 (0.3%) | 1 | 483 (0.8%) | 8 |
| HCL | 77 (0.3%) | 1 | 716 (1%) | 10 | |
| ≥101 | BCL | 418 (1%) | 3 | 1,173 (2%) | 5 |
| HCL | 0 (0%) | 0 | 5,599 (9%) | 19 | |
| Sum | BCL | 8,478 (29%) | 1,213 | 27,103 (45%) | 2,144 |
| HCL | 7,334 (25%) | 131 | 34,041 (57%) | 305 | |
Table V.
Examples of organism-restricted single-domain clusters
HCL clusters are listed that have zero members in one of the two organisms. All provided clusters are also organism restricted according to the BCL clustering and contain only one Pfam domain.
| Organism | Description | Pfam ID | HCL Size |
|---|---|---|---|
| Arabidopsis restricted | |||
| Self-incompatibility protein S1 | PF05938 | 39 | |
| Protein of unknown function | PF03384 | 26 | |
| Protein of unknown function | PF05617 | 18 | |
| Trypsin inhibitor | PF00537 | 7 | |
| Mildew resistance protein RPW8 | PF05659 | 4 | |
| Rice restricted | |||
| Ribosome inactivating protein | PF00161 | 21 | |
| Bowman-Birk protease inhibitor | PF00228 | 11 | |
| MerR family regulatory domain | PF00376 | 9 | |
| ABA/WDS induced protein | PF02496 | 6 | |
| Common central domain of tyrosinase | PF00264 | 5 |
As outlined above, comprehensive clustering of entire proteomes is a very complex process. Although automated computational approaches provide very efficient solutions to this problem, it is currently not possible to generate perfect cluster information for all families, even with two different approaches. This is largely due to the often very specialized requirements for extremely diverse candidates and incomplete proteome knowledge. Therefore, the provided family information has its limitations, and users are asked to critically assess the quality of a family of interest with their expert knowledge before they base critical research decisions on the results.
Database and Interface Design of GCD
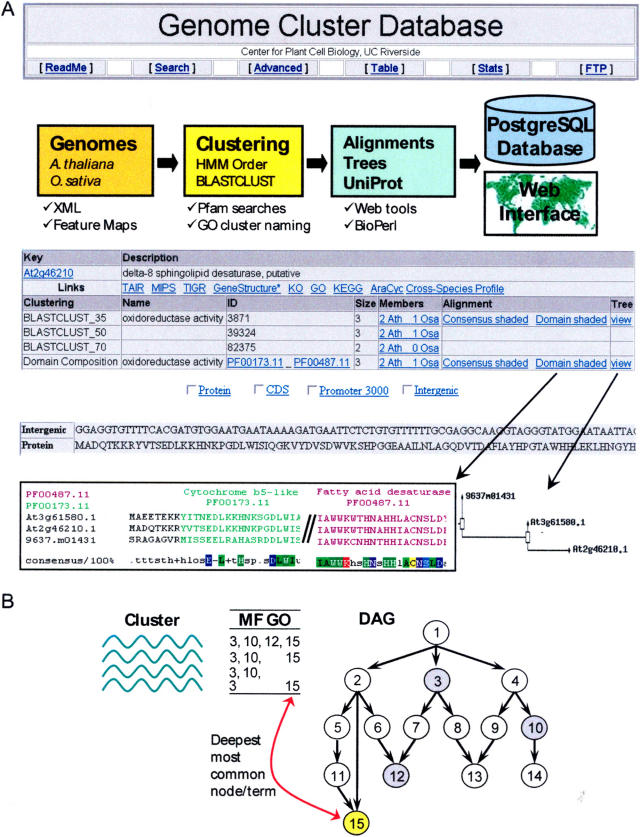
The generated information of this study was organized in the public GCD that is equipped with many powerful query, visualization, and download features for flexible interspecies analyses of gene and protein families. A multifunctional entry page allows users to search the database in single or batch mode by querying with gene/protein IDs, functional descriptions, cluster IDs, cluster names, or gene ontology keys. Combinatorial queries of scalable complexity can be generated through a separate Advanced Query page. Alternatively, a search and sortable cluster table enables navigation by cluster sizes, family names, and other criteria. All of the above query options return a result list with rich information on the specified gene/protein entries from Arabidopsis and rice. This includes the statistics of a query containing the number of its returned loci, gene models, and clusters. The corresponding protein, gene model, untranslated region, intergenic, and putative promoter sequences for any cluster or query can be displayed on the same page. This versatile sequence batch retrieval system allows efficient download of almost all types of Arabidopsis and rice sequences in a single step. The provided annotations for individual members contain detailed cluster information, including cluster names, total cluster sizes, organism distribution within clusters, and many links to external resources. Subclusters of higher similarity can be easily identified through BCL results with more stringent thresholds of 50% and 70% sequence identity. To quickly retrieve all members of a family on the result page, users can activate a hyperlinked subquery system for any given cluster in the database. This action will send the correct query syntax back to the main page and return all of the members of a family of interest. A sortable list of related sequences from all other organisms represented in the UniProt database (Leinonen et al., 2004) is accessible via a link menu. The multiple alignments for the individual clusters can be viewed and downloaded in two different color modes: the consensus shading highlights its conserved residues, while the domain shading localizes the detected Pfam domains for its members. An online HMMALIGN tool can generate custom multiple alignments against a chosen Pfam domain model. Many additional batch analysis functions are available on the same page: gene structure viewing, chromosome mapping, and Gene Ontology (GO) pie chart plotting.
Functional names for all clusters are available. Those were assigned by a computational method that is based on the GO annotations for the two organisms (Ashburner et al., 2000; Berardini et al., 2004). This approach uses the deepest and most common molecular function (MF) GO term for all proteins in a cluster (Fig. 1B). The deepest MF node was chosen since it typically provides the most detailed functional information. The developed method generates in most cases very useful cluster names that will automatically improve with future updates of the available GO annotations of the two species.
Figure 1.
Design Overview of GCD. A, Outline of data flow and graphical interface. B, Automated cluster naming strategy based on GO annotations that assigns the deepest and most common MF term to a cluster. To identify those consensus terms, the available molecular function annotations of all proteins of a cluster are mapped to the GO network that consists of a directed acyclic graph (DAG).
An automated download and reclustering pipeline has been implemented for the database backend to ensure up-to-date clustering and sequence information upon major changes in the genome annotations of the two organisms. GCD will be further maintained and improved by including additional well annotated plant genomes in the future and adding new features to enhance its functionality for the community. We will also continue to work on data interoperability and sharing of data with various protein family resources, TIGR, The Arabidopsis Information Resource (TAIR; Rhee et al., 2003), and other databases.
CONCLUSION
The comprehensive protein family information of this project and the associated GCD Web service both provide many new and unique opportunities for efficient comparative studies between Arabidopsis and rice. Those resources are expected to be of broad interest to researchers who are interested in exploring the molecular and structural diversities within and across the two plant species.
MATERIALS AND METHODS
Sequence Clustering
The plant proteome and genome sequences used for this project were downloaded from TIGR's ftp site (ftp://ftp.tigr.org). The latest genome annotation versions 5.0 and 2.0 were retrieved for Arabidopsis (Arabidopsis thaliana) and rice (Oryza sativa) spp. japonica, respectively. The nucleotide sequences and annotation data that are provided through the GCD interface were extracted with internal Bioperl parsers from the corresponding pseudochromosome files in XML format. Orthologs in other species were identified through local BLASTP (Altschul et al., 1990) searches against the UniProt database (Leinonen et al., 2004).
Protein sequence similarity clustering was performed with the BLASTCLUST program (ftp://ftp.ncbi.nlm.nihi.gov/blast/executables) using 50% overlap and 35% identity as cutoff values for family assembly. Two additional cluster sets with 50% and 70% identity were generated for the Web page. Prior to this clustering, low-complexity regions of the proteins were masked with the freely available CAST program from Promponas et al. (2000).
Domain composition clustering was performed in two steps. First, the Pfam domains were identified in the proteins with HMMPFAM searches against the latest Pfam HMM library (Pfam_ls). Second, the proteins were clustered with a custom Perl script based on their order of identified domains using an HMM E value of ≤0.1 as cutoff.
Alignments, Trees, and GO Annotations
Multiple alignments for clusters were calculated using the MultAlin program from Corpet (1988) that is capable of generating complex alignments of several thousand proteins. To visualize conserved residues in color, the alignments were reformatted with the MView software (Brown et al., 1998). Distance trees for the alignments were calculated with the PHYLIP package using a robust distance-based neighbor-joining approach for tree construction and the midpoint method for defining root positions (Felsenstein, 2004).
The GO annotations, used for protein family naming, were retrieved from TIGR's pseudochromosome files. Based on consistency considerations of GO categories between the two species, the TIGR annotations were used for both organisms. The more comprehensive Arabidopsis GO annotations from TAIR are not included at this point. For consensus mapping of terms, the current GO tree was downloaded from the GO Consortium page (http://www.geneontology.org/). The developed naming strategy is divided into two steps. First, a single molecular function GO identifier is assigned to each protein by using the deepest one in the network. If several GOs with the same depth are determined, then only the first one is used. Second, the GO term appearing most often in a cluster is chosen to be the cluster name. If the GO count ends in two or more groups of identical size, then the first one is used.
Database, Interface, and Update Strategy
The generated protein family cluster information was uploaded into a relational PostgreSQL database (http://www.postgresql.org/). To provide unlimited data access to the public through the Internet, a user-friendly Java-based Web interface was developed that integrates several open-source applications and Bioperl modules (Stajich et al., 2002). More detailed information on its design and usage can be retrieved through GCD's ReadMe page. Most data upload and clustering steps have been automated with a Perl script, so that GCD can be quickly updated on an available 64 CPU LINUX cluster when new versions of the genome annotations and Pfam domain database are released in the future.
Acknowledgments
We thank Brian Haas and Shu Ouyang from TIGR for keeping us informed about new updates in the annotations of the two plant genomes.
This work was supported by the Center for Plant Cell Biology at the University of California, Riverside, and by the National Science Foundation (grant no. IOB–0420152 to J.B.-S. and T.G., and grant no. MCB–0296080 to N.R.).
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25: 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman A, Coin L, Durbin R, Finn RD, Hollich V, Griffiths-Jones S, Khanna A, Marshall M, Moxon S, Sonnhammer EL, et al (2004) The Pfam protein families database. Nucleic Acids Res (Database issue) 32: D138–D141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beisson F, Koo AJ, Ruuska S, Schwender J, Pollard M, Thelen JJ, Paddock T, Salas JJ, Savage L, Milcamps A, et al (2003) Arabidopsis genes involved in acyl lipid metabolism. A 2003 census of the candidates, a study of the distribution of expressed sequence tags in organs, and a web-based database. Plant Physiol 132: 681–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berardini TZ, Mundodi S, Reiser L, Huala E, Garcia-Hernandez M, Zhang P, Mueller LA, Yoon J, Doyle A, Lander G, et al (2004) Functional annotation of the Arabidopsis genome using controlled vocabularies. Plant Physiol 135: 745–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolten E, Schliep A, Schneckener S, Schomburg D, Schrader R (2001) Clustering protein sequences—structure prediction by transitive homology. Bioinformatics 17: 935–941 [DOI] [PubMed] [Google Scholar]
- Briggs WR, Christie JM (2002) Phototropins 1 and 2: versatile plant blue-light receptors. Trends Plant Sci 7: 204–210 [DOI] [PubMed] [Google Scholar]
- Brown NP, Leroy C, Sander C (1998) MView: a web-compatible database search or multiple alignment viewer. Bioinformatics 14: 380–381 [DOI] [PubMed] [Google Scholar]
- Corpet F (1988) Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res 16: 10881–10890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy SR (1996) Hidden Markov models. Curr Opin Struct Biol 6: 361–365 [DOI] [PubMed] [Google Scholar]
- Enright AJ, Kunin V, Ouzounis CA (2003) Protein families and TRIBES in genome sequence space. Nucleic Acids Res 31: 4632–4638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright AJ, Ouzounis CA (2000) GeneRAGE: a robust algorithm for sequence clustering and domain detection. Bioinformatics 16: 451–457 [DOI] [PubMed] [Google Scholar]
- Enright AJ, Van Dongen S, Ouzounis CA (2002) An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res 30: 1575–1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faik A, Price NJ, Raikhel NV, Keegstra K (2002) An Arabidopsis gene encoding an alpha-xylosyltransferase involved in xyloglucan biosynthesis. Proc Natl Acad Sci USA 99: 7797–7802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J (2004) PHYLIP (Phylogeny Inference Package) Version 3.6. Distributed by the author. Department of Genetics, University of Washington, Seattle
- Fowler TJ, Bernhardt C, Tierney ML (1999) Characterization and expression of four proline-rich cell wall protein genes in Arabidopsis encoding two distinct subsets of multiple domain proteins. Plant Physiol 121: 1081–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girke T, Lauricha J, Tran H, Keegstra K, Raikhel N (2004) The Cell Wall Navigator database. A systems-based approach to organism-unrestricted mining of protein families involved in cell wall metabolism. Plant Physiol 136: 3003–3008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi S, Satoh K, Nagata T, Kawagashira N, Doi K, Kishimoto N, Yazaki J, Ishikawa M, Yamada H, Ooka H, et al (2003) Collection, mapping, and annotation of over 28,000 cDNA clones from japonica rice. Science 301: 376–379 [DOI] [PubMed] [Google Scholar]
- Koonin EV, Wolf YI, Karev GP (2002) The structure of the protein universe and genome evolution. Nature 420: 218–223 [DOI] [PubMed] [Google Scholar]
- Krause A, Haas SA, Coward E, Vingron M (2002) SYSTERS, GeneNest, SpliceNest: exploring sequence space from genome to protein. Nucleic Acids Res 30: 299–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriventseva EV, Fleischmann W, Zdobnov EM, Apweiler R (2001) CluSTr: a database of clusters of SWISS-PROT+TrEMBL proteins. Nucleic Acids Res 29: 33–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leinonen R, Diez FG, Binns D, Fleischmann W, Lopez R, Apweiler R (2004) UniProt archive. Bioinformatics 20: 3236–3237 [DOI] [PubMed] [Google Scholar]
- Mohseni-Zadeh S, Louis A, Brezellec P, Risler JL (2004) PHYTOPROT: a database of clusters of plant proteins. Nucleic Acids Res (Database issue) 32: D351–D353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DR, Schuler MA, Paquette SM, Werck-Reichhart D, Bak S (2004) Comparative genomics of rice and Arabidopsis. Analysis of 727 cytochrome P450 genes and pseudogenes from a monocot and a dicot. Plant Physiol 135: 756–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okushima Y, Overvoorde PJ, Arima K, Alonso JM, Chan A, Chang C, Ecker JR, Hughes B, Lui A, Nguyen D, et al (2005) Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: unique and overlapping functions of ARF7 and ARF19. Plant Cell 17: 444–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pipenbacher P, Schliep A, Schneckener S, Schonhuth A, Schomburg D, Schrader R (2002) ProClust: improved clustering of protein sequences with an extended graph-based approach. Bioinformatics (Suppl 2) 18: S182–S191 [DOI] [PubMed] [Google Scholar]
- Promponas VJ, Enright AJ, Tsoka S, Kreil DP, Leroy C, Hamodrakas S, Sander C, Ouzounis CA (2000) CAST: an iterative algorithm for the complexity analysis of sequence tracts. Complexity analysis of sequence tracts. Bioinformatics 16: 915–922 [DOI] [PubMed] [Google Scholar]
- Qin C, Wang X (2002) The Arabidopsis phospholipase D family. Characterization of a calcium-independent and phosphatidylcholine-selective PLD zeta 1 with distinct regulatory domains. Plant Physiol 128: 1057–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee SY, Beavis W, Berardini TZ, Chen G, Dixon D, Doyle A, Garcia-Hernandez M, Huala E, Lander G, Montoya M, et al (2003) The Arabidopsis Information Resource (TAIR): a model organism database providing a centralized, curated gateway to Arabidopsis biology, research materials and community. Nucleic Acids Res 31: 224–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond TA, Bleecker AB (1999) A defect in beta-oxidation causes abnormal inflorescence development in Arabidopsis. Plant Cell 11: 1911–1924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann JL, Ratcliffe OJ (2000) A genomic perspective on plant transcription factors. Curr Opin Plant Biol 3: 423–434 [DOI] [PubMed] [Google Scholar]
- Sarria R, Wagner TA, O'Neill MA, Faik A, Wilkerson CG, Keegstra K, Raikhel NV (2001) Characterization of a family of Arabidopsis genes related to xyloglucan fucosyltransferase1. Plant Physiol 127: 1595–1606 [PMC free article] [PubMed] [Google Scholar]
- Sperling P, Zahringer U, Heinz E (1998) A sphingolipid desaturase from higher plants. Identification of a new cytochrome b5 fusion protein. J Biol Chem 273: 28590–28596 [DOI] [PubMed] [Google Scholar]
- Stajich JE, Block D, Boulez K, Brenner SE, Chervitz SA, Dagdigian C, Fuellen G, Gilbert JG, Korf I, Lapp H, et al (2002) The Bioperl toolkit: Perl modules for the life sciences. Genome Res 12: 1611–1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov RL, Galperin MY, Natale DA, Koonin EV (2000) The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res 28: 33–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov SL, Nikolskaya AN, et al (2003) The COG database: an updated version includes eukaryotes. BMC Bioinformatics 4: 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov RL, Koonin EV, Lipman DJ (1997) A genomic perspective on protein families. Science 278: 631–637 [DOI] [PubMed] [Google Scholar]
- Tchieu JH, Fana F, Fink JL, Harper J, Nair TM, Niedner RH, Smith DW, Steube K, Tam TM, Veretnik S, et al (2003) The PlantsP and PlantsT functional genomics databases. Nucleic Acids Res 31: 342–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Harper JF, Gribskov M (2003) Systematic trans-genomic comparison of protein kinases between Arabidopsis and Saccharomyces cerevisiae. Plant Physiol 132: 2152–2165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Tischner R, Gutierrez RA, Hoffman M, Xing X, Chen M, Coruzzi G, Crawford NM (2004) Genomic analysis of the nitrate response using a nitrate reductase-null mutant of Arabidopsis. Plant Physiol 136: 2512–2522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wootton JC, Federhen S (2003) Statistics of local complexity in amino acid sequences and sequence databases. Comput Chem 17: 149–163 [Google Scholar]
- Wortman JR, Haas BJ, Hannick LI, Smith RK Jr, Maiti R, Ronning CM, Chan AP, Yu C, Ayele M, Whitelaw CA, et al (2003) Annotation of the Arabidopsis genome. Plant Physiol 132: 461–468 [DOI] [PMC free article] [PubMed] [Google Scholar]