Abstract
Exposure to microgravity (μg) results in a range of systemic changes in the organism, but may also have beneficial cellular effects. In a previous study we detected increased proliferation capacity and upregulation of genes related to proliferation and survival in boundary cap neural crest stem cells (BC) after MASER14 sounding rocket flight compared to ground-based controls. However, whether these changes were due to μg or hypergravity was not clarified. In the current MASER15 experiment BCs were exposed simultaneously to μg and 1 g conditions provided by an onboard centrifuge. BCs exposed to μg displayed a markedly increased proliferation capacity compared to 1 g on board controls, and genetic analysis of BCs harvested 5 h after flight revealed an upregulation, specifically in μg-exposed BCs, of Zfp462 transcription factor, a key regulator of cell pluripotency and neuronal fate. This was associated with alterations in exosome microRNA content between μg and 1 g exposed MASER15 specimens. Since the specimens from MASER14 were obtained for analysis with 1 week’s delay, we examined whether gene expression and exosome content were different compared to the current MASER15 experiments, in which specimens were harvested 5 h after flight. The overall pattern of gene expression was different and Zfp462 expression was down-regulated in MASER14 BC μg compared to directly harvested specimens (MASER15). MicroRNA exosome content was markedly altered in medium harvested with delay compared to directly collected samples. In conclusion, our analysis indicates that even short exposure to μg alters gene expression, leading to increased BC capacity for proliferation and survival, lasting for a long time after μg exposure. With delayed harvest of specimens, a situation which may occur due to special post-flight circumstances, the exosome microRNA content is modified compared to fast specimen harvest, and the direct effects from μg exposure may be partially attenuated, whereas other effects can last for a long time after return to ground conditions.
Keywords: microgravity, proliferation, delayed effect, neural stem cell, gene expression, exosomes, microRNA
1. Introduction
Exposure to microgravity (μg) results in a range of systemic changes in the organism, reflecting the physiological stress and adaptation associated with this extreme physical environment (Corydon et al., 2023). Space flight conditions have also been shown to alter brain structure and function with more severe effects after long-term space missions (Roy-O'Reilly et al., 2021; Shirah et al., 2022). Microgravity induces stress-related changes in cellular structure and gene expression (Corydon et al., 2023). However, μg also has some beneficial cellular effects and was shown to promote neuronal differentiation of mesenchymal stem cells (Chen et al., 2011), adipose stem cells (Zarrinpour et al., 2017; Graziano et al., 2018) and to promote cardiomyocyte development (Camberos et al., 2019).
Boundary cap neural crest stem cells (BCs) are a transient group of cells located at spinal root exit and entry points during embryonic development and can differentiate into neurons and glia (Hjerling-Leffler et al., 2005; Aldskogius et al., 2009; Trolle et al., 2014; Radomska and Topilko, 2017). In addition to their broad differentiation potential, BCs display a remarkable ability to promote survival and support the function of other cells (Olerud et al., 2009; Grouwels et al., 2012; Ngamjariyawat et al., 2013; Aggarwal et al., 2017). We previously showed that BCs analyzed 1 week after exposure to μg condition on the MASER14 sounding rocket flight markedly increased their post-flight proliferation capacity compared to ground controls, and showed activation of genes associated with survival and differentiation (Han et al., 2021). However, whether these changes were induced by exposure to μg or to hypergravity remain to be clarified. Furthermore, since space flown BCs were harvested after 1 week’s delay, the observed effects could be indirect, i.e., mediated through factors released by BCs to the medium as a result of the space flight conditions.
In the current experiment with MASER15 sounding rocket we asked (i) whether hypergravity or μg induces increased proliferation and altered gene expression in BCs; (ii) whether these effects are detectable also in BCs analyzed directly after the flight; and (iii) whether exosome content in the medium differs after direct harvest (MASER15 experiment) compared to delayed harvest (MASER14 experiment). To distinguish between the influence of μg and hypergravity, BCs were placed in two separate sections in the sounding rocket: one group was exposed to μg, whereas another group was placed in an onboard centrifuge that provided 1 g control condition.
The experiments revealed that exposure specifically to μg induces the increased BC proliferation capacity, which can be detected in specimens harvested shortly after flight and are associated with specific alterations in gene expression. Furthermore, distinct differences were identified in exosome microRNA content between MASER14 (1 week delayed harvest) and MASER15 (5 h delayed harvest).
2. Methods
2.1. Preparation and culture of boundary cap neural crest stem cells
The Regional Ethics Committee for Research on Animals approved all animal procedures. BCs were prepared from transgenic mice harboring red fluorescent protein (RFP) under the universal actin promoter as previously described and cultured under the same conditions (Aldskogius et al., 2009). Briefly, the spinal cord was exposed and dorsal root ganglia, including their attachment with the spinal cord, gently separated and mechano-enzymatically dissociated using collagenase/dispase (1 mg/mL) and DNase (0.5 mg/mL) for 30 min at room temperature. Cells were plated at 0.5–1 × 105 cells/cm2 in N2 medium containing B27 (Gibco) as well as EGF and bFGF (R&D Systems; 20 ng/mL, respectively). After 12 h of culture, cells that had not adhered were removed together with half of the medium, and a fresh medium was added. The medium was changed every second day, and neurospheres could be observed after about 2 weeks of culture.
2.2. Assembly of specimens for insertion to the space module
The final preparations before launch of MASER15 were performed in the Esrange Space Center bio-laboratory.1 SIOUX Technologies 2 provided the special hardware for storing the cells during space flight. For the assembly, a special metal tripod-table was used. All cellular materials were prepared as triplicates at around 0.3 M cells/ml density, placed to membranes within the hard-plastic building blocks. The membranes were sealed airtight with respective membrane lids and remaining bubbles were aspirated by injecting needles (G25, G27) through the membrane lids. Cassettes were installed in the “Late-access unit” (LAU), which functioned as an airtight, pressure-, temperature-controlled incubator aboard the rocket. The center part of the LAU was built as a centrifuge to mimic regular gravitational acceleration (1G), while the outer parts of the LAU were exposed to microgravity (weightlessness) during the flight. The ground control group was cultured in ambient temperature condition on the bench at Esrange throughout the flight until all materials returned to the lab.
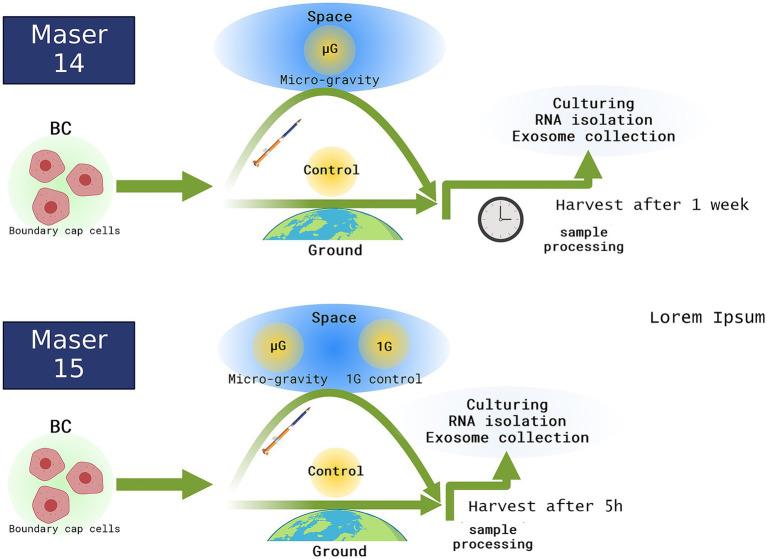
All samples for further analysis were divided into three groups; one part was placed in a centrifuge installed in the experimental module onboard to keep 1 g condition, another part was subjected to μg, and the third part remained on the ground as a control group (Figure 1, overview of the experiment).
Figure 1.
The overview and time line of the experiments in Space. BCs as floating spheres were prepared in the laboratory and placed to rocket 3 h before launch. In Maser 15 experiment just before entering space microgravity the centrifuge was started, maintaining 1G throughout flight whereas the rest of the cells were subjected to microgravity condition. After landing the cells were delivered to the laboratory and after Maser 15 cells were processed directly for analysis, whereas after Maser 14 the cells were retrieved for analysis 1 week after landing.
Microgravity conditions with sounding rocket were achieved between 75 s (at 100 km altitude) and 438.1 s (at 110 km altitude) after lift-off, providing 363 0.4 s of microgravity (source: SSC document “S1X-3 M15 post flight report,” ref.: S1XM-384989335-2407, 24 November 2021).
Microgravity conditions are identified by measuring residual accelerations in the vehicle’s Service Module. For this, a set of calibrated and flight-qualified 3-axis accelerometers (Honeywell Q-Flex® QA-1400 series) were used. Measurement range in fine mode were minus 60.0 mG to 60.0 mG with 4 μG resolution. Sampling frequency was 2,500 Hz (source: SSC document “MASM-2B design report,” ref.: SCIPROJ-1835638381-8199, 20 December 2022).
2.3. Post-flight culture and morphological assessment
Part of the cells subjected to μg or 1 g condition on MASER15 was retrieved from the membranes 5 h after landing for RNA extraction, whereas another group of cells received fresh medium and was placed into 24 -well plates for extension. The cells exposed to μg required every day split due to their extreme rate of proliferation, whereas cells in 1 g group and ground control cells were split every second day according to the standard protocol. This situation resembled the postflight conditions from the previous MASER14 BC experiment (Han et al., 2021) and prompted a comparison of the rate of BC proliferation between these two flights. For the comparison of BC proliferation rate, the neurospheres from Maser 14, Maser 15 and non-flight groups were split to single cells, stained with trypan blue, counted in Burker chamber and seeded to low-affinity 6 well-dishes with the equal concentrations. After 3 days the all cells were collected, split to the single cells and counted in the Burker chamber.
2.4. Transcriptomics analysis of BCs exposed to space flight
2.4.1. AmpliSeq sequencing
For transcriptomic analysis, the medium was replaced immediately after cells were accessible for analysis (5 h after landing) with RNAlater® (#R0901, Sigma-Aldrich), and samples were transported at room temperature and subsequently frozen at −80°C until processed for total RNA extraction using the Aurum™ Total RNA Mini Kit (#7326820, Bio-Rad) according to manufacturer’s instruction. Concentration was determined using NanoDrop™ 1000 (Thermo Fisher). Before sequencing, RNA was quality-controlled using a 2100 Bioanalyzer (Agilent). For sequencing, 10 ng of RNA from each sample was used. Sequencing was performed using an Ion S5™ XL system (Thermo Fisher). The data was processed through the ampliSeqRNA plugin in the Torrent Suite Software.
2.4.2. Differential gene expression analysis
Analysis of gene expression data was performed using R 4.3.1 (Team, 2023) with the DESeq2 1.40.2 (Love et al., 2014) package. Differential expression analysis was performed on raw read counts from AmpliSeq, normalized with the median-of-ratios method in DESeq2. Differentially expressed genes (DEGs) with an absolute log2 fold change (log2FC) above 2.0 and false discovery rate (FDR) adjusted p-value < 0.001 were used for further analysis. The MASER15 cells exposed to μg were compared to ground control and 1 g control. For MASER14, cells exposed to space flight were compared to ground control.
Pathway enrichment analysis was performed for differentially expressed genes in MASER 15 and MASER 14 compared to the respective ground control group using the REACTOME database (Fabregat et al., 2017). Up- and downregulated genes were assessed separately. The top 100 pathways for each analysis were considered. Because similar sets of genes generated hits on multiple related pathways in REACTOME, the genes triggering these hits were manually combined in four non-redundant genes of interest lists (Tables 1–4).
Table 1.
Genes of interest from MASER15, upregulated in cells exposed to space flight μg.
| Gene symbol | Gene description | Assigned function |
|---|---|---|
| Antxr2 | ANTXR Cell Adhesion Molecule 2 | Cell adhesion |
| Camkk2 | Calcium/Calmodulin Dependent Protein Kinase Kinase 2 | Neuronal signaling |
| Ccnd1 | Cyclin D1 | Cell division |
| Chst7 | Carbohydrate Sulfotransferase 7 | Proteoglycan |
| Dph2 | Diphthamide Biosynthesis 2 | Cell cycle |
| Il20rb | Interleukin 20 Receptor Subunit Beta | Immune system |
| Kcng1 | Potassium Voltage-Gated Channel Modifier Subfamily G Member 1 | Neuronal signaling |
| Map3k11 | Mitogen-Activated Protein Kinase Kinase Kinase 11 | Cell division |
| Myc | MYC Proto-Oncogene, BHLH Transcription Factor | Cell division |
| Ptger4 | Prostaglandin E Receptor 4 | Immune system |
| Slc35b3 | Solute Carrier Family 35 Member B3 | Cell division |
| Snx9 | Sorting Nexin 9 | Vesicle biogenesis |
| Srxn1 | Sulfiredoxin 1 | Stress response |
| Tor1aip2 | Torsin 1A Interacting Protein 2 | Stress response |
| Tyw3 | TRNA-YW Synthesizing Protein 3 Homolog | Cell division |
| Uroc1 | Urocanate Hydratase 1 | Histidine metabolism |
Table 4.
Genes of interest from MASER14, down-regulated in cells exposed to space flight.
| Gene symbol | Gene description | Assigned function |
|---|---|---|
| Alox5 | Arachidonate 5-Lipoxygenase | Immune system |
| Bsn | Bassoon Presynaptic Cytomatrix Protein | Vesicle biosynthesis |
| Casp1 | Caspase 1 | Immune system |
| Cftr | CF Transmembrane Conductance Regulator | Other |
| Cnih3 | Cornichon Family AMPA Receptor Auxiliary Protein 3 | Neuronal signaling |
| Cntn6 | Contactin 6 | Cell adhesion |
| D2hgdh | D-2-Hydroxyglutarate Dehydrogenase | Mitochondria |
| Fzd6 | Frizzled Class Receptor 6 | Cell division |
| Gdap1 | Ganglioside Induced Differentiation Associated Protein 1 | Mitochondria |
| Gja10 | Gap Junction Protein Alpha 10 | Cell–Cell adhesion |
| Gja4 | Gap Junction Protein Alpha 4 | Cell–Cell adhesion |
| Grb10 | Growth Factor Receptor Bound Protein 10 | Cell division |
| Grik1 | Glutamate Ionotropic Receptor Kainate Type Subunit 1 | Neuronal signaling |
| Hif3a | Hypoxia Inducible Factor 3 Subunit Alpha | Hypoxia |
| Igf2 | Insulin Like Growth Factor 2 | Cell division |
| L3mbtl1 | L3MBTL Histone Methyl-Lysine Binding Protein 1 | Cell division |
| Loxl4 | Lysyl Oxidase Like 4 | Collagen synthesis |
| Lpar3 | Lysophosphatidic Acid Receptor 3 | Differentiation |
| Mag | Myelin Associated Glycoprotein | Cell–Cell adhesion |
| Mboat4 | Membrane Bound O-Acyltransferase Domain Containing 4 | Other |
| Myh11 | Myosin Heavy Chain 11 | Other |
| Mylk | Myosin Light Chain Kinase | Other |
| Myt1 | Myelin Transcription Factor 1 | Development of nervous system |
| Nkx2-2 | NK2 Homeobox 2 | Development of nervous system |
| Osr1 | Odd-Skipped Related Transcription Factor 1 | Development of nervous system |
| Pgr | Progesterone Receptor | Other |
| Pla2g4a | Phospholipase A2 Group IVA | Immune system |
| Rab33a | RAB33A, Member RAS Oncogene Family | Vesicle biosynthesis |
| Rhd | Rh Blood Group D Antigen | Other |
| Rspo3 | R-Spondin 3 | Cell division |
| Sftpc | Surfactant Protein C | Other |
| Slc17a8 | Solute Carrier Family 17 Member 8 | Neuronal signaling |
| Slc30a10 | Solute Carrier Family 30 Member 10 | Other |
| Slc7a8 | Solute Carrier Family 7 Member 8 | Metabolism |
| Slc9a2 | Solute Carrier Family 9 Member A2 | Other |
| Spn | Sialophorin | Immune system |
| Sptbn2 | Spectrin Beta, Non-Erythrocytic 2 | Cell division |
| Stx3 | Syntaxin 3 | Neuronal signaling |
| Trpm3 | Transient Receptor Potential Cation Channel Subfamily M Member 3 | Neuronal signaling |
| Vtn | Vitronectin | Cell adhesion |
| Wnt2b | Wnt Family Member 2B | Development of nervous system |
| Zp1 | Zona Pellucida Glycoprotein 1 | Other |
The genes of interest were assigned a categorical classification based on functional classification in gene ontology using the DAVID tool (Huang da et al., 2009a,b) and the NCBI gene info database. 3 Further, the predicted cellular localization was obtained using the WegoLoc tool (Chi and Nam, 2012). Data were visualized using MS Excel and Graph Pad Prism V5.0 (GraphPad Software Inc., Boston, MA, United States).
2.5. Exosome analysis
2.5.1. Isolation of BC-derived exosomes
BCs were maintained in culture in DMEM/F12 medium supplemented with N2 and B27 (see above). After the specimens were delivered to the Esrange laboratory, the medium was collected for exosome analysis. Exosome isolation was performed using Amicon® Ultra-15 Centrifugal Filter Unit with Ultracel-100 regenerated cellulose membrane (UFC910024, Millipore, Massachusetts, United States). The cellular medium was centrifuged at 2000 rcf for 30 min at 4°C and washed with PBS at 2000 rcf for 30 min at 4°C. Exosomes kept by the filter were then collected and stored at −20°C.
2.5.2. Exosome observations—transmission electron microscopy
Exosome fixation for TEM was performed using 2% glutaraldehyde solution in phosphate buffer (ratio 1:1). After the fixation phase, exosomes were deposited, rinsed, and stained with heavy metal compounds onto a gridded slide following standard protocols. The visualization of the slide was performed using a TEM Zeiss EM 910 instrument (Zeiss, Oberkochen, Germany).
2.5.3. Exosome characterization with tunable resistive pulse sensing
BC exosome distribution and diameter size were analyzed with the qNano platform (iZON Science, UK). NP150 nanopores and CPC200 calibration particles were used to analyze at 20 mbar pressure. The output was then analyzed with the Izon control suite v3.4 software, which allows for comparing the sample and calibration particles as a comparative reference.
2.5.4. Total exosome RNA extraction and miRNA sequencing
Total RNA extraction from the BC-derived exosomes was performed using the Cell Culture Media Exosome Purification and RNA Isolation Mini Kit (Norgen Biotek Corp., Thorold, Ontario, Canada), following the manufacturer’s instructions. All RNA samples were then stored at −80°C.
Illumina sequencing was used to realize miRNA profiling, which was carried out by Area Science Park (ASP, Trieste, Italy). MiRNA-Seq libraries were realized using the QIAseq miRNA Library Kit (QIAGEN; Hilden, Germany). The sequencing was performed using Novaseq 6000 (Illumina; San Diego, CA, United States) in the 2 × 150 paired-end mode. The identification of miRNAs in the samples was done using the QIAseq miRNA-NGS data analysis software V5, considering single read as the read type and Read 1 Cycles 75 as the read cycles.
2.5.5. Bioinformatic and statistical analysis
MiRNAs from QIAseq miRNA-NGS data analysis software were selected based on read number. The final list of miRNAs obtained was used in enrichment analysis using miRNet software (Chang and Xia, 2023). Functional enrichment analysis of miRNA was realized using miRTareBase v8.0 database as reference. The software was exploited to perform a Gene Ontology Biological Process Enrichment. A p-value < 0.05 was chosen to select data, and Prism 8.03 software graphical view (GraphPad Software Inc., Boston, MA, United States) was used to report enrichment analysis.
For evaluation of BC proliferation, statistics such as One-Way ANOVA and Tukey’s HSD post-hoc test were calculated in RStudio Version 4.0.5. Additionally, the package “ggplot2” was used to plot the graph (Figure 2).
Figure 2.
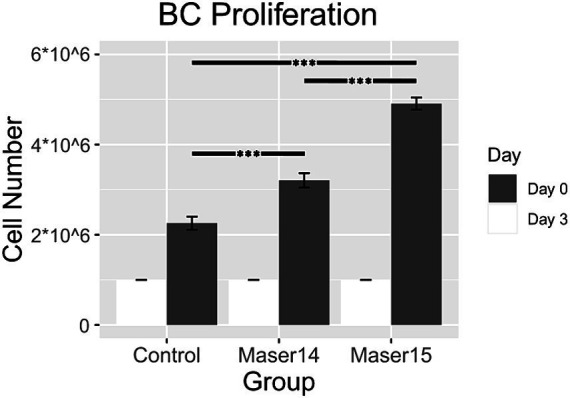
BC proliferation between the ground control group, the cells that were aboard the MASER14, and BC aboard the MASER15 on the seeding day (Day 0) and Day 3 of culture (*p < 0.05, **p < 0.01, ***p < 0.001, as per One-Way ANOVA and Tukey’s HSD post-hoc test).
3. Results
3.1. Space flown BCs show enhanced proliferation
We previously reported that BCs increased their proliferation capacity after a space flight with the sounding rocket MASER14 (Han et al., 2021), for which experimental material was harvested 1 week after landing (delayed harvest). All material from the MASER15 experiment was collected directly after the flight (immediate harvest). Control BCs and BCs from MASER14 and MASER15 μg groups were split into single cells, seeded at 0.3 M cells/mL, and cultured for 3 days, and the number of cells was assessed at the end of the experiment. The results show that 5 h after flight harvested BCs proliferated significantly faster than BCs harvested after a 1 week delay, as well as control BCs (Figure 2).
3.2. Space flown BCs show altered gene expression
Whole transcriptome profiles obtained using the AmpliSeq method for BCs exposed to μg on MASER14 and MASER15 were compared to their corresponding ground control groups. For MASER15, 104 genes were differentially expressed (log2FC > 2.0, FDR adjusted p < 0.001) in μg compared to ground control (42 upregulated and 62 downregulated genes). For MASER14, 479 genes met the fold change limit (334 upregulated and 145 downregulated). BCs from MASER14 were harvested 7 days after μg, while BCs from MASER15 were harvested directly after the flight (5–6 h after μg exposure). Hence, potential differences in the effect on gene expression in relation to time after μg exposure could be evaluated.
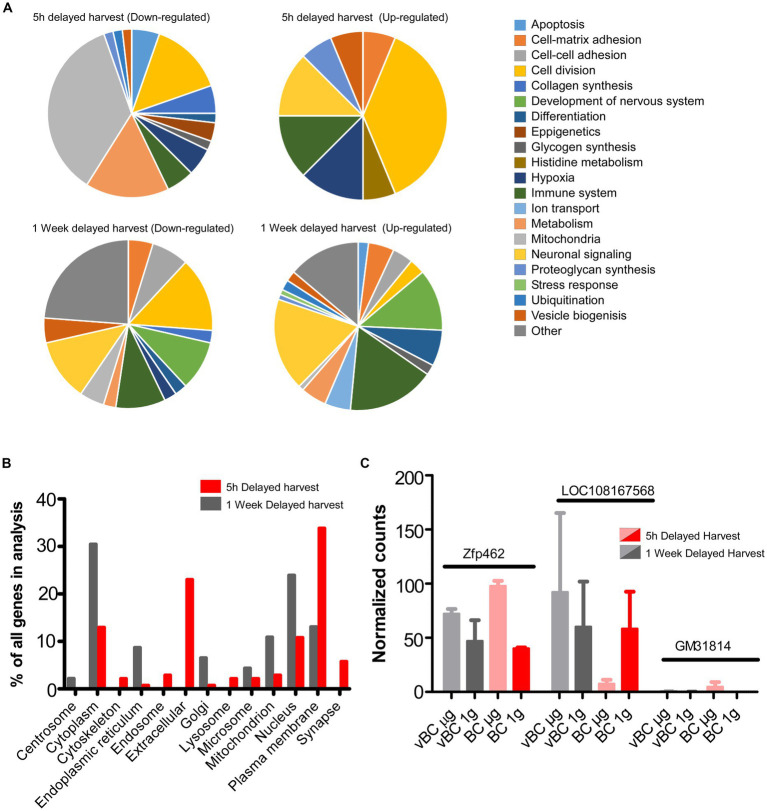
The differentially expressed genes were assessed using the REACTOME pathway database (Fabregat et al., 2017). In MASER15 samples, pathways related to disease, gene expression, signal transduction, cell cycle, and programmed death were found to be enriched (Tables 1, 2). At the same time, in MASER14, enrichment of pathways related to the immune system, transport of small molecules, cellular response to stimuli, and metabolism of proteins were detected (Tables 3, 4). Whether these differences in gene expressions are due to the delayed effect of μg in MASER14 or the changes in stem cells due to the prolonged effect of μg may be addressed in future space experiments with BCs. Genes defining the enrichment of the listed pathways were extracted and used for further analysis (Tables 1–4). The genes were further grouped into 21 classes, including one class named “Other” for singular genes with more deviant categorization, based on the REACTOME enrichment analysis (Figure 3A).
Table 2.
Genes of interest from MASER15, down-regulated in cells exposed to space flight μg.
| Gene symbol | Gene description | Assigned function |
|---|---|---|
| Aldoa | Aldolase, Fructose-Bisphosphate A | Metabolism |
| Bbc3 | BCL2 Binding Component 3 | Apoptosis |
| Bmt2 | Base Methyltransferase Of 25S RRNA 2 Homolog | Metabolism |
| Cbx4 | Chromobox 4 | Cell division |
| Cbx8 | Chromobox 8 | Cell division |
| Ccng2 | Cyclin G2 | Cell division |
| Cdkn1b | Cyclin Dependent Kinase Inhibitor 1B | Cell division |
| Chuk | Component Of Inhibitor Of Nuclear Factor Kappa B Kinase Complex | Immune system |
| Copb1 | COPI Coat Complex Subunit Beta 1 | Vesicle biogenesis |
| Crebrf | CREB3 Regulatory Factor | Cell division |
| Ddit4 | DNA Damage Inducible Transcript 4 | Hypoxia |
| Egln3 | Egl-9 Family Hypoxia Inducible Factor 3 | Hypoxia |
| Ero1l | Endoplasmic Reticulum Oxidoreductase 1 Alpha | Immune system |
| Gadd45b | Growth Arrest And DNA Damage Inducible Beta | Epigenetics |
| Gbe1 | 1,4-Alpha-Glucan Branching Enzyme 1 | Proteoglycan synthesis |
| Gys1 | Glycogen Synthase 1 | Metabolism |
| Herpud1 | Homocysteine Inducible ER Protein With Ubiquitin Like Domain 1 | Apoptosis |
| Higd1a | HIG1 Hypoxia Inducible Domain Family Member 1A | Hypoxia |
| Hk2 | Hexokinase 2 | Metabolism |
| Hoxb2 | Homeobox B2 | Cell division |
| Mmaa | Metabolism Of Cobalamin Associated A | Metabolism |
| Nampt | Nicotinamide Phosphoribosyltransferase | Metabolism |
| Nfkbia | NFKB Inhibitor Alpha | Immune system |
| P4ha1 | Prolyl 4-Hydroxylase Subunit Alpha 1 | Collagen synthesis |
| Pfkl | Phosphofructokinase, Liver Type | Metabolism |
| Pgm2 | Phosphoglucomutase 2 | Metabolism |
| Plod2 | Procollagen-Lysine,2-Oxoglutarate 5-Dioxygenase 2 | Collagen synthesis |
| Pole3 | DNA Polymerase Epsilon 3, Accessory Subunit | Collagen synthesis |
| Ppp1r3c | Protein Phosphatase 1 Regulatory Subunit 3C | Glycogen synthesis |
| Sap30 | Sin3A Associated Protein 30 | Epigenetics |
| Slc2a1 | Solute Carrier Family 2 Member 1 | Metabolism |
| Trib3 | Tribbles Pseudokinase 3 | Apoptosis |
| Ypel5 | Yippee Like 5 | Cell division |
Table 3.
Genes of interest from MASER14, upregulated in cells exposed to space flight.
| Gene symbol | Gene description | Assigned function |
|---|---|---|
| A2m | Alpha-2-Macroglobulin | Immune system |
| Abca1 | ATP Binding Cassette Subfamily A Member 1 | Immune system |
| Adm2 | Adrenomedullin 2 | Other |
| Ank3 | Ankyrin 3 | Cell–Cell contact |
| Anxa2 | Annexin A2 | Neuronal signaling |
| Apobr | Apolipoprotein B Receptor | Metabolism |
| Appl2 | Adaptor Protein, Phosphotyrosine Interacting With PH Domain And Leucine Zipper 2 | Neuronal signaling |
| Asic4 | Acid Sensing Ion Channel Subunit Family Member 4 | Neuronal signaling |
| Atp11b | ATPase Phospholipid Transporting 11B (Putative) | Ion transport |
| Atp1a4 | ATPase Na+/K+ Transporting Subunit Alpha 4 | Ion transport |
| Atp8a2 | ATPase Phospholipid Transporting 8A2 | Ion transport |
| Atxn3 | Ataxin 3 | Cell division |
| C3 | Complement C3 | Immune system |
| C4b | Complement C4B (Chido Blood Group) | Development of nervous system |
| Camp | Cathelicidin Antimicrobial Peptide | Immune system |
| Cd68 | CD68 Molecule | Immune system |
| Chrm2 | Cholinergic Receptor Muscarinic 2 | Neuronal signaling |
| Cldn10 | Claudin 10 | Cell–Cell contact |
| Cldn4 | Claudin 4 | Cell–Cell contact |
| Cldn7 | Claudin 7 | Cell–Cell contact |
| Clu | Clusterin | Metabolism |
| Cox6a2 | Cytochrome C Oxidase Subunit 6A2 | Mitochondria |
| Cp | Ceruloplasmin | Ion transport |
| Cpz | Carboxypeptidase Z | Metabolism |
| Crebrf | CREB3 Regulatory Factor | Cell division |
| Crispld2 | Cysteine Rich Secretory Protein LCCL Domain Containing 2 | Immune system |
| Ctsh | Cathepsin H | Other |
| Cyp26b1 | Cytochrome P450 Family 26 Subfamily B Member 1 | Other |
| Cyp7b1 | Cytochrome P450 Family 7 Subfamily B Member 1 | Other |
| Cystm1 | Cysteine Rich Transmembrane Module Containing 1 | Other |
| Dapk2 | Death Associated Protein Kinase 2 | Apoptosis |
| Dhrs3 | Dehydrogenase/Reductase 3 | Other |
| Dok3 | Docking Protein 3 | Immune system |
| Ecm1 | Extracellular Matrix Protein 1 | Cell adhesion |
| Efemp1 | EGF Containing Fibulin Extracellular Matrix Protein 1 | Cell adhesion |
| Gbe1 | 1,4-Alpha-Glucan Branching Enzyme 1 | Proteoglykan synthesis |
| Gngt2 | G Protein Subunit Gamma Transducin 2 | Other |
| Golga4 | Golgin A4 | Development of nervous system |
| Gria1 | Glutamate Ionotropic Receptor AMPA Type Subunit 1 | Neuronal signaling |
| Grik4 | Glutamate Ionotropic Receptor Kainate Type Subunit 4 | Neuronal signaling |
| Hkdc1 | Hexokinase Domain Containing 1 | Glycogen synthesis |
| Hmox1 | Heme Oxygenase 1 | Other |
| Hrc | Histidine Rich Calcium Binding Protein | Other |
| Htr1b | 5-Hydroxytryptamine Receptor 1B | Neuronal signaling |
| Icam5 | Intercellular Adhesion Molecule 5 | Immune system |
| Il12rb1 | Interleukin 12 Receptor Subunit Beta 1 | Immune system |
| Il23a | Interleukin 23 Subunit Alpha | Immune system |
| Il33 | Interleukin 33 | Immune system |
| Inpp5j | Inositol Polyphosphate-5-Phosphatase J | Development of nervous system |
| Kcnip1 | Potassium Voltage-Gated Channel Interacting Protein 1 | Ion transport |
| Lbp | Lipopolysaccharide Binding Protein | Immune system |
| Lcat | Lecithin-Cholesterol Acyltransferase | Other |
| Lcn2 | Lipocalin 2 | Immune system |
| Lrp4 | LDL Receptor Related Protein 4 | Neuronal signaling |
| Ltbp2 | Latent Transforming Growth Factor Beta Binding Protein 2 | Cell adhesion |
| Matn3 | Matrilin 3 | Cell adhesion |
| Met | MET Proto-Oncogene, Receptor Tyrosine Kinase | Cellular growth |
| Mmp19 | Matrix Metallopeptidase 19 | Cell adhesion |
| Mt1 | Metallothionein 1 | Stress response |
| Myl4 | Myosin Light Chain 4 | Other |
| Ndn | Necdin, MAGE Family Member | Development of nervous system |
| Nedd4l | NEDD4 Like E3 Ubiquitin Protein Ligase | Ubiquitine |
| Nod2 | Nucleotide Binding Oligomerization Domain Containing 2 | Immune system |
| Npas2 | Neuronal PAS Domain Protein 2 | Other |
| Nt5e | 5’-Nucleotidase Ecto | Immune system |
| Ntn1 | Netrin 1 | Differentiation |
| Osmr | Oncostatin M Receptor | Cell division |
| P2rx1 | Purinergic Receptor P2X 1 | Neuronal signaling |
| P2rx6 | Purinergic Receptor P2X 6 | Neuronal signaling |
| Padi2 | Peptidyl Arginine Deiminase 2 | Cell differentiation |
| Pak1 | P21 (RAC1) Activated Kinase 1 | Cell differentiation |
| Pdgfb | Platelet Derived Growth Factor Subunit B | Cellular growth |
| Plch2 | Phospholipase C Eta 2 | Neuronal signaling |
| Pou3f1 | POU Class 3 Homeobox 1 | Cell differentiation |
| Pou3f2 | POU Class 3 Homeobox 2 | Cell differentiation |
| Ptpn6 | Protein Tyrosine Phosphatase Non-Receptor Type 6 | Cell differentiation |
| Qpct | Glutaminyl-Peptide Cyclotransferase | Neuronal signaling |
| Rab18 | RAB18, Member RAS Oncogene Family | Neuronal development |
| Rap2c | RAP2C, Member Of RAS Oncogene Family | Cell differentiation |
| Rarres2 | Retinoic Acid Receptor Responder 2 | Immune system |
| Rgs6 | Regulator Of G Protein Signaling 6 | Other |
| Rims1 | Regulating Synaptic Membrane Exocytosis 1 | Vesicular exocytosis |
| Ripk3 | Receptor Interacting Serine/Threonine Kinase 3 | Apoptosis |
| Scn1b | Sodium Voltage-Gated Channel Beta Subunit 1 | Neuronal signaling |
| Scube3 | Signal Peptide, CUB Domain And EGF Like Domain Containing 3 | Development of nervous system |
| Sema3e | Semaphorin 3E | Neuronal development |
| Slc17a7 | Solute Carrier Family 17 Member 7 | Neuronal signaling |
| Slc22a3 | Solute Carrier Family 22 Member 3 | Neuronal signaling |
| Slc7a11 | Solute Carrier Family 7 Member 11 | Metabolism |
| Slit1 | Slit Guidance Ligand 1 | Neuronal development |
| Sncb | Synuclein Beta | Development of nervous system |
| Sparcl1 | SPARC Like 1 | Neuronal development |
| Stat6 | Signal Transducer And Activator Of Transcription 6 | Immune system |
| Thrsp | Thyroid Hormone Responsive | Metabolism |
| Tnni1 | Troponin I1, Slow Skeletal Type | Other |
| Trpm2 | Transient Receptor Potential Cation Channel Subfamily M Member 2 | Neuronal signaling |
| Trpm7 | Transient Receptor Potential Cation Channel Subfamily M Member 7 | Neuronal signaling |
| Trpv4 | Transient Receptor Potential Cation Channel Subfamily V Member 4 | Neuronal signaling |
| Txnip | Thioredoxin Interacting Protein | Glycogen synthesis |
| Vps37a | VPS37A Subunit Of ESCRT-I | Ubiquitine |
| Vps41 | VPS41 Subunit Of HOPS Complex | Vesicular exocytosis |
Figure 3.
(A) Classification of significantly enriched genes according to Reactome pathways. (B) Classification of significantly changed genes into predicted cellular localization according to WegoLoc classification. (C) Genes with significantly changed expression between 5 h harvest at 1G and 5 h harvest at μg.
Following immediate harvest, the largest groups of upregulated genes were related to proliferation, hypoxia, and immune signaling. Further, genes related to metabolism and proliferation were also most downregulated, emphasizing the impact of space flight on proliferation. Interestingly, we also detected downregulation of genes that prevent epigenetic changes. After delayed harvest, upregulation of genes involved in stress response and development of the nervous system and downregulation of genes involved in cell division and development of the nervous system were detected.
When genes were classified according to cellular localization, there was a higher number of genes localized to the cytoplasm and nucleus in immediately harvested BCs, while the number of genes localized to the plasma membrane and synapses was increased in delayed harvested BCs, compared to ground controls (Figure 3B). Interestingly, a relatively large number of genes on the MASER14 gene lists were found to be extracellular, e.g., neuropeptides, growth factors and cytokines (Table 3).
The MASER15 μg group was further compared to the MASER15 1 g control, which was subjected to all aspects of the flight except μg (Figure 2). Here, only five differentially expressed genes were found (FDR < 0.05, Log2FC > 1.0): 9530082P21Rik, Uox, Zfp462, Malat1, Gm31814, and LOC108167568. Three genes (9530082P21Rik, Uox, and Gm31814) were excluded from further assessment due to expression in only one sample. Upregulated genes were Zfp462, that encodes a zink-finger protein known to regulate survival in early development, and Malat1, which produces a precursor to a non-coding RNA. LOC108167568, encoding a transcription factor without known function but with an active binding site, was found to be downregulated (Figure 3C).
3.3. BC exosome number and morphology differ after direct compared to delayed harvest
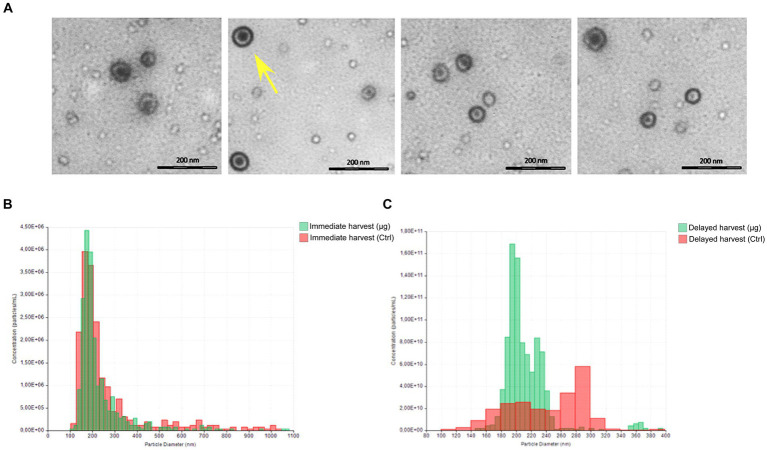
Exosomes were isolated from BC μg and BC ground control medium and characterized. First, they were observed using transmission electron microscopy (TEM). The resulting extracellular vesicles showed a typical bilayer cup-shaped membrane structure, appearing like rounded structures in TEM (Figure 4A). Tunable resistive pulse sensing analysis was used to measure the dimension of the vesicles (Table 5; Figures 4B,C). The analysis of exosome sizes shows similarity between MASER14 and MASER15 μg groups, similar to the control group of MASER15 (directly collected medium after μg exposure). In the MASER14 group, when the control medium was collected with a delay of 1 week, we detected fewer, but larger exosomes (Figure 4C).
Figure 4.
Total BC-derived exosomes characterization. (A) Representative TEM image for each samples. (B,C) Size distribution and concentration using tunable resistive pulse sensing instrument qNano (iZON Science, Oxford, UK).
Table 5.
Average exosome size and concentration in MASER14 and MASER15 BC µG samples, and in corresponding BC control samples.
| Sample | Diameter average (nm) | Concentration |
|---|---|---|
| BC μg (MASER15) | 231 | 1.07e+07 |
| BC Ctrl (MASER15) | 265 | 9.38e+06 |
| BC μg (MASER14) | 212 | 9.17e+11 |
| BC Ctrl (MASER14) | 241 | 2.26e+11 |
3.4. Exosomes from space flown BCs differ from controls in their miRNA content
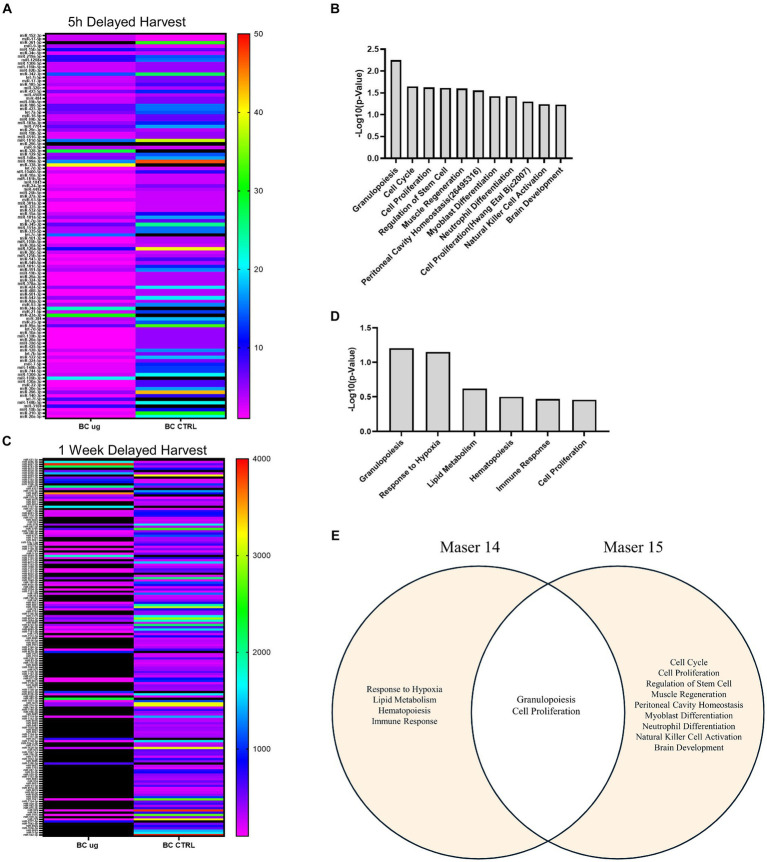
After the isolation, all the exosome content was extracted to perform miRNA sequencing analysis. From the miRNA sequencing analysis of the immediately harvested MASER15 BC μg samples compared to corresponding ground control, 110 miRNAs showed a significant fold-regulation value (cut off: < −2 or > +2) (Figure 5A). Among these significantly altered miRNAs, five were upregulated: miR-152-3p, miR-17-5p, miR-15b-5p miR-361-5p, and miR-9-3p. Of these miRNAs, the first three are involved as regulators of biological processes including proliferation, extracellular matrix production, and apoptosis (Cloonan et al., 2008; Gan et al., 2021; Pinazo-Duran et al., 2023), while miR-361-5p and miR15b-5p are reported to be tumor suppressors and brain-specific miRNA (Ji et al., 2016; Ma et al., 2017; Zhou et al., 2022).
Figure 5.
miRNAs expression profiling analysis. (A) HeatMap on the reads of 110 significant miRNAs of BC μg vs. BC Ctrl (immediate harvest) comparison. (B) MiRNet biological function enrichment on up-regulated of BC μg vs. BC Ctrl (immediate harvest) miRNAs [significant value reported as −Log10(p-value)]. (C) HeatMap on the reads of significant miRNAs of BC μg vs. BC 1G on board (immediate harvest) comparison. (D) MiRNet biological function enrichment on up-regulated of BC μg vs. BC 1G on board (immediate harvest) miRNAs [significant value reported as −Log10(p-value)]. (E) Venn diagram describing the common pathways between Maser14 and Maser15 experiments.
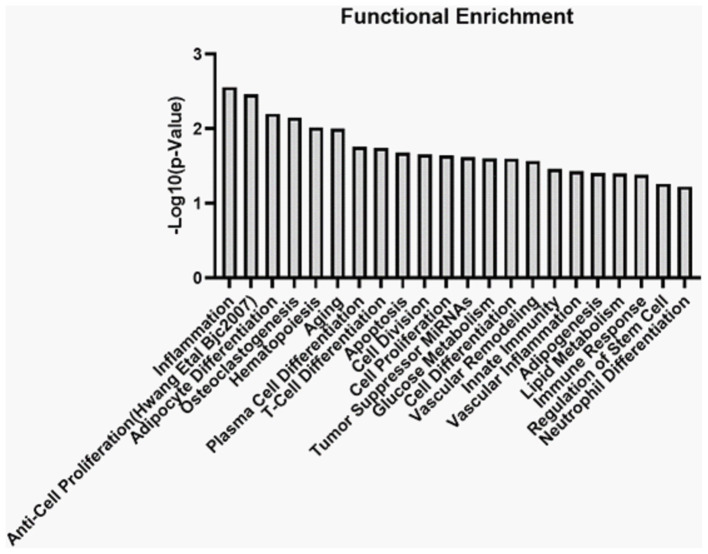
To further explore the function of upregulated, an enrichment analysis was performed using miRNet software. From the miRNet output, only the biological processes with a p-value < 0.05 were selected and reported in a bar graph (Figure 5B). The analysis performed on up-regulated miRNAs shows enrichment in immuno-modulating functions, cell cycle/proliferation mechanism, regulation of stem cells, and brain development. The miRNA sequencing analysis was also performed for MASER15 μg sample compared with the 1 g on board control sample. This analysis yielded 48 significantly altered miRNAs (cut-off: < −1.5 or > +1.5). Of these, 12 were down-regulated and 36 up-regulated in MASER15 μg sample compared to 1 g sample. Among the up-regulated miRNAs is reported let-7b-5p, that represents a regulator of Zfp462 gene resulted as a significant gene in the gene expression analysis.
An identical miRNA analysis performed on BC μg and related control samples following delayed harvest from MASER14, showed a total of 169 significantly altered miRNAs based on fold-regulation value (cut off: < −2 or > +2) (Figure 5C). Eighteen of these miRNAs (miR-651-3p, miR-6886-5p, miR-6867-3p, miR-6761-3p, miR-6879-3p, miR-6828-3p, miR-6890-3p, miR-6869-5p, miR-6877-3p, miR-3192-3p, miR-6826-3p, miR-196b-3p, miR-575, miR-4322, miR-570-3p, miR-4683, miR-5703, miR-579-3p) were upregulated in BC μg condition compared to control. An enrichment analysis of these 18 upregulated miRNAs revealed their involvement granulopoiesis, response to hypoxia, lipid metabolism, hematopoiesis, immune response, and cell proliferation (Figure 5D). A visual Venn Diagram was exploited to highlight the common biological pathways between MASER 14 and MASER 15 (Figure 5E).
Furthermore, overlap analysis performed on the list of miRNAs from MASER14 and MASER15 BC μg condition resulted in the identification of 46 common miRNAs in the samples analyzed (Table 6). An enrichment analysis on these common miRNAs to verify a possible biological process enrichment pattern shows their involvement in immune system mechanisms, differentiation, proliferation and regenerative processes, as well as in glucose and lipid metabolisms (Figure 6).
Table 6.
Common miRNAs in MASER14 and MASER15 BC µg samples.
| Common miRNAs | |
|---|---|
| let-7b-5p | miR-20a-5p |
| let-7c-5p | miR-23a-3p |
| let-7d-5p | miR-24-3p |
| let-7f-5p | miR-25-3p |
| let-7 g-5p | miR-26a-5p |
| let-7i-5p | miR-296-3p |
| miR-101-3p | miR-29a-3p |
| miR-103a-3p | miR-301a-3p |
| miR-10a-5p | miR-335-5p |
| miR-125a-5p | miR-342-3p |
| miR-125b-5p | miR-34a-5p |
| miR-1268a | miR-361-5p |
| miR-128-3p | miR-378a-3p |
| miR-130a-3p | miR-423-3p |
| miR-148a-3p | miR-424-5p |
| miR-148b-3p | miR-532-5p |
| miR-151a-3p | miR-744-5p |
| miR-152-3p | miR-92a-3p |
| miR-16-5p | miR-93-5p |
| miR-181a-5p | miR-9-3p |
| miR-181b-5p | miR-9-5p |
| miR-191-5p | miR-99a-5p |
| miR-196b-5p | miR-99b-5p |
Figure 6.
MiRNet biological function enrichment on 46 common miRNAs [significant value reported as −Log10(p-value)].
4. Discussion
We show that a short flight with sounding rocket enhances the proliferation capacity by BCs and alters their gene expression and exosome miRNA content. The different patterns of gene expression in samples collected 5 h or 1 week after the exposure to μg compared to ground control groups, show that some post-microgravity effects can be detected directly after the flight, whereas other effects can appear later.
We previously showed a markedly increased proliferation capacity in BCs flown with MASER14 sounding rocket (Han et al., 2021). MASER15 BCs showed a remarkably high proliferation rate compared to 1 g samples on board, indicating that this feature was a result specifically of μg exposure during sounding rocket flight. Genetic analysis of the same two groups of MASER15 BCs revealed significant changes in three genes. The Zfp462, which encodes a zink-finger protein known to regulate survival in early development and contribute to cell proliferation (Yelagandula et al., 2023), was up-regulated in μg exposed BCs. Zfp462 secures neural lineage specification of mouse embryonic stem cells (ESCs) by silencing mesoendodermal genes due to targeting histone methyltransferase complex and direct epigenetic regulation.
The other two genes, Malat1 and LOC108167568, are encoding a precursor to a non-coding RNA and a transcription factor without known function, respectively. The change in expression of these genes could initiate increased proliferation and survival in μg-exposed cells at later stages after μg. The role of non-coding RNAs is now under extensive investigation by several groups, and their potential role in gene activation and protein synthesis is suggested (Kehl et al., 2017; Balashanmμgam et al., 2019; Hwang et al., 2023). It has been shown that overexpression of Zfp462 is essential for the development of the nervous system (Chang et al., 2007; Laurent et al., 2009). Zfp462 has also been shown to be crucial for maintaining stemness (Masse et al., 2010, 2011; Yelagandula et al., 2023).
The exosome-derived miRNAs profile from the MASER15 experiment showed up-regulation of the miRNA let-7b-5p in the μg sample compared to 1 g according to the miRTareBase, a reference database for target genes of miRNAs, Zfp462it is one of the target genes of let-7b-5p (Huang et al., 2020). The up-regulation of this miRNA might be expected to decrease or block the expression of Zfp462. However, given the complex effects of μg exposure, with alterations in the activity of multiple genes and changes in miRNA exosomal content, the precise influence of this miRNA is speculative.
When gene activity was compared between MASER14 and MASER15, it was found that protein activity locations are distributed differently. After the flight in MASER15 cells, the primary localization was detected mostly inside the nuclei and the cytosol. In contrast, after the MASER14 flight, the activity of the proteins was localized in the cell membrane and associated with the secretion. This distribution suggests that cells concentrate their activity on their survival during flight. Still, after the flight, they possess new features related to cell–cell communication, which may reflect their supporting characteristics in co-culture (Grouwels et al., 2012; Ngamjariyawat et al., 2013; Aggarwal et al., 2017) and co-transplantation with other cells (Olerud et al., 2009; Grapensparr et al., 2015; Aggarwal et al., 2017).
The differences in gene expression between MASER15 and MASER14 showed that early gene activity in MASER15 was associated with cell division and downregulation of cell adhesion, whereas in MASER14 a gene activity related to pro-inflammatory response and, to a lesser extent, cell division was enriched. In MASER14, an upregulation of genes related to the immune system was detected. However, this designation was a result of Toll-Like Receptor (TLR) activation, which may be associated with cell defense mechanisms in non-immune cells (Song et al., 2019). The extreme resistance of BCs to external stress factors may partly reflect the upregulation of these genes.
The analysis of the number and size of exosomes from MASER14 and MASER15 medium revealed that compared to control cells cultured on the ground, the size in the MASER15 group did not differ. In contrast, in the control group of MASER14, the size of exosomes increased, whereas the number of exosomes was reduced. These findings indicate that the size of exosomes exposed to μg did not change and remained similar to the ground control exosomes, as shown after direct harvest. In contrast, exosomes from MASER14 medium, when cells were in post-flight condition for 1 week, underwent significant modification, suggesting that μg supports maintenance of baseline exosome production, similar to ground control.
We also analyzed miRNA content in the exosomes from the medium of the MASER14 and MASER15 groups. MiRNAs identified in MASER14 exosomes are involved in processes like proliferation, cell cycle, and regulation of stem cell fate. In contrast miRNA exosomes from MASER15 were found to be associated with protection from hypoxia and, to a lesser extent, with cell proliferation (Yan et al., 2020; Wei et al., 2022). These results correlate with the significantly increased proliferation of cells after space flight, as well as the pathway enrichment analysis of the transcriptomic data showing an altered gene expression related to cell proliferation. The mechanisms underlying possible delayed emergence of space flight-induced changes in cellular properties are most likely due to altered gene regulation, e.g., DNA methylation or histone modifications. These alterations may, in turn, lead to a long-lasting change in cell properties, either as a lowered cell intrinsic threshold for entering the cell cycle or by inducing the release of factors that operate in an autocrine or paracrine manner to stimulate proliferation.
There are numerous reports, based on different types of actual and simulated μg exposure, of long-term up- and downregulation of genes and of alterations in the expression of regulatory molecules such as miRNAs in a range of cell types (Corydon et al., 2023). These alterations reflect adaptations associated with cellular stress, but a correlation with beneficial effects has also been demonstrated. Our previous study on BCs exposed to sounding rocket MASER14 flight showed an upregulation of genes related to proliferation and survival (Han et al., 2021). Remarkably, MASER14-flown BCs still showed an increased proliferation rate compared to control BCs 3 years after space flight, though not as high as MASER15 BCs. This agrees with the exosome analysis of MASER14, where miRNAs related to proliferation were altered after delayed harvest, indicating a lingering effect on cell growth.
We conclude that neural crest stem cells increase their proliferation capacity after space flight due to exposure to μg, an outcome that can be detected immediately after space flight, as well as in specimens harvested after a delay. This effect is associated with alterations in gene expression, among which upregulation of the transcription factor Zfp462 may be particularly relevant for the observed increased proliferation capacity. We find a complex pattern of regulation of additional genes, as well as exosomal miRNAs, including regulators involved in cell stress response. Further studies in ground-based simulated and prolonged space μg experiments will help to elucidate the mechanisms of direct and delayed effects of μg and elucidate the metabolic characteristics of BCs during flight conditions, which underlie their remarkable survival capacity in stress conditions. This will improve our understanding of the impact of μg on neural stem cells or other type of cells, and contribute to potential clinical application, such as approaches for controlled and rapid cell renewal for cell replacement therapy and tissue engineering.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics statement
The animal study was approved by Uppsala Regional Committee for the Care and Use of Animals in Research. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
YH: Data curation, Formal analysis, Investigation, Methodology, Validation, Visualization, Writing – review & editing. PB: Data curation, Formal analysis, Investigation, Methodology, Validation, Visualization, Writing – review & editing. LZ: Data curation, Formal analysis, Investigation, Validation, Writing – review & editing. SS: Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Software, Validation, Visualization, Writing – review & editing. FZ: Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Validation, Visualization, Writing – review & editing. ME: Conceptualization, Formal analysis, Resources, Validation, Writing – review & editing. BZ: Data curation, Formal analysis, Investigation, Resources, Supervision, Validation, Visualization, Writing – review & editing. MT: Data curation, Formal analysis, Investigation, Methodology, Writing – review & editing. GF: Data curation, Formal analysis, Validation, Visualization, Writing – review & editing. AV: Investigation, Software, Validation, Writing – review & editing. HA: Data curation, Formal analysis, Project administration, Validation, Writing – review & editing. RF: Conceptualization, Investigation, Methodology, Software, Visualization, Writing – original draft. EK: Conceptualization, Funding acquisition, Methodology, Resources, Supervision, Visualization, Writing – original draft.
Acknowledgments
We are grateful for technical support at Esrange by Stefan Krämer (Swedish Space Corporation) and to Leonardo Surdo and Philippe DeGieter from ESA for help with design and implementation of the study. The authors would also like to acknowledge the support of the National Genomics Infrastructure (NGI)/Uppsala Genome Center and UPPMAX for assisting in massive parallel sequencing and computational infrastructure. Work performed at NGI/Uppsala Genome Center has been funded by RFI/VR and Science for Life Laboratory, Sweden.
Funding Statement
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. We were supported by the Swedish National Space Agency (Dnr 2020-00163, 2021-00089) and Åhlens Foundation (Dnr 2022-230).
Footnotes
Conflict of interest
GF and AV were employed by Swedish Space Corporation.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- Aggarwal T., Hoeber J., Ivert P., Vasylovska S., Kozlova E. N. (2017). Boundary cap neural crest stem cells promote survival of mutant SOD1 motor neurons. Neurotherapeutics 14, 773–783. doi: 10.1007/s13311-016-0505-8, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldskogius H., Berens C., Kanaykina N., Liakhovitskaia A., Medvinsky A., Sandelin M., et al. (2009). Regulation of boundary cap neural crest stem cell differentiation after transplantation. Stem Cells 27, 1592–1603. doi: 10.1002/stem.77, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balashanmμgam M. V., Shivanandappa T. B., Nagarethinam S., Vastrad B., Vastrad C. (2019). Analysis of differentially expressed genes in coronary artery disease by integrated microarray analysis. Biomol. Ther. 10:35. doi: 10.3390/biom10010035, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camberos V., Baio J., Bailey L., Hasaniya N., Lopez L. V., Kearns-Jonker M. (2019). Effects of spaceflight and simulated microgravity on YAP1 expression in cardiovascular progenitors: implications for cell-based repair. Int. J. Mol. Sci. 20:2742. doi: 10.3390/ijms20112742, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y. S., Stoykova A., Chowdhury K., Gruss P. (2007). Graded expression of Zfp462 in the embryonic mouse cerebral cortex. Gene Expr. Patterns 7, 405–412. doi: 10.1016/j.modgep.2006.11.009, PMID: [DOI] [PubMed] [Google Scholar]
- Chang L., Xia J. (2023). MicroRNA regulatory network analysis using miRNet 2.0. Methods Mol. Biol. 2594, 185–204. doi: 10.1007/978-1-0716-2815-7_14, PMID: [DOI] [PubMed] [Google Scholar]
- Chen J., Liu R., Yang Y., Li J., Zhang X., Li J., et al. (2011). The simulated microgravity enhances the differentiation of mesenchymal stem cells into neurons. Neurosci. Lett. 505, 171–175. doi: 10.1016/j.neulet.2011.10.014, PMID: [DOI] [PubMed] [Google Scholar]
- Chi S. M., Nam D. (2012). WegoLoc: accurate prediction of protein subcellular localization using weighted gene ontology terms. Bioinformatics 28, 1028–1030. doi: 10.1093/bioinformatics/bts062, PMID: [DOI] [PubMed] [Google Scholar]
- Cloonan N., Brown M. K., Steptoe A. L., Wani S., Chan W. L., Forrest A. R., et al. (2008). The miR-17-5p microRNA is a key regulator of the G1/S phase cell cycle transition. Genome Biol. 9:R127. doi: 10.1186/gb-2008-9-8-r127, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corydon T. J., Schulz H., Richter P., Strauch S. M., Bohmer M., Ricciardi D. A., et al. (2023). Current knowledge about the impact of microgravity on gene regulation. Cells 12:1043. doi: 10.3390/cells12071043, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabregat A., Sidiropoulos K., Viteri G., Forner O., Marin-Garcia P., Arnau V., et al. (2017). Reactome pathway analysis: a high-performance in-memory approach. BMC Bioinformatics 18:142. doi: 10.1186/s12859-017-1559-2, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan S., Mao J., Pan Y., Tang J., Qiu Z. (2021). Hsa-miR-15b-5p regulates the proliferation and apoptosis of human vascular smooth muscle cells by targeting the ACSS2/PTGS2 axis. Exp. Ther. Med. 22:1208. doi: 10.3892/etm.2021.10642, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grapensparr L., Vasylovska S., Li Z., Olerud J., Jansson L., Kozlova E., et al. (2015). Co-transplantation of human pancreatic islets with post-migratory neural crest stem cells increases β-cell proliferation and vascular and neural regrowth. J. Clin. Endocrinol. Metab. 100, E583–E590. doi: 10.1210/jc.2014-4070, PMID: [DOI] [PubMed] [Google Scholar]
- Graziano A. C. E., Avola R., Perciavalle V., Nicoletti F., Cicala G., Coco M., et al. (2018). Physiologically based microenvironment for in vitro neural differentiation of adipose-derived stem cells. World J. Stem Cells 10, 23–33. doi: 10.4252/wjsc.v10.i3.23, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grouwels G., Vasylovska S., Olerud J., Leuckx G., Ngamjariyawat A., Yuchi Y., et al. (2012). Differentiating neural crest stem cells induce proliferation of cultured rodent islet beta cells. Diabetologia 55, 2016–2025. doi: 10.1007/s00125-012-2542-0, PMID: [DOI] [PubMed] [Google Scholar]
- Han Y., Zeger L., Tripathi R., Egli M., Ille F., Lockowandt C., et al. (2021). Molecular genetic analysis of neural stem cells after space flight and simulated microgravity on earth. Biotechnol. Bioeng. 118, 3832–3846. doi: 10.1002/bit.27858, PMID: [DOI] [PubMed] [Google Scholar]
- Hjerling-Leffler J., Marmigere F., Heglind M., Cederberg A., Koltzenburg M., Enerback S., et al. (2005). The boundary cap: a source of neural crest stem cells that generate multiple sensory neuron subtypes. Development 132, 2623–2632. doi: 10.1242/dev.01852, PMID: [DOI] [PubMed] [Google Scholar]
- Huang da W., Sherman B. T., Lempicki R. A. (2009a). Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13. doi: 10.1093/nar/gkn923, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W., Sherman B. T., Lempicki R. A. (2009b). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57. doi: 10.1038/nprot.2008.211, PMID: [DOI] [PubMed] [Google Scholar]
- Huang H. Y., Lin Y. C., Li J., Huang K. Y., Shrestha S., Hong H.-C., et al. (2020). miRTarBase 2020: updates to the experimentally validated microRNA-target interaction database. Nucleic Acids Res. 48, D148–D154. doi: 10.1093/nar/gkz896, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang J., Jang S., Kim C., Lee S., Jeong H. S. (2023). Role of stem cell-derived exosomes and microRNAs in spinal cord injury. Int. J. Mol. Sci. 24:13849. doi: 10.3390/ijms241813849, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Q., Ji Y., Peng J., Zhou X., Chen X., Zhao H., et al. (2016). Increased brain-specific MiR-9 and MiR-124 in the serum exosomes of acute ischemic stroke patients. PLoS One 11:e0163645. doi: 10.1371/journal.pone.0163645, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehl T., Backes C., Kern F., Fehlmann T., Ludwig N., Meese E., et al. (2017). About miRNAs, miRNA seeds, target genes and target pathways. Oncotarget 8, 107167–107175. doi: 10.18632/oncotarget.22363, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent A., Masse J., Omilli F., Deschamps S., Richard-Parpaillon L., Chartrain I., et al. (2009). ZFPIP/Zfp462 is maternally required for proper early Xenopus laevis development. Dev. Biol. 327, 169–176. doi: 10.1016/j.ydbio.2008.12.005, PMID: [DOI] [PubMed] [Google Scholar]
- Love M. I., Huber W., Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. doi: 10.1186/s13059-014-0550-8, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma F., Zhang L., Ma L., Zhang Y., Zhang J., Guo B. (2017). MiR-361-5p inhibits glycolytic metabolism, proliferation and invasion of breast cancer by targeting FGFR1 and MMP-1. J. Exp. Clin. Cancer Res. 36:158. doi: 10.1186/s13046-017-0630-1, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masse J., Laurent A., Nicol B., Guerrier D., Pellerin I., Deschamps S. (2010). Involvement of ZFPIP/Zfp462 in chromatin integrity and survival of P19 pluripotent cells. Exp. Cell Res. 316, 1190–1201. doi: 10.1016/j.yexcr.2010.02.024, PMID: [DOI] [PubMed] [Google Scholar]
- Masse J., Piquet-Pellorce C., Viet J., Guerrier D., Pellerin I., Deschamps S. (2011). ZFPIP/Zfp462 is involved in P19 cell pluripotency and in their neuronal fate. Exp. Cell Res. 317, 1922–1934. doi: 10.1016/j.yexcr.2011.04.015, PMID: [DOI] [PubMed] [Google Scholar]
- Ngamjariyawat A., Turpaev K., Vasylovska S., Kozlova E. N., Welsh N. (2013). Co-culture of neural crest stem cells (NCSC) and insulin producing beta-TC6 cells results in cadherin junctions and protection against cytokine-induced beta-cell death. PLoS One 8:e61828. doi: 10.1371/journal.pone.0061828, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olerud J., Kanaykina N., Vasylovska S., King D., Sandberg M., Jansson L., et al. (2009). Neural crest stem cells increase beta cell proliferation and improve islet function in co-transplanted murine pancreatic islets. Diabetologia 52, 2594–2601. doi: 10.1007/s00125-009-1544-z, PMID: [DOI] [PubMed] [Google Scholar]
- Pinazo-Duran M. D., Zanon-Moreno V., Garcia-Villanueva C., Martucci A., Peris-Martinez C., Vila-Arteaga J., et al. (2023). Biochemical-molecular-genetic biomarkers in the tear film, aqueous humor, and blood of primary open-angle glaucoma patients. Front. Med. (Lausanne) 10:1157773. doi: 10.3389/fmed.2023.1157773, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radomska K. J., Topilko P. (2017). Boundary cap cells in development and disease. Curr. Opin. Neurobiol. 47, 209–215. doi: 10.1016/j.conb.2017.11.003, PMID: [DOI] [PubMed] [Google Scholar]
- Roy-O'Reilly M., Mulavara A., Williams T. (2021). A review of alterations to the brain during spaceflight and the potential relevance to crew in long-duration space exploration. NPJ Microgravity 7:5. doi: 10.1038/s41526-021-00133-z, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirah B. H., Ibrahim B. M., Aladdin Y., Sen J. (2022). Space neuroscience: current understanding and future research. Neurol. Sci. 43, 4649–4654. doi: 10.1007/s10072-022-06146-0, PMID: [DOI] [PubMed] [Google Scholar]
- Song Y., Shou L. M., Ai L. Y., Bei Y., Chen M. T. (2019). Mini-review: the non-immune functions of toll-like receptors. Crit. Rev. Eukaryot. Gene Expr. 29, 37–45. doi: 10.1615/CritRevEukaryotGeneExpr.2018027399, PMID: [DOI] [PubMed] [Google Scholar]
- Team , R.C. (2023). R: A language and environment for statistical computing. Vienna: R Foundation for Statistical Computing. [Google Scholar]
- Trolle C., Konig N., Abrahamsson N., Vasylovska S., Kozlova E. N. (2014). Boundary cap neural crest stem cells homotopically implanted to the injured dorsal root transitional zone give rise to different types of neurons and glia in adult rodents. BMC Neurosci. 15:60. doi: 10.1186/1471-2202-15-60, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei G., Li S., Wang P., Wang S., Zhao Y. (2022). Altered expression of miR-575 in glioma is related to tumor cell proliferation, migration, and invasion. NeuroMolecular Med. 24, 224–231. doi: 10.1007/s12017-021-08679-7, PMID: [DOI] [PubMed] [Google Scholar]
- Yan G., Zhao H., Hong X. (2020). LncRNA MACC1-AS1 attenuates microvascular endothelial cell injury and promotes angiogenesis under hypoxic conditions via modulating miR-6867-5p/TWIST1 in human brain microvascular endothelial cells. Ann. Transl. Med. 8:876. doi: 10.21037/atm-20-4915, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yelagandula R., Stecher K., Novatchkova M., Michetti L., Michlits G., Wang J., et al. (2023). ZFP462 safeguards neural lineage specification by targeting G9A/GLP-mediated heterochromatin to silence enhancers. Nat. Cell Biol. 25, 42–55. doi: 10.1038/s41556-022-01051-2, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarrinpour V., Hajebrahimi Z., Jafarinia M. (2017). Expression pattern of neurotrophins and their receptors during neuronal differentiation of adipose-derived stem cells in simulated microgravity condition. Iran. J. Basic Med. Sci. 20, 178–186. doi: 10.22038/ijbms.2017.8244, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Xu C., Chao D., Chen Z., Li S., Shi M., et al. (2022). Acute cerebral ischemia increases a set of brain-specific miRNAs in serum small extracellular vesicles. Front. Mol. Neurosci. 15:874903. doi: 10.3389/fnmol.2022.874903, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.