Abstract
The covalent modification of eukaryotic DNA by methylation of the 5′ carbon of cytosine residues is frequently associated with transcriptional silencing. In mammals, a potential mechanism for transducing DNA methylation patterns into altered transcription levels occurs via binding of methyl-CpG-binding domain (MBD) proteins. Mammalian MBD-containing proteins bind specifically to methylated DNA and recruit chromatin-modifying complexes containing histone deacetylase activities. Sequence similarity searches reveal the presence of multiple proteins in plants containing a putative MBD. Outside of the MBD itself, there is no sequence relationship between plant and mammalian MBD proteins. The plant MBD proteins can be divided into eight classes based on sequence similarity and phylogenetic analyses of sequences obtained from two complete genomes (rice [Oryza sativa] and Arabidopsis [Arabidopsis thaliana]) and from maize (Zea mays). Two classes of MBD proteins are only represented in dicot species. The striking divergence of plant and animal MBD-containing proteins is in stark contrast to the amino acid conservation of DNA methyltransferases across plants, animals, and fungi. This observation suggests the possibility that while plants and mammals have retained similar mechanisms for the establishment and maintenance of DNA methylation patterns, they may have evolved distinct mechanisms for the interpretation of these patterns.
Regulation of gene expression is achieved through the combined actions of sequence-specific regulators and nonsequence-specific chromatin-modifying complexes. DNA methylation is one of the most studied modifications to chromatin. In plants and many animal species, a significant fraction of the cytosine residues are covalently modified by the addition of a methyl group to the 5′ carbon. The presence of extensive 5-methylcytosine is strongly correlated with reduced transcriptional activity.
DNA methylation influences transcription through several possible mechanisms. Eden and Cedar (1994) provided evidence that DNA methylation can directly interfere with the binding of sequence-specific transcriptional activators. Another mechanism involves the recruitment of transcriptional repressors by DNA methylation (Kass et al., 1997; Bird and Wolffe, 1999). There is significant evidence in animals that the latter is the predominant mechanism through which DNA methylation is translated to reduced transcription.
Several proteins with the capability to bind specifically to methylated DNA were purified from mammalian systems (Meehan et al., 1989, 1992). Sequence analysis revealed the presence of a common domain, the methyl-CpG-binding domain (MBD), which is sufficient to provide binding to methylated DNA (Nan et al., 1993). The human genome encodes five proteins containing a canonical MBD (MBD1, MBD2, MBD3, MBD4, and MeCP2; Bird and Wolffe, 1999; Hendrich et al., 1999b), as well as an additional six genes containing a closely related TAM domain (Hendrich and Tweedie, 2003).
The mammalian MBD proteins perform multiple functions. MBD1 and MeCP2 act as transcriptional repressors via the recruitment of silencing complexes, specifically recruiting histone deacetylase activity (Nan et al., 1998; Fujita et al., 1999; Ng et al., 2000; Yu et al., 2000). However, MeCP2 can also repress transcription via a separate pathway that does not require histone deacetylation (Yu et al., 2000). Mutations in the human MeCP2 gene result in Rett syndrome, a progressive neurological disorder (Amir et al., 1999). The MBD2 and MBD3 proteins have been identified as part of the Mi2/NURD deacetylase complex (Zhang et al., 1989; Wade et al., 1999). MBD4, which contains a DNA glycosylase domain in addition to the MDB domain, performs DNA repair activities by binding to the sites of spontaneous deamination of methylated bases and repairing the damage (Hendrich et al., 1999a).
Relatively little is known about the molecular mechanisms for interpreting DNA methylation in plants. Several groups have biochemically characterized proteins that bind to methylated DNA (Zhang et al., 1989; Ehrlich, 1993; Pitto et al., 2000), but the molecular identity of these proteins is not known. Our database searches have identified a small family of proteins in plants containing a putative MBD, and these genes have been cataloged and curated by ChromDB (www.chromdb.org). Zemach and Grafi (2003) tested the ability of six Arabidopsis (Arabidopsis thaliana) MBD proteins to bind to methylated DNA and demonstrated specific binding for three of these proteins: AtMBD5, AtMBD6, and AtMBD7. Ito et al. (2003) tested seven Arabidopsis MBD proteins for the ability to specifically bind methylated DNA and found specific binding only for AtMBD5. Scebba et al. (2003) tested a partially overlapping set of six Arabidopsis MBD proteins and found specific binding for three of the proteins: AtMBD5, AtMBD6, and AtMBD11. In combination, these studies present evidence for the ability for AtMBD5, AtMBD6, AtMBD7, and AtMBD11 to specifically bind to methylated DNA, whereas AtMBD1, AtMBD2, AtMBD4, AtMBD8, and AtMBD9 have not shown specific binding abilities in these assays. The AtMBD3, AtMBD10, AtMBD12, and AtMBD13 proteins have not been biochemically tested for methyl-CpG-binding activity. Berg et al. (2003) presented a bioinformatics analysis of the Arabidopsis MBD genes and provided evidence that AtMBD11 is important for proper regulation of development.
We have searched for genes encoding proteins with an MBD in Arabidopsis, maize (Zea mays), and rice (Oryza sativa). Four groups have published reports describing the Arabidopsis MBD genes (Berg et al., 2003; Ito et al., 2003; Scebba et al., 2003; Zemach and Grafi, 2003). There has been some confusion regarding the identity of AtMBD8 and AtMBD9 as these two names have been assigned to different sequences in different manuscripts. Our nomenclature is consistent with that of ChromDB and Zemach and Grafi (2003). We have extended the analysis of previous publications by including the sequences of MBD genes from two monocot species, maize and rice. We have characterized expression profiles of the maize MBD genes using reverse transcription (RT-PCR), and have classified the plant MBD genes by phylogenetic analysis and domain analysis of the encoded proteins. Comparative analysis of monocot and dicot MBD genes reveals extensive duplication as well as the presence of dicot-specific classes of genes.
RESULTS
The MBDs from MBD1 and MeCP2 were used as queries to perform BLASTP and TBLASTN (Altschul et al., 1997) searches against the Arabidopsis and rice nonredundant databases. The similarity of the MBD in the resulting proteins to current domain models was evaluated by performing National Center for Biotechnology Information (NCBI) conserved domain searches (Altschul et al., 1997) and HMMER-based searches using SMART (Schultz et al., 2000). Thirteen Arabidopsis proteins and 16 rice proteins containing a domain with similarity to the MBD were found (Tables I and II). Iterative TBLASTN searches were performed between these species using all MBD proteins to find the complete complement of MBD proteins in the Arabidopsis and rice genomes. The E value for presence of an MBD is indicated in Tables I and II using two different alignment methods (BLAST and HMMER) to either the PFAM MBD or the CDD MBD.
Table I.
Arabidopsis MBD proteins
NA, No score available.
| Gene | Length (Amino Acids) | Protein Accession No.a | AGI Locus No. | Chromosome | Class | HMMER (PFAM MBD) | BLAST (PFAM MBD) | BLAST (CDD MBD) |
|---|---|---|---|---|---|---|---|---|
| MBD1 | 204 | AL161557 | At4g22745 | 4 | II | 4.3E-23 | 6.0E-07 | 3.0E-11 |
| MBD2 | 270 | BAB11480 | At5g35330 | 5 | III | 6.2E-10 | 4.0E-05 | 3.0E-10 |
| MBD3 | 163 | AF013293 | At4g00416 | 4 | II | 0.0012 | 0.044 | NA |
| MBD4 | 186 | CAB87748 | At3g63030 | 3 | II | 6.7E-18 | 4.0E-08 | 1.0E-11 |
| MBD5 | 182 | AAL36205 | At3g46580 | 3 | IV | 1.5E-47 | 7.0E-10 | 1.0E-06 |
| MBD6 | 225 | BAA97474 | At5g59380 | 5 | IV | 4.1E-17 | 4.0E-11 | 6.0E-07 |
| MBD7 | 306 | BAB08347 | At5g59800 | 5 | VI | 0.018 | 0.006 | 9.0E-05 |
| MBD8 | 425, 524b | AAM91660, BAC43440 | At1g22310 | 1 | VII | NA | 0.096 | NA |
| MBD9 | 2,176 | AAF24616 | At3g01460 | 3 | V | 9.3E-46 | NA | NA |
| MBD10 | 384 | AAM19811 | At1g15340 | 1 | I | 3.6E-19 | 0.005 | NA |
| MBD11 | 254 | BAB02310 | At3g15790 | 3 | I | 6.4E-12 | 5.0E-05 | 9.0E-07 |
| MBD12 | 155 | BAB11482 | Not assigned | 5 | III | 4.9E-05 | 4.0E-04 | 4.0E-06 |
| MBD13 | 746 | BAA97466 | At5g52230 | 5 | VIII | 0.84 | NA | NA |
For some MBD proteins, a model different from the predicted annotation is available at www.chromdb.org.
MBD8 is alternatively spliced, resulting in proteins of two different lengths.
Table II.
Maize MBD proteins
N.D., Not determined since available sequence lacks MBD. –, Domain was not predicted.
| Gene | Length (Amino Acids) | Accession No.a | Class | HMMER (PFAM MBD) | BLAST (PFAM MBD) | BLAST (CDD MBD) |
|---|---|---|---|---|---|---|
| MBD101 | 177 | AAK40305 | II | 0.015 | 0.003 | 3.0E-06 |
| MBD105 | 433 | AAK40307 | I | 5.8E-04 | 0.004 | – |
| MBD106 | 428 | AAK40308 | I | 0.0029 | 0.001 | – |
| MBD108 | 297 | AAK40309 | III | 1.2E-07 | 5.0E-05 | 3.0E-09 |
| MBD109 | 400 | AAM93219 | I | 6.6E-10 | 3.0E-04 | 4.0E-08 |
| MBD110 | 215 | AI901690EST | VIII | 0.0044 | 0.011 | – |
| MBD111 | 301 | AAK40310 | III | 6.6E-08 | 2.0E-05 | 4.0E-10 |
| MBD113 | 292 | AY863050 | VIII | 1.2E-25 | 0.021 | 0.25 |
| MBD114 | 336 | BG268492EST | VIII | 0.0015 | 0.011 | – |
| MBD115 | 308 | AY863051 | I | 0.00792 | 0.002 | 1.0E-05 |
| MBD116b | 1,853 | CF034340EST | V | – | – | – |
| MBD117 | 1,176 | CC343391GSS | VII | 1.4 | – | – |
| MBD119c | N.D. | CD441002EST | VII | N.D. | N.D. | N.D. |
| MBD120 | 165 | CC735189GSS | II | 0.011 | 0.016 | 5.0E-05 |
| MBD121c | N.D. | CC414396GSS | VII | N.D. | N.D. | N.D. |
| MBD122 | 432 | CL983464GSS | VIII | 3.6E-05 | 0.009 | 9.0E-04 |
| MBD123 | N.D. | CG233380GSS | VIII | 1.4E-07 | 5.0E-06 | 5.0E-06 |
Accession is for the full-length protein when available. If no full-length sequence is available, a representative EST or GSS is indicated. Contig assemblies for most genes are available at www.chromdb.org.
MBD116 does not contain a MBD but was discovered based on overall sequence similarity to MBD9.
These genes are not fully sequenced, and the current partial sequence lacks a MBD.
The MBDs from MBD1 and MeCP2 and the complete sequences of the Arabidopsis and rice MBD proteins were used to search available maize sequences. Sixteen maize MBD genes were identified (Table III). The full-length sequences of 13 of the 16 maize genes have been obtained by random amplification of cDNA ends or RT-PCR, and these sequences have been deposited at GenBank.
Table III.
Rice MBD proteins
*, Contains multiple MBD domains. –, MBD not predicted.
| Gene | Length (Amino Acids) | BAC Accession No.a | cDNA Accession | Chromosome | Class | HMMER (PFAM MBD) | BLAST (PFAM MBD) | BLAST (CDD MBD) |
|---|---|---|---|---|---|---|---|---|
| MBD701 | 315 | AC099403 | AK101317 | 9 | III | 3.0E-09 | 3.0E-06 | 7.0E-10 |
| MBD703 | 410 | AP003726 | CB097036b | 6 | III | 2.9E-10 | 6.0E-07 | 7.0E-11 |
| MBD704* | 385 | AL663008 | CA759629b | 4 | VIII | 1.2E-08 | 0.065 | – |
| MBD705* | 470 | AL731611 | AK065632 | 4 | VIII | 1.7E-17 | 0.013 | 0.069 |
| MBD706 | 157 | AC104708 | AU057642 | 5 | II | 0.0057 | 0.001 | 8.0E-07 |
| MBD707 | 303 | AL732532 | AK059006 | 12 | I | 2.6E-06 | 0.004 | 1.0E-06 |
| MBD708 | 930 | AP006169 | AK058514 | 9 | VII | 1.3 | – | – |
| MBD709 | 885 | AP004376 | AK069097 | 8 | VII | – | – | 0.93 |
| MBD710c | 1,979 | AP005785 | AK072184 | 2 | V | – | – | – |
| MBD711 | 164 | AC104708 | None | 5 | II | 3.6E-04 | 9.0E-03 | 3.0E-07 |
| MBD712 | 281 | AL662965 | None | 4 | VIII | 0.048 | – | – |
| MBD713* | 378 | AL606989 | None | 4 | VIII | 0.027 | – | – |
| MBD714* | 491 | AL606989 | None | 4 | VIII | 0.0028 | 0.54 | – |
| MBD715 | 728 | AP005918 | AK068243 | 8 | VII | – | – | 0.75 |
| MBD716* | 411 | AL606989 | None | 4 | VIII | 0.0073 | – | – |
| MBD717 | 438 | AL731610 | AK072038 | 4 | VIII | 1.1E-06 | 3.0E-05 | 1.0E-06 |
| MBD718 | 422 | AP003875 | AK071442 | 8 | VIII | 1.1E-06 | 3.0E-05 | 1.0E-04 |
Accession number is for a BAC containing the gene.
Accession number references an EST because no cDNA clone sequences are available.
MBD710 does not actually contain an MBD but was discovered based on overall sequence similarity to MBD9.
This article generally uses the nomenclature of ChromDB, in which genus and species are designated by a number rather than an alphabetical prefix. According to the ChromDB nomenclature, Arabidopsis genes are numbered between 1 and 99, maize genes are numbered between 101 and 199, and rice genes are numbered 701 to 799. However, since some mammalian and Arabidopsis MBD genes have the same name, an alphabetical prefix of At is used for clarity in distinguishing among Arabidopsis and mammalian genes.
In comparison to domains of proteins that carry out biochemical reactions, such as the SET or MET domains, the structural MBD is often more divergent and can have low similarity to consensus domains. For our analysis, we chose to be inclusive and to analyze sequences containing putative MBDs with low similarity to consensus domains, such as AtMBD13. While the sequences in this report are named based on the presence of a putative MBD, it is not expected that all of these proteins will display specific binding to methylated DNA. Only four of nine Arabidopsis genes that have been tested for in vitro activity have displayed the ability to specifically bind to methylated DNA (Ito et al., 2003; Scebba et al., 2003; Zemach and Grafi, 2003).
Phylogenetic Analysis of Plant MBD Proteins
The MBD from the human, Arabidopsis, maize, and rice sequences was aligned using ClustalW (Fig. 1). Several sequences with homology to MBD proteins in other species are not represented in this alignment due to the absence of a detectable MBD (MBD116 and MBD710) or due to the fact that only partial sequence is currently available (MBD119). Several plant proteins have multiple MBDs, and each of these domains was included in the alignment (for example, MBD7d1, d2, or d3), despite the fact that some of these domains are quite divergent. The alignment shows that many of the regions indicated as important by structural and mutational analysis of the mammalian proteins are well conserved in the plant MBD proteins (Ohki et al., 1999; Wakefield et al., 1999). However, as shown by the mammalian MBD3 protein that contains a highly conserved MBD but lacks the ability to specifically bind to methylated DNA (Hendrich and Bird, 1998), this conservation may not lead to specific binding to methylated DNA.
Figure 1.
Alignment of the MBD. The MBDs from human, Arabidopsis, maize, and rice proteins were aligned using ClustalW. The mammalian proteins are indicated by the brackets. The plant MBD proteins were assigned a number based on their species of origin, 1 to 99 for Arabidopsis, 101 to 199 for maize, and 701 to 799 for rice. Within the species designation, the numbers were assigned based on order of discovery. This alignment was shaded using Boxshade. Above and below the alignment are schematic diagrams indicating the structural regions determined for the MeCP2 and MBD1 proteins (Ohki et al., 1999; Wakefield et al., 1999). The position of three β-sheets, an α-helix, and a loop region are shown. The positions of five amino acids thought to be critical for the ability to bind methylated DNA are indicated by the arrows. The first 11 sequences in the alignment are the human proteins MeCP2 (CAA68001), MBD1 (AAD51442), MBD2 (NP_003918), MBD3 (NP_003917), MBD4 (NP_003916), BAZ2A (NP_038477), BAZ2B (NP_038478), TAM1 (XP_002359), TAM2 (BAB71176), CLLL8 (NP_114121), and SETDB1 (AAH28671). The accession numbers for the plant proteins are in Tables I to III.
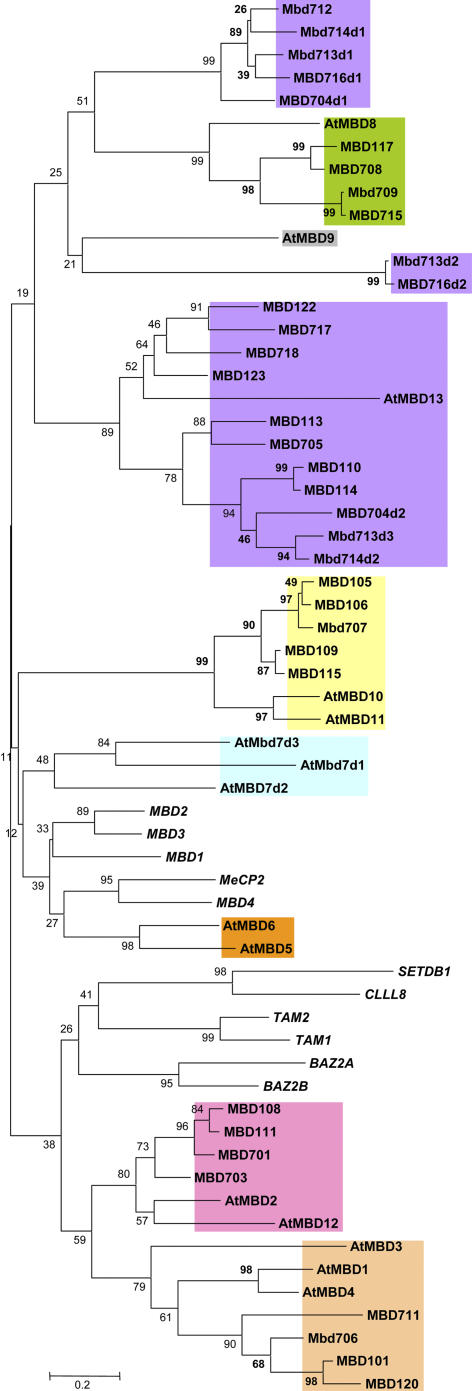
The alignment of the plant and mammalian MBDs was used to perform a neighbor-joining phylogenetic analysis using MEGA (Kumar et al., 2001; Fig. 2). The tree indicates a lack of support for clustering any of the plant MBD proteins with the animal proteins. This finding, based on the MBD alone, is bolstered by the fact that there is no detectable homology between plant and animal MBD proteins outside the MBD itself. The plant proteins were classified into eight different classes according to the phylogenetic analysis and, in addition, consideration of the protein structure and sequence similarity outside of the MBD (Table IV). Interestingly, two of the eight classes (class IV and class VI) are only represented by dicot sequences.
Figure 2.
Phylogenetic analysis of the MBD. The alignment of the MBD, shown in Figure 1, was analyzed by MEGA using neighbor joining (“Materials and Methods”). The classification of each of the proteins is indicated by color. The human sequences are indicated by the use of italics. A bootstrap value is provided for each node. These values indicate the percentage of bootstrap replicates that support this node out of 1,000 samples.
Table IV.
Classes of plant MBD proteins
| Class | Arabidopsis Genes | Maize Genes | Rice Genes |
|---|---|---|---|
| I | Mbd105 | Mbd707 | |
| Mbd10 | Mbd106 | ||
| Mbd11 | Mbd109 | ||
| Mbd115 | |||
| Mbd1 | Mbd101 | Mbd711 | |
| II | Mbd3 | Mbd120 | Mbd706 |
| Mbd4 | |||
| III | Mbd2 | Mbd108 | Mbd701 |
| Mbd12 | Mbd111 | Mbd703 | |
| IV | Mbd5 | – | – |
| Mbd6 | |||
| V | Mbd9 | Mbd116 | Mbd710 |
| VI | Mbd7 | – | – |
| Mbd117 | Mbd708 | ||
| VII | Mbd8 | Mbd119 | Mbd709 |
| Mbd121 | Mbd715 | ||
| Mbd704 | |||
| Mbd110 | Mbd705 | ||
| Mbd113 | Mbd712 | ||
| VIII | Mbd13 | Mbd114 | Mbd713 |
| Mbd122 | Mbd714 | ||
| Mbd123 | Mbd716 | ||
| Mbd717 | |||
| Mbd718 |
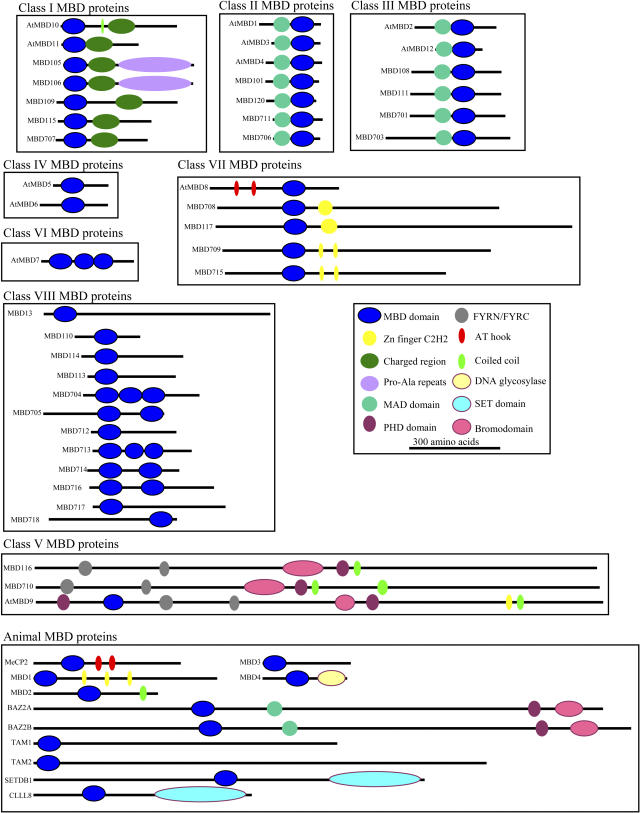
Features of Each Class of Plant MBD Proteins
Class I MBD Genes
The MBD is located within 100 amino acids of the N terminus in all class I MBD proteins (Fig. 3). A region of 80 amino acids that is highly conserved across maize, rice, and Arabidopsis immediately follows the MBD (Supplemental Fig. 1). This domain is conserved across monocot and dicot proteins in this class but does not have homology to other known protein domains. The C-terminal region of the remaining class I MBD proteins contains a charged, Lys/Gln-rich region. The C termini of the class I MBD proteins MBD105 and MBD106 also contain a low-complexity Pro/Ala-rich region that has weak similarity to neurofilament-like repeats, and that is not present in the dicot members of this family.
Figure 3.
Domain organization of the MBD proteins in plants and animals. The protein is represented as a black line; the N terminus is on the left for each representation. The location and size of domains are shown by the use of colored ovals as indicated in the key. The proteins belonging to each class are grouped together.
Class II MBD Genes
The domain architecture of the class II MBD proteins is relatively simple. The proteins are short (163–204 amino acids) with a centrally located MBD. There is significant conservation of the sequence in the N-terminal portion of these proteins that is also found in the class III MBD proteins and was named the MBD-associated domain by Berg et al. (2003; Supplemental Fig. 2). The class II sequences do contain the five residues thought to be critical for specific binding to methylated DNA (Fig. 1) but lack the highly conserved GW located in the β-2 region of the domain. Biochemical assays of AtMBD1 and AtMBD4 failed to detect any specific in vitro binding of methylated DNA (Ito et al., 2003; Scebba et al., 2003; Zemach and Grafi, 2003).
The phylogeny suggests that the presence of multiple class II genes in maize and rice is due to recent duplication events. The genome localization of these genes suggests different mechanisms for the duplication event in rice and maize, as the two maize genes Mbd101 and Mbd120 are mapped to colinear genomic regions of chromosomes 8.04 and 6.05 (K. Cone, unpublished data) and the two rice genes Mbd706 and Mbd711 occur as a tandem duplication within the same bacterial artificial chromosome (BAC).
Class III MBD Genes
The class III MBD genes were grouped together with the class II genes by Berg et al. (2003), but the results of our analysis of phylogeny and sequence similarity across the entire protein have led us to separate class II and class III proteins, as they represent two lineages that apparently have diverged prior to the divergence of monocots and dicots. However, the similarity in these two groups does suggest a more ancient common ancestor. Class III MBD proteins all contain a MBD-associated domain in addition to an MBD (Fig. 3). The MBD of the class III sequences contains all of the residues thought to be critical for specific binding to methylated DNA (Ohki et al., 1999; Wakefield et al., 1999). However, the Arabidopsis AtMBD2 protein did not display specific binding to methylated DNA in vitro (Ito et al., 2003; Scebba et al., 2003; Zemach and Grafi, 2003).
Class IV MBD Genes
The class IV MBD proteins, AtMBD5 and AtMBD6, contain an atypical MBD (Fig. 1). Alignments of the class IV MBDs with the MBDs from other plant and animal proteins indicate the presence of a 30- to 33-amino acid insertion in the loop2 region relative to the other MBDs (the loop2 region is defined according to the alignment shown in Fig. 1 and was reported by Ohki et al. [1999] and Wakefield et al. [1999]). AtMBD5 and AtMBD6 also lack a conserved Lys residue that was implicated in binding to methylated DNA (Ohki et al., 1999; Wakefield et al., 1999). In spite of these differences, the Arabidopsis class IV MBD proteins both preferentially bind methyl-CpG in vitro (Ito et al., 2003; Scebba et al., 2003; Zemach and Grafi, 2003).
The AtMBD5 and AtMBD6 sequences were used to perform BLAST searches of the available maize and rice sequences. No orthologous sequences were found that had more similarity to class IV MBDs than to other classes of MBD proteins. The AtMBD5 and AtMBD6 sequences were also used to perform BLAST searches of expressed sequence tag (EST) databases for all plant species. ESTs closely related to class IV MBD genes were detected in Brassica napus (CD829904), Populus petioles (BU891883), Ipomoea nil (BJ570214), Solanum tuberosum (BQ112257), and Lycopersicon esculentum (AW442004), but none were detected in the ESTs from any monocot species or in the rice genomic sequence. This provides evidence that class IV MBD genes are found only in dicots, not in monocots.
Class V MBD Genes
Our phylogenetic analysis and sequence searches indicate that there is not a sequence closely related to AtMBD9 in rice, maize, or humans. The AtMBD9 protein is a long protein (2,176 amino acids) with the MBD located near the N terminus (Fig. 3). In addition to the MBD, AtMBD9 also contains two PHD domains, a bromodomain and a weak FYRC domain (Fig. 3). The PHD domain is frequently found in proteins that are part of chromatin remodeling complexes, and recent evidence suggests that it may function as a nuclear phosphoinositide receptor (Gozani et al., 2003). The bromodomain is thought to bind to specifically modified histones (Dhalluin et al., 1999). The FYRC domain is found along with the FYRN domain in homologs of the SET domain protein Trithorax (Alvarez-Venegas and Avramova, 2001; Springer et al., 2003).
The full-length AtMBD9 sequence was used to perform BLAST searches to further search for orthologs in maize or rice. Sequences with significant similarity were found in both maize and rice and were named Mbd116 and Mbd710, although the monocot proteins do not contain a detectable MBD. Detailed searches failed to identify an MBD in the genomic sequence surrounding either of these genes (the MBD116 genomic sequence was obtained by a BAC skim of BAC B0265K23 by The Institute for Genomic Research).
The domains found in MBD710 and MBD116 include a PHD domain, a bromodomain, and an FYRC/FYRN domain, similar to the AtMBD9 protein. The overall sequence similarity between AtMBD9, MBD710, and MBD116 suggests that these are orthologous proteins. The class V MBD genes contain many domains commonly associated with chromatin-associated proteins and appear to have lost the MBD during the evolution of rice and maize, or, alternatively, an MBD was gained during the evolution of Arabidopsis.
Class VI MBD Genes
The relatively short AtMBD7 protein (306 amino acids) contains three MBDs. Each of these MBDs was used for the phylogenetic analysis. The MBDs found in AtMBD7 do show evidence of relationships with each other but not with the MBD from any other plant or animal proteins. The close relationship of the three multiple AtMBD7 domains with each other suggests intergenic duplication as a potential source of the multiple MBDs. The AtMBD7 protein was shown to have methyl-CpG-binding activity in vitro by Zemach and Grafi (2003), although Ito et al. (2003) did not find evidence for specific binding.
The AtMBD7 protein sequence was used to search for ESTs from other species of plants related to AtMBD7. Several ESTs from dicot species, including Vitis vinifera (CB008695), I. nil (BJ572810), and S. tuberosum (BQ506651), were identified. However, there were no monocot genes identified with more similarity to AtMBD7 than to other MBD genes. Thus, class VI is likely to represent another dicot-specific class of MBD proteins that possess methyl-CpG-binding activity.
Class VII MBD Genes
The Arabidopsis AtMbd8 is alternatively spliced (Berg et al., 2003), producing two distinct isoforms that both contain two AT-hook domains in addition to the MBD. The MBD of the class VII sequences lacks three of the five residues indicated as critical by Wakefield et al. (1999) and Ohki et al. (1999) for specific binding to methylated DNA by structural or mutational studies (Fig. 1).
The rice predicted protein sequences are much longer than those of the Arabidopsis genes. The MBD708, MBD709, and MBD715 proteins each contain zinc-finger C2H2 domains in addition to the MBD (Fig. 3). The Mbd709 and Mbd715 cDNA sequences are very closely related (96% identical), suggesting that these genes are the result of a recent duplication in the rice genome. Maize contains three class VII MBD genes, Mbd117, Mbd119, and Mbd121. Full-length sequence was obtained for Mbd117 but not Mbd119 or Mbd121. Alignments of the sequence available for MBD119 (287 amino acids) and MBD121 (520 amino acids) with the other class VII MBD proteins suggests that MBD119 and MBD121 are recent duplicates most closely related to MBD709 and MBD715, while MBD117 is most closely related to MBD708 (data not shown). The phylogeny of this group suggests that a single class VII MBD gene existed prior to the divergence of monocots and dicots and duplication events have occurred both before and after the divergence of maize and rice.
Class VIII MBD Genes
Class VIII includes a single Arabidopsis gene, five maize genes, and eight rice genes. The class VIII MBD genes are all relatively short proteins that do not contain any characterized domains except the MBD. Several of the rice genes contain multiple MBDs (Fig. 3). The phylogeny supports conservation of two separate types of MBDs within the rice genes of this class (Fig. 2). The MBD found within these proteins has low homology to MBD consensus domains but is well conserved among members of this group (Tables I–III). The AtMBD13 protein does not align well with the monocot sequences within this class and has a distinct domain architecture relative to the monocot genes. It appears that significant duplication and divergence has occurred within this class of MBD proteins. Alignments of a subset of the monocot sequences identify a region of significant conservation outside of the MBD that does not have homology to any other type of protein in monocots (Supplemental Fig. 3).
Expression of the MBD Genes
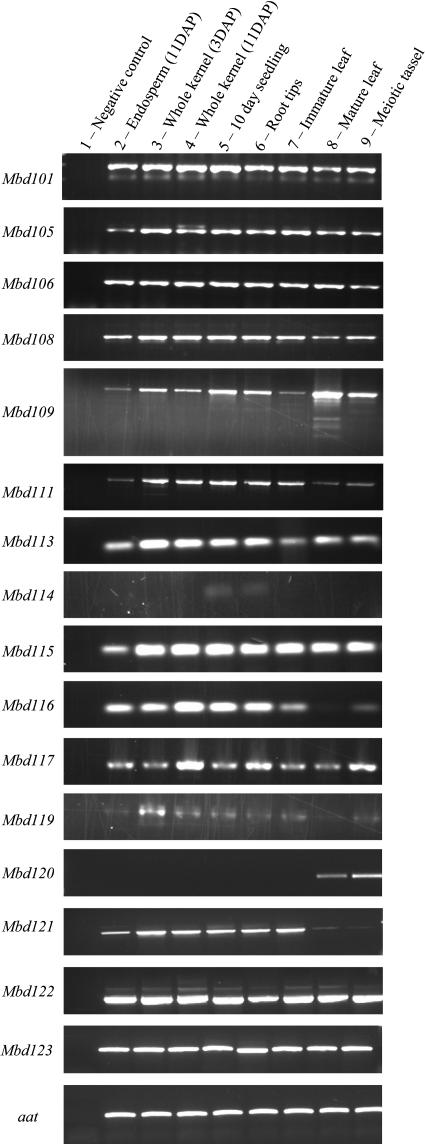
RT-PCR was used to test expression of the 16 maize MBD genes (Fig. 4) in a variety of tissues. Expression was observed for all 16 genes. Twelve genes (Mbd101, Mbd105, Mbd106, Mbd108, Mbd109, Mbd111, Mbd113, Mbd115, Mbd117, Mbd121, Mbd122, and Mbd123) had detectable levels of transcript in all samples tested. There was little evidence for differential regulation of these genes in the developmental stages analyzed, although we have not determined whether there are cell- or tissue type-specific expression patterns of these genes that would not be discernable by our sampling strategy. There were additional bands present in the 11-d-after-pollination whole-kernel tissue amplified with Mbd105 primers and in the mature leaf sample amplified with Mbd109 primers. These may represent tissue-specific alternative splicing products. Mbd110 expression was not detected within these tissues, although we were able to amplify a full-length cDNA from seedling tissue using different protocols. This may indicate that the expression level of Mbd110 is relatively low. Mbd114 transcripts were only detected in 10-d seedlings and root tips. As these were the only samples with root tissue, this suggests and may indicate a root-specific expression pattern for Mbd114. Mbd120 was only detectable in mature leaf and meiotic tissue. Mbd116 and Mbd119 were detected in all tissue tested, although there appears to be a lower expression level of each of these genes in mature leaves. Mbd111 and Mbd121 both displayed lower levels of expression from the mature leaf and meiotic tassel tissue. Southern and northern blots for several of the Arabidopsis and maize MBD genes are available at www.chromdb.org.
Figure 4.
Expression analysis of the maize MBD genes. cDNA was made from RNA isolated from tissues corresponding to the tissue and developmental stage listed above each lane. PCR was performed on these cDNA samples to test for expression of the maize MBD genes. Primers were designed to uniquely amplify cDNA from the gene indicated to the left of the gel image. Controls were performed to test for the presence of genomic DNA. No genomic DNA contamination was detected in any of the eight cDNA samples (data not shown).
DISCUSSION
Plants and Mammals Contain Distinct MBD Proteins
Database searches have identified the complement of MBD proteins in several plant species. Comparisons of the MBD proteins present within plants and mammals reveal no evidence for a common origin of any subgroups of the MBD proteins. There are no examples of plant and mammalian MBD proteins that display conserved sequence outside of the MBD. In several of the mammalian MBD proteins, the regions of the protein outside the MBD contain transcriptional repressive activities or interact with chromatin-modifying complexes such as Mi-2/NURD (Zhang et al., 1989; Wade et al., 1999). The lack of conservation within these regions of the plant proteins suggests that the molecular events initiated by methylation of DNA could be distinct in plants and animals. The lack of conservation in these genes suggests that much of the knowledge regarding the action of mammalian MBD proteins may not be directly transferable to studies of the plant MBD proteins.
In general, the plant MBD proteins do not contain the other domains found in mammalian proteins, including SET domains, bromodomains, CXXC domains, or DNA glycosylase domains. While mammals have evolved a mechanism for the repair of spontaneous deamination of methylated cytosines, there is no evidence for a MBD protein that provides this function in plants. There may be plant MBD proteins that interact with a DNA glycosylase protein to provide this function in plants. With the exception of the class V and VII proteins, the plant MBD proteins do not contain other previously characterized domains. Despite the relative lack of identifiable domains within these sequences, there are a number of regions of significant conservation within classes of these proteins that may represent novel domains or sites for conserved protein-protein interactions (Supplemental Figs. 1–3).
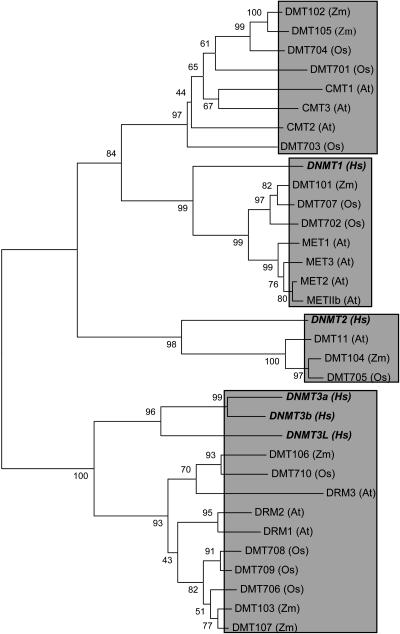
The lack of relationships between subgroups of plant and animal MBD proteins suggests that a single MBD protein may have been present at the time of divergence of plants and animals, and that this protein has undergone independent duplication and divergence events in the two kingdoms. This is distinct from the evolutionary pattern observed for the DNA methyltransferase enzymes (Fig. 5). There is evidence for at least three types of DNA methyltransferase enzyme that were present prior to the divergence of plants and animals. One interpretation is that the mechanisms for creating and maintaining DNA methylation patterns have been preserved in plants and animals, but the mechanisms for interpreting DNA methylation patterns independently evolved in plants and animals.
Figure 5.
Phylogenetic analysis of plant and animal DNA methyltransferase enzymes. A partial sequence of the catalytic domains of the DNA methyltransferases, bound by L(S/D)(L/I)(Y/F) on the N terminus and PPC on the C terminus, was aligned using ClustalX. This alignment was then analyzed by MEGA using neighbor joining (“Materials and Methods”). Bootstrap values (expressed as the percentage of 1,000 samples) are shown at the nodes of the tree. Gray shading is used to indicate the four classes of DNA methyltransferases. The human sequences are indicated by the use of italics. The accession numbers for the sequences are as follows: DNMT1 (AAF23609), DNMT2 (AAC39764), DNMT3a (AAD33084), DNMT3b (AAD53063), DNMT3L (BAA95556), MET1 (AAA32829), MET2 (NP_193150), MET3 (NP_193097), METIIb (NP_192638), DRM1 (CAB89348), DRM2 (AAF66129), DRM3 (AAN12982), CMT1 (NP_565245), CMT2 (AAK69757), CMT3 (AAK69756), DMT11 (NP_568474), DMT101 (ZMET1, AAM28226), DMT102 (ZMET2, AAK11516 ), DMT103 (ZMET3, AAF68437), DMT104 (ZMET4, AAK40306), DMT105 (ZMET5, AAK15805), DMT106 (ZMET6, AAM93211), DMT107 (ZMET7, CA399729), DMT701 (AAN60988), DMT702 (AAP44671), DMT703 (XP_476210), DMT704 (AAP51784), DMT705 (NP_917641), DMT706 (AAN61474), DMT707 (DAA01513), DMT708 (no protein accession available, BAC sequence accession is BX000492), DMT709 (no protein accession available, BAC sequence accession is BX000497), and DMT710 (AAT85176).
Evolution of the Plant MBD Proteins
Within plant species there is ongoing duplication and divergence of the MBD proteins. There is evidence of domain shuffling within the plant MBDs. In dicots, the class V MBD proteins contain an MBD, but the closest related proteins in monocots lack any evidence of an MBD. Two possibilities exist. Either the MBD has been inserted into the dicot gene, or it has been lost in the monocot lineage. The monocot genes are well conserved in other regions of the protein and are expressed, indicating that these genes are likely to still be functional. Genetic analysis of the function of the dicot and monocot class V genes will determine if these proteins have retained similar functions during evolution.
The duplication of the MBD within class VI and VIII proteins provides further evidence for shuffling of the MBD. The phylogeny suggests that the duplication of the MBDs in class VI and in class VIII were distinct events. There is evidence that multiple MBDs within the Arabidopsis class VI protein MBD7 have retained function (Zemach and Grafi, 2003). The phylogenetic analysis suggests that the duplication and triplication of the MBD within the rice class VIII sequences occurred prior to several of the whole-gene duplication events that have occurred.
One of the most interesting findings in our comparative analysis of MBD proteins in maize and rice relative to Arabidopsis was that two classes were specific to dicots. The finding of dicot-specific classes of MBD proteins and highly divergent domain structures (as in class VIII) is in contrast to the high degree of conservation of the DNA methyltransferases and SDG (SET domain group) proteins in monocots and dicots (Cao et al., 2000; Springer et al., 2003). Our analysis did not include a sufficient number of species to determine whether the lack of these classes in monocots is caused by loss of a progenitor class during monocot evolution or by duplication and divergence to create a new class in one lineage. It is striking that the two dicot-specific classes contain three of the four Arabidopsis MBD genes that have demonstrated MBD activity (Ito et al., 2003; Scebba et al., 2003; Zemach and Grafi, 2003). Further research is necessary to determine whether the functions provided by these classes are specific to dicots or whether other MBD proteins in monocots are able to perform the functions of this class. To date, AtMBD11 is the only protein with the ability to specifically bind methylated DNA that has orthologs in monocot species.
The differences in gene content and organization of small gene families between rice and maize did not fit the expectations. Often, it is assumed that the maize genome will contain approximately twice the number of genes as present in the rice genome, with two maize paralogs for each gene present in the rice genome. Our data rarely supported this simplified view of the maize and rice genomes. In some cases, such as classes II, III, V, and VII, rice and maize contained equal numbers of genes. Within these families there were differing organizations, however. The class III sequences from rice occur within the same BAC and are the result of tandem duplication, while the maize genes are likely to be paralogs resulting from the tetraploid origin of maize. The class II sequences in rice are located on different chromosomes and are both expressed, and the maize genes are likely paralogs resulting from the tetraploid ancestry of maize. In class VII, there are three rice and three maize genes, and it appears that there were at least two class VII genes prior to the divergence of maize and rice. Class I contains four maize genes and one rice gene. Class VII has five rice genes and a single maize gene. Based on the data from the MBD genes, it appears that, while maize and rice contain orthologous groups of genes, there have been substantial duplications within each lineage and the actual gene number for any gene family can be quite different in the two species.
If the plant MBD proteins are required for interpretation of DNA methylation patterns correlated with the silencing of gene expression, then it would be expected that mutations in these genes should be recovered in genetic screens for reactivation of silenced transgenes or endogenous genes such as SUP or PAI that are methylated (Lindroth et al., 2001; Malagnac et al., 2002). However, to date, there have been no reports of the isolation of mutations in MBD proteins that affect gene silencing. There are several possible explanations. One potential explanation is that it is less probable to identify loss-of-function mutations in the MBD proteins due to their relatively short length and lack of catalytic motifs. None of the plant MBD proteins contain catalytic motifs, which could make it difficult to recover missense mutations that affect the function of these proteins. A second possible explanation for the failure to recover mutations in MBD proteins in genetic screens is the apparent genetic redundancy of these proteins. The high degree of sequence similarity within the classes of MBD proteins and the overlapping expression patterns suggest that there could be genetic redundancy in the functions performed by the plant MBD proteins. Only three of the classes of MBD proteins in Arabidopsis, two in rice, and one in maize are represented by a single gene. For all of the other classes, there are multiple genes within each class. It is also possible that MBD proteins within different classes may have overlapping functions and may be able to substitute for one another. A third possibility is that there are other types of domains not yet recognized that have the ability to bind to methylated DNA. For example, there is evidence that the mammalian protein Kaiso, which lacks a canonical MBD, has the ability to bind specifically to methylated DNA (Prokhortchouk et al., 2001; Daniel et al., 2002).
We have documented the MBD-containing genes present in three plant species, Arabidopsis, maize, and rice. There are many remaining questions about the biological function of these genes, their genetic redundancy, and their biochemical activities. Currently, we are pursuing an RNAi-based approach to study the functions of these genes in Arabidopsis and maize.
MATERIALS AND METHODS
MBD Gene Discovery and Annotation in Arabidopsis and Rice
The Arabidopsis (Arabidopsis thaliana) and rice (Oryza sativa) MBD protein sequences used in this study were identified by nucleic acid and protein BLAST analysis using MeCP2 (CAA68001) and MBD1 (AAD51442) as queries. The resulting MBD proteins were then used to query the Arabidopsis and rice genomes to find other MBD proteins. The Arabidopsis MBD proteins were assigned numbers between 1 and 13, while the rice proteins are arbitrarily named MBD701 to MBD718. The gene models used for this study and expression data for the Arabidopsis and rice MBD genes are available at www.chromdb.org.
MBD Gene Discovery and Sequencing in Maize
The MBD protein sequences from Arabidopsis and rice were used to search all maize (Zea mays) ESTs and genome survey sequences (GSSs) present in GenBank (last searched November 15, 2004). Putative MBD proteins, identified by automated searching, were arbitrarily named MBD101 to MBD120. In some cases, further sequencing revealed that two ESTs actually corresponded to the same gene and one name was dropped. Full-length sequence for Mbd101, Mbd105, Mbd106, Mbd108, Mbd109, Mbd110, Mbd111, and Mbd113 was obtained by RACE. RACE reactions were performed using the Marathon cDNA kit (CLONTECH, Palo Alto, CA) on cDNA produced from 10-d-old B73 seedlings. Advantage2 polymerase (CLONTECH) was used in the RACE reactions. RACE products were gel purified and cloned into pCR-BluntII (Invitrogen, Carlsbad, CA). Further sequence, mapping, and expression data are available at www.chromdb.org for the many of the maize MBD genes.
Domain Predictions
The protein sequences of all MBD proteins were analyzed for additional recognizable domains using BLAST-based NCBI conserved domain searches (ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi; Marchler-Bauer et al., 2003). The low-complexity filter was turned off and the expect value was set at 1 in order to detect short domains or regions of less conservation in this analysis. The E value determined by these searches that indicates the strength of the alignment to the Pfam version 11.0 (pfam01429) or CDD version 2.02 (cdd1396) MBD is reported in Tables I to III. Domains were also verified using the HMMER-based SMART Web site (http://smart.embl-heidelberg.de/) to search both SMART and PFAM domains (Schultz et al., 2000).
Phylogenetic Analysis
The complete group of nonredundant human, Arabidopsis, maize, and rice MBD proteins was aligned using the MBD with ClustalX 1.83 (Thompson et al., 1997). For all proteins analyzed, the region of the MBD used for the alignment began at the conserved PXGW motif and ended at the FXF motif (not all proteins contain this motif, but the conserved region is used). Neighbor joining was then performed with a bootstrap analysis of 1,000 replicates using MEGA 2.0 (Kumar et al., 2001). The consensus tree was then displayed with bootstrap values.
RT-PCR Analysis
RT-PCR was used to assess expression patterns due to the fact that most of the genes were duplicated. RNA extraction, cDNA synthesis, and PCR conditions were as described by Springer et al. (2003). Primers used for the RT-PCR reactions were FlMbd101F1, TGT GCC CCT CTG CCA CCT CG, and FlMbd101R1, GGA ATT GAC ACG CAG GGG CTT C, for Mbd101; ZmMBD1F1, CGA GAG CGA GAG CAA AGA GCT GAG C, and ZmMBD1R1, CTC TGC CTC CTT GCC AGT TTC AGC, for Mbd105; ZmMBD2F1, GGG CAG AGC AAG AGC TAG GGA TAA CC, and ZmMBD2R1, ATC TCC ACG TCA GTC TCC TTT GTG C, for Mbd106; FlMbd107F1, TAC TAG TGC GGC GTG GAG GTG G, and Mbd107R2, CGG TCC TCT TTA GTA TGC AGG TCC CC, for Mbd107; FlMbd108F1, CGG ACT TCG ATA TCT TCG GAG ACC, and FlMBD108R1, GAT TAG ATC CGT GGT GCA GCA GAA C, for Mbd108; FlMbd109F2, GGA AAC TCG AAA GCC CGG CG, and FlMbd109R2, CGT CAC GTT ACA ACA GTT GGA GAC AG, for Mbd109; FlMbd111F1, CTC CAT TTG GAC CAC CGG GAC C, and FlMbd111R1, GAC ATT TCA AAA CCT TTG CTA CTG CC, for Mbd111; Mbd113F1, GTT TAC CAG ATG GAT GGG TGA AAG, and Mbd113R3, CCC ACC ACA GAT ATC AAC TTC CTC, for Mbd113; Mbd114F1, TCA ATC ACT GGT CTA CGA TTG CTG, and Mbd114R3, TGA ACT GTC AAG TCT TGC AAT GTG, for Mbd114; Mbd115F1, TGT AGA TGC AGC AGA GAA GAC TGG, and Mbd115R1, CAA ATG CGA GGT ATC GTC CTA AAG, for Mbd115; Mbd116F1, TGT ATT CTG GGG TAC TTT TGT ACGG, and Mbd116R1, TTA GCT GTT TCT TCC ATG AGT GG, for Mbd116; Mbd117F1, GCG ATA GCG AGT TCC TCT CTC C, and Mbd117R1, GCT GAC GTA GCT CTT CCC CAT AC, for Mbd117; Mbd118F1, CTA ATG ATG ACA CGG CTT GTA AGG, and Mbd118R1, TGA CAT TAC TCA ACT GGG CAA GAC, for Mbd118; Mbd119F1, ATT CTG TAC CCA CTG AAC CCT CAC, and Mbd119R1, CTA TCT TTA CAG GTG GGG CAA ATG, for Mbd119; Mbd120F1, CCC CGC ATC GCC TCT ATC G, and Mbd120R1, GGC CTT GGC AAC CTT GCA G for Mbd120; Mbd121F1, AGT GCT AGC CAG AAT GCC AAT AGT C, and Mbd121R1, TTT GAC TGG GCA TGT TAA CAA ACT G, for Mbd121; Mbd122F1, GGC GTA ATT ATG GAT TCT TTT GAG G, and Mbd122R1, GTG TCT GTC TGT GTG CCA ATA TGT C, for Mbd122; Mbd123F1, CAA GAC TGT AAG CAA GGA CAA AAG G, and Mbd123R1, TCA AGT TCT CAG GCT CTG GTA ACA C, for Mbd123; and AatF1, ATG GGG TAT GGC GAG GAT, and AatR1, TTG CAC GAC GAG CTA AAG ACT, for Ala aminotransferase (AF055898). Conditions of the PCR were as follows: 94°C for 2 min, 35 cycles of 94°C for 30 s, 63°C for 30 s, 72°C for 2 min, followed by 72°C for 7 min. Amplified products were separated in a 1% agarose Tris borate EDTA gel and visualized by ethidium bromide staining.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers AAK40305, AAK40307, AAK40308, AAK40309, AAM93219, AAK40310, AY863050, and AY863051.
Acknowledgments
We thank Sarah Kerns, Virginia Zaunbrecher, and Laura Schmitt for help with cloning and sequencing; and Karen Cone, Dean Bergstrom, and Miriam Hankins for generating DNA gel-blot data and northern blots for several of the maize MBD genes. The curation of the MBD genes has been performed by Carolyn Napoli at Chromdb.org. We are thankful for suggestions and editing by Carolyn Napoli, Vicki Chandler, Karen McGinnis, Karen Cone, Heidi Kaeppler, and several anonymous reviewers.
This work was supported by the National Science Foundation (DBI–9975930).
The online version of this article contains Web-only data.
References
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Venegas R, Avramova Z (2001) Two Arabidopsis homologs of the animal trithorax genes: a new structural domain is a signature feature of the trithorax gene family. Gene 271: 215–221 [DOI] [PubMed] [Google Scholar]
- Amir RE, Van den Veyver I, Wan M, Tran CQ, Francke U, Zoghbi HY (1999) Rett syndrome is caused by mutations in X-linked MeCP2, encoding methyl-CpG-binding protein 2. Nat Genet 23: 185–188 [DOI] [PubMed] [Google Scholar]
- Berg A, Meza TJ, Mahic M, Thorstensen T, Kristiansen K, Aalen RB (2003) Ten members of the Arabidopsis gene family encoding methyl-CpG-binding domain proteins are transcriptionally active and at least one, AtMBD11, is crucial for normal development. Nucleic Acids Res 31: 5291–5304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird AP, Wolffe AP (1999) Methylation-induced repression—belts, braces and chromatin. Cell 99: 451–454 [DOI] [PubMed] [Google Scholar]
- Cao X, Springer NM, Muszynski MG, Phillips RL, Kaeppler S, Jacobsen SE (2000) Conserved plant genes with similarity to mammalian de novo DNA methyltransferases. Proc Natl Acad Sci USA 97: 4979–4984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel JM, Spring CM, Crawford HC, Reynolds AB, Baig A (2002) The p120(ctn)-binding partner Kaiso is a bi-modal DNA-binding protein that recognizes both a sequence-specific consensus and methylated CpG dinucleotides. Nucleic Acids Res 30: 2911–2919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhalluin C, Carlson JE, Zeng L, He C, Aggarwal AK, Zhou MM (1999) Structure and ligand of a histone acetyltransferase bromodomain. Nature 399: 491–496 [DOI] [PubMed] [Google Scholar]
- Eden S, Cedar H (1994) Role of DNA methylation in the regulation of transcription. Curr Opin Genet Dev 4: 225–259 [DOI] [PubMed] [Google Scholar]
- Ehrlich KC (1993) Partial purification of a pea seed DNA-binding protein that specifically recognizes 5-methylcytosine. Prep Biochem 23: 423–438 [DOI] [PubMed] [Google Scholar]
- Fujita N, Takebayashi S, Okumura K, Kudo S, Chiba T, Saya H, Najao M (1999) Methylation-mediated transcriptional silencing in euchromatin by methyl-CpG binding protein MBD1 isoforms. Mol Cell Biol 19: 6415–6426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gozani O, Karuman P, Jones DR, Ivanov D, Cha J, Lugovskoy AA, Baird CL, Zhu H, Field SJ, Lessnick SL, et al (2003) The PHD finger of the chromatin-associated protein ING2 functions as a nuclear phosphoinositide receptor. Cell 114: 99–111 [DOI] [PubMed] [Google Scholar]
- Hendrich B, Abbott C, McQueen H, Chambers D, Cross S, Bird A (1999. b) Genomic structure and chromosomal mapping of the murine and human Mbd1, Mbd2, Mbd3 and Mbd4 genes. Mamm Genome 10: 906–912 [DOI] [PubMed] [Google Scholar]
- Hendrich B, Bird A (1998) Identification and characterization of a family of mammalian methyl-CpG-binding protein. Mol Cell Biol 18: 6538–6547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrich B, Hardeland U, Ng H, Jiricny J, Bird A (1999. a) The thymine glycosylase MBD4 can bind to the product of deamination at methylated CpG sites. Nature 401: 301–304 [DOI] [PubMed] [Google Scholar]
- Hendrich B, Tweedie S (2003) The methyl-CpG binding domain and the evolving role of DNA methylation in animals. Trends Genet 19: 269–277 [DOI] [PubMed] [Google Scholar]
- Ito M, Koike A, Koizumi N, Sano H (2003) Methylated DNA-binding proteins from Arabidopsis. Plant Physiol 133: 1747–1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kass SU, Pruss D, Wolffe AP (1997) How does DNA methylation repress transcription? Trends Genet 13: 444–449 [DOI] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Jakobsen IB, Nei M (2001) MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17: 1244–1245 [DOI] [PubMed] [Google Scholar]
- Lindroth AM, Cao X, Jackson JP, Zilberman D, McCallum CM, Henikoff S, Jacobsen SE (2001) Requirement of CHROMOMETHYLASE3 for maintenance of CpXpG methylation. Science 292: 2077–2080 [DOI] [PubMed] [Google Scholar]
- Malagnac F, Bartee L, Bender J (2002) An Arabidopsis SET domain protein is required for maintenance but not establishment of DNA methylation. EMBO J 21: 6842–6852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, Anderson JB, DeWeese-Scott C, Fedorova ND, Geer LY, He S, Hurwitz DI, Jackson JD, Jacobs AR, Lanczycki CJ, et al (2003) CDD: a curated Entrez database of conserved domain alignments. Nucleic Acids Res 31: 383–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meehan RR, Lewis JD, Bird AP (1992) Characterization of MeCP2, a vertebrate DNA binding protein with an affinity for methylated DNA. Nucleic Acids Res 20: 5085–5092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meehan RR, Lewis JD, McKay S, Kleiner EL, Bird AP (1989) Identification of a mammalian protein that specifically binds to DNA containing methylated CpGs. Cell 58: 499–507 [DOI] [PubMed] [Google Scholar]
- Nan X, Meehan RR, Bird AP (1993) Dissection of the methyl-CpG-binding domain from the chromosomal protein MeCP2. Nucleic Acids Res 21: 4886–4892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nan X, Ng H, Johnson CA, Laherty CD, Turner BM, Eisenman RN, Bird AP (1998) Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature 393: 386–389 [DOI] [PubMed] [Google Scholar]
- Ng HH, Jeppesen P, Bird A (2000) Active repression of methylated genes by the chromosomal protein MBD1. Mol Cell Biol 20: 1394–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohki I, Shimotake N, Fujita N, Nakao M, Shirakawa M (1999) Solution structure of the methyl-CpG-binding domain of the methylation dependent transcriptional repressor MBD1. EMBO J 18: 6653–6661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitto L, Cernilogar F, Evangelista M, Lombardi L, Miarelli C, Rocchi P (2000) Characterization of carrot nuclear proteins that exhibit specific binding affinity towards conventional and non-conventional DNA methylation. Plant Mol Biol 44: 659–673 [DOI] [PubMed] [Google Scholar]
- Prokhortchouk A, Hendrich B, Jorgensen H, Ruzov A, Wilm M, Georgiev G, Bird A, Prokhortchouk E (2001) The p120 catenin partner Kaiso is a DNA methylation-dependent transcriptional repressor. Genes Dev 15: 1613–1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scebba F, Bernacchia G, De Bastiani M, Evangelista M, Cantoni RM, Cella R, Locci MT, Pitto L (2003) Arabidopsis MBD proteins show different binding specificities and nuclear localization. Plant Mol Biol 53: 715–731 [DOI] [PubMed] [Google Scholar]
- Schultz J, Copley RR, Doerks T, Ponting CP, Bork P (2000) SMART: a Web-based tool for the study of genetically mobile domains. Nucleic Acids Res 28: 231–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer NM, Napoli CA, Selinger DA, Pandey R, Cone KC, Chandler VL, Kaeppler HF, Kaeppler SM (2003) Comparative analysis of SET domain proteins in maize and Arabidopsis reveals multiple duplications preceding the divergence of monocots and dicots. Plant Physiol 132: 907–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25: 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade PA, Gegonne A, Jones PI, Ballestar E, Aubry F, Wolffe AP (1999) Mi-2 complex couples DNA methylation to chromatin remodelling and histone deacetylation. Nat Genet 23: 62–66 [DOI] [PubMed] [Google Scholar]
- Wakefield RID, Smith BO, Nan X, Free A, Soteriou A, Uhrin D, Bird AP, Barlow PN (1999) The solution structure of the domain from MeCP2 that binds to methylated DNA. J Mol Biol 291: 1055–1065 [DOI] [PubMed] [Google Scholar]
- Yu F, Thiesen J, Stratling WH (2000) Histone deactylase-independent transcriptional repression by methyl-CpG-binding protein2. Nucleic Acids Res 28: 2201–2206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemach A, Grafi G (2003) Characterization of Arabidopsis thaliana methyl-CpG-binding domain (MBD) proteins. Plant J 34: 565–572 [DOI] [PubMed] [Google Scholar]
- Zhang DL, Ehrlich KC, Supakar PC, Ehrlich M (1989) A plant DNA-binding protein that recognizes 5-methylcytosine residues. Mol Cell Biol 9: 1351–1356 [DOI] [PMC free article] [PubMed] [Google Scholar]