Abstract
Over recent years, a number of initiatives have proposed standard reporting guidelines for functional genomics experiments. Associated with these are data models that may be used as the basis of the design of software tools that store and transmit experiment data in standard formats. Central to the success of such data handling tools is their usability. Successful data handling tools are expected to yield benefits in time saving and in quality assurance. Here, we describe the collection of datasets that conform to the recently proposed data model for plant metabolomics known as ArMet (architecture for metabolomics) and illustrate a number of approaches to robust data collection that have been developed in collaboration between software engineers and biologists. These examples also serve to validate ArMet from the data collection perspective by demonstrating that a range of software tools, supporting data recording and data upload to central databases, can be built using the data model as the basis of their design.
A recent proposal (Jenkins et al., 2004; http://www.armet.org) describes a framework, called architecture for metabolomics (ArMet), for the description of plant metabolomics experiments and their results. ArMet is a data model that describes plant metabolomics datasets together with their experimental context. Its publication represents the first step in an initiative whose long-term aim is the establishment of data standards for the field. It is hoped that discussion ensuing from its publication will lead to development and enhancement of the model and its eventual adoption by the community. In parallel with the development of the ArMet data model, other groups have produced reporting requirements for metabolomics experiments with the aim of standardizing experiment descriptions, particularly within publications. Output from this work comprises minimum information about a metabolomics experiment (Bino et al., 2004), which is a checklist of the information necessary to provide context for metabolomics data that is to be published, and a standard metabolic reporting structure policy document (SMRS Group, 2004) that is currently under development by a group from industry and academia.
Similar initiatives exist for other functional genomics approaches. The DNA microarray community has developed minimum information about a microarray experiment (MIAME; Brazma et al., 2001) as a definition of what should be recorded for a transcriptome experiment. MAGE-OM (microarray gene expression-object model; Spellman et al., 2002) is an associated data model. Proteomics experiment data repository (PEDRo; Taylor et al., 2003) is a Unified Modeling Language (Booch et al., 1999) model that seeks to meet the requirements of the proteomics field. PEDRo has been adopted by the Human Proteome Organization's Proteomics Standards Initiative (PSI; Orchard et al., 2003) as the basis for its own object model PSI-OM (Orchard et al., 2004; Taylor, 2004).
Once accepted, data standards, and the formal data descriptions that underlie them, may yield a range of benefits including the following. (1) Consideration and development of best practice and standard operating procedures, which, in turn, enable proper interpretation of experiment results, principled dataset comparison, and experiment repetition. (2) Standardized reporting of experiments and deposition and archiving of data associated with publications or other standard pieces of work. An example of this in practice is the requirement by many journals for MIAME-compliant experiment descriptions (where appropriate) in the papers that they publish and the submission of the related data to a public MIAME/MAGE-compliant data repository (e.g. ArrayExpress, Brazma et al., 2003; Gene Expression Omnibus, NCBI, 2004) with accession numbers forming part of the publication submission (Anonymous, 2002; Glueck and Dzau, 2003; Oliver, 2003). (3) Development of databases and verifiable transmission mechanisms for storage, collection, and dissemination of results. For example, downloadable implementations of database and data collection tools that are compliant with the ArMet, MIAME/MAGE, and PEDRo definitions are available (Killion et al., 2003; Taylor et al., 2003; Jenkins et al., 2004). Implementation independent data models such as ArMet, MAGE, PEDRo, and PSI-OM allow for multiple implementations tailored for use in different settings. As these implementations have a common design, they also promote fault-free interoperability.
ArMet as proposed, therefore, is not a publicly available data repository, but a data model that provides both a starting point for discussion of data standards for plant metabolomics and a basis for the design of data storage facilities and associated data handling tools for plant metabolomics datasets. Central to the success of data handling tools is their usability. In unregulated environments, the ease of use of interfaces to data handling tools that conform to proposed data standards may play a key role in their eventual adoption.
The nature of user interfaces for databases and data handling software tools can vary widely. For example, the ArrayExpress and Gene Expression Omnibus public repositories use Web-based forms and hypertext for submission and retrieval of datasets. A number of reference databases of chemical information also employ this approach (Kanehisa and Goto, 2000; Mueller et al., 2003; Krieger et al., 2004). While this type of interface is suited to fairly simple input and querying, more complicated programmatic access to remote relational database implementations may be provided through specialist programs that employ the de facto open database connectivity (ODBC; Microsoft, 2004) standard. Indeed, most modern statistical and data mining packages provide ODBC functionality for data import. Alternatively, offline database implementations can be customized with a specific look and feel to support data recording in particular environments for later upload to central databases. Similarly, where an extensible markup language (XML; Bray et al., 2004a, 2004b) schema implementation of a data model exists, this can be used to generate specialized applications such as the PEDRo data collection tool (Taylor et al., 2003). Microsoft Excel 2003 also provides a facility for mapping XML schemas to workbooks, thereby enabling spreadsheets to be customized to enforce recording of required data.
In this paper, we illustrate a number of approaches to the development of software tools to support the collection of ArMet-compliant datasets. We will show how the ArMet data model contributes to the rapid development of data collection tools that support the experimental process, ensuring that complete and error-free datasets are recorded. We also aim to validate ArMet from the data collection perspective through the demonstration of a range of data collection tools built using the data model as the basis of their design.
DATA COLLECTION
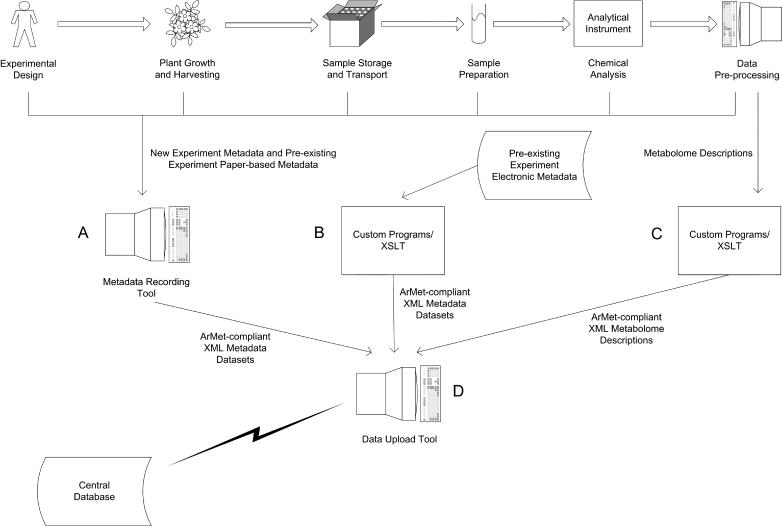
Figure 1 illustrates the timeline of a metabolomics experiment (the icons across the top) and the associated data collection activities (the icons at bottom). From this diagram, it is possible to identify two distinct aspects of data collection: gathering the metadata and results for an experiment (data recording) and the subsequent submission of the dataset to a central database (data upload). Here, we will look at the key issues to be addressed when building software tools for each of these processes.
Figure 1.
Data collection. The timeline of a metabolomics experiment (the icons across the top) and the associated data collection activities (the icons at bottom). Arrows denote data flow between people and software tools within a laboratory. The lightning flash denotes data transfer between remote computer systems across a data network. A, Software tool for recording ArMet-compliant metadata for new experiments and preexisting experiments with paper-based records. B, Custom software programs for translating electronic metadata for preexisting experiments into ArMet-compliant XML documents. C, Custom software programs for translating the datasets that result from metabolomics experiments into ArMet-compliant XML documents. D, Software tool for uploading locally held ArMet-compliant data to remote databases.
Data Recording
In practice, data that comprise a metabolomics dataset are recorded at various points during the experimental timeline. For example, plant growth and harvesting protocols will be available before information about the handling of samples and their preparation for analysis. Software tools for recording these data should reflect the experimental activities that are being performed and hide the structure of the underlying data model where that conflicts.
In our experience, experimentalists do not enjoy data input. Data recording tools that are specifically designed to support experimental procedures may go some way toward making the data input process more palatable. In addition to reflecting the experimental activities, a key way in which such software tools can make data input less onerous is to ensure that data needs to be entered only once; for example, genotype, protocol, and analytical instrument information that may be applicable to many experiments should be entered only once and then linked to additional datasets.
Finally, software tools for data recording should enforce the collection of logically correct datasets; i.e. they should ensure that data recording errors are automatically detected and flagged for correction as they are made. Such errors may go unnoticed for some time when paper-based records are created first and used to create an electronic record later.
Data Upload
The upload of datasets to central repositories, i.e. public or project-based databases, involves a number of technologies that are used to bundle the data for transmission, provide the necessary network access, and interpret the data for submission to the database at the remote site. In the same way that software tools for data recording should hide the structure of the underlying data model, tools for data upload should hide the details of these technologies from the user.
In situations where data is recorded offline and uploaded to a central database as a separate process once a complete dataset is available, it is necessary for the local data recording tool to collect all of the data that are required by the central database. Therefore, data upload tools must ensure that complete and coherent datasets are available before attempting dataset export and transmission.
Furthermore, if a local data recording tool exports data that has a different structure from that in the central database, the upload mechanism should perform the necessary transformations of the data to enable its submission to the database. Similarly, when multiple implementations of a single data model exist, as is the case when there is a central database with a number of independent data recording tools that are populating it, movement between versions of the data model can be problematic. There will inevitably be situations in which the various implementations support different versions of the model. Here again, a data upload tool should perform the necessary alterations to the dataset prior to its submission to the database.
HOW ARMET SUPPORTS DATA COLLECTION
ArMet is a data model that provides a formal specification of the data required to describe plant metabolomics experiments. As such, it may be implemented in a variety of ways to produce databases and data handling tools to support plant metabolomics experiments. Here, we will discuss the ways in which ArMet inherently supports the development of data collection tools that fulfill the requirements described above.
ArMet is an object-oriented data model. It represents data about the domain to be modeled by way of interrelated objects that contain attributes for individual data items. Essential parts of such a model are its type constraints and referential integrity (see below). It is these rules that mean that all ArMet-based data collection tools will ensure the collection of logically correct datasets and enable the collection of standard data, e.g. genotype descriptions, once only.
Referential integrity governs the relationships between objects in a model. If an attribute in one object refers to a second object, then the second object must exist. In data recording implementations of the ArMet data model, therefore, standard details, e.g. genotype definitions and protocol descriptions, need to be provided only once. The user may then rely on their existence and refer to them in multiple datasets with the knowledge that these links will not be broken by the removal of the standard data.
Each individual attribute within an object will have type constraints that specify the set of allowable values that it may contain. For numeric attributes, allowable values may be specified as a set of valid values or a valid range, whereas for text attributes, controlled vocabularies may be used to govern which entries are legal. As these constraints form part of any compliant implementation of the data model, data recording tools that are based on ArMet can automatically detect erroneous values in a dataset. These can then be flagged with the user for correction as they are made (and when they are easiest to rectify). In addition, data models are typically accompanied by additional business rules that describe further constraints on the way that a compliant software tool may be implemented; for example, a business rule that accompanies the ArMet data model is: “The date/time for the harvest of an item of source material must be later than the date/time that identifies when its cultivation started.” These additional constraints also form part of any compliant implementation of ArMet and, therefore, provide a further mechanism for detecting and flagging errors in data input.
The specification of the allowable values for an attribute includes identifying whether it may remain empty (null) within a dataset. The type constraints within ArMet, therefore, also ensure that the notion of a complete dataset is the same in all of its implementations. The issue of ensuring that a dataset is complete prior to export and upload is addressed, therefore, when the data is held locally in an ArMet-compliant data recording tool.
As the ArMet data model provides a formal specification of the data items required to describe plant metabolomics experiments and their structure, it may be used to define a format for data transmission. Such formats lend themselves to automatic manipulation. In this way, ArMet-compliant datasets that have been recorded locally and that require restructuring or alteration prior to submission to a central database may be accommodated.
In the following two sections we will discuss, and illustrate with examples, the contribution that ArMet makes to the development of data recording and upload tools.
DATA RECORDING SUPPORT
ArMet comprises nine components that, together, encompass the entire timeline of a plant metabolomics experiment. Each of these components specifies a core set of data items that are relevant to all such experiments and that serve as a minimal description for each component. Each component may also be associated with one or more subcomponents that define in detail particular experimental methodologies or technologies. It is anticipated that, as metabolomics matures and the techniques employed are standardized, the subcomponent definitions may be incorporated into the core or become standardized in their own right.
A range of data collection tools may be used to populate a single ArMet-compliant database. To collect ArMet core datasets, general purpose data collection tools, which are applicable to multiple projects, may be built. Similarly, collection tools may be built for particular subcomponents, and these too would be applicable to multiple projects that employ the techniques that the subcomponent describes. Alternatively, customized tools, for capturing either core or subcomponent data, may be built that are tailored to specific projects.
As the approach to data recording differs depending on the type of data to be recorded we have structured this discussion to address the nature of the recording mechanisms depicted in Figure 1, icons A to C.
Creating Electronic ArMet-Compliant Metadata Datasets
For new experiments, ArMet-compliant data recording can be planned from the start. Given the existence of ArMet, compliant recording tools can be developed that will ensure that the datasets produced are complete and logically correct. Tools such as these are also appropriate for assembling ArMet-compliant data from preexisting experiments where the experimental records are not already in an electronic format. Here, we illustrate two ArMet-compliant software tools for recording experimental metadata (data about the data, e.g. genotype descriptions and protocols) as they are generated in the greenhouse, field, or laboratory. These tools are examples of the “Metadata Recording Tool,” icon A in Figure 1. The two examples are as follows: (1) database example, a customized tool for capturing subcomponent data, tailored for a specific project; and (2) spreadsheet example, a general-purpose spreadsheet implementation of core data (and optional subcomponent data) that is appropriate for multiple projects.
The Database Example
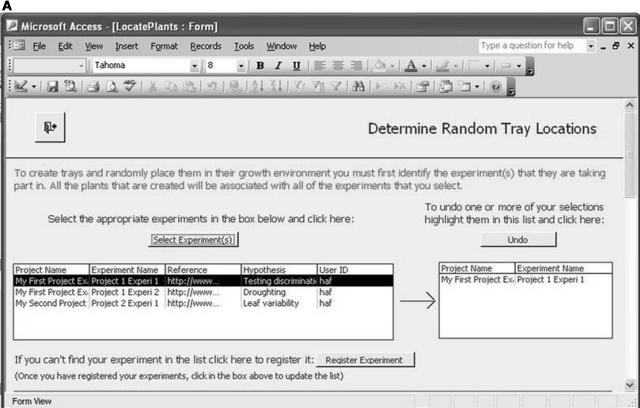
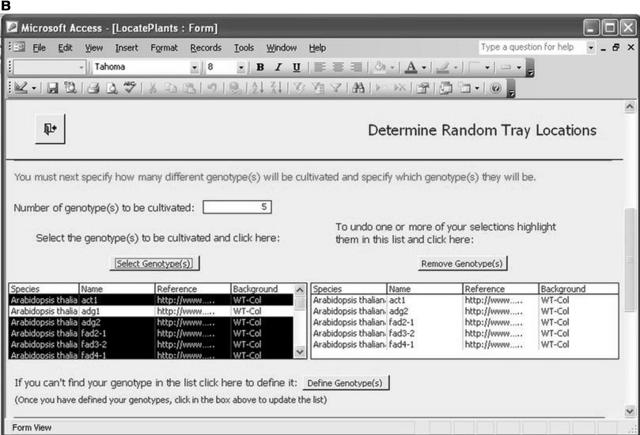
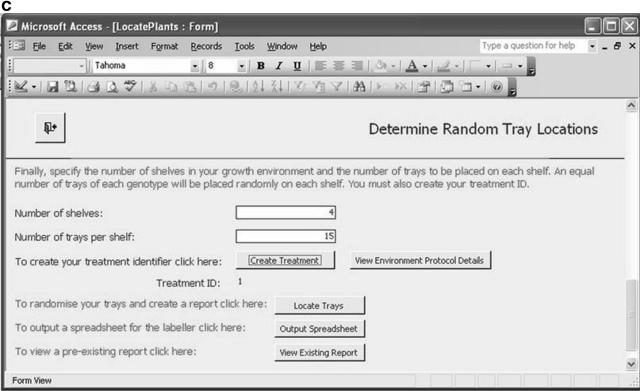
Our first example is a database implementation (in Microsoft Access) of the first five ArMet components. (This example is available as database_example.zip in the supplemental material on the Web). The choice of Microsoft Access was made for reasons of availability and ease of prototyping. There are many other database management systems that may be used to the same effect, and, clearly, custom implementations following other nondatabase approaches are feasible. This software tool was designed to be used on a touchscreen laptop computer in the laboratory and greenhouse in real-time and provides two main functions as follows: (1) recording trays of plants of specified genotypes (that are associated with one or more experiments and cultivated according to a particular treatment regime) and locating those trays within a particular growth environment in a random block design (Figure 2 depicts the form for specifying this information); and (2) support for harvesting the trays (or subsets of the plants that they contain) via forms containing clickable buttons that represent the individual trays within the growth environment.
Figure 2.
The Microsoft Access database implementation of the first five ArMet components. A, Specifying the experiments with which the trays should be associated. B, Specifying the genotypes for the trays. C, Defining the growth environment and treatment regime and locating the trays within the environment.
This software tool tackles all of the key issues that should be addressed by data recording tools as described above. First, it reflects the activities of the experimentalists and hides the component structure of the underlying data model. Second, it builds on the referential integrity inherent in the ArMet data model by providing facilities for entering standard data once and linking it to multiple datasets. For example, Figure 2B, which illustrates genotype selection, shows a list of previously defined genotypes that may be selected to describe the trays in each new experiment, while the “Define Genotype(s)” button enables the user to define additional genotypes that do not appear in the list but would then be available to all subsequent experiments. Finally, it implements ArMet's type constraints and business rules and thereby provides mechanisms for flagging data recording errors as they occur. For example, unless the experiments and genotypes to be associated with a set of trays are specified, the “Locate Trays” functionality will not work and an error message will be displayed. Software tools such as this save experimentalists' time by reducing the two-stage process of manual data recording on data recording sheets followed by transcription of the data to an electronic format to a one-stage process of real-time electronic recording of experimental data. In addition, the provision of reports at the various stages of an experiment, e.g. via the “View Existing Reports” button in Figure 2C, provides hardcopy backups and may be customized to help satisfy institutional recording and reporting requirements.
Currently, this tool has been used to record the growth and harvest of plant material for two experiments. It was developed as a result of a previous experiment in which the data were recorded on data recording sheets and subsequently transcribed to a spreadsheet. Errors in data recording meant that attempts to upload the data to an ArMet-compliant database failed. Using the error messages generated by the database, efforts were made to correct the data which, in turn, uncovered further errors. After three iterations of corrections and upload failure the process was abandoned. Of course, the results of this experiment are now useless and the time spent on growth, harvest, and error correction has been wasted. In comparison, a similar experiment, recorded using the software tool, required 2 weeks to implement the tool but saved experimentalists' time in data recording and transcription and produced a logically correct dataset. The software tool has been used at no additional cost on a subsequent experiment and will be used on further experiments of the same design. To adapt this tool for a different experimental design would entail programming effort, illustrating the fact that highly customized tools are best suited to situations where there are stable methodologies being followed routinely in a project or laboratory. However, within such situations, and given the preexistence of a data model, tools such as these are cheap in the context of a metabolomics experiment and can be built on a “throw-away” basis.
The Spreadsheet Example
In our second example, a spreadsheet (Microsoft Excel) is customized to facilitate ArMet-compliant data recording. (This example is available as spreadsheet_example.zip in the supplementary material on the Web). Spreadsheets are a commonly used tool for producing electronic records of experimental details and are generally liked by experimentalists. They have the apparent benefits of conceptual simplicity and familiarity. In general, they do not provide a robust tool with constraints to ensure logically correct data collection unless they are customized. This software tool was designed to record genotype descriptions and log items of source material within a growth environment.
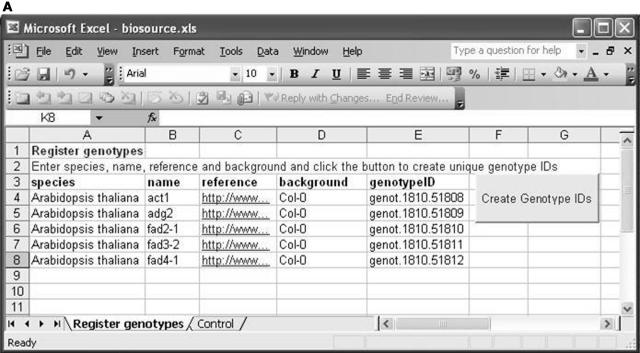
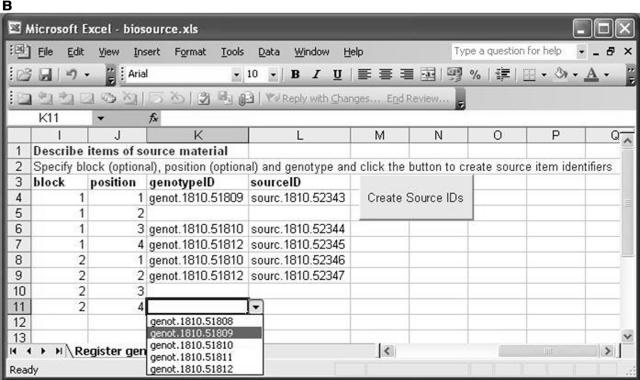
Figure 3A shows the worksheet for genotype description. The user is required to enter values for species, name, reference, and ecotype background to define their genotypes. The “Create Genotype IDs” button will then create a unique identifier for each one. Figure 3B shows the worksheet for logging items of biological source material. This is achieved by specifying their genotype (using the unique identifiers generated at genotype description that are available as a pull down list) and optionally describing their position within the growth environment. The “Create Source IDs” button creates unique identifiers for the required items of source material.
Figure 3.
The Microsoft Excel implementation of the second ArMet component. A, Genotype description. B, Logging items of biological source material.
Like the database-based tool, the spreadsheet-based tool does enforce the collection of logically correct datasets. For example, should any of the required genotype description fields be omitted from a row, and the Create Genotype IDs button pressed, a unique genotype ID will not be generated for that row and the genotype will, therefore, be unavailable for logging items of source material as only previously defined genotypes are available for entry in the genotype ID column in Figure 3B. It also facilitates the provision of standard data once that may then be linked to multiple datasets. For example, if the workbook already contains information for a previous experiment when it is opened, the user is given the option of deleting the experiment specific data (with appropriate warnings about ensuring that the data has been uploaded and/or backed up first). Using this facility, the user can make a copy of an existing workbook to use for a new experiment, choose to delete the information that it contains relating to the previous experiment, and create a new workbook that already contains a list of genotype definitions. Of course when using a spreadsheet-based approach, the well-recognized options of copy and paste and methods for creating links between workbooks may also be used to the same end. Similar spreadsheets could be created for other parts of the experimental timeline and used in real-time to log experimental activity such as harvesting and sample preparation, thereby yielding the benefits described for the database example of one-stage data collection. In addition, all of the spreadsheets may be printed to produce hardcopy backups of the data and institutional records.
In comparison with the database example, this example is less specific to a particular experimental design (for example, it refers to “items of source material” rather than “trays”; an item of source material being any basic unit of plant material used in an experiment, e.g. individual plant, tray, field plot) and, as such, may be used to support experiments in a number of projects or laboratories. Spreadsheet-based data recording is inherently adaptable and extensible and its minimal reliance on programming makes this a simple and quick process. However, this flexibility necessarily results in a software tool that is based less on the activities of the experimentalist and more on the nature of the data that is required. For example, most unique identifiers were hidden from the user in the database example, whereas they are necessary for linking different types of data, e.g. genotypes to items of source material, in the spreadsheet example. This type of tool is, perhaps, more appropriate in environments in which methodologies are being developed and are, therefore, changing rapidly.
Both of these examples rely on the ability to take a computer into the environment of an experiment. Where this is not possible due to practical limitations or institutional regulations, these software tools can be adapted to produce ArMet-compliant data recording sheets and the interfaces altered to support the process of data transcription (including checking for typing errors and logical correctness).
Creating ArMet-Compliant Metadata Datasets from Electronic Records for Preexisting Experiments
Data for preexisting experiments may be recorded in a wide variety of formats. For example, experimental metadata may have been recorded in laboratory notebooks, collected on institutional data recording sheets or alternatively deposited in a laboratory information management system or stored in another electronic format. Before the metadata for a preexisting experiment can be uploaded to an ArMet-compliant database, it must be located and used to create an electronic dataset that complies with, at least, the ArMet core specification. Here, we discuss how this may be achieved for preexisting electronic records. This relates to the “Custom Tools/XSLT” icon (B) in Figure 1.
Where metadata are already held in an electronic storage system, such as a laboratory information management system or non-ArMet database, the options for export to structured text-based formats, e.g. XML, comma-, or tab-delimited files, may be assessed. Once all of the metadata are in a text-based format, they may be merged to produce a single ArMet-compliant dataset. Although this may sound daunting, readily available technologies exist, e.g. XML, which may be used within software tools that hide the underlying complexities and permit automation.
XML is a markup language that is used to produce documents that contain textual data and markup to reflect structure. The markup takes the form of tags delimited by the characters < and > that will be familiar from hypertext markup language (HTML; W3C, 2004). XML may be used to represent semistructured data (Garcia-Molina et al., 2002), i.e. data that is encoded in XML without any constraints on the tags that may be used and therefore on its structure. Alternatively, the legal and required structures for a dataset may be specified and the resulting XML may be checked for correctness or validated against this specification. An example of such a specification is the XML Schema (Sperberg-McQueen and Thompson, 2004) implementation of ArMet (available at http://www.armet.org) that defines a markup language for ArMet-compliant plant metabolomics datasets. It is important to note that this is only one particular schema. It was designed to facilitate the transfer of data to and from relational database implementations of ArMet. Other implementations with other objectives (perhaps to be more succinct or to support Web page display of datasets) are possible.
The contents of an XML document may be transformed into another format using extensible stylesheet language transformations (XSLT; Clark, 1999). Following this approach, the extensible stylesheet language (XSL; Adler et al., 2001) is used to create stylesheets that are applied to XML documents using a software tool known as an XSLT processor. The stylesheets govern the content and structure of the output from this processing.
XML export functionality is provided by a number of widely used software packages. For example, Microsoft Office Access 2003 contains a simple menu option to save XML documents containing the data from a single database table. Alternatively, it provides functionality to generate, through computer code, an XML document containing the data from a number of tables or the results of a database query. The structure of the XML documents generated in this way depends on the decisions made by the programmer. Using Microsoft Office Excel 2003, an XML representation of an entire workbook can be saved, or alternatively, and to enable more selective data export, XML schemas can be loaded into workbooks and their fields mapped to columns in the spreadsheet. The user may then save the data in the mapped columns to an XML document. The structure of this document will conform to the schema that was mapped to the spreadsheet. The example implementations available in the supplementary material on the Web provide examples of this functionality.
In some situations, therefore, preexisting experimental metadata that are stored in electronic formats may be exported directly to ArMet-compliant XML. Where this is not an option, export to non-ArMet XML or other structured text-based formats is often available. Once all of the required data is in XML or another structured text format, XSLT or custom programs can be developed to perform any necessary merging or manipulation to produce a complete ArMet-compliant XML dataset, ensuring that it is logically correct at the same time. These transformations and/or routines can be bundled into a single program and thereby hidden from the data provider who simply has to produce the electronic records in an understood format and pass them to the program.
Metabolome Description Recording
Having addressed experimental metadata recording, we now turn our attention to the output from analytical instruments. ArMet supports this output after it has been processed from raw data to produce a metabolome description, i.e. a peak list or fingerprint or both, together with metadata about how raw machine output was processed to produce the description. Here, we discuss how this data may be captured using examples from our experience of peak lists generated from gas chromatography-mass spectrometry (GC-MS) output and Fourier transform-infrared (FT-IR) fingerprints. Our discussion applies to the analytical technology output from both new and preexisting experiments and relates to the “Custom Tools/XSLT,” icon C in Figure 1.
The extent of data processing required to transform the raw data produced by an analytical instrument into an ArMet-compliant metabolome description depends on the type of instrument and the analytical approach being taken. For example, the production of a quantified peak list from the output of GC-MS involves processing to eliminate noise, locate peaks in the chromatogram, separate out convoluted peaks, and calculate the quantity of sample material that contributes to each peak, whereas to produce an FT-IR fingerprint only background removal is necessary. This processing may be carried out in a number of ways: by the software provided with the analytical instrument, through the use of custom written programs, or by manual processing of the data.
Some analytical instruments will only export data, whether it is raw or processed, in a proprietary format. Where this is the case, it may be possible to write custom programs to convert the proprietary format to a text-based format. For example, a program written in-house may be used to convert the proprietary data format that may be saved from an FT-IR instrument into a file containing tab-delimited ASCII values. This issue will become less important, hopefully, as emerging standards for XML and other representations of analytical data develop, e.g. AnIML (Julian, 2004), JCAMP-DX (Lampen et al., 1999), and as more analytical instrument manufacturers provide output in these formats. However, in the immediate term, and with analytical instruments typically being long-term investments, it seems likely that this problem will persist.
It is evident, therefore, that the structure and format of metabolome descriptions will depend upon one or all of the following: the instruments used to generate them; how they were generated from the raw analytical instrument output; and how they are used and stored at the laboratories at which they are generated. As with electronic metadata records for preexisiting experiments, it may be possible to export them directly to ArMet-compliant XML. Alternatively, custom programs may be written to generate ArMet-compliant XML from other formats or XSLT may be used to transform other non-ArMet XML to ArMet-compliant XML.
DATA UPLOAD SUPPORT
The approaches to data recording described above all result in ArMet-compliant datasets that are held locally at the laboratory or institution that generated them. Assuming that these datasets are to be uploaded to institutional or public ArMet-compliant databases, mechanisms are required to facilitate the dataset transfer and its subsequent submission. This relates to the “Data Upload Tool,” icon D in Figure 1.
XML schemas may be used as definitions of transmission formats and, therefore, the XML schema implementation of ArMet provides a format for transmission of ArMet-compliant datasets between ArMet-compliant data handling tools. This is analogous to the use of HTML as a transmission format for Web documents that is accepted and understood by the range of available HTML-compliant Web browsers from which users may choose. Here, we describe an XML-based approach for uploading ArMet-compliant datasets to a remote ArMet-compliant Oracle 9i relational database. The decision to use the Oracle database management systems was based on the availability of central support for the service. In practice, none of the technologies that we describe are Oracle-specific.
Our approach to data upload is to provide a simple Web page containing a file upload dialog. An example of such a page is available via either of the example implementations provided in the supplementary data on the Web. Once a file is submitted, a simple button click transfers the data to the remote site and submits it to the central database. Thereby, this approach hides all of the underlying technologies from the user.
Datasets recorded locally that have been generated following the approaches described above will all be uploaded as ArMet-compliant XML. This means that the datasets will be complete and further that any necessary manipulation of the data that are uploaded, that is required to translate between ArMet versions, may be carried out by XSLT. These transformations are applied automatically when the file is uploaded, as is the submission of the data to the database tables, thereby addressing the issue of provision of version manipulation in data upload tools.
Underlying this example is Java servlet technology (Bodoff, 2004), which provides a mechanism for extending the functionality of the Web server on which it is hosted, in this case by providing XML and XSLT document handling functionality and database access via ODBC.
The upload of metabolome descriptions often involves multiple files. Usually a number of samples will be run through an analytical instrument at the same time, e.g. by way of a GC-MS autosampler or an FT-IR well plate, resulting in many datasets. To save the user the arduous job of uploading these datasets individually, our approach has been to adapt the servlet to accept and extract a single .zip file that is expected to contain a “control” file and a file for each individual dataset. The control file contains information about the individual dataset files that is used to process them, i.e. apply any necessary transformations, and submit them to the database.
When data is being uploaded from multiple sites to the same central database, the need for unique names to identify objects such as units of source material, samples, and datasets becomes evident. Obviously, if all of the data producing institutions that are populating a single database label samples with whole numbers starting from one, it will be impossible to distinguish between samples once they have been uploaded. Often, institutions have their own complicated naming conventions that produce labels that comprise a number of distinct data items. For example, “UWA_experi1_adg1_1_FT-IR” could mean “The sample of genotype adg1 that was grown in block 1 in the growth environment and analyzed using FT-IR for the experiment called ‘experi1’ carried out at the University of Wales, Aberystwyth.” This type of label has local meaning only, duplicates information that is held in other ArMet fields, and may suffer from incorrect interpretation by other users of the database. A possible solution to this problem is to use simple, but essentially meaningless, unique values to label the data items within the database and ensure that these are never presented to the end user by building or storing names that satisfy local naming conventions for presentation to the users within their interfaces. Temporary values can be generated, as shown in Figure 3A, where unique labels are required for objects within a local data collection software tool for which there are no naming conventions, such as for the genotypes in the spreadsheet example software tool described above. These can be automatically converted to the central database naming system on data upload. Such labels are necessary within the data submission interface as they allow the user to link objects (in this example, genotypes with items of source material), but are unnecessary in other interfaces in which the user would probably be presented with the complete genotype definition. Alternatively, wider naming schemes may be used to produce unique names within the context of a single database. This is analogous to the creation of the “object identification” element of a Life Sciences Identifier (OMG, 2004) or the “id” attribute of a BioMOBY (Wilkinson et al., 2003) triplet. It is a housekeeping job within a single repository, which itself may have identity to ensure global uniqueness.
SUMMARY
We have described how, by using the ArMet data model as a design template, data collection software tools that support the experimental process may be developed. The benefits of such software tools are expected to include: (1) time savings through faster input and less need for checking and correction of input errors; (2) transparent application of quality assurance procedures at the level of data values and the integrity of data sets; and (3) greater acceptance of software tools by the user because they can be quickly and cheaply customized to reflect and support working practices.
Through examples, we have discussed and illustrated a range of approaches to the development of such software tools, covering different requirements. Data collection tools, as described here, are software artifacts that must be specified, designed, built, and tested before they can be used. A data model, such as ArMet, provides a very significant proportion of that effort. Production of a new tool is made much cheaper and quicker and has a much greater likelihood of correct integration with other tools if such a model is used.
We have performed validation of the ArMet data model from a data collection perspective through the development of these tools. We have shown that it is possible to build a customized software tool that implements the first five components of ArMet (Admin, BiologicalSource, Growth, Collection, and SampleHandling) and provides support for staff performing plant growth and harvesting. We have further shown that it is possible to build a general purpose software tool that implements the second ArMet component (BiologicalSource) and provides support for experimental design.
Acknowledgments
The authors acknowledge the United Kingdom Biotechnology and Biological Sciences Research Council for funding further development of ArMet under the Exploiting Genomics Initiative.
The online version of this article contains Web-only data.
References
- Adler S, Berglund A, Caruso J, Deach S, Graham T, Grosso P, Gutentag E, Milowski A, Parnell S, Richman J, et al (2001) Extensible Stylesheet Language (XSL). http://www.w3.org/TR/xsl/
- Anonymous (2002) Microarray standards at last. Nature 419: 323. [DOI] [PubMed] [Google Scholar]
- Bino RJ, Hall RD, Fiehn O, Kopka J, Saito K, Draper J, Nikolau BJ, Mendes P, Roessner-Tunali U, Beale MH, et al (2004) Potential of metabolomics as a functional genomics tool. Trends Plant Sci 9: 418–425 [DOI] [PubMed] [Google Scholar]
- Bodoff S (2004) Java servlet technology. http://java.sun.com/j2ee/tutorial/1_3-fcs/doc/Servlets.html
- Booch G, Rumbaugh J, Jacobson I (1999) The unified modeling language user guide. Addison-Wesley, Reading, MA
- Bray T, Paoli J, Sperberg-McQueen CM, Maler E, Yergeau F (2004. a) Extensible Markup Language (XML) 1.0, Ed 3. http://www.w3.org/TR/2004/REC-xml-20040204/
- Bray T, Paoli J, Sperberg-McQueen CM, Maler E, Yergeau F, Cowan J (2004. b) Extensible Markup Language (XML) 1.1. http://www.w3.org/TR/xml11/
- Brazma A, Hingamp P, Quackenbush J, Sherlock G, Spellman P, Stoeckert C, Aach J, Ansorge W, Ball CA, Causton HC, et al (2001) Minimum information about a microarray experiment (MIAME): toward standards for microarray data. Nat Genet 29: 365–371 [DOI] [PubMed] [Google Scholar]
- Brazma A, Parkinson H, Sarkans U, Shojatalab M, Vilo J, Abeygunawardena N, Holloway E, Kapushesky M, Kemmeren P, Lara GG, et al (2003) ArrayExpress: a public repository for microarray gene expression data at the EBI. Nucleic Acids Res 31: 68–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark J (1999) XSL Transformations (XSLT). http://www.w3.org/TR/xslt
- Garcia-Molina H, Ullman JD, Widom J (2002) Database Systems: The Complete Book. Prentice Hall, Upper Saddle River, NJ
- Glueck SB, Dzau VJ (2003) Our new requirement for MIAME standards. Physiol Genomics 13: 1–2 [Google Scholar]
- Jenkins H, Hardy N, Beckmann M, Draper J, Smith AR, Taylor J, Fiehn O, Goodacre R, Bino RJ, Hall R, et al (2004) A proposed framework for the description of plant metabolomics experiments and their results. Nat Biotechnol 22: 1601–1606 [DOI] [PubMed] [Google Scholar]
- Julian RK (2004) Analytical information markup language. http://animl.sourceforge.net
- Kanehisa M, Goto S (2000) KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res 28: 27–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killion PJ, Sherlock G, Iyer VR (2003) The Longhorn Array Database (LAD): an open-source, MIAME compliant implementation of the Stanford Microarray database (SMD). BMC Bioinformatics 4: 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieger CJ, Zhang P, Mueller LA, Wang A, Paley S, Arnaud M, Pick J, Rhee SY, Karp PD (2004) MetaCyc: a multiorganism database of metabolic pathways and enzymes. Nucleic Acids Res 32: D438–D442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampen P, Lambert J, Lancashire RJ, McDonald RS, McIntyre PS, Rutledge DN, Frohlich T, Davies AN (1999) An extension to the JCAMP-DX standard file format, JCAMP-DX V.5.01 (IUPAC Recommendations 1999). Pure Appl Chem 71: 1549–1556 [Google Scholar]
- Microsoft (2004) Welcome to the MSDN library. http://msdn.microsoft.com/library/
- Mueller LA, Zhang P, Rhee SY (2003) AraCyc: a biochemical pathway database for Arabidopsis. Plant Physiol 132: 453–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- NCBI (2004) Gene expression omnibus. http://www.ncbi.nlm.nih.gov/projects/geo/
- Oliver S (2003) On the MIAME standards and central repositories, of microarray. Comp Funct Genomics 4: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OMG (2004) Life sciences identifiers specification. http://www.omg.org/docs/dtc/04-05-01.pdf
- Orchard S, Hermjakob H, Apweiler R (2003) The proteomics standards initiative. Proteomics 3: 1374–1376 [DOI] [PubMed] [Google Scholar]
- Orchard S, Hermjakob H, Julian RK, Runte K, Sherman D, Wojak J, Zhu WM, Apweiler R (2004) Common interchange of standards for proteomics data: public availability of tools and schema. Proteomics 4: 490–491 [DOI] [PubMed] [Google Scholar]
- SMRS Group (2004) The standard metabolic reporting structure. http://www.smrsgroup.org/
- Spellman PT, Miller M, Stewart J, Troup C, Sarkans U, Chervitz S, Bernhart D, Sherlock G, Ball C, Lepage M, et al (2002) Design and implementation of microarray gene expression markup language (MAGE-ML). Genome Biol 3: RESEARCH0046 [DOI] [PMC free article] [PubMed]
- Sperberg-McQueen CM, Thompson H (2004) XML Schema. http://www.w3.org/XML/Schema
- Taylor C (2004) General proteomics standards. http://psidev.sourceforge.net/gps/index.html
- Taylor CF, Paton NW, Garwood KL, Kirby PD, Stead DA, Yin ZK, Deutsch EW, Selway L, Walker J, Riba-Garcia I, et al (2003) A systematic approach to modeling, capturing, and disseminating proteomics experimental data. Nat Biotechnol 21: 247–254 [DOI] [PubMed] [Google Scholar]
- Wilkinson MD, Gessler D, Farmer A, Stein L (2003) The BioMOBY project explores open-source, simple, extensible protocols for enabling biological database interoperability. Proc Virt Conf Genom and Bioinf 3: 17–27 [Google Scholar]