Abstract
The Maize Genetics and Genomics Database (MaizeGDB) serves the maize (Zea mays) research community by making a wealth of genetics and genomics data available through an intuitive Web-based interface. The goals of the MaizeGDB project are 3-fold: to provide a central repository for public maize information; to present the data through the MaizeGDB Web site in a way that recapitulates biological relationships; and to provide an array of computational tools that address biological questions in an easy-to-use manner at the site. In addition to these primary tasks, MaizeGDB team members also serve the community of maize geneticists by lending technical support for community activities, including the annual Maize Genetics Conference and various workshops, teaching researchers to use both the MaizeGDB Web site and Community Curation Tools, and engaging in collaboration with individual research groups to make their unique data types available through MaizeGDB.
MISSION AND SCOPE
The Maize Genetics and Genomics Database (MaizeGDB) is the community resource for maize (Zea mays) data and can be accessed online at http://www.maizegdb.org. Data types stored at MaizeGDB include (but are not limited to) sequence, locus, variation, probe, map, metabolic pathway, phenotype, quantitative trait locus (QTL) experiment, stock, and contact information for hundreds of maize researchers worldwide (for review, see Lawrence et al., 2004). Data visualization is facilitated by unique views such as the highly popular genome browser (http://www.maizegdb.org/cgi-bin/bin_viewer.cgi) that displays data within their chromosomal context. Furthermore, data analysis tools such as BLAST (Altschul et al., 1997) and GeneSeqer (Brendel et al., 2004) are available for researchers to carry out their own data analyses directly via MaizeGDB Web services.
The team of people who work at MaizeGDB seek to serve the community of maize geneticists not only by making data generated by maize researchers available through the MaizeGDB site, but also by engaging in various community service projects. The MaizeGDB team supports the annual Maize Genetics Conference by maintaining the conference Web site and facilitating the abstract collection process, provides technical assistance for training workshops (e.g. the Maize Genetics, Genomics and Bioinformatics Workshop, which took place in March 2004 at the International Maize and Wheat Improvement Center [CIMMYT] in Mexico City), and sends out announcements to the community of maize geneticists via e-mail as directed by the Maize Genetics Executive Committee (http://www.maizegdb.org/mgec.php). In addition, by providing data descriptions for the general public, which can be found on each data center page at MaizeGDB (e.g. a description for gene product can be viewed at http://www.maizegdb.org/gene_product.php#dld), the MaizeGDB team works to educate the general public about the importance of maize genetic research.
It is the aim of this article to illustrate the breadth of information made available through MaizeGDB, to convey the method by which the information is curated and made accessible, and to relate how the database infrastructure was built and is currently maintained. Detailed information regarding historical aspects of the MaizeGDB project and a review of various data types made available through the MaizeGDB site are described elsewhere (Lawrence et al., 2004).
DISCUSSION AND FUTURE DIRECTIONS
In September 2004, under the guidance of the National Plant Genome Initiative (NPGI), the National Science Foundation (NSF), the U.S. Department of Energy (DOE), and the U.S. Department of Agriculture (USDA) announced that research funds would be made available to sequence the maize genome, and a solicitation for grant proposals was circulated (see http://www.nsf.gov/pubs/2004/nsf04614/nsf04614.txt). It was noted in this announcement that proposals should utilize existing, previously funded resources, one of the resources listed explicitly being the maize community genome database.
Currently, MaizeGDB provides a portal to maize genome sequencing information (which can be viewed at http://www.maizegdb.org/genome). Displayed on that page are documents outlining details concerning the maize genome sequencing endeavor, links to maize sequence repositories, information outlining the diversity of sequencing strategies currently employed, and a list of relevant publications germane to the maize genome sequencing project. As the maize genome gets sequenced, MaizeGDB will adapt to provide a sequence-centered portal to all maize data similar to the one provided by The Arabidopsis Information Resource (TAIR; http://www.arabidopsis.org; Rhee et al., 2003).
Presently, the number of people working at MaizeGDB is quite small when compared to the personnel associated with other database projects like TAIR (http://www.arabidopsis.org), Gramene (http://www.gramene.org), and the Solanaceae Genomics Network (SGN; http://www.sgn.cornell.edu). In order to serve as a central site for making large numbers of sequences, contigs, assemblies, and, eventually, a fully sequenced maize genome available at MaizeGDB, it is necessary that partnerships be forged between MaizeGDB and other databases, sequencing projects, and large-scale generators of maize data. In an initial effort to engage in such a collaboration, a pipeline for getting sequence data into MaizeGDB has been developed wherein all available maize sequences are downloaded from GenBank (http://www.ncbi.nih.gov/Genbank) by staff working at the Plant Genome Database (PlantGDB; http://www.plantgdb.org; Dong et al., 2004). Once the sequences have been cleaned, analyzed, and assembled into contigs at PlantGDB, they are delivered to MaizeGDB for long-term storage and display. By creating intuitive links to PlantGDB and facilitating database interoperability between MaizeGDB and PlantGDB, the weight of the burden to deliver high-quality sequence-based products to the community of maize geneticists is shared among mutually benefiting parties. Such collaborations are useful and are facilitated by initiatives like the Plant Ontology Consortium (http://www.plantontology.org; Bruskiewich et al., 2002), a group working to create controlled vocabularies for describing plant structures and growth and developmental stages to facilitate database interoperability among resources serving plant biologists working on a broad range of species. Forging new, useful partnerships with other databases and helping to develop methods to facilitate database interoperability are the highest priority tasks facing the MaizeGDB team in the coming year.
Over the course of the past year, cytological map images generated by the Cytogenetic Map of Maize project (http://www.cytomaize.org; Koumbaris and Bass, 2003) were added to the database, and displays for cytological map data were developed (e.g. http://www.maizegdb.org/cgi-bin/displayfishrecord.cgi?id=12098&map=892372). Community and Professional Curation Tools were developed for most data types, and the MaizeGDB Editorial Board began selecting noteworthy maize primary literature for rigorous professional data curation (see http://www.maizegdb.org/editorial_board.php). During the coming months, work will focus on associating Plant Ontology (http://www.plantontology.org) terms of type Plant Structure and Trait (an ontology currently under development) with records, and the raw data generated by QTL experiments will be made available. A Community Curation Tool module for QTL data is slated for development in the very near future.
MATERIALS AND METHODS
Web Interfaces Allow Access to Data within a Biological Context
The data stored at MaizeGDB are made available through a series of interconnected Web pages. Researchers can also contact the MaizeGDB team at mgdb@iastate.edu to request access to Web-based read-only SQL tools allowing direct queries on the curation copy of the database. These pages are coded in HTML, and most are automatically generated by PHP and Perl scripts. Through the Web interface (accessible at http://www.maizegdb.org), each data display page shows detailed information on a specific biological entity (e.g. a locus), as well as basic information about data associated with it (e.g. maps, variations, probes, and citations are among data types associated with loci), and links to related off-site resources (e.g. locus pages link directly to Gramene; Ware et al., 2002). Access to individual data displays is made possible through a number of different mechanisms, including a text search tool (available at the top right corner of each page) and a genome browser (located at the bottom left of the site's main page). These tools and interconnected pages allow researchers to easily navigate from point to point as they investigate research topics of interest. The interface design attempts to maximize the information available, requiring only a minimal amount of input from the researcher.
MaizeGDB's method of data delivery is aimed at making information available within the framework of its scientific meaning. Data displays place specific pieces of data within a biological context. For example, if you arrive at a map page by way of a locus page, the locus that was last visited is highlighted within the map display. Not only does using the biological relatedness of data types in conjunction with following a researcher's clickstream enable the interface to reflect real biological relationships, it also aids researchers by causing the site to seem to follow their actual train of thought. The following usage case demonstrates the interrelatedness of different types of biological information, reveals MaizeGDB's method of recapitulating those interrelationships, and highlights the placement of links to off-site resources that can help researchers gain access to related information that is not stored at MaizeGDB.
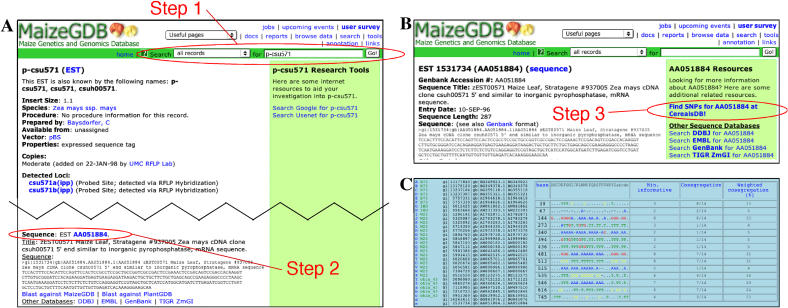
An intrepid researcher visits MaizeGDB in the hopes of finding information that would help her to design multiplex PCR primers to genotype F2 plants. She is working to determine the transmission of different variants of chromosome 10, and wishes to develop a protocol for diagnosing which chromosome 10 variants are present in any given plant growing in a half-acre field. Because the researcher knows that the expressed sequence tag (EST) probe p-csu571 labels bands that migrate differentially on Southern blots between the two backgrounds of interest (Mroczek, 2003), she decides to start by investigating sequence data for that EST. In Step 1, by searching “all records” from the top of any MaizeGDB page (see Fig. 1A) using “p-csu571” as her query, she locates the probe record for p-csu571 (http://www.maizegdb.org/cgi-bin/displayestrecord.cgi?id=118621) and discovers that the sequence AA051884 is associated with p-csu571. In Step 2, she follows the sequence link to view the MaizeGDB copy of that sequence record. In Step 3, in the right bar on the sequence record page (http://www.maizegdb.org/cgi-bin/displayseqrecord.cgi?id=1531734; Fig. 1B), a link to “Find SNPs for AA051884 at CerealsDB” is displayed. By clicking that link to automatically search CerealsDB (http://www.cerealsdb.uk.net/discover.htm; Barker et al., 2003) for single-nucleotide polymorphisms (SNPs) that include AA051884 (gi 1531734), she identifies a putative SNP cluster (snip_1563; Fig. 1C) demonstrating that, among sequences similar to AA051884, multiple polymorphisms exist. This information will allow her to design a multiplex PCR experiment protocol that could enable her to genotype plants in the field without resorting to performing hundreds of time-consuming Southern blots.
Figure 1.
The name of a probe can be used at MaizeGDB to locate SNP data available at CerealsDB. The researcher first searches “all records” from the top of any MaizeGDB page (A; step 1, circled in red) using “p-csu571” as her query; she locates the probe record. From the probe page (A), the researcher follows a link (step 2, circled in red) to view the MaizeGDB sequence display page. From the sequence display page (B), she follows a link to CerealsDB (step 3, circled in red) to check for SNP contigs that include the sequence. Data describing a putative SNP contig (C) are presented directly via the CerealsDB site (http://www.cerealsdb.uk.net/maize_snips/snip_1563.htm). The sequence of interest (EST AA051884) is listed at the bottom of the group of sequences shown.
Most of the continued development of the MaizeGDB interface is guided by members of the maize (Zea mays) genetics research community: Community members have sent hundreds of suggestions and requests concerning methods to find and display data. To aid in encouraging community feedback, a highly utilized context-sensitive feedback tool appears at the bottom of every page. The needs of researchers serve as the major impetus for interface development, and addressing those needs directly allows for tools to be developed that are both timely and germane to the needs of maize geneticists.
An example of a research need that guided interface development comes from Bill Sheridan, a maize geneticist working in the Department of Biology at the University of North Dakota. Dr. Sheridan contacted the MaizeGDB team seeking an easy method to summarize which simple sequence repeats (SSRs) detected bacterial artificial chromosomes that were also associated with genetically mapped markers. Dr. Sheridan worked with the MaizeGDB team to design a table-generating tool that provides approximate map locations for markers, the SSRs for those markers, and bacterial artificial chromosomes detected by the SSRs (see the links to each of the 10 maize chromosomes beneath the heading “Mapped & Anchored SSRs” at http://www.maizegdb.org/ssr.php). Dr. Sheridan was able to use this tool for his research and was pleased that his input guided the development of such a useful tool. This sort of interface development to address specific research needs typifies the method by which members of the MaizeGDB team work alongside researchers to help them gain access to complex relationships documented in the database. In summary, MaizeGDB's interface was initially designed to provide a context for interpreting maize data, and continued interface development is driven by specific input from and collaborative design with members of the maize research community at large.
Data Curation: Driven by the Community of Maize Geneticists
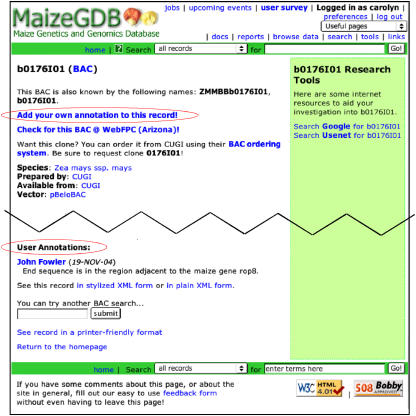
At present, the MaizeGDB team does not have any individual member dedicated solely to data curation. Instead, all team members curate data as the need arises and in accordance with his or her particular knowledge base. Most data are added to the curation copy of the database (see below for a description of how each copy of the database is utilized) in bulk and are contributed by community members directly. Feedback from researchers often guides individual data additions and edits. By describing which data to associate with existing records or by explaining why mistakenly associated information should be updated, the community of maize geneticists contributes directly to curating the data stored at MaizeGDB. Moreover, community members can add annotations to records at MaizeGDB by logging in through the annotation link at the top of any MaizeGDB page. Once logged in, researchers can add notes like the one shown in Figure 2 (http://www.maizegdb.org/cgi-bin/displaybacrecord.cgi?id=424644) by clicking the link to “Add your own annotation to this record” shown at the top of virtually all data displays.
Figure 2.
Researchers can add annotation to records at MaizeGDB. After having logged in to the site, the researcher's username appears in the upper right corner of the window, and links appear on pages allowing the researcher to add annotations (top red circle). Once an annotation has been added, it appears toward the bottom of the page (bottom red circle) along with the contributor's name and the date the annotation was submitted.
For researchers interested in contributing data directly to the database, a set of Java-based Community Curation Tools has been developed and is available for general use. Data types accessible through these tools include clone library, gel pattern, gene product, linkage group, locus, map, map scores, panel of stocks, person, phenotype, primer/enzyme, probe, recombination data, reference, species, stock, term, and variation. By creating data records, researchers become Community Curators who own the records they create and retain the ability to edit owned records directly.
To ensure that records entered by community members are complete, a curation level system has been implemented. Newly entered records are considered “submitted” and are checked by a professional curator. Once checked, the records are marked “approved” or “failed,” and only “approved” records become publicly accessible through the Web interface. Each time a community member edits a record he or she owns, the record is reassigned the “submitted” curation level and must be reapproved to regain accessibility through the Web interface.
Workshops demonstrating the use of the Community Curation Tools were taught at Iowa State University (ISU) in August 2004 and at the Plant and Animal Genome Conference in San Diego, January 2005. To schedule an on-site training session for your research group, contact the MaizeGDB team at mgdb@iastate.edu.
Professional curators have access to a set of Java-based Professional Curation Tools that were originally created to interact with the Maize Genetics Cooperation Stock Center (MGCSC) MySQL copy of the database and that subsequently were adapted to interact with the ISU Oracle-based curation database. Whereas the Community Curation Tools were designed specifically to allow researchers from the community of maize geneticists to gain limited and controlled access to the database, the Professional Curation Tools allow less restricted access to data, enabling professional curators to create and edit records in an efficient and authoritative manner.
Standard Operating Procedures, Accessibility, and Machine Architecture
Three copies of the MaizeGDB database exist at ISU: a production copy, a curation (staging) copy, and a test copy. Each copy is housed on a separate machine. The production copy of the database is accessible through http://www.maizegdb.org. This copy is not edited and is accessible by the public through the Web interface. The curation copy of the database is accessible by both community and professional curators via curation tools: It is the copy of the database to which new data are added and within which existing data are edited when the need to do so arises. The curation copy of the database is dumped in a compressed form to file each day. Compressed daily dumps of the curation database are formatted for Oracle and can be accessed at http://goblin1.zool.iastate.edu/∼oracle. A typical dump file is currently approximately 750 MB in size (approximately 2 GB when uncompressed). Dumps from the curation database are housed on a different machine than the curation database itself. Individuals can request copies of the curation database (or individual tables contained therein; see http://www.maizegdb.org/MaizeGDBSchema.pdf for access to the current MaizeGDB schema) formatted for Oracle, MySQL, or Microsoft Access by e-mailing the MaizeGDB team at mgdb@iastate.edu. On the first Tuesday of each month, a duplicate of the curation copy of the database replaces the production copy. Scheduled replacements are announced at the bottom right of the main page (http://www.maizegdb.org). The test copy of the database serves as a testing ground for tool development and improvements to both Community and Professional Curation Tools and is also used as a training site for community curators to gain familiarity with the functionality of the Community Curation Tools before using them to access the curation copy of the database.
The servers that support MaizeGDB run Oracle 9i, which is licensed every 2 years. The machines that house the various copies of the database are Dell PowerEdge servers (Round Rock, TX) with 2 × 2.0 GHz Xeon processors, 4 GB of RAM, 5 × 73 GB Ultra 320 10K RPM drives with Red Hat Advanced Server 2.1 (Raleigh, NC) installed. All servers are nearly identically configured.
In addition to the copies of the database housed at ISU, a MySQL copy exists at the MGCSC in Urbana/Champaign, Illinois, enabling the staff of the MGCSC to keep track of data associated with maize stocks directly (a service described in detail in Scholl et al., 2003). The MGCSC copy of the database is accessible through the Professional Curation Tools and is synchronized with the ISU curation database at regular intervals.
Acknowledgments
We are indebted to Darwin A. Campbell for his work as the MaizeGDB database administrator; Qunfeng Dong for his work as the database manager at PlantGDB, the source for all sequence data made available through MaizeGDB; and Marty Sachs, Director of the MGCSC, for his work curating stock and associated data types. We also thank Michael Brekke, systems support specialist; Sanford B. Baran, contract Web developer and creator of the Community Curation Tools; and Jason Carter, information technology specialist for the MGCSC and creator of the Professional Curation Tools that enable the MGCSC and MaizeGDB to maintain data synchronization. MaizeGDB would not have been possible without the legacy work of Ed Coe and Mary Polacco on the original MaizeDB resource. We are grateful for their continued interest and contributions.
This work was supported by a Specific Cooperative Agreement with the Agricultural Research Service, U.S. Department of Agriculture (no. 58–3625–1–192 to V.B.).
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker G, Batley J, O'Sullivan H, Edwards KJ, Edwards D (2003) Redundancy based detection of sequence polymorphisms in expressed sequence tag data using autoSNP. Bioinformatics 19: 421–422 [DOI] [PubMed] [Google Scholar]
- Brendel V, Xing L, Zhu W (2004) Gene structure prediction from consensus spliced alignment of multiple ESTs matching the same genomic locus. Bioinformatics 20: 1157–1169 [DOI] [PubMed] [Google Scholar]
- Bruskiewich R, Coe EH, Jaiswal P, McCouch S, Polacco M, Stein L, Vincent L, Ware D (2002) The Plant OntologyTM Consortium and plant ontologies. Comp Funct Genomics 3: 137–142 [Google Scholar]
- Dong Q, Schlueter SD, Brendel V (2004) PlantGDB, plant genome database and analysis tools. Nucleic Acids Res 32: D354–D359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koumbaris GL, Bass HW (2003) A new single-locus cytogenetic mapping system for maize (Zea mays L.): overcoming FISH detection limits with marker-selected sorghum (S. propinquum L.) BAC clones. Plant J 35: 647–659 [DOI] [PubMed] [Google Scholar]
- Lawrence CJ, Dong Q, Polacco ML, Seigfried TE, Brendel V (2004) MaizeGDB, the community database for maize genetics and genomics. Nucleic Acids Res 32: D393–D397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mroczek RJ (2003) Molecular, genetic, and cytogenetic analysis of the structure and organization of the abnormal chromosome 10 of maize. PhD dissertation. University of Georgia, Athens, GA
- Rhee S, Beavis W, Berardini T, Chen G, Dixon D, Doyle A, Garcia-Hernandez M, Huala E, Lander G, Montoya M, et al (2003) The Arabidopsis Information Resource (TAIR): a model organism database providing a centralized, curated gateway to Arabidopsis biology, research materials and community. Nucleic Acids Res 31: 224–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholl R, Sachs MM, Ware D (2003) Maintaining collections of mutants for plant functional genomics. In E Grotewold, ed, Plant Functional Genomics, Vol 236. Humana Press, Totowa, NJ, pp 311–326 [DOI] [PubMed]
- Ware DH, Jaiswal P, Ni J, Yap IV, Pan X, Clark KY, Teytelman L, Schmidt SC, Zhao W, Chang K, et al (2002) Gramene, a tool for grass genomics. Plant Physiol 130: 1606–1613 [DOI] [PMC free article] [PubMed] [Google Scholar]