Abstract
While most of the efforts to uncover mechanisms contributing to bipolar disorder (BD) focused on phenotypes at the mature neuron stage, little research has considered events that may occur during earlier timepoints of neurodevelopment. Further, although aberrant calcium (Ca2+) signaling has been implicated in the etiology of this condition, the possible contribution of store-operated Ca2+ entry (SOCE) is not well understood. Here, we report Ca2+ and developmental dysregulations related to SOCE in BD patient induced pluripotent stem cell (iPSC)-derived neural progenitor cells (BD-NPCs) and cortical-like glutamatergic neurons. First, using a Ca2+ re-addition assay we found that BD-NPCs and neurons had attenuated SOCE. Intrigued by this finding, we then performed RNA-sequencing and uncovered a unique transcriptome profile in BD-NPCs suggesting accelerated neurodifferentiation. Consistent with these results, we measured a slower rate of proliferation, increased neurite outgrowth, and decreased size in neurosphere formations with BD-NPCs. Also, we observed decreased subventricular areas in developing BD cerebral organoids. Finally, BD NPCs demonstrated high expression of the let-7 family while BD neurons had increased miR-34a, both being microRNAs previously implicated in neurodevelopmental deviations and BD etiology. In summary, we present evidence supporting an accelerated transition towards the neuronal stage in BD-NPCs that may be indicative of early pathophysiological features of the disorder.
Subject terms: Neuroscience, Bipolar disorder
Introduction
Bipolar disorder (BD) is a chronic mood disorder characterized by severe symptomatic cycling between periods of depression and mania/hypomania [1]. Twin studies support a strong genetic component to the etiology of BD [2, 3], and genome-wide association studies (GWAS) coupled with exome sequencing analyses have identified more than 30 distinct genetic variations to date that confer disease susceptibility [4–6]. Notably, specific ion channel components and signaling effectors have been implicated as risk genes like CACNA1C (Calcium Voltage-Gated Channel Subunit Alpha1 C), which encodes a subunit of the L-type voltage gated Ca2+ channel and is involved in neurodevelopment and synaptic activity [7] and NCAN (Neurocan) that codes for a secreted extracellular matrix-forming protein that modulates axon guidance and cell migration [8]. However, despite those tantalizing genetic connections specific cellular processes responsible for the manifestation and progression of BD remain elusive.
The discovery of disease mechanisms responsible for BD has progressed slowly in part due to the heterogeneity of clinical symptoms in patients, the complexity of the underlying genetics, and the lack of comprehensive animal models [9, 10]. Fortunately, the growing availability of patient-derived induced pluripotent stem cells (iPSCs) and methods to culture those as differentiated neurons in a monolayer fashion, or as three-dimensional (3D) brain organoids, is now offering new options to face this challenge [11, 12]. Research using iPSC-based models have uncovered several cellular processes suspected to play a role in BD, including impaired migration and neurogenesis [13, 14], altered expression of membrane receptor and ion channel subunits [15], and differentially expressed genes that associate with differentiation, axonogenesis, and developmental maturation [16, 17]. Together these studies suggest that deviations in neuronal excitability and connectivity, which are suspected to be part of BD pathophysiology and disease progression, may be in fact rooted in neurodevelopment.
Some of the most reproducible cellular results concerning BD are those relating to Ca2+ signaling [16], a crucial component of central nervous system development and various aspects of neuroplasticity [18–20]. Apart from specialized voltage-gated Ca2+ channels and glutamatergic receptors, neurons also employ store-operated Ca2+ entry (SOCE [21, 22]; Fig. 1A). Recent evidence found that resting neurons have the most active SOCE [23], which functions as a key mediator of cell survivability [24] and neural differentiation [25]. Given the influence SOCE exerts on neurodevelopment and cellular excitability, we formulated the idea that aberrant activity could play a part in BD neuropathology.
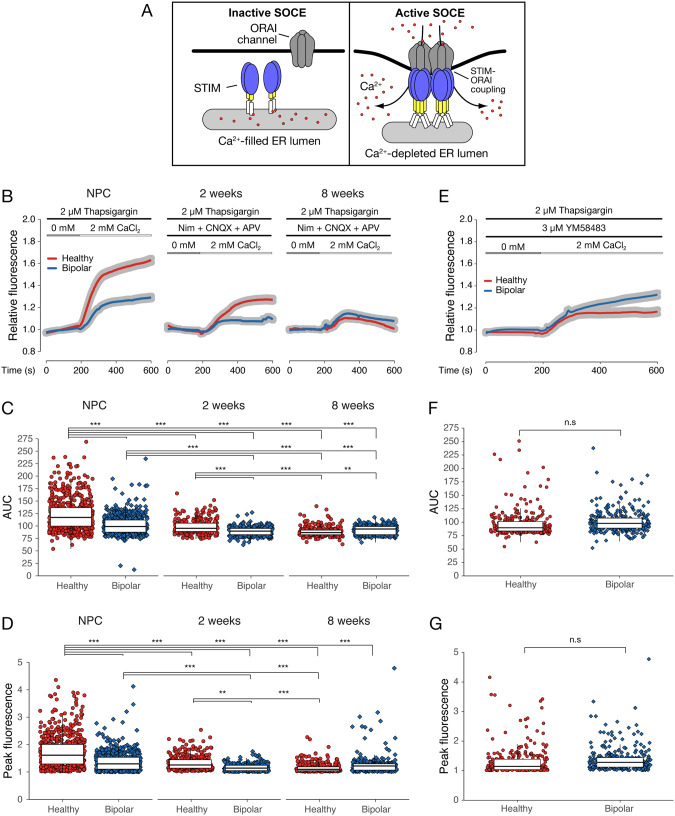
Fig. 1. Ca2+ imaging reveals SOCE deficit in BD-NPCs and neurons.
A A graphical representation of inactive (left) and active (right) SOCE. B Average increase in relative fluorescence (proportional to a baseline measurement from 0–180 s) of HC (red)- and BD (blue)-NPCs and neurons after Ca2+ re-addition. Data is presented as mean (red and blue curve) ± s.e.m (grey ribbon). C Area-under-the-curve (AUC) analysis from t = 185. Significant two-way ANOVA (F7,2966 = 139, p < 0.001) demonstrates that HC-NPCs (n = 598) had significantly higher AUCs than BD-NPCs (n = 595, p < 0.001), HC neurons differentiated for 2 weeks (n = 300, p < 0.001), BD neurons at 2 weeks (n = 287, p < 0.001), HC neurons at 8 weeks (n = 300, p < 0.001), and BD neurons at 8 weeks (n = 300, p < 0.001). BD-NPCs had significantly higher AUCs than BD neurons at 2 weeks (p < 0.001), HC neurons at 8 weeks, (p < 0.001), and BD neurons at 8 weeks (p < 0.001). HC neurons at 2 weeks had significantly higher AUCs than BD neurons at 2 weeks (p < 0.001), HC neurons at 8 weeks (p < 0.001), and BD neurons at 8 weeks (p = 0.007). D Peak fluorescence obtained per cell. Significant two-way ANOVA (F7,2966 = 66.4, p < 0.001) demonstrates that HC-NPCs had a higher peak fluorescence than BD-NPCs (p < 0.001), HC neurons differentiated for 2 weeks (p < 0.001), BD neurons at 2 weeks (p < 0.001), HC neurons at 8 weeks (p < 0.001), and BD neurons at 8 weeks (p < 0.001). BD-NPCs had significantly higher peak fluorescence than BD neurons at 2 weeks (p < 0.001), and HC neurons at 8 weeks (p < 0.001). HC neurons at 2 weeks had significantly higher peak fluorescence than BD neurons at 2 weeks (p = 0.001) and HC neurons at 8 weeks (p < 0.001). E Ca2+ re-addition assay after treatment with the ORAI channel blocker YM58483 compared to influx from (C). Data is presented as mean (red and blue curves) ± s.e.m (grey ribbon). F AUC analysis was conducted using the ANOVA described in (C) and demonstrates no significant difference between HC- and BD-NPCs (p = 0.20). G Peak fluorescence analysis was conducted using the ANOVA described in (D) and demonstrates no significant difference between HC- and BD-NPCs (p = 0.06). *p < 0.05, **p < 0.01, ***p < 0.001; ANOVA with Tukey’s post hoc test when appropriate. Each of the 6 cell lines are represented in the Ca2+ imaging where n > 198 cells per line in NPCs and n = 100 per line for differentiated neurons.
Here, we used 6 iPSC-derived NPC lines from 3 patients with type I BD (BD-NPCs) and 3 healthy controls (HC-NPCs) to study SOCE activity at the progenitor and differentiated neuron stages. Our approach allowed us to uncover a difference in the structure and function of BD-NPCs and neurons differentiated for 2 weeks compared to the HC-NPCs and neurons, namely a reduction of SOCE-dependent Ca2+ influx. Due to the range of processes affected by Ca2+ dynamics, we chose to employ RNA-sequencing (RNA-seq) and found that HC- and BD-NPCs contained a distinct transcriptome including increases in genes regulating neuronal differentiation. Next, functional assays revealed a consistent tendency towards differentiation due to decreased proliferation and increased neurite outgrowth, and characterization of iPSC-derived cerebral organoids revealed marked differences in 3D spatial patterning between HC and BD specimens. Finally, we measured an increase in the expression of the let-7 family of microRNAs (miRNAs) in BD-NPCs and of miR-34a in BD neurons—a result that is consistent with previous observations [26, 27]. Together, our results showcase a developmental component to BD that may be the key to uncovering new etiological factors responsible for the disease.
Material and methods
iPSC reprogramming, NPC derivation, and differentiation to cortical glutamatergic projection neurons
Human fibroblasts from healthy control and BD subjects enrolled in research studies at the Massachusetts General Hospital (Department of Psychiatry, Center for Genomic Medicine) were grown from dermal skin punches and reprogrammed as previously described [28].
When differentiating as 2D monolayers and neurospheres, cells were cultured in media containing 69.25% DMEM (Gibco), 29.25% Ham’s F12 (Gibco), 2% B27 (Gibco), 50 units/mL penicillin and 50 µL/mL streptomycin (Gibco) with a cortical glutamatergic projection neuron-like track, as characterized previously (29). Once in differentiation media, the cells were maintained without passage and media was changed every 2-3 days. Use of the patient-derived iPSC lines and generation of cerebral organoids for this study was performed at the University of Guelph and was approved by the University of Guelph Research Ethics Board (REB 17-11-012). All methods were carried out in accordance with relevant guidelines and regulations.
Please see Supplemental Information for detail about Cell culture, Ca2+ imaging, Puncta formation assay, Seahorse mitochondrial stress test, EdU assay, TUNEL assay, Neurosphere assay, Immunocytochemistry, Immunoblotting, RNA-seq, Cerebral organoid generation and staining, NanoString miRNA profiling, Real-time quantitative PCR, and Statistics.
Results
BD-NPCs and neurons have attenuated SOCE-specific Ca2+ influx
Most research to date about neuronal Ca2+ dynamics in relation to BD focuses on glutamatergic receptors and voltage-gated channels in differentiated neurons, whereas little attention has been directed towards possible Ca2+ mobilization differences at the NPC stage via SOCE. So, our first goal was to assess Ca2+ influx from SOCE in HC- and BD-NPCs, neurons differentiated for 2 weeks, and neurons developed to synaptic maturity at 8 weeks (Fig. S1; [29]). Using the Ca2+ indicator Fluo-4, Ca2+ imaging was conducted in NPCs, 2-week immature neurons, and 8-week mature neurons under a cocktail of inhibitors designed to abolish the activity of alternative Ca2+ pathways, namely NMDARs, AMPARs, and L-type voltage-gated Ca2+ channels [23]. In the presence of the sarcoendoplasmic reticulum Ca2+ transport ATPase (SERCA) pump inhibitor thapsigargin (Tg, a potent activator of SOCE), and after a brief incubation in Ca2+-free media, the cells were exposed to 2 mM Ca2+ and the relative increase in fluorescence from baseline (an average of fluorescence between t = 0 s and t = 180 s) was recorded (Fig. 1B; [30]). Curves are shown as a single average for all HC-NPCs or neurons from 50–100 cells in each of the 3 lines (red) and a separate, similarly obtained average for BD-NPCs or neurons. Area-under-the-curve (AUC; Fig. 1C) analysis from t = 185 s to t = 600 s and peak fluorescence analysis (Fig. 1D) revealed two key results. First, the AUC of HC-NPCs was 1.2x bigger, and the peak fluorescence 1.3x higher than BD-NPCs (p < 0.001), suggesting an attenuation in SOCE at this stage of neural differentiation. This was confirmed ratiometrically with Fura-2, which also showed that this disparity is not due to a difference in baseline Ca2+ concentrations between HC- and BD-NPCs (Fig. S2). Interestingly, at the 2-week timepoint this disparity was reduced to 1.1x for the AUC and 1.2x for the peak fluorescence measures, but still significant (p < 0.001). Finally, at the 8-week timepoint neither AUC nor peak fluorescence were statistically different between HC and BD neurons (p = 0.22). Importantly, significance was achieved even with inter- and intra-patient variability, highlighting the strength of this trend (Fig. S2).
To determine if these differences in Ca2+ influx were due to a dysregulation in SOCE, we repeated this experiment in the NPCs under SOCE-inhibiting conditions with the ORAI channel blocker YM58483 (Fig. 1E; [31]). As expected, the disparity in AUC (Fig. 1F) and peak fluorescence (Fig. 1G) between HC- and BD-NPCs was reduced and not statistically significant (p = 0.20 and p = 0.06, respectively) in the presence of YM58483. Interestingly, though, a small increase in Fluo-4 fluorescence remained following Tg application in both the BD-NPCs and the YM58483-treated HC-NPCs. We believe that these residual increases in SOCE are due to incomplete suppression since it has been previously demonstrated that germline deletion of SOCE effectors is more efficient in abolishing SOCE signal than pharmacological inhibition [32]. Of note, this residual increase could also result from the activity of Transient Receptor Potential Canonical (TRPC) channels since this pathway has been shown to influence SOCE activity in a number of contexts, including promoting the switch from proliferation to differentiation in A2B5+ NPCs [33, 34]. Therefore, to ensure that the influx we measured here is ORAI-dependent, and that the residual increase is not caused by other key Ca2+ channels, we inhibited TRPC4/5 and the activity-dependent Ca2+ channel NDMAR in succession in a representative HC and BD line to assess any effects those other players may have on the disparity between HC- and BD-NPCs (Fig. S3). Interestingly, difference in Ca2+ influx persisted (p < 0.001) through all our inhibition attempts. Altogether, these Ca2+ imaging results support the hypothesis that SOCE induced by Tg application is dysregulated in BD-NPCs and persists through the 2-week stage of differentiation but is resolved by the time the neurons reach 8-week differentiation.
BD-NPCs have decreased STIM2 puncta formation during SOCE activation
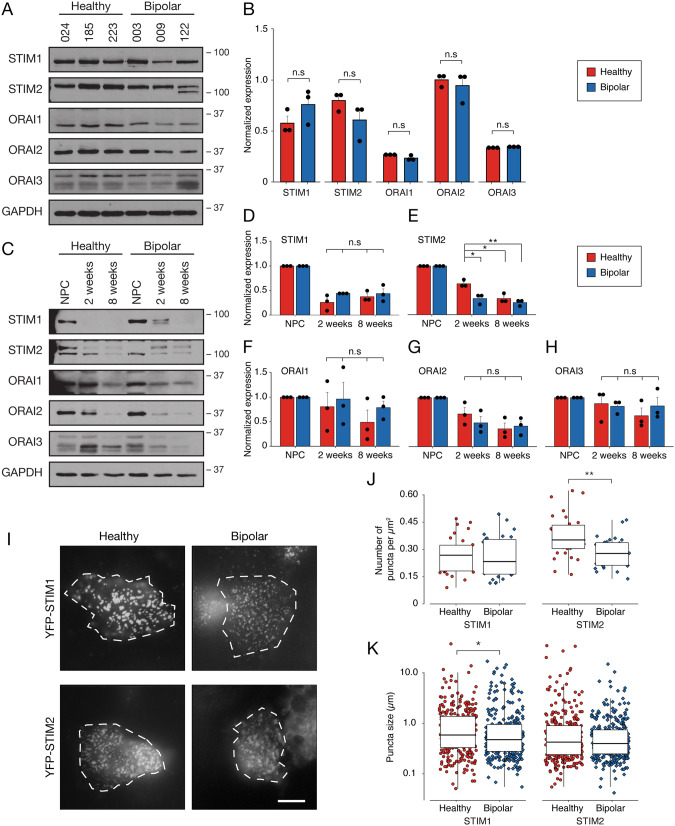
Next, we aimed to test whether the attenuation of SOCE measured in the BD-NPCs could be attributable to differences in the abundance of core effectors (STIM1/2, ORAI1/2/3). As shown in Fig. 2A, immunoblot analysis for STIMs and ORAIs revealed no apparent difference between HC- and BD-NPCs for all tested proteins. This was confirmed by quantification of the immunoblot results showing that intensity for all SOCE effectors at the NPC stage were not statistically significant (Fig. 2B), namely STIM1 (p = 0.21), STIM2 (p = 0.18), ORAI1 (p = 0.12), ORAI2 (p = 1), and ORAI3 (p = 0.37). In light of this result, we conclude that a different factor, such as membrane lipid composition [35] or physiological activation of ORAI channels [36], must be responsible for the attenuated Ca2+ influx at this stage of differentiation. It is interesting to note here that there is a qualitative, though statistically insignificant, decrease in STIM1 for BD lines 009 and 122. These are lithium-insensitive lines where 003 is lithium sensitive, and so future research with high sample sizes should consider differences in SOCE effector expression between lithium-sensitive and lithium-insensitive patient-derived cell lines. Additionally, a double band can be observed in STIM2 for patient line 122 and STIM2 is known to have several isoforms that differ slightly in weight. STIM2.2 is the canonical protein that can exist at 2 different sizes: a larger, more phosphorylated protein at 115 kDa and a smaller, less phosphorylated protein at 105 kDa [37]. Additionally, STIM2.1 and STIM2.3 have been discussed in the literature. STIM2.1 has found to be a larger, SOCE-inhibiting isoform while STIM2.3 is smaller, but little is known about its function [38]. In this case, we speculate that this band is the STIM2 isoform, STIM2.3.
Fig. 2. SOCE effector expression and puncta quantification are unique between HC- and BD-NPCs and neurons.
A SOCE effector immunoblotting on all 6 cell lines at the NPC stage with GAPDH used as a loading control. B Subsequent quantification of the western blot shows no significant differences in the expression of any effector (STIM1: t4 = 1.5, p = 0.21; STIM2: t4 = 1.63, p = 0.18; ORAI1: t4 = 1.96, p = 0.12; ORAI2: W = 5, p = 1; ORAI3: t4 = 1.0, p = 0.37, n = 3 for all effectors). C SOCE effector immunoblotting using representative cell lines at the NPC, 2-week, and 8-week stage of differentiation. Intensity was divided by intensity of GAPDH for normalization. Quantification was done for all 5 SOCE effectors using all 6 lines (n = 3 per condition with 1 replicate per cell line) and normalization was done by dividing protein band intensity by intensity of GAPDH. No interaction effect was observed for STIM1 (D; F3,8 = 1.35, p = 0.32), ORAI1, (F; F3,8 = 0.56, p = 0.66), ORAI2 (G; F3,8 = 1.19, p = 0.37), or ORAI3 (H; F3,8 = 0.49, p = 0.70). A significant interaction was observed for STIM2 (E; F3,8 = 11.47, p = 0.0028) showing a difference between HC and BD neurons at 2 weeks (p = 0.012), HC neurons at 2 weeks and BD neurons at 8 weeks (p = 0.0027), and HC neurons at 2 weeks and HC neurons at 8 weeks (p = 0.012). I All 3 cell lines from each condition (healthy, left; BD, right) were transiently transfected with YPF-STIM and imaged for YFP-STIM1 (top) and YFP-STIM2 (bottom) puncta formation 10 mins after the addition of 2 µM Tg. Cell bodies are outlined with a dotted white line. Scale bar represents 10 µm. J Subsequent quantification shows significantly more YFP-STIM2 puncta formation in HC-NPCs than BD counterparts (W = 667, p = 0.0011, n = 30 per condition, 10 per cell line). K Quantification of puncta size revealed a significant increase for HC-NPCs with YFP-STIM1 (W = 49328, p = 0.042, n = 300 per condition, 100 per cell line, 10 per cell). Data is presented as mean±s.e.m. n.s. signifies p > 0.05, *p < 0.05, **p < 0.01; Student’s t-test or ANOVA with Tukey’s HSD post hoc test was used for parametric data, Mann Whitney U test was used for non-parametric data.
We also immunoblotted SOCE effectors over the course of differentiation (Fig. 2C–H). While there were no significant differences in the expression of STIM1 (p = 0.32), ORAI1 (p = 0.66), ORAI2 (p = 0.37), or ORAI3 (p = 0.70), we found that HC neurons differentiated for 2 weeks had significantly higher expression of STIM2 than BD neurons after 2 weeks of differentiation (p = 0.012) and after 8 weeks of differentiation (p = 0.012). Importantly, 2 bands were present around the molecular weight of STIM2, which we suspect to be the differentially phosphorylated versions (39). The differences in expression presented here are concerning the less phosphorylated, smaller STIM2 protein. This suggests that the SOCE attenuation in the BD neurons over differentiation may be attributed, at least in part, to a relative decrease in the expression of STIM2.
To initiate SOCE, STIM proteins move to the plasma membrane and interact with ORAI channels to open them [39]. This event can be visualized by overexpressing STIM tagged to a fluorescent protein and monitoring the formation of discrete puncta [23, 40]. Observing a lower number of STIM puncta or smaller formations would support our finding of reduced Ca2+ influx through SOCE. To test this, we transfected our NPC lines with YFP-tagged STIM1 or YFP-tagged STIM2 (Fig. 2I) and treated the cells with 2 µM Tg for 10 min. Live NPCs from each group were then imaged and YFP-STIM puncta counted and measured. For YFP-STIM1, no difference between HC- and BD-NPCs was found for the number of puncta formed (p = 0.89; Fig. 2J); however, we observed that puncta size in HC-NPCs was on average 0.08 µm larger (p = 0.042; Fig. 2K). For YFP-STIM2, we counted 0.75x less puncta on BD-NPC membranes than HC (p = 0.0011; Fig. 2J), but no difference in size of puncta between the two groups. Importantly, it has been observed that STIM puncta can be static and, therefore, persistent even in the absence of store depletion [41]. In our NPCs, however, these static puncta were too sparse to quantify (Fig. S4). Based on those results, we propose that the lower Ca2+ influx from SOCE in BD-NPCs is primarily due to a decreased ability to form puncta on the cell surface in the NPCs.
Mitochondria function normally between HC- and BD-NPCs
Mitochondria play a key role in sequestering and shaping cytosolic Ca2+ rises [42], and mitochondrial dysfunction has been previously implicated as a contributing factor to BD pathophysiology [43]. Moreover, there is continuous Ca2+ exchange between ER and mitochondria [44]. Therefore, to ensure that mitochondria biology is consistent between the HC- and BD-NPCs, we assessed this fundamental cellular process using the Seahorse Mito Stress Test. No significant differences were observed between HC- and BD-NPCs for any parameters measured by the (Fig. S5): non-mitochondrial respiration (p = 0.4206), basal respiration (p = 0.6041), maximal respiration (p = 0.2201), ATP production (p = 0.4570), spare respiratory capacity (p = 0.3854) and coupling efficiency (p = 0.4348). This analysis shows that the differing Ca2+ dynamics that we found in our BD-NPC lines cannot be attributed to mitochondrial dysfunction.
Gene expression profiles are unique between HC- and BD-NPCs
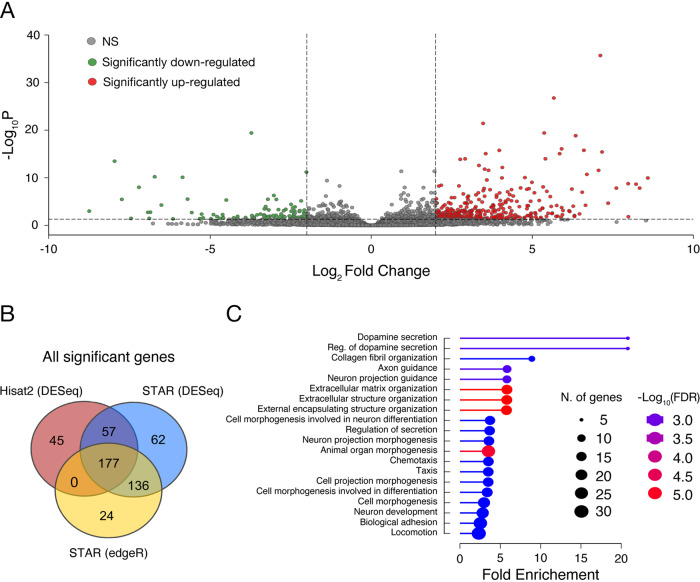
Given the finding that SOCE is dysregulated in the BD-NPCs, our next goal was to track what cellular differences this dysregulation yields. To first pinpoint transcriptional differences in a non-biased and high-throughput manner, we employed RNA-seq on the 6 iPSC-derived NPC lines using a multi-procedural approach. From STAR-DESeq2, we discovered that BD-NPCs had 283 upregulated genes and 77 downregulated genes with a log2 fold change ≥2 or ≤ -2 (p < 0.05) compared to HC-NPCs (Fig. 3A, Supplementary File 1). Of those, the top 20 most differentially expressed include the upregulated genes CFI, TGFBI, and CXCL12, and the downregulated gene TBX1. As any single method of differentially expressed gene (DEG) quantification has bias, we re-conducted the analysis with Hisat2-DESeq2 and STAR-edgeR. Commonly between the 3 analysis methods there were 148 upregulated and 29 downregulated genes (Fig. 3B). Gene Ontology (GO) analysis from geneontology.org powered by the PANTHER classification system from revealed that the top enriched biological processes from the common upregulated genes include processes implicated in neurodevelopment, such as “neuron projection guidance”, “extracellular matrix organization”, “cell morphogenesis involved in neuron differentiation”, and “neuron development”. Other enriched processes include “regulation of secretion”, “chemotaxis”, “biological adhesion”, and “locomotion” (Fig. 3C). Taken together, these results suggest that attenuation of SOCE in BD-NPCs could be responsible for wide-scale changes in gene expression affecting neurodevelopmental processes such as neurite extension, migration, and differentiation.
Fig. 3. RNA-seq demonstrates a distinct transcriptome between HC- and BD-NPCs.
A Volcano plot of the fold change of all differentially expressed genes in BD-NPCs including those with a log2 fold change significantly greater (red) or less than (green) 2 (p < 0.05) using STAR-DESeq2. 283 genes were found to be upregulated and 77 were downregulated. B To avoid bias from the analysis software, we re-quantified DEGs using 2 other methods, Hisat2-DESeq2 and STAR-edgeR. The Venn diagram represents the common DEGs found using each technique. C Gene Ontology analysis of the common DEGs ordered by category fold enrichment (ratio of the number of genes over/underexpressed compared to total number of genes associated with that biological function). Circle size illustrates number of DEGs involved in each function. Color demonstrates false discovery rate (FDR) from -log103.0 (blue) to - log105.0 (red).
BD-NPCs exhibit lower rates of proliferation, increased neurite outgrowth, and smaller neurosphere formation
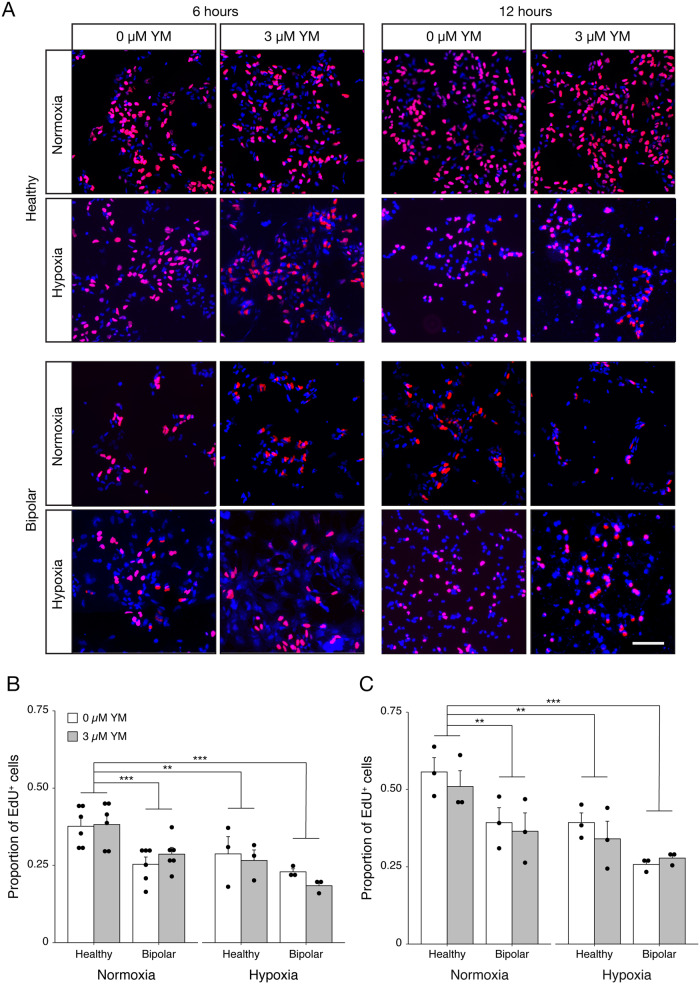
Our RNA-seq data hints at developmental differences between HC- and BD-NPCs. This, along with SOCE contributing to proliferation and differentiation [25], led us to conduct experiments to compare neurodevelopmental phenotypes between our two conditions. To assay proliferation, HC- and BD-NPCs were incubated with the thymidine analog 5-ethynyl-2’-deoxyuridine (EdU) for 6 and 12 h with and without YM58483 inhibition. To test how the NPCs responded to environmental stress, we compared cells under normoxic and hypoxic (O2 = 1%) conditions (Fig. 4). Due to a lack of significance between the untreated and treated groups, 0 µM and 3 µM YM58483 data points were pooled together for analysis. At both 6 and 12 h, HC-NPCs in normoxia had the fastest rate of proliferation compared to BD-NPCs in normoxia (p < 0.001 and p = 0.003), HC-NPCs in hypoxia (p = 0.007 and p = 0.006), and BD-NPCs in hypoxia (p < 0.001). The foremost conclusion from this data is that BD-NPCs have a slower rate of proliferation than their healthy counterparts, though this difference cannot be attributed to SOCE dysregulation. Of note, this significance is abolished in hypoxic conditions, meaning both HC- and BD-NPCs have a similar rate of proliferation under low levels of O2. This suggests a higher tolerance to stress in BD-NPCs compared to healthy counterparts, an interesting and unexpected finding. Alternatively, BD-NPCs already have a low rate of proliferation in normoxia and so any further reduction may be difficult to detect. Instead, oxidative stress present in BD-NPCs under basal conditions may be the source of this reduction in proliferation [45]. To ensure that this difference in proliferation was not due to increased cell death in the BD-NPCs, a TUNEL assay was conducted under normoxia and hypoxia between our 2 conditions (Fig. S6). There were no significant differences between cell death in any treatment group.
Fig. 4. EdU assay demonstrates lower rates of proliferation and higher resilience to stress in BD-NPCs.
A Representative immunofluorescent images of HC- and BD-NPCs without and with SOCE inhibition via 3 µM YM58483 (YM) after 6 and 12 h of incubation under normoxic and hypoxic conditions with the thymidine analog EdU (red). DAPI is shown in blue. Scale bar represents 100 µm. B Quantification of EdU+ cells / DAPI after 6 h of incubation under normoxia and hypoxia to measure proliferation rate. There was a significant interaction between condition and oxygen level (F3,32 = 13.7, p = 6.3 × 10−6, n = 12 per condition, 4 per cell line) that demonstrates a higher rate of proliferation in HC-NPCs in normoxia compared to BD-NPCs in normoxia (p = 0.00039), HC-NPCs in hypoxia (p = 0.0072), and BD-NPCs in hypoxia (p = 8.6 × 10-6). C These results were consistent after 12 h in the different oxygen conditions: a significant interaction effect (F3,20 = 14.4, p = 3.1 × 10-5, n = 6 per condition, 2 per cell line) demonstrates a higher rate of proliferation in the HC-NPCs in normoxia compared to BD-NPCs in normoxia (p = 0.0059), HC-NPCs in hypoxia (p = 0.0030), and BD-NPCs in hypoxia (p = 1.37 ×10-5). Model testing was used to determine a lack of evidence to include YM58483 treatment to the model, and so results at 0 µM and 3 µM YM treatment were pooled for both the 6- and 12-hour analyses. Data is presented as mean ± s.e.m. **p < 0.01, ***p < 0.001; ANOVA with Tukey’s post hoc test when appropriate.
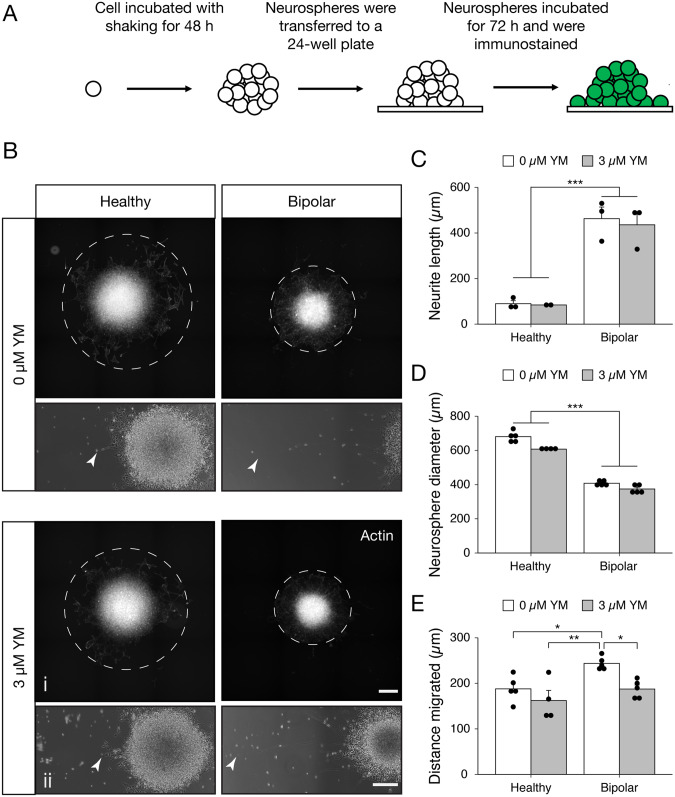
To assay migration and differentiation capacity, we employed a neurosphere assay in a representative HC- and BD-NPC line with and without pharmacological SOCE inhibition. HC and BD neurospheres were generated and plated onto Geltrex-coated plates. After a 72 h incubation period in differentiation media [29], the neurospheres were fixed and stained for visualization (Fig. 5A, B) and quantification in Image J (Fig. 5C–E). A striking result was uncovered in the amount of neurite outgrowth in the BD neurospheres. The distinction between HC and BD neurospheres is apparent both qualitatively (Fig. 5B) and quantitatively (Fig. 5C–E). Neurites developed from BD neurospheres were on average 362 nm longer than HC counterparts, regardless of YM58483 treatment condition (p = 0.002). An additional observation was the size of the neurospheres, a parameter that has been linked to the self-renewal capacity of NPCs [25]. BD neurospheres were 255 nm smaller in diameter than HC neurospheres, again regardless of YM58483 treatment condition (p < 0.001; Fig. 5D). In terms of migration, untreated BD-NPCs migrated 55.8 nm farther than untreated HC-NPCs on average, and 81.6 nm farther than treated HC-NPCs (p = 0.028 and p = 0.0027, respectively). Interestingly, they also migrated 56.4 nm farther than YM58483-treated BD-NPCs (p = 0.027; Fig. 5E). Taken together, these data suggest a tendency towards differentiation in the BD-NPCs.
Fig. 5. Neurosphere assay uncovers longer neurites, farther migration, and smaller sized spheres in BD-NPCs.
A A schematic depicting the methodology of the assay. B Representative fluorescent (i) and phase contrast (ii) micrographs for actin staining and neurite extension of HC and BD- derived neurospheres without (top) and with (bottom) SOCE inhibition (0 µM or 3 µM YM58483 (YM), respectively). Dotted lines migration of NPCs from the neurospheres. Arrow heads highlight the tip of the longest neurite imaged. Scale bars represent 200 µm. C Average length of neurites. 0 µM and 3 µM YM were pooled due to a lack of significance (W = 18, p = 1, n = 3), after which BD neurospheres had significantly longer extensions (W = 36, p = 0.002, n = 6). D Average size (diameter) of neurospheres measured. 0 µM and 3 µM YM data were pooled due to insignificance (W = 72, p = 0.11, n = 5), after which BD neurospheres were significantly smaller (W = 0, p = 1.1 ×10-5, n = 10). E Average distance cell bodies migrated from the neurosphere. Significant interaction (F3,15 = 7.21, p = 0.0032) demonstrates farther migration in untreated BD neurospheres (n = 5) compared to untreated HC (p = 0.028; n = 5), treated HC (p = 0.0027, n = 4) and treated BD- (p = 0.027, n = 5) derived neurospheres. Due to inadequate neurosphere formation with the other cell lines, data used only healthy line PSC-01-223 and BD line PSC-01-003 and is presented as mean ± s.e.m. *p < 0.05, **p < 0.01, ***p < 0.001; Mann–Whitney U test.
Cerebral organoid model reveals differences in subventricular zone (SVZ) formation between HC and BD organoids
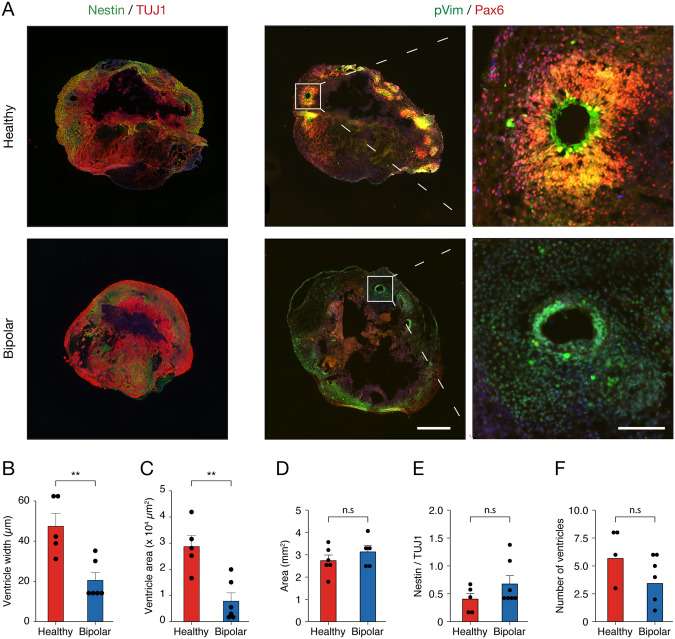
To further support the differences found between the HC and BD conditions above, we prepared 3D cerebral organoids with iPSCs from the same individuals as our NPC lines to characterize SVZ regions. Specifically, we focused on overall differentiation and size at the 50-day maturation timepoint (Fig. 6A). Comparison between the HC and BD groups revealed that SVZs were 2.3x thinner (p = 0.0046; Fig. 6B) and occupied 3.7x smaller area (p = 0.0028; Fig. 6C) in BD cerebral organoids. As SVZs are the sites of ongoing neurogenesis in a developing and adult brain, this result suggests that development in BD brains may be affected. Area (Fig. 6D), relative expression of Nestin (Fig. 6E), and number of ventricular zones (Fig. 6F) were not significantly different. In addition, a set of HC cerebral organoids was also treated with 3 µM and 6 µM YM to test SOCE-specific processes in this model, though no differences were observed between untreated and YM58483-treated organoids for number of ventricles, width of the SVZ, or subventricular area (Fig. S7). This result is not surprising, in our opinion, considering that deep pharmacological penetration into organoid tissue is difficult to achieve [46]. Overall, these results strengthen the idea that aspects of neurodevelopment are dysregulated in BD by introducing SVZ morphology as a specific phenotypic difference.
Fig. 6. BD organoids have smaller and thinner SVZs compared to HC organoids.
A Micrographs of a representative HC and BD organoid matured to 50 days. Organoids were co-stained for radial glia marker Nestin and immature neuron marker TUJ1 or for migration marker pVim and proliferation marker PAX6. Inlet shows an example SVZ. Scale bars represent 500 µm and 100 µm. B, C Average ventricular width and area per organoid. There was a significant decrease in width (t9 = 3.74, p = 0.0046) and area (t9 = 4.07, p = 0.0028) in the BD organoids (n = 3, healthy: n = 6). At least 1 organoid per cell line was used in each analysis. Data is presented as mean ± s.e.m; **p < 0.01, n.s signifies p > 0.05; Student’s t-test. D Average area of organoids. No significant differences were found between HC (n = 6) and BD (n = 4) organoids (t8 = 1.05, p = 0.32). E Ratio of Nestin/TUJ1 used to measure organoid maturity. Compared to HC (n = 5), BD (n = 4) organoids show no difference in relative Nestin expression (W = 22, p = 0.53). F Average number of ventricles per organoid. No significant differences were observed in BD- (n = 4) compared to HC (n = 6) organoids (t11 = 1.35, p = 0.20).
let-7 family and miR-34a are differentially expressed in BD-NPCs and neurons
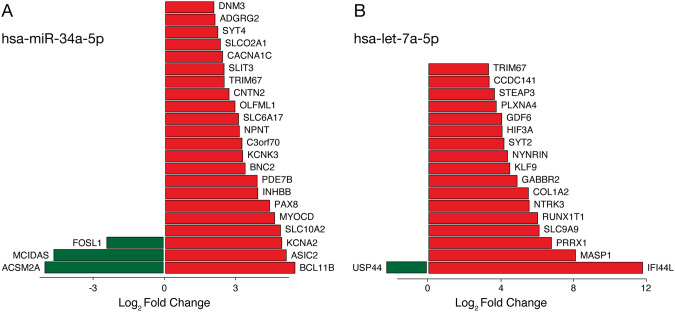
Finally, miRNAs are linked to numerous cellular processes, including neurodevelopment, and have been implicated as biomarkers for neuropsychiatric conditions like schizophrenia and major depression [47, 48]. Consequently, we sought to probe for differences in miRNA expression between our HC- and BD-NPC lines in addition to two others, GM08330 and GM05440, as described previously by Bavamian and colleagues [26]. Of special interest, previous studies found that miR-34a overexpression is associated with significant reduction of STIMATE (also known as TMEM110), which is a crucial protein in SOCE regulation [26, 49]. Moreover, miR-34a is also known to influence proliferation, morphogenesis, and differentiation of newborn neurons [50, 51]. Using Nanostring miRNA profiling of our NPC lines, we found higher counts of miR-34a at 4 weeks (3.24x higher) and 6 weeks (2.4x higher) of neuronal differentiation in BD lines, but not in NPCs or 2-week differentiated neurons (Supplementary File 2). This was further confirmed in our cell lines using qPCR (Fig. S8). When comparing the targets of this miRNA to the DEGs from our STAR-DESeq2 RNA-seq analysis, we identified 22 upregulated and 3 downregulated genes considered to be target of this miRNA (Fig. 7A). Of note, the downregulated gene FOSL1 is part of the FOS family, which are genes canonically known to influence proliferation and differentiation in neurons through their dimerization with JUN proteins [52]. Above all, our results concerning miR-34a support previous data from Bavamian and colleagues that used a different BD patient-derived NPC line (5440). While our findings suggest accelerated differentiation, they claim that this overexpression stunts neuronal morphology and synaptogenesis [26]. Further research is needed to consolidate these converse results.
Fig. 7. Cross referencing predicted targets of has-miR-34a-5p and hsa-let-7a-5p with DEGs from RNA-seq between HC- and BD-NPCs.
A miR-34a was shown to be upregulated in BD neurons at 4 and 6 weeks of differentiation in a NanoString analysis, so its predicted targets were compared to DEGs from the RNA-seq presented earlier. The plot shows putative targets and their respective expression levels. B hsa-let-7a and its family members with similar putative targets were shown to be upregulated at the NPC stage in BD lines in a NanoString analysis, so predicted targets of hsa-let-7a-5p were compared to DEGs from the RNA-seq presented earlier. The plot shows putative targets and their respective expression levels. Green represents downregulated genes while red represents upregulated genes.
Also of interest, the entire family of let-7 was upregulated in BD-NPCs including 7a (76x higher), b (8.2x higher), c (10x higher), d (30x higher), e (5.7x higher), f (15x higher), g (18x higher), and i (25x higher; Supplementary File 2). Since the predicted targets of all these family members are highly similar, we compared the targets of let-7a-5p to the DEGs in our RNA-seq analysis and found 17 upregulated and 1 downregulated gene in common (Fig. 7B). Ubiquitin-specific protease 44 (USP44) regulates the spindle assembly checkpoint and has been implicated in intellectual disability, but never BD [53]. Future research is needed to establish its role in BD.
While this comparison between the RNA-seq and Nanostring analysis demonstrates that DEGs may be due to translational regulation by miRNAs, a more comprehensive analysis on non-coding RNAs must be done to fully understand this process. Firstly, alternative polyadenylation alters miRNA binding capabilities to the 3’UTR of transcripts [54]. Additionally, crosstalk between miRNAs and other non-coding RNAs contribute to translational regulation more than miRNAs alone [55]. Finally, miRNAs have been found to increase translational efficiency in addition to inhibiting it [56]. Overall, processes of differentiation rely on careful manipulation of gene expression and dysregulation in these mechanisms contribute greatly to pathology of disease. Here we uncovered a new potential mechanism of dysregulation through differential expression of miRNAs crucial for neurodevelopment.
Discussion
BD is a complex disorder for which various theories about its etiology, like mitochondrial dysfunction (reviewed in [45]) and inositol depletion (reviewed in [57]), have been proposed. None of the models presented to date, however, have been able to adequately explain the pathology nor the cause of the disease. Alternative efforts in recent years have been to examine the role of neurodevelopment in NPCs, with much of that research suggesting that aberrant Ca2+ dynamics play a central part [15, 16]. The role SOCE plays in these aberrant Ca2+ dynamics has been explored in non-neuronal surrogate cell models of BD such as lymphoblastoid cells [58, 59], but not NPCs or neuronal models of the condition. Furthermore, few studies have examined the contribution of individual SOCE effectors and more often focused on a single isoform, in particular STIM1 and ORAI1. Here, our goal was to explore SOCE-related developmental defects in BD-NPCs in a more holistic manner and our conclusions are summarized in Table 1.
Table 1.
Summary of results.
| Experiment | Outcome |
|---|---|
| Ca2+ imaging | BD-NPCs have reduced Ca2+ influx from SOCE. |
| Effector quantification | NPCs have no differences in effector expression. Immature neurons have reduced STIM2 expression. |
| Puncta formation assay | BD-NPCs form smaller STIM1 puncta and less STIM2 puncta. |
| RNA-sequencing | BD-NPCs have a distinct transcriptome with enriched neurodevelopmental processes. |
| Proliferation assay | BD-NPCs have slower rates of proliferation. |
| Neurosphere generation | BD-NPCs form smaller neurospheres, have more migration, and longer neurite extension. |
| Organoid generation | BD organoids have subventricular zones with smaller width and area. |
| Nanostring miRNA analysis | BD-NPCs have increased expression of the let-7 miRNA family and BD immature neurons have increased expression of miR-34a. |
Our Ca2+ imaging analysis revealed that SOCE was indeed dysregulated in BD-NPCs as Ca2+ influx via this pathway was barely measurable in response to Tg application (Fig. 1). Further supporting this claim, pre-treatment with the ORAI channel blocker YM58483 significantly reduced the SOCE signal in the HC-NPCs but not the BD-NPCs (Fig. 1). From this, we conclude that the BD-NPCs have minimal SOCE activity. Importantly, this disparity persists at 2 weeks of differentiation but is abolished after 8 weeks, when the neurons reach maturity (Fig. 1). This decrease in activity over differentiation was expected since previous studies have shown that SOCE promotes self-renewal in progenitors, and the decrease of this activity coincides with differentiation [25, 32, 60]. Furthermore, as the neurons begin to express more specialized Ca2+ channels, SOCE becomes less vital for Ca2+ influx. When examining protein expression, the effectors also showed an overall decrease over differentiation. Of note, STIM2 protein expression was significantly decreased in a pattern that exactly matched that of the differences in Ca2+ influx. STIM2 in BD neurons was consistent with that of HC neurons at an early stage of differentiation, highlighting a premature differentiation phenotype dependent on its expression. Following this discovery in the neurons, we sought to understand why a disparity in Ca2+ influx existed between HC- and BD-NPCs. Immunoblot analysis of SOCE effectors revealed no differences between the 2 conditions at the NPC stage (Fig. 2). Since another cause for decreased influx could be due to aberrant puncta formation, we tested this scenario with transiently transfecting YFP-STIM1/2 constructs and found that the BD-NPCs could not form as many STIM2-specific puncta. This is a finding that may have relevant implications for the pathogenesis and progression of BD, especially with respect to STIM2. Specifically, of the two effectors, STIM2 is more widely expressed and is the most prominent effector of SOCE in the mouse hippocampus [61]. A key feature of STIM2 is its role in regulating basal Ca2+ concentrations in the cytosol due to its higher dissociation constant (KD) and sensitivity to smaller Ca2+ fluctuations [41, 62–64]. While baseline Ca2+ concentrations could not be measured in this study due to the non-ratiometric dye used, it would be of interest to examine differences in these levels over differentiation given that mature neurons from BD and schizophrenic patients have increased basal cytosolic Ca2+ concentrations [65]. Further, while STIM1 has been shown to be pivotal for self-renewal and proliferation [25], the specific effects of STIM2 on differentiation have not been explored.
Since intact mitochondria are required for proper SOCE function, we ensured that the resulting phenotypes observed are due to SOCE dysfunction and not mitochondrial effect [44]. Upon completion of a Seahorse Mito Stress Test, we have concluded that the HC- and BD-NPCs have similar mitochondrial function and no defects (Fig. S5), which is in line with previous studies using IPSC-derived HC- and BD-NPCs [66]. While this may seem inconsistent with other findings using hippocampal DG granule cell-like neurons derived from BD patients showing mitochondrial deficits [67], we would like to note the changes that are occurring in mitochondria during neurogenesis—including mitochondria morphology, RNA expression profiles, and metabolic shift from glycolysis to OXPHOS [68]—may account for these differences. Further research comparing mitochondrial functionality on different stages of neurogenesis is needed to explain this phenomenon.
Next, we looked for possible differences in functionality of the BD-NPCs to explain the tendency observed in the RNA-seq towards differentiation. For this aim we used EdU proliferation assay, in which BD-NPCs exhibited a significantly reduced rate of proliferation compared to the HC-NPCs (Fig. 4). To take it one step forward, we also tested the proliferation under a hypoxic environment since stress can unleash specific responses that could exacerbate cellular deficits. We found that while the rate of proliferation significantly decreased for HC-NPCs under hypoxia, it stayed the same for BD-NPCs, resulting consequently in similar rates of proliferation between HC- and BD-NPCs (Fig. 4). This suggests that either the BD-NPCs were less susceptible to cellular stress, or effect in proliferation cannot be observed after addition of stressors due to the intrinsic stress the BD-NPCs are already under. While the literature has not directly connected resilience to hypoxia with BD, hypoxia is connected to the Wnt/ß-catenin pathway via TLX expression and hypoxia-inducible factor 1 [69, 70]. This pathway activates neurogenesis in NPCs, including proliferation [71]. One possible explanation for this stress resilience is the reduced capacity for STIM2 puncta formation in the BD-NPCs. Loss of STIM2 is known to promote apoptosis resistance by reducing SOCE-dependent Ca2+ influx [72]. Additionally, mice with neuron-specific deficiencies in STIM2 are protected from apoptosis under oxidative stress, which is elevated during hypoxia [63]. These data support that a reduction of SOCE from decreased STIM2 expression is neuroprotective under hypoxia. It could be that a reduction in STIM2 puncta formation elicits a similar protection. Importantly, any changes in rate of proliferation could not be due to increased cell death in the cell populations, as a TUNEL assay showed no significant differences in rate of cell death between any of the above conditions (Fig. S6).
Exploring more differentiation-associated phenotypes, neurospheres generated by BD-NPCs were significantly smaller in size with significantly longer neurites, further supporting a trend towards differentiation and away from a symmetric progenitor proliferation ([25]; Fig. 5). The elongated neurites phenotype is consistent with research suggesting that CRMP2, a GSK3β-dependent microtubule-associated protein involved in axonal growth, could be responsible [73]. Transcripts of this protein are not expected to differ as its mechanism of action depends on the ratio of pCRMP2:CRMP2, which is why our RNA-seq did not yield significant differences in this transcript. In addition to decreased size and increased neurite outgrowth, migration was also affected by condition: untreated BD-NPCs migrated farther away from the neurosphere body compared to HC-NPCs and YM58483-treated BD-NPCs. Migration is partly regulated in cells by stromal cell-derived factor 1 (SDF-1, or otherwise known as CXCL12; see [74]). In agreement with our RNA-sequencing data, CXCL12 was one of the top 3 overexpressed genes in the STAR-DESeq2 analysis (6.7 log2-fold change in BD-NPCs, F = 7.7, p = 1.86 × 10−11), suggesting a role of CXCL12 for enhanced migration in the BD-NPCs. In addition to speed of migration, another recent study has shown that BD-NPCs have quasi-Brownian migration patterns instead of the typical linear migration patterns in a 2D cell culture model [75]. This further highlight aberrant migration patterns in BD-NPCs.
Some parameters are unable to be probed for in 2D cell culture. Neurogenesis and the development of the human brain is an extremely complex process in which spatio-temporal factors are crucial [12]. To model BD brain growth in a 3D model, cerebral organoids were generated from both HC- and BD-iPSCs and differences in protein expression were analyzed using immunohistochemistry. While testing for the effects that SOCE inhibition has on development, no differences were observed between untreated and treated organoids (Fig. S7). It has been shown, however, that penetrance of certain compounds into organoids is weak [76]. So, the lack of significance may be due to inadequate treatment across the entire organoid. In the future, the generation of sliced neocortical organoids or germline deletion of SOCE effectors STIM1 and STIM2 may uncover a significant role for SOCE in organoid development [46]. The BD organoids, on the other hand, had a reduced subventricular width and area (Fig. 6). This can be the result of an excess of asymmetric divisions which reduce the stem cell pool in the area [12]. Connecting this claim to the reduced capacity of SOCE in our BD-NPCs, Domenichini and colleagues found that pharmacological inhibition of SOCE impaired the self-renewal capabilities of NPCs and shifted cell division to a more asymmetric paradigm, therefore reducing the stem cell population [60]. All of this evidence suggests that BD-NPCs differentiate prematurely.
SOCE was inhibited by blocking ORAI channels with YM throughout all experiments, however this never mimicked the phenotypes observed in the BD-NPCs as we predicted it would. From Fig. 2, it is apparent that reduced capacity for puncta formation is the reason for reduced Ca2+ influx, not reduced influx from the ORAI channel. This difference in the method used to block SOCE could explain why we could not replicate the phenotype. Alternatively, SOCE attenuation may be occurring independently of the accelerated differentiation shown here. A closer investigation into the functional impact of attenuated Ca2+ influx from SOCE is needed to elucidate this further.
Aside from transcriptional regulation of neurodevelopmental genes, these processes can also be regulated post-transcriptionally by miRNAs. To examine if these types of noncoding RNAs could play a role in the phenotypes shown in our study, we probed for differentially expressed transcripts in an unbiased manner with NanoString technology. From this we found several miRNAs of interest that have differential expressions between HC- and BD-NPCs, namely the let-7 family of miRNAs. Most interestingly, one of the top predicted targets of let-7a according to the online database miRDB (www.mirdb.org; [77]) is STIMATE—a mRNA for the STIMATE protein active at the ER and plasma membrane junction where it regulates STIM-ORAI signaling [49, 78]. Although we found no difference in the expression of STIMATE mRNA between HC- and BD-NPCs, the possibility exists that translational repression of the protein due to let-7 miRNAs could help explain the aberrant formation of STIM2 puncta we measure in this study [79]. Beyond this connection, the let-7 family of miRNAs is also known to regulate neuronal differentiation: increased expression of let 7a, let-7b, and let-7d promote cell cycle exit by repressing proliferative and cell cycle progression genes such as TLX and CCND1, which is consistent with the reduced proliferation in our BD-NPCs and smaller SVZs in the organoids [80–83]. The family is also hypothesized to contribute to an intrinsic clock that regulates the balance between proliferation and differentiation [83]. Overall, we speculate that the overexpression of let-7 in the BD-NPC lines contributes to the premature differentiation phenotype due to the dysregulation of this intrinsic clock.
Several other miRNAs were upregulated over the late stages of differentiation, one of these being miR-34a. Bavamian and colleagues have similarly found that miR-34a is overexpressed in BD patient postmortem cerebellum tissue and, more interestingly, in BD-NPCs from separate patient lines than ours [26]. Interestingly, miR-34a is known to decrease SOCE activity in human Jurkat T-cells, though in part through STIM1 downregulation [84]. No miRNAs have thus far been identified to target STIM2, though miRNA expression may further contribute to the attenuated Ca2+ influx shown here.
In conclusion, we discovered a deficit of SOCE activity in BD neurons and subsequently uncovered several fundamental differences compared to HC-NPCs including a unique transcriptome and miRNA pool, reduced proliferation, enhanced differentiation, and unique 3D development. These data open avenues into exploring BD as a neurodevelopmental disorder and upon further research may elucidate novel mechanism susceptible to environmental factors molecular underpinnings of this complex disease.
Resource availability
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact Jasmin Lalonde (jlalon07@uoguelph.ca)
Supplementary information
Acknowledgements
This study was funded by the Brain & Behavior Research Foundation (NARSAD Young Investigator Grant 24990), the Canadian Foundation for Innovation (CFI Grant 034455), and generous start-up funding from the University of Guelph (all to Jasmin Lalonde). We thank Dr. Michaela Strüder-Kypke (Advanced Analysis Centre, University of Guelph) for assistance with microscopy, Dr. Anthony Mutsaers (Biomedical Sciences, University of Guelph) for access to his Agilent Seahorse Analyzer, and Dr. Jim Uniacke (Molecular and Cellular Biology, University of Guelph) for access to his hypoxic workstation.
Author contributions
TH, BA, and JL conceived the hypothesis, supervised the experiments, and prepared the manuscript. TH, BA, NB, EC, JW, and JL performed experiments. TH, BA, MT, JW, MP, SJH, NM, and JL analyzed data. SDS and RHP derived and supplied the cell lines. All authors reviewed the manuscript and provided feedback.
Data availability
Any additional information required to reanalyze the data reported in this paper is available form the lead contact upon request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Tristen Hewitt, Begüm Alural.
Supplementary information
The online version contains supplementary material available at 10.1038/s41380-023-02152-6.
References
- 1.Askland K, Read C, Moore J. Pathways-based analyses of whole-genome association study data in bipolar disorder reveal genes mediating ion channel activity and synaptic neurotransmission. Hum Genet. 2008;125:63. doi: 10.1007/s00439-008-0600-y. [DOI] [PubMed] [Google Scholar]
- 2.Barnett JH, Smoller JW. The genetics of bipolar disorder. Neuroscience. 2009;164:331–43. doi: 10.1016/j.neuroscience.2009.03.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van der Schot AC, Vonk R, Brans RG, van Haren NE, Koolschijn PC, Nuboer V, et al. Influence of Genes and Environment on Brain Volumes in Twin Pairs Concordant and Discordant for Bipolar Disorder. Arch Gen Psychiat. 2009;66:142–51. doi: 10.1001/archgenpsychiatry.2008.541. [DOI] [PubMed] [Google Scholar]
- 4.Hou L, Bergen SE, Akula N, Song J, Hultman CM, Landén M, et al. Genome-wide association study of 40,000 individuals identifies two novel loci associated with bipolar disorder. Hum Mol Genet. 2016;25:3383–94. doi: 10.1093/hmg/ddw181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Orrù G, Carta MG. Genetic Variants Involved in Bipolar Disorder, a Rough Road Ahead. Clin Pract Epidemiol Ment Heal. 2018;14:37–45. doi: 10.2174/1745017901814010037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kushima I, Nakatochi M, Aleksic B, Okada T, Kimura H, Kato H, et al. Cross-Disorder Analysis of Genic and Regulatory Copy Number Variations in Bipolar Disorder, Schizophrenia, and Autism Spectrum Disorder. Biol Psychiat. 2022;92:362–74. doi: 10.1016/j.biopsych.2022.04.003. [DOI] [PubMed] [Google Scholar]
- 7.Green EK, Hamshere M, Forty L, Gordon-Smith K, Fraser C, Russell E, et al. Replication of bipolar disorder susceptibility alleles and identification of two novel genome-wide significant associations in a new bipolar disorder case–control sample. Mol Psychiatr. 2013;18:1302–7. doi: 10.1038/mp.2012.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cichon S, Mühleisen TW, Degenhardt FA, Mattheisen M, Miró X, Strohmaier J, et al. Genome-wide Association Study Identifies Genetic Variation in Neurocan as a Susceptibility Factor for Bipolar Disorder. Am J Hum Genetics. 2011;88:396. doi: 10.1016/j.ajhg.2011.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Charney AW, Ruderfer DM, Stahl EA, Moran JL, Chambert K, Belliveau RA, et al. Evidence for genetic heterogeneity between clinical subtypes of bipolar disorder. Transl Psychiat. 2017;7:e993–e993. doi: 10.1038/tp.2016.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coombes BJ, Markota M, Mann JJ, Colby C, Stahl E, Talati A, et al. Dissecting clinical heterogeneity of bipolar disorder using multiple polygenic risk scores. Transl Psychiat. 2020;10:314. doi: 10.1038/s41398-020-00996-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, et al. Induction of Pluripotent Stem Cells from Adult Human Fibroblasts by Defined Factors. Cell. 2007;131:861–72. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 12.Lancaster MA, Renner M, Martin C-A, Wenzel D, Bicknell LS, Hurles ME, et al. Cerebral organoids model human brain development and microcephaly. Nature. 2013;501:373–9. doi: 10.1038/nature12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Muñoz‐Estrada J, Benítez‐King G, Berlanga C, Meza I. Altered Subcellular Distribution of the 75‐kDa DISC1 Isoform, cAMP Accumulation, and Decreased Neuronal Migration in Schizophrenia and Bipolar Disorder: Implications for Neurodevelopment. Cns Neurosci Ther. 2015;21:446–53. doi: 10.1111/cns.12377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Najt P, Wang F, Spencer L, Johnston JAY, Lippard ETC, Pittman BP, et al. Anterior Cortical Development During Adolescence in Bipolar Disorder. Biol Psychiat. 2016;79:303–10. doi: 10.1016/j.biopsych.2015.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Madison JM, Zhou F, Nigam A, Hussain A, Barker DD, Nehme R, et al. Characterization of Bipolar Disorder Patient-Specific Induced Pluripotent Stem Cells from a Family Reveals Neurodevelopmental and mRNA Expression Abnormalities. Mol Psychiatr. 2015;20:703–17. doi: 10.1038/mp.2015.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen HM, DeLong CJ, Bame M, Rajapakse I, Herron TJ, McInnis MG, et al. Transcripts involved in calcium signaling and telencephalic neuronal fate are altered in induced pluripotent stem cells from bipolar disorder patients. Transl Psychiat. 2014;4:e375–e375. doi: 10.1038/tp.2014.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kathuria A, Lopez-Lengowski K, Vater M, McPhie D, Cohen BM, Karmacharya R. Transcriptome analysis and functional characterization of cerebral organoids in bipolar disorder. Genome Med. 2020;12:34. doi: 10.1186/s13073-020-00733-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rosenberg SS, Spitzer NC. Calcium Signaling in Neuronal Development. Csh Perspect Biol. 2011;3:a004259. doi: 10.1101/cshperspect.a004259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wegierski T, Kuznicki J. Neuronal calcium signaling via store-operated channels in health and disease. Cell Calcium. 2018;74:102–11. doi: 10.1016/j.ceca.2018.07.001. [DOI] [PubMed] [Google Scholar]
- 20.Horigane S, Ozawa Y, Yamada H, Takemoto-Kimura S. Calcium signalling: a key regulator of neuronal migration. J Biochem. 2019;165:401–9. doi: 10.1093/jb/mvz012. [DOI] [PubMed] [Google Scholar]
- 21.Hartmann J, Karl RM, Alexander RPD, Adelsberger H, Brill MS, Rühlmann C, et al. STIM1 Controls Neuronal Ca2+ Signaling, mGluR1-Dependent Synaptic Transmission, and Cerebellar Motor Behavior. Neuron. 2014;82:635–44. doi: 10.1016/j.neuron.2014.03.027. [DOI] [PubMed] [Google Scholar]
- 22.Samtleben S, Wachter B, Blum R. Store-operated calcium entry compensates fast ER calcium loss in resting hippocampal neurons. Cell Calcium. 2015;58:147–59. doi: 10.1016/j.ceca.2015.04.002. [DOI] [PubMed] [Google Scholar]
- 23.Lalonde J, Saia G, Gill G. Store-Operated Calcium Entry Promotes the Degradation of the Transcription Factor Sp4 in Resting Neurons. Sci Signal. 2014;7:ra51. doi: 10.1126/scisignal.2005242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Secondo A, Petrozziello T, Tedeschi V, Boscia F, Vinciguerra A, Ciccone R, et al. ORAI1/STIM1 Interaction Intervenes in Stroke and in Neuroprotection Induced by Ischemic Preconditioning Through Store-Operated Calcium Entry. Stroke. 2019;50:1240–9. doi: 10.1161/STROKEAHA.118.024115. [DOI] [PubMed] [Google Scholar]
- 25.Gopurappilly R, Deb BK, Chakraborty P, Hasan G. Stable STIM1 Knockdown in Self-Renewing Human Neural Precursors Promotes Premature Neural Differentiation. Front Mol Neurosci. 2018;11:178. doi: 10.3389/fnmol.2018.00178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bavamian S, Mellios N, Lalonde J, Fass DM, Wang J, Sheridan SD, et al. Dysregulation of miR-34a links neuronal development to genetic risk factors for bipolar disorder. Mol Psychiatr. 2015;20:573–84. doi: 10.1038/mp.2014.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chua CEL, Tang BL. miR-34a in Neurophysiology and Neuropathology. J Mol Neurosci. 2019;67:235–46. doi: 10.1007/s12031-018-1231-y. [DOI] [PubMed] [Google Scholar]
- 28.Zimmerman AJ, Hafez AK, Amoah SK, Rodriguez BA, Dell’Orco M, Lozano E, et al. A psychiatric disease-related circular RNA controls synaptic gene expression and cognition. Mol Psychiatr. 2020;25:2712–27. doi: 10.1038/s41380-020-0653-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shi Y, Kirwan P, Smith J, Robinson HPC, Livesey FJ. Human cerebral cortex development from pluripotent stem cells to functional excitatory synapses. Nat Neurosci. 2012;15:477–86. doi: 10.1038/nn.3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bird GS, DeHaven WI, Smyth JT, Putney JW. Methods for studying store-operated calcium entry. Methods. 2008;46:204–12. doi: 10.1016/j.ymeth.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu B, Fivaz M. Neuronal SOCE: Myth or Reality? Trends Cell Biol. 2016;26:890–3. doi: 10.1016/j.tcb.2016.09.008. [DOI] [PubMed] [Google Scholar]
- 32.Somasundaram A, Shum AK, McBride HJ, Kessler JA, Feske S, Miller RJ, et al. Store-Operated CRAC Channels Regulate Gene Expression and Proliferation in Neural Progenitor Cells. J Neurosci. 2014;34:9107–23. doi: 10.1523/JNEUROSCI.0263-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Louis M, Zanou N, Schoor MV, Gailly P. TRPC1 regulates skeletal myoblast migration and differentiation. J Cell Sci. 2008;121:3951–9. doi: 10.1242/jcs.037218. [DOI] [PubMed] [Google Scholar]
- 34.Shin HY, Hong YH, Jang SS, Chae HG, Paek SL, Moon HE, et al. A Role of Canonical Transient Receptor Potential 5 Channel in Neuronal Differentiation from A2B5 Neural Progenitor Cells. Plos One. 2010;5:e10359. doi: 10.1371/journal.pone.0010359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cao X, Choi S, Maléth JJ, Park S, Ahuja M, Muallem S. The ER/PM microdomain, PI(4,5)P2 and the regulation of STIM1–Orai1 channel function. Cell Calcium. 2015;58:342–8. doi: 10.1016/j.ceca.2015.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yoast RE, Emrich SM, Zhang X, Xin P, Johnson MT, Fike AJ, et al. The native ORAI channel trio underlies the diversity of Ca2+ signaling events. Nat Commun. 2020;11:2444. doi: 10.1038/s41467-020-16232-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Williams RT, Manji SSM, Parker NJ, Hancock MS, Stekelenburg LV, Eid J-P, et al. Identification and characterization of the STIM (stromal interaction molecule) gene family: coding for a novel class of transmembrane proteins. Biochem J. 2001;357:673. doi: 10.1042/0264-6021:3570673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Berna-Erro A, Jardin I, Salido GM, Rosado JA. Role of STIM2 in cell function and physiopathology: STIM2 in cell function and physiopathology. J Physiol. 2017;595:3111–28. doi: 10.1113/JP273889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qiu R, Lewis RS. Structural features of STIM and Orai underlying store-operated calcium entry. Curr Opin Cell Biol. 2019;57:90–98. doi: 10.1016/j.ceb.2018.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang H, Bryson VG, Wang C, Li T, Kerr JP, Wilson R, et al. Desmin interacts with STIM1 and coordinates Ca2+ signaling in skeletal muscle. Jci Insight. 2021;6:e143472. doi: 10.1172/jci.insight.143472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gruszczynska-Biegala J, Pomorski P, Wisniewska MB, Kuznicki J. Differential Roles for STIM1 and STIM2 in Store-Operated Calcium Entry in Rat Neurons. Plos One. 2011;6:e19285. doi: 10.1371/journal.pone.0019285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Contreras L, Drago I, Zampese E, Pozzan T. Mitochondria: The calcium connection. Biochimica Et Biophys Acta Bba - Bioenergetics. 2010;1797:607–18. doi: 10.1016/j.bbabio.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 43.Cataldo AM, McPhie DL, Lange NT, Punzell S, Elmiligy S, Ye NZ, et al. Abnormalities in Mitochondrial Structure in Cells from Patients with Bipolar Disorder. Am J Pathology. 2010;177:575–85. doi: 10.2353/ajpath.2010.081068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Deak AT, Blass S, Khan MJ, Groschner LN, Waldeck-Weiermair M, Hallström S, et al. IP3-mediated STIM1 oligomerization requires intact mitochondrial Ca2+ uptake. J Cell Sci. 2014;127:2944–55. doi: 10.1242/jcs.149807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kato T. Neurobiological basis of bipolar disorder: Mitochondrial dysfunction hypothesis and beyond. Schizophr Res. 2017;187:62–66. doi: 10.1016/j.schres.2016.10.037. [DOI] [PubMed] [Google Scholar]
- 46.Qian X, Su Y, Adam CD, Deutschmann AU, Pather SR, Goldberg EM, et al. Sliced Human Cortical Organoids for Modeling Distinct Cortical Layer Formation. Cell Stem Cell. 2020;26:766. doi: 10.1016/j.stem.2020.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Alural B, Genc S, Haggarty SJ. Diagnostic and therapeutic potential of microRNAs in neuropsychiatric disorders: Past, present, and future. Prog Neuro-Psychopharmacol Biol Psych. 2017;73:87–103. doi: 10.1016/j.pnpbp.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miao C, Chang J. The important roles of microRNAs in depression: new research progress and future prospects. J Mol Med. 2021;99:619–36. doi: 10.1007/s00109-021-02052-8. [DOI] [PubMed] [Google Scholar]
- 49.Jing J, He L, Sun A, Quintana A, Ding Y, Ma G, et al. Proteomic mapping of ER–PM junctions identifies STIMATE as a regulator of Ca2+ influx. Nat Cell Biol. 2015;17:1339–47. doi: 10.1038/ncb3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Aranha MM, Santos DM, Solá S, Steer CJ, Rodrigues CMP. miR-34a Regulates Mouse Neural Stem Cell Differentiation. Plos One. 2011;6:e21396. doi: 10.1371/journal.pone.0021396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mollinari C, Racaniello M, Berry A, Pieri M, de Stefano MC, Cardinale A, et al. miR-34a regulates cell proliferation, morphology and function of newborn neurons resulting in improved behavioural outcomes. Cell Death Dis. 2015;6:e1622–e1622. doi: 10.1038/cddis.2014.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Raivich G, Bohatschek M, Costa CD, Iwata O, Galiano M, Hristova M, et al. The AP-1 Transcription Factor c-Jun Is Required for Efficient Axonal Regeneration. Neuron. 2004;43:57–67. doi: 10.1016/j.neuron.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 53.Koprulu M, Shabbir RMK, Zaman Q, Nalbant G, Malik S, Tolun A. CRADD and USP44 mutations in intellectual disability, mild lissencephaly, brain atrophy, developmental delay, strabismus, behavioural problems and skeletal anomalies. Eur J Med Genet. 2021;64:104181. doi: 10.1016/j.ejmg.2021.104181. [DOI] [PubMed] [Google Scholar]
- 54.Fu Y, Chen L, Chen C, Ge Y, Kang M, Song Z, et al. Crosstalk between alternative polyadenylation and miRNAs in the regulation of protein translational efficiency. Genome Res. 2018;28:1656–63. doi: 10.1101/gr.231506.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mehta SL, Chokkalla AK, Vemuganti R. Noncoding RNA crosstalk in brain health and diseases. Neurochem Int. 2021;149:105139. doi: 10.1016/j.neuint.2021.105139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vasudevan S, Tong Y, Steitz JA. Switching from Repression to Activation: MicroRNAs Can Up-Regulate Translation. Science. 2007;318:1931–4. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- 57.Yu W, Greenberg ML. Inositol depletion, GSK3 inhibition and bipolar disorder. Futur. Neurology. 2016;11:135–48. doi: 10.2217/fnl-2016-0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kato T, Ishiwata M, Mori K, Washizuka S, Tajima O, Akiyama T, et al. Mechanisms of altered Ca2+ signalling in transformed lymphoblastoid cells from patients with bipolar disorder. Int J Neuropsychoph. 2003;6:379–89. doi: 10.1017/S1461145703003717. [DOI] [PubMed] [Google Scholar]
- 59.Wasserman MJ, Corson TW, Sibony D, Cooke RG, Parikh SV, Pennefather PS, et al. Chronic Lithium Treatment Attenuates Intracellular Calcium Mobilization. Neuropsychopharmacol. 2004;29:759–69. doi: 10.1038/sj.npp.1300400. [DOI] [PubMed] [Google Scholar]
- 60.Domenichini F, Terrié E, Arnault P, Harnois T, Magaud C, Bois P, et al. Store‐Operated Calcium Entries Control Neural Stem Cell Self‐Renewal in the Adult Brain Subventricular Zone. Stem Cells. 2018;36:761–74. doi: 10.1002/stem.2786. [DOI] [PubMed] [Google Scholar]
- 61.Moccia F, Zuccolo E, Soda T, Tanzi F, Guerra G, Mapelli L, et al. Stim and Orai proteins in neuronal Ca2+ signaling and excitability. Front Cell Neurosci. 2015;9:153. doi: 10.3389/fncel.2015.00153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brandman O, Liou J, Park WS, Meyer T. STIM2 Is a Feedback Regulator that Stabilizes Basal Cytosolic and Endoplasmic Reticulum Ca2+ Levels. Cell. 2007;131:1327–39. doi: 10.1016/j.cell.2007.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Berna-Erro A, Braun A, Kraft R, Kleinschnitz C, Schuhmann MK, Stegner D, et al. STIM2 Regulates Capacitive Ca2+ Entry in Neurons and Plays a Key Role in Hypoxic Neuronal Cell Death. Sci Signal. 2009;2:ra67. doi: 10.1126/scisignal.2000522. [DOI] [PubMed] [Google Scholar]
- 64.Zhou Y, Mancarella S, Wang Y, Yue C, Ritchie M, Gill DL, et al. The Short N-terminal Domains of STIM1 and STIM2 Control the Activation Kinetics of Orai1 Channels*. J Biol Chem. 2009;284:19164–8. doi: 10.1074/jbc.C109.010900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Harrison PJ, Hall N, Mould A, Al-Juffali N, Tunbridge EM. Cellular calcium in bipolar disorder: systematic review and meta-analysis. Mol Psychiatr. 2021;26:4106–16. doi: 10.1038/s41380-019-0622-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Osete JR, Akkouh IA, de Assis DR, Szabo A, Frei E, Hughes T, et al. Lithium increases mitochondrial respiration in iPSC-derived neural precursor cells from lithium responders. Mol Psychiatr. 2021;26:6789–805. doi: 10.1038/s41380-021-01164-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mertens J, Wang Q-W, Kim Y, Yu DX, Pham S, Yang B, et al. Differential responses to lithium in hyperexcitable neurons from patients with bipolar disorder. Nature. 2015;527:95–99. doi: 10.1038/nature15526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Son G, Han J. Roles of mitochondria in neuronal development. Bmb Rep. 2018;51:549–56. doi: 10.5483/BMBRep.2018.51.11.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mazumdar J, O’Brien WT, Johnson RS, LaManna JC, Chavez JC, Klein PS, et al. O2 regulates stem cells through Wnt/β-catenin signalling. Nat Cell Biol. 2010;12:1007–13. doi: 10.1038/ncb2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Qu Q, Sun G, Li W, Yang S, Ye P, Zhao C, et al. Orphan nuclear receptor TLX activates Wnt/β-catenin signalling to stimulate neural stem cell proliferation and self-renewal. Nat Cell Biol. 2010;12:31–40. doi: 10.1038/ncb2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chavali PL, Saini RKR, Matsumoto Y, Ågren H, Funa K. Nuclear Orphan Receptor TLX Induces Oct-3/4 for the Survival and Maintenance of Adult Hippocampal Progenitors upon Hypoxia*. J Biol Chem. 2011;286:9393–404. doi: 10.1074/jbc.M110.167445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Frisch J, Angenendt A, Hoth M, Roma LP, Lis A. STIM-Orai Channels and Reactive Oxygen Species in the Tumor Microenvironment. Cancers. 2019;11:457. doi: 10.3390/cancers11040457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tobe BTD, Crain AM, Winquist AM, Calabrese B, Makihara H, Zhao W, et al. Probing the lithium-response pathway in hiPSCs implicates the phosphoregulatory set-point for a cytoskeletal modulator in bipolar pathogenesis. Proc Natl Acad Sci. 2017;114:E4462–E4471. doi: 10.1073/pnas.1700111114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bagley JA, Reumann D, Bian S, Lévi-Strauss J, Knoblich JA. Fused cerebral organoids model interactions between brain regions. Nat Methods. 2017;14:743–51. doi: 10.1038/nmeth.4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sukumaran SK, Paul P, Guttal V, Holla B, Vemula A, Bhatt H, et al. Abnormalities in the migration of neural precursor cells in familial bipolar disorder. Dis Model Mech. 2022;15:dmm049526. doi: 10.1242/dmm.049526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Srikanth P, Lagomarsino VN, Muratore CR, Ryu SC, He A, Taylor WM, et al. Shared effects of DISC1 disruption and elevated WNT signaling in human cerebral organoids. Transl Psychiat. 2018;8:77. doi: 10.1038/s41398-018-0122-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chen Y, Wang X. miRDB: an online database for prediction of functional microRNA targets. Nucl Acids Res. 2019;48:D127–D131. doi: 10.1093/nar/gkz757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Quintana A, Rajanikanth V, Farber-Katz S, Gudlur A, Zhang C, Jing J, et al. TMEM110 regulates the maintenance and remodeling of mammalian ER–plasma membrane junctions competent for STIM–ORAI signaling. Proc National Acad Sci. 2015;112:E7083–E7092.. doi: 10.1073/pnas.1521924112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Humphreys DT, Westman BJ, Martin DIK, Preiss T. MicroRNAs control translation initiation by inhibiting eukaryotic initiation factor 4E/cap and poly(A) tail function. Proc National Acad Sci. 2005;102:16961–6. doi: 10.1073/pnas.0506482102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhao C, Sun G, Li S, Lang M-F, Yang S, Li W, et al. MicroRNA let-7b regulates neural stem cell proliferation and differentiation by targeting nuclear receptor TLX signaling. Proc National Acad Sci. 2010;107:1876–81. doi: 10.1073/pnas.0908750107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhao C, Sun G, Ye P, Li S, Shi Y. MicroRNA let-7d regulates the TLX/microRNA-9 cascade to control neural cell fate and neurogenesis. Sci Rep-Uk. 2013;3:1329. doi: 10.1038/srep01329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Song J, Cho KJ, Oh Y, Lee JE. Let7a involves in neural stem cell differentiation relating with TLX level. Biochem Bioph Res Co. 2015;462:396–401. doi: 10.1016/j.bbrc.2015.05.004. [DOI] [PubMed] [Google Scholar]
- 83.Fairchild CLA, Cheema SK, Wong J, Hino K, Simó S, Torre AL. Let-7 regulates cell cycle dynamics in the developing cerebral cortex and retina. Sci Rep-Uk. 2019;9:15336. doi: 10.1038/s41598-019-51703-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Diener C, Hart M, Alansary D, Poth V, Walch-Rückheim B, Menegatti J, et al. Modulation of intracellular calcium signaling by microRNA-34a-5p. Cell Death Dis. 2018;9:1008. doi: 10.1038/s41419-018-1050-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Any additional information required to reanalyze the data reported in this paper is available form the lead contact upon request.