Abstract
Glycosylation is a posttranslational modification occurring in many secreted and membrane-associated proteins in eukaryotes. It plays important roles in both physiological and pathological processes. Most of these protein modifications depend on UDP-N-acetylglucosamine. In this study, a T-DNA insertional rice (Oryza sativa) mutant exhibiting a temperature-sensitive defect in root elongation was isolated. Genetic and molecular analysis indicated that the mutated phenotype was caused by loss of function of a gene encoding a glucosamine-6-P acetyltransferase (designated OsGNA1), which is involved in de novo UDP-N-acetylglucosamine biosynthesis. The aberrant root morphology of the gna1 mutant includes shortening of roots, disruption of microtubules, and shrinkage of cells in the root elongation zone. Our observations support the idea that protein glycosylation plays a key role in cell metabolism, microtubule stabilization, and cell shape in rice roots.
UDP-N-acetylglucosamine (UDP-GlcNAc) is an essential metabolite that plays an important role in protein and lipid glycosylation in eukaryotes. The glycans on cell surface glycoproteins are instrumental for cell-cell or cell-matrix interactions, immune reactions, and tumor development, while sugar modifications on secreted proteins are important for their transport, biological activity, and clearance from circulation (Varki et al., 1999). Asn (N)-linked glycans (N-glycans) are components of most membrane-associated and secreted proteins in eukaryotic cells. N-glycans are assembled by the initial step of the transfer of a synthesized dolichol-linked oligosaccharide precursor en bloc to a nascent polypeptide chain in the endoplasmic reticulum. Maturation of the oligomannose precursor requires the action of glucosidases, mannosidases, and Golgi-resident glycosyltransferases. Synthesis of this oligosaccharide precursor initiates with the donor of UDP-GlcNAc and the enzyme of UDP-GlcNAc: dolichol phosphate GlcNAc-1-P transferase (Lowe and Marth, 2003; Haltiwanger and Lowe, 2004). The other proteins that pass through the secretory apparatus are O-glycan modified by the addition of carbohydrates to the OH group of Ser or Thr. The synthesis of O-glycans is initiated with N-acetylgalactosamine (GalNAc) modification of Ser or Thr and employs sequential addition of monosaccharides. The branch synthesis of many O-glycans requires Golgi-routed UDP-GlcNAc (for review, see Lowe and Marth, 2003).
UDP-GlcNAc also involves the generation of glycosylphospatidylinositol linkers for anchoring a variety of cell surface molecules to the plasma membrane. The transfer of GlcNAc from UDP-GlcNAc to phosphatidylinositol is the initial reaction of the glycosylphospatidylinositol assembly (Lowe and Marth, 2003; Haltiwanger and Lowe, 2004). Furthermore, many cytosolic and nuclear proteins, such as transcription factors, nuclear pore proteins, and cytoskeletal components, are O-glycosylated on Ser or Thr with a single GlcNAc residue (O-GlcNAc). This type of modification is reversible. The protein modification by O-GlcNAc affects protein stability, subcellular localization, and interaction with other proteins (Hanover, 2001; Wells and Hart, 2003).
In plants, glycans modulate many biological processes, such as somatic embryogenesis (van Hengel et al., 2001), gametophytic self-incompatibility (Ishimizu et al., 1999), cell expansion (Benfey et al., 1993; Schindelman et al., 2001; Sedbrook et al., 2002), and programmed cell death (Majewska-Sawka and Nothnagel, 2000). The reversible posttranslational modification of proteins by O-GlcNAc was also found in plants (Heese-Peck et al., 1995; Heese-Peck and Raikhel, 1998). In this pathway, O-GlcNAc transferases (OGTs) catalyze the transfer of GlcNAc from UDP-GlcNAc to Ser or Thr residues. Arabidopsis (Arabidopsis thaliana) SPINDLY (SPY) and SECRET AGENT proteins play similar roles as the animal OGT. Arabidopsis SPY acts as both a repressor of GA responses and a positive regulator of cytokinin signaling (Jacobsen et al., 1996; Swain and Olszewski, 1996; Greenboim-Wainberg et al., 2005). OGT activity is also required during gametogenesis and embryogenesis, with lethality occurring when protein modification by O-GlcNAc becomes limited in Arabidopsis (Hartweck et al., 2002). Orthologs of Arabidopsis SPY have been isolated in barley (cv Himalaya; HvSPY), petunia (Petunia hybrida; PhSPY), and tomato (Lycopersicon esculentum; LeSPY; Robertson et al., 1998; Izhaki et al., 2001; Greb et al., 2002). The specific involvement of HvSPY as a negative regulator of GA response has also been demonstrated (Robertson et al., 1998). These results indicate that the reversible protein modification by O-GlcNAc is likely ubiquitous in plants.
Generation of UDP-GlcNAc is possible by both de novo synthesis and salvage pathways, and the respective enzymes have been identified (for review, see Schachter, 1978). In eukaryotic cells, the sequence of intermediates in the de novo synthesis pathway is fructose-6-P (Fru-6-P) → glucosamine-6-P (GlcN-6-P) → N-acetyl glucosamine-1-P (GlcNAc-1-P) → UDP-GlcNAc. GlcN-6-P, as the recipient of the acetyl moiety, is converted into GlcNAc-1-P by the enzyme of GlcN-6-P acetyltransferase (encoded by GNA1 in yeast and EMeg32 in mice; Mio et al., 1999; Boehmelt et al., 2000a).
In this study, we identified and characterized a temperature-sensitive short-root mutant in a T-DNA insertional rice (Oryza sativa) library. The genetic and molecular analyses indicated that a yeast (Saccharomyces cerevisiae) GNA1 homology gene was disrupted by the insertion of T-DNA in this mutant. Downstream routes emanating from UDP-GlcNAc in the gna1 mutant were affected differentially with predominant aberrations in O-GlcNAc modification and partly in N-glycan modification of cellular proteins. Our results support the idea that protein glycosylation plays an important role in root cell metabolism and microtubule (MT) stabilization in rice.
RESULTS
Isolation of the Short-Root Rice Mutant in Rice
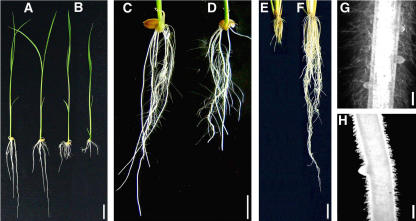
To study the genetic mechanism controlling root growth in plants, we screened a rice T-DNA insertional library (Chen et al., 2003) for rice mutants defective in root elongation. One mutant, designated NB208 (constructed from the Japonica cultivar Nipponbare and containing the bar gene marker phosphinothricin [PPT]), showed a defect in the elongation of roots, including primary roots, adventitious roots, and root hairs at room temperature (25°C; Fig. 1, B, G, and H). The shoot of the mutant, however, was not so prominently affected (Fig. 1, A and B).
Figure 1.
A and B, Root growth phenotypes. Seven-day-old wild-type (A) and NB208 mutant (B) seedlings grown in rice nutrient solution at 25°C. C and D, Seven-day-old wild type (C) and NB208 mutant (D) seedlings grown in rice nutrient solution at 32°C. E, NB208 mutant seedling grown for 35 d in rice nutrient solution at 25°C; F, NB208 mutant seedlings grown for 25 d in rice nutrient solution at 25°C and then at 32°C for an additional 10 d. G and H, Lateral roots and root hairs on primary roots of wild-type (G) and NB208 mutant (H) seedlings grown for 5 d in rice nutrient solution at 25°C. Bars = 2 cm (A–F) and 200 μm (G and H).
The defect in root elongation of the NB208 mutant proved to be temperature sensitive. The root lengths of 1-week-old mutant seedlings were about 70% of those of wild-type seedlings at 32°C (Fig. 1, C and D), while showing very short roots at 25°C (Fig. 1E). The defect in root elongation at 25°C could be restored when seedlings were moved to 32°C (Fig. 1F).
Cloning of the Gene
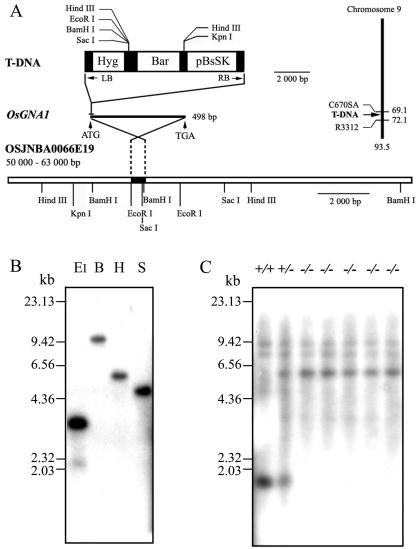
Two flanking sequences of the left border and right border of the T-DNA tag were obtained using the plasmid rescue strategy and the thermal asymmetric interlaced (TAIL)-PCR method (Manuel et al., 1980). The obtained sequences match a clone of the rice bacterial artificial chromosome (OSJNBA0066E19), which is located on chromosome 9. The insertion locus contains a putative GlcN-6-P AT gene (OsGNA1). Its coding product is homologous to the GlcN-6-P AT-1 proteins from yeast, Candida albicans, and Mus musculus (EMeg32). A cDNA sequence (accession no. AK063214), including the complete coding sequences (CDS), was obtained from the National Center for Biotechnology Information (NCBI) database by BLAST search with the putative GNA gene sequence. A 96-bp sequence from −83 to +13 of the GNA CDS (the A site of the first ATG signed as +1) was replaced by the T-DNA sequence in the NB208 mutant plants (Fig. 2A). Twenty-seven short-root seedlings were identified out of 111 T2 mutant seedlings, indicating that the defect in root elongation is controlled by a single recessive nuclear locus. Southern-blot analysis of the genomic DNA extracted from homozygous NB208 mutant plants, using a hygromycin phosphotransferase fragment as a probe, revealed that the mutant plants contained only one copy of the T-DNA insertion (Fig. 2B). The insertion was confirmed by Southern-blot analysis using the OsGNA1 probe for the wild-type, the gan1 mutant, and the heterozygous genotype (Fig. 2C).
Figure 2.
Cloning of the OsGNA1 gene. A, Restriction sites of the 13-kb genomic fragment and T-DNA. OsGNA1 is located on chromosome 9. The T-DNA insertion location was indicated. LB, Left border of T-DNA; RB, right border of T-DNA; Hyg, hygromycin phosphotransferase expression cassette; BAR, BASTA-resistance expression cassette; pBsSK, the pBluescript II SK(+). B, Southern blot of the gan1 (NB208) mutant using the Hyg probe. DNA was digested by EcoRI (EI), BamHI (B), HindIII (H), and SacI (S), respectively. HindIII λ-DNA size markers are indicated on the left. C, Southern blot of GNA1/GNA1 (+/+), GNA1/gna1 (+/−), and gna1/gna1 (−/−) using the OsGNA1 probe. The DNA was digested with EcoRI.
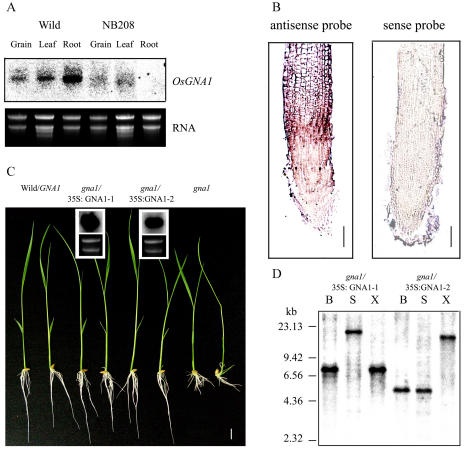
Northern-blot analysis and RNA in situ hybridization indicate that the OsGNA1 gene was mainly expressed in roots, particularly in the elongation zone of roots in wild-type plants, whereas OsGNA1 transcripts were scarcely detected in the NB208 mutant roots (Fig. 3, A and B). This result indicates that NB208 is a GNA1 null mutant. OsGNA1 cDNA was cloned into the binary vector pCAMBIA2301. The cauliflower mosaic virus 35S promoter-driven OsGNA1 was then introduced into gna1 mutant plants via Agrobacterium tumefaciens-mediated transformation. The root length of nine independent T2 families (20–50 seedlings each) was evaluated. A 3:1 segregation ratio of wild-type to short-root mutants was observed among two independent T2 families. OsGNA1 overexpression in transgenic plants was determined by northern-blot analysis (Fig. 3C). Southern-blot analysis showed that these two transgenic events contain a single copy of the insertion (Fig. 3D). These results confirmed that overexpression of OsGNA1 complemented the short-root phenotype. Thus, the mutant is designated gna1.
Figure 3.
The expression of OsGNA1 and complementation of the gan1 mutant by the OsGNA1 gene. A, Northern blot of OsGNA1 in different plant tissues. B, OsGNA1 expression in rice root revealed by RNA in situ hybridization with the antisense (left) or sense (right) probes for OsGNA1. C, Seven-day-old seedlings of two representative T2 lines of gna1 harboring the OsGNA1 gene grown at 25°C and the northern blot of the OsGNA1 transcripts in the two lines. The first two are wild-type plants and the last two are gna1 mutants. D, Southern blot of the pCAMBIA2301 insertion in the two transformed lines (gna1 + 35S∷OsGNA1-1 and gna1+35S∷OsGNA1-2 using the NPTII probe). DNA was digested by BamHI (B), SacI (S), and XhoI (X), respectively. The positions of HindIII λ-DNA size markers are indicated on the left.
Structural and Functional Characterization of OsGNA1
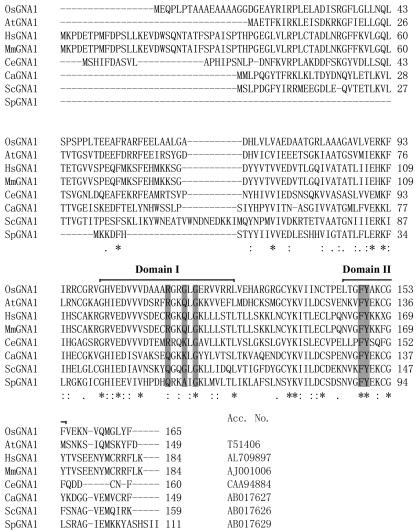
The OsGNA1 gene contains an open reading frame of 165 amino acids. The C terminus of the OsGNA1 polypeptide matches the AT motifs (Mio et al., 1999; Fig. 4). Two AT signature motifs were identified in OsGNA1 between amino acids 101 and 125 (domain I), and between amino acids 145 and 154 (domain II). The consensus motif, (R/Q)-X-X-G-X-G, has been demonstrated to be important to acetyl-CoA binding and could be established for the whole GCN5-related N-AT family of proteins (Lu et al., 1996). The corresponding motif in OsGNA1 is R-G-R-G-L-G in domain I. Furthermore, a conserved Phe-Tyr in domain II in ScGNA1 (Mio et al., 1999) is also found in OsGNA1 (Fig. 4).
Figure 4.
Structure of OsGNA1. Alignment of the deduced amino acid sequence of OsGNA1 with Caenorhabditis elegans (CeGNA1), yeast (YBF7/ScGNA1), C. albicans (CaGNA1), Schizosaccharomyces pombe (YDR3/SpGNA1), M. musculus (EMeg32/MmGNA1), Homo sapiens (HsGNA1), and Arabidopsis (AtGNA1) homologs using ClustalW software. Asterisks (*), Matching amino acids; double dots (:), strong similarities; single dots (.), weak similarities; dashes (-), gaps. Numbers refer to amino acids. The AT signature motifs are indicated as domains I and II. The conserved key amino acids for AT activity are shaded.
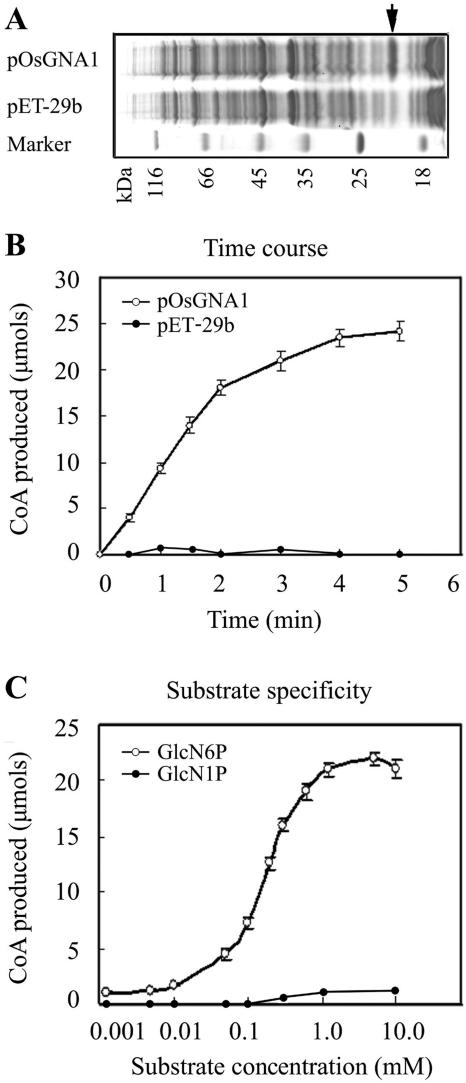
To verify whether OsGNA1 can acetylate GlcN-6-P, bacterium-expressed His-OsGNA1 (Fig. 5A) and a control protein, His-S-tag/thrombin, were purified under identical conditions. With 0.1 μg of purified protein, 200 μm of acetyl-CoA, and different amounts of GlcN-6-P per reaction, His-OsGNA1 exerted GlcN-6-P AT activity, while the control proteins did not (Fig. 5B). Since GlcN-1-P did not lead to the release of CoA from acetyl-CoA in the assay, OsGNA1 prefers to use GlcN-6-P as a substrate (Fig. 5C). The average Michaelis-Menten Km value for GlcN-6-P and acetyl-CoA are 145 and 180 μm, respectively. Therefore, it is likely that OsGNA1 exerts GlcN-6-P AT activity but not GlcN-1-P AT activity.
Figure 5.
GlcN-6-P AT activity analysis. A, Coomassie-stained gel showing bacterially expressed OsGNA1 proteins. The arrow indicates the pOsGNA1 protein. Protein standards are indicated in kilodaltons. B, Kinetics of pOsGNA1 using 200 μm of acetyl-CoA, 200 μm of GlcN-6-P, and 0.1 μg of purified recombinant proteins. At the indicated time points, aliquots of the reaction were stopped. Released CoA was measured by incubating terminated duplicate reactions with DTNB and measuring optical densities at 412 nm. C, Purified recombinant protein (0.1 μg) was incubated with 200 μm of acetyl-CoA and increasing concentrations of GlcN-6-P/GlcN-1-P for 10 min at 30°C.
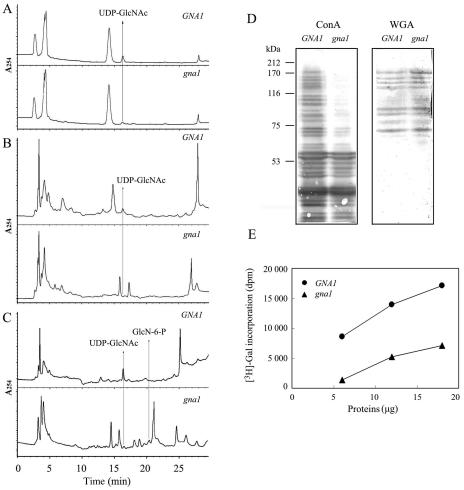
Analysis of UDP-GlcNAc steady-state levels in roots showed that the level of UDP-GlcNAc in gna1 mutants was less than 10% of that in wild-type plants (Table I; Fig. 6, A and B), suggesting that the GNA1-dependent pathway is the major source of UDP-GlcNAc in rice roots. After incubation for 6 h with GlcN, the UDP-GlcNAc level increased in the roots of wild-type plants, but not in the roots of the gna1 mutant (Table I; Fig. 6C). The results indicate that GlcN-6-P AT activity is impaired in gna1 root cells.
Table I.
The amount (ng g−1 FW) of UDP-GlcNAc in the gna1 mutant and wild-type seedlings under different temperatures and GlcN-6-P supplementation conditions (n = 3)
Six 10-d-old seedlings grown at 32°C or 25°C were sampled for analysis of the amount of UDP-GlcNAc using HPLC. The amount of UDP-GlcNAc was analyzed with the chromatography software program Chemistation 2170AA (Agilent 1100, LC system), according to the peak area. The data represent averages of three repeats of root samples with the mean sd. FW, Fresh weight; +GlcN-6-P, supplemented with 10 mm GlcN-6-P for 6 h before being sampled.
| Growth Conditions
|
GNA1
|
gna1
|
||||
|---|---|---|---|---|---|---|
| 32°C | 25°C | 25°C + GlcN-6-P | 32°C | 25°C | 25°C + GlcN-6-P | |
| UDP-GlcNAc (ng g−1 FW) | 16.2 ± 2.9 | 11.8 ± 3.4 | 38.5 ± 6.4 | 1.5 ± 1.4 | 0.8 ± 0.8 | 1.4 ± 1.0 |
Figure 6.
Decrease of UDP-GlcNAc levels and the differential impairment of downstream routes of UDP-GlcNAc in root cells of the gan1 mutant. A to C, HPLC elution profile at A254 nm. The rice root cells used were from 10-d-old seedlings grown at 32°C (A) or 25°C (B). C, Ten-day-old seedlings supplemented with 10 mm GlcN-6-P for 6 h. Arrows indicate elution positions of UDP-GlcNAc and GlcN-6-P peaks. D, Normalized plant cell extracts were blotted and analyzed using peroxidase-conjugated lectin ConA or WGA. Protein standards are in kilodaltons at the left. E, Cytosolic extracts prepared from roots labeled by β-1,4-galactosyltransferase-mediated [3H]Gal.
Glycosylation in gna1 Mutant Roots
To determine whether endoplasmic reticulum/Golgi-mediated glycosylation is impaired in root cells of the gna1 mutant, extracts of root cells were subjected to immunoblot analysis using the peroxidase-conjugated lectins, concanavalin A (ConA) and wheat germ agglutinin (WGA). ConA is used to bind to branches of oligomannose chains on N-glycoproteins, while WGA recognizes salic acid and GlcNAc on N- and O-linked oligosaccharides. The results showed that the amount of higher molecular weight glycoproteins was decreased dramatically in mutant roots compared to wild-type roots for lectin ConA. No significant difference in the overall amount of glycoproteins between the gna1 mutant and the wild-type root cells was detected for WGA used (Fig. 6D).
To assess the extent of cytosolic O-linked GlcNAc modifications of proteins in the gna1 mutant, cytosolic extracts were labeled with UDP-[3H]Gal using β-1,4-galactosyltransferase. This enzyme transfers Gal residues specifically to unsubstituted GlcNAc residues on proteins, the majority of which are O-linked GlcNAc (Torres and Hart, 1984). The result showed that levels of cytosolic O-linked GlcNAc were much lower in lysates of gna1 mutant root cells than in wild-type root cells (Fig. 6E).
Reduced Root Viability and Cell Division in Root Tips of the gna1 Mutant
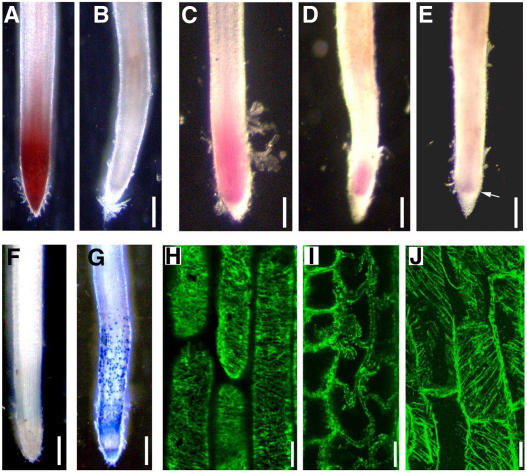
To investigate the relationship between glycoproteins and root elongation in rice, physiological and anatomical analyses were performed. Triphenyltetrazolium chloride (TTC) reduction has been widely used as a general measurement of vitality in plant tissues. In plant tissues, TTC is mainly reduced by dehydrogenase enzymes, most of which are associated with mitochondrial function (Comas et al., 2000). The wild-type rice root tips turned red when incubated with TTC solution for 2 h (Fig. 7A), while gna1 mutant root tips did not (Fig. 7B). These results suggest that normal respiration metabolism is arrested in the cells of gna1 root tips.
Figure 7.
Cell viability, cell division, and MT in primary root tips. The germinated seeds were grown at 32°C for 3 d and then at 25°C for an additional 2 to 5 d. A and B, TTC-incubated roots. Wild-type roots grown at 25°C for 2 d (A) showed TTC reduction in root tips (red stain); gna1 roots (B) exhibited no staining in root tips. C to E, Feulgen-stained roots. Wild-type roots grown at 25°C for 5 d (C) showed high DNA content in root tips (red stain). The gna1 roots exhibited dramatic reduced DNA content in root tips after growth at 25°C for 2 d (D) and 5 d (E). The arrow in E indicates the restricted high-DNA content zone. F and G, Evans blue-stained root tips of wild-type (F) and gna1 mutant (G) seedlings grown at 25°C for 2 d. The blue patches of dead cell zones can be easily detected. H to J, Antitubulin immunofluorescence analysis. H, The regular cortical MTs in wild-type root tip cells after growth at 25°C for 2 d. I, The MT was disrupted in the gna1 mutant root tip cells after growth at 25°C for 2 d. J, The cortical MTs in the gna1 mutant root tip cells grown at 32°C. Bars = 200 μm (A–G) and 20 μm (H–J).
Feulgen staining, which selectively stains DNA, has been widely used as a measure of DNA content and reveals distinctive tissue with high DNA content (Demchenko et al., 2004). A red region in the meristematic zone was observed in wild-type root tips after Feulgen stain (Fig. 7C) but not in mutant root tips (Fig. 7, D and E). Compared with wild-type roots, the cell division region of the mutant roots is smaller when grown at room temperature for 2 and 5 d (Fig. 7E), indicating that cell division in the mutant roots was inhibited.
Evans blue dye staining of cells with a completely permeable plasma membrane is used to identify the dead cells (Gaff and Okong'O-gola, 1971). With Evans blue dye, many large patches of cells were found to be stained along the roots of gna1 mutant plants (Fig. 7, F and G), but some cells, especially the meristem zone cells in the root tips of the gna1, were still alive after growth at room temperature for 2 d.
MT Disruption in Root Tips of the gna1 Mutant
Immunolocalization of cortical MTs was performed to examine their status in the mutant plants. In wild-type root tips, cortical MTs at the elongation zone were oriented transversely along the elongation axis (Fig. 7H), whereas no cortical MT bundles were detected in gna1 roots grown at room temperature. The disrupted MTs associated with the plasma membrane exhibited condensed spots in the cortex cells spreading into the endodermal cells of gna1 roots (Fig. 7I). The gna1 root cells grown at high temperature (32°C), however, showed normal cortical MTs (Fig. 7J).
Abnormal Cell Shape
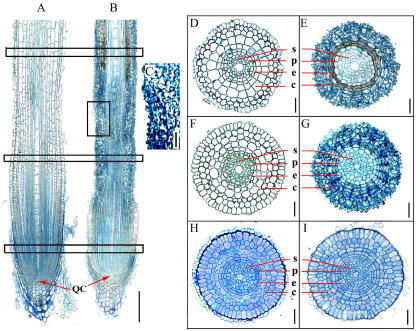
To observe cell size and shape, longitudinal and transverse sections were prepared from root maturation, elongation, and meristem zones. Roots of wild-type seedlings exhibited highly organized cell files and defined cell shapes after 2 d of growth at room temperature (Fig. 8A). The root tips of gna1 seedlings at the same temperatures displayed abnormal cell shape and length under the same growth conditions (Fig. 8, B and C). From the epidermis to the cortical layer, the mutant root cells were shrunken and the adjacent cell walls were inlaid on each other in maturation, elongation, and meristem zones (Fig. 8, E, G, and I), compared with wild-type roots (Fig. 8, D, F, and H). The stele cells also became distorted after 5 d of growth at room temperature (Fig. 8F).
Figure 8.
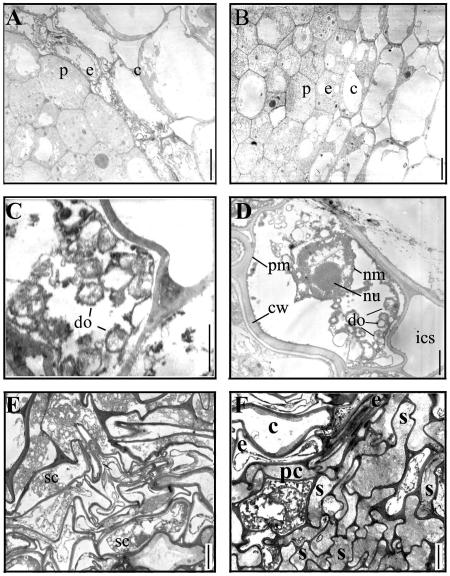
Longitudinal and transverse sections of primary roots of wild-type and gna1 plants. The germinated seeds were grown at 32°C for 3 d, and then at 25°C for an additional 2 d. A and B, Longitudinal sections of wild-type (A) and gna1 mutant (B) seminal roots. C, Detailed examination of longitudinal section of the root elongation zone of gna1. Squares indicate the approximate regions from where the transverse sections were prepared. D to I, Transverse sections of different zones. Maturation zones of wild type (D) and gna1 (E); elongation zones of wild type (F) and gna1 (G); and meristem zones of wild type (H) and gna1 (I). Bars = 200 μm (A and B) and 20 μm (C–I). QC, Quiescent center; c, cortex; e, endodermis; p, pericycle; s, stele.
Using a transmission electron microscope (H-600; Hitachi, Tokyo), it was observed that the endodermis, cortex, and stele cells of the gna1 root were characterized by cytoplasmic shrinkage and distorted cell shape after being grown at room temperature (Fig. 9, A, C, E, and F). Further examination of the shrunken cells revealed that organelles in the cells were disaggregated and empty. The nucleolus and nuclear membrane were intact before the cell became empty, although the chromatin in the nuclei had been disaggregated (Fig. 9, C and D).
Figure 9.
Ultrastructure of wild-type and gna1 root cells. A and B, Transverse sections of the elongation zones of the seminal root tips of gna1 (A) grown at 25°C for 1 d and the wild type (B) grown at 25°C for 5 d. C and D, Endodermis cells in gna1 root tips after growth at 25°C for 1 d (C) or 5 d (D). E and F, Transverse sections of the cortex (E) and stele (F) regions of the elongation zone of the gna1 root after growth at 25°C for 5 d. c, Cortex; cw, cell wall; do, degenerated organelle; e, endodermis; ics, intercellular space; nm, nuclear membrane; nu, nucleolus; pc, pericycle; pm, plasma membrane; s, stele; sc, shrunken cytoplasm. Bars = 20 μm (A, B, E, and F), 2 μm (C), and 5 μm (D).
DISCUSSION
A novel short-root gene for a GlcN-6-P AT in rice (OsGNA1) was identified from a T-DNA insertional rice mutant library. Our results indicate that OsGNA1 is required for maintaining normal root cell metabolism and cell shape. Northern-blot analysis and RNA in situ hybridization revealed that the OsGNA1 is expressed preferentially in the root elongation zone, suggesting its participation in UDP-GlcNAc synthesis in roots.
Using purified recombinant His-tagged OsGNA1 expressed in Escherichia coli (Fig. 5A), we determined that the Michaelis-Menten Km value for GlcN-6-P and acetyl-CoA was 145 and 180 μm, respectively, which is comparable to the Km value of ScGNA1 for GlcN-6-P and acetyl-CoA (124.13 and 228.78 μm). These results support the idea that the AT motif and the enzymatic character of the GNA family were conserved in yeast, animals, and plants (Mio et al., 1999; Boehmelt et al., 2000a). The UDP-GlcNAc content in root cells of the gna1 mutant is less than 10% of that in root cells of the wild type (Fig. 6). The dramatically reduced UDP-GlcNAc concentration in root cells of the gna1 mutant suggests that the de novo synthesis pathway dependent on GNA1 synthesizes the most UDP-GlcNAc in rice root cells.
The limited UDP-GlcNAc level in root cells of the gna1 mutant was insufficient to provide UDP-GlcNAc for protein N-glycosylation (Fig. 6). N-Glycosylation is essential for the proper folding, targeting, and functioning of secreted proteins. N-Glycoproteins play important roles in plant developmental processes, including cell wall synthesis (Lerouge et al., 1998; Lukowitz et al., 2001). Therefore, a decrease of N-glycoprotein levels in root cells of the gna1 mutant could be a major factor accounting for cell shrinkage. In addition, the insufficient substrate of UDP-GlcNAc also reduced the cellular O-linked GlcNAc modifications as measured by the β-1,4-galactosyltransferase assay. In murine EMeg32−/− cells, removal of O-GlcNAc modification makes the eukaryotic initiation factor 2-associated protein p67 and the transcription factor SP1 susceptible to degradation, whereas in the case of p53 and c-Myc, transcriptional activities are affected (Boehmelt et al., 2000b). In plants, the genes encoding O-GlcNAc transferase were cloned and functioned in many developmental processes (Hartweck et al., 2002). Therefore, the regulatory functions of O-GlcNAc modification and their absence in root cells of the gna1 mutant may be another reason accounting for the observed aberrant cells.
MTs take part in governing the formation of cell walls and, consequently, cell shape in plants. Many MT-associated proteins regulate the organization and dynamics of MTs in cells. In animals, MT-binding proteins, such as Tau, MAP2, and MAP4, are modified by O-GlcNAc. It has been suggested that acetylation increases the stability of MTs. The acetylation of rabbit skeletal troponin C increases its ability to interact with other troponin components (Maruta et al., 1986; Grabarek et al., 1995). In root cells of the gna1 mutant, O-GlcNAc modification of these proteins is reduced due to lack of cytosolic O-linked GlcNAc modifications. Therefore, it is likely that the loss of function of OsGNA1 gives rise to an unstable structure of MT-associated proteins and, consequently, results in MT disruption in root cells of the gna1 mutant grown at room temperature (Fig. 7). Yeast Pat1/GNA1 functions in a process where tubulin or related proteins are involved. Under low-temperature conditions, e.g. 23°C, pat1 mutants showed increased benomyl (an anti-MT drug) sensitivity compared to wild-type cells (Lin et al., 1996). GNA1 functions in plant MT dynamics in a way similar to the above yeast process at lower temperature conditions, which may partly explain the abnormal root growth of the gna1 mutant. High temperatures can repair the defect in root elongation and cell shape in gna1. It is noted that, at higher temperatures (32°C), the UDP-GlcNAc level in the gna1 mutant was about 1 time higher than at lower temperatures (25°C; Table I), which may, at least partially, make a contribution to the repair of the short-root phenotype at higher temperatures. It is possible that cytoskeleton proteins such as MTs are made more stable by other components at the higher temperatures.
Root length is determined by cell division in the root meristem and cell elongation in the root elongation zone. Our observations suggest that the gna1 mutant primarily reduced cell elongation in the elongation zone and above by causing cell shrinkage (Fig. 8). The effects on cell division in the mutant may be secondary to the effects on elongating cells. Perhaps cell-cell signaling between the cells in the elongation zone and the meristem are required for meristem maintenance, which is disrupted in the mutant. The reduction of respiration in the mutant may result from the disaggregation of the organelle. Cell shrinkage in root cells of the gna1 mutant may be attributed to the reduction of respiration metabolism and the disruption of MTs. The reduced respiration results in the lack of energy to support ion absorption to maintain cell osmotic potential and prevent loss of cellular water. The disrupted MT could not support the cell shape and intracellular transportation. It is possible that the loss of function of many unknown essential proteins, such as ion transporters and cellulose synthase, due to the lack of glycosylation, could also be important in determining cell shrinkage.
Proteomic analysis is required to identify the glycosylation-modified proteins, especially the O-GlcNAc-modified proteins related to the defect in the gna1 mutant at room temperature. The collection of O-GlcNAc-modified proteins would greatly facilitate the elucidation of the regulatory mechanism of responding proteins and the specific consensus sequence directing the O-GlcNAc modification.
MATERIALS AND METHODS
Mutant Screening
One rice (Oryza sativa) short-root mutant, designated NB208 (Nipponbare containing a bar gene marker), was isolated from about 1,000 T-DNA insertion-mutagenized lines (Chen et al., 2003) in rice culture solution (Yoshida et al., 1976). The bar gene (PPT AT) was used as a selection marker in the vector of T-DNA insertion, which allows the use of a PPT resistance assay for primary analysis for cosegregation of mutated phenotypes and T-DNA insertions. A PPT resistance assay was performed by spraying 250 mg L−1 glufosinate solution (Aventis, Strasbourg, France) on the middle leaf blades of 10-d-old seedlings grown in solution culture. The scoring for herbicide resistance or sensitivity was carried out 5 d after spraying.
Cloning of the Short-Root Gene
DNA fragments flanking the T-DNA insertions were isolated using plasmid rescue strategy (Manuel et al., 1980) and amplified using TAIL-PCR from the mutant plant genomic DNA. For plasmid rescue, 5 μg of genomic DNA were digested overnight with HindIII. TAIL-PCR amplification was performed as described by Chen et al. (2003). Plasmids were isolated from positive clones and sequenced.
BLAST analysis indicates that the gene obtained from the NB208 mutant encodes a putative GlcN-6-P AT (OsGNA1) on chromosome 9. The OsGNA1 cDNA was cloned into a pUC-T vector by reverse transcription-PCR using the primers of 5′-AACCGAAAACCCTAACTGAGTACG-3′ and 5′-TCCATCCATACAAGGAGCAATAAC-3′. For the complementation test, the OsGNA1 cDNA clone with the complete CDS was digested with KpnI and XbaI and subcloned into the corresponding site of pCAMBIA2301. The cauliflower mosaic virus 35S promoter was then inserted by EcoRI/SacI sites. Agrobacterium strain EHA105 harboring the above vector was used to transfer the mutant calli via Agrobacterium tumefaciens-mediated transformation (Chen et al., 2003).
DNA and RNA Gel-Blot Analysis
Five micrograms of genomic DNA was digested with appropriate restriction enzymes, separated on a 0.8% agarose gel, and blotted onto a Hybond-N+ nylon membrane (Amersham Pharmacia Biotech, Piscataway, NJ). The probe was prepared using a random priming method. Hybridization and washing were conducted according to the manufacturer's instructions. After washing, the blots were analyzed using a Typhoon-8600 phosphorimager system. Twenty micrograms of total RNA was isolated using Trizol (Gibco-BRL, Cleveland) following the manufacturer's protocol, separated on 1% agarose gels containing 1× MOPS and 0.66 m formaldehyde, and transferred onto a Hybond-N+ nylon membrane (Amersham Pharmacia Biotech). The probe was prepared using a random priming method and was hybridized following the manufacturer's instructions.
In Situ Hybridization
RNA in situ hybridization was performed as described by Harding et al. (2002). Briefly, the OsGNA1 cDNA was cloned into a pBSSK vector and used as a template to generate RNA probes. Transverse sections (10 μm thick) were probed with a digoxigenin-labeled antisense probe or sense probe (control; DIG Northern starter kit; Roche, Indianapolis). The slides were observed using a microscope (Zeiss Axiovert 200 with color CCD camera; Zeiss, Jena, Germany) and photographed.
Expression and Purification of Recombinant Proteins
To make the OsGNA1∷6-His fusion, the OsGNA1 gene was amplified by PCR using the primers of 5′-AAAAAACATATGGAACAGCCGCTCCCCAC-3′ and 5′-AAAAAACTCGAGGAAGTAGAGCCCCATCTGGACG-3′. The reconstructed sequences of OsGNA1 were subcloned into the NdeI and XhoI sites of pET-29b (Novagen, Madison, WI) and defined as pOsGNA1.
pOsGNA1 and pET-29b (negative control) were transformed into Escherichia coli BL21 (DE3; Stratagene, La Jolla, CA), and grown on Luria-Bertani medium. Overnight cultures were inoculated into fresh medium at a 1:100 dilution and grown at 37°C until the density was 0.6 A600/mL, and 0.5 mm IPTG was added to the cultures, followed by another 5 h of incubation at 28°C. The cells were collected by centrifugation, suspended in one-twentieth culture volume of sonication buffer (50 mm Tris-acetate, 10 mm EDTA, and 5 mm DTT, pH 7.5), and sonicated on ice for 4 min (1 s on, 2 s off). Cell lysates were centrifuged at 15,000g for 30 min and analyzed using SDS-PAGE. The OsGNA1-6-His fusion proteins were purified under natural conditions using a Ni-NTA column according to the manufacturer's instructions (Qiagen, Hilden, Germany).
AT Assay
The enzyme assay was performed according to the description by Mio et al. (1999). The assay for GlcN-6-P AT was performed in 50 μL of a reaction mixture containing 50 mm Tris-HCl, pH 7.5, 5 mm MgCl2, 200 μm GlcN-6-P, 200 μm acetyl-CoA, 10% (v/v) glycerol, and approximately 0.1 μg of the recombinant His-Gna1p fusion protein. After incubation at 30°C for 10 min or the indicated time, the reaction was terminated by adding 50 μL of stop solution containing 50 mm Tris-HCl, pH 7.5, and 6.4 m guanidine hydrochloride, and then 50 μL of a solution containing 50 mm Tris-HCl, pH 7.5, 1 mm EDTA, and 20 μm 2-nitrobenzoic acid. The amount of CoA produced by the GNA1 protein was estimated by 4-nitrothiophenolate and measured at the optimal density of 412 nm.
Western-Blot Analysis
Proteins were separated by SDS-PAGE using a 7.5% acrylamide gel and electroblotted onto a polyvinylidene difluoride membrane (Amresco, Solon, OH). Blots were incubated in Tris-buffered saline (TBS) plus Tween (TBST; 20 mm Tris-HCl, pH 7.5, 150 mm NaCl, and 0.05% [v/v] Tween 20) solution containing 3% (w/v) bovine serum albumin for 60 min at 30°C, and further incubated for 6 h at 4°C with peroxidase-conjugated ConA and WGA diluted with TBST solution containing 1% (w/v) bovine serum albumin. The membranes were washed three times in TBST for 10 min each, and rinsed with TBS for 5 min. The reaction was performed in the reaction buffer with 10 mL 100 mm Tris-HCl, pH 7.5, 100 μL 40 mg/mL diaminobenzidine, 25 μL 80 mg/mL NiCl2, and 15 μL 3% H2O2.
β-1,4-Galactosyltransferase Assay
β-1,4-Galactosyltransferase assay was performed according to Boehmelt et al. (2000b). Briefly, root cell extracts were prepared in phosphate-buffered saline containing 1% Triton X-100 and centrifuged at 15,000g for 10 min at 4°C. Aliquots of lysate were incubated at 37°C for 20 min in 50 μL of the assay mixture containing buffer (10 mm HEPES, pH 7.0, 5 mm MnCl2, and 10 mm Gal), 0.01 mm UDP-Gal, 10 mU of bovine galactosyltransferase (G5507; Sigma, St. Louis), and 0.5 μCi UDP-[1-3H]Gal (U1506; Sigma). Reactions were stopped by the addition of 5 μL 100 mm EDTA. Protein reaction products were isolated in the exclusion volume from Sepharose G-50 columns and analyzed using liquid scintillation counting (Wallic 1414; Perkin-Elmer, Foster City, CA).
HPLC Analysis for Sugars
UDP-GlcNAc in distilled water (U4375; Sigma) has a high absorption at 250 to 260 nm and peaks at 254 nm, while GlcN-6-P (G5509; Sigma) also has a high absorption at 250 to 280 nm according to wavelength-scanning analysis. The samples were quantified using a UV spectrophotometer at 254 nm. The retention time and the peaks of UDP-GlcNAc and GlcN-6-P were determined with 1 μg mL−1 UDP-GlcNAc and 10 μg mL−1 GlcN-6-P in distilled water and the root extract, respectively. For rice samples, 5 g of fresh roots were homogenized with a mortar and pestle in 10 mL of distilled water. The homogenate was centrifuged at 12,000g for 10 min at 4°C. The pellet was washed twice with 10 mL of distilled water. The supernatant was filtered with an 0.2-μm membrane and vacuum dried. The dried filtrate was dissolved in 0.2 mL of distilled water and referred to as the nucleotide-sugars extract used for HPLC analysis (Agilent 1100; Agilent Technologies, Palo Alto, CA). Reverse-phase HPLC was performed on a Synchropak WAX anion-exchange column (4.6 × 250 mm; Agilent Technologies), eluted with a concave gradient of ammonium phosphate from 15 mm, pH 3.8, to 1 mm, pH 4.5, over 45 min at a flow rate of 0.8 mL min−1. The operations were repeated three times. The detector signal was recorded and integrated by a personal computer and a chromatography software program (Chemistation 2170AA; Agilent).
Microscopic Analysis
The root tips, hairs, and surface were examined by a Leica MZ95 stereomicroscope with a color CCD camera. To examine the DNA content in the root meristem, Feulgen staining was used according to Demchenko et al. (2004), with a slight modification. Root tips (8–10 mm) were fixed in Carnoy's fixative for at least 24 h. The root tips were rinsed in 1 m HCl, transferred to 1 m HCl prewarmed to 60°C for 12 min, and stained with Schiff's reagent until the tip of the root showed distinct coloring (about 10 min). Afterward, the root tips were rinsed twice for 2 min in a rinse solution (50 mm HCl, 0.5% Na2S2O5). The samples were then placed in 45% (v/v) HOAc, and checked and photographed under a stereomicroscope. To observe root vitality, root tips (8–10 mm) were submerged in 3 mL of 0.2% (w/v) TTC in 50 mm Na2HPO4-KH2PO4, pH 7.4, with 0.05% Triton X-100, and were vacuum infiltrated for 5 min to ensure infiltration of TTC (Steponkus and Lanphear, 1967). Samples were incubated at 37°C for 2 h, rinsed twice with distilled water, and photographed immediately. Evans blue enters dead cells due to freely permeable plasma membranes (Gaff and Okong'O-gola, 1971). To examine whether the cells were dead, root tips were submerged in 20 mL of 1% (w/v) Evans blue water solution for 10 min, washed with distilled water for 2 h, and photographed.
For light and electromicroscopic analysis, root tips were fixed overnight at 4°C in 2.5% glutaraldehyde in 0.1 m sodium phosphate buffer, pH 7.2, and washed three times for 30 min in the same buffer. Root samples were then refixed for 4 h in 1% OsO4 in 0.1 m sodium phosphate buffer, pH 7.2, and washed for 30 min in the same buffer. The samples were dehydrated in a gradient ethanol and embedded in Spurr resin. Semithin sections (3 μm thick) were made using glass knives on a Power Tome XL (RMC-Boeckeler Instruments, Tucson, AZ) microtome and stained in 0.1% methylene blue for 3 to 5 min at 70°C. The samples were rinsed with distilled water and visualized with a Zeiss Axiovert 200 microscope with a color CCD camera (Zeiss).
For ultrathin sections, root tips were fixed in 2.5% glutaraldehyde and 1% OsO4, dehydrated in an acetone series, and embedded in Spurr resin. Ultrathin sections (0.1 μm thick) were stained with uranyl acetate for 45 min at room temperature and observed with an electron microscope.
Immunolocalization of MTs
The protocol used for fixing and immunostaining seedling roots has been described (Vitha et al., 1997). MTs were visualized using immunolabeling β-tubulins according to Sugimoto et al. (2000). Roots from 5-d-old seedlings were used for MT localization. Samples were probed first with mouse monoclonal antibody against chicken β-tubulin (1:200 dilution; Sigma) and then incubated with Alexa Fluor 488 goat anti-mouse IgG antibody (1:300 dilution; Molecular Probes, Eugene, OR). MT sections were viewed with a Zeiss confocal laser-scanning microscope (LSM 510; Zeiss) equipped with a 40× N.A. 0.75 water-immersion objective and a 100× N.A. 1.30 oil objective. Alexa-FITC was excited at 488 nm with emission from 505 to 530 nm. Confocal images are a projection of 10 optical sections taken at approximately 0.3- to 1-μm thickness, with each plane being averaged two times. All images were adjusted using Zeiss LSM image examiner software.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession number AY722189.
This work was supported by the Natural Science Foundation of China (project no. 30230220) and the Bureau of Science and Technology of Zhejiang Province, China.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.058248.
References
- Benfey PN, Linstead PJ, Roberts K, Schiefelbein JW, Hauser MT, Aeschbacher RA (1993) Root development in Arabidopsis: four mutants with dramatically altered root morphogenesis. Development 119: 57–70 [DOI] [PubMed] [Google Scholar]
- Boehmelt G, Fialka I, Brothers G, McGinley MD, Patterson SD, Mo R, Hui CC, Huber LA, Mak TW, Iscove NN (2000. a) Cloning and characterization of the murine glucosamine-6-phosphate acetyltransferase EMeg32. J Biol Chem 275: 12821–12832 [DOI] [PubMed] [Google Scholar]
- Boehmelt G, Wakeham A, Elia A, Sasaki T, Plyte S, Potter J, Yang Y, Tsang E, Ruland J, Iscove NN, et al (2000. b) Decreased UDP-GlcNAc levels abrogate proliferation control in EMeg32-deficient cells. EMBO J 19: 5092–5104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SY, Jin WZ, Wang MY, Zhang F, Zhou J, Jia QJ, Wu YR, Liu FY, Wu P (2003) Distribution and characterization of over 1000 T-DNA tags in rice genome. Plant J 36: 105–113 [DOI] [PubMed] [Google Scholar]
- Comas LH, Eissenstat DM, Lakso AN (2000) Assessing root death and root system dynamics in a study of grape canopy pruning. New Phytol 147: 171–178 [Google Scholar]
- Demchenko K, Winzer T, Stougaard J, Parniske M, Pawlowski K (2004) Distinct roles of Lotus japonicus SYMRK and SYM15 in root colonization and arbuscule formation. New Phytol 163: 381–392 [DOI] [PubMed] [Google Scholar]
- Gaff DF, Okong'O-gola O (1971) The use of non-permeating pigments for testing the survival of cells. J Exp Bot 22: 757–758 [Google Scholar]
- Grabarek Z, Mabuchi Y, Gergely J (1995) Properties of troponin C acetylated at lysine residues. Biochemistry 34: 11872–11881 [DOI] [PubMed] [Google Scholar]
- Greb T, Schmitz G, Theres K (2002) Isolation and characterization of the Spindly homologue from tomato. J Exp Bot 53: 1829–1830 [DOI] [PubMed] [Google Scholar]
- Greenboim-Wainberg Y, Maymon I, Borochov R, Alvarez J, Olszewski N, Ori N, Eshed Y, Weiss D (2005) Cross talk between gibberellin and cytokinin: the Arabidopsis GA response inhibitor SPINDLY plays a positive role in cytokinin signaling. Plant Cell 17: 92–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haltiwanger RS, Lowe JB (2004) Role of glycosylation in development. Annu Rev Biochem 73: 491–537 [DOI] [PubMed] [Google Scholar]
- Hanover JA (2001) Glycan-dependent signaling: O-linked N-acetylglucosamine. FASEB J 15: 1865–1876 [DOI] [PubMed] [Google Scholar]
- Harding SA, Leshkevich J, Chiang VL, Tsai CJ (2002) Differential substrate inhibition couples kinetically distinct 4-coumarate:coenzyme A ligases with spatially distinct metabolic roles in quaking aspen. Plant Physiol 128: 428–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartweck LM, Scott CL, Olszewski NE (2002) Two O-linked N-acetylglucosamine transferase genes of Arabidopsis thaliana L. have overlapping functions necessary for gamete and seed development. Genetics 161: 1279–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heese-Peck A, Cole RN, Borkhsenious ON, Hart GW, Raikhel NV (1995) Plant nuclear pore complex proteins are modified by novel oligosaccharides with terminal N-acetylglucosamine. Plant Cell 7: 1459–1471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heese-Peck A, Raikhel NV (1998) A glycoprotein modified with terminal N-acetylglucosamine and localized at the nuclear rim shows sequence similarity to aldose-1-epimerases. Plant Cell 10: 599–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishimizu T, Mitsukami Y, Shinkawa T, Natsuka S, Hase S, Miyagi M, Sakiyama F, Norioka S (1999) Presence of asparagine-linked N-acetylglucosamine and chitobiose in Pyrus pyrifolia S-RNases associated with gametophytic self-incompatibility. Eur J Biochem 263: 624–634 [DOI] [PubMed] [Google Scholar]
- Izhaki A, Swain SM, Tseng TS, Borochov A, Olszewski NE, Weiss D (2001) The role of SPY and its TPR domain in the regulation of gibberellin action throughout the life cycle of Petunia hybrida plants. Plant J 28: 81–90 [DOI] [PubMed] [Google Scholar]
- Jacobsen SE, Binkowski KA, Olszewski NE (1996) SPINDLY, a tetratricopeptide repeat protein involved in gibberellin signal transduction in Arabidopsis. Proc Natl Acad Sci USA 93: 9292–9296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerouge P, Cabanes-Macheteau M, Rayon C, Fischette-Laine AC, Gomord V, Faye L (1998) N-Glycoprotein biosynthesis in plants: recent developments and future trends. Plant Mol Biol 38: 1–48 [PubMed] [Google Scholar]
- Lin R, Allis CD, Elledge SJ (1996) PAT1, an evolutionarily conserved acetyltransferase homologue, is required for multiple steps in the cell cycle. Genes Cells 1: 923–942 [DOI] [PubMed] [Google Scholar]
- Lowe JB, Marth JD (2003) A genetic approach to mammalian glycan function. Annu Rev Biochem 72: 643–691 [DOI] [PubMed] [Google Scholar]
- Lu L, Berkey KA, Casero RA Jr (1996) RGFGIGS is an amino acid sequence required for acetyl coenzyme A binding and activity of human spermidine/spermine N1acetyltransferase. J Biol Chem 271: 18920–18924 [DOI] [PubMed] [Google Scholar]
- Lukowitz W, Nickle TC, Meinke DW, Last RL, Conklin PL, Somerville CR (2001) Arabidopsis cyt1 mutants are deficient in a mannose-1-phosphate guanylyltransferase and point to a requirement of N-linked glycosylation for cellulose biosynthesis. Proc Natl Acad Sci USA 98: 2262–2267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majewska-Sawka A, Nothnagel EA (2000) The multiple roles of arabinogalactan proteins in plant development. Plant Physiol 122: 3–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manuel P, Douhlas H, Leah L, Michael W (1980) Isolation of the chicken thymidine kinase gene by plasmid rescue. Nature 285: 207–210 [DOI] [PubMed] [Google Scholar]
- Maruta H, Greer K, Rosenbaum JL (1986) The acetylation of alpha-tubulin and its relationship to the assembly and disassembly of microtubules. J Cell Biol 103: 571–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mio T, Yamada-Okabe T, Arisawa M, Yamada-Okabe H (1999) Saccharomyces cerevisiae GNA1, an essential gene encoding a novel acetyltransferase involved in UDP-N-acetylglucosamine synthesis. J Biol Chem 274: 424–429 [DOI] [PubMed] [Google Scholar]
- Robertson M, Swain SM, Chandler PM, Olszewski NE (1998) Identification of a negative regulator of gibberellin action, HvSPY, in barley. Plant Cell 10: 995–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schachter H (1978) Glycoprotein biosynthesis. In D Pigman, MI Horowitz, eds, The Glycoconjugates, Vol. II. Academic Press, New York, pp 87–181
- Schindelman G, Morikami A, Jung J, Baskin TI, Carpita NC, Derbyshire P, McCann MC, Benfey PN (2001) COBRA encodes a putative GPI-anchored protein, which is polarly localized and necessary for oriented cell expansion in Arabidopsis. Genes Dev 15: 1115–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedbrook JC, Carroll KL, Hung KF, Masson PH, Somerville CR (2002) The Arabidopsis SKU5 gene encodes an extracellular glycosyl phosphatidylinositol-anchored glycoprotein involved in directional root growth. Plant Cell 14: 1635–1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steponkus PL, Lanphear FO (1967) Refinement of the triphenyl tetrazolium chloride method of determining cold injury. Plant Physiol 42: 1423–1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto K, Williamson RE, Wasteneys GO (2000) New techniques enable comparative analysis of microtubule orientation, wall texture, and growth rate in intact roots of Arabidopsis. Plant Physiol 124: 1493–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swain SM, Olszewski NE (1996) Genetic analysis of gibberellin signal transduction. Plant Physiol 112: 11–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres CR, Hart GW (1984) Topography and polypeptide distribution of terminal N-acetylglucosamine residues on the surfaces of intact lymphocytes. Evidence for O-linked GlcNAc. J Biol Chem 259: 3308–3317 [PubMed] [Google Scholar]
- van Hengel AJ, Tadesse Z, Immerzeel P, Schols H, van Kammen A, de Vries SC (2001) N-Acetylglucosamine and glucosamine-containing arabinogalactan proteins control somatic embryogenesis. Plant Physiol 125: 1880–1890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varki A, Cummings R, Esko J, Freeze H, Hart D, March J (1999) Essentials of Glycobiology. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp 85–195 [PubMed]
- Vitha S, Baluska F, Mews M, Volkmann D (1997) Immunofluorescence detection of F-actin on low melting point wax sections from plant tissues. J Histochem Cytochem 45: 89–95 [DOI] [PubMed] [Google Scholar]
- Wells L, Hart GW (2003) O-GlcNAc turns twenty: functional implications for post-translational modification of nuclear and cytosolic proteins with a sugar. FEBS Lett 546: 154–158 [DOI] [PubMed] [Google Scholar]
- Yoshida S, Forno DA, Cock JH, Gomez KA (1976) Laboratory Manual for Physiological Studies of Rice, Ed 3. The International Rice Research Institute, Manila, The Philippines