Abstract
Organisms exhibit a diverse set of responses when exposed to low-phosphate conditions. Some of these responses are specific for phosphorus limitation, including responses that enable cells to efficiently scavenge phosphate from internal and external stores via the production of high-affinity phosphate transporters and the synthesis of intracellular and extracellular phosphatases. Other responses are general and occur under a number of different environmental stresses, helping coordinate cellular metabolism and cell division with the growth potential of the cell. In this article, we describe the isolation and characterization of a mutant of Chlamydomonas reinhardtii, low-phosphate bleaching (lpb1), which dies more rapidly than wild-type cells during phosphorus limitation. The responses of this mutant to nitrogen limitation appear normal, although the strain is also somewhat more sensitive than wild-type cells to sulfur deprivation. Interestingly, depriving the cells of both nutrients simultaneously allows for sustained survival that is similar to that observed with wild-type cells. Furthermore, upon phosphorus deprivation, the lpb1 mutant, like wild-type cells, exhibits increased levels of mRNA encoding the PHOX alkaline phosphatase, the PTB2 phosphate transporter, and the regulatory element PSR1. The mutant strain is also able to synthesize the extracellular alkaline phosphatase activity upon phosphorus deprivation and the arylsulfatase upon sulfur deprivation, suggesting that the specific responses to phosphorus and sulfur deprivation are normal. The LPB1 gene was tagged by insertion of the ARG7 gene, which facilitated its isolation and characterization. This gene encodes a protein with strong similarity to expressed proteins in Arabidopsis (Arabidopsis thaliana) and predicted proteins in Oryza sativa and Parachlamydia. A domain in the protein contains some similarity to the superfamily of nucleotide-diphospho-sugar transferases, and it is likely to be localized to the chloroplast or mitochondrion based on programs that predict subcellular localization. While the precise catalytic role and physiological function of the putative protein is not known, it may function in some aspect of polysaccharide metabolism and/or influence phosphorus metabolism (either structural or regulatory) in a way that is critical for allowing the cells to acclimate to nutrient limitation conditions.
The predominant form of available phosphorus (P) in the environment is inorganic phosphate (Pi), which is incorporated into numerous molecules including nucleic acids, phospholipids, and proteins. Pi availability is often limiting to plant growth, and while many soils have a Pi content of between 0.5 and 1.5 mm, the majority of the Pi is present as insoluble Fe3+, Al3+, and Ca2+ salts or as esterified organic compounds; these forms may not be readily assimilated. To maintain high crop yields, Pi is included as an abundant component of commercial fertilizers. Much of this supplementary Pi may be leached from agricultural fields and deposited into nearby lakes and rivers, triggering rapid algal growth (algal blooms) that results in the proliferation of heterotrophs, eutrophication of the environment, and massive fish kills (Wetzel, 1983). Sustainability of agricultural yields and the quality of terrestrial and aquatic ecosystems will benefit from a clearer understanding of Pi assimilation and the regulatory processes that control its acquisition and utilization.
Many soil-dwelling organisms have evolved a suite of responses that enable them to survive extended exposure to conditions of low P availability. These responses can be divided into the P-specific responses and the general (or global) stress responses (Quisel et al., 1996; Davies and Grossman, 1998; Wykoff et al., 1998). P-specific responses facilitate mobilization and acquisition of Pi from both extracellular and intracellular sources (Quisel et al., 1996; Shimogawara et al., 1999), while the global responses allow for long-term survival by coordinating cellular metabolism and growth with nutrient availability.
Starvation of most plants and microorganisms for P results in elevated phosphatase activities (Juma and Tabatabai, 1988; Lee, 1988; Goldstein et al., 1989; Duff et al., 1991a, 1991b; Sachay et al., 1991; Tadano et al., 1993; Nakazato et al., 1997; Trull et al., 1997), which helps to mobilize internal Pi stores (e.g. from phytic acid and inositol phosphates in vacuoles) and to generate free Pi from external organic phosphates. Increased RNase production during P deprivation (Jost et al., 1991; Taylor and Green, 1991; Yen and Green, 1991; Löffler et al., 1992, 1993; Taylor et al., 1993; Bariola et al., 1994) may also boost Pi availability by hydrolyzing Pi from various RNA reservoirs (Green, 1994). Furthermore, the roots of specific plants such as rape (Brassica napus) may exude organic acids during P deprivation that lower the rhizosphere pH (Hoffland et al., 1992), facilitating solubilization and increased availability of soil Pi for transport into roots (Lopez-Bucio et al., 2000). P limitation can also result in the elevation of the Vmax (but not necessarily alter the Km) for Pi uptake into roots by triggering accumulation of high-affinity Pi uptake systems (Bieleski, 1973; Drew et al., 1984; Lefebvre and Clarkson, 1984; Schjorring and Jensen, 1984; McPharlin and Bieleski, 1987; Jungk et al., 1990; Lefebvre et al., 1990; Bieleski and Laluchli, 1992; Shimogawara and Usuda, 1995). While genes encoding plant Pi transporters have been isolated (Muchhal and Raghothama, 1999; Raghothama, 2000a, 2000b; Smith et al., 2000; Baek et al., 2001), the acquisition of Pi by plants can be complex because of interactions of plant roots with soil organisms such as mycorrhizal fungi (Rhizoctoni solani; Yao et al., 2002). Proliferation of mycorrhizal fungi helps mobilize soil Pi for uptake into the roots (Pfleger and Linderman, 1994; Rosewarne et al., 1999; Yao et al., 2002).
Morphological and biochemical changes in plants and microbes may also accompany P limitation. In P-starved plants, the root-to-shoot ratio may increase in conjunction with decreased lateral-root proliferation and the development of longer, more densely packed root hairs. P-starved black mustard (Brassica nigra) exhibits increased fixed carbon storage and elevated accumulation of lipids and phenolics. Furthermore, enzymes that serve a bypass function (Duff et al., 1989; Theodorou et al., 1991), helping to eliminate the need for phosphorylated adenylates, which can be limiting during P starvation, may replace other enzymes in metabolic pathways (e.g. phosphoenolpyruvate phosphatase may replace pyruvate kinase).
Like vascular plants, Chlamydomonas reinhardtii synthesizes a number of phosphatases (Lien and Knutsen, 1973; Matagne and Loppes, 1975; Loppes, 1976; Matagne et al., 1976; Patni et al., 1977; Loppes and Deltour, 1981; Quisel et al., 1996) and exhibits a change in the kinetics of Pi transport (Shimogawara et al., 1999) during P limitation. The cells synthesize an abundant extracellular phosphatase activity, with 85% to 90% associated with a 5′ nucleotidase having an alkaline pH optimum and a monomeric, apparent molecular mass of 190 kD (Dumont et al., 1990; Quisel et al., 1996). Starved cells also display increased Pi uptake potential. The Vmax for Pi increases by well over 10-fold and the Km for Pi decreases from approximately 10 μm to between 0.1 and 0.3 μm (Shimogawara et al., 1999). Other genes that appear to be activated during P starvation encode enolase, pyruvate-formate lyase, and two ribosomal proteins, although some of these genes may also be induced under other growth-limiting conditions (Dumont et al., 1993).
Currently, little is known about regulatory elements that control P limitation responses in plants and algae. A number of mutants of C. reinhardtii have been isolated that exhibit aberrant phosphatase activity during P deprivation (Loppes and Matagne, 1973; Matagne and Loppes, 1975; Loppes, 1976; Loppes et al., 1977; Loppes and Deltour, 1981; Wykoff et al., 1999), and we have identified the altered gene in one such mutant. This gene, designated PSR1 (Phosphate Starvation Response), encodes a protein of 762 amino acids that is synthesized at elevated levels during P deprivation (Wykoff et al., 1999) and is a member of the MYB-CC subfamily of regulatory elements (Pabo and Sauer, 1992). A psr1 null mutant is unable to synthesize extracellular phosphatase in response to Pi deprivation and is aberrant for the other specific responses to P deprivation that were tested. Recently, PSR1 was shown to be important for the development of some general responses and survival of the cells during P deprivation under elevated light conditions (J. Moseley, C.-W. Chang, and A.R. Grossman, unpublished data). A similar regulatory element designated PHR1 was identified in Arabidopsis (Arabidopsis thaliana; Rubio et al., 2001).
In this study, we used a genetic screen to identify C. reinhardtii mutants that die more rapidly than wild-type cells during P limitation conditions. Two allelic mutants, low-P bleaching (lpb1-1) and lpb1-2, were isolated and the LPB1 gene was identified and sequenced. The predicted gene product has limited similarity to a UDP-Glc pyrophosphorylase (UDPGPase), which is involved in the synthesis or degradation of polysaccharides.
RESULTS
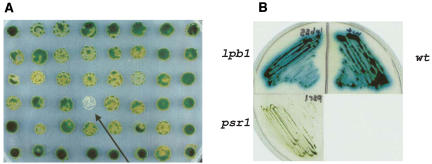
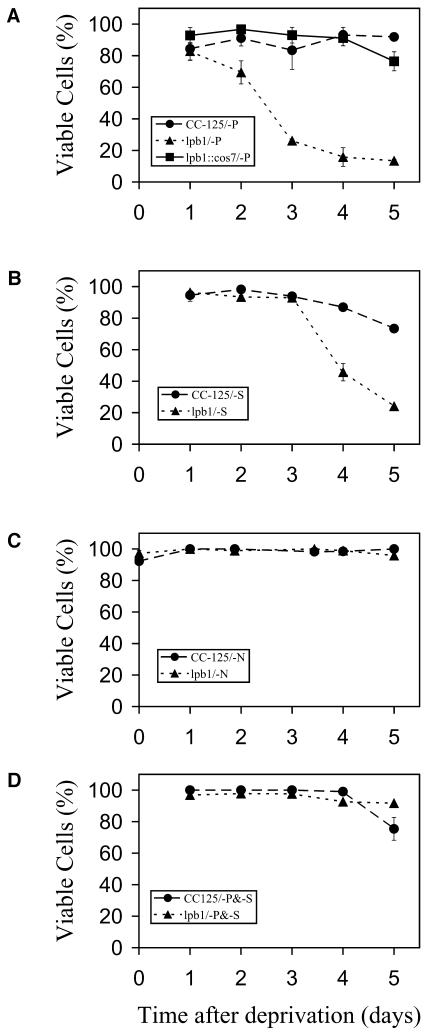
We previously isolated the PSR1 gene, which regulates the specific responses of C. reinhardtii to P starvation (Wykoff et al., 1999) but also affects at least some of the general responses to P deprivation (J. Moseley, C.-W. Chang, and A.R. Grossman, unpublished data). In this article, we report the identification of a C. reinhardtii mutant that was still able to perform specific P-deprivation responses, but was screened for rapid death on solid medium following the imposition of P deprivation; this mutant, designated lpb1, is defined by two alleles. As shown in the colony assay (Fig. 1A), the lpb1 mutant bleaches and dies rapidly on solid medium lacking P. The loss of pigmentation or bleaching of the cells parallels cell death (data not shown). Quantification of viability of the mutant and wild-type strain in liquid medium is presented in Figure 2A. Approximately 30% of lpb1 mutant cells died after 2 d of P starvation, and after 5 d, less then 20% viable mutant cells remained. In contrast, essentially 100% of the wild-type cells were viable after 5 d of P deprivation (Fig. 2A).
Figure 1.
Cell viability and phosphatase activity. A, Viability of the lpb1 mutant and wild-type cells after 1 week of growth on solid medium devoid of phosphate. The arrow denotes the bleached lpb1 colony. B, Qualitative analysis of phosphatase activity secreted by wild-type cells (wt), the lpb1 mutant, and the psr1 mutant. The psr1 strain represents a negative control; the mutant is unable to synthesize alkaline phosphatase in response to P deprivation (Wykoff et al., 1999). Cells were streaked onto TA solid medium prior to spraying the plates with the colorimetric phosphatase substrate 5-bromo-4-chloro-3-indolyl-P, which turns blue after phosphate is cleaved from the molecule by alkaline phophatase. The plates were allowed to develop for 2 h before recording the results.
Figure 2.
Viability of wild-type cells, mutant strains, and the complemented lpb1 mutant in medium lacking P, S, N, or P plus S. A, Viability of wild-type cells (CC-125), lpb1, and lpb1 complemented with cos7 (containing the wild-type LPB1 gene) following P deprivation in liquid medium. B, Viability of wild-type cells and lpb1 following S deprivation in liquid medium. C, Viability of wild-type cells and lpb1 following N deprivation in liquid medium. D, Viability of wild-type cells and lpb1 following both P and S deprivation in liquid medium. A viability dye was used to distinguish viable from nonviable cells. This was confirmed by monitoring cell viability by plating the cells onto solid TAP medium (see “Materials and Methods” for further details).
To determine whether the loss of viability of the mutant strain was specific to P deprivation or occurred under other nutrient deprivation conditions, we starved both mutant and wild-type cells for nitrogen (N), sulfur (S), and S plus P and measured viability over a 5-d period. The lpb1 mutant also showed decreased viability relative to wild-type cells during S deprivation (Fig. 2B), but not during N deprivation (Fig. 2C). These results suggest that the lesion affects acclimation of C. reinhardtii to reduced P and S concentrations, but not to a reduced N concentration. The lpb1 mutant appears to be more sensitive to P deprivation than to S deprivation since it dies more rapidly in the former. Interestingly, as shown in Figure 2D, depriving the mutant strain simultaneously for P and S rescues the lpb1 death phenotype.
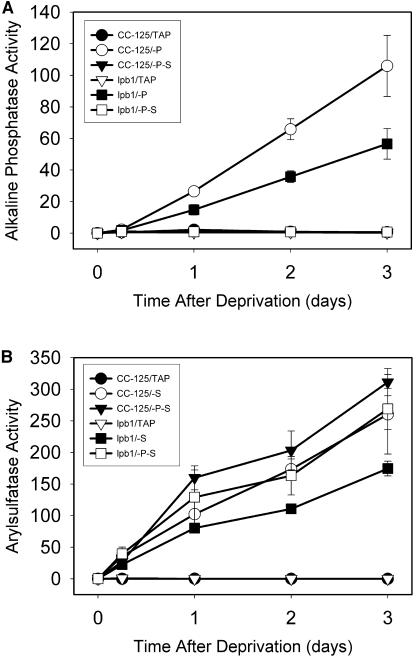
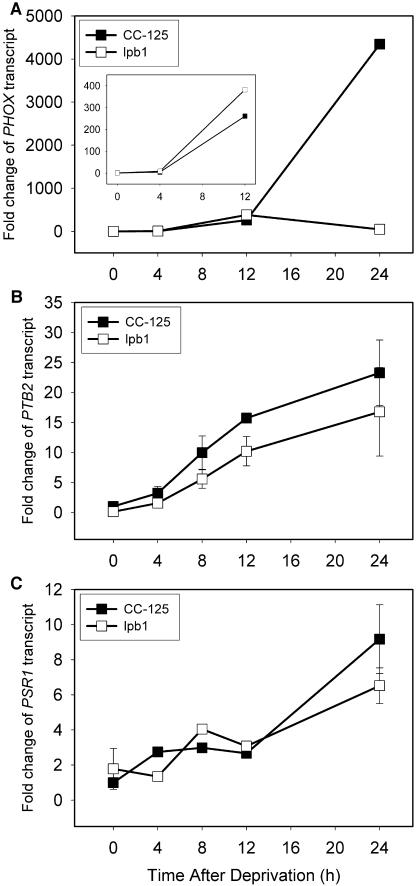
While survival of the lbp1 mutant is specifically compromised during P or S starvation, this strain can still perform P and S deprivation-specific acclimation responses. The lpb1 mutant still exhibits the synthesis of an extracellular alkaline phosphatase activity (blue halo around the plated cells) during P starvation (Fig. 1B), and the accumulation of alkaline phosphatase activity in liquid cultures of the mutant cells following elimination of P from the medium is about 60% of that of wild-type cells (Fig. 3A). The mutant strain also develops relatively high levels of arylsulfatase (ARS) activity during S deprivation (Fig. 3B). Furthermore, both the mutant and wild-type strains display increased accumulation of transcripts encoding the PHOX phosphatase (Fig. 4A), the PTB2 phosphate transporter (Fig. 4B), and the regulatory element PSR1 (Fig. 4C) after 12 h of P deprivation. These transcripts were previously shown to increase during P deprivation (Wykoff et al., 1999; J. Moseley, C.-W. Chang, and A.R. Grossman, unpublished data). The high level of PHOX transcript at 24 h after the initiation of P deprivation of wild-type cells relative to the lpb1 mutant may reflect a rapid turnover of the transcript when the mutant cells begin to die.
Figure 3.
Measurements of alkaline phosphatase and ARS activities following exposure of wild-type cells and the lpb1 mutant to P and S deprivation. A, Quantification of alkaline phosphatase activity associated with wild-type cells (CC-125) and lpb1 (lpb1) following P or P and S deprivation. B, Quantification of ARS activity associated with wild-type cells (CC-125) and lpb1 (lpb1) following S or S and P deprivation. Alkaline phosphatase and ARS activities were determined by monitoring the hydrolysis of ρ-nitrophenol phosphate and ρ-nitrophenol sulfate, respectively, at OD410. The absorbance readings were converted to nmol PNPP/mg chl/min.
Figure 4.
Change in mRNA levels of P deprivation-responsive genes following exposure of wild-type cells and the lpb1 mutant to medium lacking P. A to C, The PHOX (A), PTB2 (B), and PSR1 (C) mRNA levels over a period of 24 h of P deprivation. Levels of the mRNA were quantified by qPCR, and the threshold cycle number or CT value was normalized to the amount of constitutively expressed CBLP transcript. The fold change was calculated relative to the amount of transcript at time 0 for wild-type cells.
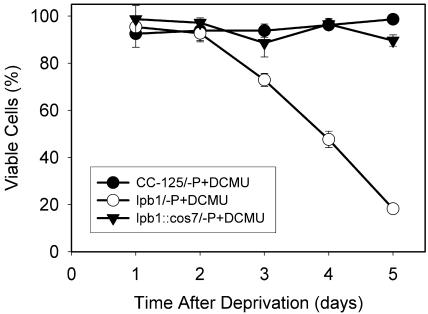
To determine whether the death of lpb1 was a consequence of its inability to modulate photosynthetic electron flow during nutrient limitation, we examined the effect of 3-(3,4-dichlorophenyl)-1,1-dimethylurea (DCMU) on lpb1 viability during P deprivation. As shown in Figure 5, although DCMU did not totally rescue P deprivation-induced death of the mutant, blocking electron flow delayed the death by approximately 1 d.
Figure 5.
Viability of P-starved wild-type cells (CC-125) and the lpb1 mutant (lpb1) in the presence of DCMU. DCMU was added to 1 μm DCMU, which eliminated all photosynthetic O2 evolution (data not shown).
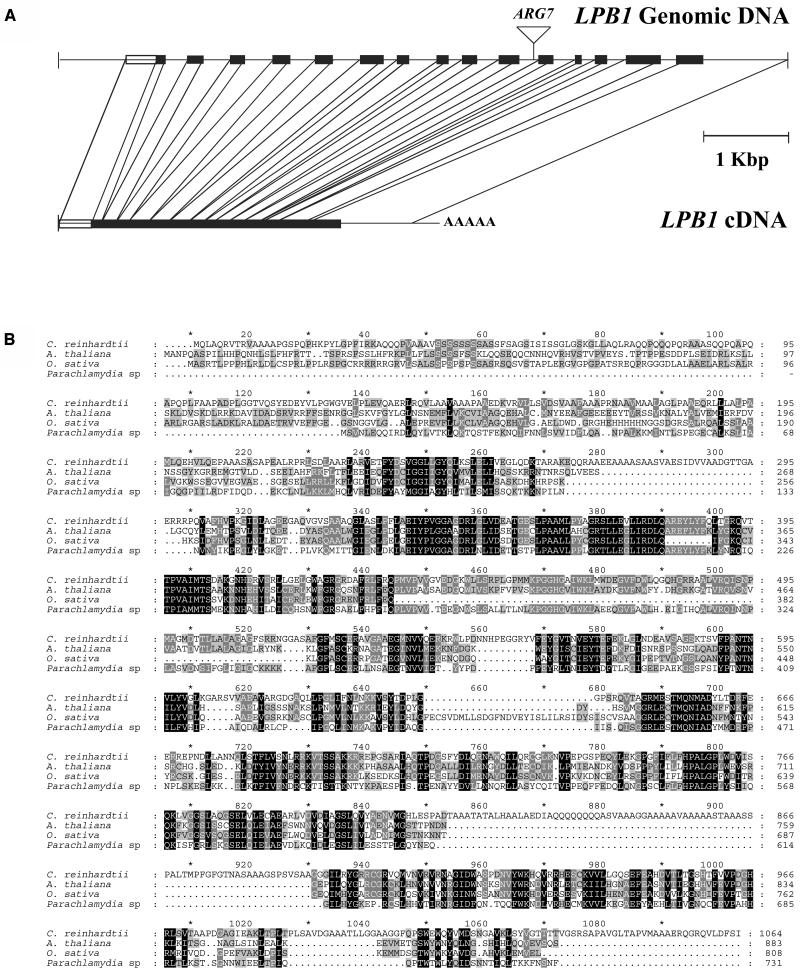
The putative LPB1 gene in the mutant strain was disrupted by the ARG7 gene; the insertion occurred within intron 10 and was not accompanied by a deletion in the genomic DNA (Fig. 6A). We isolated a C. reinhardtii bacterial artificial chromosome clone containing the site of the insertion and generated a 9.3-kb fragment that was able to complement the mutant phenotype. The complemented strain survived P deprivation (Fig. 2A). Using RACE, we obtained a near full-length cDNA sequence; the GreenGenie Program predicts a full-length cDNA with 360 additional nucleotides at the 5′ end (in the predicted 5′-untranslated region). The sequences of the LPB1 cDNA and genomic clones have been deposited in the National Center for Biotechnology Information (NCBI) GenBank database with accession numbers AY946347 and AY946348, respectively. A comparison of the cDNA with the genomic sequence demonstrates that the gene contains 15 exons and 14 introns; the latter range in size from about 150 to 400 nucleotides. The predicted polypeptide (Fig. 6B) has considerable similarity to a putative Arabidopsis protein (NCBI accession no. NP567031) and hypothetical proteins of Oryza sativa (NCBI accession no. AC112159) and Parachlamydia sp. UWE25 (NCBI accession no. YP007323). Interestingly, the C. reinhardtii sequence has several insertions relative to the other two sequences; one of these insertions is high in both Gln and Ala residues and has limited sequence similarity to the nucleotide-binding domain (P-loop) of nucleoside triphosphate hydrolases (e-value 0.004). The overall identity and similarity along the entire 1,064-amino acid region of the putative polypeptides of Arabidopsis, O. sativa, and Parachlamydia sp. that match the C. reinhardtii open reading frame ranges from 38% to 44%.
Figure 6.
The LPB1 gene and predicted gene product. A, Structure of LPB1 cDNA and genomic clones. The black rectangles represent the exons, while the lines represent introns. The rectangle at the beginning of the gene sequence represents the part of the cDNA that was deduced by RNA ligase-mediated RACE (see “Materials and Methods”). The site of insertion of the ARG7 gene is marked above the genomic sequence. B, Alignment of predicted LPB1 polypeptide sequence with homologous polypeptides encoded by an expressed sequence of Arabidopsis and hypothetical proteins of O. sativa and Parachlamydia sp. Amino acids in a blackened background represent identical or conserved residues for all four sequences, while amino acids in a gray background represent identical or conserved residues for two or three of the four sequences.
The C. reinhardtii LPB1 protein and the putative LPB1-like proteins of Arabidopsis, O. sativa, and Parachlamydia sp. have domains similar to those that are present in the enzymes UDP-acetylhexosamine pyrophosphorylase and UDPGPase. The former enzyme catalyzes the formation of UDP-N-acetyl-d-glucosamine/hexosamine (UDPGlcNAc) and pyrophosphate from N-acetyl-d-glucosamine-1-P and UTP, while the latter enzyme catalyzes the reversible formation of UDP-Glc (UDPG) and pyrophosphate from UTP and Glc-1-P. UDPGlcNAc is an important precursor for peptidoglycan, chitin, and lipopolysaccharide synthesis (Yarema and Bertozzi, 2001), while UDPG is critical for the synthesis of various carbohydrates including Suc, cellulose, starch, glycogen, and β-glucans (Kleczkowski, 1994; Kleczkowski et al., 2004; Kotake et al., 2004). However, the conservation is not extensive enough to draw strong conclusions concerning the potential catalytic activity of the polypeptide (see “Discussion”).
LPB1 mRNA was not detected by RNA-blot hybridization using RNA isolated from wild-type cells grown under either nutrient-replete or P starvation conditions. However, we were able to detect low-level constitutive expression from the gene using quantitative PCR (qPCR; data not shown); the level of RNA did not appear to change during P deprivation.
DISCUSSION
In this study, we identified a C. reinhardtii mutant, lpb1, that still performs the specific P-deprivation responses, as evaluated by the induction of alkaline phosphatase activity and the accumulation of mRNAs encoding the PTB2 phosphate transporter, the PHOX phosphatase, and the regulatory protein PSR1, but dies much more rapidly than wild-type cells during P starvation. The mutant also accumulates ARS activity following the initiation of S starvation, but was more sensitive than wild-type cells to conditions of S depletion. The accumulation of the ARS and alkaline phosphatase activities were somewhat lower in the mutant than in the wild-type strains, but the reduced levels may reflect the inability of the cells to survive extended periods of P or S stress relative to the wild-type strain. When lpb1 mutant cells were starved simultaneously for P and S, the development of ARS activity, but not alkaline phosphatase activity, was observed. It was previously demonstrated that acclimation of C. reinhardtii to S deprivation occurs more rapidly than acclimation to P deprivation. Generally, it takes longer for cells to respond to P than to S deprivation; this is exemplified by the less rapid decline in photosynthetic O2 evolution during P deprivation, as was previously reported (Wykoff et al., 1998). Therefore, the more rapid response of the cells to S deprivation leads to domination of S-deprivation over P-deprivation responses. However, the dual-stress conditions did enable the mutant to survive for a longer time than either of the single-stress conditions, suggesting that there is some combination of S and P-deprivation responses occurring in the cells; the combination of these responses must have a synergistic role in maintaining cell viability.
One of the general responses to nutrient deprivation in C. reinhardtii is a marked decline in photosynthetic activity (Wykoff et al., 1998), and it was previously demonstrated that this decline was critical for sustaining the viability of the cells during nutrient deprivation (Davies et al., 1996). Therefore, the death of lpb1 during P deprivation may be a consequence of the inability of the mutant strain to properly modulate photosynthetic electron flow. However, the loss of photosynthetic activity during P deprivation is similar in the two strains (data not shown), and while DCMU, an inhibitor of photosynthetic electron flow, did delay of the death of lpb1 to some extent, it did not entirely rescue the viability phenotype. These results suggest that there is some other cause(s) for the death of the lpb1 mutant during P deprivation.
Genes encoding proteins similar to the predicted LPB1 polypeptide were identified in other organisms; one is an expressed protein of Arabidopsis and the other two are hypothetical proteins, one in O. sativa and the other in Parachlamydia sp. (UWE25). Furthermore, these sequences have domains with some similarity to sequences of UDP-acetylhexosamine pyrophosphorylase and UDPGPase, enzymes that catalyze the formation of UDPGlcNAc and UDPG, respectively. Indeed, LPB1 appears to be part of a larger family of proteins that includes UDPGPase, UDP-acetylhexosamine pyrophosphorylase (Peneff et al., 2001), and Suc phosphate synthase (Eimert et al., 1996). The UDPGlcNAc and UDPG sugar donors are key metabolites required for glycoprotein and glycolipid synthesis (UDPGlcNAc) as well as for the synthesis of Suc and various polysaccharides associated with carbon storage, cell wall structure, and protein modification (UDPG; Kleczkowski, 1994; Kleczkowski et al., 2004). Furthermore, in Arabidopsis, both expression from the UGP gene (gene encoding UDPGPase) and UDPGPase activity were shown to be up-regulated by conditions leading to P deprivation (Ciereszko et al., 2001; Repetto et al., 2003); the activity may be involved in optimizing the use of P resources (by generating pyrophosphate that could be hydrolyzed by pyrophosphatases to Pi) in times of need.
A number of observations suggest that it is unlikely that LPB1 has either UDP-acetylhexosamine pyrophosphorylase or UDPGPase activity. UDPGPase is 97% to 99.6% identical among mammalian proteins and 33% to 54% identical between mammalian and nonmammalian proteins (Flores-Diaz et al., 1997). Gene models with much greater conservation with the nucleotide-diphospho-sugar transferases are found in the draft C. reinhardtii genome (http://genome.jgi-psf.org/chlre2/chlre2.home.html; model C_40043 is the best match to UDPGP, and model C_100106 is the best match to UDP-acetylhexosamine pyrophosphorylase). The identity between LBP1 and potential UDPGPase homologs of C. reinhardtii and other organisms, including those of plants, is only 8% to 12% over the entire protein sequence and 25% to 29% over the regions that align among the proteins (Supplemental Fig. 1). Furthermore, there are three clearly defined, conserved motifs of UDPGPase (Bork and Koonin, 1996; Flores-Diaz et al., 1997). These motifs are defined as regions of 30 amino acids, in which at least 11 amino acids are strictly conserved in all homologs (Flores-Diaz et al., 1997). LPB1 has 8/24 identical amino acids for conserved motif 1 and 9/21 identical amino acids for conserved motif 2 (Fig. 7). Also, LPB1 and its Arabidopsis, O. sativa, and Parachlamydia sp. putative homologs lack the five conserved Lys residues that were previously shown to be important for substrate binding or catalysis (Katsube et al., 1991; Kazuta et al., 1991). Finally, UDPGPase in both plant and animal systems occurs almost exclusively in the cytosol, with activity sometimes associated with Golgi membranes (Simcox et al., 1977; Azzar et al., 1978; Nishimura and Beevers, 1979; Persat et al., 1983, 1984; Kleczkowski, 1994). Based on programs designed to predict subcellular locations of nuclear-encoded polypeptides, LPB1 appears to be localized in an organelle. Predictions from a variety of different localization programs are presented in Table I. For C. reinhardtii, LPB1 is predicted to localize to the chloroplast or mitochondrion, depending on the prediction tool used. In contrast, the prediction programs place the Arabidopsis LPB1-like protein in the chloroplast. Since different prediction tools used for these analyses are based on different algorithms, it is highly probable that LPB1 of C. reinhardtii and Arabidopsis are organelle localized. Frequently, there are difficulties in predicting whether a precursor polypeptide of C. reinhardtii is targeted to the mitrochondrion or chloroplast, probably because the algorithms used have not been trained extensively enough with polypeptides previously shown to localize to each of the organelles. It is also worth noting that the predicted cleavage sites for the LPB1 polypeptides of C. reinhardtii and Arabidopsis are in the same position with respect to the sequence alignment (the cleavage site of LPB1 predicted by MitoProtII is at amino acid 86, while the cleavage site of the Arabidopsis protein predicted by ChloroP is at amino acid 72). The lack of a targeting peptide in UWE25 of Parachlamydia (but present on the predicted sequences of Arabidopsis, O. sativa, and C. reinhardtii) is apparent from the alignment shown in Figure 6B. UWE25 is an endosymbiotic Chlamydial species that is thought to have diverged from the Chamydiaceae about 700 million years ago. Interestingly, a number of putative plant genes with significant similarity to UWE25 have been localized to the chloroplast, suggesting cyanobacterial anscestry (Horn et al., 2004). Similarly, there is only limited similarity between LPB1 and UDP-acetylhexosamine pyrophosphorylase.
Figure 7.
Sequence similarity between two conserved domains of UDPGPases from human (Swissprot Q07131), Caenorhabditis elegans (NCBI accession no. U58727), barley (Hordeum vulgare; NCBI accession no. X91347), Arabidopsis (NCBI accession no. BAB10518), and gene model C_40034 from the draft C. reinhardtii genome, with the similar domains of C. reinhardtii LPB1 and the putative LPB1 homolog in Arabidopsis (locus id, At3g56040).
Table I.
Predicted subcellular targeting of C. reinhardtii LPB1 and its putative Arabidopsis homolog
| Prediction Tool
|
LPB1 Protein
|
LPB1 Homolog in Arabidopsis
|
||||
|---|---|---|---|---|---|---|
| Probability Mitochondrial | Probability Chloroplastic | Prediction Result | Probability Mitochondrial | Probability Chloroplastic | Prediction Result | |
| PCLR | NAa | 0.988 | Chloroplast | NA | 0.964 | Chloroplast |
| ChloroP | NA | 0.562 | Chloroplast | NA | 0.593 | Chloroplast |
| MitoProtII | 0.9680 | NA | Mitochondrial | 0.1368 | NA | Chloroplastb |
| Predotar | 0.995 | 0.03 | Mitochondrial | 0 | 0.927 | Chloroplast |
| TargetP | 0.690 | 0.358 | Mitochondrial | 0.075 | 0.953 | Chloroplast |
| iPSORT | NA | NA | Mitochondrial | NA | NA | Chloroplast |
NA, Not applicable (probability score not provided).
Probability is below the confidence level for mitochondrial targeting, and other programs predict chloroplast targeting.
While LPB1 is probably located in chloroplasts, the precise function of the protein is still not clear. Based on its similarity to UDP-sugar pyrophosphorylases, LPB1 appears to be a member of a family of proteins associated with sugar metabolism and perhaps the transfer of sugar moieties to other molecules in the cell. C. reinhardtii cells mutated for LPB1 show no apparent growth defect relative to wild-type cells when maintained on nutrient-replete medium, but they die rapidly when exposed to P or S limitation. This phenotype may be related to the potential function of LPB1 in the metabolism of sugars/polysaccharides, the decoration of macromolecules with sugar moieties, and/or the conservation/efficient utilization of Pi during P deprivation. Such activities may result in modification of regulatory processes and/or facilitate the storage of fixed carbon, relieving, to some extent, stress that results from the excitation of the photosynthetic reaction centers during a time when cell growth is beginning to slow. Perhaps more insights will be gained with respect to the function of the LPB1 protein with the initiation of biochemical analyses of the purified protein and the analysis of sugar and polysaccharide metabolism in both mutant and wild-type strains during nutrient deprivation. The use of DNA microarrays to compare expression patterns of wild-type C. reinhardtii with that of the lpb1 mutant following the imposition of P deprivation may also provide insights into the physiological function of this protein.
MATERIALS AND METHODS
Strains and Growth Condition
The wild strain of Chlamydomonas reinhardtii used in these studies was CC-125. The lpb1 mutant was generated in the CC-425 background by transformation with pJD67. Selected transformants were backcrossed to CC-125 five times to ensure genetic homogeneity of the two strains. The standard growth medium was Tris-acetate phosphate (TAP; Harris, 1989), and liquid cultures were grown in Erlenmeyer flasks with gentle agitation on a rotary shaker (120 rpm) and illuminated at 80 μmol photon m−2 s−1 from cool-white fluorescent lamps at 27°C. To achieve P deprivation, cells in mid-logarithmic phase of growth (1–3×106 cells/mL) were pelleted by centrifugation (3,000g, 5 min), washed twice with TAP medium lacking P (TA; Quisel et al., 1996), and then resuspended in TA to 1×106 cells/mL. This protocol was modified when performing viability measurements; the cells were resuspended in either TA or TAP medium at 2.5×104 cells/mL to allow for a longer periods of sustained growth. To prepare TA solid medium, we used 0.5% agarose instead of 1.2% agar (Shimogawara et al., 1999). To impose S and N starvation on the cells, TAP lacking sulfate (chloride salts were substituted for the sulfate salts) and TAP lacking ammonium (potassium chloride and molybdic acid were substituted for ammonium chloride and ammonium molybdate, respectively) liquid medium was used for washing the cells, in a manner similar to that described for achieving P deprivation.
Insertional Mutagenesis and Screening for Low-P-Bleaching Mutants
The plasmid pJD67, harboring the ARG7 gene encoding arginosuccinate lyase (Davies et al., 1994), was linearized by digestion with HindIII and transformed (Kindle, 1990) into the arg auxotrophic strain CC-425. Transformants that could grow on medium lacking Arg were spread at low density on solid TAP medium containing low Pi (10 μm) and grown for 7 d (Shimogawara et al., 1999). Those transformants that rapidly bleached were subjected to a secondary screen on solid TAP medium to eliminate pigment mutants and on high-salt medium lacking acetate (Harris, 1989) to eliminate those mutants that were defective for photosynthesis.
Viability and Activity Assays
Cells were counted in a hemocytometer, and cell viability was determined by staining with a solution of 0.0125% phenosafranin, 0.0125% methylene blue, 2.5 mm potassium phosphate, and 2.5% ethanol, similar to the assay previously described by Davies et al. (1996). Dead cells stained blue, whereas live cells excluded the dyes and remained green. The production of alkaline phosphatase activity by colonies growing on solid medium was evaluated by spraying colonies with an aqueous solution of 10 mm 5-bromo-4-chloro-3-indolyl-P (X-Pi; Sigma-Aldrich, St. Louis). Alkaline phosphatase activity in liquid cultures was measured as previously described (Shimogawara et al., 1999), except that the reaction was incubated at 37°C for 15 min. ARS activity was measured as described by de Hostos et al. (1988), except that the reaction was incubated at 37°C for 15 min.
In Vivo Measurements of Oxygen Evolution
Light-dependent O2 evolution and dark respiration were measured with a Clark-type oxygen electrode (Hansatech, King's Lynn, UK) and reported by the Oxygraph system, as previously described (Wykoff et al., 1998).
Transformation and Complementation of lpb1
The lpb1 mutant was generated by introduction of the ARG7 gene (Wykoff et al., 1999) into CC-425 (cw15, nit1, arg). The ARG phenotype and the introduced ARG7 gene cosegregated with the lpb1 phenotype (death during P deprivation), suggesting that the inserted pJD67 DNA was responsible for sensitivity of the mutant to P deprivation. Plasmid rescue (Davies et al., 1996) was used to isolate DNA flanking the inserted vector; genomic DNA of the transformant was digested with SacI, and the fragments generated were self-ligated, transformed into Escherichia coli, and transformants selected for kanamycin resistance. A 300-bp region of the DNA flanking the vector sequence was amplified by PCR and used to screen a wild-type cosmid library (Purton and Rochaix, 1994; Zhang et al., 1994). Three cosmids were isolated, all of which shared identical restriction fragment profiles. The lpb1 mutant strain (nit1 cw15) was cotransformed with pMN24 (NIT1) and the identified cosmids containing the putative LPB1 gene, transformants were selected for growth on nitrate, and each of the transformants was screened for growth characteristics on TA plates; several of the cotransformed strains showed growth characteristics that were similar to those observed for the wild-type strain. The cosmid was able to rescue the lpb phenotype even after it was digested with EcoRI, KpnI, or XbaI. These enzymes were used to identify fragments of the insert DNA harboring the gene responsible for rescuing the mutant phenotype; a 9-kb XbaI-EcoRI complementing fragment was further characterized.
Isolation of LPB1 cDNA
Based on information generated by the C. reinhardtii genome project (http://genome.jgi-psf.org/chlre2/chlre2.home.html), we identified four 5′ end-truncated LPB1 cDNA clones. Because the longest cDNA clone was 2.2 kb and did not contain the full-length sequence, RNA ligase-mediated RACE (GeneRacer kit, Invitrogene, Carlsbad, CA) was used to recover a full-length cDNA sequence. Total RNA was extracted from P-starved cells for 2, 4, 8, 12, and 24 h and treated with DNase I at 37°C for 1 h. The protocol described by the manufacture was followed except that we did not dephosphorylate the nucleic acid fragments by calf intestine phosphatase and, because the primers used had high-GC content, the reverse transcription was performed at 42°C for 30 min and then 50°C for 20 min. Furthermore, because the lpb1 transcript was not very abundant, nested PCR was necessary to obtain a 1.9-kb cDNA fragment containing the 5′ region of the gene.
qPCR
qPCR was performed in the Lightcycler (Roche Applied Science, Indianapolis) using the SYBR green I RNA amplification kit. Total RNA was extracted from C. reinhardtii cells deprived of P for 4, 8, 12, 24, and 48 h (Shrager et al., 2003) as described previously (Wykoff et al., 1999), and polyA RNA was purified (polyA extraction kit, Ambion, Austin, TX), quantified using the ribogreen RNA quantification kit (Molecular Probes, Eugene, OR), and 10 ng of polyA RNA was used for each real-time qPCR reaction. The reaction mixture also included carrier RNA (RNA from bacteriophage MS2). The primers used for the CBLP transcript were 5′GTCATCCACTGCCTGTGCTT and 5′CCTTCTTGCTGGTGATGTTG. An intron is positioned between the latter two primers, and amplification with these primers would detect the presence of contaminating genomic DNA (based on amplicon sizes). The primer pairs used for qPCR were 5′CTAGAATGGAAACTGGTTGATG and 5′GTTACACGGAGAAAGCTATCA, 5′CAACAGCAGCAACAAGAGCA and 5′ATCACCGAAGTCAAAGTCCC, and 5′-TTCCGTTTCCGTTCTCTGAC-3′ and 5′-CCCTGCATCTTGTTCTCCAG-3′ for PTB2 PSR1, and PHOX, respectively. In total, three pairs of primers were used for amplification of the LPB1 transcript: (1) 5′GCAGTTGCAGTGAAGGAAAAGG and 5′TGTTCCTACAGCTCCCCTATGC, (2) 5′TGATGTACCCCAATTACTGCCA and 5′CAAATGTAFCACCTCTCAGCGA, and (3) 5′CCGGCTTCATCTTCCTGTTCCA and 5′CCCATCACATTCTCCGCGTACA. The sizes of amplicons associated with each of these transcripts were approximately 100 bp, which yields optimal PCR efficiency. The annealing temperature used for amplifications was 53°C. Quantification of relative transcript levels was accomplished by the 2e−DDCt method (Livak and Schmittgen, 2001), using CBLP (Schloss, 1990) as an internal control gene (endogenous reference; the RNA level from this gene remained constant during P deprivation).
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers AY946347, AY946348, NP567031, AC112159, and YP007323.
Supplementary Material
Acknowledgments
This is a Carnegie Institution Publication (no. 1684).
This work was supported by the U.S. Department of Agriculture (grant no. 2002–35301–12178) and by the Carnegie Institution of Washington.
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.059550.
References
- Azzar G, Berthillier G, Got R (1978) Presence of enzymes catalyzing UDP-glucose biosynthesis in low density golgi fractions of cat hepatocytes. Biochimie 60: 1339–1342 [PubMed] [Google Scholar]
- Baek SH, Chung IM, Yun SJ (2001) Molecular cloning and characterization of a tobacco leaf cDNA encoding a phosphate transporter. Mol Cells 11: 1–6 [PubMed] [Google Scholar]
- Bariola PA, Howard CJ, Taylor CB, Verburg MT, Jaglan VD, Green PJ (1994) The Arabidopsis ribonuclease gene RNS1 is tightly controlled in response to phosphate limitation. Plant J 6: 673–685 [DOI] [PubMed] [Google Scholar]
- Bieleski RL (1973) Phosphate pools, phosphate transport, and phosphorus availability. Annu Rev Plant Physiol Plant Mol Biol 24: 225–252 [Google Scholar]
- Bieleski RL, Laluchli A (1992) Phosphate uptake, efflux and deficiency in the water fern, Azolla. Plant Cell Environ 15: 665–673 [Google Scholar]
- Bork P, Koonin EV (1996) Protein sequence motifs. Curr Opin Struct Biol 6: 366–376 [DOI] [PubMed] [Google Scholar]
- Ciereszko I, Johansson H, Hurry V, Kleczkowski LA (2001) Phosphate status affects the gene expression, protein content and enzymatic activity of UDP-glucose pyrophosphorylase in wild-type and pho mutants of Arabidopsis. Planta 212: 598–605 [DOI] [PubMed] [Google Scholar]
- Davies J, Yildiz F, Grossman AR (1996) Sac1, a putative regulator that is critical for survival of Chlamydomonas reinhardtii during sulfur deprivation. EMBO J 15: 2150–2159 [PMC free article] [PubMed] [Google Scholar]
- Davies JD, Grossman AR (1998) Responses to deficiencies in macronutrients. In J-D Rochaix, M Goldschmidt-Clermont, S Merchant, eds, The Molecular Biology of Chlamydomonas. Kluwer Academic Publishers, Dortrecht, The Netherlands, pp 613–635
- Davies JP, Yildiz F, Grossman AR (1994) Mutants of Chlamydomonas reinhardtii with aberrant responses to sulfur deprivation. Plant Cell 6: 53–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Hostos EL, Togasaki RK, Grossman AR (1988) Purification and biosynthesis of a derepressible periplasmic arylsulfatase from Chlamydomonas reinhardtii. J Cell Biol 106: 29–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew MC, Saker LR, Barber SA, Jenkins W (1984) Changes in the kinetics of phosphate and potassium absorption in nutrient-deficient barley roots measured by a solution-depletion technique. Planta 160: 490–499 [DOI] [PubMed] [Google Scholar]
- Duff SMG, Lefebvre DD, Plaxton WC (1991. a) Purification, characterization and subcellular localization of an acid phosphatase from Brassica nigra suspension cells: comparison with phosphoenolpyruvate phosphatase. Arch Biochem Biophys 286: 226–232 [DOI] [PubMed] [Google Scholar]
- Duff SMG, Moorhead GBG, Lefebre DD, Plaxton WC (1989) Phosphate starvation inducible ‘bypasses’ of adenylate and phosphate dependent glycolytic enzymes in Brassica nigra suspension cells. Plant Physiol 90: 1275–1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duff SMG, Plaxton WD, Lefebure DD (1991. b) Phosphate-starvation response in plant cells: de novo synthesis and degradation of acid phosphatases. Proc Natl Acad Sci USA 88: 9538–9542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont F, Joris B, Gumusboga A, Bruyninx M, Loppes R (1993) Isolation and characterization of cDNA sequences controlled by inorganic phosphate in Chlamydomonas reinhardtii. Plant Sci 89: 55–67 [Google Scholar]
- Dumont F, Loppes R, Kremers P (1990) New polypeptides and in-vitro-translatable mRNAs are produced by phosphate-starved cells of the unicellular alga Chlamydomonas reinhardtii. Planta 182: 610–616 [DOI] [PubMed] [Google Scholar]
- Eimert K, Villand P, Kilian A, Kleczkowski LA (1996) Cloning and characterization of several cDNAs for UDP-glucose pyrophosphorylase from barley (Hordeum vulgare) tissues. Gene 170: 227–232 [DOI] [PubMed] [Google Scholar]
- Flores-Diaz M, Alape-Giron A, Persson B, Pollesello P, Moos M, von Eichel-Streiber C, Thelestam M, Florin I (1997) Cellular UDP-glucose deficiency caused by a single point mutation in the UDP-glucose pyrophosphorylase gene. J Biol Chem 272: 23784–23791 [DOI] [PubMed] [Google Scholar]
- Goldstein AH, Baertlein DA, Danon A (1989) Phosphate starvation stress as an experimental system for molecular analysis. Plant Mol Biol Rep 7: 7–16 [Google Scholar]
- Green PJ (1994) The ribonucleases of higher plants. Annu Rev Plant Physiol Plant Mol Biol 45: 421–445 [Google Scholar]
- Harris EH (1989) The Chlamydomonas Sourcebook: A Comprehensive Guide to Biology and Laboratory Use. Academic Press, San Diego [DOI] [PubMed]
- Hoffland E, Vandenboogaard R, Nelemans J, Findenegg G (1992) Biosynthesis and root exudation of citric and malic acids in phosphate starved rape plants. New Phytol 122: 675–680 [Google Scholar]
- Horn M, Collingro A, Schmitz-Esser S, Beier CL, Purkhold U, Fartmann B, Brandt P, Nyakatura GJ, Droege M, Frishman D, et al (2004) Illuminating the evolutionary history of Chlamydiae. Science 304: 728–730 [DOI] [PubMed] [Google Scholar]
- Jost W, Bak H, Blund K, Terpstra P, Beintema JJ (1991) Amino acid sequence of an extracellular, phosphate-starvation-induced ribonuclease from cultured tomato (Lycopersicon esculentum) cells. Eur J Biochem 198: 1–6 [DOI] [PubMed] [Google Scholar]
- Juma NG, Tabatabai MA (1988) Comparison of kinetic and thermodynamic parameters of phosphomonoesterases of soils and corn and soybean roots. Soil Biol Biochem 20: 533–540 [Google Scholar]
- Jungk A, Asher CJ, Edwards DG, Meyers D (1990) Influence of phosphate status on phosphate uptake kinetics of maize (Zea mays) and soybean (Glycine max). Plant Soil 124: 175–182 [Google Scholar]
- Katsube T, Kazuta Y, Tanizawa K, Fukui T (1991) Expression in Escherichia coli of UDP-glucose pyrophosphorylase cDNA from potato tuber and functional assessment of the five lysyl residues located at the substrate-binding site. Biochemistry 30: 8546–8551 [DOI] [PubMed] [Google Scholar]
- Kazuta Y, Omura Y, Tagaya M, Nakano K, Fukui T (1991) Identification of lysyl residues located at the substrate-binding site in UDP-glucose pyrophosphorylase from potato tuber: affinity labeling with uridine di- and triphosphopyridoxals. Biochemistry 30: 8541–8545 [DOI] [PubMed] [Google Scholar]
- Kindle KL (1990) High-frequency nuclear transformation of Chlamydomonas reinhardtii. Proc Natl Acad Sci USA 87: 1228–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleczkowski LA (1994) Glucose activation and metabolism through UDP glucose pyrophosphorylase in plants. Phytochemistry 37: 1507–1515 [Google Scholar]
- Kleczkowski LA, Geisler M, Ciereszko I, Johansson H (2004) UDP-glucose pyrophosphorylase: an old protein with new tricks. Plant Physiol 134: 912–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotake T, Yamaguchi D, Ohzono H, Hojo S, Kaneko S, Ishida H, Tsumuraya Y (2004) UDP-sugar pyrophosphorylase with broad substrate specificity toward various monosaccharide 1-phosphate from pea sprouts. J Biol Chem 279: 45728–45736 [DOI] [PubMed] [Google Scholar]
- Lee RB (1988) Phosphatase influx and extracellular phosphatase activity in barley roots and rose cells. New Phytol 109: 141–148 [Google Scholar]
- Lefebvre DD, Clarkson DT (1984) Characterization of orthophosphate absorption by pea root protoplasts. J Exp Bot 35: 1265–1276 [Google Scholar]
- Lefebvre DD, Duff SMG, Fife CA, Julien-Inalsingh C, Plaxton WC (1990) Response to phosphate deprivation in Brassica nigra suspension cells: enhancement of intracellular, cell surface, and secreted phosphatase activities compared to increases in Pi-absorption rate. Plant Physiol 93: 504–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lien T, Knutsen G (1973) Synchronous cultures of Chlamydomonas reinhardtii: properties and regulation of repressible phosphatases. Physiol Plant 28: 291–298 [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Löffler A, Abel S, Jost W, Glund K (1992) Phosphate-regulated induction of intracellular ribonucleases in cultured tomato (Lycoperisicon esculentum) cells. Plant Physiol 98: 1472–1478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löffler A, Glund K, Irie M (1993) Amino acid sequence of an intracellular, phosphate-starvation inducible ribonuclease from cultured tomato (Lycopersicon esculentum) cells. Eur J Biochem 214: 627–633 [DOI] [PubMed] [Google Scholar]
- Lopez-Bucio J, de La Vega OM, Guevara-Garcia A, Herrera-Estrella L (2000) Enhanced phosphorus uptake in transgenic tobacco plants that overproduce citrate. Nat Biotechnol 18: 450–453 [DOI] [PubMed] [Google Scholar]
- Loppes R (1976) Genes involved in the regulation of the neutral phosphatase in Chlamydomonas reinhardtii. Mol Gen Genet 148: 315–321 [DOI] [PubMed] [Google Scholar]
- Loppes R, Braipson J, Matagne RF, Sassen A, Ledoux L (1977) Regulation of the neutral phosphatase in Chlamydomonas reinhardtii: an immunogenetic study of wild-type and mutant strains. Biochem Genet 15: 1147–1157 [DOI] [PubMed] [Google Scholar]
- Loppes R, Deltour R (1981) A pleiotropic mutant of Chlamydomonas reinhardtii showing cell wall abnormalities and altered phosphatase activities. Plant Sci Lett 21: 193–197 [Google Scholar]
- Loppes R, Matagne RF (1973) Acid phosphatase mutants in Chlamydomonas: isolation and characterization by biochemical, electrophoretic and genetic analysis. Genetics 75: 593–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matagne RF, Loppes R (1975) Isolation and study of mutants lacking a derepressible phosphatase in Chlamydomonas reinhardtii. Genetics 80: 239–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matagne RF, Loppes R, Deltour R (1976) Phosphatases of Chlamydomonas reinhardtii: biochemical and ctyochemical approach with specific mutants. J Bacteriol 126: 937–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPharlin IR, Bieleski RL (1987) Phosphate uptake by Spirodela and Lemna during early phosphorus deficiency. Aust J Plant Physiol 14: 561–572 [Google Scholar]
- Muchhal US, Raghothama KG (1999) Transcriptional regulation of plant phosphate transporters. Proc Natl Acad Sci USA 96: 5868–5872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazato H, Okamoto T, Ishikawa M, Okuyama H (1997) Purification and characterization of phosphate inducibly synthesized in Spirodela oligorrhiza grown under phosphate deficient conditions. Plant Physiol 35: 437–446 [Google Scholar]
- Nishimura M, Beevers H (1979) Subcellular distribution of gluconeogenetic enzymes in germinating castor bean endosperm. Plant Physiol 64: 31–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabo CO, Sauer RT (1992) Transcription factors: structural families and principles of DNA recognition. Annu Rev Biochem 61: 1053–1095 [DOI] [PubMed] [Google Scholar]
- Patni NJ, Dhawale SW, Aaronson S (1977) Extracellular phosphatases of Chlamydomonas reinhardtii and their regulation. J Bacteriol 130: 205–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peneff C, Ferari P, Charrier V, Taburet Y, Monnier C, Zamboni V, Winter J, Harnois M, Fassy F, Bourne Y (2001) Crystal structures of two pyrophosphorylase isoforms in complexes with UDPGlc(Gal)NAc: role of the alternatively spliced insert in the enzyme oligomeric assembly and active site architecture. EMBO J 20: 6191–6202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persat F, Azzar G, Martel MB, Got R (1983) Properties of uridine diphosphate glucose pyrophosphorylase from Golgi apparatus of liver. Biochim Biophys Acta 749: 329–332 [DOI] [PubMed] [Google Scholar]
- Persat F, Azzar G, Martel MB, Got R (1984) Evidence for coupling between transport of UDP-glucose and its synthesis by membrane-bound pyrophosphorylase in Golgi apparatus of cat liver. Biochim Biophys Acta 769: 377–380 [DOI] [PubMed] [Google Scholar]
- Pfleger FL, Linderman RG (1994) Mycorrhizae and Plant Health. APS Press, St. Paul
- Purton S, Rochaix J-D (1994) Complementation of a Chlamydomonas reinhardtii mutant using a genomic cosmid library. Plant Mol Biol 24: 533–537 [DOI] [PubMed] [Google Scholar]
- Quisel J, Wykoff D, Grossman AR (1996) Biochemical characterization of the extracellular phosphatases produced by phosphorus-deprived Chlamydomonas reinhardtii. Plant Physiol 111: 839–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghothama KG (2000. a) Phosphate transport and signaling. Curr Opin Plant Biol 3: 182–187 [PubMed] [Google Scholar]
- Raghothama KG (2000. b) Phosphorus acquisition: plant in the driver's seat! Trends Plant Sci 5: 412–413 [DOI] [PubMed] [Google Scholar]
- Repetto O, Bestel-Corre G, Dumas-Gaudot E, Berta G, Gianinazzi-Pearson V, Gianinazzi S (2003) Targeted proteomics to identify cadmium-induced protein modifications in Glomus mosseae-inoculated pea roots. New Phytol 157: 555–567 [DOI] [PubMed] [Google Scholar]
- Rosewarne GM, Barker SJ, Smith SE, Smith F, Schachtman DP (1999) A Lycopersicon esculentum phosphate transporter (lePT1) involved in phosphorus uptake from a vesicular-arbuscular mycorrhizal fungus. New Phytol 144: 507–516 [DOI] [PubMed] [Google Scholar]
- Rubio V, Linhares F, Solano R, Martin AC, Iglesias J, Leyva A, Paz-Ares J (2001) A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev 15: 2122–2133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachay JE, Wallace RL, Johns MA (1991) Phosphate stress response in hydroponically grown maize. Plant Soil 132: 85–90 [Google Scholar]
- Schjorring JK, Jensen P (1984) Phosphorus nutrition of barley, buckwheat and rape seedlings: influx and efflux of phosphorus by intact roots of different P status. Physiol Plant 61: 584–590 [Google Scholar]
- Schloss JA (1990) A Chlamydomonas gene encodes a G protein β subunit-like polypeptide. Mol Gen Genet 221: 443–452 [DOI] [PubMed] [Google Scholar]
- Shimogawara K, Usuda H (1995) Uptake of inorganic phosphate by suspension-cultured tobacco cells: kinetics and regulation of Pi starvation. Plant Cell Physiol 36: 341–351 [Google Scholar]
- Shimogawara K, Wykoff DD, Usuda H, Grossman AR (1999) Chlamydomonas reinhardtii mutants abnormal in their responses to phosphorus deprivation. Plant Physiol 120: 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrager J, Hauser C, Chang CW, Harris EH, Davies J, McDermott J, Tamse R, Zhang Z, Grossman AR (2003) Chlamydomonas reinhardtii genome project: a guide to the generation and use of the cDNA information. Plant Physiol 131: 401–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simcox PD, Reid EE, Canvin DT, Dennis DT (1977) Enzymes of the glycolytic and pentose phosphate pathways in proplastids from developing endosperm of Ricinus communis L. Plant Physiol 59: 1128–1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith FW, Rae AL, Hawkesford MJ (2000) Molecular mechanisms of phosphate and sulphate transport in plants. Biochim Biophys Acta 1465: 236–245 [DOI] [PubMed] [Google Scholar]
- Tadano T, Ozawa K, Sakai H, Osaki M, Matsui H (1993) Secretion of acid phosphatase by the roots of crop plants under phosphorus-deficient conditions and some properties of the enzyme secreted by lupin roots. Plant Soil 155–156: 95–98 [Google Scholar]
- Taylor CB, Bariola PA, del Cardayré SB, Raines RT, Green PJ (1993) RNS2: a senescence-associated RNase of Arabidopsis that diverged from the S-RNases before speciation. Proc Natl Acad Sci USA 90: 5118–5122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor CB, Green PJ (1991) Genes with homology to fungal and S-gene RNases are expressed in Arabidopsis thaliana. Plant Physiol 96: 980–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theodorou ME, Elrifi IR, Turpin DH, Plaxton WC (1991) Effects of phosphorus limitation on respiratory metabolism in the green alga Selenastrum minutum. Plant Physiol 95: 1089–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trull MC, Guiltinan MJ, Lynch JP, Deikman J (1997) The response of wild-type and ABA mutant of Arabidopsis thaliana to phosphorous starvation. Plant Cell Environ 20: 85–92 [Google Scholar]
- Wetzel RG (1983) Limnology, Ed 2. Saunders College Publishing, Philadelphia
- Wykoff D, Davies J, Grossman A (1998) The regulation of photosynthetic electron transport during nutrient deprivation in Chlamydomonas reinhardtii. Plant Physiol 117: 129–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wykoff D, Grossman A, Weeks DP, Usuda H, Shimogawara K (1999) Psr1, a nuclear localized protein that regulates phosphorus metabolism in Chlamydomonas. Proc Natl Acad Sci USA 96: 15336–15341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao MK, Tweddell RJ, Desilets H (2002) Effect of two vesicular-arbuscular mycorrhizal fungi on the growth of micropropagated potato plantlets and on the extent of disease caused by Rhizoctonia solani. Mycorrhiza 12: 235–242 [DOI] [PubMed] [Google Scholar]
- Yarema K, Bertozzi CR (2001) Characterization of glycosylation pathways. Genome Biol 2: 0004.1–0004.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen Y, Green PJ (1991) Identification and properties of the major ribonucleases of Arabidopsis thaliana. Plant Physiol 97: 1487–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Herman PL, Weeks DP (1994) Gene isolation through genomic complementation using an indexed library of Chlamydomonas reinhardtii DNA. Plant Mol Biol 24: 663–672 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.