Abstract
Herpes simplex virus type 1 (HSV-1) mutants defective for envelope glycoprotein C (gC) and gB are highly impaired in the ability to attach to cell surface heparan sulfate (HS) moieties of proteoglycans, the initial virus receptor. Here we report studies aimed at defining the HS binding element of HSV-1 (strain KOS) gB and determining whether this structure is functionally independent of gB’s role in extracellular virus penetration or intercellular virus spread. A mutant form of gB deleted for a putative HS binding lysine-rich (pK) sequence (residues 68 to 76) was transiently expressed in Vero cells and shown to be processed normally, leading to exposure on the cell surface. Solubilized gBpK− also had substantially lower affinity for heparin-acrylic beads than did wild-type gB, confirming that the HS binding domain had been inactivated. The gBpK− gene was used to rescue a KOS gB null mutant virus to produce the replication-competent mutant KgBpK−. Compared with wild-type virus, KgBpK− showed reduced binding to mouse L cells (ca. 20%), while a gC null mutant virus in which the gC coding sequence was replaced by the lacZ gene (KCZ) was substantially more impaired (ca. 65%-reduced binding), indicating that the contribution of gC to HS binding was greater than that of gB. The effect of combining both mutations into a single virus (KgBpK−gC−) was additive (ca. 80%-reduced binding to HS) and displayed a binding activity similar to that observed for KOS virus attachment to sog9 cells, a glycosaminoglycan-deficient L-cell line. Cell-adsorbed individual and double HS mutant viruses exhibited a lower rate of virus entry following attachment, suggesting that HS binding plays a role in the process of virus penetration. Moreover, the KgBpK− mutant virus produced small plaques on Vero cells in the presence of neutralizing antibody where plaque formation depended on cell-to-cell virus spread. These studies permitted the following conclusions: (i) the pK sequence is not essential for gB processing or function in virus infection, (ii) the lysine-rich sequence of gB is responsible for HS binding, and (iii) binding to HS is cooperatively linked to the process of efficient virus entry and lateral spread but is not absolutely required for virus infectivity.
Herpes simplex virus type 1 (HSV-1) is a neurotropic human pathogen capable of infection and spread in a variety of cells. Infection is mediated by the viral envelope glycoproteins, which have been assigned specific and often redundant functional roles. Of the 10 virus envelope glycoproteins, only gB, gD, gH, and gL are essential to the process of infection in cell culture, while the other six contribute to virus infectivity and spread in the host (2, 4, 5, 10, 14, 27, 29, 42, 43, 54). An additional glycoprotein, gK, has been shown to be absent from the virus envelope; however, it is required for the production of infectious virions (30, 31).
Infection involves virus attachment to the cell surface membrane followed by virus penetration and entry of the nucleocapsid into the cytoplasm (53, 57). Current evidence indicates that virus attachment is a two-step process (48) involving different glycoproteins and several receptors. Glycoprotein B (gB) and gC have been shown to be involved in the initial attachment phase through the interaction of positively charged glycoprotein structures with negatively charged heparan sulfate (HS) moieties located on cell surface proteoglycans (44, 56). This HS-dependent attachment may facilitate a second attachment in which gD binds to a cellular receptor, one of them recently reported to be a member of the tumor necrosis factor-nerve growth factor receptor family (50). Following attachment, the virus penetrates the cell by fusion of the virus envelope with the cell plasma membrane (57). Genetic studies have shown that gB, gD, and gH are required to carry out the fusion-penetration process (4, 10, 32, 42) and that gL is essential for proper processing and insertion of gH into the virus envelope (29). These studies have demonstrated that virus penetration is a highly complex process involving the cooperative activities of multiple viral glycoproteins.
Different lines of evidence have identified HS as an initial receptor for HSV infection. First, HS proteoglycans are commonly found on the surface of most vertebrate cell types (15), including those susceptible to HSV infection (16, 21, 44, 58, 64). Second, removal of HS from the cell surface, either by enzymatic treatment or by selection of cell lines defective in the pathway of HS (3, 17, 41, 56), renders the cells at least partially resistant to HSV infection by reducing virus attachment to the cell surface. Third, heparin, a molecule chemically similar to HS (35), has been shown to inhibit viral infection by masking the HS binding domain on the virus envelope (21, 22, 55), and immobilized heparin columns bind to the principal mediators of virus attachment, gB and gC, either derived from HSV-1-infected cells or produced in a baculovirus expression system (24, 59). Fourth, construction of deletion mutants for the glycoproteins involved in HS binding, namely gC and gB, impairs virus binding to the cell surface (23).
Glycoprotein C interactions with HS have been extensively studied (6, 12, 16, 23, 24, 48), and HS binding domains have been identified (62). HSV-1 gC-negative mutants (23, 24, 28, 59) are impaired in binding and slightly impaired in penetration but remain highly infectious, presumably because the HS binding function of gB is sufficient to mediate virus attachment to HS (24). A deletion mutant lacking both gC and gB coding sequences was demonstrated to be substantially impaired in binding compared with a gC null mutant virus, suggesting that gB also contributes to HS binding (23). The residual attachment ability of this mutant may indicate either that other viral glycoproteins possess limited HS binding activity or that additional receptors are recognized in the absence of HS binding. Mutants deleted for gB cannot be used to determine whether gB binding to HS is essential to the process of penetration and cell-to-cell spread, since gB is required for both functions.
In this report, experiments were performed to (i) identify the HS binding domain of gB and quantify its contribution to HS binding, (ii) determine whether the HS binding domain of gB is required for gB’s essential role in virus penetration and lateral spread, and (iii) compare the contributions of both gB- and gC-mediated HS binding to the efficiency of virus attachment, rate of penetration, and lateral spread in plaque formation. To achieve these goals, a lysine-rich (pK) sequence (amino acids 68 to 76) was deleted from the N terminus of gB (gBpK−) and analyzed for its ability to bind heparin-acrylic beads. The reduction in gBpK− binding to heparin confirmed that an HS binding domain had been removed. A mutant virus carrying this altered gB molecule showed reduced adsorption to the cell membrane and reduced capacity to spread from cell to cell but remained infectious, demonstrating that gB binding to HS is not essential for gB function. A double mutant virus (KgBpK−gC−) deleted for gC in addition to the pK domain of gB, was constructed and shown to be highly impaired in binding and to have a reduced rate of penetration. Taken together, these data demonstrate that HS binding is required for efficient penetration by both extra- and intercellular virus.
MATERIALS AND METHODS
Cells and viruses.
Vero cells were used to propagate the KOS strain of HSV-1, from which all recombinant viruses were derived. The Vero cell line A1 (60) (kindly provided by Fred Homa, Kalamazoo, Mich.), stably transfected with the HSV-1 genes encoding gB and ICP 18.5, was used to propagate a double-mutant virus, KΔ4BX (9), deleted for both genes. The Vero cell line C1 (60) (kindly provided by Fred Homa), stably transfected with the HSV-1 gene encoding ICP 18.5, was used to passage the double-mutant virus (KgBpK−gC−) to generate a virus particle deleted for gB and gC only. Mouse L cells and their glycosaminoglycan (GAG)-negative derivatives (sog9 [3]) were kindly provided by Frank Tufaro, Vancouver, British Columbia, Canada. All cell lines were maintained at 37°C in Dulbecco’s modified minimum essential medium (DMEM) (Gibco BRL, Grand Island, N.Y.) supplemented with 10% fetal bovine serum.
Construction of gB mutant and gC-deleted plasmids.
The HSV-1 gB coding sequence (UL27) was excised by enzymatically digesting the BamG fragment of the viral genome with KpnI and SalI endonucleases and subcloned with the KpnI-XhoI restriction sites of the pTZ18U vector (pTZ18UgB1). Site-specific mutagenesis was performed to delete 27 nucleotides encoding the putative HS binding domain of gB (amino acids 68 to 76 [KPKKNKKPK]), and a BamHI recognition site was inserted in frame at the site of the mutation. The resulting gBpK− mutation was subsequently inserted into the pKBXX vector (4, 25) to create pgBpK−, and both plasmids (pKBXX and pgBpK−) were further modified by inserting the human cytomegalovirus immediate early promoter (HCMV-IEp) at their 5′ ends to create HCMV-BXX and HCMV-gBpK−.
The HSV-1 gC coding sequence (UL44) contained in the pgC1 plasmid (27) was deleted by digestion with the restriction endonucleases XhoI and EcoRV, and the 1,622-bp fragment was replaced with the SalI/BamHI fragment encoding the HCMV-lacZ cassette of the pIEP-lacZ plasmid (12) in order to create pΔgC:lacZ.
Construction and isolation of recombinant viruses.
Mutant HSV-1 viruses were constructed by standard methods for transfer by using LipofectAmine (Gibco BRL) for cotransfection, and mutant or recombinant virus plaques were thrice plaque purified by limiting dilution prior to characterization. To construct the KgBpK− recombinant virus, plasmid pgBpK− DNA was cotransfected with viral DNA from KΔ4BX on the complementing A1 cell line. The KgBpK− recombinant virus was selected for growth on Vero cells and screened by Southern hybridization with a gB probe that hybridized to the gB coding sequence (see Fig. 3A). The pΔgC:lacZ plasmid was used to cotransfect Vero cells with viral DNA from KOS. A virus recombinant (KCZ) deleted for 1,622 bp of the gC coding sequence was selected by complement-dependent neutralization with a pool of gC-specific monoclonal antibodies (MAbs) (47). The viral plaques formed by the neutralization escape mutants were further screened for the “blue-plaque” phenotype in presence of X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) substrate. The same pΔgC:lacZ plasmid was used to cotransfect A1 cells with KΔ4BX viral DNA in order to create the KΔ4BXCZ virus, which was deleted for genes encoding ICP 18.5, gB, and gC. The recombinant virus was selected on A1-complementing cells, as described above for KCZ on Vero cells. Southern blot hybridization with a gC probe was used to confirm the absence of the gC coding sequence in the KCZ (see Fig. 3B) and KΔ4BXCZ viruses. The double mutant KgBpKgB−gC− was produced by cotransfection of pgBpK− plasmid DNA with viral DNA from KΔ4BXCZ on A1 cells. Viruses from the cotransfection were plated on Vero cells, and the recombinant virus deleted for both the pK region of gB and the gC coding sequence (KgBpK−gC−) was selected for the presence of the gBpK− sequence, as described above for the KgBpK− recombinant virus, and for the absence of the gC coding sequence, as described above for KCZ. KgBpK−gC− recombinant virus was rescued for the full-length gB and gC coding sequences in order to produce KgBpKRgC− and KgBpK−gCR, respectively. Viral DNA from KgBpK−gC− was cotransfected with wild-type KOS gB DNA (pKBXX [4]) or wild-type KOS gC DNA (pgC1 [27]). Vero cell plaques from both cotransfections were isolated and tested for the rescue of each respective gene by Southern blot hybridization, as shown in Fig. 3. The rescue of gC was also confirmed by complement-dependent neutralization of the recombinant virus with gC-specific MAbs and by a “clear-plaque” phenotype following X-Gal staining.
FIG. 3.
Southern blot characterization of the recombinant viruses. Viral DNAs from KOS (lane 1), KgBpK− (lane 2), KCZ (lane 3), and KgBpK−gC− (lane 4), as well as viral DNAs from KgBpK−gCR and KgBpKRgC−, were digested with the restriction endonuclease BamHI (A) or NcoI (B and C) and subjected to Southern blot analysis. (A) A 32P-labeled gB probe hybridized to a 7,774-bp fragment of wild-type gB sequence encoded by KOS (lane 1) and KCZ (lane 3), and KgBpKRgC− (lane 6) hybridized with a smaller fragment of 3,009 bp in the recombinant viruses KgBpK− (lane 2), KgBpK−gC− (lane 4), and KgBpK−gCR (lane 5) due to the introduction of a BamHI restriction endonuclease site within the gB sequence at the site of the deletion of amino acids 68 to 76. (B) A 32P-labeled gC probe (642-bp NcoI fragment of pgC1) encoding the deleted gC sequence hybridized to a 642-bp fragment containing wild-type gC in KOS (lane 7), KgBpK− (lane 8), and KgBpK−gCR (lane 11). (C) A different gC probe (828-bp NcoI fragment of pgC1 undeleted in all viruses) hybridized with an 11.2-kbp fragment in KCZ (lane 15), KgBpK−gC− (lane 16), and KgBpKRgC− (lane 18), in which the gC coding sequence was deleted and replaced with the human cytomegalovirus immediate early promoter driving the lacZ gene, and it hybridized with an 828-bp fragment in KOS (lane 13), KgBpK− (lane 14), and KgBpK−gCR (lane 17), in which the wild-type gC coding sequence was present.
Immunofluorescence.
Thirty hours posttransfection with HCMV-BXX or HCMV-gBpK−, the Vero cell monolayers were incubated for 1 h at 4°C with a pool of gB MAbs (46), rinsed with cold Tris-buffered saline (TBS), pH 7.4, and incubated for an additional hour with a cy3-conjugated anti-mouse antibody (Jackson Immunoresearch Laboratories, West Grove, Pa.); the monolayer was then fixed with 2% paraformaldehyde. Immunofluorescence-positive cells were photographed with a Nikon TMS microscope-camera (model 211910).
Binding of gB and gBpK− to heparin-acrylic beads.
Plasmids pKBXX (4) and pgBpK− were used individually to transfect Vero cells. Twenty-four hours posttransfection, the cell monolayers were infected at a multiplicity of infection (MOI) of 3 with KΔ4BX virus followed by overlay with DMEM containing [35S]methionine/cysteine. Ten hours postinfection (p.i.), the cell monolayers were scraped, harvested, and resuspended in 200 μl of lysis buffer (0.5 M Tris-HCl, 150 mM NaCl, 1% Triton X-100, 10 mM phenylmethylsulfonyl fluoride, and 1 mM TLCK [Nα-p-tosyl-l-lysine chloromethyl ketone]). The Triton-soluble extracts were incubated for 2 h at 4°C in the presence of heparin-acrylic beads (Sigma, St. Louis, Mo.). The beads were rinsed three times with lysis buffer or lysis buffer supplemented with 10 mg of heparin (Sigma) per ml. The Triton-soluble extract and lysis buffer washes or lysis buffer with heparin washes were immunoprecipitated with a pool of gB-specific MAbs (46) and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Quantification of precipitated radiolabeled proteins was determined with the 1-D Scan and Report program (Biomed Instruments, Fullerton, Calif.).
Purification of radiolabeled virus.
Virions used for binding and immunoprecipitation assays were labeled and purified as follows. Confluent Vero cell monolayers in T150s flasks (Falcon; Becton Dickinson, Franklin Lakes, N.J.) were infected with viruses at an MOI of 10. Four hours p.i., 16 ml of DMEM without methionine and cysteine (Gibco BRL) and supplemented with 1% fetal calf serum was added to the infected cell monolayers. [35S]methionine/cysteine (ExpreSS; Dupont, NEN, Boston, Mass.) having a specific activity of 50 μCi/ml was added after 4 h. Twenty-four hours p.i., medium containing radiolabeled virus was harvested and virions were purified by centrifugation (SWTi-40 Beckman rotor) through sucrose gradients (30 to 65% sucrose). The fractions containing the radiolabeled virus were pooled, diluted in sterile TBS, and centrifuged at 20,000 × g for 1 h at 4°C in a SWTi-40 rotor. The virus pellet was resuspended in DMEM containing 10% fetal bovine serum, and radioactivity was determined with a beta counter (Beckman, Fullerton, Calif.).
Immunoprecipitation analysis of surface glycoproteins.
Aliquots of radiolabeled virus were immunoprecipitated with a pool of gB-specific (46), gC-specific (47), or gD-specific (26) MAbs. Each virus aliquot was diluted in 200 μl of lysis buffer containing 2 μl of antibody and incubated at 4°C for a minimum of 4 h. The quantity of antibody used in this assay was in 10-fold excess of the amount needed for complete immunoprecipitation of gB from wild-type virus-infected cells. The immune complexes were incubated with protein A-Sepharose (Sigma) for 1 h, centrifuged at 500 × g, and washed five times with 600 μl of lysis buffer. The protein A-Sepharose complexes were resuspended in Laemmli loading buffer (36), boiled for 2 min, and subjected to SDS-PAGE. After electrophoresis, the gels were fixed, treated with En3Hance solution (Dupont, NEN), vacuum dried, and exposed to X-Omat-XAR film (Kodak, Rochester, N.Y.). Quantification of radiolabeled proteins precipitated was determined with the 1-D Scan and Report program (Biomed Instruments).
Binding of radiolabeled virus to cells.
Monolayers of confluent Vero, murine L, or sog9 cells in 24-well plates were incubated at 4°C with radiolabeled purified virions. The viruses were allowed to bind to the cell surface for 10 to 320 min, after which the unbound viruses were removed and the cell monolayer was washed three times with cold TBS. Cell monolayers with bound virions were resuspended in 100 μl of lysis buffer and transferred to vials for liquid scintillation counting. Quantification of cell-associated labeled virions was determined with a beta counter.
Elution of bound virus with heparin.
Confluent monolayers of Vero cells in 6-well plates were infected with 200 PFU of the KOS, KCZ, KgBpK−, KgBpK−gC−, KgBpKRgC−, or KgBpK−gCR virus. Following a 4-h incubation at 4°C, the cells were washed three times with complete medium supplemented with 500 μg of heparin per ml (or without heparin in control wells). The cell monolayers were then overlaid with medium containing 0.5% methyl-cellulose at 37°C to allow virus plaques to form. Cells were then fixed and stained with crystal violet for plaque quantification.
Virus titration in the presence of heparin.
Confluent monolayers of Vero cells were infected for 2 h at different dilutions with all mutant viruses or with vesicular stomatitis virus (VSV) (kindly provided by Patricia A. Dowling, University of Pittsburgh) in the absence or presence of 50 or 500 and 10,000 μg of heparin per ml. Virus inoculates were removed, and the cell monolayer was rinsed twice with medium or heparin-supplemented medium. Two additional washes were performed with medium only in all samples to remove the presence of heparin that could reduce the efficiency of plaque formation. The cell monolayers were then overlaid with medium containing 0.5% methyl-cellulose at 37°C to allow virus plaques to form. Cells were fixed and stained with crystal violet to quantify plaque numbers.
Virus penetration assay.
The rate of virus penetration was assessed by determining the rate at which adsorbed virus became resistant to inactivation by a low-pH glycine buffer (0.1 M glycine [pH 3.0] [8]). Confluent Vero, murine L, and sog9 cells in 6-well plates were incubated at 4°C for 2 h with 300 PFU of wild-type KOS, HS binding mutants, or rescued viruses. Following virus adsorption the cells were rinsed three times, overlaid with complete medium, and shifted to 37°C to allow virus penetration. At selected times after temperature shift, wells were treated with 2 ml of glycine buffer for 1 min while control wells were treated with 2 ml of TBS for 1 min. The monolayers were then washed three times with complete medium, overlaid with DMEM containing 0.5% methyl-cellulose, and incubated at 37°C, allowing virus plaques to form. Cells were then fixed and stained with crystal violet to visualize and count viral plaques.
In vitro cell-to-cell spread of virus.
Vero and sog9 cell monolayers were infected with KOS, KCZ, KgBpK−, and KgBpK−gC− viruses for 2 h, the medium was aspirated, and the cells were washed twice with TBS and overlaid with fresh medium supplemented with 0.2% human gamma globulin (HGG) (Bayer Corporation, Elkhart, Ind.) containing anti-HSV neutralizing antibody. Twenty-four, 36, and 48 h p.i., cell monolayers were fixed with methanol, washed three times with TBS, and processed for immunofluorescence as described above by using rabbit anti-HSV-1 antibodies combined with secondary cy3-conjugated antibody (Jackson Immunoresearch Laboratories). Plaque sizes were determined by a Zeiss Axiophot microscope linked to a Xillix digital camera with personal computer-based image analysis (MCID) from Imaging Resources Incorporated (Brock, Ontario, Canada).
RESULTS
Construction of a gB mutant molecule impaired in heparin binding.
The role of gB in HS proteoglycan binding may be distinguished from its essential role in penetration if an HS binding domain can be identified and removed from the molecule without inactivating its ability to function in membrane fusion. If so, then a double mutant virus also lacking gC could be constructed since gC is nonessential for virus infectivity (27). Polylysine inhibits HSV infection (38) presumably by interacting with HS, a property shared with other cationic substances such as polyarginine and neomycin (37, 38). Accordingly, a lysine-rich (pK) sequence (residues 68 to 76) was deleted from the N-terminal region of gB (gBpK−) for studies of its ability to be processed and bind heparin, an HS-like molecule.
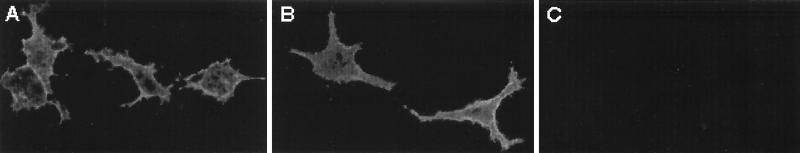
Deletion or substitution of peptide sequences from HSV-1 glycoproteins can result in altered glycoprotein processing and prevent analysis of the effect of the mutation on the glycoprotein’s functional role in virus infection. For example, modified glycoproteins can be retained within the endoplasmic reticulum and fail to be transported to the cell surface membrane and incorporated into virus particles (9). In order to evaluate the posttranslational processing of the mutated gB molecule, Vero cells were transfected with expression vectors encoding wild-type (HCMV-BXX) (Fig. 1A) or mutant (HCMV-gBpK−) (Fig. 1B) gB; mock-transfected Vero cells were used as a negative control (Fig. 1C). Twenty-four hours posttransfection, the cells were analyzed by immunofluorescence for the presence and localization of gB molecules. Untreated cell monolayers were incubated at 4°C with a pool of gB-specific MAbs (46) to determine whether the mutant gB molecule was processed and transported to the cell surface membrane. As demonstrated in Fig. 1, both wild-type (panel A) and mutant (panel B) gB (gBpK−) were recognized by a pool of specific MAbs, and both proteins were incorporated into the cell surface membrane. No fluorescence was detected on mock-transfected cells (panel C). These data demonstrate that mutant gB (gBpK−) was transported to the cell membrane of transfected cells, showing that deletion of the pK domain of gB permits endoplasmic reticulum and Golgi processing of the mutant molecule. Moreover, the molecular size of the fully processed mutant molecule was similar to the wild-type gB molecule, revealing no obvious differences in the addition of complex carbohydrates (see Fig. 4).
FIG. 1.
Cell surface detection of gBpK− mutant glycoprotein. Vero cells were transfected with expression plasmids encoding the wild-type gB protein (HCMV-BXX) (A) and the pK-deleted gB protein (HCMV-gBpK−) (B) or were mock transfected (C). After 30 h, unfixed transfected monolayers were incubated with a pool of monoclonal anti-gB antibody for 1 h at 4°C, washed with cold TBS, and incubated for 1 h at 4°C with a cy3 anti-mouse antibody. Monolayers were visualized with a Nikon microscope (model 211910) and photographed.
FIG. 4.
Comparative incorporation of glycoproteins B, C, and D into mutant virus envelopes. [35S]methionine/cysteine-labeled sucrose-purified KOS (A), KgBpK− (B), KCZ (C), KgBpK−gC− (D), KgBpK−gCR (E), and KgBpKRgC− (F) viruses were solubilized with detergent and immunoprecipitated with a pool of MAbs directed against gB (lanes 1), gC (lanes 2), or gD (lanes 3). The immune complexes were captured on protein A-Sepharose, resuspended in Laemmli buffer, and separated by SDS-PAGE. Asterisks indicate the positions of the glycoproteins designated on the left, when present.
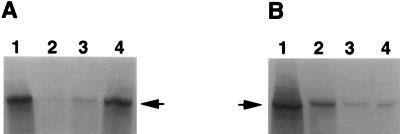
Since the mutant form of gB was apparently normally processed it could be compared with the wild-type molecule for its reactivity with heparin, an HS-like molecule, to determine whether the pK deletion had removed the HS binding activity attributed to the gB molecule. Expression plasmids encoding either wild-type gB (pKBXX) or gBpK− (pgBpK−) were used to transfect Vero cells in order to produce wild-type and mutant proteins, respectively, after induction of gB expression by infection with a gB–ICP 18.5-deleted virus (KΔ4BX). The transfected/infected cells were radiolabeled with [35S]methionine/cysteine, and the cell monolayers were harvested and solubilized prior to analysis of binding to heparin-acrylic beads. As shown in Fig. 2, approximately the same quantity of gB or gBpK− molecules was incubated with the heparin beads (lanes 1). However, as demonstrated by the quantity of unbound glycoprotein (lanes 2), gBpK− displayed a lower affinity for heparin-acrylic beads than did wild-type gB; more than 40% of the gBpK− molecules were recovered in the unbound fraction compared to less than 3% of the gB wild-type molecules. Figure 2, lanes 3, demonstrates that similar amounts of gB molecules were bound nonspecifically to the heparin beads and could be eluted with lysis buffer. In order to show that wild-type gB molecules were bound specifically to heparin-coated beads, the bound glycoprotein was eluted by washing the beads with lysis buffer supplemented with heparin. The results revealed that bound wild-type gB was efficiently eluted by heparin washes (Fig. 2A, lane 4), while a small amount of gBpK− was released by heparin wash due to the small quantity of gBpK− nonspecifically associated with the heparin beads (Fig. 2B, lane 4). Together, these studies confirmed that the gBpK− mutant molecule was fully glycosylated, incorporated into the cell surface membrane, and that a domain responsible for heparin binding had been inactivated by deletion of the pK region of gB.
FIG. 2.
Heparin binding capacity of gBpK− mutant glycoprotein. Vero cells were transfected with plasmids encoding wild-type gB (A) and gBpK− (B). Twenty-four hours posttransfection, the cell monolayers were infected with KΔ4BX virus in the presence of [35S]methionine/cysteine, as described in Materials and Methods. Ten hours p.i., the monolayers were harvested and solubilized with 0.1% Triton X-100-containing buffer and equivalent amounts of detergent-extracted protein (lanes 1) and were incubated for 2 h at 4°C with heparin-acrylic beads. The unbound proteins (lanes 2) and proteins eluted from the heparin-acrylic beads with detergent buffer (lanes 3) or detergent buffer supplemented with 10 mg of heparin per ml (lanes 4) were immunoprecipitated with a pool of gB-specific MAbs. Each sample was analyzed by SDS-PAGE and autoradiography. The arrows indicate the positions of gB wild-type or gBpK− mutant glycoproteins.
Construction of HS attachment-defective mutant viruses.
An HSV-1 viral mutant carrying the gBpK− coding sequence (KgBpK−) was constructed to evaluate the effect of the pK deletion on virus infection. The gBpK− gene was inserted together with the UL28 gene (ICP 18.5) by marker rescue of the gB and UL28 double-deletion virus (KΔ4BX) (9). The recombinant viruses (KgBpK−) were selected by growth on noncomplementing Vero cells. The DNA sequence encoding the major HSV-1 HS binding protein, gC, was deleted from wild-type KOS virus and replaced with the human cytomegalovirus immediate early gene promoter (HCMV-IEp) lacZ gene expression cassette in order to create KCZ. The virus recombinants were selected by complement-dependent neutralization escape with gC-reactive MAbs and identified by their blue-plaque phenotype following X-Gal staining. A double HS attachment-defective virus encoding gBpK− and deleted for the gC coding sequence (KgBpK−gC−) was created to determine the role of virus binding to HS in virus infection. The gBpK− gene, together with the UL28 gene (ICP 18.5), was used to marker rescue KΔ4BXCZ, a virus deleted for gC, gB, and UL28. The recombinant KΔ4BXCZ mutant virus was created by replacing the gC coding sequence of KΔ4BX with the HCMV-IEp-lacZ cassette, as described above for the generation of KCZ. Both gB and gC wild-type genes were used subsequently to rescue either the gB or gC mutation of the KgBpK−gC− mutant virus to produce the KgBpKRgC− and KgBpK−gCR viruses, respectively.
The mutant and recombinant virus genotypes were analyzed by Southern blotting to confirm their genotypes. Viral DNAs were extracted from purified virions, digested with BamHI (Fig. 3A) or NcoI (Fig. 3B and C) endonuclease, and Southern blotted with a 32P-labeled gB (Fig. 3A) or gC (Fig. 3B and C) probe. As shown in Fig. 3A, the 32P-labeled gB probe hybridized to the BamG fragment (7,774 bp) of HSV-1 viral DNA containing the wild-type gB coding sequence encoded by the KOS, KCZ, and KgBpKRgC− viruses (lanes 1, 3, and 6, respectively). The same probe hybridized to a 3,009-bp fragment in the recombinant KgBpK−, KgBpK−gC−, and KgBpK−gCR viruses (lanes 2, 4, and 5, respectively). This confirmed the presence of the mutant gB gene because a BamHI recognition sequence was introduced at the site of the pK mutation in the mutant viruses, resulting in the production of two subfragments (3,009 and 4,738 bp) after digestion. Replacement of the gC coding sequence by the HCMV-IEp-lacZ cassette was confirmed by Southern blot hybridization of NcoI-digested viral DNA using a 32P-labeled gC probe (642-bp NcoI fragment of pgC1) that hybridized to a gC sequence deleted in the gC-negative recombinant viruses. As shown in Fig. 3B, the 32P- labeled gC probe hybridized to a 642-bp fragment in KOS, KgBpK−, and KgBpK−gCR digested viral DNA (lanes 7, 8, and 11, respectively) containing the wild-type gC coding sequence. The same probe, however, failed to hybridize with KCZ, KgBpK−gC−, and KgBpKRgC− viral DNA (lanes 9, 10, and 12, respectively), demonstrating that the gC coding sequence was deleted. The presence of the HCMV-IEp-lacZ cassette was identified with an 828-bp NcoI 32P-labeled gC probe that hybridized to the wild-type gC gene (KOS, KgBpK−, and KgBpK−gCR viruses [Fig. 3C, lanes 13, 14, and 17, respectively]) but with an 11.2-kb fragment derived from viral DNA containing the HCMV-IEp-lacZ cassette (KCZ, KgBpK−gC−, and KgBpKRgC− viruses [Fig. 3C, lanes 15, 16, and 18, respectively]). This hybridization pattern confirmed the absence of the NcoI endonuclease restriction site within the HCMV-IEp-lacZ cassette and its presence within the wild-type gC sequence. Together, these data confirm the isolation of single- and double-mutant viruses deleted for one or both HS binding elements (pK of gB and gC), as well as both mutant virus rescuants.
Evaluation of the envelope glycoprotein composition of the mutant viruses.
Since the quantity of viral glycoproteins in the virus envelope could influence the ability of individual glycoproteins to carry out virus attachment, penetration, and lateral spread, the mutant virus envelopes were analyzed for glycoprotein content to determine whether the amounts of the individual HSV-1 glycoproteins were similar to that of wild-type KOS virus. The level of incorporation of these glycoproteins into mutant virions was determined by specific immunoprecipitation of solubilized radiolabeled envelopes from wild-type virus (KOS), the mutants KCZ, KgBpK−, and KgBpK−gC−, and the rescued KgBpK−gCR and KgBpKRgC− viruses by using pools of monoclonal antibodies specific for gB, gC, and gD. The protein A-captured immune complexes were then analyzed by SDS-PAGE and autoradiography. As shown in Fig. 4, wild-type gB was immunoprecipitated from the KOS (panel A, lane 1), KCZ (panel C, lane 1), and KgBpKRgC− (panel F, lane 1) viruses and the mutant gBpK− was immunoprecipitated from the KgBpK− (panel B, lane 1), KgBpK−gC− (panel D, lane 1), and KgBpK−gCR (panel E, lane 1) viruses, demonstrating that the mutant gB molecule was incorporated into the envelopes of all viruses under study. As expected, gC was present in KOS (Fig. 4A, lane 2), KgBpK− (Fig. 4B, lane 2), and KgBpK−gCR (Fig. 4E, lane 2) and absent in KCZ (Fig. 4C, lane 2), KgBpK−gC− (Fig. 4D, lane 2), and KgBpKRgC− (Fig. 4F, lane 2) virus envelopes. Glycoprotein D (Fig. 4, lanes 3) was detected in all virion envelope preparations, and the ratio of the quantity (analyzed by densitometry) of immunoprecipitated gB or gBpK− to gD demonstrated that the pK mutation in gB did not affect the level of incorporation of the mutant gBpK− molecules into the virion envelopes and that the absence of gC did not increase the relative amount of gB incorporation into virus envelopes compared with gD.
Attachment properties of HS binding-deficient mutant viruses.
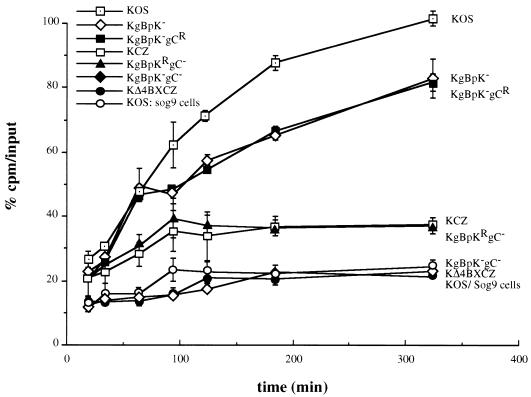
The attachment efficiency of the HS binding mutant viruses was assessed on mouse L cells and compared to the binding capacity of KOS on sog9 cells, a GAG-deficient L-cell-line derivative (3). As shown in Fig. 5, the binding capacity of the single-mutant KgBpK− and KgBpK−gCR viruses after 5.5 h of incubation with cell monolayers was reduced by approximately 20% compared to the binding capacity of the wild-type KOS virus. However, the binding capacity of the KgBpK− and KgBpK−gCR viruses was higher than that of both gC-deleted viruses (KCZ and KgBpKRgC−), which displayed a 65% reduction in binding compared to wild-type KOS virus. This result demonstrates that the pK region of gB contributes less to virus binding to HS than does gC. Analysis of the binding capacity of the double-mutant virus KgBpK−gC− demonstrates that the ability of this mutant to attach to cells was reduced by 80% and was similar to the binding activity displayed by wild-type KOS virus on GAG-deficient sog9 cells. These results demonstrate that the double KgBpK−gC− recombinant virus was greatly impaired in its ability to attach to the cell surface, which could be attributed to the removal of the HS binding domains of gB and the deletion of gC. Moreover, the removal of the pK domain from gB and deletion of gC abrogated all detectable HS binding activity, since the double-mutant virus KgBpK−gC− bound to mouse L cells to an extent similar to wild-type KOS virus binding to the GAG-deficient sog9 cells. The binding capacity of a double mutant deleted for gB and gC (KΔ4BXCZ) showed a binding activity on L cells comparable to that of the KgBpK−gC− virus, demonstrating that removal of the pK domain of gB eliminated all of its detectable HS binding activity and confirmed studies described above (Fig. 2) which determined that the pK sequence of gB was solely responsible for the HS binding function of gB. In similar studies using Vero cells, the ratios of bound virus to input were higher for all viruses on Vero than on L cells; however, a proportional reduction in binding for all recombinant viruses compared to wild-type KOS virus was observed, confirming studies using L cells (data not shown).
FIG. 5.
Cell surface binding capacity of viruses altered in HS proteoglycan binding domains. Vero cells infected with KOS, KCZ, KgBpK−, KgBpK−gC−, KgBpKRgC−, or KgBpK−gCR viruses and C1 cells infected with KΔ4BXCZ virus were labeled with [35S]methionine/cysteine, and the virions from cell supernatants were subsequently purified on sucrose gradients. The binding capacity of each purified virion was determined on mouse L cells and compared to the binding of wild-type KOS virus on sog9 cells (GAG-deficient L-cell derivatives). Aliquots from the different virus preparations were incubated on the two cell lines at 4°C for up to 320 min and washed with cold TBS, and the cells were scraped, harvested, and counted for virus-associated radioactivity. The percentage of bound virus was determined as radioactive counts representing the bound fraction divided by the total counts per minute (input). The binding capacity of KOS virus on mouse L cells after 320 min was designated 100% binding. Error bars indicate results determined for triplicate wells.
Although removal of the pK domain of gB and deletion of gC greatly impaired the adsorption of the recombinant virus KgBpK−gC− to L cells (Fig. 5), there remained a 20% residual binding activity. This residual binding capacity was also observed for wild-type KOS virus on sog9 cells (Fig. 5), confirming that it was HS independent. In order to determine whether this residual binding was HS independent on cell types other than L cells, the HS binding-deficient recombinant viruses were titrated on Vero cells in the presence or absence of heparin (50 μg/ml), an HS-like molecule. The data presented in Table 1 demonstrate that the wild-type KOS and mutant KgBpK− and KCZ viruses infected Vero cells more efficiently in the absence than in the presence of heparin, confirming that these viruses contained HS binding activity. However, the ratio of the titers of the double-mutant KgBpK−gC− virus was similar on Vero cells in the presence or absence of heparin. The small observed difference (relative ratio of 2.1) could be accounted for by the nonspecific inhibitory effect of heparin on virus binding, since this effect also was observed when an HS binding independent virus, VSV, was titrated in the presence of heparin (data not shown). Similar results were also obtained at different concentrations (500 μg/ml and 10 mg/ml) of heparin, and the rescued viruses KgBpKRgC− and KgBpK−gCR showed results similar to the KCZ and KgBpK− mutant viruses, respectively (data not shown). In experiments in which the viruses were bound at 4°C in the presence or absence of heparin rather than titrated in heparin-supplemented media (Table 1), similar results were observed (data not shown), demonstrating that the inhibitory effect of heparin was at the adsorption level. Moreover, similar titers for the double-mutant KgBpK−gC− virus were obtained on L and sog9 cells (relative ratio of 1.1), demonstrating that this virus was deficient for HS binding (data not shown).
TABLE 1.
Effects of heparin on infection of Vero cells by HSV
| Virus | Titer (PFU/ml)
|
Ratio | |
|---|---|---|---|
| Without heparin | With heparin | ||
| KOS | 4.8 × 109 | 1.1 × 108 | 44.6 |
| KgBpK− | 1.9 × 109 | 1.0 × 108 | 19.0 |
| KCZ | 2.2 × 108 | 2.0 × 107 | 11.0 |
| KgBpK−gC− | 8.8 × 108 | 4.2 × 108 | 2.1 |
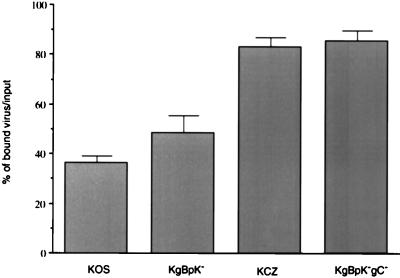
To confirm that the impaired binding of KgBpK−gC− was due to the loss in virus-specific HS recognition sites, viruses bound to the cell surface were washed with heparin and the quantity of released virus particles was taken as a measure of virus specifically bound to HS. Vero cells were incubated for 4 h at 4°C with the KOS, KCZ, KgBpK−, and KgBpK−gC− viruses, and washes were performed in the presence or absence of heparin to elute the fraction of virus bound to HS. As shown in Fig. 6, the binding of the KgBpK− virus was less sensitive (12%) to heparin washes than that of KOS, consistent with other evidence showing that the HS binding region of gB had been deleted. Moreover, the binding of the double-mutant KgBpK−gC− virus was highly resistant to heparin washes but only to an extent slightly higher (2.4%) than the resistance demonstrated by KCZ, again showing that the majority of HS binding activity was attributable to gC. The residual bound virus that was resistant to heparin washes depended on binding to a non-HS receptor, most likely due to the binding of gD to its specific receptor (50).
FIG. 6.
Contribution of a viral glycoprotein(s) other than gB and gC to HS binding. Confluent monolayers of Vero cells in 12-well plates were inoculated with 100 PFU of each test virus per well for an adsorption period of 2 h at 4°C. The cells were then rinsed three times with complete medium in the absence or presence of 500 μg of heparin per ml, overlaid with medium containing methyl-cellulose, and shifted to 37°C to allow viral plaque formation. The cells were then fixed and stained with crystal violet, and plaques were counted. At each time point, the average number of plaques produced on heparin-washed monolayers was expressed as a percentage of the average number of plaques produced on complete medium-washed monolayers. Error bars indicate results determined for triplicate wells.
Virus mutants altered in HS binding domains also showed a reduced capacity for viral entry and infectivity.
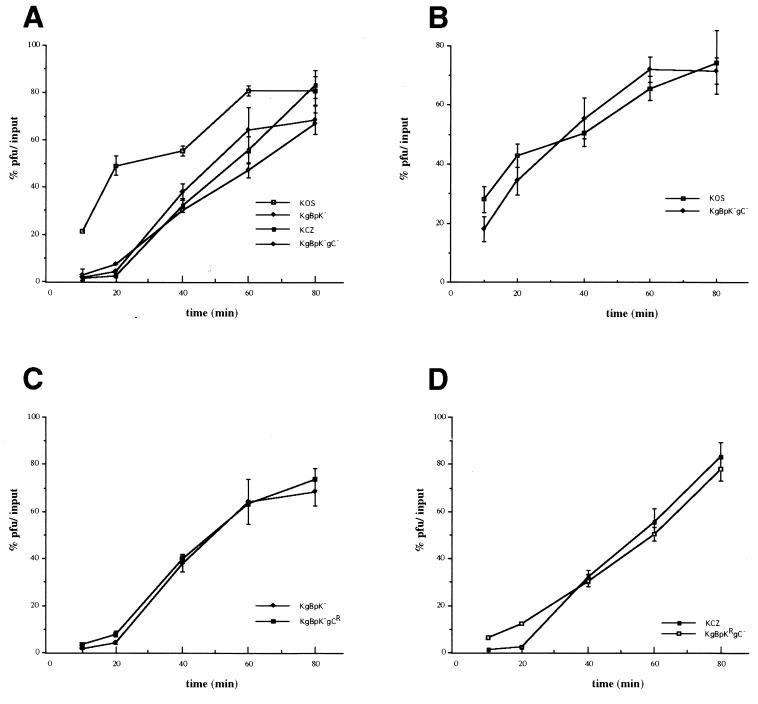
The entry of HSV-1 capsids into the host cell is believed to involve a two-step process requiring the binding of the virus to the cell surface prior to fusion of the viral envelope with the cellular plasma membrane. Whether the binding and fusion events are independent is unknown. In particular, does binding to HS contribute to the efficiency of entry of bound virus or does HS binding-defective virus enter cells with the same kinetics as bound wild-type virus? Assays of the rate of virus entry were performed to answer this question. The rate of virus entry into host cells was determined as the rate at which a virus bound to the cell surface becomes resistant to acid treatment as a consequence of penetration of the virus into the host cell, compared with untreated virus controls. The results presented in Fig. 7A show that the single-mutant KgBpK− and KCZ viruses displayed reduced kinetics of penetration compared to wild-type KOS virus. The rate of entry of the double-mutant KgBpK−gC− virus was slightly lower than each individual single mutant virus but equivalent to the additive effect of each individual mutation. Similar data were also obtained with L cells (data not shown). These data suggest either that stable attachment to the cell surface HS receptor is required for efficient virus entry or that deletion of the pK domain of gB reduces its fusion capacity in addition to its contribution to virus adsorption to the cell surface. In order to discriminate between these two possibilities, the rates of penetration of the wild-type and double-mutant KgBpK−gC− viruses were determined on GAG-deficient sog9 cells (Fig. 7B). We reasoned that if the fusion function of gB was altered by deletion of the pK domain, the rates of penetration of both viruses should be different on HS-deficient sog9 cells, while if the differences in the rates of penetration were due to the absence of binding to HS then both viruses should have the same rate of penetration on these cells. The data presented in Fig. 7B show that the rate of penetration of the double-mutant virus was similar to that of the wild-type virus on sog9 cells, confirming that the fusion function of gB was not altered by deletion of the pK domain of gB. Taken together, these data demonstrate that stable attachment to cell surface HS is required for efficient virus entry. The rate of entry of KCZ virus was compared to that of KgBpKRgC− (Fig. 7C), and the rate of entry of KgBpK− virus was compared to that of KgBpK−gCR (Fig. 7D). Both combinations gave similar results, confirming that the double-mutant viruses rescued for gB or gC showed rates of penetration similar to the corresponding single-mutant viruses.
FIG. 7.
Rate of penetration of viruses altered in HS proteoglycan binding domains. Confluent monolayers of Vero cells (A, C, and D) or sog9 cells (B) in 6-well plates were inoculated with 250 PFU of each indicated virus per well for an adsorption period of 2 h at 4°C. The cells were then rinsed three times, overlaid with complete medium, and shifted to 37°C to allow virus penetration. At selected times after the temperature shift, the cells were treated with 2 ml of glycine buffer (0.1 M glycine [pH 3.0]) or TBS for 1 min. The cell monolayers were then washed three times with complete medium, overlaid with medium containing methyl-cellulose, and incubated at 37°C to allow viral plaques to form. After plaque formation, cells were fixed and stained with crystal violet, and plaques were counted. At each time point, the average number of plaques produced on glycine-treated monolayers was expressed as a percentage of the average number of plaques produced on TBS-treated monolayers. Results are averages from triplicate wells.
The infectivity of the pK-deleted viruses (KgBpK− and KgBpK−gC−) compared to the wild-type KOS virus was measured on Vero cells by calculating the capacity of the 35S-labeled sucrose-purified recombinant viruses to form plaques on Vero cells and expressed as a ratio of PFU per counts per minute, where counts per minute was used to normalize the number of input particles. As demonstrated in Table 2, the single-mutant virus KgBpK− demonstrated 1.56-fold-less infectivity than wild-type KOS virus, while the double-mutant KgBpK−gC− virus demonstrated 3.55-fold-less infectivity than KOS virus. These data confirm results reported previously for gC deletion mutants (23) where a decrease in infectivity corresponded with a reduction in virus binding to the cell surface.
TABLE 2.
Infectivity of pK-deleted versus wild-type viruses for Vero cells
| Virus | cpm/ml | PFU/ml | PFU/cpm | Ratio to KOS |
|---|---|---|---|---|
| KOS | 4.299 × 107 | 7.50 × 107 | 1.74 | 1.00 |
| KgBpK− | 1.027 × 107 | 1.14 × 107 | 1.11 | 1.56 |
| KgBpK−gC− | 7.768 × 107 | 3.80 × 107 | 0.49 | 3.55 |
Virus mutants altered in gB-dependent HS binding also showed reduced cell-to-cell virus spread.
Virus penetration and cell-to-cell spread of viruses have been uncoupled through isolation of virus mutants deleted for glycoprotein gE and gI (11) that are defective in cell-to-cell spread but competent for virus entry. These mutants retained the ability to initiate infection of cells but could not form plaques in the presence of virus-neutralizing antibody. Glycoprotein B is required for both penetration and cell-to-cell spread; however, the role of its HS binding domain in lateral virus transmission has not been examined. To determine whether gB binding to HS influences lateral virus spread, Vero cells were infected with KOS, KgBpK−, KCZ, and KgBpK−gC− in the presence of HGG, which neutralized extracellular virus particles. As demonstrated in Fig. 8A, at 24, 36, and 48 h p.i., the average plaque size of mutant KgBpK− virus was significantly smaller than that of wild-type KOS virus at all time points, showing that deletion of the HS binding function of gB reduced lateral virus spread. These data demonstrate that the HS binding activity of gB is important to this mode of virus infection, an activity which could not be compensated for by the HS binding activity of gC. However, deletion of gC from wild-type KOS virus increased the efficiency of the mutant virus (KCZ) in spreading from cell to cell, as reported by Manservigi et al. (45), producing significantly larger plaques at 36 and 48 h p.i. These findings demonstrate that unlike virus penetration (Fig. 7A), the presence of gC is not required for efficient virus transmission to neighboring cells. However, similar experiments carried out with the double-mutant virus (KgBpK−gC−) that was deleted for the HS binding activity of gB and gC did not show significant reduction in plaque size compared with wild-type virus, suggesting that the opposing effects of each mutation on plaque development mask their individual phenotypes. In order to determine if the reduction in plaque size observed with the KgBpK− virus was related to an additional alteration in gB function that resulted from the deletion of the HS binding sequence, similar experiments were performed on GAG-deficient sog9 cells in the presence of HGG (Fig. 8B). The virus plaque sizes measured at 24, 36, and 48 h p.i. showed that plaque formation on sog9 cells was similar for each virus at all time points, demonstrating that the altered function of gB in Vero cell-to-cell spread was due to the loss in HS binding and was not a change in the ability of gB to mediate lateral virus spread independent of its HS binding functions. Together, these observations demonstrate that gB contributes to virus infection across cell membranes in an HS binding-dependent manner and that the loss of HS binding activity results in a small-plaque phenotype that can be masked through further deletion of gC.
FIG. 8.
Effect of HS binding mutations on lateral mutant virus spread. Confluent monolayers of Vero and sog9 cells were infected with 300 PFU of KOS, KCZ, KgBpK−, and KgBpK−gC− viruses per well. Twenty-four, 36, and 48 h p.i., Vero cell (A) and sog9 cell (B) monolayers were fixed with ice-cold methanol and processed for immunofluorescence with an anti-rabbit HSV-1 polyclonal antibody for 1 h at 37°C, washed with TBS, and incubated for 1 h at 37°C with a cy3 anti-rabbit antibody. Plaque sizes were determined with a Zeiss Axiophot microscope linked to a Xillix digital camera. For Vero cells, statistically significant differences (P < 0.05) in plaque sizes between viruses are marked by like symbols above the measured values; for sog9 cells, the plaque sizes were not statistically different (P > 0.39).
DISCUSSION
Previous studies have demonstrated that the initial attachment phase of infection by many herpesviruses (pseudorabies virus [PrV] [34, 34b, 49, 52, 61], bovine herpesvirus [13, 39, 40, 51], equine herpesvirus [1], cytomegalovirus [8], and HSV-1 and HSV-2 [16, 21, 44, 58, 64]) is mediated primarily by binding of one or more envelope glycoproteins to HS proteoglycans on the surface of susceptible cells. The HS binding activity of HSV-1 is attributable primarily to gC and gB (23), an observation confirmed in the present study. Mutants deleted for both gB and gC are severely impaired for binding to HS, to an extent similar to the highly impaired adsorption of wild-type virus to mutant cells defective for HS expression (17, 23, 55). Glycoprotein C accounts for the most avid binding, since mutations that result in gC-deficient enveloped particles show at least a 60% reduction in the efficiency of virus attachment compared to gB null virion particles (4, 24). Moreover, mutant viruses deleted for gC penetrate cells at a reduced rate (24), suggesting that attachment to HS may be required either to increase binding to a second receptor potentially recognized by gD or to initiate changes in the bound particle envelope that trigger the process of envelope fusion with the cell surface membrane (33). The interdependence of attachment to HS with virus entry and spread previously could not be studied in HSV with either gB or gB and gC null viruses, since gB is essential for both processes and may itself initiate fusion.
Initial experiments were aimed at determining whether an HS binding domain could be identified within gB based on analysis of the predicted amino acid sequence of the gB gene product. Studies of other HS binding molecules indicate that the negatively charged proteoglycan molecules would be recognized by peptide domains rich in positively charged residues, particularly lysine (38). Adenovirus vectors modified to contain a polylysine sequence within the fiber protein, for example, bind to HS and increase virus infection of cells low in adenovirus receptor density (63). Glycoprotein B contains a polylysine region (pK) that lies within amino acid residues 68 to 76 and has the sequence KPKKNKKPK. This sequence is similar to the consensus sequences predicted for the HS binding domain of proteins (7, 62) and was deleted from gB to produce a mutant form designated gBpK−. SDS-PAGE analysis of the transiently expressed mutant gene revealed a molecular size consistent with the predicted sequence. The processed glycoprotein was transported to the cell surface membrane in amounts similar to wild-type gB, and the gBpK− molecule was reactive with gB-specific monoclonal antibodies, indicating that these conformational epitopes were preserved. The gBpK− transiently expressed molecule was highly impaired in its ability to bind to heparin-conjugated acrylic beads, confirming that at least the major HS binding domain of gB was localized to this lysine-rich sequence. These results showed that although the HS binding domain of gB was deleted, its removal did not appear to significantly alter its conformational structure, glycosylation, or intracellular trafficking. The mutant gB molecule also proved to be functional for virus penetration by rescue of gB null mutant virus. Biochemical analysis of purified mutant virus particles indicated that normal levels of the mutant molecules were incorporated into virions, ruling out the possibility that any phenotypic change in mutant virus behavior was due to a reduction of gB in virus particles.
The KgBpK− mutant virus showed a decreased capacity for attachment to susceptible cells in culture. However, previous studies showed that gB-negative mutant viruses demonstrated the same binding capacity as wild-type virus to susceptible cells (4, 23). The discrepancy between our data and these reports could be explained on the basis of altered capacity for incorporation of the remaining glycoproteins into the virus envelope (9, 18, 19). For example, deletion of gB from the virus envelope might provide added space for increasing the amount of other glycoproteins such as gC in the envelope of mature particles, which in turn compensates for and thereby masks the HS binding activity of gB. Removal of both the HS binding function of gB and gC by construction of a double-mutant virus (KgBpK−gC−) had a profound effect on HS binding; however, the mutant virus still retained the ability to attach to murine L cells to an extent similar to the binding of wild-type virus to sog9 cells, a GAG-deficient mutant derivative (3). Moreover, the binding of the double mutant virus (KgBpK−gC−) was similar to the binding of the double deletion mutant gB/gC virus (K4BXCZ), demonstrating that notwithstanding the sensitivity of our binding assay gB and gC are the only HS binding proteins encoded by HSV-1 and that the pK domain of gB is the only binding domain of gB. The ratios of the titer of the double-mutant (KgBpK−gC−) virus on Vero cells in the absence or presence of heparin on L cells compared to sog9 cells were close to unity, demonstrating that all detectable HS-dependent binding activity had been deleted from the double-mutant virus. This conclusion was also supported by results demonstrating that the double-mutant virus was resistant to heparin washes, indicating that attachment had occurred through recognition of a non-HS receptor, presumably gD attachment to its specific receptor (50). We cannot rule out the possibility, however, that binding to the non-HS receptor is more avid in the absence of HS binding, making bound virus resistant to heparin washes and thus masking additional HS binding activity associated with gB and/or other glycoproteins. The ability of heparin to release bound virus to the HS receptor where, presumably, the non-HS receptor is also recognized might be explained by the order of receptor recognition. That is, HS receptor binding may preceed binding to the non-HS receptor; thus, heparin may release those particles in transition. Comparison of the infectivity of the double mutant with the wild-type virus demonstrated that in the absence of HS binding more input mutant virus was required to obtain infectivity comparable to the wild-type virus, indicating that the non-HS receptor was less abundant or less accessible. Similar studies have been performed with a gC-deleted PrV mutant where gC is solely responsible for the virus HS binding function. The PrV HS binding-deficient mutant showed reduced attachment to HS-bearing cells but not to HS-deficient mutant cells, a finding consistent with our studies of HSV mutants defective for HS binding. Both the HSV and PrV mutants remain active, demonstrating that HS binding is not essential for virus infectivity (34a).
The reduction in double-mutant virus attachment to the cell surface might also reflect a change in the ability of the virus particle to “neutralize” a repulsive charge on the cell surface, resulting in a change in the kinetics of recognition of the non-HS virus receptor. Such repulsive forces may not be present on HS-deficient sog9 cells; thus, the kinetics of virus attachment would appear similar to the wild-type virus. However, studies of the specificity of HSV-1 and HSV-2 for different locations of virus transmission may be related to the ability of the two serotypes to recognize different HS moieties on cell surface proteoglycans, and the HS specificity and recognition sequence of gC of PrV is apparently different from that of HSV-1 (24a, 61). In the present study, we show that the HS recognition sequence of gB differs from earlier reports for the HS binding element of gC (12a, 62). Thus, although HS recognition involves a charge interaction, the specific HS receptors recognized by different viruses may vary, suggesting that the interaction between the virus envelope and cell surface HS species are specific (3a, 12a).
Because gBpK− was functional in virus infection, it could be directly determined whether HS binding was also essential to the process of virus penetration. The rate of cell-bound KgBpK− mutant virus penetration was reduced compared with the wild-type virus, indicating that gB recognition of HS was required in this process. Moreover, the gC null virus (KCZ) also entered cells more slowly, in agreement with an earlier report (24), whereas the double-mutant (KgBpK−gC−) virus did not differ significantly from either single-mutant virus, suggesting that HS recognition by both glycoproteins is required for efficient virus entry and that they may cooperate in this process. This conclusion was supported by the finding that the KgBpK−gC− mutant entered cells with kinetics similar to the wild-type virus on sog9 cells. Moreover, this result suggests that deletion of the HS binding function of gB does not alter the fusion function of gB. Since the double-mutant KgBpK−gC− virus was capable of binding to the cell surface through a non-HS receptor but demonstrated a lower rate of penetration than wild-type virus, we conclude that virus binding to the inherent non-HS receptor can substitute for HS binding in promoting virus entry but with less efficiency. The biochemical mechanism which underlies these cooperative functions is unknown but may relate to the possibility that HS binding brings the virus in closer proximity to the cell membrane, thereby enhancing the ability of gB to contribute to virus envelope cell membrane fusion. The binding to HS, for example, may alter the conformation of gB or other viral fusogens in a manner that allows invasion of the fusogenic peptide into the cell membrane (20).
The role of HS receptor binding glycoproteins in entry of extracellular virus may be different when virus infection proceeds from cell to cell. For example, mutants deficient in gC form larger plaques in the presence of neutralizing antibodies than does the wild-type virus, suggesting that gC reduces the efficiency of virus spread. In our studies we have made similar observations, indicating that the HS binding activity of gC may not be required for intercellular virus entry into neighboring cells. Shieh and Spear (55) reported that binding to HS is involved in virus spread from cell to cell since wild-type HSV-1 cannot spread laterally in CHO cells deficient for HS but can be enhanced in such cells if soluble heparin is provided. In contrast, Gruenheid et al. (17) showed that HS may be dispensable for lateral virus spread, since virus plaques were of similar size on murine L cells or their HS-deficient derivatives, gro2C cells. These mutant cells retained condroitin sulfate moieties that were subsequently demonstrated to be recognized by HSV in virus attachment to murine L cells (3). In the present study, we compared the ability of both single and double HS binding mutant viruses to form plaques on Vero cells in the presence of virus-neutralizing antiserum where plaque formation depended exclusively on cell-to-cell virus spread. The results showed that the KgBpK− mutant virus produced significantly smaller plaques than wild-type virus on Vero cells, while the gC null mutant produced larger-than-wild-type virus plaques in the presence of antiserum. However, deletion of both the pK domain of gB and the gC gene created a mutant virus (KgBpK−gC−) that formed plaques of similar size to the wild-type virus, suggesting that only gB binding to the HS receptor is required for efficient intercellular infection and indicating that the roles of HS binding are different in extra- and intercellular infection. These conclusions were strengthened by comparisons of the virus plaque sizes on GAG-deficient cells in which all mutants tested formed plaques of comparable size to wild-type KOS virus. Because the presence of gC appeared to inhibit lateral virus spread, deletion of both the gB and gC HS binding activity resulted in a normal-size plaque phenotype in which the plaque-size-reducing effect of mutant gB appeared to be masked by the plaque-size-enhancing phenotype associated with the deletion of gC. The reason(s) for these opposing effects is unknown but might be explained by gB and gC recognition of different HS structures such as, for instance, those that might be associated with cell junctions where virus transmission occurs. Alternatively, these differences could be due to a change in the stoichiometry of gC-deleted virus envelope components, some of which may also contribute to lateral virus spread. While these observations distinguished between the roles of gB and gC in plaque size development, deletion of either HS binding activity reduced the binding of virus to cell surfaces and slowed the rate of extracellular virus entry. Further studies are in progress to determine whether HS binding is important to the pathogenesis of HSV where virus spread occurs from cell to cell. These studies should determine whether the HS binding function of gB is essential to the process of virus infection in vivo.
ACKNOWLEDGMENTS
We thank Frank Tufaro for providing murine L cells and derivative sog9 cells, Patricia Dowling for VSV, and David Fink for assistance in measuring plaque size areas. We also thank Thomas Holland for critical reading of the manuscript.
This work was supported by Public Health Service grant R01 CA66141-07 from the National Institutes of Health (J.C.G.) and by grants from Telethon A100 and AIRC (R.M.). Rafaela Argnani was supported by an AIRC fellowship, and Sylvie Laquerre was supported by L’Association Française contre les Myopathies (AFM).
REFERENCES
- 1.Allen G P, Coogle L D. Characterization of an equine herpesvirus type 1 gene encoding a glycoprotein (gp13) with homology to herpes simplex virus glycoprotein C. J Virol. 1988;62:2850–2858. doi: 10.1128/jvi.62.8.2850-2858.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baines J D, Roizman B. The UL10 gene of herpes simplex virus 1 encodes a novel glycoprotein, gM, which is present in the virion and in the plasma membrane of infected cells. J Virol. 1993;67:1441–1452. doi: 10.1128/jvi.67.3.1441-1452.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Banfield B W, Leduc Y, Esford L, Schubert K, Tufaro F. Sequential isolation of proteoglycan synthesis mutants by using herpes simplex virus as a selective agent: evidence for a proteoglycan-independent virus entry pathway. J Virol. 1995;69:3290–3298. doi: 10.1128/jvi.69.6.3290-3298.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3a.Bergström T, Trybala E, Spillmann D. Heparan sulfate and viral tropism. Nat Med. 1997;3:1177. doi: 10.1038/nm1197-1177b. [DOI] [PubMed] [Google Scholar]
- 4.Cai W, Gu B, Person S. Role of glycoprotein B of herpes simplex virus type 1 in viral entry and cell fusion. J Virol. 1988;62:2596–2604. doi: 10.1128/jvi.62.8.2596-2604.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cai W, Person S, Warner S C, Zhou J, DeLuca N A. Linker-insertion nonsense and restriction-site deletion mutations of the gB glycoprotein gene of herpes simplex virus type 1. J Virol. 1987;61:714–721. doi: 10.1128/jvi.61.3.714-721.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campadelli-Fiume G, Stirpe D, Boscaro A, Avitabile E, Foá-Tomasi L, Barker D, Roizman B. Glycoprotein C-dependent attachment of herpes simplex virus to susceptible cells leading to productive infection. Virology. 1990;178:213–222. doi: 10.1016/0042-6822(90)90396-9. [DOI] [PubMed] [Google Scholar]
- 7.Cardin A D, Weintraub H J R. Molecular modeling of protein-glycosaminoglycan interactions. Arteriosclerosis. 1989;9:21–32. doi: 10.1161/01.atv.9.1.21. [DOI] [PubMed] [Google Scholar]
- 8.Compton T, Nowlin D M, Cooper N R. Initiation of human cytomegalovirus infection requires initial interaction with cell surface heparan sulfate. Virology. 1993;193:834–841. doi: 10.1006/viro.1993.1192. [DOI] [PubMed] [Google Scholar]
- 9.Desai P, Homa F L, Person S, Glorioso J C. A genetic selection method for the transfer of HSV-1 glycoprotein B mutations from plasmid to the viral genome: preliminary characterization of transdominance and entry kinetics of mutant viruses. Virology. 1994;204:312–322. doi: 10.1006/viro.1994.1536. [DOI] [PubMed] [Google Scholar]
- 10.Desai P J, Schaffer P A, Minson A C. Excretion of non-infectious virus particles lacking glycoprotein H by a temperature-sensitive mutant of herpes simplex virus type 1: evidence that gH is essential for virion infectivity. J Gen Virol. 1988;69:1147–1156. doi: 10.1099/0022-1317-69-6-1147. [DOI] [PubMed] [Google Scholar]
- 11.Dingwell K S, Brunetti C R, Hendricks R L, Tang Q, Tang M, Rainbow A J, Johnson D C. Herpes simplex virus glycoproteins E and I facilitate cell-to-cell spread in vivo and across junctions of cultured cells. J Virol. 1994;68:834–845. doi: 10.1128/jvi.68.2.834-845.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dolter K E, Goins W F, Levine M, Glorioso J C. Genetic analysis of type-specific antigenic determinants of herpes simplex virus glycoprotein C. J Virol. 1992;66:4864–4873. doi: 10.1128/jvi.66.8.4864-4873.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12a.Feyzi E, Trybala E, Bergström T, Lindahi U, Spillmann D. Structural requirement of heparan sulfate for interaction with herpes simplex virus type 1 glycoprotein C. J Biol Chem. 1997;272:24850–24857. doi: 10.1074/jbc.272.40.24850. [DOI] [PubMed] [Google Scholar]
- 13.Fitzpatrick D R, Babiuk L A, Zamb T J. Nucleotide sequence of bovine herpesvirus type 1 glycoprotein gIII, a structural model of gIII as a new member of the immunoglobulin superfamily, and implications for the homologous glycoproteins of other herpesviruses. Virology. 1989;173:46–57. doi: 10.1016/0042-6822(89)90220-1. [DOI] [PubMed] [Google Scholar]
- 14.Fuller A O, Lee W-C. Herpes simplex virus type 1 entry through a cascade of virus-cell interactions requires different roles of gD and gH in penetration. J Virol. 1992;66:5002–5012. doi: 10.1128/jvi.66.8.5002-5012.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gallagher J T, Lyon M, Steward W P. Structure and function of heparan sulfate proteoglycans. Biochem J. 1986;236:313–325. doi: 10.1042/bj2360313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gerber S I, Belval B J, Herold B C. Differences in the role of glycoprotein C of HSV-1 and HSV-2 in viral binding may contribute to serotype differences in cell tropism. Virology. 1995;214:29–39. doi: 10.1006/viro.1995.9957. [DOI] [PubMed] [Google Scholar]
- 17.Gruenheid S, Gatzke L, Meadows H, Tufaro F. Herpes simplex virus infection and propagation in a mouse L-cell mutant lacking heparan sulfate proteoglycans. J Virol. 1993;67:93–100. doi: 10.1128/jvi.67.1.93-100.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Handler G C, Eisenberg R J, Cohen G H. Oligomeric structure of glycoproteins in herpes simplex virus type 1. J Virol. 1996;70:6067–6075. doi: 10.1128/jvi.70.9.6067-6070.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Handler G C, Cohen G H, Eisenberg R J. Oligomeric structure of glycoproteins in herpes simplex virus type 1. J Virol. 1996;70:6076–6082. doi: 10.1128/jvi.70.9.6067-6070.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haywood A M. Virus receptors: binding, adhesion strengthening, and changes in viral structure. J Virol. 1994;68:1–5. doi: 10.1128/jvi.68.1.1-5.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Herold B C, Gerber S I, Belval B J, Siston A M, Shulman N. Differences in the susceptibility of herpes simplex virus types 1 and 2 to modified heparin compounds suggest serotype differences in viral entry. J Virol. 1996;70:3461–3469. doi: 10.1128/jvi.70.6.3461-3469.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Herold B C, Gerber S I, Polonsky T, Belval B J, Shaklee P N, Holme K. Identification of structural features of heparin required for inhibition of herpes simplex virus type 1 binding. Virology. 1995;206:1108–1116. doi: 10.1006/viro.1995.1034. [DOI] [PubMed] [Google Scholar]
- 23.Herold B C, Visalli R J, Sumarski N, Brandt C R, Spear P G. Glycoprotein C-independent binding of herpes simplex virus to cells requires cell surface heparan sulfate and glycoprotein B. J Gen Virol. 1994;75:1211–1222. doi: 10.1099/0022-1317-75-6-1211. [DOI] [PubMed] [Google Scholar]
- 24.Herold B C, WuDunn D, Soltys N, Spear P G. Glycoprotein C of herpes simplex virus type 1 plays a principal role in the adsorption of virus to cells and in infectivity. J Virol. 1991;65:1090–1098. doi: 10.1128/jvi.65.3.1090-1098.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24a.Herold B C, Gerber S I, Belval B J, Siston A M, Scrulman N. Differences in the susceptibility of herpes simplex virus types 1 and 2 to modified heparin compounds suggest serotype differences in viral entry. J Virol. 1996;70:3461–3469. doi: 10.1128/jvi.70.6.3461-3469.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Highlander S L, Goins W F, Person S, Holland T C, Levine M, Glorioso J C. Oligomer formation of the gB glycoprotein of herpes simplex virus type 1. J Virol. 1991;50:805–812. doi: 10.1128/jvi.65.8.4275-4283.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Highlander S L, Sutherland S L, Gage P J, Johnson D C, Levine M, Glorioso J C. Neutralizing monoclonal antibodies specific for herpes simplex virus glycoprotein D inhibit virus penetration. J Virol. 1987;61:3356–3364. doi: 10.1128/jvi.61.11.3356-3364.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Holland T C, Homa F L, Marlin S D, Levine M, Glorioso J C. Herpes simplex virus type 1 glycoprotein C-negative mutants exhibit multiple phenotypes, including secretion of truncated glycoproteins. J Virol. 1984;52:566–574. doi: 10.1128/jvi.52.2.566-574.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Homa F L, Purifoy D J M, Glorioso J C, Levine M. Molecular basis of the glycoprotein C-negative phenotypes of herpes simplex virus type 1 mutants selected with a virus-neutralizing monoclonal antibody. J Virol. 1986;58:281–289. doi: 10.1128/jvi.58.2.281-289.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hutchinson L, Browne H, Wargent V, Davis-Poynter N, Primorac S, Goldsmith K, Minson A C, Johnson D C. A novel herpes simplex glycoprotein, gL, forms a complex with glycoprotein H (gH) and affects normal folding and surface expression of gH. J Virol. 1992;66:2240–2250. doi: 10.1128/jvi.66.4.2240-2250.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hutchinson L, Goldsmith K, Snoddy D, Ghosh H, Graham F L, Johnson D C. Identification and characterization of a novel herpes simplex virus glycoprotein, gK, involved in cell fusion. J Virol. 1992;66:5603–5609. doi: 10.1128/jvi.66.9.5603-5609.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hutchinson L, Johnson D C. Herpes simplex virus glycoprotein K promotes egress of virus particles. J Virol. 1995;69:5401–5413. doi: 10.1128/jvi.69.9.5401-5413.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnson D C, Ligas M W. Herpes simplex viruses lacking glycoprotein D are unable to inhibit viral penetration: quantitative evidence for virus-specific cell surface receptors. J Virol. 1988;62:4605–4612. doi: 10.1128/jvi.62.12.4605-4612.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johnson D C, McDermott M R, Chrisp C, Glorioso J C. Pathogenicity in mice of herpes simplex virus type 2 mutants unable to express glycoprotein C. J Virol. 1986;58:36–42. doi: 10.1128/jvi.58.1.36-42.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karger A, Mettenleiter T C. Glycoproteins gIII and gp50 play dominant roles in the biphasic attachment of pseudorabies virus. Virology. 1993;194:654–664. doi: 10.1006/viro.1993.1305. [DOI] [PubMed] [Google Scholar]
- 34a.Karger A, Saalmüller A, Tufaro F, Banfield B W, Mettenleiter T C. Cell surface proteoglycans are not essential for infection by pseudorabies virus. J Virol. 1995;69:3482–3489. doi: 10.1128/jvi.69.6.3482-3489.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kjellén L, Lindahl U. Proteoglycans: structures and interactions. Annu Rev Biochem. 1991;60:443–475. doi: 10.1146/annurev.bi.60.070191.002303. [DOI] [PubMed] [Google Scholar]
- 36.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 37.Langeland N, Holmsen H, Lillehaug J R, Haarr L. Evidence that neomycin inhibits binding of herpes simplex virus type 1 to the cellular receptor. J Virol. 1987;61:3388–3393. doi: 10.1128/jvi.61.11.3388-3393.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Langeland N, Moore L J, Holmsen H, Haarr L. Interaction of polylysine with the cellular receptor for herpes simplex virus type 1. J Gen Virol. 1988;69:1137–1145. doi: 10.1099/0022-1317-69-6-1137. [DOI] [PubMed] [Google Scholar]
- 39.Liang X, Babiuk L A, van Drunen Littel-van den Hurk S, Fitzpatrick D R, Zamb T J. Bovine herpesvirus 1 attachment to permissive cells is mediated by its major glycoproteins gI, gIII, and gIV. J Virol. 1991;65:1124–1132. doi: 10.1128/jvi.65.3.1124-1132.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liang X, Babiuk L A, Zamb T J. Mapping of heparin-binding structures of bovine herpesvirus 1 and pseudorabies virus gIII glycoproteins. Virology. 1993;194:233–243. doi: 10.1006/viro.1993.1254. [DOI] [PubMed] [Google Scholar]
- 41.Lidholt K, Weinke J L, Kiser C S, Lugemwa F N, Bame K J, Cheifetz S, Massague J, Lindahl U, Esko J D. A single mutation affects both N-acetylglucosaminyltransferase and glucoronosyltransferase activities in a Chinese hamster ovary cell mutant defective in heparan sulfate biosynthesis. Proc Natl Acad Sci USA. 1992;89:2267–2271. doi: 10.1073/pnas.89.6.2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ligas M W, Johnson D C. A herpes simplex virus mutant in which glycoprotein D sequences are replaced by β-galactosidase sequences binds to but is unable to penetrate into cells. J Virol. 1988;62:1486–1494. doi: 10.1128/jvi.62.5.1486-1494.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Little S P, Jofre J T, Courtney R J, Schaffer P A. A virion-associated glycoprotein essential for infectivity of herpes simplex virus type 1. Virology. 1981;115:149–160. doi: 10.1016/0042-6822(81)90097-0. [DOI] [PubMed] [Google Scholar]
- 44.Lycke E, Johansson M, Svennerholm B, Lindahl U. Binding of herpes simplex virus to cellular heparan sulfate, an initial step in the adsorption process. J Gen Virol. 1991;72:1131–1137. doi: 10.1099/0022-1317-72-5-1131. [DOI] [PubMed] [Google Scholar]
- 45.Manservigi R, Spear P G, Buchan A. Cell fusion induced by herpes simplex virus is promoted and suppressed by different viral glycoproteins. Proc Natl Acad Sci USA. 1977;74:3913–3917. doi: 10.1073/pnas.74.9.3913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marlin S D, Highlander S L, Holland T C, Levine M, Glorioso J C. Antigenic variation (mar mutations) in herpes simplex virus glycoprotein B can induce temperature-dependent alterations in gB processing and virus production. J Virol. 1986;59:142–153. doi: 10.1128/jvi.59.1.142-153.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marlin S D, Holland T C, Levine M, Glorioso J C. Epitopes of herpes simplex virus type 1 glycoprotein gC are clustered in two distinct antigenic sites. J Virol. 1985;53:128–136. doi: 10.1128/jvi.53.1.128-136.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McClain D S, Fuller A O. Cell-specific kinetics and efficiency of herpes simplex virus type 1 are determined by two distinct phases of attachment. Virology. 1994;198:690–702. doi: 10.1006/viro.1994.1081. [DOI] [PubMed] [Google Scholar]
- 49.Mettenleiter T C, Zsak L, Zuckermann F, Sugg N, Kern H, Ben-Porat T. Interaction of glycoprotein gIII with a cellular heparinlike substance mediates adsorption of pseudorabies virus. J Virol. 1990;64:278–286. doi: 10.1128/jvi.64.1.278-286.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Montgomery R I, Warner M S, Lum B J, Spear P G. Herpes simplex virus-1 entry into cells mediated by a novel member of the TNF/NGF receptor family. Cell. 1996;87:427–436. doi: 10.1016/s0092-8674(00)81363-x. [DOI] [PubMed] [Google Scholar]
- 51.Okazaki K, Matsuzaki T, Sugahara Y, Okada J, Hasebe M, Iwamura Y, Ohnishi M, Kanno T, Shimizu M, Honda E, Kono Y. BHV-1 adsorption is mediated by the interaction of glycoprotein gIII with heparinlike moiety on the cell surface. Virology. 1991;181:666–670. doi: 10.1016/0042-6822(91)90900-v. [DOI] [PubMed] [Google Scholar]
- 52.Robbins A K, Watson R J, Whealy M E, Hays W W, Enquist L W. Characterization of a pseudorabies virus glycoprotein gene with homology to herpes simplex virus type 1 and type 2 glycoprotein C. J Virol. 1986;58:339–347. doi: 10.1128/jvi.58.2.339-347.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Roizman B, Sears A E. Herpes simplex viruses and their replication. In: Fields B N, Knipe D M, Howley P M, et al., editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincot-Raven Publishers; 1996. pp. 2231–2295. [Google Scholar]
- 54.Roop C, Hutchinson L, Johnson D C. A mutant herpes simplex virus type 1 unable to express glycoprotein L cannot enter cells, and its particles lack glycoprotein H. J Virol. 1993;67:2285–2297. doi: 10.1128/jvi.67.4.2285-2297.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shieh M-T, Spear P G. Herpesvirus-induced cell fusion that is dependent on cell surface heparan sulfate or soluble heparin. J Virol. 1994;68:1224–1228. doi: 10.1128/jvi.68.2.1224-1228.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shieh M-T, WuDunn D, Montgomery R I, Esko J D, Spear P G. Cell surface receptors for herpes simplex virus are heparan sulfate proteoglycans. J Cell Biol. 1992;116:1273–1281. doi: 10.1083/jcb.116.5.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Spear P G. Entry of alphaherpesviruses into cells. Semin Virol. 1993;4:167–180. [Google Scholar]
- 58.Subramanian G, McClain D S, Perez A, Fuller A O. Swine testis cells contain functional heparan sulfate but are defective in entry of herpes simplex virus. J Virol. 1994;68:5667–5676. doi: 10.1128/jvi.68.9.5667-5676.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tal-Singer R, Peng C, Ponce de Leon M, Abrams W R, Banfield B W, Tufaro F, Cohen G H, Eisenberg R J. Interaction of herpes simplex virus glycoprotein gC with mammalian cell surface molecules. J Virol. 1995;69:4471–4483. doi: 10.1128/jvi.69.7.4471-4483.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tengelsein L A, Pederson N E, Shaver P R, Wathen M W, Homa F L. Herpes simplex virus type 1 DNA cleavage and encapsidation require the product of the UL28 gene: isolation and characterization of two UL28 deletion mutants. J Virol. 1993;67:3470–3480. doi: 10.1128/jvi.67.6.3470-3480.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Trybala E, Bergström T, Spillmann D, Svennerholm B, Olofsson S, Flynn S J, Ryan P. Mode of interaction between pseudorabies virus and heparan sulfate/heparin. Virology. 1996;218:35–42. doi: 10.1006/viro.1996.0163. [DOI] [PubMed] [Google Scholar]
- 62.Trybala E, Bergström T, Svennerholm B, Jeansson S, Glorioso J C, Olofsson S. Localization of the functional site on herpes simplex virus type 1 glycoprotein C involved in binding to cell surface heparan sulfate. J Gen Virol. 1994;75:743–752. doi: 10.1099/0022-1317-75-4-743. [DOI] [PubMed] [Google Scholar]
- 63.Wickham T J, Roelvink P W, Brough D E, Kovesdi I. Adenovirus targeted to heparan containing receptors increases its gene delivery efficiency to multiple cell types. Nat Biotechnol. 1996;14:1570–1573. doi: 10.1038/nbt1196-1570. [DOI] [PubMed] [Google Scholar]
- 64.WuDunn D, Spear P G. Initial interaction of herpes simplex virus with cells is binding to heparan sulfate. J Virol. 1989;63:52–58. doi: 10.1128/jvi.63.1.52-58.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]