Abstract
We identified four genes for potential equilibrative nucleoside transporters (ENTs) from rice (Oryza sativa; designated OsENT1 through OsENT4). Growth analysis of budding yeast (Saccharomyces cerevisiae) cells expressing OsENTs showed that OsENT2 transported adenosine and uridine with high affinity (adenosine, Km = 3.0 μm; uridine, Km = 0.7 μm). Purine or pyrimidine nucleosides and 2′-deoxynucleosides strongly inhibited adenosine transport via OsENT2, suggesting that OsENT2 possesses broad substrate specificity. OsENT2-mediated adenosine transport was resistant to the typical inhibitors of mammalian ENTs, nitrobenzylmercaptopurine ribonucleoside, dilazep, and dipyridamole. The transport activity was maximal at pH 5.0 and decreased slightly at lower as well as higher pH. In competition experiments with various cytokinins, adenosine transport by OsENT2 was inhibited by isopentenyladenine riboside (iPR). Direct measurements with radiolabeled cytokinins demonstrated that OsENT2 mediated uptake of iPR (Km = 32 μm) and trans-zeatin riboside (Km = 660 μm), suggesting that OsENT2 participates in iPR transport in planta. In mature plants, OsENT2 was predominantly expressed in roots. The OsENT2 promoter drove the expression of the β-glucuronidase reporter gene in the scutellum during germination and in vascular tissues in germinated plants, suggesting a participation of OsENT2 in the retrieval of endosperm-derived nucleosides by the germinating embryo and in the long-distance transport of nucleosides in growing plants, respectively.
Nucleotides participate in various biochemical processes in plants (Stasolla et al., 2003). They are essential precursors for nucleic acids, as well as metabolites participating in bioenergetic processes and in the synthesis of polysaccharides, phospholipids, and glycolipids (Ross, 1981). Furthermore, they are essential constituents of cytokinin (CK), a plant hormone controlling growth and development (Mok, 1994). During de novo nucleotide biosynthesis, nucleosides do not occur as intermediates, but appear during nucleotide turnover in the salvage pathway (Stasolla et al., 2003). Plant cells harbor enzymes allowing nucleoside salvage, and some of the genes encoding these enzymes have been identified and characterized (Stasolla et al., 2003). The enzymes that act on adenine and adenosine have also attracted the attention of researchers interested in CKs because these enzymes are involved in the metabolism of this group of phytohormones (Mok and Mok, 2001). For instance, adenine phosphoribosyltransferase (APT) converts free-base CKs into the corresponding nucleotides (Schnorr et al., 1996). Arabidopsis (Arabidopsis thaliana) mutants lacking an APT gene are male sterile and are impaired in CK metabolism (Moffatt et al., 1991; Gaillard et al., 1998).
Nucleosides are readily taken up into plant cells. Pollen cells of petunia (Petunia hybrida) are able to uptake exogenous nucleosides via both an active transport system and a facilitated diffusion (Kamboj and Jackson, 1984, 1985, 1987). Cotyledons separated from castor bean (Ricinus communis) efficiently import endosperm-derived secretion products, including purine and pyrimidine bases, nucleosides, and AMP, but not ATP (Kombrink and Beevers, 1983). Furthermore, when [3H]dihydrozeatin riboside and [3H]trans-zeatin riboside (tZR) were supplied to soybean (Glycine max) explants, they were incorporated and immediately metabolized into the corresponding nucleotides (Singh et al., 1988).
Although plant cells are capable of absorbing nucleosides, our knowledge of the mechanism of the process is limited. In general, nucleoside transporters have been categorized into concentrative nucleoside transporters (CNTs) and equilibrative nucleoside transporters (ENTs), which share no structural similarity (Hyde et al., 2001; Acimovic and Coe, 2002; Cabrita et al., 2002). The CNT system catalyzes the active transport of nucleosides against their concentration gradients. Typical CNT proteins possess 12 to 13 predicted transmembrane (TM) helices and are either sodium or proton symporters (Cabrita et al., 2002). Genes for CNTs have been identified in a variety of bacteria, Caenorhabditis elegans, Drosophila melanogaster, and several mammalian species (Cabrita et al., 2002). However, structurally homologous genes have not been found in Arabidopsis (Arabidopsis thaliana) and rice (Oryza sativa). On the other hand, ENT proteins typically exhibit 11 predicted TMs and catalyze the transport of nucleosides down their concentration gradients (Hyde et al., 2001; Cabrita et al., 2002). Genes for ENT family proteins have been identified in a variety of eukaryotes such as C. elegans and yeast (Saccharomyces cerevisiae), various parasites such as Trypanosoma brucei brucei and Leishmania donovani, mammals, and higher plants (Hyde et al., 2001; Acimovic and Coe, 2002; Cabrita et al., 2002). The Arabidopsis genome harbors eight ENT homologous genes (AtENTs), out of which AtENT1, AtENT3, AtENT4, AtENT6, and AtENT7 have been biochemically characterized (Li and Wang, 2000; Möhlmann et al., 2001; Li et al., 2003; Wormit et al., 2004). When heterologously expressed in yeast cells, these five AtENTs transport purine or pyrimidine nucleosides with broad substrate specificity. The AtENTs commonly have low sensitivity against typical inhibitors of mammalian ENT proteins such as nitrobenzylmercaptopurine ribonucleoside (NBMPR), dilazep, and dipyridamole (Möhlmann et al., 2001; Li et al., 2003; Wormit et al., 2004). The pH dependency of the adenosine transport activity differs between the AtENTs (Wormit et al., 2004). It has been suggested that AtENT1, AtENT3, and AtENT6 are localized in the plasma membrane (Li and Wang, 2000; Li et al., 2003; Wormit et al., 2004). In Arabidopsis suspension cells, the transcripts of AtENT1, AtENT3, AtENT4, AtENT6, and AtENT8 accumulated under nitrogen deprivation and following application of fluorouracil and methotrexate, two inhibitors of de novo nucleotide synthesis. This suggested that some AtENTs may be involved in the supply of substrates to the salvage pathway of nucleotide synthesis (Li et al., 2003). Although valuable information on the transport properties and the expression patterns of AtENTs has become available recently, their physiological roles remain to be elucidated, especially regarding their possible involvement in CK transport. Moreover, it is unfortunate that current knowledge of plant ENTs is restricted to Arabidopsis.
Here, we report the isolation of four ENT genes from rice designated OsENT1 through OsENT4. We demonstrate that OsENT2 is predominantly expressed in the vasculature and that its gene product can transport a wide spectrum of nucleosides, including nucleoside-type CKs. Potential functions of OsENT2 during rice development and participation in CK nucleoside transport are discussed.
RESULTS
Isolation of Members of the ENT Gene Family in Rice
To identify rice ENT genes, a BLAST search was performed in rice genome databases using the amino acid sequence of AtENT1 as a query. Four ENT genes were found and designated OsENT1, OsENT2, OsENT3, and OsENT4. The genes are distributed over two chromosomes (chromosome 8, OsENT1; chromosome 7, OsENT2, OsENT3, and OsENT4). The bacterial artificial chromosome clone (accession no. AP005125) on chromosome 7 contains a tandem repeat of three related OsENTs (OsENT2, OsENT3, and OsENT4). The cDNA clones of all OsENTs were found in the database of full-length cDNA clones from japonica rice at the Knowledge-based Oryza Molecular biological Encyclopedia (KOME; http://cdna01.dna.affrc.go.jp/cDNA): OsENT1 (accession no. AK059439), OsENT2 (AK102045, AK058524), OsENT3 (AK101098), and OsENT4 (AK065096). We obtained the cDNA clones of OsENT2 and OsENT4 from the Rice Genome Resource Center (www.rgrc.dna.affrc.go.jp). The OsENT1 cDNA clone deposited in the database at KOME appeared truncated, while the OsENT3 cDNA clone was not available from any of the above sources. Therefore, we isolated the cDNA clones of OsENT1 and OsENT3 by reverse transcription-PCR.
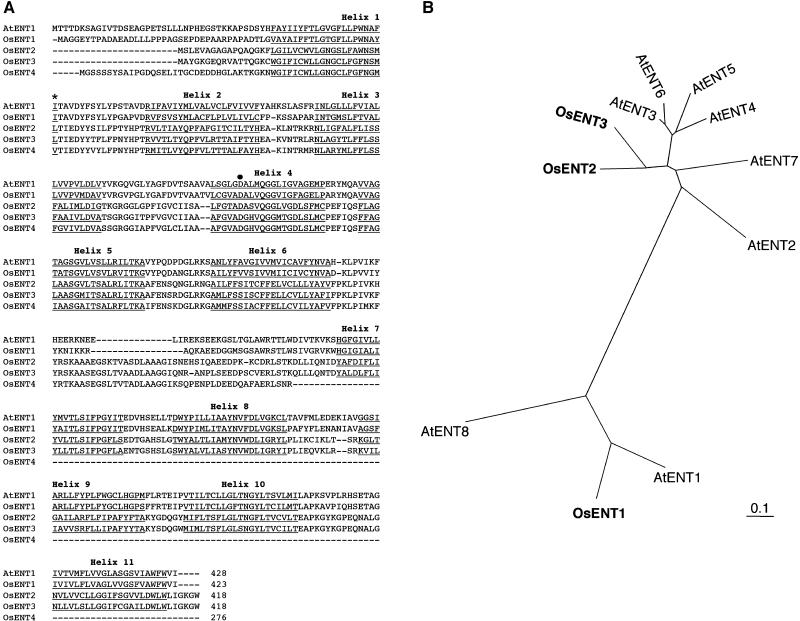
The cDNA clones of OsENT1, OsENT2, and OsENT3 contained reading frames of 423, 418, and 418 amino acids, respectively (Fig. 1A). These three OsENTs possessed 11 putative TMs (Fig. 1A). On the other hand, the OsENT4 cDNA clone contained a reading frame of 276 amino acids (Fig. 1A). Comparisons of the nucleotide sequence of the OsENT4 cDNA clone with that of the bacterial artificial chromosome clone (AP005125) revealed that OsENT4 carried a stop codon in the eighth exon. Thus, OsENT4 may be a pseudogene, at least in the Nipponbare cultivar, or it may encode a truncated form of typical ENT. OsENT1 exhibited 65% and 45% amino acid sequence identity with AtENT1 and AtENT8, respectively. OsENT2 and OsENT3 shared 71% amino acid sequence identity. Phylogenetic analysis indicated that OsENT2 and OsENT3 are more highly homologous to a group of Arabidopsis ENTs consisting of AtENT2 through AtENT7 than to AtENT1 or AtENT8 (Fig. 1B).
Figure 1.
Structural features of OsENTs. A, Multiple alignment of the predicted amino acid sequences of four OsENTs with that of AtENT1. Putative transmembrane helices are underlined. The asterisk highlights the amino acid in plant ENTs that corresponds to residue 33 of hENT1 and hENT2. The black dot marks the Gly residue at the 154 position of hENT1. B, Phylogenetic relationship of OsENT1, OsENT2, OsENT3, and AtENTs. Amino acid alignment of ENTs was performed using the ClustalW program at the DDBJ Web site. Bar = 0.1 amino acid substitutions per site.
Expression Patterns of OsENTs in Mature Rice Plants
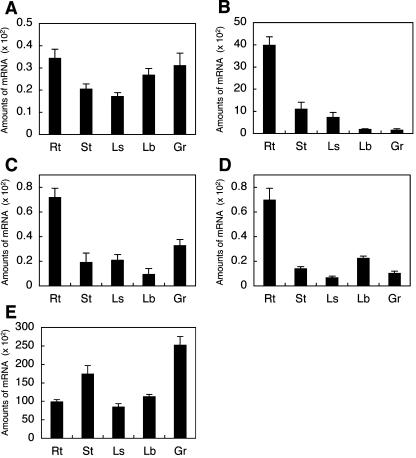
To evaluate levels of OsENT expression in different organs, the accumulation of OsENT transcripts was analyzed by quantitative real-time PCR using RNA samples extracted from various organs of mature rice plants. The transcripts of OsENTs were detected in all organs tested, but their distribution patterns differed (Fig. 2). Transcripts of OsENT2, OsENT3, and OsENT4 accumulated predominantly in roots, whereas that of OsENT1 did not show any pronounced preference. Accumulation levels of the OsENT2 transcript were significantly higher than those of the other OsENTs in all organs tested, suggesting that OsENT2 is the dominant form of the rice ENT family.
Figure 2.
Accumulation patterns of OsENT transcripts in various rice organs. Total RNA prepared from various organs was subjected to quantitative real-time PCR. A, OsENT1; B, OsENT2; C, OsENT3; D, OsENT4; E, Act1. Rt, Root; St, stem; Ls, leaf sheath; Lb, leaf blade; Gr, grain. Amounts of mRNA are given as the copy number of OsENT mRNA/ng total RNA. Real-time PCR was performed three times; values shown are means ± sd. Actin1 was used as an internal standard.
Growth Analysis of Yeast Expressing OsENTs
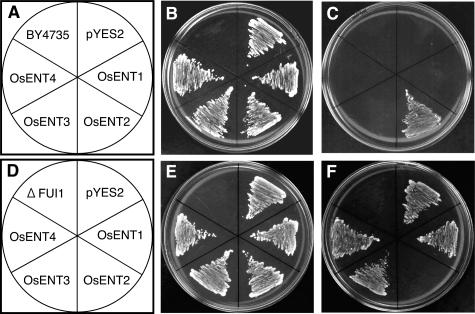
Under normal conditions, yeast cells do not uptake exogenous adenosine because of the lack of an endogenous transport system for the nucleoside and cannot use it as a purine source (Mässer et al., 1999). Exploiting this property, adenine auxotroph ade2 mutant strains, which are unable to synthesize adenine, have been successfully employed to evaluate the uptake of adenosine via AtENTs (Möhlmann et al., 2001; Li et al., 2003; Wormit et al., 2004). Either the presence of adenine in the medium or the endowment with a transgenic adenosine uptake system enables the yeast strain to grow in a minimal medium. The transport properties of AtENTs have also been evaluated in the yeast mutant ΔFUI1, which lacks the plasma membrane-localized uridine transporter FUI1 (Vickers et al., 2000). To examine whether OsENTs transport the nucleosides, we analyzed growth in the yeast mutants equipped with OsENT genes in the presence of either adenosine (a purine nucleoside) or fluorouridine (a toxic pyrimidine nucleoside analog). Although OsENT4 appears to be truncated, we expressed all OsENTs. Figure 3, A to C, shows the growth patterns of the BY4735 (ade2) strain, while Figure 3, D to F, shows those of the ΔFUI1 (fui1) strain. In both cases, the plain recipient cells (BY4735 and ΔFUI1) could not grow on media lacking uracil, a selection substance for the pYES2 expression vector (Fig. 3, B and E). OsENT2 was the only OsENT able to complement the ade2 mutation on an adenosine-containing medium (Fig. 3C). On the other hand, although ΔFUI1 cells with the empty pYES2 vector, as well as ΔFUI1 cells expressing OsENT1, OsENT3, or OsENT4, were able to grow in the presence of the toxic compound fluorouridine (Fig. 3F), expression of OsENT2 proved lethal. These results showed that OsENT2 mediated adenosine and uridine transport in the yeast system.
Figure 3.
Growth analysis of yeast cells expressing OsENTs. Growth of yeast cells harboring expression plasmids containing OsENT1, OsENT2, OsENT3, OsENT4, or controls (BY4735; BY4735 plain recipient cells, ΔFUI1; ΔFUI1 plain recipient cells, pYES2; cells transformed with empty vector pYES2). A, Schematic representation of BY4735 strains used in B and C. B and E, Growth on synthetic minimal medium lacking uracil but containing Gal (20 g L−1). C, Growth on synthetic minimal medium lacking uracil and adenine but containing adenosine (150 μm) and Gal. D, Schematic representation of ΔFUI1 strains used in E and F. F, Growth on synthetic minimal medium lacking uracil but containing fluorouridine (1 mm) and Gal. Cells were grown for 2 to 3 d at 30°C.
Transport Properties of Yeast Cells Expressing OsENT2
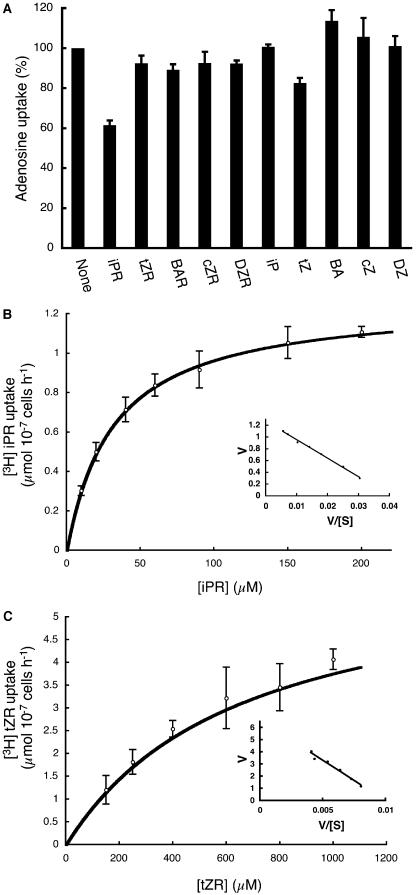
The properties of OsENT2-mediated adenosine and uridine transport were examined using the ade2 and ΔFUI1 mutants, respectively. In initial time course experiments with [3H]adenosine and [3H]uridine, the uptake rate of both radiochemicals was practically constant for the first 10 min (data not shown). Therefore, subsequent quantifications of uptake kinetics were based on a 2-min period. Figure 4 shows the Michaelis-Menten kinetics of adenosine and uridine uptake mediated by OsENT2. The deduced Eadie-Hofstee plots (Fig. 4, insets) indicate Km values for adenosine and uridine of 3.0 and 0.7 μm, respectively. The Vmax values were 3.9 μmol (107 cells)−1 h−1 for adenosine, and 1.6 μmol (107 cells)−1 h−1 for uridine. Thus, OsENT2 exhibited higher affinity for uridine than for adenosine, whereas the Vmax was lower. In competition experiments, [3H]adenosine uptake via OsENT2 was significantly and consistently reduced by a series of purine and pyrimidine nucleosides and 2′-deoxynucleosides (Table I), suggesting that OsENT2 possesses broad substrate specificity for nucleosides. In addition, nucleotides slightly inhibited [3H]adenosine uptake, while none of the nucleobases tested exerted any significant effects (Table I).
Figure 4.
Characterization of [3H]adenosine and [3H]uridine uptake by yeast cells expressing OsENT2. [3H]adenosine uptake was examined in BY4735 ade2 cells expressing OsENT2 (A), whereas [3H]uridine uptake was tested in the yeast mutant ΔFUI1 expressing OsENT2 (B). Cells were incubated for 2 min at the indicated substrate concentrations at pH 6.0. Background uptake rates (empty vector pYES2) were subtracted. Affinity constants were calculated on the basis of Eadie-Hofstee plots (insets). Data shown are means ± se (n = 3).
Table I.
Inhibition of OsENT2-mediated [3H]adenosine uptake in BY4735 ade2 yeast cells by nucleosides, nucleobases, and nucleotides
[3H]Adenosine uptake was examined at a substrate concentration of 2.0 μm for 2 min at pH 6.0. The concentrations of various additives were 20 μm. Mean ± se (n = 3).
| Additive | [3H]Adenosine Uptake (% of Control) |
|---|---|
| None | 100 |
| Nucleosides: | |
| Adenosine | 23.8 ± 2.6 |
| Cytidine | 35.1 ± 4.0 |
| Guanosine | 20.4 ± 3.5 |
| Uridine | 13.3 ± 2.2 |
| Inosine | 16.6 ± 2.4 |
| 2′-Deoxyadenosine | 37.5 ± 5.6 |
| 2′-Deoxycytidine | 25.3 ± 3.8 |
| 2′-Deoxyguanosine | 29.7 ± 3.3 |
| 2′-Deoxythymidine | 6.7 ± 1.6 |
| 2′-Deoxyinosine | 24.4 ± 3.2 |
| Nucleobases: | |
| Adenine | 100.5 ± 0.7 |
| Cytosine | 94.6 ± 0.9 |
| Guanine | 98.8 ± 2.4 |
| Thymine | 89.7 ± 2.6 |
| Uracil | 85.7 ± 2.8 |
| Hypoxanthine | 79.7 ± 2.1 |
| Nucleotides: | |
| dATP | 77.2 ± 2.8 |
| ATP | 73.5 ± 1.5 |
| ADP | 71.9 ± 0.3 |
| AMP | 72.3 ± 2.7 |
| CTP | 69.4 ± 2.8 |
| GTP | 62.9 ± 0.1 |
| UTP | 64.4 ± 1.0 |
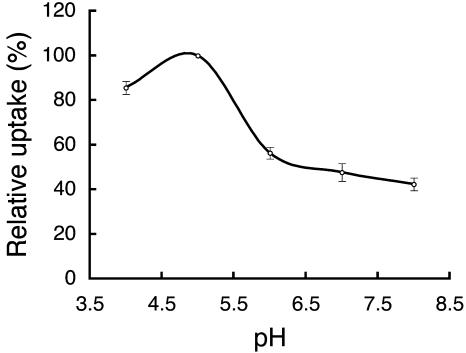
The sensitivity of OsENT2-mediated uptake of [3H]adenosine uptake to typical inhibitors of mammalian ENT proteins, such as NBMPR, dilazep, dipyridamole, and the protonophore, carbonyl cyanide m-chlorophenyl-hydrazone (CCCP), was tested. The effector concentrations and duration of the treatment in our experiments were almost identical to those in previous studies on AtENTs (Möhlmann et al., 2001; Li et al., 2003; Wormit et al., 2004). All three inhibitors of mammalian ENTs tested did not affect the transport activity of OsENT2 (Table II). On the other hand, CCCP reduced [3H]adenosine uptake by about 30% (Table II). To characterize the pH dependency of the transport in more detail, we examined OsENT2-mediated [3H]adenosine transport at different extracellular pH values. The maximum transport activity was observed at pH 5.0 (Fig. 5), but 40% of the activity still was recorded at pH 8.0. These characteristics of OsENT2 transport, namely, low sensitivity to CCCP and slight dependency on pH, resemble those of AtENT3 (Li et al., 2003; Wormit et al., 2004).
Table II.
Inhibition of OsENT2-mediated [3H]adenosine uptake in BY4735 ade2 yeast cells
[3H]Adenosine uptake was examined at a substrate concentration of 2.0 μm for 2 min at pH 6.0. The concentration of the inhibitor applied is shown in parenthesis. Mean ± se (n = 3).
| Additive | [3H]Adenosine Uptake (% of Control) |
|---|---|
| None | 100 |
| NBMPR (20 nm) | 97.2 ± 3.7 |
| NBMPR (20 μm) | 86.6 ± 1.1 |
| Dilazep (20 nm) | 98.2 ± 0.2 |
| Dilazep (20 μm) | 95.9 ± 3.9 |
| Dipyridamole (20 nm) | 96.9 ± 4.1 |
| Dipyridamole (20 μm) | 91.4 ± 4.0 |
| CCCP (5 μm) | 70.7 ± 3.7 |
Figure 5.
pH dependency of [3H]adenosine uptake into BY4735 ade2 yeast cells expressing OsENT2. [3H]adenosine uptake was tested at a substrate concentration of 2.0 μm for 2 min at various pH. Data shown are means ± se (n = 3).
OsENT2 Transports Nucleoside-Type CKs in Yeast
Recently, the Arabidopsis purine transporters AtPUP1 and AtPUP2 have been shown to translocate adenine and its various derivatives, such as caffeine, and as well as CK nucleobases (Gillissen et al., 2000; Bürkle et al., 2003). In this context, we speculated that OsENT2 might play a role as a CK nucleoside transporter. Competition experiments using various CKs showed that the transport of [3H]adenosine mediated by OsENT2 was partially inhibited by isopentenyladenine riboside (iPR; Fig. 6A). The main characteristics of iPR and trans-zeatin riboside (tZR) transport through OsENT2 were examined using the yeast ade2 mutant. Since [3H]iPR and [3H]tZR were chemically unstable (data not shown), we prepared them just prior to the transport experiments as described previously (Takei et al., 2003). In initial time course experiments, the uptake rate of both radiochemicals was found to be practically constant for the first 10 min (data not shown). Consequently, uptake periods of 5 min formed the basis of kinetic analyses in subsequent experiments. Eadie-Hofstee plots (Fig. 6, inset) deduced from the Michaelis-Menten kinetics of OsENT2-mediated uptake of iPR and tZR (Fig. 6) indicated Km values for iPR and tZR of 32 and 660 μm, respectively. The Vmax values for uptake were 1.3 μmol (107 cells)−1 h−1 for iPR and 6.2 μmol (107 cells)−1 h−1 for tZR. We concluded that OsENT2 transported CK nucleosides and had a higher affinity for iPR than for tZR.
Figure 6.
Uptake of nucleoside-type CKs by yeast cells expressing OsENT2. A, Inhibition of OsENT2-mediated [3H]adenosine uptake in BY4735 ade2 yeast cells by various CKs. [3H]adenosine uptake was tested at a substrate concentration of 2.0 μm for 2 min at pH 6.0. The concentrations of the various additives were 20 μm. [3H]iPR (B) and [3H]tZR (C) uptake was examined in BY4735 ade2 cells expressing OsENT2. Cells were incubated for 5 min at the substrate concentrations indicated at pH 6.0. Background uptake rates (empty vector pYES2) were subtracted. Affinity constants were calculated on the basis of an Eadie-Hofstee plot (inset). Data shown are means ± se (n = 3). BAR, benzyladenine riboside; cZR, cis-zeatin riboside; DZR, dihydrozeatin riboside; BA, benzyladenine; cZ, cis-zeatin; DZ, dihydrozeatin.
OsENT2 Is Predominantly Expressed in the Vasculature and during Germination
To study the tissue specificity of OsENT2 expression throughout plant development, the promoter region was isolated and fused to the Escherichia coli β-glucuronidase (GUS) gene. Twenty-five independent T1 transgenic lines were analyzed for reporter gene activity. All lines analyzed showed a very similar pattern of expression, varying only in the intensity of GUS staining. Four representative lines were used to determine the expression pattern in more detail.
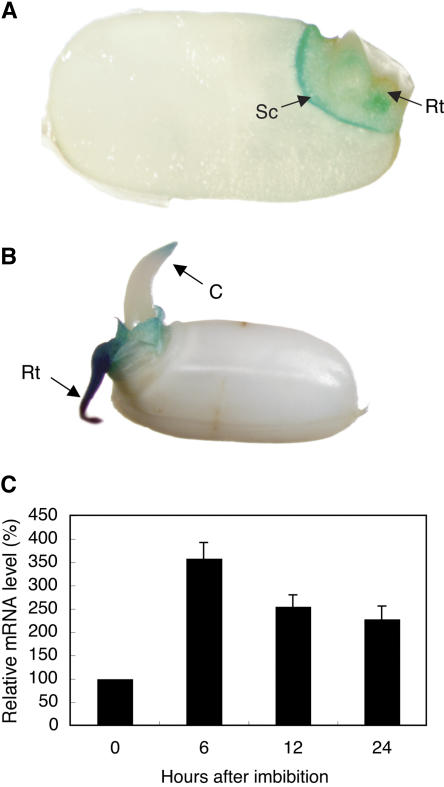
In 1-d-old seedlings, a strong signal for OsENT2 promoter activity was detected predominantly in the root primordium and in the scutellum (Fig. 7A). Intense staining was also observed in the root and in the coleoptile tip of 2-d-old seedlings (Fig. 7B). OsENT2 mRNA accumulation was induced within 6 h after the initiation of germination by imbibition of the dry seeds, indicating that OsENT2 gene expression is up-regulated at the earliest stages of germination (Fig. 7C). The GUS staining in the scutellum was observed for at least 2 weeks after the start of imbibition (data not shown).
Figure 7.
Expression of the OsENT2 gene during germination. Histochemical localization of OsENT2-promoter-controlled GUS activity in seedlings at 1 d (A) and 2 d (B) after the start of imbibition. A longitudinal section of a representative grain is shown in A. C, Coleoptile; Rt, root; Sc, scutellum. Relative amount of OsENT2 mRNA during germination is shown in C. The OsENT2 mRNA level was normalized with respect to the corresponding actin transcript level. The data are given as relative values with respect to the value determined at 0 h. Data shown are means ± se (n = 3).
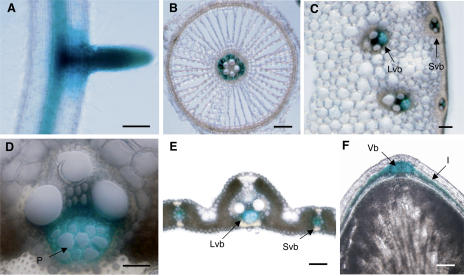
In adult plants, the OsENT2 promoter was active in the vasculature throughout the whole plant (roots, stems, leaves, and grains; Fig. 8, A–F). In roots, GUS staining derived from the OsENT2 promoter activity was observed within the stele and lateral roots, but not in epidermal, exodermal, or cortical cells (Fig. 8, A and B). In leaf sheaths, GUS activity was detected exclusively in the phloem (Fig. 8D). In 10-DPA-old grains, the GUS signal was found in the vascular bundle on the dorsal side and in the inner integument (Fig. 8F). Taken together, these results suggested that OsENT2 participates in the long-distance transport of nucleosides.
Figure 8.
Tissue specificity of the activity of the OsENT2 promoter. Histochemical localization of OsENT2 promoter-controlled GUS activity in a representative root (A), a root cross-section (B), a stem cross-section (C), a vascular bundle in a leaf sheath (D), a leaf blade cross-section (E), and a longitudinal section of a grain (F). I, Inner integument; Lvb, large vascular bundle; P, phloem; Svb, small vascular bundle; Vb, vascular bundle on the dorsal side of grain. Bar = 500 μm for A; 100 μm for B, C, E and F; 50 μm for D.
DISCUSSION
In contrast to the Arabidopsis genome, which harbors eight ENT genes, the rice genome contains only four (Fig. 1A). OsENT1, OsENT2, and OsENT3 possessed the 11 putative TMs that are typical of ENTs. On the other hand, OsENT4 appeared to be carboxy-terminally truncated. In this context, it is interesting that the human ENT2 gene (hENT2) is alternatively spliced to produce a number of variants, including a hydrophobic nucleolar family of proteins (Sankar et al., 2002). An amino-terminally truncated form of hENT2 failed to stimulate uridine transport in transiently transfected cultured cells (Crawford et al., 1998). Additional studies are required to determine whether OsENT4 actually is translated as a truncated ENT in rice and whether it has a role in nucleoside transport.
Growth analysis of yeast cells expressing OsENTs and competition tests indicated that OsENT2 can transport both purine and pyrimidine nucleosides, while OsENT1, OsENT3, and OsENT4 did not exhibit any transport competence (Fig. 3; Table I). Not all ENTs may be functionally expressed in this yeast system: For instance, AtENT8, an Arabidopsis ENT, also did not exhibit transport activity when it was expressed in ade2 and fui1 mutants (Wormit et al., 2004). Further studies will establish whether the lack of transport activity we observed in OsENT1 and OsENT3 was due to ineffective plasma membrane targeting in the heterologous expression system and whether these two OsENTs transport substrates other than nucleosides.
OsENT2 resembles AtENT1, AtENT3, AtENT4, AtENT6, and AtENT7 (Möhlmann et al., 2001; Li et al., 2003; Wormit et al., 2004) in its low sensitivity to NBMPR, dilazep, and dipyridamole (Table II). Amino acids at positions 33 and 154 have been identified as the residues responsible for the difference in the sensitivity to vasodilator drugs that is observed between hENT1 and hENT2 (Hyde et al., 2001; Visser et al., 2002, 2005). Comparing the sequences of AtENTs with that of hENT1, Li et al. (2003) found that Met-33 of hENT1 is replaced by Ile or Leu and that Gly-154 of hENT1 is exchanged for Asp in all AtENTs. The authors speculated that these two exchanges are responsible for the low sensitivity of AtENTs to vasodilator drugs. In OsENT2, Leu and Asp occupied the positions corresponding to residues 33 and 154 of hENT1, respectively (Fig. 1A). Since OsENT2 is also resistant to vasodilator drugs, as demonstrated in this investigation, these two residues appear likely to be involved in the resistance to vasodilators of all plant ENTs tested so far.
Our analysis of cloned OsENT2 implicates a role of the ENT as a CK nucleoside transporter. At first sight, the affinity constant for the CK nucleosides, especially that of tZR, may seem high (Fig. 6) compared to the predicted concentration of CKs in plant tissues in the nanomolar range. In some enzymes of the purine salvage pathway and of CK metabolism, the affinity constants and specificity constants have been determined. As for cloned Arabidopsis adenosine kinases, Km values for iPR and adenosine were 3 to 5 μm and 0.3 to 0.5 μm, respectively, while the Vmax/Km for iPR was 100-fold lower than for adenosine (Moffatt et al., 2000). The Km value of cloned APT2 for isopentenyladenine (iP) was 110 μm and that of APT3 for trans-zeatin (tZ) was 76 μm. In both cases, the Vmax/Km was significantly lower than for adenine (Allen et al., 2002). Km values of cloned maize (Zea mays) O-glucosyltransferases for cis-zeatin were between 46 and 95 μm (Veach et al., 2003), and the Km values of cloned Arabidopsis N-glucosyltransferases for iP and tZ ranged from 70 to 240 μm (Hou et al., 2004). In cloned maize CK oxidase/dehydrogenase, a CK-degrading enzyme, the Km values for iP and tZ ranged from 1.5 to 14 μm (Bilyeu et al., 2001). Thus, the substrate affinities of enzymes involved in CK metabolism and that of OsENT2 seem to be in a compatible range (at least for iPR). Moreover, the Km value of a CK nucleobase transporter, AtPUP1, for tZ was calculated to be around 40 μm (Bürkle et al., 2003). In this context, we speculated that OsENT2 functions as an iPR transporter in planta.
Analysis of OsENT2 promoter:GUS transgenic rice enabled us to deduce the physiological function of OsENT2 (Figs. 7 and 8). OsENT2 was predominantly expressed in the scutellum of the germinating seed (Fig. 7A). The epithelium layer in the dorsal portion of the scutellum elongates and develops during germination and functions as the absorptive tissue of storage reserves from the endosperm (Hoshikawa, 1989). Significant levels of nucleosides such as adenosine and guanosine were released into the incubation medium when halved endosperm of castor been seeds was incubated in buffer (Kombrink and Beevers, 1983). Active nucleotide biosynthesis is needed to provide the embryos with sufficient purine and pyrimidine nucleotides to support nucleic acid synthesis (Deltour, 1985). In this context, we assumed that OsENT2 might be involved in the uptake of nucleosides derived from the endosperm, which are subsequently metabolized to nucleotides in developing embryos. Strong GUS staining was detected in the roots of 2-d-old seedlings (Fig. 7B) and in lateral roots of mature plants (Fig. 8A), reflecting the high demand for nucleotides in young sink organs. Real-time PCR analysis also indicated that OsENT2 generally is at high levels in roots of mature plants (Fig. 2B).
Predominant expression of OsENT2 in vascular tissues throughout the whole plant (Fig. 8, A–F), and particularly in the phloem of leaf sheaths (Fig. 8D), suggests an involvement of OsENT2 in the long-distance transport of nucleosides by loading and unloading into and from the phloem, respectively. OsENT2 was expressed in the vascular tissues of grains during the period of seed loading, pointing to a role of OsENT2 in providing grains with nucleosides, possibly derived from senescing tissues where nucleosides are liberated by the nucleolytic breakdown of RNA and DNA. Long-distance translocation of nucleoside-type CKs is believed to occur in the xylem and phloem (Letham, 1994). The highest concentrations of CKs were found in the developing rice seed (Yang et al., 2001, 2002). It has been suggested that cell number and cell division activity in rice endosperm are regulated by CK levels in this tissue (Yang et al., 2002). OsENT2 might contribute to the retrieval of CKs transported through the vascular systems in developing grains.
Distinct distributions of iPR and tZR in phloem and xylem, respectively, were reported in Sinapis alba (Lejeune et al., 1994). We found similarly distinct distributions of CK nucleosides in rice as well (N. Hirose and H. Sakakibara, unpublished data). Considering that OsENT2 has different affinities for iPR and tZR, vascular-localized OsENT2 might play an important role in the selective transport of nucleoside-type CKs. The sites of CK biosynthesis recently have been revealed in Arabidopsis (Miyawaki et al., 2004; Takei et al., 2004), but not yet in rice. To more precisely deduce the possible roles of OsENT2 in CK transport, future studies will have to focus on the identification of the sites of CK biosynthesis in rice. Furthermore, detailed analyses of knockout or RNA interference mutants will enable us to further elucidate the physiological roles of OsENT2 in the transport of nucleosides, including nucleoside-type CKs.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
For the determination of organ distribution of OsENT mRNA and histochemical analysis, rice (Oryza sativa) L. cv Nipponbare plants were hydroponically grown in an environment-controlled greenhouse at a 12-h-light (30°C)/12-h-dark (25°C) photoperiod as described by Kamachi et al. (1991). For the propagation of transgenic rice plants, single plants were grown in 0.3-L pots with 150 g of synthetic culture soil (Mitsui-Toatsu No. 3; Mitsui-Toatsu, Tokyo) in a greenhouse under the conditions described above. For germination experiments, rice grains were surface sterilized and incubated on wet filter paper at 30°C in the dark.
Cloning of OsENT cDNAs and Comparative Analysis of Amino Acid Sequence of OsENTs
The OsENT genes were identified by BLAST queries in GenBank (www.ncbi.nlm.nih.gov/BLAST), The Institute for Genomic Research (TIGR rice genome project; http://www.tigr.org/tdb/e2k1/osa1/index.shtml), the DNA Data Bank of Japan (DDBJ; http://www.ddbj.nig.ac.jp/search/blast-j.html), and the rice cDNA database at KOME (http://cdna01.dna.affrc.go.jp/cDNA) using the amino acid sequence of AtENT1 as a search sequence. To amplify cDNAs of OsENT1 and OsENT3 using reverse transcription-PCR, the following primers were used: 5′-AAGATCCCCACCCAAATCCACCTC-3′ and 5′-AGCATGGCACTATGATACACTGAC-3′ for OsENT1; 5′-GGGTCATCCTAGTTGACTACAAAC-3′ and 5′-TGGGAAAAAGACATGTAATGCAACTATG-3′ for OsENT3. Total RNA was prepared from 3-week-old seedlings with the RNeasy plant mini kit (Qiagen, Valencia, CA) with RNase-free DNase I (Qiagen). cDNA was synthesized by SuperScriptII reverse transcriptase (Invitrogen, Carlsbad, CA) with oligo(dT)12–18 primers. The amplified fragment was cloned into pCR-Blunt-TOPO (Invitrogen) and sequenced to confirm that no substitution had occurred during PCR. Analysis of cDNA and amino acid sequences was carried out using GENETYX-MAC version 11 (Software Development, Tokyo). To investigate phylogenetic relationships among OsENTs and AtENTs, the amino acid sequences were aligned using the ClustalW program (Thompson et al., 1994) at the DDBJ Web site with default settings; phylogenetic trees were developed with Tree View (Page, 1996). The locations of transmembrane helices in OsENTs were predicted by the program TMHMM 2.0 (http://www.cbs.dtu.dk/services/TMHMM) with manual adjustment aided by the comparison of the amino acid sequences of OsENTs with that of hENT1 for which structural information is currently available (Sundaram et al., 2001).
Quantitative Real-Time PCR
Total RNA was prepared from the roots, stems, leaves, and young grains (10 DPA) of ripening rice plants with the RNeasy plant mini kit (Qiagen) and from mature grains by the method of Shirzadegan et al. (1991). Total RNA extracted was treated with RNase-free DNase I (Qiagen). cDNA was synthesized using SuperScriptII reverse transcriptase (Invitrogen) with oligo(dT)12–18 primers. Accumulation levels of the transcripts of interest were analyzed by a real-time PCR method, with the ABI Prism 7000 sequence detection system (PE-Applied Biosystems, Foster City, CA). The primers for PCR were designed using Primer Expression software (PE-Applied Biosystems) and checked for the specific product formation by polyacrylamide gel analysis. The sequences of the primers used for PCR were 5′-GCTCAGAAGGCAGAGGAAGATG-3′ and 5′-CAACAATGCTCCACAAGGTTGA-3′ for OsENT1; 5′-CATCGTGAAGCACTACCGATCA-3′ and 5′-CCTGCAGCAGCAAGATCACTT-3′ for OsENT2; 5′-AGCTGAAAACACTGGATCACACA-3′ and 5′-ATACGTTATAGCTCGCGATCAAGA-3′ for OsENT3; 5′-TGCATCGGGTGCAATAACC-3′ and 5′-GGCCATCTTTGGAGTTCTCAAA-3′ for OsENT4; 5′-CGAGGCGCAGTCCAAGAG-3′ and 5′-CCCAGTTGCTGACGATACCA-3′ for actin1 (ACT1; McElroy et al., 1990). In each case, plasmid DNA containing the corresponding cDNAs was used as a template to generate a calibration curve.
Expression of OsENTs in the Yeast Mutant
The coding regions of OsENT cDNAs were amplified by PCR using the primers 5′-TGGATCCAAGCGAAAGGCTCAAGAAAGC-3′ and 5′-GTCTAGATCAAATGACCCAAAACCAAGC-3′ for OsENT1; 5′-GAAGCTTATCATGAGCCTCGAGGTCGCA-3′ and 5′-GTCTAGATCACCAGCCTTTTCCTATC-3′ for OsENT2; 5′-AGGTACCATGGCATATGGTAAAGGAG-3′ and 5′-GTCTAGATCACCACCCTTTACCTATC-3′ for OsENT3; 5′-AGGTACCATGGGCAGCTCTTCCTCC-3′ and 5′-ACTCGAGCTATCTGTTGCTTAATCG-3′ for OsENT4. The amplified fragments were cloned into pCR-Blunt-TOPO (Invitrogen) and sequenced. The OsENT1 cDNA was cut out as a BamHI-XbaI fragment and was cloned into BamHI-XbaI sites of pYES2 (Invitrogen). The OsENT2 cDNA was cut out as a HindIII-XbaI fragment and was ligated into HindIII-XbaI sites of pYES2. The OsENT3 cDNA was cut out as a KpnI-XbaI fragment and was cloned into KpnI-XbaI sites of pYES2. The OsENT4 cDNA was cut out as a KpnI-XhoI fragment and was ligated into KpnI-XhoI sites of pYES2. The resulting plasmids were transformed into a yeast (Saccharomyces cerevisiae) ade2 strain (BY4735, Mat α, ade2Δ∷hisG, his3Δ200, leu2Δ0, met15Δ0, trp1Δ63, ura3Δ0, purchased from America Type Culture Collection, Manasas, VA) or the yeast mutant ΔFUI1 (BY4742, Mat α, his3Δ1, leu2Δ0, lys2Δ0, ura3Δ0, YBL042C∷KanMX4, purchased from Open Biosystems, Huntsville, AL) by the lithium acetate method (Gietz et al., 1992), and the transformants were selected on synthetic minimal medium lacking uracil (BD Biosciences Clontech, Palo Alto, CA) as described in Yeast Protocols Handbook (Clontech). For plate assays, the cells were grown on three types of medium: minimal medium lacking uracil but containing Gal (20 g L−1), minimal medium lacking both uracil and adenine but containing adenosine (150 μm) and Gal, and minimal medium lacking uracil but containing fluorouridine (1 mm) and Gal.
The uptake of tritiated adenosine, iPR, and tZR was evaluated by using the BY4735 ade2 yeast strain, while that of [3H]uridine was assessed in the yeast mutant ΔFUI1. Gal (20 g L−1) was applied to culture media as a sole carbon source to activate the GAL1 promoter on pYES2. Cells were cultured to an A600 of 0.8 to 1.2. They were harvested by centrifugation (4,000g, 10 min), and washed twice with 25 mm phosphate buffer, pH 6.0. Washed cells were resuspended in the same buffer to an A600 of 5. For uptake experiments at different pH, cells were first suspended in distilled water, distributed, and resuspended in 25 mm phosphate buffer of the given pH. The [3H]nucleoside uptake was measured by the method of Smith et al. (1995), with minor modification (Kataoka et al., 2004). The uptake was terminated by centrifuging (10,000g, 1 min) 100 μL of the yeast cell suspension through 150 μL of 1:1 silicone oil (SH-150):dinonylphthalate into 14 μL of 60% perchloric acid (Nakalai Tesque, Kyoto) placed at the bottom of 250-μL microcentrifuge tubes. After centrifugation, the tips of these tubes were cut off and the contents were transferred into 2 mL of Ultima Gold scintillation cocktail (Perkin-Elmer, Boston) to determine the radioactivity with a liquid scintillation counter (Aloka, Tokyo). For investigating the sensitivity of OsENT2-mediated transport to inhibitors, vasodilator drugs (dilazep or dipyridamole; Sigma, St. Louis), a nucleoside analog (NBMPR; Research Biochemicals, Natick, MA), or a protonophore (CCCP; Sigma) were added to the yeast cell suspensions 30 min before the addition of [3H]adenosine (Visser et al., 2002).
Transformation of Rice
Genomic sequences containing putative promoter regions of OsENT2 (−2,012 to −1 bp from the translational initiation codon) were amplified by PCR with genomic DNA. Primers were forward, 5′-GAAGCTTCTTCTTTGATTTTTATGGTCATC-3′ and reverse, 5′-AGTCGACGATAAAGTTCACCTGCGCCTATTC-3′. The resulting PCR product was cloned into pCR-Blunt-TOPO (Invitrogen) and verified by complete sequencing. The verified fragments were introduced into the upstream region of the open reading flame of the GUS gene that is inserted into the SmaI-SpeI site of the pCAMBIA1390 vector (CAMBIA, Canberra, Australia).
Agrobacterium tumefaciens (EHA101) carrying the above construct was used to transform rice following the method of Hiei et al. (1994). Hygromycin B-resistant plants from callus, defined as transgenic plants of the T0 generation, were transplanted into soil and grown in a greenhouse. The primary transformants were self-pollinated and the resulting seeds (T1) were collected.
Histochemical Analysis
Histochemical analysis of GUS activity was performed by the method of Jefferson (1987), modified by Kosugi et al. (1990). The tissues of transgenic plants were infiltrated with GUS reaction buffer: 1 mm 5-bromo-5-chloro-3-indolyl-β-d-glucuronide, 0.5% Triton X-100, 50 mm sodium phosphate buffer, pH 7.0, and 20% (v/v) methanol. For fresh sections, the tissues of transgenic plants were cut with a scalpel into approximately 0.5-cm sections. These sections were embedded in 5% agar and then cut into 80- to 130-μm sections using a DTK-100 microslicer (Dosaka EM, Kyoto). The sections were incubated at 37°C for between 30 min and overnight in GUS reaction buffer. After staining, the sections were washed in 70% ethanol overnight to remove chlorophyll and stored in 70% ethanol until further analysis. GUS staining was observed using an Olympus BX100 microscope (Olympus, Tokyo) following the manufacturer's instructions.
Chemicals
iP, tZ, benzyladenine, dihydrozeatin, iPR, tZR, and dihydrozeatin riboside were purchased from Sigma, cis-zeatin from ICN Biomedicals (Irvine, CA), benzyladenine riboside from Fluka (Buchs, Switzerland), and [2-3H]adenosine (TRK423; 814 GBq/mmol) and [5-3H]uridine (TRK178; 1.04 TBq/mmol) from Amersham Biosciences (Uppsala). The preparations and quality checks of [3H]iPR and [3H]tZR were performed as described previously (Takei et al., 2003).
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers AP005125, AK059439, AK102045, AK058524, AK101098, AK065096, and AB201113.
Acknowledgments
We thank the Rice Genome Project of the National Institute of Agrobiological Sciences and the Rice Genome Resource Center, Japan, for providing us with full-length cDNAs of OsENT2 and OsENT4. We are grateful to Dr. T. Sakamoto, University of Tokyo, for the kind gift of the pCAMBIA1390-GUS vector and the Agrobacterium EHA101 strain. We also thank Dr. J. Zuo for sharing unpublished data.
This work was supported by the Ministry of Education, Culture, Sports, Science and Technology and by a Grant-in-Aid for Scientific Research on functional analysis of genes relevant to agriculturally important traits in rice genome (grant no. IP–3003 to H.S.) from the Ministry of Agriculture, Forestry and Fisheries, Japan.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.060137.
References
- Acimovic Y, Coe IR (2002) Molecular evolution of the equilibrative nucleoside transporter family: identification of novel family members in prokaryotes and eukaryotes. Mol Biol Evol 19: 2199–2210 [DOI] [PubMed] [Google Scholar]
- Allen M, Qin W, Moreau F, Moffatt B (2002) Adenine phosphoribosyltransferase isoforms of Arabidopsis and their potential contributions to adenine and cytokinin metabolism. Physiol Plant 115: 56–68 [DOI] [PubMed] [Google Scholar]
- Bilyeu KD, Cole JL, Laskey JG, Riekhof WR, Esparza TJ (2001) Molecular and biochemical characterization of a cytokinin oxidase from maize. Plant Physiol 125: 378–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bürkle L, Cedzich A, Döpke C, Stransky H, Okumoto S, Gillissen B, Kühn C, Frommer WB (2003) Transport of cytokinins mediated by purine transporters of the PUP family expressed in phloem, hydathodes, and pollen of Arabidopsis. Plant J 34: 12–26 [DOI] [PubMed] [Google Scholar]
- Cabrita MA, Baldwin SA, Young JD, Cass CE (2002) Molecular biology and regulation of nucleoside and nucleobase transporter proteins in eukaryotes and prokaryotes. Biochem Cell Biol 80: 623–638 [DOI] [PubMed] [Google Scholar]
- Crawford CR, Patel DH, Naeve C, Belt JA (1998) Cloning of the human equilibrative, nitrobenzylmercaptopurine riboside (NBMPR)-insensitive nucleoside transporter ei by functional expression in a transport-deficient cell lines. J Biol Chem 273: 5288–5293 [DOI] [PubMed] [Google Scholar]
- Deltour R (1985) Nuclear activation during early germination of the higher plant embryo. J Cell Sci 75: 43–83 [DOI] [PubMed] [Google Scholar]
- Gaillard C, Moffat BA, Blacker M, Laloue M (1998) Male sterility associated with APRT deficiency in Arabidopsis thaliana results from a mutation in the gene APT1. Mol Gen Genet 257: 348–353 [DOI] [PubMed] [Google Scholar]
- Gietz D, Jean AS, Woods DA, Schiestl RH (1992) Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res 20: 1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillissen B, Bürkle L, André B, Kühn C, Rentsch D, Brandl B, Frommer WB (2000) A new family of high-affinity transporters for adenine, cytosine, and purine derivatives in Arabidopsis. Plant Cell 12: 291–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiei Y, Ohta S, Komari T, Kumashiro T (1994) Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of T-DNA. Plant J 6: 271–282 [DOI] [PubMed] [Google Scholar]
- Hoshikawa K (1989) XI. Ripening 1. Development of brown rice (hulled rice). The Growing Rice Plant. Nobunkyo, Tokyo, pp 28–29
- Hou B, Lim E-K, Higgins GS, Bowles DJ (2004) N-glucosylation of cytokinins by glycosyltransferases of Arabidopsis thaliana. J Biol Chem 279: 47822–47832 [DOI] [PubMed] [Google Scholar]
- Hyde RJ, Cass CE, Young JD, Baldwin SA (2001) The ENT family of eukaryote nucleoside and nucleobase transporters: recent advances in the investigation of structure/function relationships and the identification of novel isoforms. Mol Membr Biol 18: 53–63 [PubMed] [Google Scholar]
- Jefferson RA (1987) Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol Biol Rep 5: 387–405 [Google Scholar]
- Kamachi K, Yamaya T, Mae T, Ojima K (1991) A role for glutamine synthetase in the remobilization of leaf nitrogen during natural senescence in rice leaves. Plant Physiol 96: 411–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamboj RK, Jackson JF (1984) Divergent transport mechanisms for pyrimidine nucleosides in Petunia pollen. Plant Physiol 75: 499–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamboj RK, Jackson JF (1985) Pyrimidine nucleoside uptake by Petunia pollen. Plant Physiol 79: 801–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamboj RK, Jackson JF (1987) Purine nucleoside transport in Petunia pollen is an active, carrier-mediated system not sensitive to nitrobenzylthioinosine and not renewed during pollen tube growth. Plant Physiol 84: 688–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka T, Hayashi N, Yamaya T, Takahashi H (2004) Root-to-shoot transport of sulfate in Arabidopsis. Evidence for the role of SULTR3;5 as a component of low-affinity sulfate transport system in the root vasculature. Plant Physiol 136: 4198–4204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kombrink E, Beevers H (1983) Transport of purine and pyrimidine bases and nucleosides from endosperm to cotyledons. Plant Physiol 73: 370–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosugi S, Ohashi Y, Nakajima K, Arai Y (1990) An improved assay for β-glucuronidase in transformed cells: Methanol almost completely suppresses a putative endogenous β-glucuronidase activity. Plant Sci 70: 133–140 [Google Scholar]
- Lejeune P, Bernier G, Requier MC, Kinet JM (1994) Cytokinins in phloem and xylem saps of Sinapis alba during floral induction. Physiol Plant 90: 522–528 [Google Scholar]
- Letham DS (1994) Cytokinins as phytohormones: sites of biosynthesis, translocation, and function of translocated cytokinin. In DWS Mok, MC Mok, eds, Cytokinins: Chemistry, Activity, and Function. CRC Press, Boca Raton, FL, pp 57–80
- Li G, Liu K, Baldwin SA, Wang D (2003) Equilibrative nucleoside transporters of Arabidopsis thaliana: cDNA cloning, expression pattern and analysis of transport activities. J Biol Chem 278: 35732–35742 [DOI] [PubMed] [Google Scholar]
- Li J, Wang D (2000) Cloning and in vitro expression of the cDNA encoding a putative nucleoside transporter from Arabidopsis thaliana. Plant Sci 157: 23–32 [DOI] [PubMed] [Google Scholar]
- McElroy D, Rothenberg M, Wu R (1990) Structural characterization of a rice actin gene. Plant Mol Biol 14: 163–171 [DOI] [PubMed] [Google Scholar]
- Mässer P, Sütterlin C, Kralli A, Kaminsky R (1999) A nucleoside transporter from Trypanosoma brucei involved in drug resistance. Science 285: 242–244 [DOI] [PubMed] [Google Scholar]
- Miyawaki K, Matsumoto-Kitano M, Kakimoto T (2004) Expression of cytokinin biosynthetic isopentenyltransferase genes in Arabidopsis: tissue specificity and regulation by auxin, cytokinin, and nitrate. Plant J 37: 128–138 [DOI] [PubMed] [Google Scholar]
- Moffatt B, Pethe C, Laloue M (1991) Metabolism of benzyladenine is impaired in a mutant of Arabidopsis thaliana lacking adenine phosphoribosyltransferase activity. Plant Physiol 95: 900–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffatt BA, Wang L, Allen MS, Stevens YY, Qin W, Snider J, von Schwartzenberg K (2000) Adenosine kinase of Arabidopsis. Kinetic properties and gene expression. Plant Physiol 124: 1775–1785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Möhlmann T, Mezher Z, Schwerdtfeger G, Neuhaus HE (2001) Characterization of a concentrative type of adenosine transporter from Arabidopsis thaliana (ENT1, At). FEBS Lett 509: 370–374 [DOI] [PubMed] [Google Scholar]
- Mok DW, Mok MC (2001) Cytokinin metabolism and action. Annu Rev Plant Physiol Plant Mol Biol 52: 89–118 [DOI] [PubMed] [Google Scholar]
- Mok MC (1994) Cytokinins and plant development: an overview. In DWS Mok, MC Mok, eds, Cytokinins: Chemistry, Activity, and Function. CRC Press, Boca Raton, FL, pp 155–166
- Page RD (1996) Tree View: an application to display phylogenetic trees on personal computers. Comput Appl Biosci 12: 357–358 [DOI] [PubMed] [Google Scholar]
- Ross CW (1981) Biosynthesis of nucleotides. In PK Stump, EE Conn, eds, The Biochemistry of Plants, Vol 6. Academic Press, New York, pp 169–205
- Sankar N, Machado J, Abdulla P, Hilliker AJ, Coe IR (2002) Comparative genomic analysis of equilibrative nucleoside transporters suggests conserved protein structure despite limited sequence identity. Nucleic Acids Res 30: 4339–4350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnorr KM, Gaillard C, Biget E, Nygaad P, Laloue M (1996) A second form of adenine phosphoribosyltransferase in Arabidopsis thaliana with relative specificity towards cytokinins. Plant J 9: 891–898 [DOI] [PubMed] [Google Scholar]
- Shirzadegan M, Christie P, Seemann JR (1991) An efficient method for isolation of RNA from tissue cultured plant cells. Nucleic Acids Res 19: 6055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S, Letham DS, Jameson PE, Zhang R, Parker CW, Bandenoch-Jones J, Nooden LD (1988) Cytokinin biochemistry in relation to leaf senescence. IV. Cytokinin metabolism in soybean explants. Plant Physiol 88: 788–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith FW, Ealing PM, Hawkesford MJ, Clarkson DT (1995) Plant members of a family of sulfate transporters reveal functional subtypes. Proc Natl Acad Sci USA 92: 9373–9377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stasolla C, Katahira R, Thorpe TA, Ashihara H (2003) Purine and pyrimidine nucleotide metabolism in higher plants. J Plant Physiol 160: 1271–1295 [DOI] [PubMed] [Google Scholar]
- Sundaram M, Yao SYM, Ingram JC, Berry ZA, Abidi F, Cass CE, Baldwin SA, Young JD (2001) Topology of a human equilibrative, nitrobenzylthioinosine (NBMPR)-sensitive nucleoside transporter (hENT1) implicated in the cellular uptake of adenosine and anti-cancer drugs. J Biol Chem 276: 45270–45275 [DOI] [PubMed] [Google Scholar]
- Takei K, Dekishima Y, Eguchi T, Yamaya T, Sakakibara H (2003) A new method for enzymatic preparation of isopentenyladenine-type and trans-zeatin type cytokinins with radioisotope-labeling. J Plant Res 116: 259–263 [DOI] [PubMed] [Google Scholar]
- Takei K, Ueda N, Aoki K, Kuromori T, Hirayama T, Shinozaki K, Yamaya T, Sakakibara H (2004) AtIPT3 is a key determinant of nitrate-dependent cytokinin biosynthesis in Arabidopsis. Plant Cell Physiol 45: 1053–1062 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ (1994) ClustalW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veach YK, Martin RC, Mok DW, Malbeck J, Vankova R, Mok MC (2003) O-glucosylation of cis-zeatin in maize. Characterization of genes, enzymes, and endogenous cytokinins. Plant Physiol 131: 1374–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vickers MF, Yao SYM, Baldwin SA, Young JD, Cass CE (2000) Nucleoside transporter proteins of Saccharomyces cerevisiae. J Biol Chem 275: 25931–25939 [DOI] [PubMed] [Google Scholar]
- Visser F, Baldwin SA, Isaac RE, Young JD, Cass CE (2005) Identification and mutational analysis of amino acid residues involved in dipyridamole interactions with human Caenorhabditis elegans equilibrative nucleoside transporters. J Biol Chem 280: 11025–11034 [DOI] [PubMed] [Google Scholar]
- Visser F, Vickers MF, Ng AML, Baldwin SA, Young JD, Cass CE (2002) Mutation of residue 33 of the human equilibrative nucleoside transporters 1 and 2 alters sensitivity to inhibition of transport by dilazep and dipyridamole. J Biol Chem 277: 395–401 [DOI] [PubMed] [Google Scholar]
- Wormit A, Traub M, Flörchinger M, Neuhaus HE, Möhlmann T (2004) Characterization of three novel members of the Arabidopsis thaliana equilibrative nucleoside transporter (ENT) family. Biochem J 383: 19–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Zhang J, Huang Z, Wang Z, Zhu Q, Liu L (2002) Correlation of cytokinin levels in the endosperms and roots with cell number and cell division activity during endosperm development. Ann Bot (Lond) 90: 369–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Zhang J, Wang Z, Zhu Q, Wang W (2001) Hormonal changes in the grains of rice subjected to water stress during grain filling. Plant Physiol 127: 315–323 [DOI] [PMC free article] [PubMed] [Google Scholar]