Abstract
The chloroplast contains many iron (Fe)-sulfur (S) proteins for the processes of photosynthesis and nitrogen and S assimilation. Although isolated chloroplasts are known to be able to synthesize their own Fe-S clusters, the machinery involved is largely unknown. Recently, a cysteine desulfurase was reported in Arabidopsis (Arabidopsis thaliana; AtCpNifS) that likely provides the S for Fe-S clusters. Here, we describe an additional putative component of the plastid Fe-S cluster assembly machinery in Arabidopsis: CpIscA, which has homology to bacterial IscA and SufA proteins that have a scaffold function during Fe-S cluster formation. CpIscA mRNA was shown to be expressed in all tissues tested, with higher expression level in green, photosynthetic tissues. The plastid localization of CpIscA was confirmed by green fluorescent protein fusions, in vitro import, and immunoblotting experiments. CpIscA was cloned and purified after expression in Escherichia coli. Addition of CpIscA significantly enhanced CpNifS-mediated in vitro reconstitution of the 2Fe-2S cluster in apo-ferredoxin. During incubation with CpNifS in a reconstitution mix, CpIscA was shown to acquire a transient Fe-S cluster. The Fe-S cluster could subsequently be transferred by CpIscA to apo-ferredoxin. We propose that the CpIscA protein serves as a scaffold in chloroplast Fe-S cluster assembly.
Iron (Fe)-sulfur (S) clusters are cofactors of proteins that perform a number of biological roles, including electron transfer; redox and nonredox catalysis; regulation of gene expression; and as sensors for Fe and oxygen within all living organisms, prokaryotes, and eukaryotes (Beinert, 2000). Although Fe-S clusters can be assembled in proteins in vitro with ferrous Fe and sulfide, it is now clear that the process is not spontaneous in vivo, and proteins have been shown to be required for the biological formation of these clusters (for review, see Lill and Kispal, 2000; Frazzon et al., 2002). Genetic and biochemical studies in microorganisms initially led to identification of two types of Fe-S cluster machinery, termed NIF (nitrogen fixation) and ISC (iron-sulfur cluster; Zheng et al., 1993, 1998). A third machinery (SUF) with homologs in a wide range of organisms was characterized more recently (Takahashi and Tokumoto, 2002).
The proposed mechanism of cluster formation is as follows: (1) S is mobilized from Cys by the action of a Cys desulfurase enzyme (Zheng et al., 1993, 1998); (2) the S atoms are transferred to a scaffold protein (Urbina et al., 2001), Fe atoms are supplied to the scaffold protein, and the transient Fe-S cluster is assembled; and (3) the Fe-S cluster is inserted into various apo-proteins to form the Fe-S proteins (Yuvaniyama et al., 2000; Agar et al., 2000a; Krebs et al., 2001; Wu et al., 2002; Tong et al., 2003). In addition to Cys desulfurase and scaffold proteins, other factors such as Hsp70- and Hsp40-type chaperones as well as ferredoxin (Fd)/Fd reductase systems may be involved (Lill and Kispal, 2000; Frazzon et al., 2002). There are indications that molecular chaperone proteins interact with the scaffold protein and keep the scaffold protein in a conformation that facilitates the Fe-S cluster assembly and the transfer to an apo-Fe-S protein (Hoff et al., 2000; Silberg et al., 2001; Muhlenhoff et al., 2003).
In the nitrogen-fixing bacterium Azotobacter vinelandii, NifU was shown to provide a scaffold for Fe-S cluster formation (Zheng et al., 1993, 1994; Agar et al., 2000a; Yuvaniyama et al., 2000). In non-nitrogen-fixing bacteria and eukaryotic mitochondria, an IscU scaffold protein is essential and the main protein for Fe-S cluster biosynthesis; this protein shows a high sequence identity with the N-terminal domain of A. vinelandii NifU (Agar et al., 2000a, 2000b). IscU proteins accept S from a NifS-like Cys desulfurase and are also the binding site for Fe to build an Fe-S cluster (Agar et al., 2000b). By contrast, plastids and most non-nitrogen-fixing cyanobacteria whose genome sequence is known do not possess any homolog of IscU. Instead, proteins, termed NFUs, with sequence similarity to the C-terminal domain of A. vinelandii NifU were found to be scaffolds for Fe-S cluster assembly (Leon et al., 2003; Yabe et al., 2004). SyNifU, the NifU of cyanobacterium Synechocystis, serves as a scaffold for Fe-S cluster assembly and delivery (Nishio and Nakai, 2000).
IscA is another protein that may function as a scaffold protein for Fe-S cluster synthesis in Escherichia coli (Takahashi and Nakamura, 1999; Tokumoto and Takahashi, 2001), yeast (Saccharomyces cerevisiae; Jensen and Culotta, 2000; Kaut et al., 2000), and cyanobacteria (Morimoto et al., 2002; Wollenberg et al., 2003). It is proposed that the IscA family of proteins provide alternative scaffolds to the NifU and IscU proteins for mediating Nif-specific and general Fe-S cluster assembly (Krebs et al., 2001). In addition to a scaffold function in cluster formation in E. coli, it was proposed that IscA can provide Fe for the assembly of the transient Fe-S cluster in IscU in the presence of IscS and Cys in vitro (Ding and Clark, 2004). In Synechocystis PCC 6803, IscA1, the product of SLR1417, predominantly binds the Fe ion alone, whereas IscA2, the product of SLR1565, binds a 2Fe-2S cluster (Morimoto et al., 2002). IscA2 forms a complex with a HEAT-repeat-containing protein, IaiH, which stabilizes the Fe-S cluster in IscA2 (Morimoto et al., 2002).
In addition, the SUF gene clusters of E. coli and Erwinia chrysanthemi have genes that encode components similar to IscS and IscA, named SufS and SufA, respectively (Takahashi and Tokumoto, 2002). These proteins are thought to be involved in Fe-S cluster formation under Fe limitation and oxidative stress conditions (Ollagnier-de-Choudens et al., 2001; Outten et al., 2003, 2004).
In plant cells, mitochondria and chloroplasts are believed to originate from endosymbiotic bacteria and therefore are predicted to have their own Fe-S cluster biosynthesis machinery. The structural and physiological differences between mitochondria and chloroplasts suggest the existence of two distinct Fe-S assembly machineries. Mitochondria are a site of oxygen consumption, whereas chloroplasts produce oxygen through the photosynthetic process. Based on sequence homology of putative components, the mitochondrial Fe-S machinery may be most similar to the bacterial ISC machinery, where IscU is regarded as the major and essential scaffold protein for the cluster assembly. In chloroplasts, the components of the Fe-S machinery are starting to be identified. A NifS-like protein (AtCpNifS) with Cys desulfurase activity was localized in the chloroplast (Leon et al., 2002; Pilon-Smits et al., 2002). This protein is similar in sequence to a cyanobacterial l-Cys/l-cystine C-S lyase (C-DES; Lang and Kessler, 1999; Kessler, 2004). Other putative components include three NFU proteins (NFU1–3), similar to cyanobacterial NFU-like scaffold proteins and to the C terminus of A. vinelandii NifU (Leon et al., 2003; Yabe et al., 2004). Nfu2 purified from E. coli contained a transient Fe-S cluster (Leon et al., 2003) that could be passed on to apo-Fd in vitro (Yabe et al., 2004). Nfu2 insertion mutants have a dwarf phenotype and are deficient in some but not all plastid Fe-S proteins (Touraine et al., 2004; Yabe et al., 2004). Other potential Fe-S candidate proteins in plastids are the Arabidopsis homologs of SufBCD and E and the high chlorophyll fluorescence 101 protein (HCF101). The putative SufC homolog AtNAP7 is confirmed to be in the chloroplast, and mutants have phenotypes that indicate a role in plastidic Fe-S cluster maintenance and repair during Arabidopsis embryogenesis (Xu and Moller, 2004). HCF101 encodes a NifH-related P-loop ATPase that seems required for 4Fe-4S but not 2Fe-2S clusters in chloroplasts (Lezhneva et al., 2004). So far, a link of either NFUs or CpSufBCDE with CpNifS has not been made. Here, we report the cloning, purification, and characterization of a chloroplast-localized IscA-like protein (AtCpIscA) from Arabidopsis. CpIscA is shown to act as a molecular scaffold in Fe-S cluster biosynthesis in vitro and to be able to transfer the cluster to apo-Fd producing active holo-Fd.
RESULTS
Cloning of CpIscA and Sequence Analysis
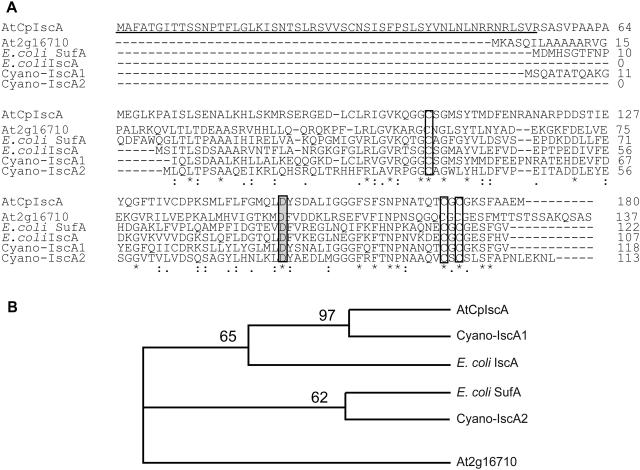
In earlier studies, the NifS-like protein AtCpNifS from Arabidopsis was shown to have Cys desulfurase activity and to be located in the chloroplast (Leon et al., 2002; Pilon-Smits et al., 2002). To identify another Arabidopsis protein(s) that might be involved in Fe-S cluster formation in plastids, a database search was performed to identify homologs of the cyanobacterial Synechocystis PCC 6803 IscA proteins. BLAST searches in the Munich Information Center for Protein Sequences (http://mips.gsf.de/proj/thal/db/) and The Arabidopsis Information Resource (TAIR; http://www.arabidopsis.org) databases revealed the presence of one IscA-like protein (At1g10500). TargetP predictions suggest that this protein is a chloroplast protein with a 55-amino-acid-long chloroplast transit sequence. The corresponding gene was named AtCpIscA, and its cDNA was cloned using reverse transcription-PCR. Alignment of the AtCpIscA primary sequence with IscA- and SufA-like proteins from cyanobacteria and E. coli reveals the presence of three conserved Cys residues (residues 104, 170, and 172), including the characteristic motif CXC and a conserved Asp (residue 149) in the C-terminal region (Fig. 1A). These residues are conserved among all IscA-type proteins from prokaryotic and eukaryotic organisms. In addition to the three Cys conserved in all IscA-type proteins, AtCpIscA contains two other Cys, at residues 94 and 135, which are also present in cyanobacterial IscA1 from Synechocystis PCC 6803. AtCpIscA exhibits higher sequence similarity to IscA1 (Slr1417) from Synechocystis PCC 6803 (58% identity) and IscA from E. coli (47% identity) compared to the mitochondrial IscA-like protein from Arabidopsis (At2g16710; 24%) and SufA from E. coli (25%). Consequently, in a phylogenetic analysis, AtCpIscA was grouped with cyanobacterial IscA1 and E. coli IscA, separate from E. coli SufA and the Arabidopsis mitochondrial NifA-like protein (Fig. 1B).
Figure 1.
Sequence alignment and phylogenetic tree of IscA-like proteins from various organisms. A, Sequence alignment. The predicted chloroplast targeting sequence is underlined. Conserved amino acids are marked by asterisks, and the conserved Cys residues are boxed. The conserved Asp residue is shown in gray. Sequences are: AtCpIscA, At1g10500 (this article); At2g16710, encoding the putative Arabidopsis mitochondrial IscA-like protein; E. coli SufA (NC_004431); E. coli IscA (NC_002655.2); cyano-IscA1, Synechocystis IscA Slr1417; and cyano-IscA2, Synechocystis IscA Slr1565. B, Phylogenetic tree of IscA-like proteins used in the sequence alignment.
Expression Analysis of CpIscA
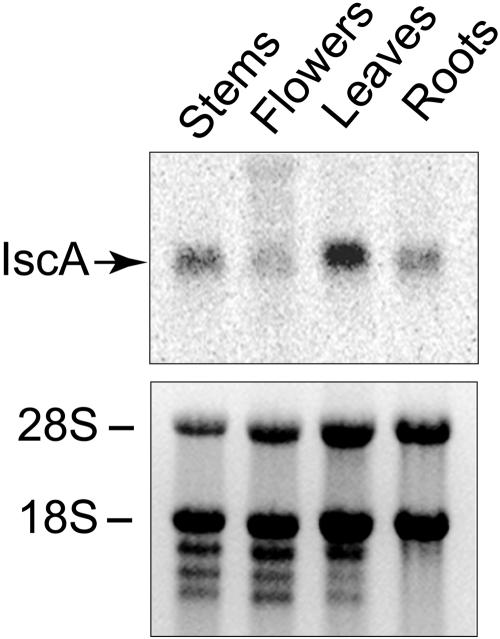
The AtCpIscA gene expression pattern in different tissues was analyzed using RNA-blot analysis (Fig. 2). Ten micrograms of total RNA extracted from roots, stems, leaves, and flowers was electrophoresed, and ethidium bromide staining was used to verify equal loading. Northern-blot analysis reveals that AtCpIscA is expressed in all tested tissues, with higher expression level in green photosynthetic tissues (leaves and stems) than non-green tissues (roots and flowers; Fig. 2).
Figure 2.
Expression analysis of CpIscA in different tissues. Total RNA from roots, leaves, stems, and flowers of 4-week-old Arabidopsis plants was isolated. Ten micrograms of total RNA was separated by electrophoresis, transferred to a Hybond-N membrane, and probed with 33P-labeled CpIscA cDNA (top). Ethidium bromide-stained agarose gel was used to show loading (bottom). 28S and 18S are ribosomal RNA subunits.
Intracellular Localization of CpIscA
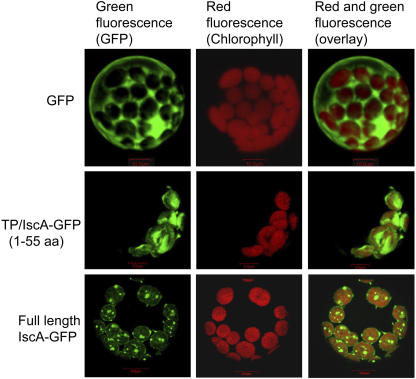
The TargetP program (Emanuelsson et al., 2000) predicted a chloroplast localization of AtCpIscA and a cleavable transit sequence of 55 amino acids. To examine the subcellular localization, we constructed fusions with the green fluorescent protein (GFP). GFP was fused to the coding region for the predicted chloroplast transit sequence of CpIscA (TP-IscA-GFP) and to the full-length IscA including its transit sequence (Full length-IscA-GFP). GFP alone expressed from the same constitutive promoter was used as a control. The localization in cells was analyzed using confocal laser microscopy. Fluorescence corresponding to GFP expressed without a transit sequence was excluded from the chloroplasts as expected (Fig. 3, top). By contrast, green fluorescence from TP-IscA-GFP was localized to the chloroplast stroma, as indicated by the overlay of green fluorescence and red autofluorescence (Fig. 3, middle). Because chloroplast transit sequences effectively mediate translocation of a passenger protein across the envelope (Keegstra and Froehlich, 1999), this is the expected location for a transit sequence fusion. Interestingly, green fluorescence from the full-length CpIscA coupled to GFP (Full length-IscA-GFP) was localized to discrete locations in the chloroplast stroma, which may be indicative of the inclusion in discretely localized complexes (Fig. 3, bottom).
Figure 3.
Subcellular localization of AtCpIscA. Arabidopsis protoplasts were transformed with plasmids that express the indicated gene constructs under control of the constitutive 35S cauliflower mosaic virus promoter. The nonfused GFP protein-coding sequence was used as a control. TP/IscA-GFP encodes the fusion of the CpIscA TP (1–55) to the N terminus of GFP, and full-length IscA GFP contains the full coding sequence of the IscA precursor fused to GFP. After 16 h of expression, cells were observed using a confocal laser-scanning microscope. Green fluorescence signals, chlorophyll red autofluorescence, and an overlay of green and red signals are shown.
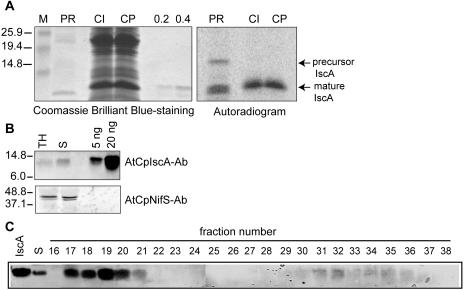
To investigate the chloroplast localization of CpIscA by an alternative method and to determine the size of the mature CpIscA protein in plastids, we performed an in vitro chloroplast uptake experiment (Fig. 4A). Radiolabeled precursor protein was produced by in vitro transcription of the cloned cDNA and subsequent translation of the synthetic mRNA in the presence of 35S-Met. The translation reaction (Fig. 4A, right, PR) resulted in a radiolabeled protein band with a size expected for the precursor. In addition to the precursor, two translation products with a size smaller than the expected mature protein were present that may result from initiation at either of the two downstream AUG codons corresponding to residues 65 and 85. The translation mixture including the precursor was incubated with purified intact chloroplasts in the light and in the presence of ATP. After import and treatment with protease, the chloroplasts were recovered and proteins were analyzed by SDS-PAGE (Fig. 4A). The E. coli-expressed recombinant mature CpIscA was electrophoresed in the same gel to allow a direct comparison of the molecular mass with the imported protein. Chloroplasts incubated with the precursor encoded by CpIscA and treated with protease accumulated a protein of about 14 kD in size that has the same electrophoretic mobility as the purified recombinant mature protein. Quantitation of the precursor and the mature bands indicated that 32% of the added CpIscA precursor was imported in this assay, which was comparable to the plastocyanin control (data not shown). We conclude that the CpIscA precursor contains plastid-targeting information and that the protein is active in the chloroplast. The recombinant protein purified from E. coli corresponds in size to the imported mature CpIscA.
Figure 4.
Chloroplast targeting of AtCpIscA and gel filtration analysis. A, In vitro import of radiolabeled CpIscA precursor. 35S-Met-labeled Precursor CpIscA protein (PR) was obtained by in vitro translation and incubated with isolated pea chloroplasts. Chloroplasts were reisolated, and proteins in the intact chloroplasts were analyzed either directly (CI) or after treatment of intact chloroplast with protease (CP). Proteins were separated by 15% SDS-PAGE. The gel was stained with Coomassie Brilliant Blue (left), and the radiolabeled protein bands were visualized using a PhosphorImager (right). B, Immunoblot analysis of 20 μg of total leaf homogenate protein (TH) and 20 μg of chloroplast stroma (S). Purified CpIscA (5 ng and 20 ng) was applied for comparison in the two right lanes of Figure 4B. AtCpNifS antibody (bottom) was used as a control for stromal protein. C, Gel filtration analysis of AtCpIscA in the stroma. One milligram of stromal protein was separated on a Superdex-S200 gel filtration column. Elution was monitored by the A280 (data not shown) and by immunoblotting of collected fractions with CpIscA-specific antibodies. The void volume of this column corresponds to fraction 11.
To analyze localization and expression levels in Arabidopsis plants, we raised an antibody (see “Materials and Methods”) that was used in immunoblots to analyze the presence of CpIscA in total leaf homogenate and chloroplast stroma fractions. The antibody recognized a band corresponding to the size of mature CpIscA in total homogenate and isolated stroma, confirming the stromal localization of AtCpIscA (Fig. 4B). The mobility of the detected CpIscA protein band in plant fractions is the same as that of purified recombinant CpIscA. Control experiments showed that these bands were not detected with preimmune serum (data not shown). Based on comparisons of the CpIscA staining intensities in stromal fractions and purified protein, we estimate the abundance of CpIscA in stroma to be approximately 0.01% to 0.02% of protein, which is slightly lower than the measured abundance of CpNifS (Ye et al., 2005). To investigate the oligomeric state of CpIscA in the stroma, a gel filtration experiment was performed using a high-resolution column and detection of AtCpIscA using immunoblotting (Fig. 4C). Approximately 90% of the stromal IscA was eluted in a single peak at high molecular mass (approximately 600 kD, fractions 17–20). In addition, a smaller amount of IscA was eluted as a dimer. This result indicates that CpIscA present in stroma interacts with other proteins in vivo and may form a transient complex with them. Interestingly, a fraction of CpNifS was also detected in a complex of approximately 600 kD (Ye et al., 2005), and coelution of CpIscA and CpNifS was confirmed by gel filtration (data not shown).
Expression and Purification of AtCpIscA
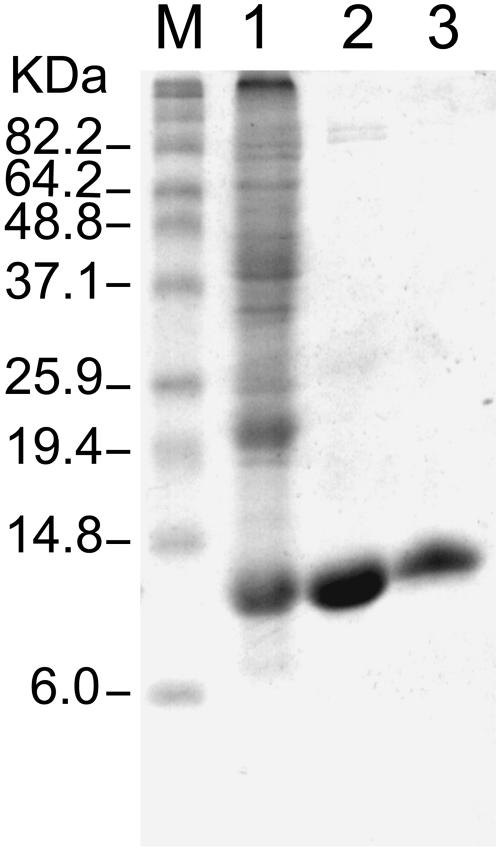
The coding sequence for mature CpIscA (without transit sequence) was cloned into the expression vector pET11d, and the mature-sized protein was expressed in BL21 codon+ E. coli cells. Three hours after induction with isopropyl-β-d-thiogalactopyranoside (0.4 mm), a protein with a molecular mass of about 14 kD accumulated to about 5% to 10% of the soluble protein, as shown by Coomassie Brilliant Blue staining (Fig. 5, lane 1). This molecular mass is in agreement with the molecular mass calculated from the DNA sequence (13,971 D). Soluble IscA was purified using cation exchange chromatography and a gel filtration step (Fig. 5, lanes 2 and 3). The protein was eluted from the calibrated S200 gel filtration column with an apparent molecular mass of 54 kD, suggesting that the protein is purified as a tetramer (data not shown). N-terminal sequencing (Macromolecular Resources, Colorado State University) by Edmann degradation yielded the sequence RNRLSV, corresponding to the predicted N-terminal sequence of mature CpIscA. The pure CpIscA protein was colorless, indicating it did not contain an Fe-S cluster while in E. coli.
Figure 5.
SDS-PAGE of IscA at different stages of its purification. Lane 1, Molecular mass marker; lane 2, total lysate from E. coli BL21 (DE3)/codon+ expressing IscA; lane 3, IscA after cation exchange column; lane 4, IscA after HPLC gel filtration column.
CpIscA with CpNifS Stimulates Fe-S Cluster Formation in Fd
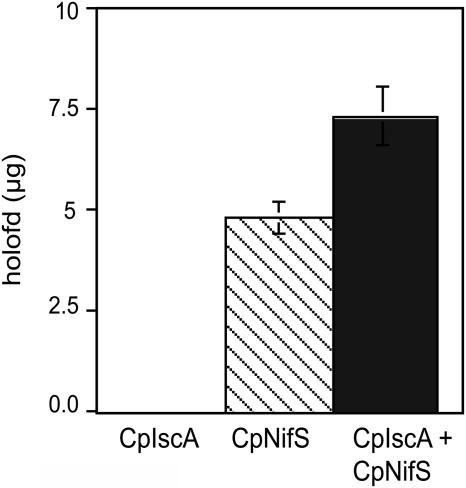
To study the role of CpIscA in Fe-S cluster formation, an in vitro reconstitution assay was developed. In this assay, apo-Fd was reconstituted to the holo-form by acquiring an Fe-S cluster, which was synthesized in vitro from Cys S and a ferrous Fe salt. Holo-Fd was separated from apo-Fd and other proteins and quantified by HPLC using an ion-exchange column. The activity of CpNifS proved to be sufficient and required for the Fe-S cluster formation in Fd (Ye et al., 2005). However, other proteins may promote the efficiency of CpNifS-mediated holo-Fd formation in a plant cell. In the presence of all necessary substrates for Fe-S cluster formation, except CpNifS, CpIscA alone did not show any reconstitution activity. However, preincubation of CpIscA with CpNifS significantly increased subsequent Fd reconstitution activity compared to that shown by CpNifS alone (P < 0.05; Fig. 6). The observed modest increase in reconstitution activity was a first indication of a possible role of CpIscA in Fe-S cluster formation.
Figure 6.
Purified CpIscA enhances the Fd reconstitution activity of CpNifS. Purified CpNifS (7.5 μg), CpIscA (30 μg), or a combination of both CpNifS (7.5 μg) and CpIscA (30 μg) were incubated with 50 mm Tricine-NaOH, pH 7.5, 5 mm DTT, 1 mm l-Cys, 1 mm ferrous ammonium sulfate, 20 μm PLP for 30 min, followed by addition of 30 μg of apo-Fd. After 20 min, the reconstituted holo-Fd was assayed by HPLC. Data represent the average and se of three replicates.
Incorporation of an Fe-S Cluster into CpIscA
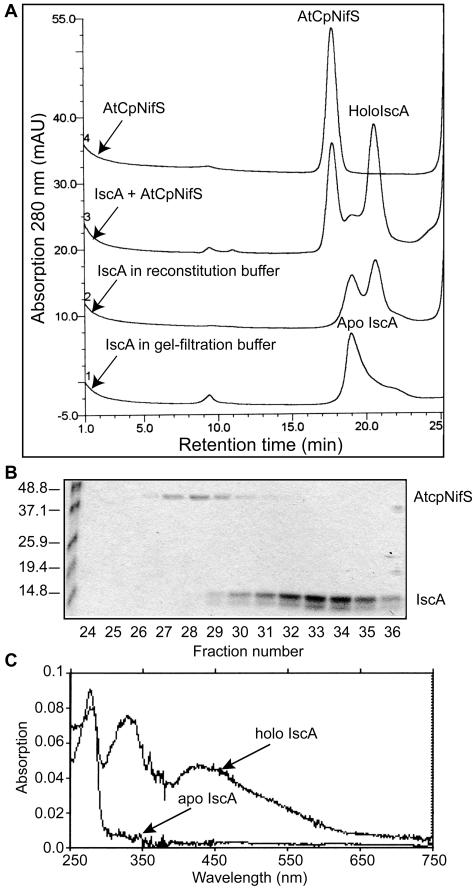
The stimulation of CpNifS-dependent Fd reconstitution activity by CpIscA suggested possible interactions between CpIscA and CpNifS in the Fe-S cluster formation where CpIscA may be a scaffold for cluster assembly and an intermediate in Fe-S insertion. To investigate this possibility in more detail, CpIscA (300 μg) was incubated with CpNifS (75 μg) in reconstitution buffer (5 mm dithiothreitol [DTT], 1 mm l-Cys, 1 mm ferrous ammonium sulfate, 20 μm pyridoxal 5′ phosphate [PLP]) for 30 min and then applied to a gel filtration column (Fig. 7A, line 3). As controls, apo-CpIscA (300 μg) incubated in gel filtration buffer or in reconstitution buffer, as well as CpNifS alone (75 μg), were incubated and run through the same column (Fig. 7A, lines 1, 2, and 4, respectively). Proteins were detected by A280 (Fig. 7) and 420 nm (data not shown). Purified untreated apo-CpIscA eluted from the column in a peak with a retention time expected for the tetramer but a shoulder was present, suggesting the existence of lower-molecular mass IscA species (Fig. 7A, line 1). Incubation of ApoIscA with DTT-containing buffer resulted in mainly the dimeric form of the protein (line 2). No absorbance was detected at 420 nm, indicating the absence of Fe-S clusters in the apo-IscA fractions (data not shown). As shown in Figure 7A (line 3), incubation of CpIscA with CpNifS resulted in two major peaks corresponding to CpNifS and a CpIscA dimer, according to its calculated molecular mass. SDS-PAGE (Fig. 7B) indicated that the peak of CpNifS eluted at 17.5 min (fraction 28) and the dimeric CpIscA at 20.5 min (fraction 34). Interestingly, the peak corresponding to reconstituted dimeric CpIscA could also be detected by A420 (data not shown), suggesting the presence of an Fe-S cluster. Apo-CpIscA (line 2) and reconstituted dimeric CpIscA (line 3) were collected from gel filtration runs, and their absorption spectra were measured (Fig. 7C). Unreconstituted CpIscA showed only one peak at 280 nm, characteristic for aromatic residues in proteins. Reconstituted CpIscA (fraction 34), however, showed extra maxima at 330 nm, 420 nm (with a shoulder at 470), and 580 nm, indicative of a 2Fe-2S cluster (Fig. 7C). In a typical 2Fe-2S cluster, absorption from 420 nm to 460 nm is attributed to the vibration between Fe and bridging inorganic S (Morimoto et al., 2002).
Figure 7.
Purified CpIscA can acquire an Fe-S cluster. A, Gel filtration analysis. CpIscA (300 μg) was incubated with CpNifS (75 μg) in reconstitution buffer (50 mm Tricine-NaOH, pH 7.5, 5 mm DTT, 1 mm l-Cys, 1 mm ferrous ammonium sulfate, 20 μm PLP; line 3) for 30 min. CpIscA (300 μg) in gel filtration buffer (25 mm Tricine/KOH, pH 7.9, 50 mm KCl; line 1) or in reconstitution buffer (reaction 2) and CpNifS (75 μg) in reconstitution buffer (reaction 4) were used as controls. After incubation at 37°C, proteins were separated on a Superdex S200 gel filtration column equilibrated in 25 mm Tricine/KOH, pH 7.9, 50 mm KCl, and elution was monitored by the A280. NifS eluted at 17.5 min (fraction 28), apo-IscA (tetramer) at 19 min (fraction 31), and holo-IscA (dimer) at 20.5 min (fraction 34). B, SDS-PAGE of gel filtration fractions of treatments shown in line 3. C, Absorption spectrum of apo-CpIscA and holo-CpIscA. The protein concentration was 300 μg in 25 mm Tricine/KOH, pH 7.9.
The presence of Fe in CpIscA was determined chemically (see “Materials and Methods”) using spinach (Spinacia oleracea) holo-Fd as control, which contains one 2Fe-2S cluster per protein. We measured 2.0 ± 0.2 Fe/protein for spinach holo-Fd. Reconstituted CpIscA contained 1.4 ± 0.2 Fe/dimer, while no Fe was detected in the apo-form. Because the reconstituted fraction obtained after incubation with CpNifS (line 3) still may contain some inactive apo-CpIscA, a number slightly lower than two is expected for the number of Fe atoms/dimer. These data indicate that the isolated and reduced CpIscA is a clusterless apo-protein, while the reconstituted dimer may contain a 2Fe-2S cluster. The Fe-S cluster formed in IscA is stable since treatment with Fe chelator (1 mm EDTA) or overnight incubation at 4°C did not induce destruction of the Fe-S cluster, as indicated by the reconstitution efficiency (data not shown).
Fe-S Cluster Transfer from CpIscA to Apo-Fd
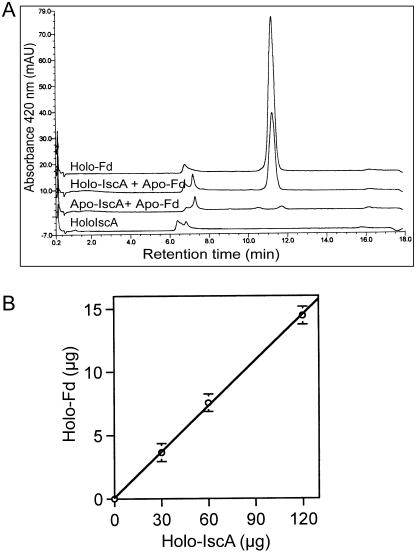
As CpIscA appeared to have an Fe-S cluster, the potential transfer of the Fe-S cluster to apo-Fd was investigated. Both apo- and holo-CpIscA (150 μg) were collected from the gel filtration column and incubated with 30 μg of apo-Fd in the presence of 5 mm DTT at 37°C for 30 min. As controls, holo-Fd (30 μg) alone and holo-IscA (150 μg) alone were incubated. Incubation mixtures were used for holo-Fd analysis by HPLC. As shown in Figure 8A, the apo-CpIscA fraction did not lead to any formation of holo-Fd, while 50% (15 μg) of apo-Fd was reconstituted to holo-Fd by acquiring an Fe-S cluster from the dimeric holo-CpIscA. To test if the amount of holo-Fd formation depends on the amount of holo-IscA, various amounts of holo-IscA were incubated with apo-Fd (30 μg) for 30 min and the amount of holo-Fd was assayed (Fig. 8B). The amount of holo-Fd formed increased with increasing the amount of holo-IscA. Importantly, in the same condition when holo-IscA (60 μg) was incubated with apo-Fd (30 μg) in the presence of 1 mm EDTA, reconstitution was still observed with 70% activity of the reaction without EDTA indicating direct transfer from IscA to Apo-Fd (data not shown). Thus, it is proposed that holo-CpIscA can be formed from Apo-CpIscA by acquisition of a transient Fe-S cluster, which it can subsequently deliver to apo-Fd. This suggests a role for CpIscA as a scaffold during Fe-S cluster formation in plastids and in transfer of the formed cluster to apo-Fe-S proteins.
Figure 8.
Fe-S cluster transfer from CpIscA to Fd apo-protein. A, Fe-S cluster transfer from holo-IscA to apo-Fd. Apo-Fd (30 μg) was incubated with 150 μg of holo-CpIscA (collected from gel filtration column, reaction 3, Fig. 7) or 150 μg of apo-CpIscA purified by gel filtration column. After 30 min, the Fd was assayed by HPLC and quantified. Holo-Fd (30 μg) and holo-IscA (150 μg) were used as controls. B, The amount of holo-Fd formed depends on the amount of holo-IscA. Thirty micrograms of apo-Fd was incubated with various amounts of holo-IscA for 30 min, and the amounts of holo-IscA were assayed by HPLC and quantified as before.
DISCUSSION
Recently, it was demonstrated that AtCpNifS, a chloroplastic NifS-like Cys desulfurase of Arabidopsis, is responsible for the release of S from Cys for the biogenesis of Fe-S clusters in vitro (Ye et al., 2005). In this study, we report that Arabidopsis chloroplasts contain an IscA-like protein, CpIscA, which enhances the CpNifS-dependent Fe-S cluster formation in vitro. In addition, we provide evidence that recombinant apo-CpIscA, upon incubation with AtCpNifS and appropriate substrates, can acquire an Fe-S cluster, which in turn can be donated to apo-Fd in vitro. From these observations, we hypothesize that CpIscA functions as a scaffold protein for the assembly of transient Fe-S clusters from Fe and elemental S, which can be donated to Fe-S apo-proteins in the plastids. These results are of significance since Fe-S cluster proteins play crucial roles in essential plant processes, such as photosynthesis, which determine plant productivity and nutritional value. Although plastids are known to be able to synthesize their own Fe-S clusters (Takahashi et al., 1986), the machinery involved in Fe-S cluster formation in plastids remains largely to be elucidated. The results presented here contribute to the understanding of this important process.
CpIscA has three conserved Cys residues and a conserved Asp residue (Fig. 1) with the sequence arrangement C-X42-44-D-X20-C-G-C. These residues are conserved among all IscA-type proteins from prokaryotic and eukaryotic organisms, including nif-specific IscA proteins (Krebs et al., 2001). This sequence motif of IscA scaffold proteins is different from the primary sequence arrangement of Cys in NifU/IscU scaffold proteins (C-G-D-X22-24-C-X43-C; Krebs et al., 2001). Individual amino acid substitutions for each of the three conserved Cys residues of the yeast Isa1p and Isa2p proteins (IscA homologs) yield the same phenotype as the gene knockout (Jensen and Culotta, 2000). When any one of three conserved Cys residues in Synechocystis IscA2 was replaced with Ser, the amount of assembled 2Fe-2S was significantly reduced (Morimoto et al., 2003). It has been suggested that two of the three conserved Cys residues are involved in cluster binding, whereas the third one provides an electron during cluster assembly for the reduction of the Cys persulfide at NifS/IscS (Krebs et al., 2001). Further studies are required to investigate the role of these Cys residues in CpIscA.
Two scaffold proteins for Fe-S cluster formation in nitrogenase, called NifU and nifIscA, were discovered in the Nif operon of A. vinelandii. NifU consists of three domains: an N-terminal region that can accept a transient cluster, a central region with stable cluster, and a C-terminal thioredoxin-like domain known as the Nfu region (Agar et al., 2000a; Yuvaniyama et al., 2000). Homologs of the N-terminal domain of NifU as well as of NifA seem to function in the general Fe-S cluster machinery found in mitochondria (Lill and Kispal, 2000) and are also encoded by the Isc-gene cluster of bacteria such as E. coli (Frazzon et al., 2002). This Isc-type machinery was reported to be sensitive to oxygen in E. coli, A. vinelandii, and Schizosaccharomyces pombe (Krebs et al., 2001; Ollagnier-de-Choudens et al., 2001; Wu et al., 2002). In E. coli, a second Fe-S assembly machinery is encoded by the SUF operon and the gene cluster includes SufA, a homolog of IscA/NifA. The Suf operon may function in Fe-S assembly under conditions of oxidative stress and Fe limitation (Outten et al., 2004).
The CpIscA-mediated formation of the [2Fe-2S] cluster in apo-Fd proceeds under normal atmospheric (oxygenic) conditions in the presence of a thiol reductant (DTT) in the assembly cocktail. By comparison, the reconstituted Fe-S cluster in either E. coli or A. vinelandii IscA proteins was fairly labile (Ollagnier-de-Choudens et al., 2001; Krebs et al., 2001), and the cluster formed in CpIscA was stable and insensitive to oxygen, as indicated by the persistence over several days at 4°C. Since photosynthetic activity produces oxygen, this reaction condition may be appropriate for a chloroplast enzyme. By contrast, the mitochondrial Isc-type Fe-S protein maturation machinery requires the exclusion of oxygen in vitro (Muhlenhoff et al., 2002). Chloroplasts are proposed to be derived from an ancestral cyanobacterium-like endosymbiont (Martin et al., 1998). No IscU-type scaffold proteins have been identified in the nine completely sequenced genomes of cyanobacteria (http://www.ncbi.nlm.nih.gov/blast/Blast/.cgi). However, IscA scaffold proteins were identified in Synechocystis PCC 6803 (Wollenberg et al., 2003). Genes encoding IscU homologs with putative chloroplast-targeting sequences were not found in the Arabidopsis genome (TAIR; www.arabidopsis.org). This suggests that the Arabidopsis chloroplast, like cyanobacteria, does not express an IscU scaffold homolog. Therefore, the CpIscA protein may provide an alternative scaffold to the IscU proteins for Fe-S cluster formation and delivery in chloroplasts.
In addition to CpIscA, three chloroplast-localized CpNfu proteins were identified that may function as molecular scaffolds for Fe-S cluster biosynthesis (Leon et al., 2003; Touraine et al., 2004; Yabe et al., 2004). An interesting question that remains to be answered is whether the CpIscA and CpNfu scaffold proteins function complementary, overlapping, or parallel to each other in chloroplasts. Some plausible reasons for the need of alternative scaffolds are that each scaffold protein is optimized for assembly of either [2Fe-2S] or [4Fe-4S] clusters in vivo, that they preferentially transfer clusters to different acceptor proteins, or that each functions optimally under different physiological conditions, e.g. conditions that are more reducing or oxidizing. Complementary or partially overlapping roles in the delivery of Fe-S clusters to various substrate apo-proteins in different plastids and/or under different growth conditions were postulated for AtCpNfus (Touraine et al., 2004; Yabe et al., 2004). A scaffold role for CpIscA in plastid Fe-S cluster formation is similar to the finding that IscA from E. coli (Ollagnier-de-Choudens et al., 2001) and cyanobacterium Synechocystis PCC 6803 (Wollenberg et al., 2003) can serve as a scaffold for formation of a [2Fe-2S] cluster, which can be donated to the Fe-S apo-protein Fd. The homolog from A. vinelandii NifIscA can also serve as a scaffold for 4Fe-4S clusters (Krebs et al., 2001). Once a suitable in vitro plant system for reconstitution of a 4Fe-4S apo-protein has been established, it will be interesting to investigate whether CpIscA serves as a scaffold for both types of Fe-S cluster.
MATERIALS AND METHODS
Cloning and Plasmid Construction
The Arabidopsis (Arabidopsis thaliana) CpIscA coding sequence was amplified by PCR using cDNA as a template. cDNA was prepared from DNase-treated total RNA prepared from 2-week-old seedlings as described (Pilon-Smits et al., 2002). Primers used for IscA amplification were 5′-GCTCTAGACAGAAGATTATGGCTTTCGC-3′ (forward primer) and 5′-TCCCCCGGGTGTGAAACTGGTTCACATCT-3′ (reverse primer). Underlined bases indicate XbaI and SmaI sites, respectively. The PCR product was digested with XbaI and SmaI and then ligated into a pBS (KS+) vector (Stratagene, La Jolla, CA), digested with the same restriction enzymes to produce plasmid pPrIscA. To subclone the mature sequence of IscA in pET11d for expression, PCR was performed with another set of nested primers, 5′-CATGCCATGGCTGTTCGATCCGCTTCGGTTC-3′ (forward primer) and 5′-CGGGATCCTCACATCTCGGCAGCAAAAGA-3′ (reverse primer). Underlined bases indicate NcoI and BamHI sites, respectively. The PCR product was digested with NcoI and BamHI and subcloned in pET11d to produce plasmid pMIscA.
Construction of the plasmid for expression of the transit peptide (TP; amino acids 1–55) of IscA fused to GFP was performed as follows. The sequence encoding the predicted TP of IscA was amplified by PCR using flanking primers SalI-N (5′-GAATGGTCGACATGGGCTTTCGCTACTGGAATCACG-3′) and NcoI-C-TP (5′-CATGCCATGGACATCTCGGCAGCAAAAGACTTCC-3′). To make a fusion of GFP with the full-length protein, the entire coding sequence of IscA was amplified by PCR using flanking primers SalI-N and NcoI-C-Fl (5′-CATGCCATGGCTGATCTCATCTTACTCAGATGCTTAA-3′). pPrIscA plasmid was used as a template for PCR. PCR products were digested and inserted into the SalI/NcoI-digested GFP reporter plasmid, 35Ω-SGFP(S65T), to create the TP/IscA-GFP and Full length-IscA-GFP, respectively. The inserts in all plasmids were sequenced by the dideoxy dye termination method to ensure that no error was introduced during PCR reactions. The plasmids used for protoplast transformation were prepared using the Plasmid Midi kit (QIAGEN, Valencia, CA).
Sequence Analysis and Alignments
Sequence analysis was performed using the Mac Vector sequence analysis software (International Biotechnologies, New Haven, CT). Searches for sequence similarity were performed using the BLAST network service provided by the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov). Sequence alignment was performed using ClustalW at European Bioinformatics Institute, ExPASY Proteomics tools (http://www.ebi.ac.uk/clustalw). Phylogenetic analysis was performed using the PAUP (Sinauer Associates, Sunderland, MA; version 4.0b10) heuristic search method with tree bisection-reconnection branch swapping (Swofford, 1993). Bootstrap analysis with 1,000 random replicates was performed using the heuristic method.
In Vitro Chloroplast Import Assay
The pPrIscA plasmid was linearized with KpnI and transcribed in vitro using T7 polymerase (Epicenter Technologies, Madison, WI) according to the manufacturer's instructions. Radiolabeled precursors were synthesized in a wheat germ lysate system in the presence of 35S-Met (25 μCi/50 μL reaction; Amersham/Pharmacia, Piscataway, NJ) according to suggested protocols (Promega, Madison, WI). Chloroplasts for import experiments were isolated from 10-d-old pea (Pisum sativum) seedlings (cv Little Marvel) and incubated with radiolabeled precursor as described (Pilon et al., 1992). The postimport thermolysin treatment and reisolation of intact chloroplasts were performed as described (Smeekens et al., 1986). Proteins from import experiments equivalent to 10 μg of chlorophyll were separated by 15% SDS-PAGE, stained with Coomassie Brilliant Blue, fixed in 7% (v/v) acetic acid, 25% (v/v) methanol, dried, and the radiolabeled proteins visualized and quantified using a STORM PhosphorImager (Molecular Dynamics, Sunnyvale, CA).
Immunoblotting, Gel Filtration, and Antibody Production
Total homogenate and intact chloroplasts were isolated from rosette leaves of Arabidopsis plants as described by Rensink et al. (1989). A stromal protein fraction was obtained from chloroplasts as described by Smeekens et al. (1986). Gel filtration analysis was performed as described (Ye et al., 2005). Protein samples were separated by SDS-PAGE, transferred to cellulose membrane, and probed with CpIscA-specific antibody as described by Pilon-Smits et al. (2002). Polyclonal antibodies against bacterially expressed CpIscA were raised in rabbits at a commercial facility (Pocono Rabbit Farm and Laboratory, Canadensis, PA). Two rabbits were immunized after collection of preimmune sera. Preimmune serum at dilutions between 1/500 and 1/2,000 in phosphate-buffered saline did not recognize either purified CpIscA or CpIscA in plant homogenates. Antiserum from both rabbits detected CpIscA, and the optimal antiserum dilutions in phosphate-buffered saline were found to be 1/2,000 for immunoblots. The antibody for CpNifS has been described (Pilon-Smits et al., 2002).
RNA-Blot Analysis
Total RNA from different tissues (roots, leaves, stems, and flowers) was isolated by the TRIzol method (Invitrogen, Carlsbad, CA). Ten micrograms of total RNA was electrophoresed on a 1% (w/v) agarose gel containing 4% (w/v) formaldehyde, transferred to a nylon membrane, and probed with a 33P-labeled AtCpIscA cDNA synthesized with a random primer labeling kit from Amersham. Prehybridization, hybridization, washing, and detection were done as described previously (Pilon-Smits et al., 2002).
Subcellular Localization of GFP-Fusion Proteins
GFP fusions were expressed in Arabidopsis protoplast-derived cells from the 35S promoter of cauliflower mosaic virus in the GFP reporter plasmid 35Ω-SGFP(S65T). For protoplast preparation, Arabidopsis plants (ecotype Columbia) were grown on Murashige and Skoog medium (Murashige and Skoog, 1962) for 3 weeks. Two grams fresh weight of leaf tissue was placed in 30 mL of a buffer containing 1% (w/v) cellulase Onozuka R-10, 0.25% Macerozyme R-10 (Karlan Research Products, Santa Rosa, CA), 8 mm CaCl2, 0.5 m mannitol, 5 mm MES, pH 5.5, vacuum infiltrated for 1 min, and incubated for 3 h at room temperature with gentle shaking. The clear digest was filtered through a 37- to 70-μm nylon mesh (Carolina Biological Supply, Burlington, NC), and the protoplasts were harvested by centrifugation for 5 min at 1,000 rpm and washed twice in 10 mL of cold W5 wash solution (154 mm NaCl, 125 mm CaCl2, 5 mm KCl, 5 mm Glc, 0.5 m mannitol, adjusted to pH 5.8 with KOH). The pellet was suspended in about 2 mL of mannitol/magnesium solution (15 mm MgCl2, 0.4 m mannitol, 0.1% MES, pH 5.6). Protoplasts were counted using a hemocytometer, and their concentration was adjusted to 3 × 106 cells mL−1 with mannitol/magnesium solution. Fifty micrograms of plasmid DNA, 100 μg of salmon sperm DNA, and 300 μL of polyethylene glycol (PEG) solution [40% PEG 6000/0.4 m mannitol/0.1 m Ca(NO3)2, adjusted to pH 8.0 with KOH] were added to 300 μL of the protoplast solution, very gently mixed, and left for 30 min at room temperature. The solution was very slowly diluted with 10 mL of W5 solution and then pelleted by centrifugation for 5 min at 1,000 rpm. Protoplasts were further resuspended in 5 mL of protoplast culture medium composed of Murashige and Skoog medium supplemented with 0.4 m Glc, 0.4 m mannitol, 1 mg/L 2,4-dichlorophenoxyacetic acid, 0.15 mg/L kinetin, pH adjusted to 5.8 with KOH, and left at 23°C for 16 h under continuous light. Confocal images were obtained using an Olympus Fluoview 300 inverted confocal laser-scanning microscope (1X70) equipped with an argon ion laser system and a 1.4 numerical aperture 60× objective lens (Olympus America, Melville, NY). The fluorescence signals were detected at 530 nm for GFP and 660 nm for chlorophyll. Sections of 1-μm thickness were scanned, and images were captured by the Fluoview software as Tiff files.
Preparation of Proteins
Fd was extracted from fresh spinach (Spinacia oleracea) that was obtained at a local health-food store, essentially as described (Yocum, 1982), and further purified as described (Pilon et al., 1992). The apo-Fd for reconstitution assays was prepared from holo-Fd by removal of the Fe-S cluster as described (Kato et al., 2000). Apo-Fd (at 0.5 mg/mL) was stored in 100 mm Tris-HCl, pH 8.0, under nitrogen in small aliquots at −80°C. CpNifS was expressed and purified as described (Pilon-Smits et al., 2002). For overexpression of AtCpIscA, Escherichia coli BL21 (DE3) codon+ (Stratagene) was transformed with pMIscA vector, and 2 L of LB medium containing 50 μg mL−1 ampicillin was inoculated with 1/100 volume of overnight culture. Cells were grown at 37°C to an OD600 of 0.5, and expression was induced with 0.4 mm isopropyl-β-d-thiogalactopyranoside for 3 h at 37°C. The bacterial pellet was washed with 150 mm NaCl and resuspended in buffer A (25 mm potassium phosphate buffer, pH 7.5, 1 mm EDTA) and passed twice through a French press (8,000 psi) at 4°C to disrupt the cells. All further manipulations were at 4°C unless indicated otherwise. The lysate was centrifuged for 20 min at 12,500g, and the cleared supernatant was loaded onto a cation-exchange SP Sepharose 1.6- × 20-cm column (Amersham Pharmacia Biotech, Piscataway, NJ) equilibrated in 25 mm potassium phosphate, pH 7.5, 1 mm EDTA at a flow rate of 5 mL min−1. The column was washed with three volumes of the same buffer and eluted with a 500-mL linear gradient of 0 to 0.5 m KCl in the same buffer, and 6-mL fractions were collected. Peak fractions were concentrated by adding ammonium sulfate to 70%, and the precipitated protein was collected by centrifugation for 20 min at 12,500g, dissolved in 5 mL of buffer A, and dialyzed overnight in 3 L of the same buffer. The dialyzed protein was filtered through a 0.2-μm filter and applied to a Sephacryl-S200 Hiprep 16/60 column (Amersham Bioscience, Piscataway, NJ) equilibrated in 25 mm potassium phosphate, pH 7.5, 100 mm KCl, 1 mm EDTA at room temperature and connected to a Summit HPLC system (Dionex, Sunnyvale, CA). The column was eluted, and fractions of 1 mL were collected. Elution was monitored by detection of the OD at 280 nm. The purified protein was concentrated as before and dissolved in the same buffer, without EDTA, dialyzed, aliquoted, and stored at −80°C with 15% (w/v) glycerol. Typical yields were 5 to 10 mg L−1 of culture. The N-terminal sequence was determined by Edmann degradation at the Colorado State University Macromolecular Resources facility.
Enhancement of Fe-S Cluster Assembly in Fd by IscA
The role of IscA in Fe-S cluster assembly was assayed by incubating IscA (30 μg) with NifS (7.5 μg) for 30 min in reconstitution buffer containing 50 mm Tricine-NaOH, pH 7.5, 5 mm DTT (Roche Diagnostics, Palo Alto, CA), 1 mm l-Cys (Sigma, St. Louis), 1 mm ferrous ammonium sulfate, and 20 μm PLP. Apo-Fd (30 μg) was added and incubated for 20 min at 37°C. After incubation, the reaction mixture was centrifuged at 14,000g for 1 min and directly applied to a 1-mL RESOURCE Q anion-exchange column (Amersham) connected to a summit HPLC system with a UVD170 detector and controlled by Chromeleon software (Dionex). The sample loop size was 100 μL. The column had been equilibrated with 25 mm Tris-HCl, pH 7.5. The following KCl gradient was applied in this buffer at a flow rate of 1.5 mL min−1: 0 to 4.5 min, 0 m KCl; 4.5 to 5 min, 0 to 0.25 m KCl; 5 to 14 min, 0.25 to 0.55 m KCl; ramp up to 1 m KCl; 14.1 to 15.6 min, 1 m KCl; ramp down to 0 m KCl; and hold for 2 min to reequilibrate. Holo-Fd eluted at 10.8 min. Elution was monitored by absorbance at both 280 and 420 nm. Holo-Fd was quantified by signal integration at 420 nm, a characteristic absorption maximum for Fd. Spinach holo-Fd (30 μg in 150 μL) was used as a standard. The activity of the holo-Fd recovered from CpNifS/CpIscA reconstitution was assayed by examining NADP reduction (Smillie and Entsch, 1971). The ΔA340 was 0.27 mg−1 Fd min−1, and this activity was light dependent. The absorption spectrum change was the same for both reconstituted Fd and spinach holo-Fd.
Incorporation of a Transient Fe-S Cluster into CpIscA, Gel Filtration Analysis, and Cluster Transfer
Incorporation of the Fe-S cluster into CpIscA was achieved by incubating IscA (300 μg) with 75 μg of NifS in reconstitution buffer (reaction 3; Fig. 7). Separate incubations of CpIscA (300 μg) in gel filtration buffer (25 mm Tricine/KOH, pH 7.9, 50 mm KCl; reaction 1; Fig. 7), IscA (300 μg) in reconstitution buffer (reaction 2), and CpNifS (75 μg) in reconstitution buffer (reaction 4; Fig. 7) were used as controls. After incubation at 37°C for 30 min, the mixtures were centrifuged at 14,000g for 1 min and directly applied to a Superdex-200 gel filtration column (1 × 30 cm; Pharmacia) connected to a summit HPLC system with a UVD170 detector and controlled by Chromeleon software (Dionex). The column was equilibrated in 25 mm Tricine/KOH, pH 7.9, 50 mm KCl. The flow rate was 0.75 mL min−1, and fractions were collected every 0.5 min. A loop size of 500 μL was used. Elution was monitored by A280 and 420 nm. The void volume was determined with blue dextran. Standards used for calibration were IgY, bovine serum albumin, ovalbumin, chymotrypsinogen, and RNase.
Apo-Fd (30 μg) was incubated with holo-IscA, which was collected from a gel filtration column (reaction 3), for 30 min at 37°C in gel filtration buffer with 5 mm DTT. The resulting holo-Fd was assayed by HPLC as described (Ye et al., 2005). Gel filtration analysis of CpIscA in stromal fractions utilized the same column, buffer, and elution conditions as described above.
UV-Visible Spectroscopy
UV-visible absorption spectra (250–750 nm) were recorded with a Beckman DU 530 spectrophotometer (Beckman Instruments, Fullerton, CA) in a 100-μL quartz cuvette containing 20 to 30 μg of IscA protein in assay buffer (25 mm Tricine/KOH, pH 7.9, 50 mm KCl) in 1.0-nm scan steps.
General Methods
Protein was assayed according to Bradford using bovine serum albumin as a standard (Bradford, 1976). For CpIscA, the protein assay by Bradford was calibrated by measuring the protein absorption at 280 nm in 6 m guanidine HCl and calculation of absolute amounts using the molar extinction coefficient predicted by the primary sequence (Gill and von Hippel, 1989). Fe content of proteins was determined according to Fish (1988). Spinach holo-Fd was used as a control. Statistical analysis was performed with the JMP-IN software (SAS Institute, Cary, NC).
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession number AY971959.
Acknowledgments
We thank Dr. Norbert Rolland for the generous gift of the GFP-reporter plasmid. Also, we would like to thank Dr. Libing Zhang for help with the phylogenetic analysis.
This work was supported by the National Research Initiative of the U.S. Department of Agriculture Cooperative State Research, Education and Extension Service (grant no. 2003–35318–13758 to E.A.H.P.S and M.P.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.058602.
References
- Agar JN, Yuvaniyama P, Jack RF, Cash VL, Smith AD, Dean DR, Johnson MK (2000. a) Modular organization and identification of a mononuclear iron-binding site within the NifU protein. J Biol Inorg Chem 5: 167–177 [DOI] [PubMed] [Google Scholar]
- Agar JN, Krebs C, Frazzon J, Huynh BH, Dean DR, Johnson MK (2000. b) IscU as a scaffold for iron-sulfur cluster biosynthesis: sequential assembly of [2Fe-2S] and [4Fe-4S] clusters in IscU. Biochemistry 39: 7856–7862 [DOI] [PubMed] [Google Scholar]
- Beinert H (2000) Iron-sulfur proteins: ancient structures, still full of surprises. J Biol Inorg Chem 5: 2–15 [DOI] [PubMed] [Google Scholar]
- Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Ding H, Clark RJ (2004) Characterization of iron binding in IscA, an ancient iron-sulfur cluster assembly protein. Biochem J 379: 433–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol 300: 1005–1016 [DOI] [PubMed] [Google Scholar]
- Fish W (1988) Rapid colorimetric micromethod for the quantitation of complexed iron in biological samples. Methods Enzymol 158: 357–364 [DOI] [PubMed] [Google Scholar]
- Frazzon J, Fick JR, Dean DR (2002) Biosynthesis of iron-sulphur clusters is a complex and highly conserved process. Biochem Soc Trans 30: 680–685 [DOI] [PubMed] [Google Scholar]
- Gill SC, von Hippel PH (1989) Calculation of protein extinction coefficients from amino acid sequence data. Anal Biochem 182: 319–326 [DOI] [PubMed] [Google Scholar]
- Hoff KG, Silberg JJ, Vickery LE (2000) Interaction of the iron-sulfur cluster assembly protein IscU with the Hsc66/Hsc20 molecular chaperone system of Escherichia coli. Proc Natl Acad Sci USA 97: 7790–7795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen LT, Culotta VC (2000) Role of Saccharomyces cerevisiae ISA1 and ISA2 in iron homeostasis. Mol Cell Biol 20: 3918–3927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato S, Mihara H, Kurihara T, Yoshimura T, Esaki N (2000) Gene cloning, purification, and characterization of two cyanobacterial NifS homologs driving iron-sulfur cluster formation. Biosci Biotechnol Biochem 64: 2412–2419 [DOI] [PubMed] [Google Scholar]
- Kaut A, Lange H, Diekert K, Kispal G, Lill R (2000) Isa1p is a component of the mitochondrial machinery for maturation of cellular iron-sulfur proteins and requires conserved cysteine residues for function. J Biol Chem 275: 15955–15961 [DOI] [PubMed] [Google Scholar]
- Keegstra K, Froehlich JE (1999) Protein import into chloroplasts. Curr Opin Plant Biol 2: 471–476 [DOI] [PubMed] [Google Scholar]
- Kessler D (2004) Slr0077 of Synechocystis has cysteine desulfurase as well as cystine lyase activity. Biochem Biophys Res Commun 320: 571–577 [DOI] [PubMed] [Google Scholar]
- Krebs C, Agar JN, Smith AD, Frazzon J, Dean DR, Huynh BH, Johnson MK (2001) IscA, an alternate scaffold for Fe-S cluster biosynthesis. Biochemistry 40: 14069–14080 [DOI] [PubMed] [Google Scholar]
- Lang T, Kessler D (1999) Evidence for cysteine persulfide as reaction product of L-cyst(e)ine C-S lyase (C-DES) from Synechocystis. J Biol Chem 274: 189–195 [DOI] [PubMed] [Google Scholar]
- Leon S, Touraine B, Ribot C, Briat JF, Lobreaux S (2003) Iron-sulphur cluster assembly in plants: distinct NFU proteins in mitochondria and plastids from Arabidopsis thaliana. Biochem J 371: 823–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leon S, Touraine B, Briat JF, Lobreaux S (2002) The AtNFS2 gene from Arabidopsis thaliana encodes a NifS-like plastidial cysteine desulphurase. Biochem J 366: 557–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lezhneva L, Amann K, Meurer J (2004) The universally conserved HCF101 protein is involved in assembly of [4Fe-4S]-cluster-containing complexes in Arabidopsis thaliana chloroplasts. Plant J 37: 174–185 [DOI] [PubMed] [Google Scholar]
- Lill R, Kispal G (2000) Maturation of cellular Fe-S proteins: an essential function of mitochondria. Trends Biochem Sci 25: 352–356 [DOI] [PubMed] [Google Scholar]
- Martin W, Stoebe B, Goremykin V, Hapsmann S, Hasegawa M, Kowallik KV (1998) Gene transfer to the nucleus and the evolution of chloroplasts. Nature 393: 162–165 [DOI] [PubMed] [Google Scholar]
- Morimoto K, Nishio K, Nakai M (2002) Identification of a novel prokaryotic HEAT-repeats-containing protein which interacts with a cyanobacterial IscA homolog. FEBS Lett 519: 123–127 [DOI] [PubMed] [Google Scholar]
- Morimoto K, Sato S, Tabata S, Nakai M (2003) A HEAT-repeats containing protein, IaiH, stabilizes the iron-sulfur cluster bound to the cyanobacterial IscA homologue, IscA2. J Biochem (Tokyo) 134: 211–217 [DOI] [PubMed] [Google Scholar]
- Muhlenhoff U, Gerber J, Richhardt N, Lill R (2003) Components involved in assembly and dislocation of iron-sulfur clusters on the scaffold protein Isu1p. EMBO J 22: 4815–4825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlenhoff U, Richhardt N, Ristow M, Kispal G, Lill R (2002) The yeast frataxin homolog Yfh1p plays a specific role in the maturation of cellular Fe/S proteins. Hum Mol Genet 11: 2025–2036 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol Plant 15: 437–497 [Google Scholar]
- Nishio K, Nakai M (2000) Transfer of iron-sulfur cluster from NifU to apoferredoxin. J Biol Chem 275: 22615–22618 [DOI] [PubMed] [Google Scholar]
- Ollagnier-de-Choudens S, Mattioli T, Takahashi Y, Fontecave M (2001) Iron-sulfur cluster assembly: characterization of IscA and evidence for a specific and functional complex with ferredoxin. J Biol Chem 276: 22604–22607 [DOI] [PubMed] [Google Scholar]
- Outten FW, Djaman O, Storz G (2004) A suf operon requirement for Fe-S cluster assembly during iron starvation in Escherichia coli. Mol Microbiol 52: 861–872 [DOI] [PubMed] [Google Scholar]
- Outten FW, Wood MJ, Munoz FM, Storz J (2003) The SufE protein and the SufBCD complex enhance SufS cysteine desulfurase activity as part of a sulfur transfer pathway for Fe-S cluster assembly in Escherichia coli. J Biol Chem 278: 45713–45719 [DOI] [PubMed] [Google Scholar]
- Pilon M, Rietveld AG, Weisbeek PJ, de Kruijff B (1992) Secondary structure and folding of a functional chloroplast precursor protein. J Biol Chem 267: 19907–19913 [PubMed] [Google Scholar]
- Pilon-Smits EAH, Garifullina GF, Abdel-Ghany S, Kato S, Mihara H, Hale KL, Burkhead JL, Esaki N, Kurihara T, Pilon M (2002) Characterization of a NifS-like chloroplast protein from Arabidopsis. Implications for its role in sulfur and selenium metabolism. Plant Physiol 130: 1309–1318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rensink WA, Pilon M, Weisbeek P (1989) Domains of transit sequence required for in vivo import in Arabidopsis chloroplast. Plant Physiol 118: 691–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silberg JJ, Hoff KG, Tapley TL, Vickery LE (2001) The Fe/S assembly protein IscU behaves as a substrate for the molecular chaperone Hsc66 from Escherichia coli. J Biol Chem 276: 1696–1700 [DOI] [PubMed] [Google Scholar]
- Smeekens S, Baurle C, Hageman J, Keegstra K, Weisbeek P (1986) The role of the transit peptide in the routing of pre-cursors toward different chloroplast compartments. Cell 46: 365–375 [DOI] [PubMed] [Google Scholar]
- Smillie RM, Entsch B (1971) Phytoflavin. Methods Enzymol 23: 504–514 [Google Scholar]
- Swofford DL (1993) PAUP: Phylogenetic Analysis Using Parsimony 3.1.1. Illinois Natural History Survey, Champaign, IL
- Takahashi Y, Mitsui A, Hase T, Matsubara H (1986) Formation of the iron sulfur cluster of ferredoxin in isolated chloroplast. Proc Natl Acad Sci USA 83: 2434–2437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y, Nakamura M (1999) Functional assignment of the ORF2-iscS-iscU-iscA-hscB-hscA-fdx-ORF3 gene cluster involved in the assembly of Fe-S clusters in Escherichia coli. J Biochem (Tokyo) 126: 917–926 [DOI] [PubMed] [Google Scholar]
- Takahashi Y, Tokumoto U (2002) A third bacterial system for the assembly of iron-sulfur clusters with homologs in archaea and plastids. J Biol Chem 277: 28380–28383 [DOI] [PubMed] [Google Scholar]
- Tokumoto U, Takahashi Y (2001) Genetic analysis of the isc operon in Escherichia coli involved in the biogenesis of cellular iron-sulfur proteins. J Biochem (Tokyo) 130: 63–71 [DOI] [PubMed] [Google Scholar]
- Tong WH, Jameson GN, Huynh BH, Rouault TA (2003) Subcellular compartmentalization of human Nfu, an iron-sulfur cluster scaffold protein, and its ability to assemble a [4Fe-4S] cluster. Proc Natl Acad Sci USA 100: 9762–9767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touraine B, Boutin J, Marion-Poll A, Briat J, Peltier G, Lobreaux S (2004) Nfu2: a scaffold protein required for [4Fe-4S] and ferredoxin iron-sulfur cluster assembly in Arabidopsis chloroplasts. Plant J 40: 101–111 [DOI] [PubMed] [Google Scholar]
- Urbina HD, Silberg JJ, Hoff KG, Vickery LE (2001) Transfer of sulfur from IscS to IscU during Fe/S cluster assembly. J Biol Chem 276: 44521–44526 [DOI] [PubMed] [Google Scholar]
- Wollenberg M, Berndt C, Bill E, Schwenn JD, Seidler A (2003) A dimer of the FeS cluster biosynthesis protein IscA from cyanobacteria binds a [2Fe2S] cluster between two protomers and transfers it to [2Fe2S] and [4Fe4S] apo proteins. Eur J Biochem 270: 1662–1671 [DOI] [PubMed] [Google Scholar]
- Wu G, Mansy SS, Hemann C, Hille R, Surerus KK, Cowan JA (2002) Iron-sulfur cluster biosynthesis: characterization of Schizosaccharomyces pombe Isa1. J Biol Inorg Chem 7: 526–532 [DOI] [PubMed] [Google Scholar]
- Xu X, Moller S (2004) AtNAP7 is a plastidic SufC-like ATP-binding cassette/ATPase essential for Arabidopsis embryogenesis. Proc Natl Acad Sci USA 101: 9143–9148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabe T, Morimoto K, Kikuchi S, Nishio K, Terashima I, Nakai M (2004) The Arabidopsis chloroplastic NifU-like protein CnfU, which can act as an iron-sulfur cluster scaffold protein, is required for biogenesis of ferredoxin and photosystem I. Plant Cell 16: 993–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye H, Garifullina G, Abdel-Ghany S, Zhang L, Pilon-Smits EAH, Pilon M (2005) The chloroplast NifS-like protein of Arabidopsis thaliana is required for iron-sulfur cluster formation in ferredoxin. Planta 220: 602–608 [DOI] [PubMed] [Google Scholar]
- Yocum CF (1982) Purification of ferredoxin and plastocyanin. In M Edelman, RB Hallick, N-H Chua, eds, Methods in Chloroplast Molecular Biology. Elsevier Biomedical Press, Amsterdam, pp 973–981
- Yuvaniyama P, Agar JN, Cash VL, Johnson MK, Dean DR (2000) NifS-directed assembly of a transient [2Fe-2S] cluster within the NifU protein. Proc Natl Acad Sci USA 97: 599–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L, Cash VL, Flint DH, Dean DR (1998) Assembly of iron-sulfur clusters. Identification of an iscSUA-hscBA-fdx gene cluster from Azotobacter vinelandii. J Biol Chem 273: 13264–13272 [DOI] [PubMed] [Google Scholar]
- Zheng L, White RH, Cash VL, Dean DR (1994) Mechanism for the desulfurization of L-cysteine catalyzed by the nifS gene product. Biochemistry 33: 4714–4720 [DOI] [PubMed] [Google Scholar]
- Zheng L, White RH, Cash VL, Jack RF, Dean DR (1993) Cysteine desulfurase activity indicates a role for NIFS in metallocluster biosynthesis. Proc Natl Acad Sci USA 90: 2754–2758 [DOI] [PMC free article] [PubMed] [Google Scholar]